
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 October 2017
Sec. Plant Pathogen Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01758
 Rajtilak Majumdar1†
Rajtilak Majumdar1† Kanniah Rajasekaran1†
Kanniah Rajasekaran1† Christine Sickler1
Christine Sickler1 Matthew Lebar1
Matthew Lebar1 Bryan M. Musungu2,3
Bryan M. Musungu2,3 Ahmad M. Fakhoury4
Ahmad M. Fakhoury4 Gary A. Payne5
Gary A. Payne5 Matt Geisler2
Matt Geisler2 Carol Carter-Wientjes1
Carol Carter-Wientjes1 Qijian Wei1
Qijian Wei1 Deepak Bhatnagar1
Deepak Bhatnagar1 Jeffrey W. Cary1*†
Jeffrey W. Cary1*†Aspergillus flavus is an opportunistic plant pathogen that colonizes and produces the toxic and carcinogenic secondary metabolites, aflatoxins, in oil-rich crops such as maize (Zea mays ssp. mays L.). Pathogenesis-related (PR) proteins serve as an important defense mechanism against invading pathogens by conferring systemic acquired resistance in plants. Among these, production of the PR maize seed protein, ZmPRms (AC205274.3_FG001), has been speculated to be involved in resistance to infection by A. flavus and other pathogens. To better understand the relative contribution of ZmPRms to A. flavus resistance and aflatoxin production, a seed-specific RNA interference (RNAi)-based gene silencing approach was used to develop transgenic maize lines expressing hairpin RNAs to target ZmPRms. Downregulation of ZmPRms in transgenic kernels resulted in a ∼250–350% increase in A. flavus infection accompanied by a ∼4.5–7.5-fold higher accumulation of aflatoxins than control plants. Gene co-expression network analysis of RNA-seq data during the A. flavus-maize interaction identified ZmPRms as a network hub possibly responsible for regulating several downstream candidate genes associated with disease resistance and other biochemical functions. Expression analysis of these candidate genes in the ZmPRms–RNAi lines demonstrated downregulation (vs. control) of a majority of these ZmPRms-regulated genes during A. flavus infection. These results are consistent with a key role of ZmPRms in resistance to A. flavus infection and aflatoxin accumulation in maize kernels.
Mycotoxin contamination of food and feed crops is a global problem. Exposure to mycotoxins in humans and livestock primarily occurs through ingestion of contaminated seeds or other edible plant parts. The majority of mycotoxin contamination of crop plants is the result of infection by members of the three fungal genera, Aspergillus, Fusarium, and Penicillium among which Aspergillus is responsible for the most adverse economic and health impacts (Ismaiel and Papenbrock, 2015; Kumar et al., 2016; Mitchell et al., 2016; Umesha et al., 2017). Maize is a major crop grown worldwide and is susceptible to aflatoxin contamination by Aspergillus flavus, especially during episodes of severe drought (Kebede et al., 2012; Fountain et al., 2014). Aflatoxin contamination of maize can result in economic losses as high as US$686.6 million/year in the United States based on a recent estimation of economic losses for the year 2013 (Mitchell et al., 2016). With predicted changes in the global climate, it is estimated that aflatoxin contamination could cause losses to the maize industry ranging from US$52.1 million to US$1.68 billion/year in the United States (Mitchell et al., 2016).
Production of pathogenesis-related (PR) proteins in response to biotic stressors is a defense strategy often employed by plants to resist pathogen invasion. PR proteins inhibit pathogens either by their direct antimicrobial properties or by regulating the expression of key genes involved in host defense or both. This class of defense proteins has been shown to accumulate at the site of infection upon pathogen invasion and contribute to systemic acquired resistance (SAR) (Ryals et al., 1996). Transcriptomic and proteomic studies have identified changes in protein production during maize seed infection by A. flavus and Fusarium verticillioides (reviewed by Pechanova and Pechan, 2015). Among the different PR proteins reported, PR10 (GenBank accession no. AY953127) has been extensively studied in corn in relation to A. flavus infection (Chen et al., 2010). Silencing of PR10 (under constitutive promoter) in maize resulted in a significant increase in fungal growth in the kernels accompanied by higher amounts of aflatoxin accumulation. The contribution of PR10 to aflatoxin resistance in maize has been mainly attributed to the antimicrobial property of the PR10 protein. The other PR protein that is also highly induced upon infection by necrotrophic seed pathogenic fungi is the PR maize seed (ZmPRms; AC205274.3_FG001) protein (Casacuberta et al., 1991; Murillo et al., 1999; Dolezal et al., 2014; Shu et al., 2015; Musungu et al., 2016).
