- Botanical Institute, Cologne Biocenter, Cologne University, Cologne, Germany
The GLABRA3 gene is a major regulator of trichome patterning in Arabidopsis thaliana. The regulatory regions important for the trichome-specific expression of GL3 have not been characterized yet. In this study, we used a combination of marker and rescue constructs to determine the relevant promoter regions. We demonstrate that a 1 kb 5′ region combined with the second intron is sufficient to rescue the trichome mutant phenotype of gl3 egl3 mutants. Swap experiments of the second intron suggest that it is not sufficient to generally enhance the expression level of GL3. This implies that the second intron contains regulatory regions for the temporal and spatial regulation of GL3. The corresponding GUS-marker constructs revealed trichome-specific expression in young trichomes.
Introduction
Arabidopsis trichomes are single epidermal cells that develop on the surfaces of most aerial organs. Trichomes are regularly distributed on rosette leaves, cauline leaves, sepals and the stem without any obvious reference to other morphological structures (Hulskamp et al., 1994; Hulskamp, 2004; Balkunde et al., 2010). The distribution of trichomes is regulated by gene regulatory network containing genes promoting or inhibiting trichome fate. The positive regulators include the WD40 factor TRANSPARENT TESTA GLABRA1 (TTG1) (Koornneef, 1981; Galway et al., 1994; Walker et al., 1999), the bHLH factors GLABRA3 (Koornneef et al., 1982; Payne et al., 2000; Bernhardt et al., 2003) and the redundantly acting ENHANCER OF GLABRA3 (EGL3) (Bernhardt et al., 2003), and the R2R3MYB factors GLABRA1 (GL1, trichome system) (Oppenheimer et al., 1991) and MYB23 (Kirik et al., 2001, 2005). In addition, several redundantly acting negative regulators, the R3MYB proteins, mediate cell–cell communication by moving between cells (Wada et al., 1997; Schellmann et al., 2002; Kirik et al., 2004a,b; Kurata et al., 2005; Digiuni et al., 2008; Tominaga et al., 2008; Wang et al., 2008; Wester et al., 2009). The trichome promoting genes and the negative regulators are engaged in a regulatory feed back loop that is described in the activator-inhibitor model (Hulskamp, 2004; Pesch and Hulskamp, 2009): The activators TTG1, GL3/EGL3, and GL1/MYB23 form an activator complex, in which TTG1 and the R2R3MYB protein bind to GL3 or EGL3. This complex activates the R3MYBs that in turn can move into neighboring cells where they repress the activators. In addition a so-called activator-depletion mechanism has been postulated according to which TTG1 is mobile in the epidermis and due to its binding to GL3 protein it is trapped in trichomes and depleted in the neighboring cells (Bouyer et al., 2008; Pesch and Hulskamp, 2009; Balkunde et al., 2011).
For a better understanding of the regulatory network it is important to analyze the transcriptional regulation of the key gene GL3. In rosette leaves, GL3 in situ hybridization experiments have shown that GL3 is expressed in developing trichomes. The expression analysis of a 2.5 kb 5′-promoter fragment driving the GUS reporter gene revealed a similar expression pattern (Zhang et al., 2003; Zhao et al., 2008) suggesting that the promoter fragment is sufficient for GL3 function in leaves. As a 1 kb 5′-promoter fragment can rescue gl3 mutants it is likely that this 1 kb fragment contains all regulatory sequences essential for trichome patterning (Bernhardt et al., 2005). Recently, the 2.5 kb 5′-promoter driving the GL3 cDNA was shown to rescue the trichome and root hair phenotype of gl3 egl3 double mutants though the rescue was not complete (Zhao et al., 2012).
In this study, we aimed to identify the relevant promoter regions of the GL3 gene in the context of trichome patterning. We could not confirm that the 2.5 kb 5′-promoter fragment drives GUS expression in trichomes (Zhang et al., 2003). We also show that a 1 kb 5′-promoter region is not sufficient for rescue. As this upstream region was previously shown to rescue the trichome phenotype when combined with the genomic region of GL3 containing introns and the 3′-1 kb downstream region (Bernhardt et al., 2005) we speculated that introns or the 3′ region contains additional regulatory sequences. A detailed analysis of the function of all introns revealed that intron 2 is essential for rescuing the gl3 mutant trichome phenotype and that it is sufficient for rescue in combination with the 1 kb 5′-promoter fragment. We exchanged the second intron with intron sequences leading to a generally enhanced expression without rescuing the gl3 egl3 phenotype. This implies that the intron sequences contain regulatory sequences for the temporal and spatial regulation of GL3 rather than for an un-specific up-regulation of the GL3 levels. Finally, we show that the relevant promoter sequences mediate a trichome specific expression of the GUS marker gene.
Materials and Methods
Molecular Biology
The 35S promoter cassette of the vector pAMPAT-GW (GenBank accession no. AY436765, Pesch et al., 2015) was exchanged with the 898 bp 5′ sequence immediately upstream of the start codon of GL3 gene using AscI and XhoI [pAMPAT-GW-GL3(5′-1 kb)]. The 1051 bp 3′ fragment was cloned into the PmeI site of pAMPAT-GW-GL3(5′-1 kb) to create the pAMPAT-GW- GL3(5′-1 kb):LR recombination cassette:(3′-1 kb) vector. All genomic fragments of GL3 were cloned into pDONR201 by BP reactions (Invitrogen). Deletions of single introns within the genomic sequence of GL3 were introduced by PCR based site directed mutagenesis. The entry clone carrying the GL3 gene with the second intron was generated using the following strategy: an entry clone carrying the genomic GL3 was cut with EcoRV and KpnI generating a GL3 fragment that includes the second and third intron of GL3. This fragment was exchanged against the corresponding GL3 fragment without introns in the entry clone carrying the coding sequence of GL3. Thereafter, the third intron was deleted by PCR based site directed mutagenesis. Coding and genomic sequences of GL3 were introduced into pAMPAT-GW-GL3(5′-1 kb):LR recombination cassette:(3′-1 kb) by LR recombination with the respective entry clones to generate the various intron deletion constructs. Plants were transformed using the floral dip method described previously (Clough and Bent, 1998).
