- 1College of Plant Protection, Shenyang Agricultural University, Shenyang, China
- 2Department of Life Sciences, Chung-Ang University, Seoul, South Korea
Ammonium transporter (AMT) proteins have been reported in many plants, but no comprehensive analysis was performed in wheat. In this study, we identified 23 AMT members (hereafter TaAMTs) using a protein homology search in wheat genome. Tissue-specific expression analysis showed that TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a were relatively more highly expressed in comparison with other TaAMTs. TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a-GFP were localized in the plasma membrane in tobacco leaves, and TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a successfully complemented a yeast 31019b strain in which ammonium uptake was deficient. In addition, the expression of TaAMT1;1b in an Arabidopsis AMT quadruple mutant (qko) successfully restored uptake ability. Resupply of rapidly increased cellular contents and suppressed expression of TaAMT1;3a, but not of TaAMT;1;1a and TaAMT1;1b expressions. Expression of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a was not changed in leaves after resupply. In contrast, nitrogen (N) deprivation induced TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a gene expressions in the roots and leaves. Expression analysis in the leaves of the stem rust-susceptible wheat line “Little Club” and the rust-tolerant strain “Mini 2761” revealed that TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a were specifically induced in the former but not in the latter. Rust-susceptible wheat plants grown under N-free conditions exhibited a lower disease index than plants grown with as the sole N source in the medium after infection with Puccinia graminis f. sp. tritici, suggesting that and its transport may facilitate the infection of wheat stem rust disease. Our findings may be important for understanding the potential function TaAMTs in wheat plants.
Introduction
In most soils, nitrate () and ammonium () represent the major forms of nitrogen (N) uptake in higher plants. The ions accumulate in cells either by direct uptake from the rhizosphere via ammonium transporters (AMTs) or by reduction of . The is then assimilated into glutamate via the glutamine synthetase/glutamate synthase (GS/GOGAT) cycle. Glutamine and asparagine have been identified as the major forms of organic N in the xylem and are translocated from the roots to the shoots (Fukumorita and Chino, 1982). The energy cost of reducing to involves the consumption of 12–26% of photosynthetically generated reductants. Therefore, the use of as an N source conserves a large amount of energy for plants (Bloom, 1997; Noctor and Foyer, 1998; Patterson et al., 2010). AMT genes have been identified in many plant species including Arabidopsis thaliana (Ninnemann et al., 1994; Gazzarrini et al., 1999; Sohlenkamp et al., 2000, 2002; Loqué and von Wirén, 2004; Yuan et al., 2007a, 2009), Lycopersicon esculentum (Lauter et al., 1996; Von Wirén et al., 2000), Lotus japonicus (Salvemini et al., 2001; Simon-Rosin et al., 2003; D’Apuzzo et al., 2004), Brassica napus (Pearson et al., 2002), Oryza sativa (Sonoda et al., 2003), Zea mays (Gu et al., 2013), and Sorghum bicolor (Koegel et al., 2013). In wheat, AMT TaAMT1;1 was stimulated by an acidic pH in vitro (Sogaard et al., 2009). In addition, three TaAMTs identified their transcriptional regulation under arbuscular mycorrhizal (AM) fungi infection (Duan et al., 2015). However, no further information regarding wheat AMTs has been reported. Although is an energetically favorable N source, various plants exhibit toxic symptoms in response to high concentrations (Britto and Kronzucker, 2002).
In contrast to other grasses, rice is tolerant of and the tolerance relies on an energetically favorable equilibration of influx and efflux at elevated supplies (Britto et al., 2001). The uptake of high affinity into root cells is mediated by AMT type transporters, encompassing a family of 10 AMT paralogs that have been classified into four subfamilies in rice (Suenaga et al., 2003; Loqué and von Wirén, 2004; Ye et al., 2016). Among them, AMT1;1, AMT1;2, and AMT1;3 are of particular importance; AMT1;1 is constitutively expressed in the shoots and roots, while AMT1;2 and AMT1;3 are specifically expressed in the roots (Sonoda et al., 2003). Arabidopsis is a -sensitive species, and all AtAMT1 (AtAMT1;1, AtAMT1;2, and AtAMT1;3) and AtAMT2;1 gene expression are suppressed at high concentrations (Sohlenkamp et al., 2000; Loqué et al., 2006). Furthermore, post-transcriptional and post-translational regulation in AMTs has been reported. In particular, the -mediated phosphorylation at T460 in the cytosolic tail of AMT1;1 has been found to inhibit transporter activity in A. thaliana (Yuan et al., 2007b; Lanquar et al., 2009). In rice, the transcriptional regulation of AMT genes is dependent on the N nutritional status of the plant and on the external availability of different N forms. AMT1;1 and AMT1;2 are up-regulated in response to ; however, AMT1;3 is up-regulated by N deprivation (Kumar et al., 2003; Sonoda et al., 2003). The overexpression of AMT1;1 enhanced uptake and improved plant growth and yield production in rice, at least under specialized N fertilization conditions (Ranathunge et al., 2014). In contrast, the overexpression of AMT1;3 resulted in poor growth and reduced uptake in rice (Bao et al., 2015). As rice plants use as a favorable N source, the importance of AMTs is evident, though their biological function remains unclear. The only information regarding the function of AMT genes is that -triggered lateral root branching is controlled by AMT1;3 in Arabidopsis (Lima et al., 2010).
Nitrogen status is closely associated with plant defense. In rice, treatment of the roots with glutamate induces systemic resistance to rice blast disease, partially through salicylic acid signaling (Kadotani et al., 2016). In Arabidopsis, AMT1;1 alters basal defense, generating resistance against Pseudomonas syringae and Plectosphaerella cucumerina (Pastor et al., 2014), and the expressions of AMT1;1, AMT1;2, AMT1;3, and AMT2;1 have been found to be altered by both biotic and abiotic stresses (Fagard et al., 2014). In sorghum, the expression of SbAMT3;1 and SbAMT4 was greatly induced locally in roots colonized by AM fungi (Koegel et al., 2013). Wheat stem rust is one of the most serious diseases of wheat worldwide (Pardey et al., 2013). In China, it has been effectively controlled through the development of resistant cultivars and effective resistant genes, particularly the 1B/1R translocation gene Sr31, in different epidemiological regions since the 1970s (Cao and Chen, 2010). However, a new strain of stem rust pathogen designated as Ug99 (TTKS), expressing virulence to Sr31, was first identified in Uganda in 1998 (Pretorius et al., 2000). It has since spread throughout the major wheat growing regions of the world such as Ethiopia, Zimbabwe, Mozambique, Kenya, Sudan, Yemen, and Tanzania (Singh et al., 2008). Ug99 and related strains threaten global wheat production because they are virulent on widely used cultivars that had otherwise been effective for many years (Yu et al., 2010). This has initiated renewed genetic research into wheat to identify tolerant strains and the regulatory mechanism thereof.
In the present study, we identified and characterized TaAMTs in wheat plants. The homology of 23 TaAMTs was compared with AMTs from other species and their tissue-specific or -mediated expressions were analyzed. In addition, the localization of TaAMT1;1 proteins and their functions were analyzed using a yeast uptake deficient strain and an Arabidopsis AMT qko mutant. TaAMT1;1 expression in wheat was also examined upon Puccinia graminis f. sp. tritici (Pgt) infection.
Materials and Methods
Plant Growth
Arabidopsis seeds (qko, qko+AtAMT1;1, and qko+TaAMT1;1b) (Yuan et al., 2007a) were surface sterilized and kept in a 4°C chamber for 2 days. The seeds were planted in modified 0.5× MS medium containing 1 mM KNO3 as the sole N source. Arabidopsis were cultured in the chamber at 22°C with 12 h/12 h: light/dark cycle. Three-day-old plants were transferred to the same medium containing 0 or 10 mM methyl-ammonium (MeA) and grown for another 4 days. In order to analyze the cellular contents, the Arabidopsis mutants (qko, qko+AtAMT1;1, and qko+TaAMT1;1b) were planted in 0.5× MS medium and grown for 7 days, after which their complete root systems from 30 plants were collected.
The stem rust-susceptible wheat (Triticum aestivum) line “Little Club” (LC) was used in the experiments to examine the effects of on TaAMT1 gene expression. Germinated seeds were grown in deionized water in a greenhouse for 2 weeks to consume all the nutrient solution in the endosperm. Wheat plants were cultured in the chamber at 21°C with 12/12: light/dark cycle. The seedlings were then grown for another 3 days in N-free nutrient solution (–N basal salt: 7 μM Na2HPO4, 16 μM KCl, 7 μM CaCl2.2H2O, 15 μM MgCl2.6H2O, 36 μM FeSO4.7H2O, 9 μM MnSO4.4H2O, 45 μM H3BO4, 3 μM ZnSO4.7H2O, 0.2 μM CuSO4.7H2O, 0.05 μM Na2MoO4.2H2O) (Abiko et al., 2005), after which they were transferred to a nutrient solution containing 0.5 mM (NH4)2SO4 at pH 5.5. Whole roots and leaves were harvested at 0, 1, 3, and 6 h following the provision of 0.5 mM (NH4)2SO4. Two-week-old LC plants grown in water were transferred to a nutrient solution (–N basal salt) containing 0.5 mM (NH4)2SO4. After 3 days of growth, the plants were transferred to the same nutrient solution without the N source (–N basal salt). Whole roots and leaves were collected after 0, 1, 2, and 3 days of N deprivation. Seventeen-day-old LC plant roots and leaves, as well as 2-month-old plant stems and flowers, were harvested for RNA extraction.
