- 1Section of Genetics and Plant Breeding, Department of Plant, Soil and Food Science, University of Bari Aldo Moro, Bari, Italy
- 2Plant Breeding, Wageningen University & Research, Wageningen, Netherlands
- 3Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
Specific syntaxins, such as Arabidopsis AtPEN1 and its barley ortholog ROR2, play a major role in plant defense against powdery mildews. Indeed, the impairment of these genes results in increased fungal penetration in both host and non-host interactions. In this study, a genome-wide survey allowed the identification of 21 tomato syntaxins. Two of them, named SlPEN1a and SlPEN1b, are closely related to AtPEN1. RNAi-based silencing of SlPEN1a in a tomato line carrying a loss-of-function mutation of the susceptibility gene SlMLO1 led to compromised resistance toward the tomato powdery mildew fungus Oidium neolycopersici. Moreover, it resulted in a significant increase in the penetration rate of the non-adapted powdery mildew fungus Blumeria graminis f. sp. hordei. Codon-based evolutionary analysis and multiple alignments allowed the detection of amino acid residues that are under purifying selection and are specifically conserved in syntaxins involved in plant-powdery mildew interactions. Our findings provide both insights on the evolution of syntaxins and information about their function which is of interest for future studies on plant–pathogen interactions and tomato breeding.
Introduction
In eukaryotic cells, compartmentalization through the endomembrane apparatus and exocytosis require a highly regulated transport system. Soluble N-ethylmaleimide-sensitive-factor attachment protein receptor (SNARE) proteins are involved in such a transport, as they mediate the fusion of membranes of cargo-containing small shuttles, referred to as vesicles, and target membranes (Lipka et al., 2007). According to their localization, SNARE proteins can be classified into vesicle-associated (v-SNAREs) and target-membrane-associated (t-SNAREs) (Söllner et al., 1993). Alternatively, they can be classified into Q-SNARE and R-SNARE proteins, which contain either arginine or glutamine at the center of the SNARE domain, respectively (Fasshauer et al., 1998). Typically, SNARE complexes involved in membrane fusion are tetrameric coiled-coil structures containing one R-SNARE protein anchored to the vesicle and one of each of the three Q-SNARE proteins, namely Qa- (also referred to as syntaxins or SYPs), Qb- and Qc-SNAREs (Fasshauer et al., 1998; Bock et al., 2001; Schilde et al., 2008). Based on sequence homology, syntaxins of the Qa SNARE family can be divided into five subfamilies known as SYP-1, -2, -3, -4, -8 (Sanderfoot et al., 2000; Bock et al., 2001).
Ascomycete fungi of the order of Erysiphales cause the powdery mildew disease on 1000s of plant species and lead to massive economic losses in agricultural settings (Takamatsu, 2004). Specific members of the syntaxin protein family, such as Arabidopsis AtPEN1, barley ROR2, and grapevine VvPEN1, are essential components of a SNARE-dependent antimicrobial secretion pathway that limits the penetration of powdery mildew fungi (Collins et al., 2003; Feechan et al., 2013). In compatible interactions between barley and the barley powdery mildew pathogen Blumeria graminis f. sp. hordei (Bgh), ror2 mutations lead to a significant increase of fungal entry, thus indicating a functional role in basal defense mechanisms (Collins et al., 2003). In non-host interactions, involving Arabidopsis and barley with non-adapted powdery mildew species, Atpen1 and ror2 mutations also result in increased penetration rates (Peterhänsel et al., 1997; Collins et al., 2003; Assaad et al., 2004). However, further fungal growth is prevented by the occurrence of cell hypersensitive response, which is assumed to act as a post-invasion defense mechanism (Collins et al., 2003; Trujillo et al., 2004; Lipka et al., 2005).
Syntaxin-dependent exocytosis is also required for mlo-based defense, exhibited by plants harboring loss-of-function mutations of MLO susceptibility genes and associated with the failure of fungal penetration in correspondence of thick cell wall appositions termed papillae (Pavan et al., 2010; Seifi et al., 2014). In barley, ror2 mutations break mlo resistance to Bgh, resulting in the restoration of visible disease symptoms (Freialdenhoven et al., 1996; Collins et al., 2003). In Arabidopsis, Atmlo2/Atpen1 double mutants display higher penetration rate of the fungal species Golovinomyces cichoracearum than single Atmlo2 mutants. However, they do not exhibit increased fungal sporulation and still appear phenotypically resistant (Consonni et al., 2006). The mlo-based defense and syntaxins also operate at the non-host level. Indeed, barley mlo mutants show enhanced penetration resistance to the non-adapted wheat powdery mildew fungus Blumeria graminis f. sp. tritici, while mlo/ror2 double mutants display wild-type penetration rates (Peterhänsel et al., 1997).
In this study, we exploited available genomic information to perform a genome-wide characterization of tomato syntaxins. The functional role of two tomato syntaxins closely related to Arabidopsis PEN1, barley HvROR2, and grapevine VvPEN1 was investigated using RNA interference (RNAi) technology. Evolutionary analysis and multiple alignments were performed to identify molecular features putatively important for the function of the syntaxins.
Materials and Methods
Plant and Fungal Materials
The tomato Slmlo1 line, carrying a loss-of-function mutation in the SlMLO1 susceptibility gene (Bai et al., 2008; Zheng et al., 2016), and the tomato cultivar (cv) Moneymaker (MM) were used in the disease tests as resistant and susceptible control, respectively. The tomato Slmlo1 line was also used as background genotype for RNAi experiments.
The Wageningen isolate of the tomato powdery mildew fungus Oidium neolycopersici and the Wag04 isolate of the barley powdery mildew fungus Bgh, maintained on the susceptible cvs. MM and Manchuria, respectively, were used for inoculation. Infected tomato and barley plants were kept in a greenhouse compartment at 20 ± 2°C with 70 ± 15% relative humidity at the Unifarm of Wageningen University & Research, Netherlands.
Identification and Phylogenesis of Tomato Syntaxins
The Arabidopsis syntaxin AtPEN1 amino acid sequence (GenBank ID: NP_187788.1; gene AT3G11820) was used as query for a tBLASTn search against the Tomato Genome SL2.50 CDS of Sol Genomics Network (SGN), using default settings. The resulting 21 hits were used for a ClustalW alignment of 41 sequences, also including the complete Arabidopsis Qa SNARE family, grapevine VvPEN1, and barley HvROR2. The gap open cost and the gap extension cost were set equal to 10 and 4, respectively. Afterward, an Unweighted Pair Group Method with Arithmetic Mean (UPGMA) phylogenetic tree was built setting the bootstrap value equal to 100. All the bioinformatic analyses were performed using the CLC sequence viewer software1.
Generation of Tomato RNAi Transformants
To generate RNAi constructs, the two tomato cDNA sequences showing the highest similarity to AtPEN1, Solyc10g081850.1.1 (named SlPEN1a) and Solyc01g006950.2.1 (named SlPEN1b), were used to design primer pairs for Gateway cloning (Invitrogen), using the Primer3 software (Untergasser et al., 2012). These primers, named SlPEN1a_gate__Fw/Rev (5′-caccCTGGTTGTGGACCTGGAAGT-3′/5′-TGTCCTCTTCCTTGCTCCTG-3′) and SlPEN1b_gate_Fw/Rev (5′-caccACGAGCTGAAAAACCTCGAA-3′/5′-ACA ACAGACGTCCTCGTCCT-3′), allowed the amplification of 259 and 250 bp PCR products for SlPEN1a and SlPEN1b, respectively. Amplification was conducted using PfuUltraII Fusion HS DNA Polymerase (Agilent Technologies). PCR fragments were first cloned into the Gateway-compatible vector pENTR D-TOPO (Invitrogen) and then transferred to Escherichia coli competent cells (XL10-Gold Ultracompetent Cells, Agilent Technologies). The presence of the inserts was assessed by colony PCR, restriction enzyme digestion and sequencing using the universal M13 primers. Positive clones were used in a LR reaction, which allowed the inserts to be transferred to the pHELLSGATE12 vector (Helliwell and Waterhouse, 2003), harboring the 35S Cauliflower Mosaic Virus (CaMV) promoter for constitutive expression and the marker gene NPTII for kanamycin resistance selection. Plasmids were cloned into E. coli competent cells DH5α and positive colonies were screened by colony PCR and sequencing as described before. Recombinant vectors were finally extracted and transferred to the AGL1+virG strain of Agrobacterium tumefaciens by electroporation.