Plant Materials and Growth Conditions
Plants were grown on soil at 24°C with 16 h of light per day. All Arabidopsis thaliana used in this study were of the Columbia (Col-0) ecotype. The gl3-3 mutant line has been described previously (Jakoby et al., 2008). egl3-77439 corresponds to the TAIR accession 1008704039.
Expression Analysis
Total RNA was extracted from 10-day-old true leaves using the RNeasy Mini Kit (Qiagen, Cat No./ID: 74106) and first-strand cDNA was then synthesized from the total RNA (1 μg) using the RevertAid H Minus 1st strand cDNA synthesis (Thermo) as described by manufacturer’s instruction. Real-time polymerase chain reactions (PCR) contained 1 μl of primer mix (10 μM), 1 μl cDNA template (10-fold dilution),10 μl 2 × SYBR Green master PCR mix and 8 μl water to a total of 20 μl. cDNA concentrations in different samples were normalized with reference to AtAct2. Gene-specific primers are listed in Supplementary Table S1.
Morphological and Histochemical Analysis
GUS stainings were essentially done as previously (Sessions et al., 1999; Schroeder et al., 2016). After staining for 16 h at 37°C, tissues were cleared and leaves were inspected by light microscopy and pictures taken using the DISKUS software (Carl H. Hilgers -Technisches Büro, Germany). Trichome numbers were determined on the third and fourth fully expanded leaf of soil-grown seedlings.
Results
5′-Promoter Region of GL3 Is Not Sufficient for Proper Expression and Rescue
It has been previously reported, that a 2.5 kb 5′-promoter fragment of GL3 fused to GUS reveals trichome specific expression in leaves and that a fusion to the GL3 cDNA can rescue the trichome phenotype in gl3 egl3 double mutants (Zhang et al., 2003; Zhao et al., 2012). In addition, it was shown that a genomic GL3 fragment including 1kb of the 5′-promoter was sufficient to rescue the gl3 trichome phenotype (Bernhardt et al., 2005). To test, whether the 1 kb 5′-promoter fragment is sufficient for trichome-specific expression or whether the introns are also important we created a pGL3(1 kb):GUS line. We observed ubiquitous GUS expression in young leaves (Supplementary Figures S1A–D). In older leaves, expression levels were close to background (Supplementary Figures S1E–G).