Stem Rust Infection
For inoculation of the urediniospores of Pgt (race 21C3CTHTM), the urediniospores were separately inoculated to stem rust-susceptible wheat (T. aestivum) “LC” and stem rust resistant line “Mini 2761” seedlings once the primary leaves had fully expanded while the secondary leaves were being sprouted. The urediniospores of Pgt (race 21C3CTHTM) were propagated by inoculation of LC leaves in the growth chamber. The inoculated seedlings were kept in moist conditions for 14–16 h, and then cultured at 21 ± 1°C, 12 h/12 h: light/dark cycle and a light intensity of 5.8–6.0 klx. The infection types were recorded according to six classes of standards (Supplementary Figure S1; Roelfs and Martens, 1988). To analyze N fertilization dependent stem rust disease index, seeds were grown in deionized water for 2 weeks before being transferred to –N basal salt containing 0.5 mM (NH4)2SO4 solution (pH 5.5) for another 2 weeks prior to inoculation of Pgt.
Molecular Phylogenetic Analysis Using Maximum Likelihood
To search AMT amino sequences in wheat, the rice, Arabidopsis, and potato AMT sequences were used as a bait and searched in the Uniprot database1 using BLAST with e-value cutoff of e-10 (Pearson, 2013). The retrieved sequences are listed in Supplementary Table S1. The multiple sequence alignments were performed by ClustalW and the evolutionary history was inferred using maximum likelihood based on the JTT matrix-based model. The tree with the highest log likelihood (-12,240.60) is built. Initial tree(s) for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model. The topology with the superior log likelihood value was then selected. The analysis involved 42 amino acid sequences. All positions containing gaps and missing data were eliminated. There was a total of 49 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016).
RNA Extraction and qRT-PCR
Total cellular RNA was isolated from 20 plant tissues with TRIzol (Takara, Dalian, Liaoning, China), and 2 μg of total RNA was subsequently treated with RQ1 RNase free DNase (Promega, Madison, WI, United States) to eliminate genomic DNA contamination. For cDNA synthesis, a GoScript Reverse Transcription Kit was used following the manufacturer’s instructions (Promega, Madison, WI, United States). Subsequently, qRT-PCR was performed in triplicate using the SYBR Green Mix (Bio-Rad). The three replicates of PCR in each time were analyzed for one sample and the experiments were repeated at least three times. The PCR products were quantified using the Illumina Research Quantity software Illumina Eco 3.0 (Illumina, San Diego, CA, United States). The values of each sample were first normalized against TaEF1α levels from the same samples, and next compared with indicated control group value to analyze the ratio for each gene. Changes in gene expression were calculated using the 2-ΔΔCT method (Han et al., 2006). The primers used for qRT-PCR are listed in Supplementary Table S2.
Yeast Complementation Assay
The ammonium uptake deficient yeast strain 31019b (Δmep1, Δmep2, Δmep3, ura3; Marini et al., 1997) was obtained from the Frommer Laboratory (Carnegie Institution for Science). A pDRf1-GW (Xuan et al., 2013) vector harboring TaAMT1;1a, TaAMT1;1b, or TaAMT1;3a was transformed into the yeast cells. The successful transformants were screened by growing of yeast cells in the SD/-ura solid medium. Each transformant was plated on yeast N base media containing 1 mM NH4Cl or 1 mM arginine, and yeast growth was monitored at 28°C for 3 days. The primers used for cloning the TaAMT genes are listed in Supplementary Table S1.
Arabidopsis qko Mutant Complementation
Nucleotides of TaAMT1;1b ORF were fused to 1.5 kb of the AtAMT1;1 promoter sequence and cloned into the pABind vector (Bleckmann et al., 2010). The pABind-pAtAMT1;1–TaAMT1;1b construct was transformed into the Arabidopsis qko mutant background via Agrobacterium-mediated transformation. qko mutant has mixed Col-0 and Ws-2 genomes; therefore, the qko+AtAMT1;1 was used as an control. The transgenic plant seeds were selected in the plates containing hygromycin, and around 20 individual plants were selected. For analyzing MeA uptake and cellular contents, three independent lines were further examined.
Localization of TaAMT1;1 in Plants
Nucleotides of TaAMT1;1 ORFs were cloned into a pABindGFP (35S promoter) destination plasmid (Bleckmann et al., 2010) followed by transient expression in Nicotiana benthamiana leaves using the Agrobacterium-mediated transient expression method (Kim et al., 2009). Green fluorescent protein fluorescence was detected under a confocal microscope (SP5; Leica).
Determination of Ammonium Contents
Enzymatic determination of the ammonium contents in roots was performed using an F-Kit (Roche) according to the manufacturer’s instructions (Oliveira et al., 2002).
Statistical Analysis
Statistical calculations were conducted using Prism 5 (GraphPad, San Diego, CA, United States). Significant differences between two groups were analyzed by Student’s t-test. Comparisons between more than two groups were performed by using one-way ANOVA followed by Bonferroni’s multiple comparison test. P-values of <0.05 were considered statistically significant.
Results
Identification of TaAMTs and Phylogenetic Relationships between AMTs
High affinity transport is mediated by AMT family transporters. To isolate wheat TaAMTs sequences, 10 rice, 6 Arabidopsis, and 3 potato AMT sequences were used as baits to obtain the amino sequences of AMTs in wheat. A total of 23 AMTs (among 25 identified in Uniprot database, 2 AMTs were duplicated) were identified in T. aestivum, and were designated as TaAMTs on the basis of their similarity to OsAMTs, AtAMTs, and LeAMTs (Figure 1 and Supplementary Table S1). In order to understand the evolutionary relationships between TaAMTs and AMTs from other plant species, the amino acid sequences of 10 AMTs from O. sativa, 6 AMTs from A. thaliana, and 3 AMTs from Solanum lycopersicum were collected and aligned. An unrooted phylogenetic ML tree was constructed and the results indicated that the AMTs from four species could be classified into four sub-groups (AMT1, AMT2, AMT3, and AMT4). Among these, all the AMT1 proteins clustered together with high bootstrap support (100%), while AMT2, AMT3, and AMT4 were closely associated in the phylogenetic tree. Furthermore, the tree topology was independent of the methods used for the phylogenetic reconstruction (data not shown). Rice and wheat were clustered more closely, whereas the tomato and Arabidopsis AMTs were clustered relatively distantly (Figure 1).
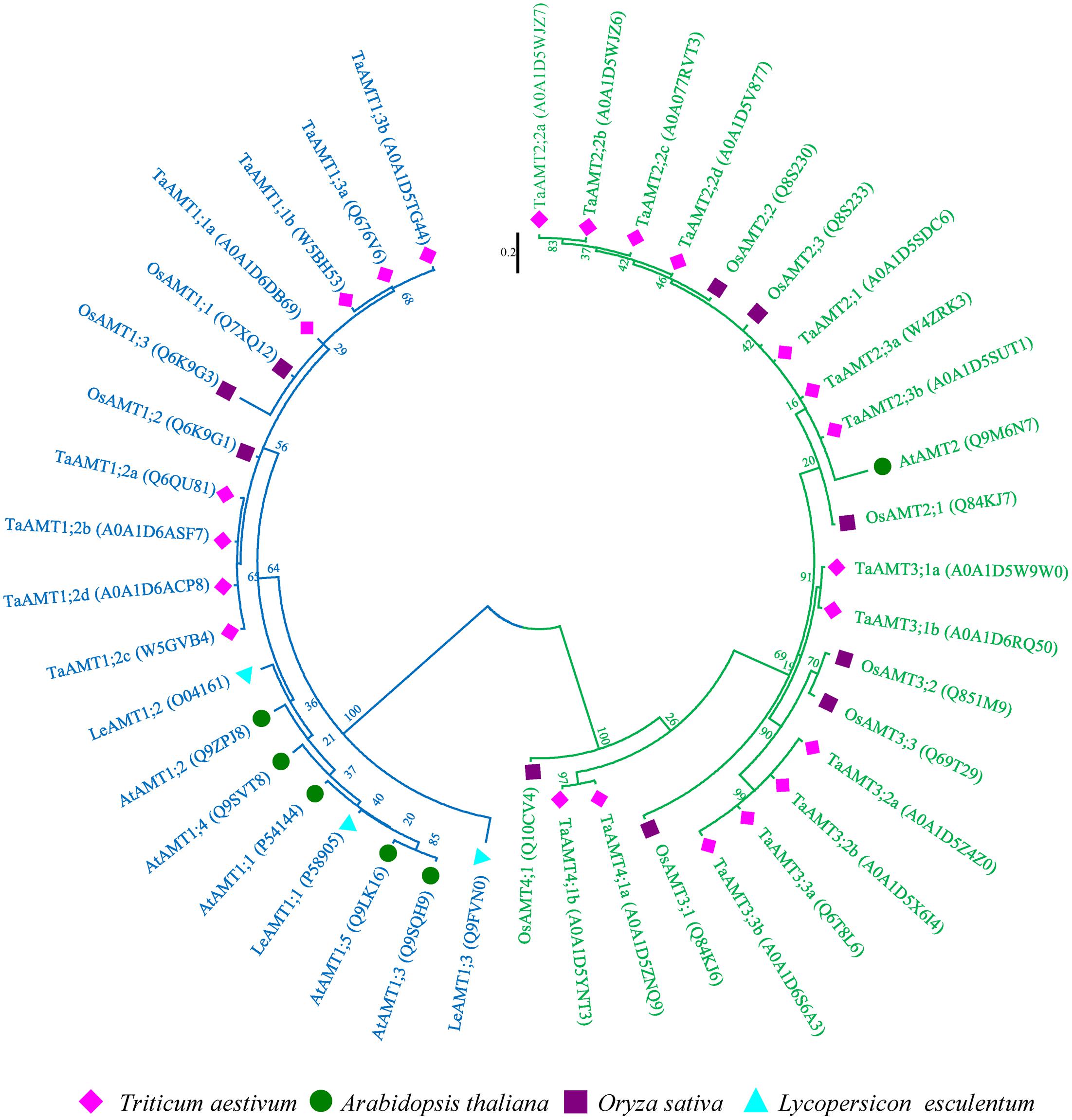
FIGURE 1. Phylogenetic tree of AMTs from Oryza sativa, Arabidopsis thaliana, Triticum aestivum, and Solanum lycopersicum. The phylogenetic tree was constructed using the maximum-likelihood method. The unrooted tree was generated using ClustalW in MEGA 7.0 using AMT amino acid sequences from O. sativa (OsAMTs; dark blue color), A. thaliana (AtAMTs; dark orange color), T. aestivum (TaAMTs; light green color), and S. lycopersicum (LeAMTs; light blue color). AMT1s are marked with blue and others are marked with green.