ZmPRms transcripts are reported to accumulate at the aleurone layer and scutellum of germinating maize seeds (Casacuberta et al., 1991) and their production is increased upon fungal infection (Casacuberta et al., 1992; Murillo et al., 1999; Shu et al., 2015). The involvement of the ZmPRms gene promoter in response to fungal elicitors was also reported by Raventós et al. (1995). ZmPRms promoter::reporter gene fusion assays demonstrated activation of the promoter by fungal elicitors (mycelial extract derived from F. verticillioides). Presence of a specific motif in the promoter region showed strong association between promoter induction and biotic stressors. Involvement of both elicitor-response element (ERE) and enhancing sequences in the PRms promoter were implicated in induction of the promoter by fungal elicitors. In another study, transgenic expression of the ZmPRms gene in rice resulted in broad-spectrum resistance against fungal (Magnaporthe oryzae, F. verticillioides, and Helminthosporium oryzae) and bacterial (Erwinia chrysanthemi) pathogens suggesting involvement of this gene in the central defense mechanism in plants (Gómez-Ariza et al., 2007). Overexpression of ZmPRms in rice also primed the expression of other pathogen-induced defense genes leading to significantly higher expression at the basal level (in absence of the pathogen) and higher induction upon pathogen exposure as compared to the wild-type rice plants.
Both marker-assisted breeding and transgenic approaches are being used to develop maize lines with enhanced resistance to A. flavus infection and aflatoxin contamination (Cary et al., 2011; Warburton and Williams, 2014; Bhatnagar-Mathur et al., 2015; Schubert et al., 2015; Brown et al., 2016; Thakare et al., 2017). Both of these approaches require identification of host genes that contribute to enhanced resistance. The objective of the current study was to determine and evaluate the relative contribution of ZmPRms against A. flavus infection and aflatoxin production during infection of maize kernels. Using an RNAi-based approach, transgenic maize plants were generated with seed-specific expression of hairpin RNAs (hpRNA) to silence the ZmPRms gene. Downregulation in expression of the native ZmPRms gene in selected transgenic RNAi maize lines allowed us to quantify the role of ZmPRms in fungal growth and aflatoxin accumulation in infected kernels. We also evaluated ZmPRms-mediated global regulation of host defense-related genes and other biochemical function-related genes associated with resistance to A. flavus in maize.
An RNAi-based binary vector for host-induced gene silencing (HIGS) of the ZmPRms gene (Figures 1A,B) was constructed using an In-fusion HD Cloning Kit (Clontech Laboratories, Inc.; Cat# 011614). Briefly, a 1115 bp Zein promoter (Shepherd and Scott, 2009), a 460 bp 5′ arm (the sense strand) and a 454 bp 3′ arm (the antisense strand) of the ZmPRms gene and a PR10 intron (Chen et al., 2010) were PCR amplified using sequence-specific primers (Supplementary Table S1) containing a 15 bp overlap at the 5′ ends. The three PCR fragments were joined together with ScaI-SpeI restriction enzyme digested pMCG1005 binary plant transformation vector (McGinnis et al., 2005) using the In-Fusion® HD Cloning Kit to generate the vector, pMCG–ZmPRms–RNAi (Figure 1C). The PR10 intron inserted between the sense and antisense ZmPRms fragments facilitated efficient splicing and formation of hpRNA in the transgenic ZmPRms–RNAi plants.
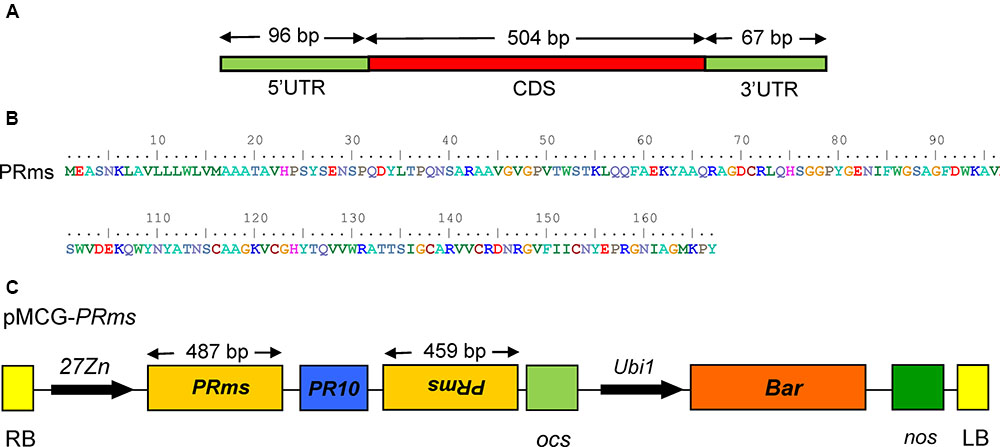
FIGURE 1. ZmPRms gene and vector construction. (A) Genomic structure of the Zea mays (Zm) PRms gene; (B) Amino acid sequence of the ZmPRms gene; (C) Vector diagram of the RNAi construct used for maize transformation to silence the ZmPRms gene (Abbreviations: 27Zn = maize endosperm-specific promoter, RB = right border, LB = left border, PR10 = intron (maize), Ubi1 = constitutive promoter, Bar = bialaphos resistance gene, ocs and nos = transcription terminators).