The tomato Slmlo1 line was used for transformation, that was carried out as described by Appiano et al. (2015). A total of 15 T1 plants were generated and, after self-pollination, T2 families were obtained.
To select transgenic plants within each segregating family, the DNA of the T2 plants was amplified with primer pairs NPTII_Fw/ NPTII_Rev (5′-TCGGCTATGACTGGGCACAAC-3′)/5′-AAGAAGGCGATAGAAGGCGA-3′), and 35S-Fw/Rev (5′-GCTCCTACAAATGCCATCA-3′)/(5′-GATAGTGG GATTG TGCGTCA-3′). The expression of the target genes in selected T2 families was assessed by real-time qPCR using the primer pairs qPEN1a_Fw/Rev (5′-CGAGATGCTTTGTGCATCAG-3′/5′-CAGTCTCCTTCAGCTCCATTTC-3′) and qPEN1b_Fw/Rev (5′-TGGTTTAGTTGTTGATGGACCTC-3′/5′-ACCCCCATCCAACTTACTTACTTCTC-3′).
Transgenic T2 plants showing high silencing level for each construct were crossed and eight F1 individuals were obtained. Four-week-old F1 plants were tested through qPCR for the expression of SlPEN1a and SlPEN1b, using the primer pairs mentioned above.
Disease Tests with O. neolycopersici and Quantification of Fungal Biomass
Three disease tests were performed using the adapted powdery mildew pathogen O. neolycopersici. The first one encompassed 20 plants for each of the 15 T2 families, seven Slmlo1 plants and five MM plants. The second one included four T2 families (two carrying the RNAi::SlPEN1a construct and two carrying the RNAi::SlPEN1b construct), each including 15 transgenic plants, five non-transgenic plants and five Slmlo1 plants. The third test included eight F1 individuals, 14 T3 transgenic individuals silenced for SlPEN1a, 19 T3 individuals silenced for SlPEN1b, and 8 Slmlo1 individuals.
In all cases, the inoculation was performed by spraying plants with a suspension of conidiospores obtained from heavily infected leaves of MM plants and adjusted to a final concentration of 2∗104 spores per milliliter, as described by Pavan et al. (2008). Inoculated plants were grown at 20 ± 2°C with 70 ± 15% relative humidity and day length of 16 h in a greenhouse of Unifarm of Wageningen University & Research, Netherlands.
Disease evaluation was carried out 15 days after inoculation. Powdery mildew symptoms were visually scored on each plant using a scale of symptoms severity ranging from 0 to 3 (Bai et al., 2008), and means and standard deviation were calculated for each T2 family. In addition, in the case of the second and third disease tests, fungal biomass was quantified by real-time qPCR as reported by Huibers et al. (2013). Briefly, plant and fungal genomic DNAs were isolated from infected tomato leaves using the Qiagen DNeasy Plant Mini Kit and amplified with the primer pairs On-Fw (5′-CGCCAAAGACCTAACCAAAA-3′)/On-Rev (5′-AGCCAAGAGATCCGTTGTTG-3′), designed on O. neolycopersici internal transcribed spacer (ITS) sequences (GenBank accession number EU047564), and Ef-Fw (5′-GGAACTTGAGAAGGAGCCTAAG-3′)/Ef-Rev (5′-CAACACCAACAGCAACAGTCT-3′), designed on the tomato Elongation Factor 1α (Ef1α) reference gene (Løvdal and Lillo, 2009). Relative fold-change of the ratio between fungus and tomato gDNAs was inferred by the 2-ΔΔCT method (Livak and Schmittgen, 2001; Pfaffl, 2001) and results were analyzed by the Student’s t-test.
Disease Test with Blumeria graminis f. sp. hordei (Bgh) and Histological Analysis
Three transgenic plants from the T2 families RNAi::SlPEN1a_I and RNAi::SlPEN1b_I were selected by PCR using the 35S and NPTII primer pairs. Around 18 days after sowing, these plants, together with three plants of the Slmlo1 line, were transferred to an infection chamber.
A dry inoculum of Bgh was brushed off heavily infected barley leaves with a paintbrush and applied on the third and fourth leaves of 4-week old tomato plants. At least three samples for each inoculated plant were collected 72 hours post-inoculation (hpi). These samples were stained with trypan blue as described by Freialdenhoven et al. (1996) and mounted on glass slides with a 1:1 (v/v) glycerol:water solution. Observation of the slides was done using a Zeiss Axiophot bright field microscope and pictures were taken with an Axiocam ERc5s. For each sample, more than thirty infection units (one infection unit representing a germinated spore) per slide were observed. The pathogen penetration rate was estimated as the percentage of units displaying hypersensitive response. Statistical analysis was carried out using the Student’s t-test.
Detection of Conserved Syntaxin Motifs and Codon Evolutionary Analysis
To detect conserved motifs in the syntaxin family, the alignment used for phylogenetic analysis was given as input to the BOXSHADE software2, setting as 1.0 the fraction of sequences that must agree. Furthermore, with the aim of identifying residues specifically conserved in syntaxins acting in powdery mildew defense, another ClustalW alignment was performed, using proteins of the SYP-1b subclade. This new alignment was also fed to the BOXSHADE software.
Moreover, the same dataset was used for a codon-based evolutionary analysis, based on the difference of nonsynonymous-to-synonymous substitutions per non-synonymous and synonymous sites (dN-dS). The single-likelihood ancestor counting (SLAC) method implemented by the Datamonkey web server3 was performed. The default p-value of 0.1 was used as threshold for statistical significance to make predictions on the kind of selection pressure (negative, neutral, or positive) acting on each codon.
Results
In Silico Identification of Tomato Syntaxins
Twenty-one putative syntaxins were retrieved in the tomato genome with a BLAST search using the Arabidopsis AtPEN1 amino acid sequence as input. These were used for a phylogenetic study, together with the 18 Arabidopsis syntaxins, barley HvROR2 and grapevine VvPEN1. The resulting phylogenetic tree was composed by five clearly distinct clades (Figure 1). The distribution of Arabidopsis syntaxins within the clades fully matched with their previous assignment to the five subfamilies SYP-1, -2, -3, -4, -8 (Sanderfoot et al., 2000). Within each clade, at least one tomato putative syntaxin was found. The most represented clade (SYP-1), harboring 21 homologs, was further partitioned into four subclades (indicated as SYP-1a to -1d in Figure 1).