In parallel, we performed rescue experiments by expressing the GL3 cDNA under the control of the 1 kb 5′-promoter and 1 kb downstream of STOP codon (termed as 3′-1 kb) in the gl3-3 egl3-77439 double mutant. The gl3-3 single mutant shows about 50% reduction in trichome number whereas the egl3-77439 mutant shows a significant reduction of about 10% similar as reported for the egl3 allele in the Ler background (Table 1 and Supplementary Figures S2A,B) (Zhang et al., 2003). The gl3-3 egl3-77439 double mutant is completely glabrous and one would expect that rescued lines should exhibit the egl3 77439 mutant phenotype. Among 80 transformed T1 plants we found no rescue. All plants were completely glabrous (Supplementary Figures S2C,D).

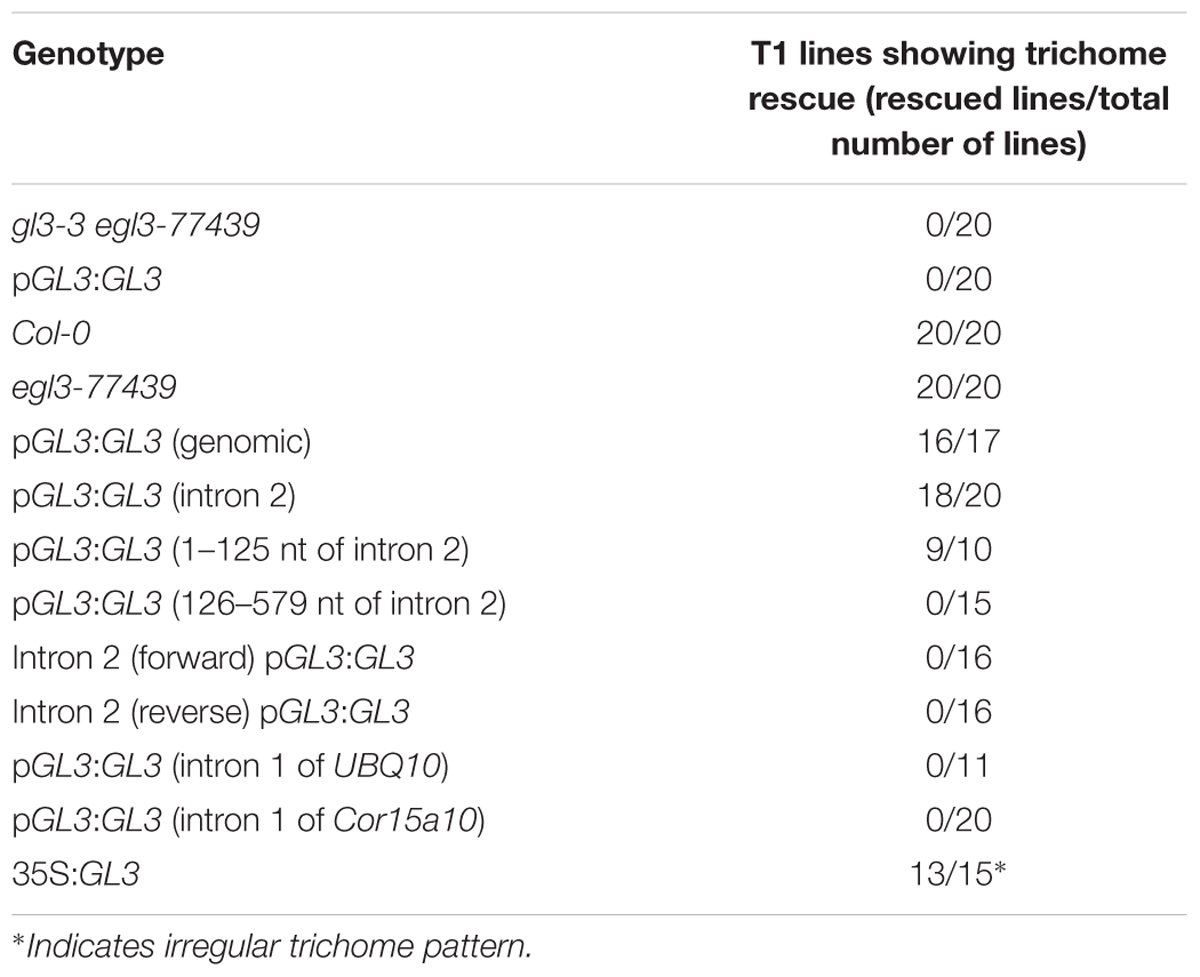
TABLE 1. Trichome number of the third and fourth true leaf in gl3-3 egl3-77349 mutants transformed with different GL3 rescue constructs.
The Second Intron of GL3 Is Essential for Rescue
Our data indicated that the 5′ promoter region and the 3′-1 kb are not sufficient for rescuing the trichome phenotype of gl3-3 egl3-77439 double mutant. We therefore tested the possibility that introns are relevant for the proper regulation of GL3 during trichome formation. Toward this end, we created a gateway construct containing 1 kb of the 5′ promoter region and 1 kb of the 3′-1 kb [called pGL3:GL3(genomic):3′-1 kb] such that the coding region can be replaced by recombination (Figure 1). This construct was used to study the rescue ability in gl3 egl3 mutants in the T1 generation. As expected, we found a range of rescue phenotypes in the T1 generation. The average rescue efficiency was used as a reference for subsequent analysis (Table 1). Next, we created a series of constructs each lacking one of the six introns (Figure 1). We found a clear rescue with constructs missing the third, fourth, or fifth intron. The deletion of the first or the sixth intron resulted in a weaker rescue of trichome formation. No rescue was observed in plants carrying the pGL3:GL3 (genomicΔintron 2):3′-1 kb construct (Table 1) indicating that the second intron is essential. We therefore focused in the following on the function of the second intron.
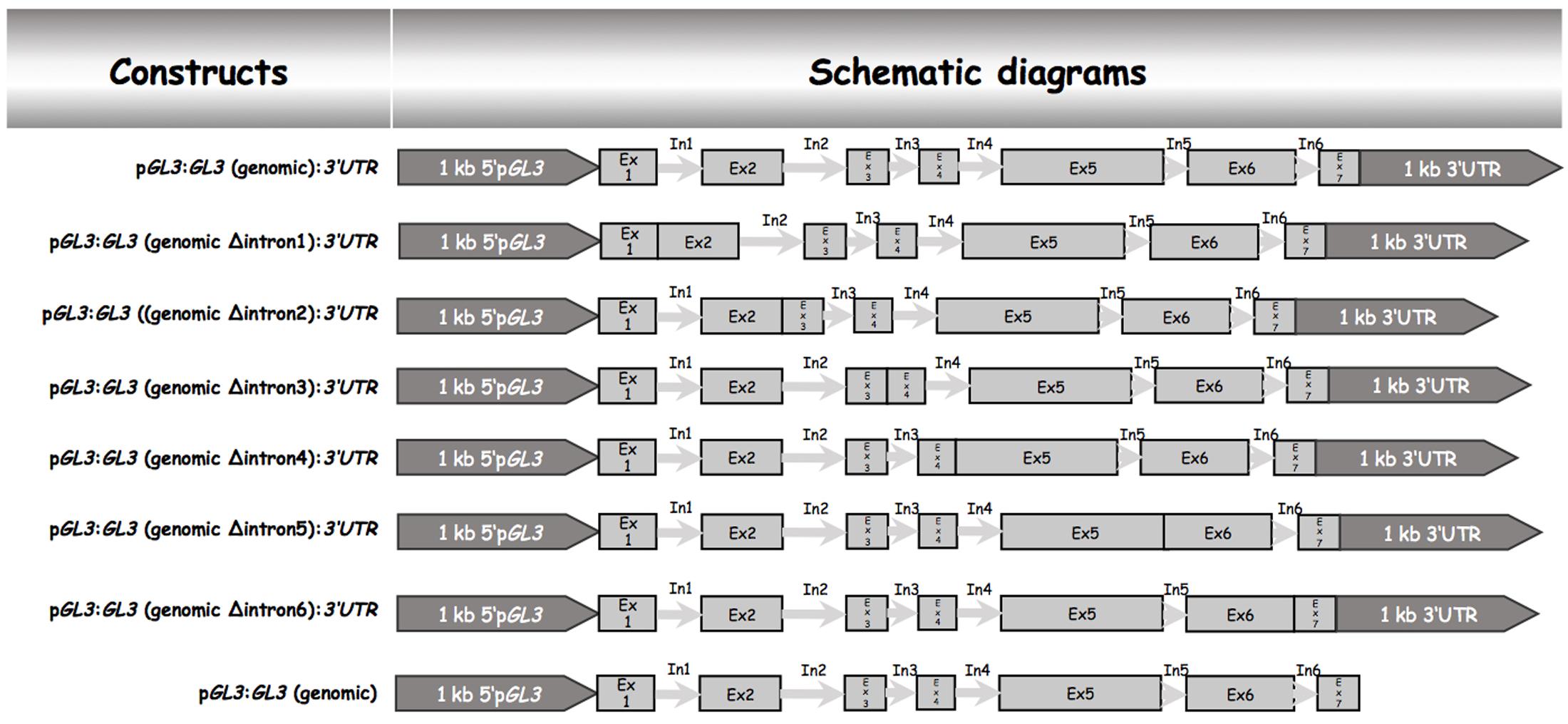
FIGURE 1. Schematic description genomic GL3 constructs and intron deletions. All constructs were cloned into the pAMPAT vector backbone. The length of promoter, exon, and intron elements are proportional to the respective sequence length. Ex, exon; In, intron.