Agrobacterium-mediated transformation of maize (inbred B104) with the pMCG–ZmPRms–RNAi vector was carried out at the Iowa State University Plant Transformation Center (Frame et al., 2011). Several independent transgenic events were regenerated for further analysis. Putative transgenic plants and their progenies were grown in moist soil mix containing three parts Scott’s 360 Metro-Mix (Scotts Company, Marysville, OH, United States) and one part perlite in 3′′ (7.6 cm) pots. Seedlings were first grown in a growth chamber at 25°C under 16-h photoperiod (80 μmol m-2 s-1) for 4 weeks prior to transfer to 5 gal (0.02 cubic meter) pots in the greenhouse (27 ± 2°C).
Plants were screened by PCR using the ‘Phire Plant Direct PCR Kit’ (ThermoFisher Scientific; Cat# F160S) according to the manufacturer’s protocol. Primers used to screen putativeZmPRms–RNAi plants for the presence of the RNAi transgene cassette were 27zn945-F 5′-ccatgaagctgcctacagc-3′ and PR10_R 5′-cggaattccgtatggcaa-3′, while primers Bar_F: 5′-caccatcgtcaaccactacatcgagac-3′ and Bar_R: 5′-cagctgccagaaacccacgtcatgc-3′ (Rajasekaran et al., 2017) were used to screen empty vector transformed control plants. A 55°C annealing temperature and 30 s elongation time were used to amplify 1156 bp and 433 bp fragments that confirmed the presence of the RNAi expression cassette and the bar gene, respectively, in the transgenic plants.
An aflatoxin-producing A. flavus 70 strain expressing GFP (AF70-GFP; Rajasekaran et al., 2008) was obtained from the SRRC fungal collection (SRRC 1436; ARS-USDA, New Orleans, LA, United States). The fungal strain was grown on 2x concentrated V8 agar media (2x V8; Cary et al., 2015) for 7 days at 30°C with illumination. Conidia were harvested by flooding each plate with 20 ml of 0.02% (v/v) sterile Triton X-100 solution and gently dislodging conidia from the surface mycelia with a sterile scraper. Conidial suspensions were adjusted to 4 × 106 spores/ml prior to the kernel inoculation.
Undamaged and roughly uniform size T1 maize kernels (B104) collected from transgenic ZmPRms–RNAi and control plant lines were randomly assigned and processed according to a kernel-screening assay (KSA; Rajasekaran et al., 2013). All kernels were surface sterilized in 70% ethanol, air dried, and stored in sterile tubes. Kernels were inoculated by immersion into a 4 × 106 spores/ml suspension of the AF70-GFP strain followed by stirring for 3 min. The inoculum was then drained off and the kernels were transferred to plastic caps that were placed in a tray for each transgenic line. The kernels were incubated under high RH ( > 90%) at 31°C for 7 days in the dark. The filter paper inside the tray was kept moist by adding extra water when needed during the incubation period.
AF70-GFP infected maize kernels were harvested 7 days post inoculation (dpi), longitudinally sectioned, and photographed using a stereomicroscope (Nikon SMZ25, Melville, NY, United States) equipped with a GFP filter and a camera to capture images of GFP fluorescence (excitation 485 nm, emission 528 nm). Individual kernels frozen in liquid nitrogen were homogenized using a SPEX SamplePrep (Geno/Grinder, Metuchen, NJ, United States) and the ground seed samples (∼25 mg fresh weight) were extracted in 500 μl of Sorenson’s phosphate buffer (pH 7.0). Samples were vortexed for 30 s followed by centrifugation at 9000 × g for 15 min. A 100 μl aliquot of the supernatant was carefully transferred to a 96-well plate and GFP fluorescence was measured at an excitation wavelength of 485 nm and an emission wavelength of 535 nm using a fluorometer (BioTek, Synergy4, Winooski, VT, United States) along with the appropriate control (buffer only blank).