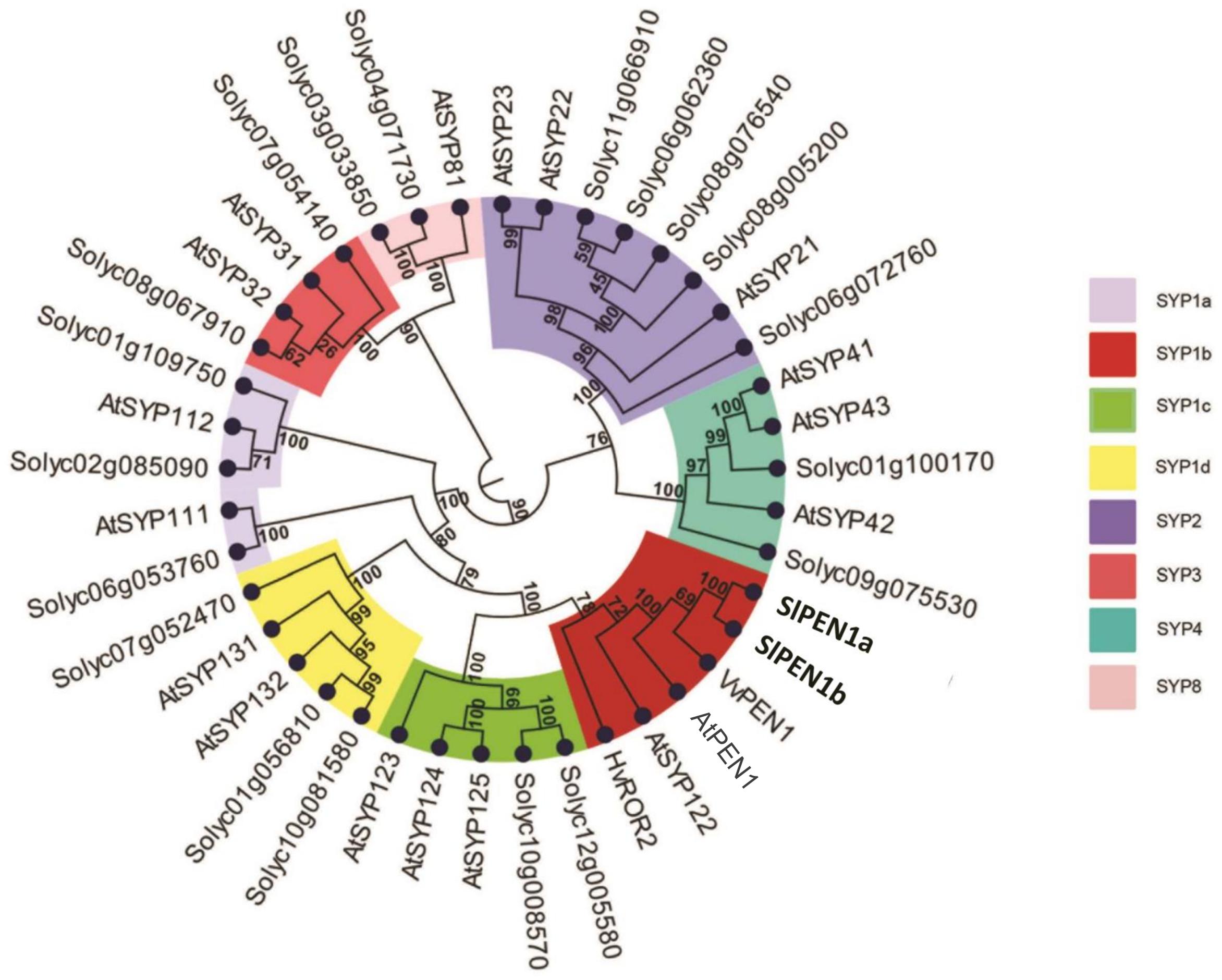
FIGURE 1. Phylogenetic relationships of 41 syntaxins. The dataset includes barley HvROR2, grapevine VvPEN1, the 18 Arabidopsis syntaxins (AtSYPs and AtPEN1) and the 21 predicted tomato syntaxins identified in this study (named with the SolGenomics Network database ID code). Numbers at nodes indicate bootstrap support values. Clades are named according to the nomenclature used for syntaxins subfamilies (SYP1, -2, -3, -4, -8). The clade SYP1 is further divided in four subclades (SYP1-a, -b, -c, and -d). The subclade SYP1b containing all the homologs known to be involved in plant–pathogen interactions (AtPEN1, AtSYP122, HvROR2, and VvPEN1) is highlighted in red.
Two putative tomato syntaxins, Solyc10g081850 and Solyc01g006950, were assigned with strong bootstrap support to the subclade SYP-1b, containing all the functional homologs shown to be involved in powdery mildew penetration resistance, i.e., AtPEN1, HvROR2, and VvPEN1 (Collins et al., 2003; Feechan et al., 2013). Therefore, Solyc10g081850 and Solyc01g006950 were renamed SlPEN1a and SlPEN1b, respectively (Figure 1). Protein sequences of SlPEN1a and SlPEN1b are highly related with each other (85.3% amino-acid conservation) and with AtPEN1 (72.3 and 71.7% amino-acid conservation, respectively).
SlPEN1a Is Required for mlo Resistance to Tomato Powdery Mildew
To gain insights into the functional role of SlPEN1a and SlPEN1b, we obtained RNAi silencing constructs for each of the two genes and transformed the tomato Slmlo1 line. Eleven RNAi::SlPEN1a and four RNAi::SlPEN1b T1 plants were obtained and self-pollinated to provide T2 families.
Two weeks after the inoculation, all the MM plants were covered by the fungal mycelium and had an average disease index (DI) score of 3 (Supplementary Table S1). Individuals of the Slmlo1 line showed no symptoms on the 4th leaf. Since weak mycelium growth was occasionally observed on the first three true leaves, an average DI score of 0.62 was assigned to them.
The 11 T2 families obtained with the RNAi::SlPEN1a construct showed various levels of symptoms corresponding to average DI scores ranging from 0.47 to 1.49. The four T2 families obtained with the RNAi::SlPEN1b construct displayed average DI scores ranging from 0.8 to 0.95.
Individuals of five selected T2 families associated with the highest average DI scores (namely RNAi::PEN1a_I, RNAi::PEN1a_II, RNAi::PEN1a_III, RNAi::PEN1b_I, and RNAi::PEN1b_II), were screened for the presence/absence of the construct. Plants carrying the RNAi constructs were identified and further referred to as RNAi::SlPEN1a(+) and RNAi::SlPEN1b(+). A higher average DI score was assigned to them compared to T2 plants not harboring the corresponding RNAi constructs (Supplementary Table S1). To verify that higher susceptibility of individuals carrying the RNAi constructs was due to reduced SlPEN1a and SlPEN1b transcripts, the expression level of these genes was evaluated. The SlPEN1a homolog was significantly silenced in transgenic individuals of the families RNAi::SlPEN1a_I(+) and RNAi::SlPEN1a_II(+) compared to Slmlo1 plants. In addition, no unwanted cross-silencing of SlPEN1b was found (Figure 2). Transgenic plants identified in family RNAi::SlPEN1a_III did not show a significant silencing of the target gene. However, this family was the least susceptible of the three T2 families carrying the RNAi::SlPEN1a construct. The SlPEN1b homolog was strongly silenced in transgenic plants of families RNAi::PEN1b_I and _II compared to Slmlo1 plants, while the expression of SlPEN1a was not significantly reduced.