Tissue-Specific Expression of TaAMT Genes
To examine the expression patterns of TaAMT genes, the roots, leaves, stems, and flowers were collected for RNA extraction. qRT-PCR was performed to analyze 12 TaAMT gene expressions among the 23 TaAMT members, and the results showed that two TaAMT1;1 group genes (1;1a and 1;1b) and TaAMT1;3a were highly expressed, while the remainder of the TaAMT genes were barely detected (Figure 2). The TaAMT1;1 group genes (1;1a and 1;1b) and TaAMT1;3a exhibited similar patterns and showed the highest expression levels in the roots, while similar expression levels were found in the leaves, stems, and flower tissues. The expression levels of TaAMT1;2a and TaAMT1;2c were relatively higher in the leaves and roots than in the stems and flowers. The expression levels of the TaAMT3;2 group genes were not as high as the TaAMT1;1 group genes, and TaAMT3;3a exhibited the highest expression levels in the roots, and the lowest in the stems and flowers. The remaining genes either showed reduced expression levels or were not detected (Figure 2).
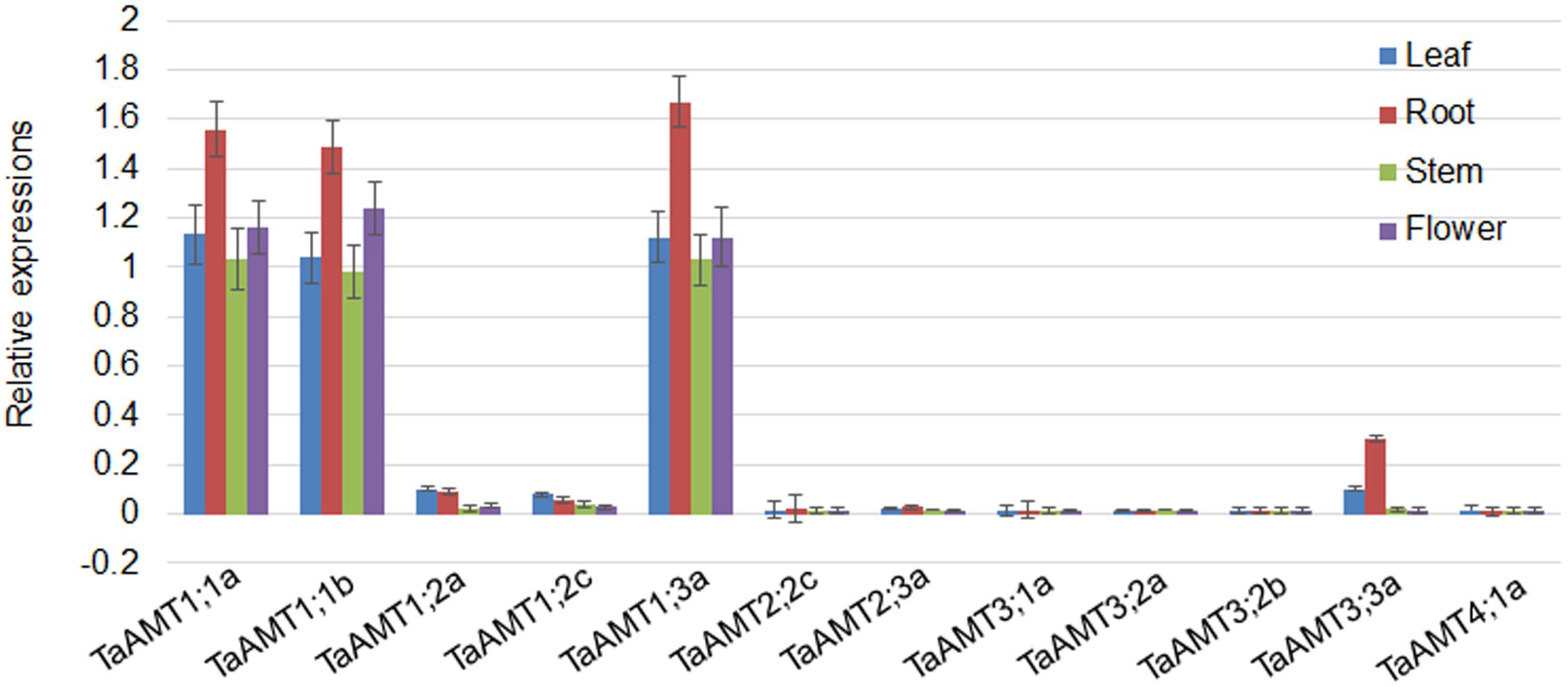
FIGURE 2. Tissue-specific expression of TaAMT genes in wheat. Expression levels of TaAMTs in the leaves, roots, stems, and flowers were analyzed. The expression patterns were analyzed by qRT-PCR. The TaEF1α gene was used as the internal control. The experiments were repeated three times.
Two TaAMT1;1 and TaATM1;3a Proteins Localize at the Plasma Membrane and Transport in Yeast
Since TaAMT1;1 group genes (1;1a and 1;1b) and TaAMT1;3a were the dominant TaAMT members, the subcellular localization of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a was monitored. GFP coding sequences were C-terminally fused to TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a and the fusion proteins were transiently expressed in N. benthamiana leaves via Agrobacterium-mediated transformation. Two days after infection, the TaAMT-GFP signal was detected on the plasma membrane (Figure 3A). TaAMTs conserved with other characterized AMTs from different plant species in a phylogenetic tree (Figure 1). Therefore, we examined their transport activity. Since the TaAMT1;1 group of genes and TaAMT1;3a were most strongly expressed in all the tissues tested, the transport activity of these three proteins was analyzed (Figure 2). We used the yeast mutant complement approach to test the transport activity of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a when was supplied. Coding sequences of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a were cloned into the yeast expression vector pDRf1-GW, and the three genes were expressed in the yeast strain 31019b (Δmep1, Δmep2, Δmep3, ura3), which is deficient in uptake (Marini et al., 1997). Yeast cell growth was monitored in the media containing 1 mM NH4Cl or 1 mM arginine. The results indicate that TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a are able to transport into yeast cells (Figure 3B).
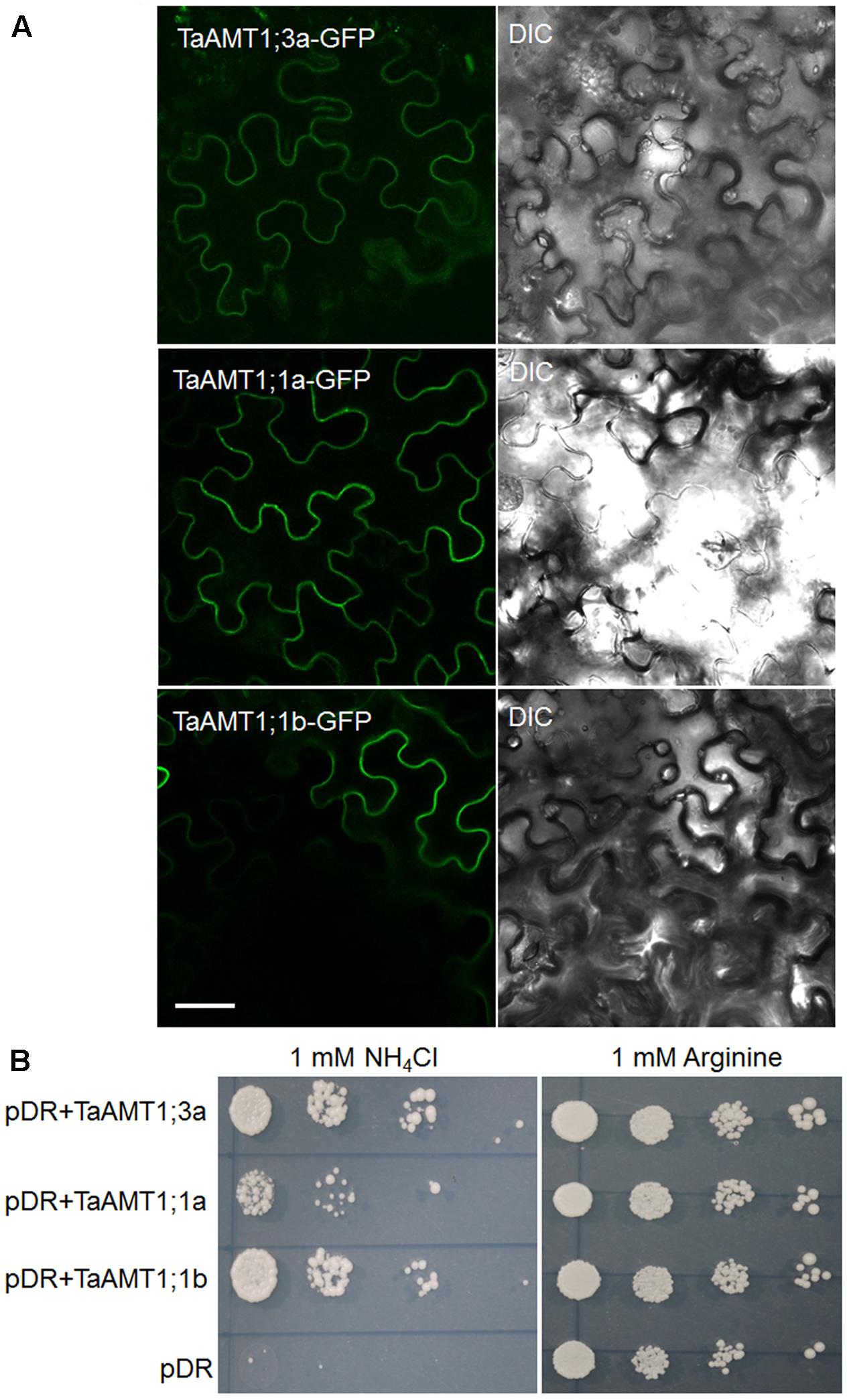
FIGURE 3. Localization and functional analysis of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a proteins. Agrobacterium-mediated transient expression of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a-GFP constructs in N. benthamiana leaves. GFP fluorescence and bright field images are shown in the left and right panels, respectively. Bar = 20 μm (A). The functions of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a genes were analyzed by complementation of an uptake defective yeast strain 31019b (Δmep1, Δmep2, Δmep3, ura3). Yeast cells were transformed expressing TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a via a pDRf1 (pDR) vector or an empty vector, and were tested for growth complementation on yeast N base plates supplemented with 1 mM NH4Cl or 1 mM arginine (Arg). The empty vector (pDRf1) was used as the control. The yeast cells were grown at 30°C for 3 days (B).