Homogenized maize kernel tissue (∼20–70 mg) was extracted with ethyl acetate/acetone (1:1)/0.1 % formic acid (1 ml) for 24 h at room temperature. The extracts were filtered through cotton plugs and the filtrates were concentrated under nitrogen to dryness. Each extract was re-dissolved in acetonitrile (1 mg/ml), filtered through a 0.22 μm Spin-X centrifuge tube filter, and analyzed on a Waters Acquity UPLC system (40% MeOH in water, BEH C18 1.7μm, 2.1 × 50 mm column) using fluorescence detection (Ex = 365 nm, Em = 440 nm). Samples were diluted 10-fold if the aflatoxin signal saturated the detector. Analytical standards (Sigma–Aldrich, St. Louis, MO, United States) were used to identify and quantify aflatoxins: aflatoxin B1 (AFB1, retention time = 4.60 min.); aflatoxin B2 (AFB2, retention time = 3.55 min.). Aflatoxin contents were expressed in ng/mg fresh weight of homogenized maize kernels.
The data used to identify potential maize genes involved in resistance to A. flavus infection and aflatoxin contamination was obtained from a set of RNA-seq data of the A. flavus–maize interaction publically available and located at NCBI [SRP082421] (Musungu et al., 2016). Additionally, data from the PiZeam interactome (Musungu et al., 2015) was utilized to detect additional gene targets by analyzing the overlap between the networks. For mining of the RNA-seq data, a two-pronged approach was utilized with PiZeaM, a Z. mays–A. flavus interactome (Musungu et al., 2015), and a published A. flavus/Z. mays gene co-expression network (Musungu et al., 2016). First, the RNA-seq data was analyzed as described in Musungu et al. (2016) and large regulatory networks inferred. The ‘R’ statistical language was then used to mine the networks for ZmPRms. After filtering by correlation strength and overlap, ZmPRms was predicted to be regulating multiple Z. mays and A. flavus genes. The sub-network was further analyzed using the PiZeaM interactome (Musungu et al., 2015) to identify protein–protein interactions in Z. mays, potentially revealing downstream targets of ZmPRms. This approach resulted in the identification of additional targets that would have been missed if differential expression had been used as the only metric cutoff to identify partners of ZmPRms.
RNA was isolated from individual homogenized maize kernels using the ‘SpectrumTM Plant Total RNA kit’ (Sigma–Aldrich, St. Louis, MO, United States) and cDNA was synthesized using iScriptTM cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR (qRT-PCR) was performed using SYBR green I chemistry and iCycler iQ5 Multicolor real-time PCR detection system (Bio-Rad). The thermocycling conditions included a pre-incubation at 95°C for 3 min, dye activation at 95°C for 10 s, primer annealing at 55°C for 30 s, elongation at 55°C for 50 s followed by a dissociation curve between 65°C and 95°C for 30 min (with 0.5°C increments). The primers used for qRT-PCR are presented in Supplementary Table S1. Gene expression was normalized by ΔΔCT analysis (Livak and Schmittgen, 2001) to Zea mays ribosomal structural gene GRMZM2G024838 expression (Shu et al., 2015) utilizing the gene expression analysis software package of the Bio-Rad iQ5.
A 700 bp upstream promoter region of the maize PRms gene was analyzed using the ‘PlantPAN 2.0’ web tool (Chow et al., 2016) to identify putative transcription factor–binding site motifs present in the promoter region.
For all experiments, typically three to five biological replicates were used per treatment. The data presented here are mean ± SE. Statistical significance between control and RNAi lines were determined by Student’s t-test. Significant difference between treatment and control was analyzed at ∗∗P ≤ 0.05 and/or ∗P ≤ 0.1 as indicated in the Figure legends.
PCR screening of genomic DNA isolated from the leaves of seven independent transgenic events of ZmPRms–RNAi plants were initially confirmed for the presence of an 1156 bp product spanning the 27 Zein promoters to the PR10 intron region of the ZmPRms–RNAi cassette (Figure 2A). Genomic DNA from both ZmPRms–RNAi and empty vector control plants amplified a region of 433 bp specific to the plant selection marker gene ‘Bar’ (Figure 2B). Based on the screening of fungal growth in the ZmPRms–RNAi silenced lines, three independent ZmPRms–RNAi lines that showed higher fungal growth were selected for further investigation to understand the impact of seed-specific ZmPRms silencing on A. flavus growth and aflatoxin production. Expression of the native ZmPRms gene in transgene positive individual seeds of RNAi lines and empty vector control plants inoculated with the AF70-GFP strain were examined by qRT-PCR. Seed-specific RNAi-mediated silencing of the native ZmPRms gene resulted in a ∼90–99% downregulation in expression of the gene (Figure 2C) in seeds of infected ZmPRms–RNAi lines compared to seeds of infected control lines. Line 1-5 showed the lowest relative expression (0.001) followed by the line 4–5 (0.104) and line 3–5 (0.100). Transgenic ZmPRms–RNAi plants were comparable to the wild type (WT) or empty vector transformed control plants with respect to plant morphology (Figure 2D).