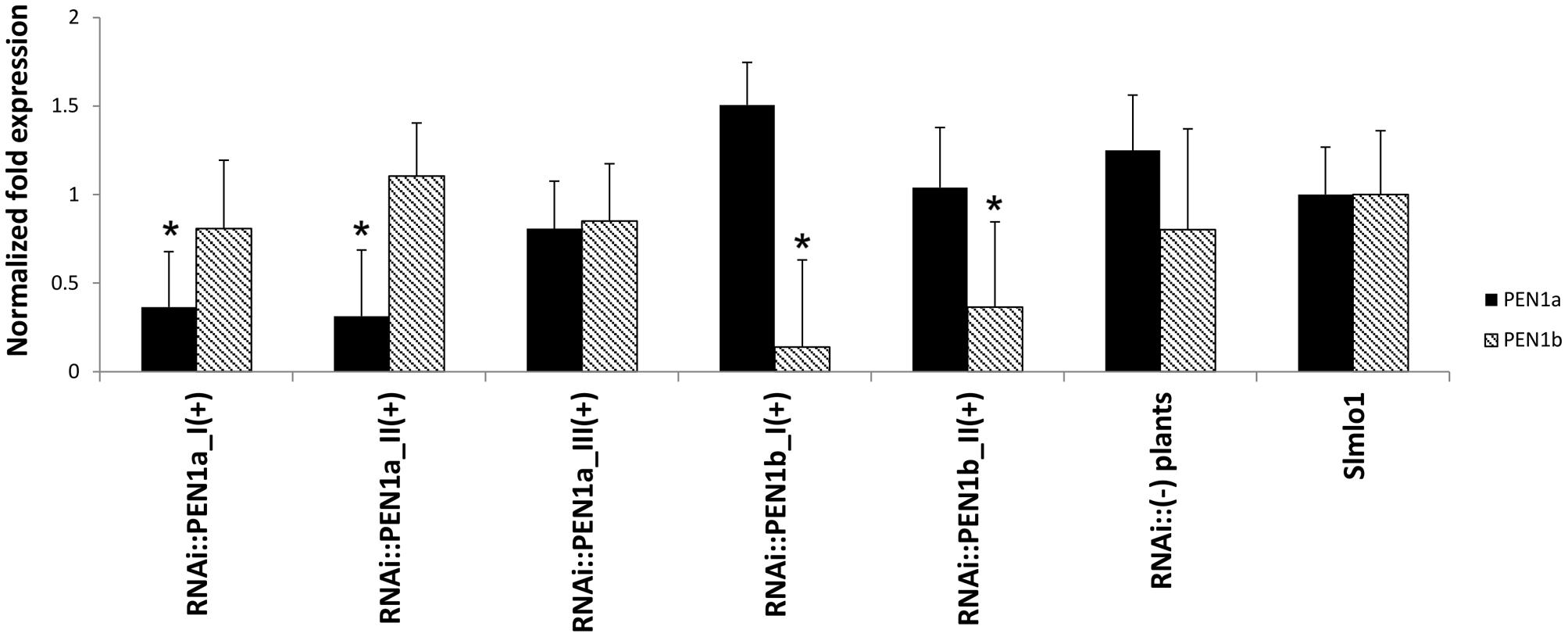
FIGURE 2. Relative expression of SlPEN1a and SlPEN1b in RNAi transformants obtained from the genetic background of the Slmlo1 line. Data refer to 15 transgenic individuals of three T2 families (I, II, and III) for the RNAi::SlPEN1a silencing construct and two T2 families (I and II) for the RNAi::SlPEN1b. The columns indicated as RNAi::(-) plants and Slmlo1 refer to five individuals, respectively. Bars indicate the standard errors. Asterisks indicate significant differences with the Slmlo1 line, inferred by Student’s t-test (p < 0.05).
A total of five non-transgenic individuals segregating from the five T2 families were grouped to evaluate the expression of SlPEN1a and SlPEN1b. The expression of these genes did not significantly change compared to Slmlo1 plants (Figure 2).
Aimed to confirm our results, the four families showing high silencing level of the target genes (RNAi::PEN1a_I and _II; RNAi::PEN1b_I and _II) were selected for a second disease test (Figure 3). Compared to the Slmlo1 plants, RNAi::SlPEN1a(+) individuals showed once more clear fungal sporulation (Figure 3A) and significantly increased fungal biomass (Figure 3B). Although RNAi::SlPEN1b(+) individuals showed slightly higher DI than Slmlo1 plants, fungal biomass did not significantly increase (Figure 3B). Non-transgenic individuals of the four T2 families displayed the same DI of Slmlo1 individuals (Figure 3).
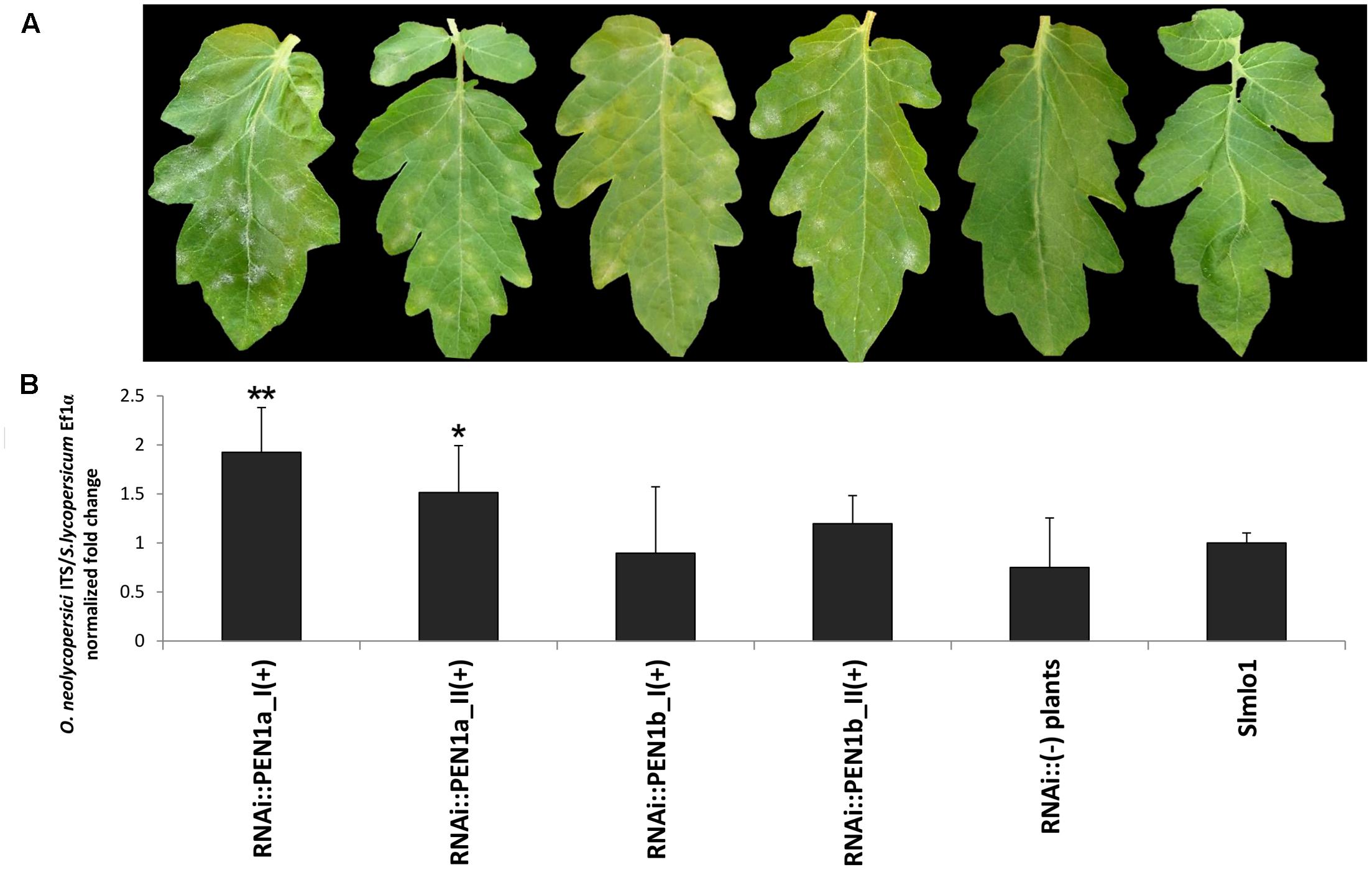
FIGURE 3. Effect of RNAi silencing of tomato SlPEN1a and SlPEN1b in the genetic background of the Slmlo1 line on the growth of Oidium neolycopersici. (A) Shows the phenotype of leaves collected 15 days after pathogen inoculation. From left to right: selected transgenic individuals of two independent T2 families carrying the SlPEN1a silencing construct [RNAi::SlPEN1a_I(+) and RNAi::SlPEN1a_II(+)], selected individuals of two independent T2 families carrying the SlPEN1b silencing construct [RNAi::SlPEN1b_I(+) and RNAi::SlPEN1b_II(+)], a non-transgenic plant segregating from one of the T2 families, and an individual of the Slmlo1 resistant line. (B) Refers to the relative quantification of the ratio between O. neolycopersici and tomato gDNAs in 15 transgenic individuals of the same families above mentioned, five non-transgenic T2 individuals, and five Slmlo1 individuals. Bars indicate the standard errors. Asterisks refer to significant differences with the Slmlo1 plants, inferred by Student’s t-test (∗p < 0.05; ∗∗p < 0.01).