Complementation of the qko Mutant by Heterologous Expression of TaAMT1;1b
Two TaAMT1;1 genes (TaAMT1;1a and TaAMT1;1b) are able to complement the yeast mutant 31019b in which uptake is deficient, and TaAMT1;1b showed higher affinity than TaATM1;1a in transport of (Figure 3B). Therefore, TaAMT1;1b function was further analyzed in plants. The Arabidopsis quadruple mutant (qko) missing AMT1;1, AMT1;2, AMT1;3, and AMT2;1 greatly reduced uptake ability (Yuan et al., 2007a). Since gene transformation in wheat is challenging, TaAMT1;1b was selected and expressed in qko under the control of a 1.5 kb AtAMT1;1 promoter. The RT-PCR results indicate that TaAMT1;1b was expressed in the three independent transgenic Arabidopsis lines (#1, #2, and #4), while no visible transcript of TaAMT1;1b was detected in the qko mutant (Figure 4A). MeA uptake was tested in qko, qko+AtAMT1;1 (in which AtAMT1;1 was expressed by its own promoter), and three independent qko+TaAMT1;1b plants. Three-day-old seedlings grown on modified 0.5× MS medium containing 1 mM KNO3 as the sole N source were transferred to the same medium with or without 10 mM MeA. qko, qko+AtAMT1;1, and qko+TaAMT1;1b exhibited similar growth patterns without the addition of MeA. However, the growth of qko+AtAMT1;1 and qko+TaAMT1;1b, but not gko, was severely affected after the addition of MeA to the growth medium (Figure 4B). Root growth and seedling fresh weight of qko+TaAMT1;1b plants was more severely affected by MeA than qko+AtAMT1;1 plants (Figures 4B,C). Additionally, cellular contents were measured from the 7-day-old qko, qko+AtAMT1;1, and qko+TaAMT1;1b plant roots, and the results indicated that the roots of qko+AtAMT1;1 and qko+TaAMT1;1b contained more than the roots of qko (Figure 4D).
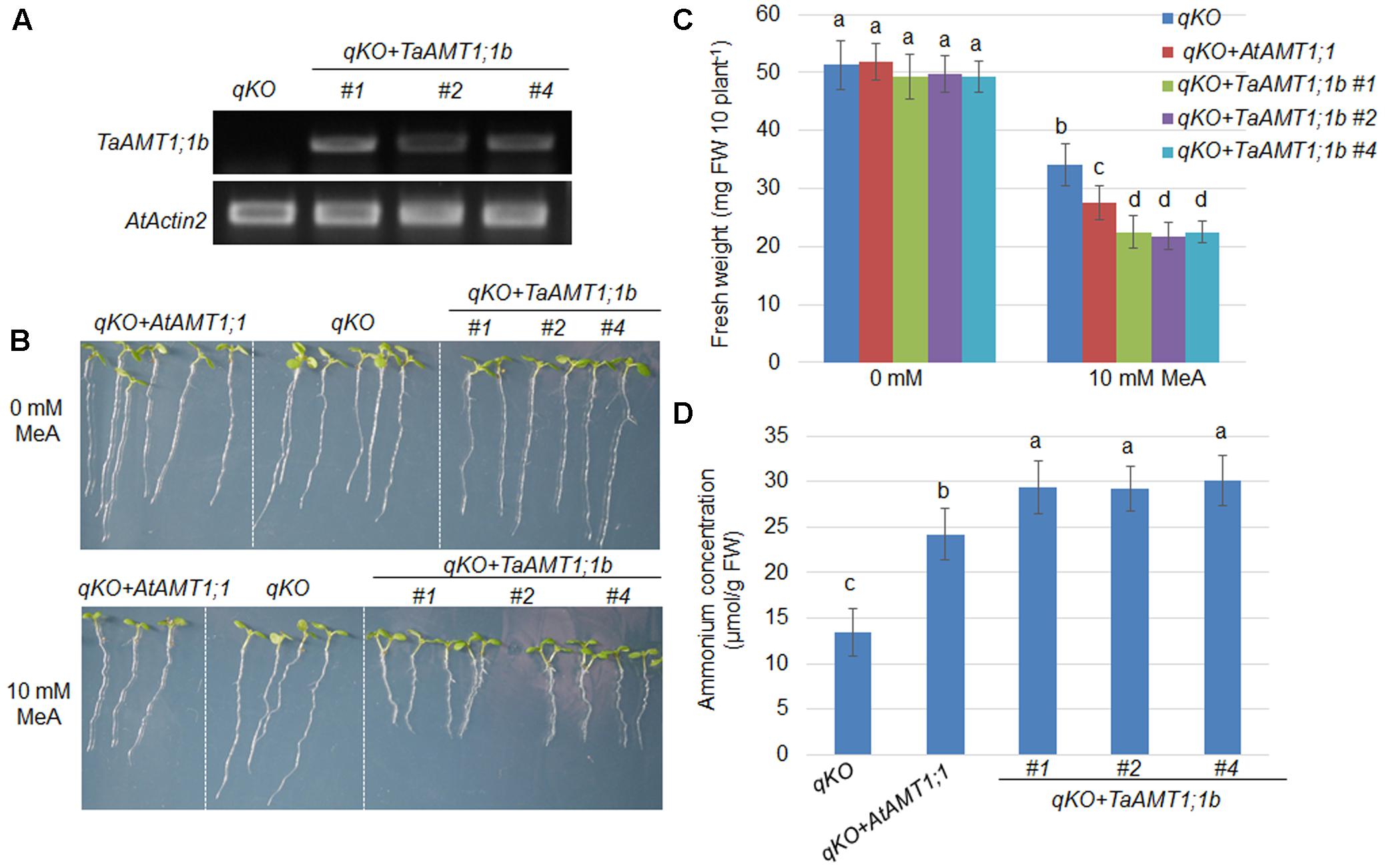
FIGURE 4. Complementation of the Arabidopsis uptake defective mutant qko by TaAMT1;1b. TaATM1;1b was expressed in the qko background under the control of the AtAMT1;1 promoter. Expression levels of TaATM1;1b were monitored in the TaATM1;1b transgenic plant roots (#1, #2, and #4), and AtActin2 was used as a loading control (A). Methyl-ammonium (MeA) dependent growth of qko, qko+AtAMT1;1, and qko+TaAMT1;1b was photographed. Three-day-old seedlings grown in 1 mM KNO3 containing modified 0.5× MS medium were transferred to the same medium containing either 0 or 10 mM MeA. Plant growth was photographed after 4 days (B). Total fresh weight of the seedlings (30 seedling for each line) shown in (B) was measured. Significant differences between qko and qko+AtAMT1;1 or qko+TaAMT1;1b were assessed (C). contents from the roots of 7-day-old qko, qko+AtAMT1;1, and qko+TaAMT1;1b plants grown in 0.5× MS medium were measured (D). Significant differences at P < 0.05 level are indicated by different letters.
N Dependent Expressions of TaAMT1;1 Genes
The expression of AMT genes is sensitive to exogenous N conditions. In Arabidopsis, the AMT1 genes were repressed upon the resupply of , while the AMT1;1 and AMT;12 genes in rice were highly induced by application (Sonoda et al., 2003; Loqué et al., 2006). To examine the -mediated expression of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a, 17-day-old wheat seedlings grown under N-free conditions were treated with 1 mM for 0, 1, 3, and 6 h, and their whole roots and leaves were sampled (Figure 5A). Prior to analyzing the -dependent expression patterns of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a, the cellular contents were measured in the roots. After transferring the wheat plants from the N-free medium to the solution containing , the contents were rapidly increased up to 6 h of treatment (Figure 5B). qRT-PCR results showed that TaAMT1;3a was suppressed while TaAMT1;1a and TaAMT1;1b were not suppressed by treatment in the roots (Figure 5C). The expression levels of the remaining TaAMT members were not altered by (data not shown). Furthermore, the contents in the leaves were monitored before and after treatment. The levels increased and reached a maximum at 6 h of treatment, but the contents were much lower than in the roots (Figure 5D). However, the -mediated expression levels of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a were unaltered in the leaves (Figure 5E).