To test the relevance of the 3′-1 kb region we studied the rescue in lines harboring the pGL3:genomic GL3 construct (Figure 1). These lines showed a rescue of the trichome phenotype. Thus, the 3′-1 kb of GL3 is not necessary for trichome rescue.
Analysis of the Function of the Second Intron of GL3
In order to demonstrate that the second intron together with the 1 kb 5′-promoter fragment is sufficient for the transcriptional regulation of GL3 in the leaf, we expressed the GL3 cDNA containing the second intron at its original site under the 1 kb 5′-promoter [pGL3:GL3(Intron 2)] in gl3 egl3 mutants (Figure 2). The majority of T1 lines showed rescue of the trichome phenotype (Table 2). In an attempt to map potential relevant regions in the second intron, we compared two deletion constructs missing either the 5′ 125 nt (pGL3:GL3(Intron 2 delta 3-125) or 454 nt at the 3′end [pGL3:GL3(Intron 2 delta 126-579)] of the second intron. Only the construct containing the 125 nt at the 5′end rescued the gl3 egl3 mutant trichome phenotype (Table 2) suggesting that this fragment contains all regulatory sequences. In a next step, we assessed whether the position of the intron 2 is important. Toward this end we placed intron 2 in front of the 5′ promoter in both directions (Figure 2). Neither construct was able to rescue the gl3 egl3 trichome mutant phenotype (Table 2) indicating that intron 2 does not act as a transcriptional enhancer element. This suggested to us that its position in the transcribed region is important for its function. One well-characterized regulatory mechanism that requires the intron