To further investigate the functional role of SlPEN1b and its relation with SlPEN1a, we performed an additional experiment using the F1 progeny obtained by crossing well-silenced RNAi::SlPEN1a_I(+) and RNAi::SlPEN1b_I(+) individuals. Significantly reduced expression level of the target gene was observed in the eight F1 individuals and the selfing progenies of their parents, which were included in the same experiment (Figure 4). Compared to the control group of eight Slmlo1 plants, the F1 progeny and RNAi::SlPEN1a(+) plants showed a significantly higher fungal biomass. However, F1 individuals were not significantly more susceptible than plants carrying the RNAi::SlPEN1a(+) construct (data not shown).
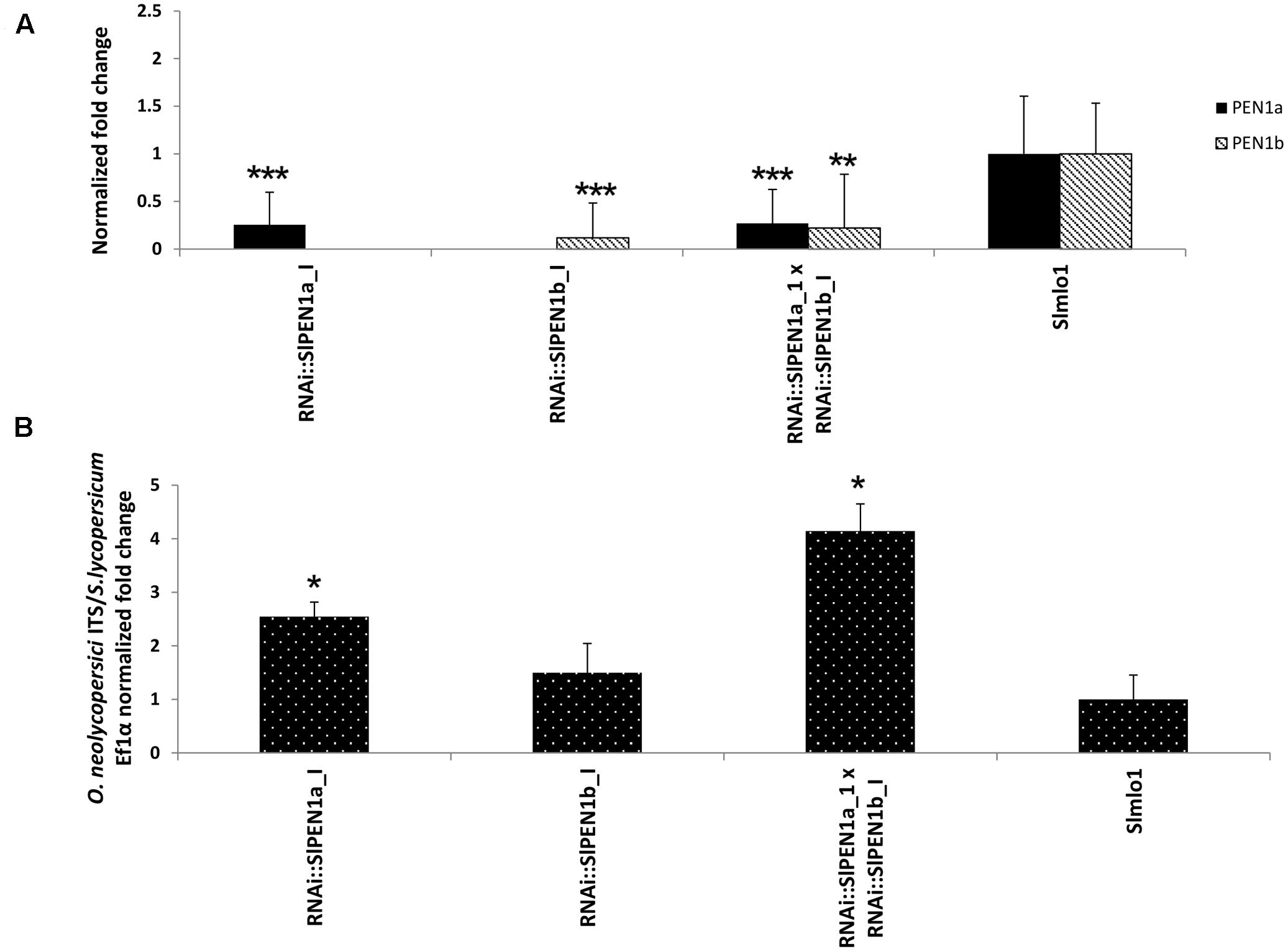
FIGURE 4. Effect of silencing both SlPEN1a and SlPEN1b [RNAi::SlPEN1a_I(+) × RNAi::SlPEN1b_I(+)] in the genetic background of the Slmlo1 line on the growth of O. neolycopersici. (A) Shows the expression of SlPEN1a and SlPEN1b evaluated on 14 individuals carrying the RNAi::SlPEN1a_I construct, 19 carrying the RNAi::SlPEN1b_I and eight individuals of the F1 progeny RNAi::SlPEN1a_I (+) × RNAi::SlPEN1b_I(+), compared to eight Slmlo1 plants. (B) Shows the quantification of fungal growth on the same dataset of plants. In both panels, bars indicate standard errors. Asterisks refer to significant differences, inferred by Student’s t-test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Functional Characterization of SlPEN1a and SlPEN1b in a Non-host Interaction
Since it has been shown that PEN genes are involved in resistance to non-adapted powdery mildew species, we set-up an assay to investigate the role of the two tomato syntaxins, SlPEN1a and SlPEN1b, in the interaction with barley powdery mildew fungus Bgh. An artificial inoculum of this pathogen was used to inoculate RNAi::SlPEN1a_I(+) and RNAi::SlPEN1b_I(+) individuals, together with Slmlo1 plants. An histological analysis was performed to observe if pathogen entry rate changed in these transgenic plants. We noticed that the percentage of infection units showing HR increased dramatically from 22 to 72% in RNAi::SlPEN1a-I(+) individuals compared to the Slmlo1 line (Figure 5). A slight increase (2.5%) of HR was also found in RNAi::SlPEN1b_I(+) plants, although this value was not statistically different from the one of the Slmlo1 line (Figure 5). The increased HR rate in RNAi::SlPEN1a_I(+) plants suggests that silencing SlPEN1a favors Bgh penetration. Indeed, HR has been assumed to act as a post-penetration defense mechanism in non-host interactions (Lipka et al., 2005).