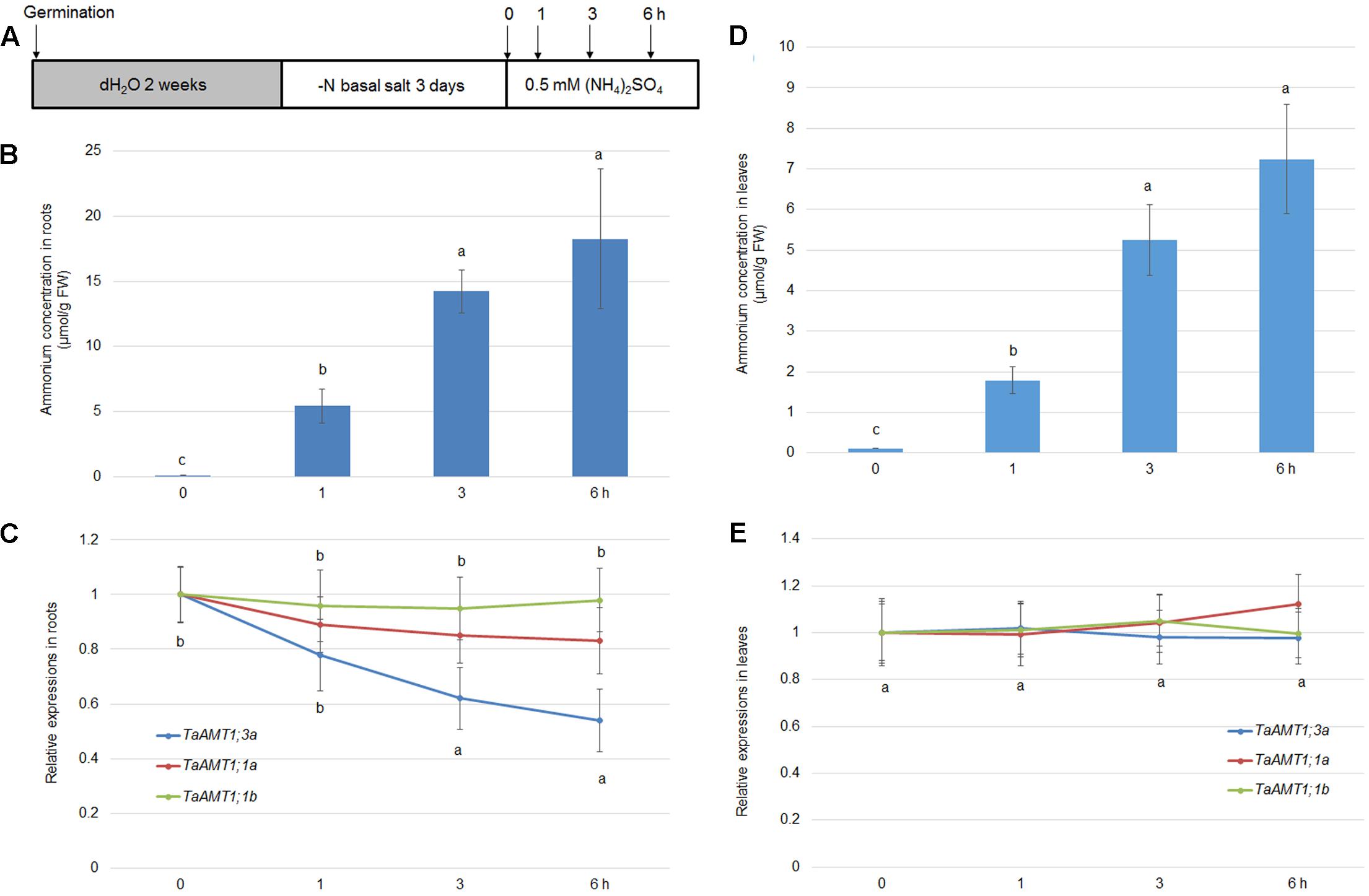
FIGURE 5. -dependent transcriptional changes in TaAMT1;1 genes. Seventeen-day-old seedlings grown as described in the section “Materials and Methods” were transferred to 0.5 mM (NH4)2SO4 containing solution (pH 5.5). Total roots and leaves of 17-day-old plants were sampled at 0, 1, 3, and 6 h after the addition of (A). Endogenous levels measured in the roots of 17-day-old plants (n > 10). Data represent means ± SE (n = 3) (B). qRT-PCR was performed to determine the expression levels of TaAMT genes in the roots. The level of TaAMT1;1 genes before treatment was defined as 1. The relative ratio shown is the ratio against 1 (C). Endogenous levels measured in the leaves of 17-day-old plants. Data represent means ± SE (n = 3) (D). qRT-PCR was performed to determine the expression levels of TaAMT genes in the leaves (E). The levels of TaAMT1;1 genes before treatment was defined as 1. The relative ratio shown is the ratio against 1. qRT-PCR was performed to determine the expression levels of TaAMT1 genes in the roots. The levels of TaAMT1;1 genes before N deprivation were defined as 1. The relative ratio shown is the ratio against 1. Significant differences at P < 0.05 level are indicated by different letters.
AMT expression is not only sensitive to conditions, but also responds to N deprivation in Arabidopsis and rice (Sonoda et al., 2003; Loqué et al., 2006). Therefore, expression of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a was analyzed upon N starvation. The 14-day-old seedlings grown in water were grown in nutrient solution containing for another 3 days. They were subsequently transferred to an N deprived solution (Figure 6A). After the transfer, the cellular levels rapidly decreased in the roots, and the lowest level was observed after 3 days (Figure 6B). Simultaneously, qRT-PCR was used to determine the N-status-dependent expressions of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a. The results indicated that TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a tested were induced under N starved conditions. The TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a were highly induced, especially on the third day in the roots (Figure 6C), and leaves (Figure 6D).
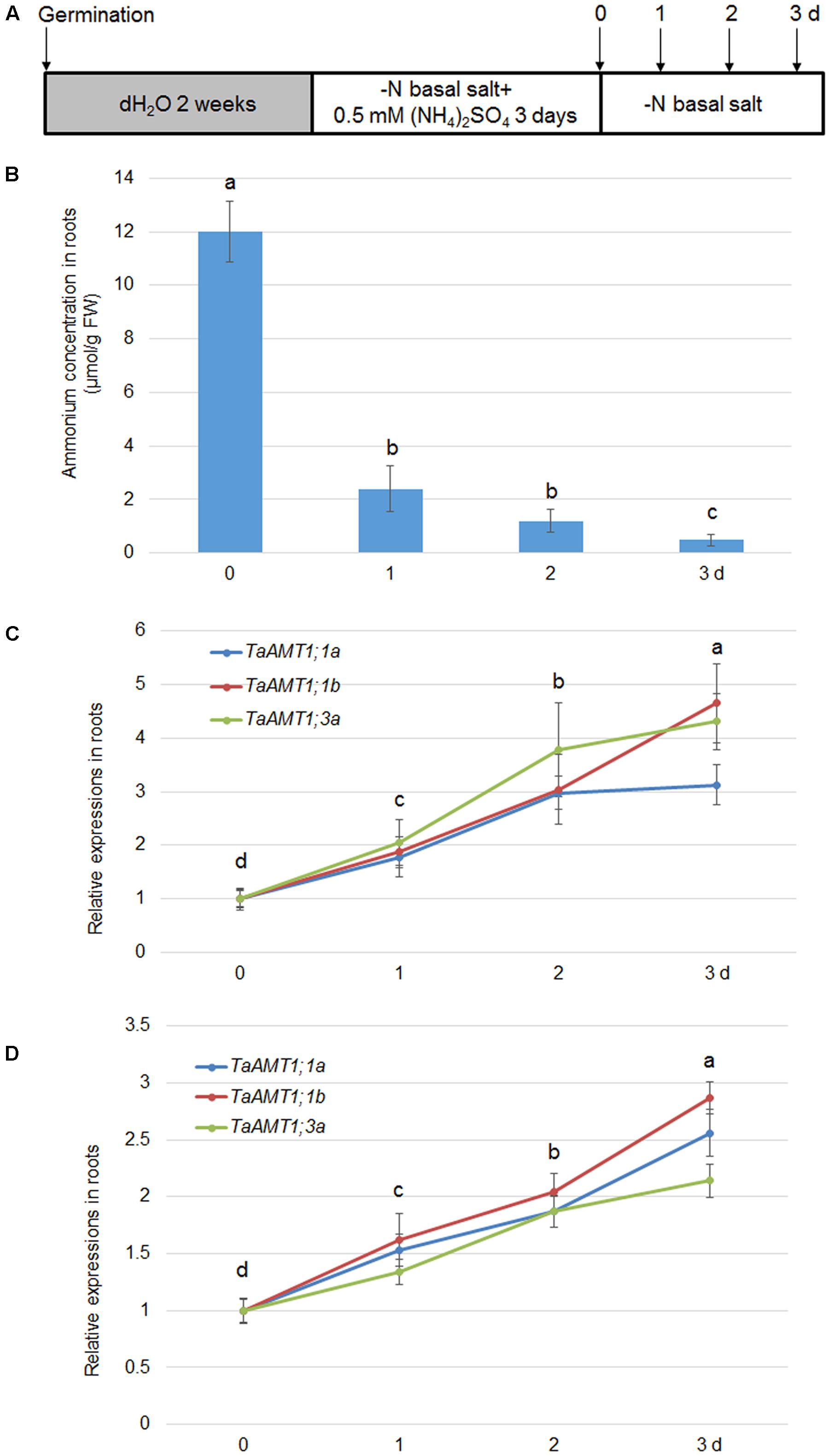
FIGURE 6. Effect of N-deprivation on TaAMT1;1 gene expressions. For testing N-deprivation dependent gene expressions, the plants were grown as described in the section “Materials and Methods” (A). Endogenous levels measured in the roots of 17-day-old plants. Data represent means ± SE (n = 3) (B). The N-deficiency dependent expression of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a were analyzed in the roots (C) and leave (D) samples. Significant differences at P < 0.05 level are indicated by different letters.
TaAMT1;1 and TaAMT1;3a Expressions Are Induced by Pgt Infection
AtAMT1;1 has been shown to alter basal defense against P. syringae and P. cucumerina (Pastor et al., 2014). In wheat, three TaAMTs levels were altered by AM fungi infection (Duan et al., 2015). To dissect whether there is a relationship between Pgt infection and expression of TaAMT1;1 genes, the stem rust-susceptible line LC (Roelfs and Martens, 1988) and resistant line Mini 2761 (Luan et al., 2013) were utilized, respectively. Following inoculation, urediniospore multiplication was detected in the leaves of the LC plants, while Mini 2761 exhibited lack of lesions in leaves (Figure 7A). Furthermore, the urediniospore inoculation-induced expression of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a genes was examined. The expression levels of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a differed insignificantly between the leaves of LC and Mini 2761. TaAMT1;3a was induced at 12 and 24 h (Figure 7B); TaAMT1;1a was induced at 36 and 48 h (Figure 7C); and TaAMT1;1b was induced at 24, 36, and 48 h after inoculation in LC only (Figure 7D). The expression levels of the three genes showed trifle difference following inoculation of the leaves of Mini 2761.