within the transcribed sequence in its correct orientation is intron mediated enhancement (IME) (Rose, 2002; Gallegos and Rose, 2017). To address this possibility, we created two constructs in which the second intron of GL3 was replaced by introns for which their ability to mediate IME is well characterized (Figure 2). The first intron of UBQ10 resulted in a 2- to 10-fold higher GL3 expression as compared to wild type Col (Figure 3). Insertion of the first intron of the Cor15a gene lead to wild-type levels or up to about two-fold increased GL3 expression (Figure 3). By comparison, constructs containing intron 2 or the first 125 nt of intron 2 could enhance GL3 expression up to three-fold (Figure 3). However, neither the UBQ10 nor the Cor15a constructs rescued the trichome phenotype (Table 2).
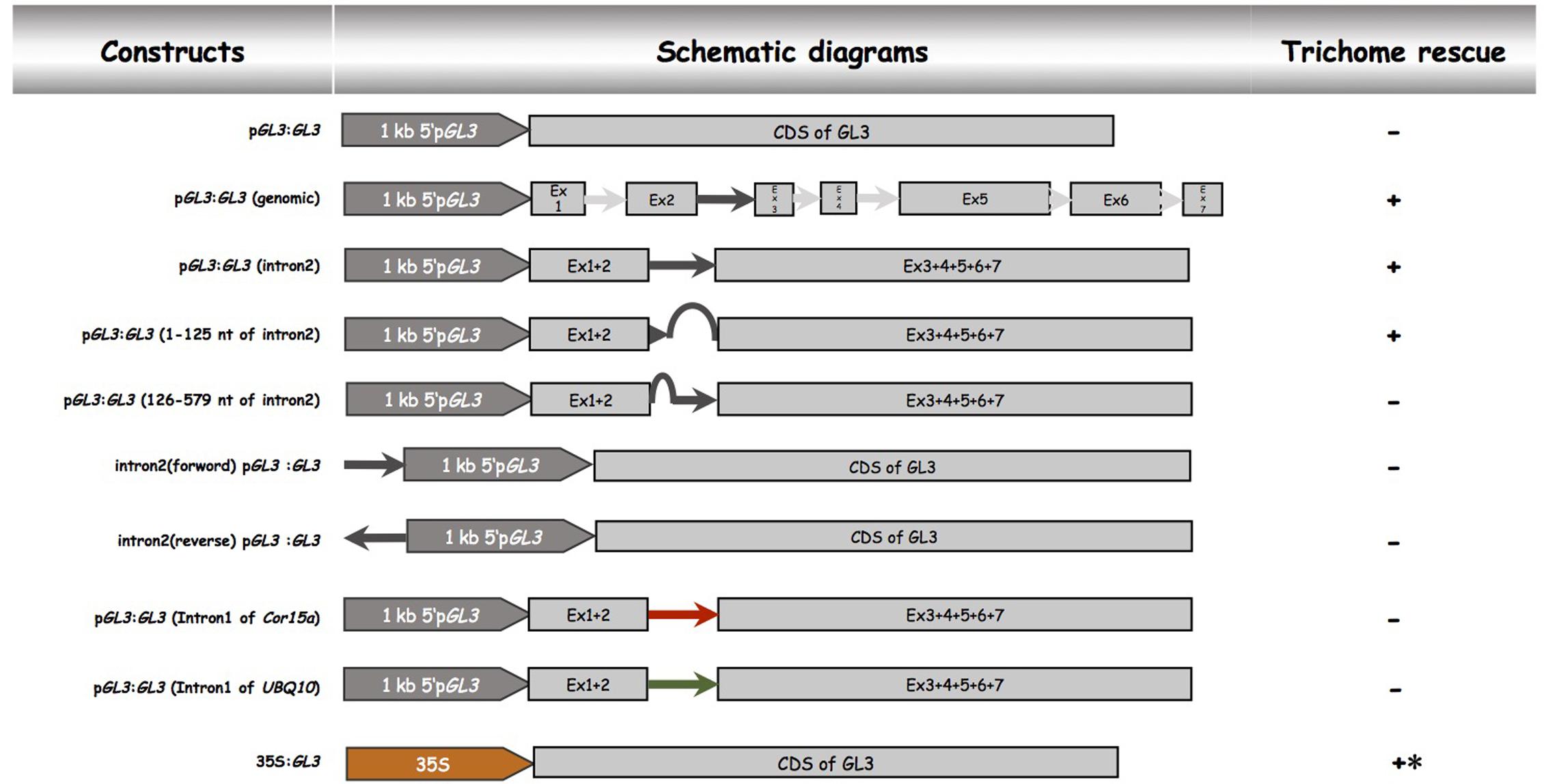
FIGURE 2. Functional analysis of the second intron of GL3. Schematic description of pGL3 driven genomic GL3 constructs. All constructs were cloned into the pAMPAT vector backbone. Promoter, exon, and intron elements are proportional to the respective sequence length. + indicates trichome rescue of the gl3 egl3 double mutant, – indicates no rescue, ∗ indicates irregular trichome pattern.
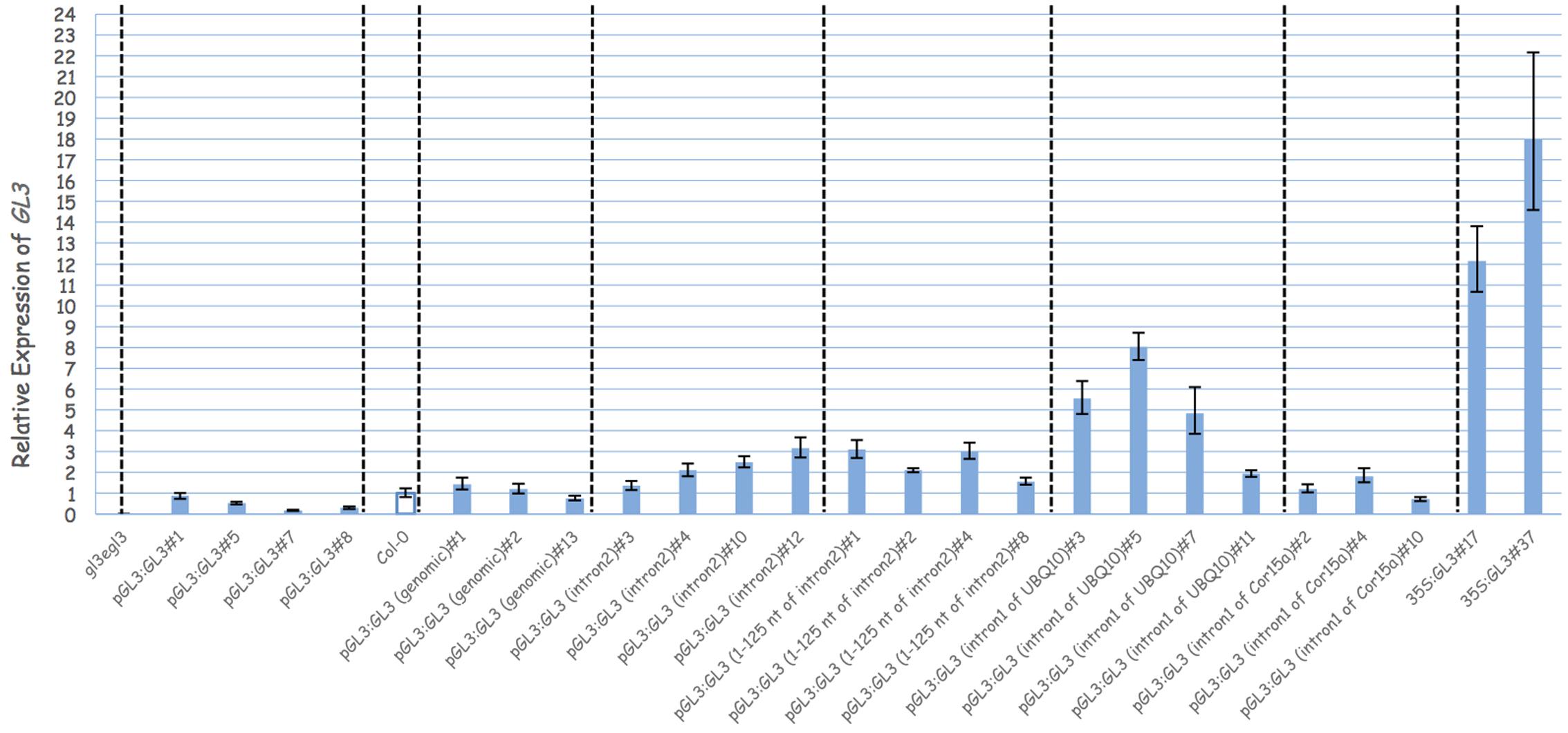
FIGURE 3. Analysis the function of the second intron of GL3. Relative expression of GL3 in different genotypes. Transcript levels were measured by quantitative real-time PCR and the expression was normalized with reference to the expression of Arabidopsis Actin2 gene. Error bars are the standard deviations of three technical replicas.