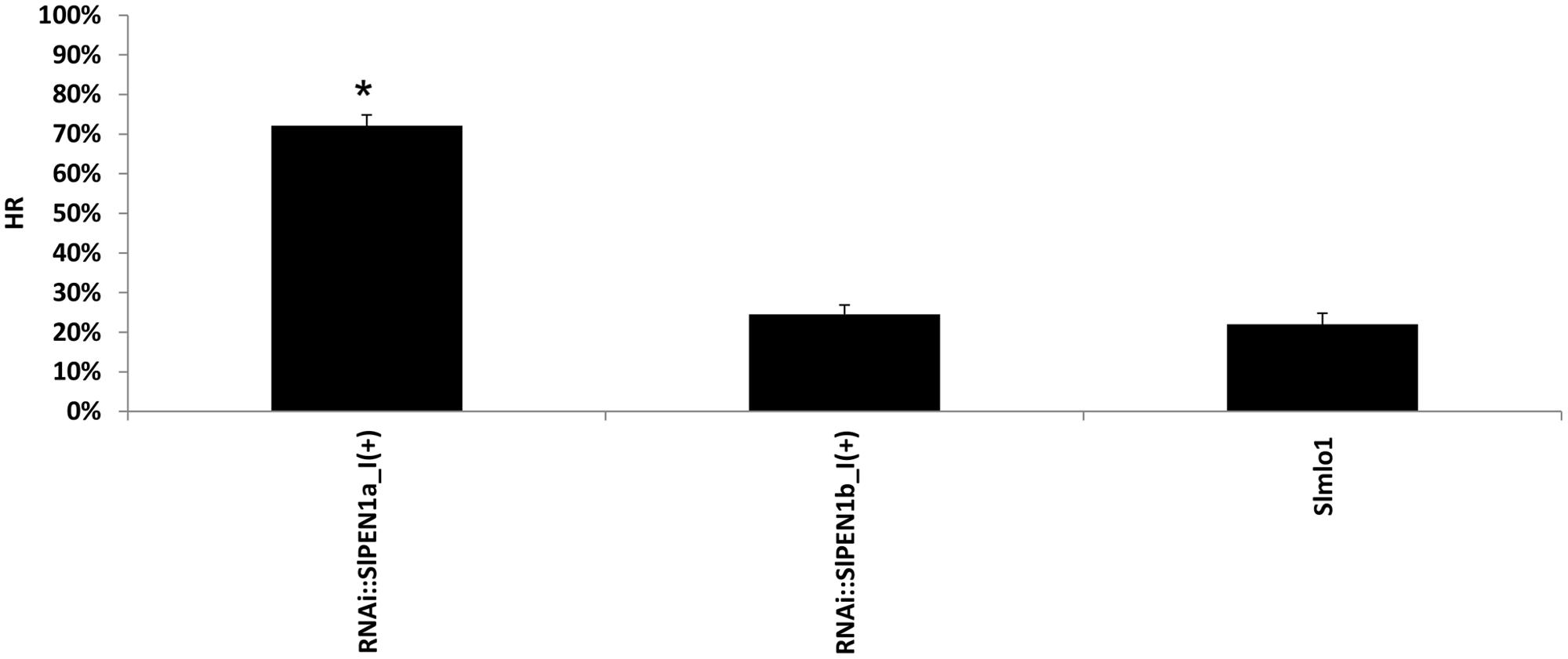
FIGURE 5. Effect of RNAi silencing of tomato SlPEN1a and SlPEN1b on the interaction with the non-adapted powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh). The graph shows the percentage of infection units causing hypersensitive response on the Slmlo1 line and on transgenic individuals silenced for SlPEN1a and SlPEN1b [RNAi::SlPEN1a_I(+) and RNAi::SlPEN1b_I(+)]. Bars refer to standard errors calculated on three biological replicates. Asterisks indicate significant difference with the Slmlo1 line inferred by the Student’s t-test (p ≤ 0.05).
Identification of Molecular Features Putatively Required for Syntaxin Function
Aiming to investigate the magnitude and direction of natural selection acting on syntaxins involved in defense against powdery mildew fungi, we used the nucleotide sequence of SYP-1b homologs for a codon-based SLAC evolutionary analysis. This analysis is based on the dissimilarity level between non-synonymous substitution (dN) and synonymous substitution (dS) values. Evidence for negative selection was found on 77 codons, associated with amino acid residues scattered in various syntaxin domains (Qa-SNARE, membrane-spanning, and the three auto-inhibitory helix domains known as Ha, Hb, Hc) (Figure 6 and Supplementary Table S2). Alignment of the SYP-1b subclade proteins revealed that 65 of these residues are invariable throughout the dataset, suggesting they might play a crucial role for protein function.
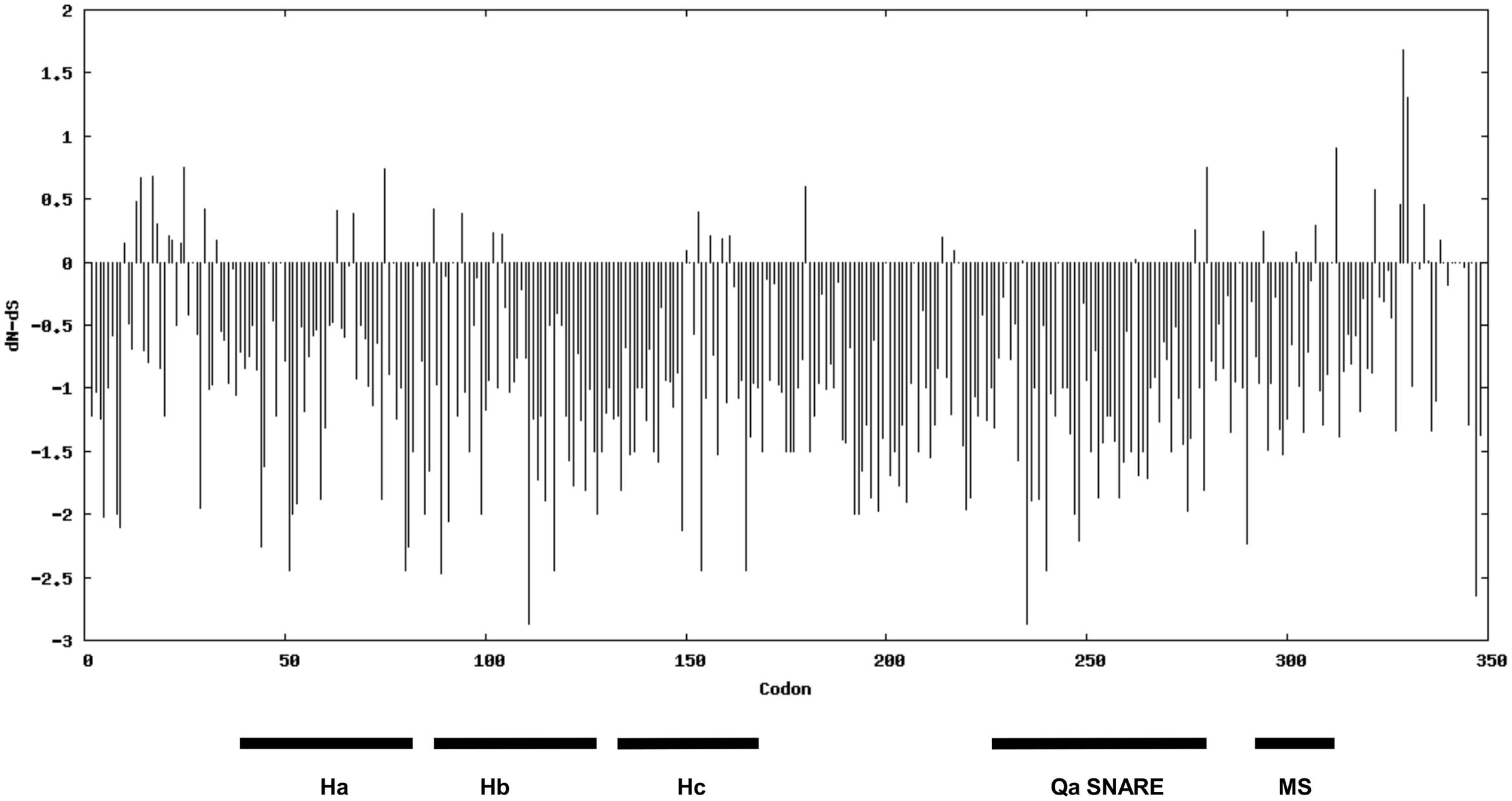
FIGURE 6. Direction and magnitude of natural selection acting on the syntaxins belonging to the SYP1b subclade. The bars indicate dN-dS values. The position of characteristic domains of syntaxins (the three helix domain Ha, Hb and Hc, the Qa-SNARE domain and the membrane-spanning (MS) domain are indicated.