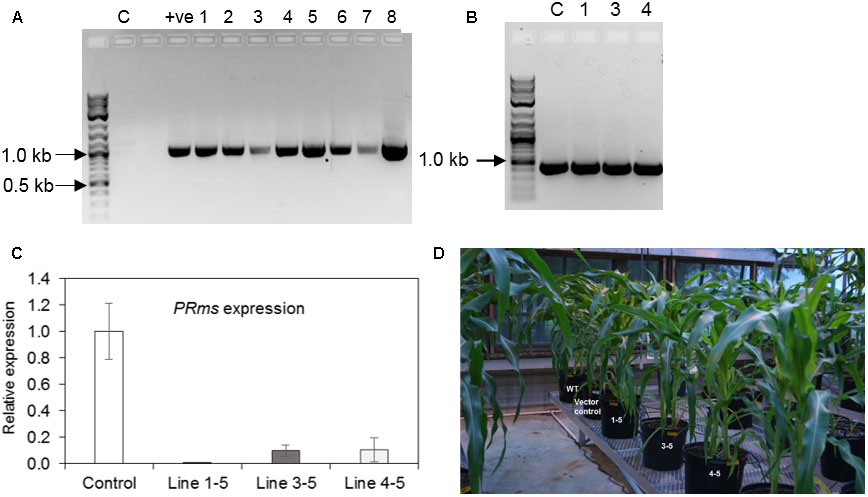
FIGURE 2. Confirmation of maize transgenic plants and plant phenotype. (A) Genomic DNA isolated from ZmPRms–RNAi lines and control plant were used to amplify a 1156 bp DNA fragment to confirm the presence of the RNAi cassette (C = empty vector transformed control plants; +ve = plasmid vector used as a PCR template; NEB 2-Log DNA ladder user as a DNA marker); (B) Genomic DNA isolated from ZmPRms–RNAi and empty vector transformed control (C) plants were used to amplify a 433 bp diagnostic DNA fragment to confirm the presence of the ‘Bar’ plant selection marker gene (Line 1 = 1–5, Line 3 = 3–5, Line 4 = 4–5; NEB 2-Log DNA ladder user as a DNA marker); (C) Relative expression of the native PRms gene in the kernels of empty vector transformed control and PRms-RNAi maize lines at 7 days post inoculation (dpi) [Gene expression was normalized to the maize ribosomal structural gene GRMZM2G024838 (Shu et al., 2015); data are mean ± SE of 3–4 biological replicates]; (D) Plant growth phenotype (uninfected) of ZmPRms–RNAi lines as compared to the wild type (WT) or empty vector transformed control transgenic maize plants.
Fungal growth in the seeds from ZmPRms–RNAi plant lines compared to control plants was qualitatively analyzed by fluorescence microscopy of longitudinally sectioned seeds (Figure 3A). Imaging of seed from the three ZmPRms–RNAi lines showed a significantly higher degree of GFP fluorescence and spread compared to seed from an empty vector control. Visual evidence of increased GFP fluorescence, primarily in the scutellar and adjacent endosperm tissue of the seed-correlated well with the data obtained from absolute quantification of GFP fluorescence. An increase of ∼250–350% in GFP fluorescence was observed in the ZmPRms–RNAi lines compared to the control with lines 1–5 showing the greatest increase followed by lines 4–5 and 3–5 (Figure 3B).