These results suggest that it is not sufficient to merely increase the GL3 expression by replacing intron 2. It is therefore conceivable that intron 2 is important for the proper regulation of the temporal and spatial expression of GL3.
Expression Analysis of pGL3: GL3(intron 2)-GUS
Our analysis revealed that a 1 kb promoter fragment combined with intron 2 in the transcribed region of GL3 is sufficient for complete rescue. To study the expression pattern mediated by this construct we fused the GUS marker gene directly after the second intron. As the signal levels were very low when using X-Gluc as a substrate, we used the more sensitive magenta-Glc-A as a substrate (Schroeder et al., 2016). We detected GL3 expression in young leaves in all stages of trichome development Figures 4A,B). In addition we noted weak expression in epidermal pavement cells in young leaves (Figure 4A). In older leaves with young trichome stages at the leaf bases and mature trichomes at the tip of the leaf trichomes exhibited much stronger expression then the mature trichomes (Figure 4B). Low levels of GL3 were maintained during further leaf growth but disappeared in fully mature leaves (Figures 4C,D).
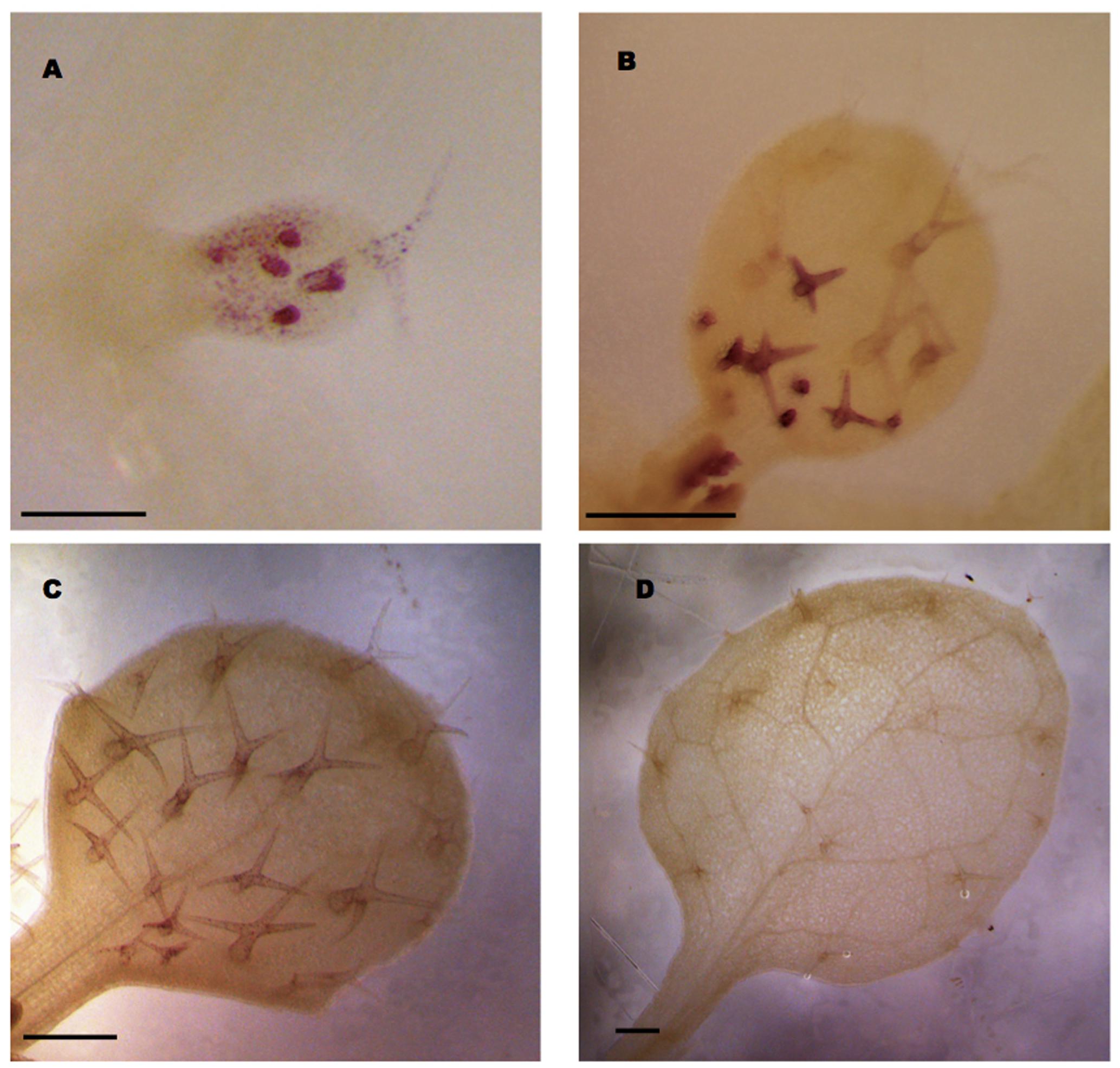
FIGURE 4. GL3 expression as revealed by pGL3:GL3(intron2)-GUS. Glucuronidase activity was detected in the first or second true leaves using magenta Glc-A as a substrate. (A) Incipient leaf with six trichomes at different stages 4 days after germination (DAG). (B) Young leaf with young stained trichomes at the leaf base and weakly stained older trichomes at the leaf tip 6 DAG. (C) Ten days old mature leaf with weakly stained trichomes. (D) Fourteen days old leaf with mature unstained trichomes. Scale bars in (A–C) are 100 μm. Scale bar in (D) is 200 μm.