We performed an additional alignment with the same set of syntaxins used for phylogenetic analysis. In total, we found that 24 out of 65 residues that are invariable in the SYP-1b subclade are not conserved in other syntaxins, thus pointing to the identification of residues that are, possibly, specifically important for pathogen defense (Supplementary Figure S1). In addition, we noticed two amino acid differences (highlighted in red) between the proteins playing a major role in penetration defense (AtPEN1/SlPEN1a/VvPEN1/HvROR2) and the other two proteins of subclade SYP-1b (AtSYP122/SlPEN1b) (Supplementary Figure S2).
Discussion
The Tomato Syntaxin Family
In this study, a genome-wide survey allowed the identification of 21 syntaxins in the tomato genome. The number of tomato homologs is consistent with similar genome-wide studies showing that A. thaliana contains 18 syntaxins, O. sativa 14 and P. trichocarpa 22 (International Rice Genome Sequencing Project, 2005; Tuskan et al., 2006; Lipka et al., 2007). The identification of 10 putative tomato SYP1 members corroborates previous findings indicating that this subfamily, containing homologs localized in the plasma membrane, is more represented than the other subfamilies SYP2, -3, -4, and -8 in plants (Lipka et al., 2007) (Supplementary Table S3).
It is known that specific SYP1 homologs in Arabidopsis are involved in other physiological processes besides defense against pathogens. Specifically, SYP111 controls cytokinesis (Lauber et al., 1997), whereas SYP123 and SYP132 mediate root hair tip growth (Ichikawa et al., 2014). Furthermore, the SYP132 ortholog in Medicago truncatula homolog (MtSYP132) and Nicotiana benthamiana (NtSYP132) were found to be involved in the symbiosome formation of Sinorhizobium meliloti and Pseudomonas syringae resistance, respectively (Catalano et al., 2007; Kalde et al., 2007; Limpens et al., 2009). Phylogenetic analysis presented in this study suggests that the tomato syntaxin Solyc06g053760 is an ortholog of Arabidopsis SYP111. Evolutionary relatedness between SYP132 and a couple of tomato syntaxins, Solyc01g056810 and Solyc10g081580, might indicate that these genes are SYP132 co-orthologs due to a recent duplication event after the split of two lineages from an Arabidopsis-tomato common ancestor.
Involvement of a Tomato Syntaxin in Plant-PM Pathogen Interaction
Two tomato syntaxins, SlPEN1a and SlPEN1b, are grouped in the phylogenetic clade also containing homologs previously reported to play a major (AtPEN1, HvROR2, and VvPEN1) or minor (AtSYP122) role in penetration resistance to powdery mildew fungi (Figure 1). Silencing of SlPEN1a compromised penetration resistance to both adapted and non-adapted powdery mildew pathogens in the Slmlo1 tomato line, showing that SlPEN1a is likely the functional ortholog of HvROR2, AtPEN1 and VvPEN1. This information might be useful for tomato breeding and for future studies on plant–pathogen interaction using tomato as model organism.
Similarly to barley, we found that the impairment of the tomato SlPEN1a syntaxin in the Slmlo1 background macroscopically restored the susceptibility to the powdery mildew disease. In contrast, the Atmlo2pen1 double mutant, although being characterized by wild-type levels of fungal penetration, still displayed a macroscopically resistant phenotype (Consonni et al., 2006). AtPEN1 was shown to act as a negative regulator of the hormone salicylic acid, known to enhance post-penetration defense mechanisms against biotrophic (Zhang et al., 2007). Possibly, differently from AtPEN1, SlPEN1a and barley HvROR2 have minor or no role in the regulation of salicylic-acid-mediated defense pathways. However, further testing needs to be done to support this hypothesis.
The Atpen1Atsyp122 double knock-out mutant displays a severely stunted and necrotic phenotype (Assaad et al., 2004). In the present study, transgenic individuals, silencing both SlPEN1a and SlPEN1b homologs, were devoid of markedly visible pleiotropic effects which might be due to residual gene expression. Alternatively, they may not affect the growth and development of the plant as their corresponding Arabidopsis orthologs. Also in this case, further tests are required to prove this hypothesis.
Amino Acids of Syntaxins Playing a Potential Role in Penetration Resistance
In this work, a codon-based evolutionary analysis allowed us to detect 77 codons under significant negative selection in the SYP-1b subclade, containing the syntaxins associated with defense against powdery mildew fungi. Moreover, alignment of all the 41 syntaxin sequences pointed out the occurrence of 24 residues specifically conserved in the SYP-1b subclade. Three of these residues are located in the Qa-SNARE domain, whereas 10 are found in the auto-inhibitory motifs Ha, Hb or Hc helices of Qa-SNARE proteins. These helices, when folded in a closed conformation, prevent the exposure of the Qa-SNARE domain and thus the formation of SNARE complexes (Collins et al., 2003). In barley ror2 mutant, a deletion of 31 amino acids (S118-E148) of the Hc helix occurred, which might have led to a constitutively open state of the protein resulting in enhanced binding to HvSNAP34 (Collins et al., 2003). Interestingly, the deletion contains four of the 24 conserved residues identified in this study, namely G123, P124, T133 and G138, in HvROR2 sequence, suggesting an important function of these residues.
Besides the above discussed 24 amino acids specifically conserved in the SYP-1b subclade, we pointed out two different amino acids present in AtSYP122 and SlPEN1b but not in any of the four SYP1b syntaxins with well-characterized role in plant–pathogen interactions, namely A111 and I286 (Supplementary Figure S2). In the study of Pajonk et al. (2008), authors obtained a chimeric syntaxin by swapping the first 175 amino acids of the AtPEN1 N-terminal domain (including the residue A111), with the corresponding sequence of AtSYP122. The construct was then used to transform pen1 mutant. The authors did not observe any difference in the level of Bgh entry rate between the obtained transformants and their corresponding genetic background. This observation suggests that N-terminal region of the AtPEN1 protein contains amino acids of critical importance for its functionality, possibly including the A111 residue mentioned above. Future functional analyses, aimed at targeting these amino acids, might unravel their importance toward the functional specialization observed between AtPEN1/ SlPEN1a/VvPEN1/HvROR2 and AtSYP122/SlPEN1b.
Author Contributions
Conceived and designed the experiments: MA, VB, SP, and YB. Performed the experiments: VB, MA, ZY, and ZZ. Analyzed the data: MA, VB, and SP. Contributed reagents/materials/ analysis tools: LR and RV. Wrote and edited the paper: VB, MA, SP, A-MW, RV, and YB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer PS-B and handling Editor declared their shared affiliation.