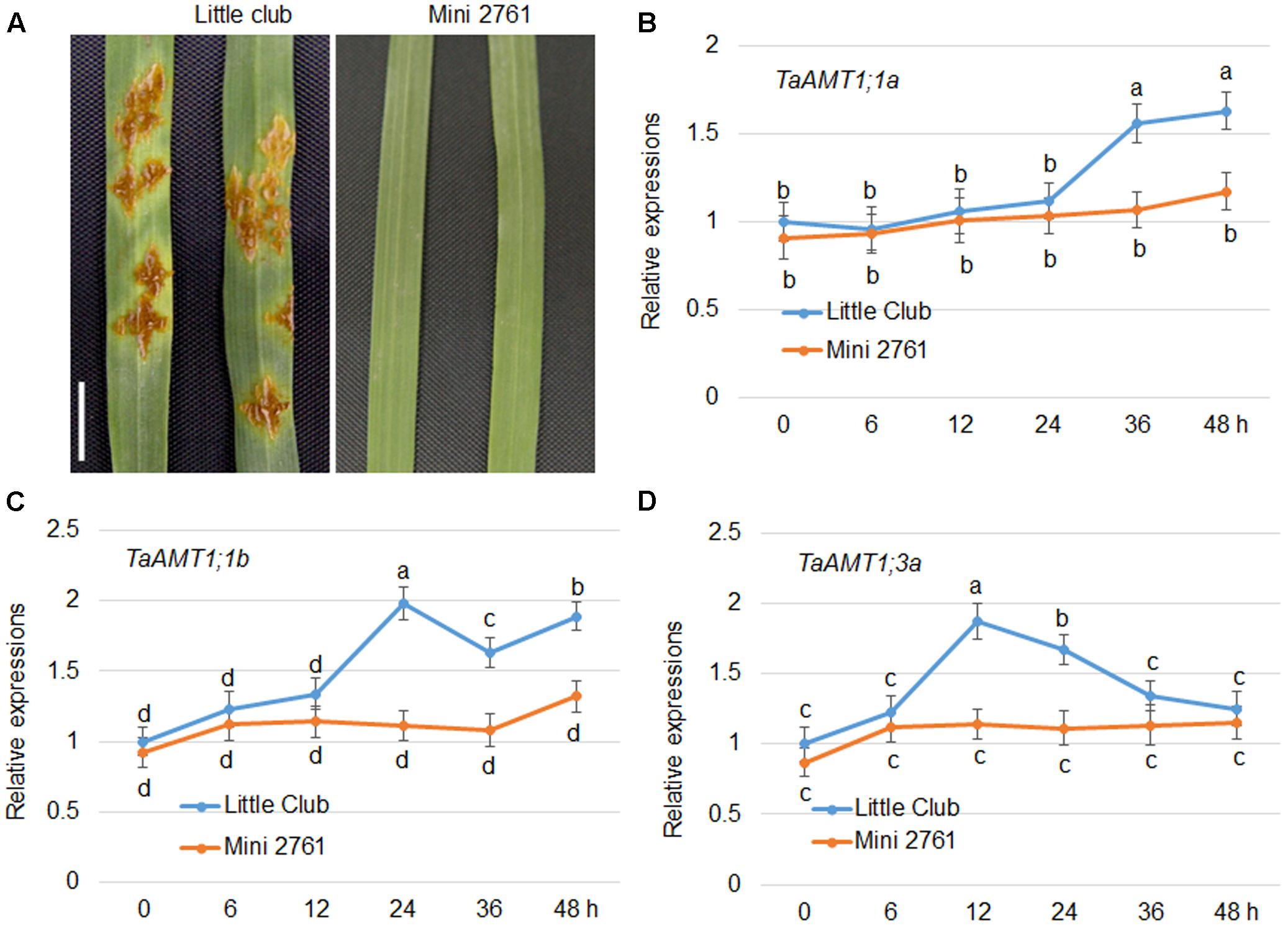
FIGURE 7. Urediniospore infection-mediated expression patterns of TaAMT1;1 genes. Phenotype of uredinia growth in two different wheat varieties. Leaves of 4-week-old LC and Mini 2761, a stem rust susceptible and immune line, respectively, were inoculated with urediniospores. After 7 days of infection, the uredinia on the surface of the leaves were photographed. Bar = 1 cm. Leaves of 4-week-old LC and Mini 2761 were inoculated with urediniospores, and the leaves were collected for RNA extraction 0, 6, 12, 24, 36, and 48 h after inoculation. TaAMT1;1 gene expressions were monitored by qRT-PCR (A). The levels of TaAMT1;1a (B), TaAMT1;1b (C), and TaAMT1;3a (D), before inoculation (0 h) was defined as 1. The relative ratio shown is the ratio against 1. The experiments were repeated three times and significant differences at P < 0.05 level are indicated by different letters.
As TaAMT genes were specifically induced in a stem rust-susceptible line (LC) after inoculation of the urediniospores, the effects of N availability in the growth medium on wheat stem rust disease were examined further. The LC plants were grown without any nutrient supply for the first 2 weeks after germination, following which they were transferred to a medium containing either 0 or 1 mM . After 2 weeks of growth, the urediniospores were inoculated evenly on the surface of the leaves (Figure 8A). The symptoms were monitored after another 2 weeks of inoculation. The results indicated that the plants grown under N-free conditions exhibited lighter colored uredinia as well as chlorosis surrounding the uredinium-formed regions (Figure 8B). Disease class standards were calculated for the symptoms shown in Figure 7B following international wheat rust disease standards (Abiko et al., 2005). The results indicate that plants grown on the medium containing were on average Class 4, while the plants grown under N-free conditions were on average Class 3 (Figure 8C). These results suggest that N deficiency inhibits stem rust disease in wheat.
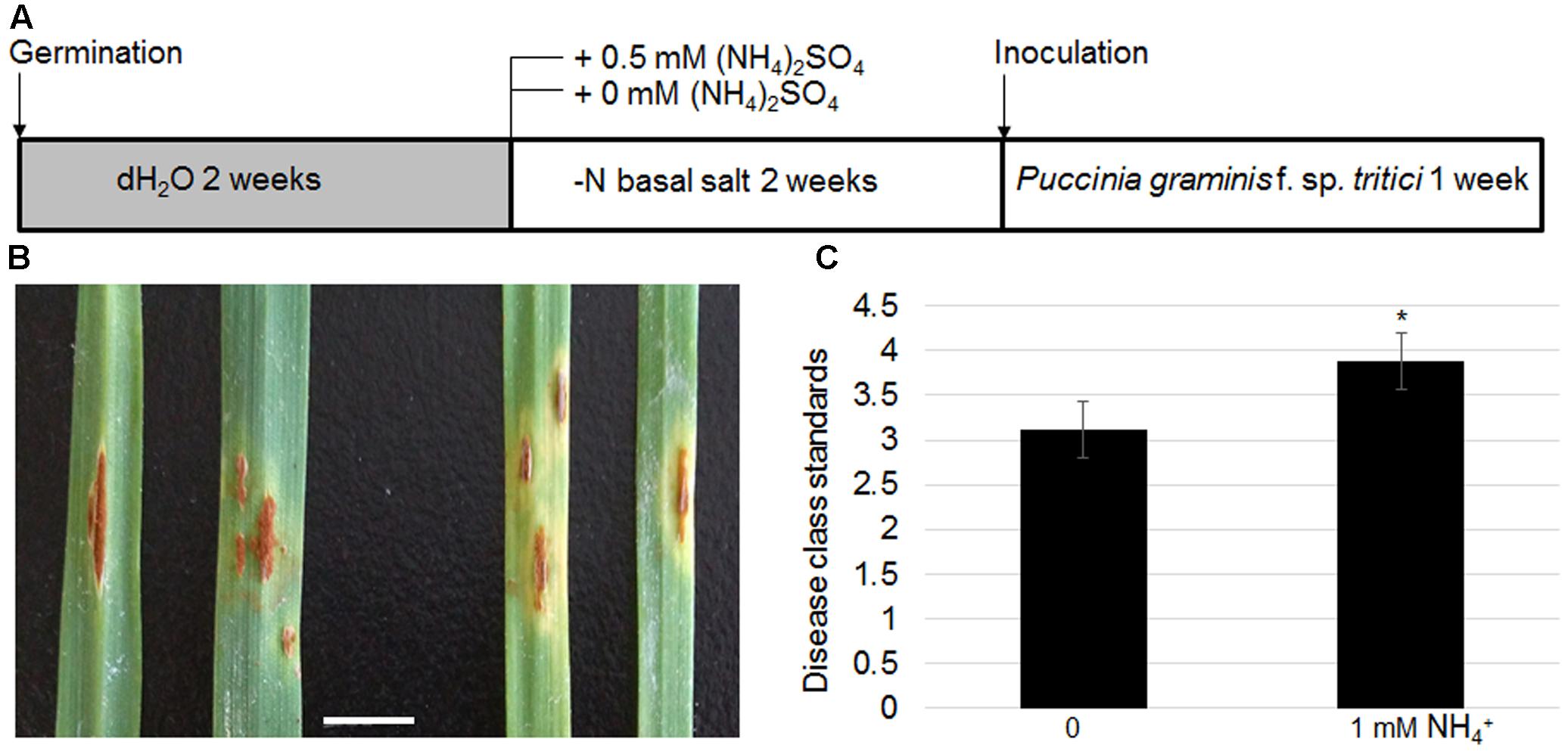
FIGURE 8. Effects of N-deficient growth conditions on wheat stem rust disease. Schematic diagram showing plant growth conditions for testing N effects on stem rust disease. Four-week-old LC seedling was inoculated with Pgt (A). Photograph of uredinia growth in the leaves of LC grown on -supplied (left panel) or N-free media (right panel). Bar = 1 cm (B). Disease class standards were calculated following the international standard for wheat stem rust disease corresponding to the phenotypes shown in (B,C). Significant differences were analyzed (∗P < 0.05).