Discussion
In this study, we examined which regions of the GL3 gene are important for the transcriptional regulation in the context of trichome development. While previous data suggested that the 1 kb 5′ sequences together with 1 kb 3′ sequences might be sufficient for rescuing the trichome phenotype (Zhang et al., 2003; Bernhardt et al., 2005; Zhao et al., 2008) we show that the presence of the second intron is essential and that the insertion of only the second intron in the coding region is sufficient for full rescue in combination with a 1 kb 5′ region. Our data also suggest that intron 2 contains regulatory sequences for the temporal and/or spatial expression of GL3 as high expression levels mediated by the UBQ10 intron cannot rescue the trichome mutant phenotype. Consistent with this, the intron 2 of the GL3 gene lies in a region that is hypersensitive to DNase digestion1. DNase hypersensitive sites are well-established to indicate regions of active transcriptional elements (Keene et al., 1981; Mcghee et al., 1981) and DNase hypersensitivity of DNA regions in intron 2 support therefore a regulatory role.
This raises the question, how and by which factors intron 2 is regulated to mediate trichome specific expression. One likely scenario would be the regulation by trichome patterning genes, in particular GL1, GL3/EGL3, and TTG1. However, previous studies had shown that the transcriptional regulation of GL3 does not seem to require any of the known trichome activator genes GL1, GL3, or TTG1. The total expression level of GL3 is not reduced or absent in gl1, gl3, or ttg1 mutants as judged by RT-PCR experiments (Payne et al., 2000). A possible negative auto-regulation was postulated because overexpression of GL3 can suppress its own expression (Morohashi et al., 2007). This regulation is likely to be direct as GL3 protein binds to the 5′ region immediately upstream of the transcriptional start site in chromatin-immunoprecipitation experiments (Morohashi et al., 2007). In contrast to the GL2 and CPC promoters the recruitment of GL3 to this promoter region was independent of GL1 (Morohashi et al., 2007). Although these data suggest that GL3 regulation does not involve GL1 and TTG1 this possibility is not ruled out as the exact temporal-spatial expression of GL3 might be important and not the overall level as measured in PCR experiments. A possible role of transcription factors that are involved in trichome patterning in the regulation of the second intron of GL3 is suggested by several conserved MYB and WRKY (WBOX) binding sites in the second intron of GL3 (Supplementary Figures S3–S5 and Table S2). Intron 2 and in particular the first 125 nt fragment is generally highly conserved (Supplementary Figure S4) in several Brassicaceae species including Arabidopsis lyrata, Capsella rubella, and Arabis alpina (Supplementary Table S2 and Figures S3–S5). Strikingly, the relative position of several MYB and WRKY binding sites (WBOXes) is conserved.
Therefore, these binding sites are potentially relevant for the regulation of GL3 in Arabidopsis. For future studies of the temporal and spatial regulation of GL3 it will be helpful that we could map one relevant region down to a fairly small fragment of only 125 nt containing conserved binding sites.
Author Contributions
AF, BZ, SH, MP, and AS designed, planned and performed the experiments and analyzed the data. MH supervised the project and wrote the manuscript.
Funding
AF was funded by the IMPRS. The project was funded from the Deutsche Forschungsgemeinschaft (DFG).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JB and handling Editor declared their shared affiliation, and the handling Editor states that the process met the standards of a fair and objective review.
Acknowledgments
We thank Dr. Swen Schellmann for critically reading the manuscript. We thank Eva-Maria Willing and Korbinian Schneeberger for access to the AaGL3 genomic sequence.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01382/full#supplementary-material
Footnotes
References
Balkunde, R., Bouyer, D., and Hulskamp, M. (2011). Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 138, 5039–5048. doi: 10.1242/dev.072454
Balkunde, R., Pesch, M., and Hulskamp, M. (2010). Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr. Top. Dev. Biol. 91, 299–321. doi: 10.1016/S0070-2153(10)91010-7
Bernhardt, C., Lee, M. M., Gonzalez, A., Zhang, F., Lloyd, A., and Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439. doi: 10.1242/dev.00880
Bernhardt, C., Zhao, M. Z., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132, 291–298. doi: 10.1242/dev.01565
Bouyer, D., Geier, F., Kragler, F., Schnittger, A., Pesch, M., Wester, K., et al. (2008). Two-dimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 6:e141. doi: 10.1371/journal.pbio.0060141
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Digiuni, S., Schellmann, S., Geier, F., Greese, B., Pesch, M., Wester, K., et al. (2008). A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol. Syst. Biol. 4, 217. doi: 10.1038/msb.2008.54
Gallegos, J. E., and Rose, A. B. (2017). Intron DNA sequences can be more important than the proximal promoter in determining the site of transcript initiation. Plant Cell 29, 843–853. doi: 10.1105/tpc.17.00020
Galway, M. E., Masucci, J. D., Lloyd, A. M., Walbot, V., Davis, R. W., and Schiefelbein, J. W. (1994). The Ttg gene is required to specify epidermal-cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754. doi: 10.1006/dbio.1994.1352