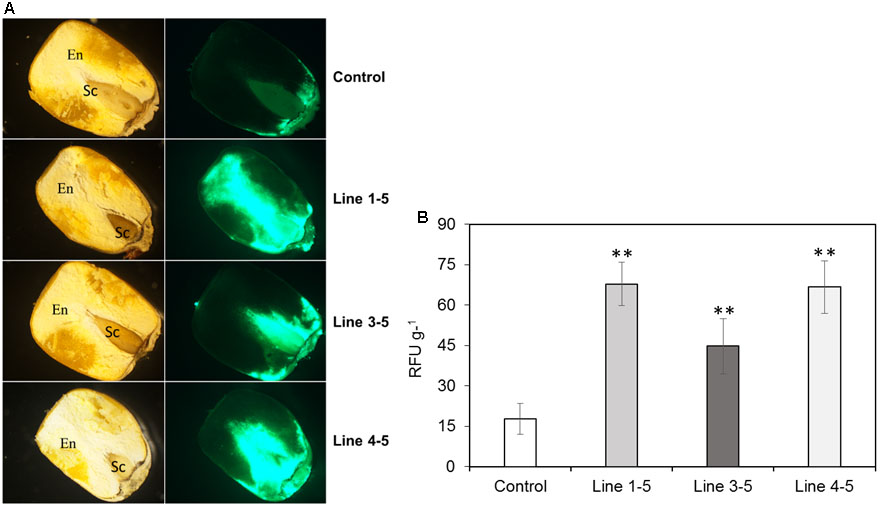
FIGURE 3. Fungal growth in the T1 generation maize kernels. (A) Growth of A. flavus-GFP at 7 dpi (as indicated by relative GFP fluorescence; Rajasekaran et al., 2013) in empty vector transformed control and ZmPRms–RNAi transgenic maize kernels (First column: light micrographs and second column: GFP fluorescence micrographs of longitudinal sections of kernels. En: endosperm; Sc: scutellum); (B) Quantification of GFP fluorescence in the kernels of empty vector transformed control and ZmPRms–RNAi maize transgenic plants at 7 days post inoculation (dpi) with A. flavus. Relative fluorescence units (RFU) is directly proportional to fungal growth in the kernels. Data are mean ± SE of 5 biological replicates. ∗∗Denotes significant difference between control and PRms silenced lines (P ≤ 0.05).
Silencing of the PRms gene in maize significantly affected aflatoxin content in the kernels of silenced lines as compared to the control. In general, the quantities of aflatoxin B1 were several folds higher than those of B2 in both control and silenced lines. A 4.6–7.4-fold increase (significant) in aflatoxin B1 content (ranging between 58 and 94 ng/mg FW) was observed in the ZmPRms–RNAi kernels (vs. control; Figure 4A) and a 4.2–6.0-fold increase (significant) in aflatoxin B2 content (ranging between 3.5 and 5 ng/mg FW) were observed in the ZmPRms–RNAi kernels (vs. control; Figure 4B).
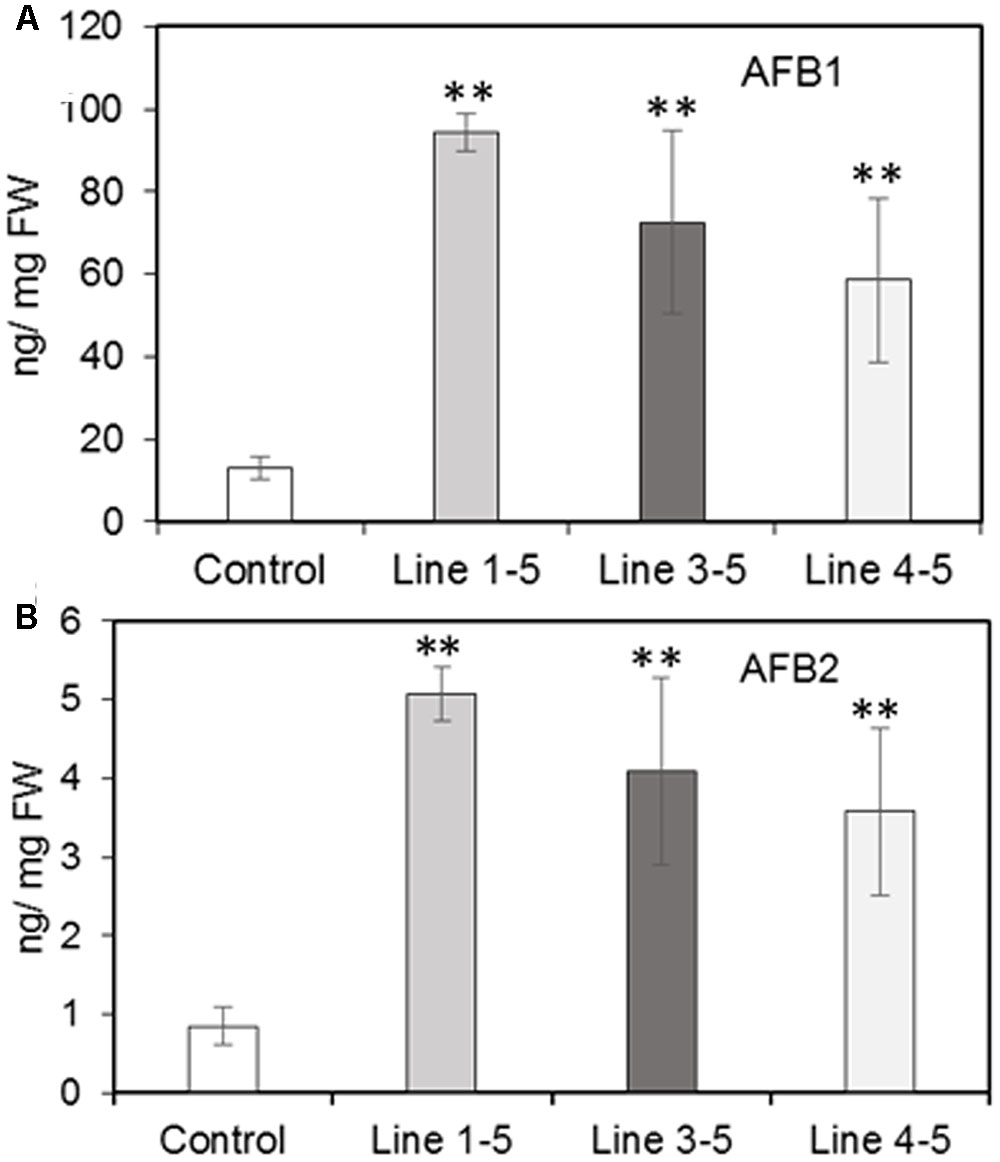
FIGURE 4. Aflatoxin (AF) contents in the control (empty vector transformed) and ZmPRms–RNAi transgenic maize kernels at 7 days post inoculation (dpi) with A. flavus. (A) AFB1; and (B) AFB2. Data are mean ± SE of 4 biological replicates. ∗∗Denotes significant difference between control and PRms silenced lines (P ≤ 0.05).