Acknowledgments
The work of ZZ is supported by the Chinese Academy of Agricultural Sciences Fundamental Research Budget Increment Project (Grant No. 2015ZL008), The Agricultural Science and Technology Innovation Program (Grant No. CAAS-ASTIP-2013-IVFCAAS) and the Merit-based Scientific Research Foundation of the State Ministry of Human Resources and Social Security of China for Returned Overseas Chinese Scholars (Grant No. 2015-192).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01573/full#supplementary-material
Footnotes
References
Appiano, M., Pavan, S., Catalano, D., Zheng, Z., Bracuto, V., Lotti, C., et al. (2015). Identification of candidate MLO powdery mildew susceptibility genes in cultivated Solanaceae and functional characterization of tobacco NtMLO1. Transgenic Res. 24, 847–858. doi: 10.1007/s11248-015-9878-4
Assaad, F., Qiu, J. L., Youngs, H., Ehrhardt, D., Zimmerli, L., Kalde, M., et al. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15, 5118–5129. doi: 10.1091/mbc.E04-02-0140
Bai, Y., Pavan, S., Zheng, Z., Zappel, N. F., Reinstädler, A., Lotti, C., et al. (2008). Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of Mlo function. Mol. Plant Microbe Interact. 21, 30–39. doi: 10.1094/MPMI-21-1-0030
Bock, J. B., Matern, H. T., Peden, A. A., and Scheller, R. H. (2001). A genomic perspective on membrane compartment organization. Nature 409, 839–841. doi: 10.1038/35057024
Catalano, C. M., Czymmek, K. J., Gann, J. G., and Sherrier, D. J. (2007). Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta 225, 541–550. doi: 10.1007/s00425-006-0369-y
Collins, N. C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J. L., et al. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. doi: 10.1038/nature02076
Consonni, C., Humphry, M. E., Hartmann, H. A., Livaja, M., Durner, J., Westphal, L., et al. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720. doi: 10.1038/ng1806
Fasshauer, D., Sutton, R. B., Brunger, A. T., and Jahn, R. (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. U.S.A. 95, 15781–15786. doi: 10.1073/pnas.95.26.15781
Feechan, A., Jermakow, A. M., Ivancevic, A., Godfrey, D., Pak, H., Panstruga, R., et al. (2013). Host cell entry of powdery mildew is correlated with endosomal transport of antagonistically acting VvPEN1 and VvMLO to the papilla. Mol. Plant Microbe Interact. 26, 1138–1150. doi: 10.1094/MPMI-04-13-0091-R
Freialdenhoven, A., Peterhansel, C., Kurth, J., Kreuzaler, F., and Schulze-Lefert, P. (1996). Identification of genes required for the function of non-race-specific mlo resistance to powdery mildew in barley. Plant Cell 8, 5–14. doi: 10.1105/tpc.8.1.5
Helliwell, C., and Waterhouse, P. (2003). Constructs and methods for high-throughput gene silencing in plants. Methods 30, 289–295. doi: 10.1016/S1046-2023(03)00036-7
Huibers, R. P., Loonen, A. E. H. M., Gao, D., Van den Ackerveken, G., Visser, R. G. F., and Bai, Y. (2013). Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLOS ONE 8:e67467. doi: 10.1371/journal.pone.0067467
Ichikawa, M., Hirano, T., Enami, K., Fuselier, T., Kato, N., Kwon, C., et al. (2014). Syntaxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana. Plant Cell Physiol. 55, 790–800. doi: 10.1093/pcp/pcu048
International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Kalde, M., Nühse, T. S., Findlay, K., and Peck, S. C. (2007). The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. U.S.A. 104, 11850–11855. doi: 10.1073/pnas.0701083104
Lauber, M. H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., et al. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485–1493. doi: 10.1083/jcb.139.6.1485
Limpens, E., Ivanov, S., van Esse, W., Voets, G., Fedorova, E., and Bisseling, T. (2009). Medicago N(2)-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 21, 2811–2828. doi: 10.1105/tpc.108.064410
Lipka, V., Dittgen, J., Bednarek, P., Bhat, R., Wiermer, M., Stein, M., et al. (2005). Plant science: Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183. doi: 10.1126/science.1119409
Lipka, V., Kwon, C., and Panstruga, R. (2007). SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 23, 147–174. doi: 10.1146/annurev.cellbio.23.090506.123529
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Løvdal, T., and Lillo, C. (2009). Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 387, 238–242. doi: 10.1016/j.ab.2009.01.024
Pajonk, S., Kwon, C., Clemens, N., Panstruga, R., and Schulze-Lefert, P. (2008). Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J. Biol. Chem. 283, 26974–26984. doi: 10.1074/jbc.M805236200
Pavan, S., Jacobsen, E., Visser, R. G., and Bai, Y. (2010). Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 25, 1–12. doi: 10.1007/s11032-009-9323-6
Pavan, S., Zheng, Z., Borisova, M., Van Den Berg, P., Lotti, C., De Giovanni, C., et al. (2008). Map- vs. homology-based cloning for the recessive gene ol-2 conferring resistance to tomato powdery mildew. Euphytica 162, 91–98. doi: 10.1007/s10681-007-9570-8
Peterhänsel, C., Freialdenhoven, A., Kurth, J., Kolsch, R., and Schulze-Lefert, P. (1997). Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. Plant Cell 9, 1397–1409. doi: 10.1105/tpc.9.8.1397
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. doi: 10.1093/nar/29.9.e45
Sanderfoot, A. A., Assaad, F. F., and Raikhel, N. V. (2000). The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124, 1558–1569. doi: 10.1104/pp.124.4.1558
Schilde, C., Lutter, K., Kissmehl, R., and Plattner, H. (2008). Molecular identification of a SNAP-25-like SNARE protein in Paramecium. Eukaryot. Cell 7, 1387–1402. doi: 10.1128/EC.00012-08
Seifi, A., Gao, D., Zheng, Z., Pavan, S., Faino, L., Visser, R. F., et al. (2014). Genetics and molecular mechanisms of resistance to powdery mildews in tomato (Solanum lycopersicum) and its wild relatives. Eur. J. Plant Pathol. 138, 641–665. doi: 10.1007/s10658-013-0314-4
Söllner, T., Whiteheart, S. W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P., et al. (1993). SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324. doi: 10.1038/362318a0
Takamatsu, S. (2004). Phylogeny and evolution of the powdery mildew fungi (Erysiphales, Ascomycota) inferred from nuclear ribosomal DNA sequences. Mycoscience 45, 147–157. doi: 10.1007/S10267-003-0159-3
Trujillo, M., Troeger, M., Niks, R. E., Kogel, K. H., and Hückelhoven, R. (2004). Mechanistic and genetic overlap of barley host and non-host resistance to Blumeria graminis. Mol. Plant Pathol. 5, 389–396. doi: 10.1111/j.1364-3703.2004.00238.x
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115. doi: 10.1093/nar/gks596
Zhang, Z., Feechan, A., Pedersen, C., Newman, M. A., Qiu, J., Olesen, K. L., et al. (2007). A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49, 302–312. doi: 10.1111/j.1365-313X.2006.02961.x
Keywords: mlo resistance, non-host resistance, Blumeria graminis f. sp. hordei, Oidium neolycopersici, tomato syntaxins
Citation: Bracuto V, Appiano M, Zheng Z, Wolters A-MA, Yan Z, Ricciardi L, Visser RGF, Pavan S and Bai Y (2017) Functional Characterization of a Syntaxin Involved in Tomato (Solanum lycopersicum) Resistance against Powdery Mildew. Front. Plant Sci. 8:1573. doi: 10.3389/fpls.2017.01573
Received: 23 May 2017; Accepted: 28 August 2017;
Published: 20 September 2017.
Edited by:
Víctor Flors, Jaume I University, SpainReviewed by:
Paloma Sanchez-Bel, Jaume I University, SpainHans Thordal-Christensen, University of Copenhagen Faculty of Science, Denmark
Copyright © 2017 Bracuto, Appiano, Zheng, Wolters, Yan, Ricciardi, Visser, Pavan and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuling Bai, bai.yuling@wur.nl
†These authors have contributed equally to this work.
 Valentina Bracuto
Valentina Bracuto Michela Appiano
Michela Appiano Zheng Zheng
Zheng Zheng Anne-Marie A. Wolters2
Anne-Marie A. Wolters2 Richard G. F. Visser
Richard G. F. Visser Stefano Pavan
Stefano Pavan Yuling Bai
Yuling Bai