Discussion
Nitrogen is an important nutrient for plant growth and production, accounting for 2% of the dry weight. Ammonium is the common N source for higher plants, and acquisition of from the rhizosphere occurs through transporters (AMTs) in plants. These AMTs have been characterized in many plant species including Arabidopsis, rice, tomato, rape, maize, and sorghum (Ninnemann et al., 1994; Lauter et al., 1996; Gazzarrini et al., 1999; Sohlenkamp et al., 2000, 2002; Von Wirén et al., 2000; Salvemini et al., 2001; Pearson et al., 2002; Simon-Rosin et al., 2003; Sonoda et al., 2003; Suenaga et al., 2003; D’Apuzzo et al., 2004; Loqué and von Wirén, 2004; Yuan et al., 2007a,b, 2009; Gu et al., 2013; Koegel et al., 2013; Ye et al., 2016). In this study, we identified AMT members in a major global crop, wheat.
Based on the sequence similarity search, 23 AMTs were retrieved from the T. aestivum genome. In phylogenetic results showed that four AMT group (AMT1, AMT2, AMT3, and AMT4) members were all present in the wheat genome (Figure 1). In Arabidopsis and rice, the AMT1 group genes are expressed either ubiquitously or tissue specifically (Sonoda et al., 2003; Loqué et al., 2006). In rice, the diverse genomic structure and transmembrane helices numbers of OsAMT members have been identified (Ye et al., 2016). We found that TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a were strongly expressed, while the other TaAMT members were weakly expressed or otherwise not detected in wheat (Figure 2). To analyze the localization of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a, GFP fusion proteins were expressed in N. benthamiana leaves. Observation of GFP signal indicated that TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a comprise plasma membrane proteins (Figure 3A). Furthermore, TaAMT1;1 activities were analyzed by complementation of a yeast strain Δmep123 in which uptake is deficient. The results indicated that TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a proteins can transport in yeast cells (Figure 3B). To analyze TaAMT1;1 transportation function in plants, TaAMT1;1b was expressed in the Arabidopsis qko mutant in which four AMT genes were mutated (Yuan et al., 2007a). Expression of TaAMT1;1b driven by the AtAMT1;1 promoter increased cellular contents in the mutant, which was similar to the levels in qko+AtAMT1;1 plants (Figure 4D). qko+TaAMT;1b was more sensitive to MeA than qko, and was slightly more sensitive than the qko+AtAMT1;1 plants. This sensitivity was exhibited in the relatively shorter roots in comparison to the control (Figures 4B,C). These results indicate that TaAMT1;1 transports , and that TaAMT1;1b exhibits similar affinity as AtAMT1;1 against . However, the data showed that MeA-dependent root growth was more severely affected in qko+TaAMT1;1b than in qko+AtAMT1;1 and qko+TaAMT1;1b accumulated more ammonium than in qko+AtAMT1;1, suggesting that TaAMT1;1b may have higher affinity than AtAMT1;1 in transport of .
The gene expression levels of AMTs are sensitive to in Arabidopsis and rice (Sonoda et al., 2003; Loqué et al., 2006). We discovered that TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a were not altered by treatment (Figure 5C). In addition, the expression of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a genes was induced in the roots and leaves of the wheat plants upon N starvation (Figure 6C). As Arabidopsis is an -sensitive species, its AMTs were suppressed under a high concentration treatment (Loqué et al., 2006). Conversely, in rice, the -tolerant species AMT1;1 and 1;2 were highly up-regulated upon supply of (Sonoda et al., 2003). Suppression of TaAMT1;3a in response to may explain why wheat is also an -sensitive species. The sensitivity to of barley, which is a close relative of wheat, was explained by the activation of futile cycling in the membrane (Britto et al., 2001).
Ammonium transporter proteins are involved in a diversity of aspects of plant growth and development. For instance, Arabidopsis AMT1;3 regulates -triggered lateral branching (Lima et al., 2010). The expression levels of AMT1;1, AMT1;2, AMT1;3, and AMT2;1, as well as other N metabolic genes, were altered by biotic stresses in Arabidopsis (Fagard et al., 2014). Furthermore, AMT1;1 was involved in P. syringae- and P. cucumerina-mediated disease in Arabidopsis (Pastor et al., 2014). In wheat, expression of three TaAMTs was changed by AM fungi infection (Duan et al., 2015), implying a potential regulation of microbe on TaAMT regulations. In addition, glutamate supply to the roots induces systemic resistance to rice blast disease in the leaves (Kadotani et al., 2016). These findings suggest that cellular N levels or N signals are closely associated with plant defense. Expression tests of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a in LC and Mini 2761 following urediniospore inoculation of Pgt showed that all three genes were induced by the infection only in LC, a wheat stem rust-susceptible line, but not in Mini 2761, a wheat stem rust immune variety. However, the inducement time points were slightly different among the three genes (Figure 7). The urediniospore inoculation of Pgt in the leaves of LC formed uredinium; however, this was not observed in the leaves of Mini 2761 (Figure 7). This implies that TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a inducement might require the successful infection of urediniospores into the plant leaves. To understand the mechanism of on wheat stem rust disease, LC plants were cultured in medium containing either 0 or 1 mM prior to urediniospore inoculation. The results indicated that the disease class standard levels were lower in the plants grown in the medium without (Figure 8).
Further genetic experiments are required to verify the role of TaAMT1;1a, TaAMT1;1b, and TaAMT1;3a in wheat stem rust disease, as well as to determine whether the lower disease class standards under N-free conditions are the result of either changes in basal immune response or disruption of N metabolism. The present study characterized the functions of AMTs in wheat plants for the first time. Identification of the interaction between TaAMTs and wheat stem rust disease will broaden our understanding of N uptake and metabolism in plant pathogenic infections.
Author Contributions
TL, BJ, and YX conceived and designed the research. KL, XZ, and XX conducted DNA and RNA isolations. All the experiments were supervised by YX, BJ performed the bioinformatics analysis, and ZW and YG provided figures and tables. YX wrote the manuscript. TL and BJ edited the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the grants from the National Natural Science Foundation (31701738) and Technology Research Project of Education Department of Liaoning (1102/01032616003) as well as a startup funding (880416008) from Shenyang Agricultural University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The qko and qko+ATM1;1 seeds were obtained from Dr. Nicolaus Von Wirén’s group at Leibniz Institute for Plant Genetics and Crop Plant Research.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01637/full#supplementary-material
Footnotes
References
Abiko, T., Obara, M., Ushioda, A., Hayakawa, T., Hodges, M., and Yamaya, T. (2005). Localization of NAD-isocitrate dehydrogenase and glutamate dehydrogenase in rice roots: candidates for providing carbon skeletons to NADH-glutamate synthase. Plant Cell Physiol. 46, 1724–1734. doi: 10.1093/pcp/pci188
Bao, A., Liang, Z., Zhao, Z., and Cai, H. (2015). Overexpressing of OsAMT1-3, a high affinity ammonium transporter gene, modifies rice growth and carbon-nitrogen metabolic status. Int. J. Mol. Sci. 16, 9037–9063. doi: 10.3390/ijms16059037
Bleckmann, A., Weidtkamp-Peters, S., Seidel, C. A., and Simon, R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176. doi: 10.1104/pp.109.149930
Bloom, A. J. (1997). “Nitrogen as a limiting factor: crop acquisition of ammonium and nitrate,” in Ecology in Agriculture, ed. L. E. Jackson (San Diego, CA: Academic Press), 145–172.
Britto, D. T., and Kronzucker, H. J. (2002). NH4+ toxicity in higher plants: a critical review. J. Plant Physiol. 159, 567–584. doi: 10.1078/0176-1617-0774
Britto, D. T., Siddiqi, M. Y., Glass, A. D., and Kronzucker, H. J. (2001). Futile transmembrane NH4(+) cycling: a cellular hypothesis to explain ammonium toxicity in plants. Proc. Natl. Acad. Sci. U.S.A. 98, 4255–4258. doi: 10.1073/pnas.061034698
Cao, Y. Y., and Chen, W. Q. (2010). Stepwise shift of differential hosts and racial designation of Puccinia graminis f. sp. tritici. J. Triticeae Crops 30, 167–172.
D’Apuzzo, E., Rogato, A., Simon-Rosin, U., El Alaoui, H., Barbulova, A., Betti, M., et al. (2004). Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol. 134, 1763–1774. doi: 10.1104/pp.103.034322
Duan, J., Tian, H., Drijber, R. A., and Gao, Y. (2015). Systemic and local regulation of phosphate and nitrogen transporter genes by arbuscular mycorrhizal fungi in roots of winter wheat (Triticum aestivum L.). Plant Physiol Biochem. 96, 199–208. doi: 10.1016/j.plaphy.2015.08.006
Fagard, M., Launay, A., Clement, G., Courtial, J., Dellagi, A., Farjad, M., et al. (2014). Nitrogen metabolism meets phytopathology. J. Exp. Bot. 65, 5643–5656. doi: 10.1093/jxb/eru323
Fukumorita, T., and Chino, M. (1982). Sugar, amino acid and inorganic contents in rice phloem sap. Plant Cell Physiol. 23, 273–283.