Analysis of the RNA-seq data resulting from the A. flavus–maize pathogenic interaction has previously identified ZmPRms as a kernel PR protein possibly involved in resistance against A. flavus (Musungu et al., 2016). Further gene regulatory network analysis (in the current study) of the previously published RNA-seq data identified downstream gene targets possibly regulated by ZmPRms during the A. flavus–maize interaction (Figure 5, Table 1 and Supplementary Figures S1–S3). These included candidates associated with pathogen responses in plants, LRR and NB-ARC domain-containing gene (GRMZM2G060714), RabGAP/TBC-domain containing gene (GRMZM2G156320), Rab28 defense-related gene (GRMZM2G472236), and F-Box gene (GRMZM2G008528). Several other genes belonging to diverse biochemical functions, namely anthocyanidin 3-O-glucosyltransferase (GRMZM2G165390), adenine nucleotide alpha hydrolases-like gene (GRMZM2G151425), inositol monophosphatase (GRMZM2G036007), plasma-membrane choline transporter (GRMZM2G330453), choline transporter (GRMZM2G330453), tRNA-His guanylyltransferase (GRMZM2G158901), and an ATP-binding microtubule motor family gene (GRMZM5G878823), were also identified as ZmPRms-regulated genes. Other transcription factor-related candidates included Myb (GRMZM2G003406) and ereb44 (GRMZM5G806839). Several other candidates referred to ‘uncharacterized’ (unknown biological function; Table 1) also appeared to be regulated by ZmPRms.
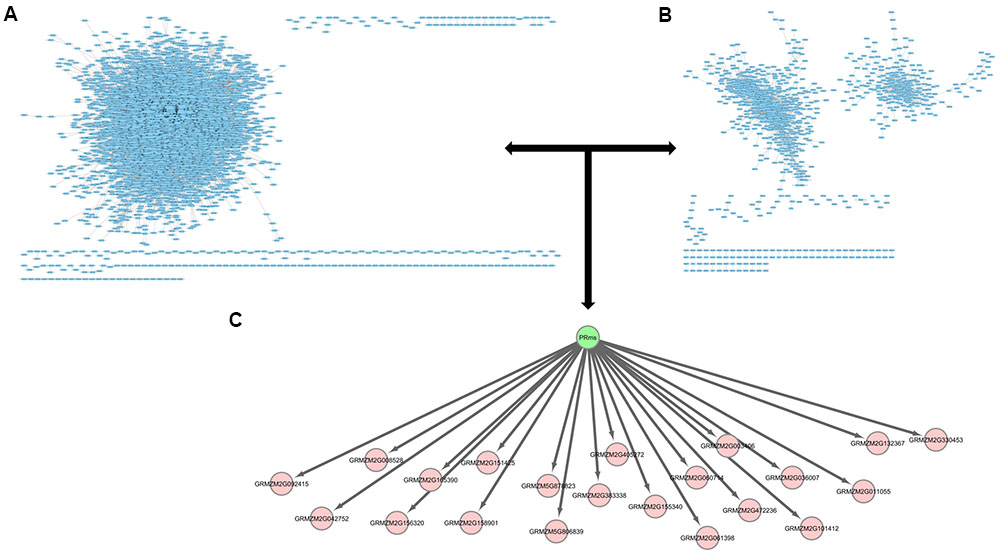
FIGURE 5. Potential downstream targets of ZmPRms. (A) The predicted maize interactome (Musungu et al., 2016); (B) a maize co-expression network (Musungu et al., 2016); (C) potential ZmPRms transcriptional targets.