Hulskamp, M. (2004). Plant trichomes: a model for cell differentiation. Nat. Rev. Mol. Cell Biol. 5, 471–480. doi: 10.1038/nrm1404
Hulskamp, M., Misra, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. doi: 10.1016/0092-8674(94)90118-X
Jakoby, M. J., Falkenhan, D., Mader, M. T., Brininstool, G., Wischnitzki, E., Platz, N., et al. (2008). Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 148, 1583–1602. doi: 10.1104/pp.108.126979
Keene, M. A., Corces, V., Lowenhaupt, K., and Elgin, S. C. (1981). DNase I hypersensitive sites in Drosophila chromatin occur at the 5’ ends of regions of transcription. Proc. Natl. Acad. Sci. U.S.A. 78, 143–146. doi: 10.1073/pnas.78.1.143
Kirik, V., Lee, M. M., Wester, K., Herrmann, U., Zheng, Z. G., Oppenheimer, D., et al. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132, 1477–1485. doi: 10.1242/dev.01708
Kirik, V., Schnittger, A., Radchuk, V., Adler, K., Hulskamp, M., and Baumlein, H. (2001). Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev. Biol. 235, 366–377. doi: 10.1006/dbio.2001.0287
Kirik, V., Simon, M., Huelskamp, M., and Schiefelbein, J. (2004a). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268, 506–513. doi: 10.1016/j.ydbio.2003.12.037
Kirik, V., Simon, M., Wester, K., Schiefelbein, J., and Hulskamp, M. (2004b). ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55, 389–398. doi: 10.1007/s11103-004-0893-8
Koornneef, M., Dellaert, L. W., and van der Veen, J. H. (1982). EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat. Res. 93, 109–123. doi: 10.1016/0027-5107(82)90129-4
Kurata, T., Ishida, T., Kawabata-Awai, C., Noguchi, M., Hattori, S., Sano, R., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132, 5387–5398. doi: 10.1242/dev.02139
Mcghee, J. D., Wood, W. I., Dolan, M., Engel, J. D., and Felsenfeld, G. (1981). A 200-base pair region at the 5’ end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell 27, 45–55. doi: 10.1016/0092-8674(81)90359-7
Morohashi, K., Zhao, M., Yang, M., Read, B., Lloyd, A., Lamb, R., et al. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145, 736–746. doi: 10.1104/pp.107.104521
Oppenheimer, D. G., Herman, P. L., Sivakumaran, S., Esch, J., and Marks, M. D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. doi: 10.1016/0092-8674(91)90523-2
Payne, C. T., Zhang, F., and Lloyd, A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362.
Pesch, M., and Hulskamp, M. (2009). One, two, three...models for trichome patterning in Arabidopsis?. Curr. Opin. Plant Biol. 12, 587–592. doi: 10.1016/j.pbi.2009.07.015
Pesch, M., Schultheiss, I., Klopffleisch, K., Uhrig, J. F., Koegl, M., Clemen, C. S., et al. (2015). TRANSPARENT TESTA GLABRA1 and GLABRA1 Compete for Binding to GLABRA3 in Arabidopsis. Plant Physiol. 168, 584–597. doi: 10.1104/pp.15.00328
Rose, A. B. (2002). Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA 8, 1444–1453. doi: 10.1017/S1355838202020551
Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036–5046. doi: 10.1093/emboj/cdf524
Schroeder, M., Tsuchiya, T., He, S. L., and Eulgem, T. (2016). Use of enhancer trapping to identify pathogen-induced regulatory events spatially restricted to plant-microbe interaction sites. Mol. Plant Pathol. 17, 388–397. doi: 10.1111/mpp.12287
Sessions, A., Weigel, D., and Yanofsky, M. F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263. doi: 10.1046/j.1365-313x.1999.00594.x
Tominaga, R., Iwata, M., Sano, R., Inoue, K., Okada, K., and Wada, T. (2008). Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135, 1335–1345. doi: 10.1242/dev.017947
Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. doi: 10.1126/science.277.5329.1113
Walker, A. R., Davison, P. A., Bolognesi-Winfield, A. C., James, C. M., Srinivasan, N., Blundell, T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1349. doi: 10.1105/tpc.11.7.1337
Wang, S., Hubbard, L., Chang, Y., Guo, J., Schiefelbein, J., and Chen, J. G. (2008). Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 8:81. doi: 10.1186/1471-2229-8-81
Wester, K., Digiuni, S., Geier, F., Timmer, J., Fleck, C., and Hulskamp, M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136, 1487–1496. doi: 10.1242/dev.021733
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681
Zhao, H. T., Wang, X. X., Zhu, D. D., Cui, S. J., Li, X., Cao, Y., et al. (2012). A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J. Biol. Chem. 287, 14109–14121. doi: 10.1074/jbc.M111.280735
Keywords: Arabidopsis, trichomes, patterning, GLABRA3, transcriptional regulation
Citation: Friede A, Zhang B, Herberth S, Pesch M, Schrader A and Hülskamp M (2017) The Second Intron Is Essential for the Transcriptional Control of the Arabidopsis thaliana GLABRA3 Gene in Leaves. Front. Plant Sci. 8:1382. doi: 10.3389/fpls.2017.01382
Received: 23 June 2017; Accepted: 25 July 2017;
Published: 08 August 2017.
Edited by:
Robert G. Franks, North Carolina State University, United StatesReviewed by:
John Larkin, Biological Sciences, Louisiana State University, United StatesJavier Brumos, North Carolina State University, United States
Copyright © 2017 Friede, Zhang, Herberth, Pesch, Schrader and Hülskamp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Hülskamp, martin.huelskamp@uni-koeln.de
†These authors have contributed equally to this work.
 Alexandra Friede†
Alexandra Friede† Bipei Zhang
Bipei Zhang Stefanie Herberth
Stefanie Herberth Andrea Schrader
Andrea Schrader Martin Hülskamp
Martin Hülskamp