Gazzarrini, S., Lejay, L., Gojon, A., Ninnemann, O., Frommer, W. B., and von Wirén, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11, 937–947. doi: 10.1105/tpc.11.5.937
Gu, R., Duan, F., An, X., Zhang, F., von Wirén, N., and Yuan, L. (2013). Characterization of AMT-mediated high-affinity ammonium uptake in roots of maize (Zea mays L.). Plant Cell Physiol. 54, 1515–1524. doi: 10.1093/pcp/pct099
Han, M. J., Jung, K. H., Yi, G., Lee, D. Y., and An, G. (2006). Rice immature pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol. 47, 1457–1472. doi: 10.1093/pcp/pcl013
Kadotani, N., Akagi, A., Takatsuji, H., Miwa, T., and Igarashi, D. (2016). Exogenous proteinogenic amino acids induce systemic resistance in rice. BMC Plant Biol. 3:60. doi: 10.1186/s12870-016-0748-x
Kim, J. G., Li, X., Roden, J. A., Taylor, K. W., Aakre, C. D., Su, B., et al. (2009). Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21, 1305–1323. doi: 10.1105/tpc.108.063123
Koegel, S., Ait Lahmidi, N., Arnould, C., Chatagnier, O., Walder, F., Ineichen, K., et al. (2013). The family of ammonium transporters (AMT) in Sorghum bicolor: two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 198, 853–865. doi: 10.1111/nph.12199
Kumar, A., Silim, S. N., Okamoto, M., Siddiqi, M. Y., and Glass, A. D. (2003). Differential expression of three members of the AMT1 gene family encoding putative high-affinity NH4+ transporters in roots of Oryza sativa subspecies indica. Plant Cell Environ. 26, 907–914. doi: 10.1046/j.1365-3040.2003.01023.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lanquar, V., Loqué, D., Hörmann, F., Yuan, L., Bohner, A., Engelsberger, W. R., et al. (2009). Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell 21, 3610–3622. doi: 10.1105/tpc.109.068593
Lauter, F. R., Ninnemann, O., Bucher, M., Riesmeier, J. W., and Frommer, W. B. (1996). Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc. Natl. Acad. Sci. U.S.A. 93, 8139–8144. doi: 10.1073/pnas.93.15.8139
Lima, J. E., Kojima, S., Takahashi, H., and von Wirén, N. (2010). Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1; 3-dependent manner. Plant Cell 22, 3621–3633. doi: 10.1105/tpc.110.076216
Loqué, D., and von Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305. doi: 10.1093/jxb/erh147
Loqué, D., Yuan, L., Kojima, S., Gojon, A., Wirth, J., Gazzarrini, S., et al. (2006). Additive contribution of AMT1; 1 and AMT1; 3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 48, 522–534. doi: 10.1111/j.1365-313X.2006.02887.x
Luan, Z. J., Cao, Y. Y., Li, T. Y., Chen, S., Chen, X. M., Zhu, G. Q., et al. (2013). cDNA-AFLP analysis of differentially expressed resistant genes of Minn2761. Sci. Agric. Sin. 46, 5058–5065.
Marini, A. M., Soussi-Boudekou, S., Vissers, S., and Andre, B. (1997). A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 4282–4293. doi: 10.1128/MCB.17.8.4282
Ninnemann, O., Jauniaux, J., and Frommer, W. (1994). Identification of a high affinity NH4+ transporter from plants. EMBO J. 13, 3464–3471.
Noctor, G., and Foyer, C. H. (1998). A re-evaluation of the ATP: NADPH budget during C3 photosynthesis: a contribution from nitrate assimilation and its associated respiratory activity? J. Exp. Bot. 49, 1895–1908. doi: 10.1093/jxb/49.329.1895
Oliveira, I. C., Brears, T., Knight, T. J., Clark, A., and Coruzzi, G. M. (2002). Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 129, 1170–1180. doi: 10.1104/pp.020013
Pardey, P. G., Beddow, J. M., Kriticos, D. J., Hurley, T. M., Park, R. F., Duveiller, E., et al. (2013). Agriculture. Right-sizing stem-rust research. Science 340, 147–148. doi: 10.1126/science.122970
Pastor, V., Gamir, J., Camanes, G., Cerezo, M., Sanchez-Bel, P., and Flors, V. (2014). Disruption of the ammonium transporter AMT1.1 alters basal defenses generating resistance against Pseudomonas syringae and Plectosphaerella cucumerina. Front. Plant. Sci. 5:231. doi: 10.3389/fpls.2014.00231
Patterson, K., Cakmak, T., Cooper, A., Lager, I., Rasmusson, A. G., and Escobar, M. A. (2010). Distinct signalling pathways and transcriptome response signatures differentiate ammonium-and nitrate-supplied plants. Plant Cell Environ. 33, 1486–1501. doi: 10.1111/j.1365-3040.2010.02158.x
Pearson, J. N., Finnemann, J., and Schjoerring, J. K. (2002). Regulation of the high-affinity ammonium transporter (BnAMT1;2) in the leaves of Brassica napus by nitrogen status. Plant Mol. Biol. 49, 483–490. doi: 10.1023/A:1015549115471
Pearson, W. R. (2013). An introduction to sequence similarity (“homology”) searching. Curr. Protoc. 42, 3.1.1–3.1.8. doi: 10.1002/0471250953.bi0301s42
Pretorius, Z., Singh, R., Wagoire, W., and Payne, T. (2000). Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis. f. sp. tritici in Uganda. Plant Dis. 84:203. doi: 10.1094/PDIS.2000.84.2.203B
Ranathunge, K., El-Kereamy, A., Gidda, S., Bi, Y. M., and Rothstein, S. J. (2014). AMT1;1 transgenic rice plants with enhanced NH4(+) permeability show superior growth and higher yield under optimal and suboptimal NH4(+) conditions. J. Exp. Bot. 65, 965–979. doi: 10.1093/jxb/ert458
Roelfs, A., and Martens, J. (1988). An international system of nomenclature for Puccinia graminis f. sp. tritici. Phytopathology 78, 526–533. doi: 10.1094/Phyto-78-526
Salvemini, F., Marini, A. M., Riccio, A., Patriarca, E. J., and Chiurazzi, M. (2001). Functional characterization of an ammonium transporter gene from Lotus japonicus. Gene 270, 237–243. doi: 10.1016/S0378-1119(01)00470-X
Simon-Rosin, U., Wood, C., and Udvardi, M. K. (2003). Molecular and cellular characterisation of LjAMT2; 1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol. Biol. 51, 99–108. doi: 10.1023/A:1020710222298
Singh, R. P., Hodson, D. P., Huerta-Espino, J., Jin, Y., Njau, P., Wanyera, R., et al. (2008). Will stem rust destroy the world’s wheat crop? Adv. Agron. 98, 271–309. doi: 10.1016/S0065-2113(08)00205-8
Sogaard, R., Alsterfjord, M., Macaulay, N., and Zeuthen, T. (2009). Ammonium ion transport by the AMT/Rh homolog TaAMT1;1 is stimulated by acidic pH. Pflugers. Arch. 458, 733–743. doi: 10.1007/s00424-009-0665-z
Sohlenkamp, C., Shelden, M., Howitt, S., and Udvardi, M. K. (2000). Characterization of Arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett. 467, 273–278. doi: 10.1016/S0014-5793(00)01153-4
Sohlenkamp, C., Wood, C. C., Roeb, G. W., and Udvardi, M. K. (2002). Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 130, 1788–1796. doi: 10.1104/pp.008599
Sonoda, Y., Ikeda, A., Saiki, S., von Wirén, N., Yamaya, T., and Yamaguchi, J. (2003). Distinct expression and function of three ammonium transporter genes (OsAMT1; 1–1; 3) in rice. Plant Cell Physiol. 44, 726–734. doi: 10.1093/pcp/pcg083
Suenaga, A., Moriya, K., Sonoda, Y., Ikeda, A., Von Wirén, N., Hayakawa, T., et al. (2003). Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 44, 206–211. doi: 10.1093/pcp/pcg017
Von Wirén, N., Laute, R. F. R., Ninnemann, O., Gillissen, B., Walch-Liu, P., Engels, C., et al. (2000). Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J. 21, 167–175. doi: 10.1046/j.1365-313x.2000.00665.x
Xuan, Y. H., Hu, Y. B., Chen, L. Q., Sosso, D., Ducat, D. C., Hou, B. H., et al. (2013). Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. U.S.A. 110, E3685–E3694. doi: 10.1073/pnas.1311244110
Ye, X., Sun, Y., Liu, P., and Lee, I. (2016). Evolutionary analysis of AMT (Ammonium Transporters) family in Arabidopsis thaliana and Oryza sativa. Mol. Soil Biol. 7, 1–7.
Yu, L. X., Liu, S., Anderson, J. A., Singh, R. P., Jin, Y., Dubcovsky, J., et al. (2010). Haplotype diversity of stem rust resistance loci in uncharacterized wheat lines. Mol. Breed. 26, 667–680. doi: 10.1007/s11032-010-9403-7
Yuan, L., Graff, L., Loqué, D., Kojima, S., Tsuchiya, Y. N., Takahashi, H., et al. (2009). AtAMT1; 4, a pollen-specific high-affinity ammonium transporter of the plasma membrane in Arabidopsis. Plant Cell Physiol. 50, 13–25. doi: 10.1093/pcp/pcn186
Yuan, L., Loqué, D., Kojima, S., Rauch, S., Ishiyama, K., Inoue, E., et al. (2007a). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19, 2636–2652.
Keywords: TaAMT, ammonium, expressions, stem rust, wheat
Citation: Li T, Liao K, Xu X, Gao Y, Wang Z, Zhu X, Jia B and Xuan Y (2017) Wheat Ammonium Transporter (AMT) Gene Family: Diversity and Possible Role in Host–Pathogen Interaction with Stem Rust. Front. Plant Sci. 8:1637. doi: 10.3389/fpls.2017.01637
Received: 03 May 2017; Accepted: 06 September 2017;
Published: 20 September 2017.
Edited by:
Gerald Alan Berkowitz, University of Connecticut, United StatesReviewed by:
Fatima Cvrckova, Charles University, CzechiaTamara Pecenkova, Institute of Experimental Botany (ASCR), Czechia
Copyright © 2017 Li, Liao, Xu, Gao, Wang, Zhu, Jia and Xuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanhu Xuan, xuanyuanhu115@syau.edu.cn Baolei Jia, jiabaolei@hotmail.com
†These authors have contributed equally to this work.
 Tianya Li
Tianya Li Kai Liao
Kai Liao Xiaofeng Xu
Xiaofeng Xu Yue Gao
Yue Gao Ziyuan Wang
Ziyuan Wang Xiaofeng Zhu
Xiaofeng Zhu Baolei Jia
Baolei Jia Yuanhu Xuan
Yuanhu Xuan