Real-time expression analyses were performed on several of the aforementioned maize genes identified from the gene network analysis, that are possibly regulated by ZmPRms during the A. flavus–maize interaction (Table 1). Several disease resistance/biotic stress response–related genes, namely the LRR and NB-ARC domain-containing gene, RabGAP/TBC-domain containing gene, and F-Box gene, were downregulated in the ZmPRms–RNAi lines (Figure 6). Other genes associated with diverse biochemical functions including an adenine nucleotide alpha hydrolase-like gene, inositol monophosphatase, plasma-membrane choline transporter, and a tRNA-His guanylyltransferase were also downregulated in the ZmPRms silenced lines. Genes that were upregulated in the ZmPRms–RNAi lines included ereb44 (transcription factor), an ATP-binding microtubule motor family gene, and a Rab28 defense-related gene among which the Rab28 gene showed the highest upregulation (∼6–10-fold) in the ZmPRms silenced lines.
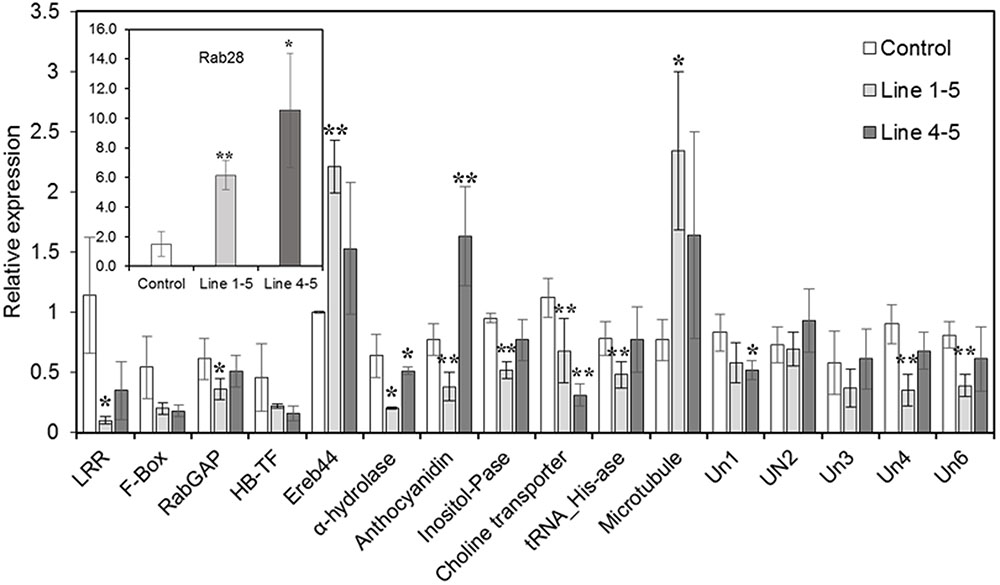
FIGURE 6. Relative expression of predicted ZmPRms-regulated downstream target (obtained from gene regulatory network analysis; Figure 5 and Table 1) candidate genes in the control (empty vector transformed) and ZmPRms–RNAi transgenic maize kernels at 7 days post inoculation (dpi) with A. flavus. Gene expression was normalized to the maize ribosomal structural gene GRMZM2G024838 (Shu et al., 2015). Data are mean ± SE of 3–4 biological replicates. ∗∗/∗Denote significant difference between control and PRms silenced lines (∗∗P ≤ 0.05, ∗P ≤ 0.1). Un1 (Uncharacterized 1; GRMZM2G042752), Un2 (Uncharacterized 2; GRMZM2G061398), Un3 (Uncharacterized 3; GRMZM2G092415), Un4 (Uncharacterized 4; GRMZM2G101412), Un6 (Uncharacterized 6; GRMZM2G383338).
Based on the physiological responses of the PRms gene in maize, we analyzed the upstream promoter region to identify any stress-related motifs that might help us better understand the biological function of this gene. Analysis of the 700 bp upstream promoter region of the ZmPRms gene revealed the presence of several transcription factor–binding motif sites associated with both biotic and abiotic stress responses in plants. These include MYBPLANT, MybSANT, WRKY, ANAERO1CONSENSUS motifs, and several others. Details of the locations, consensus sequences, and physiological functions of these cis elements are presented in Table 2.
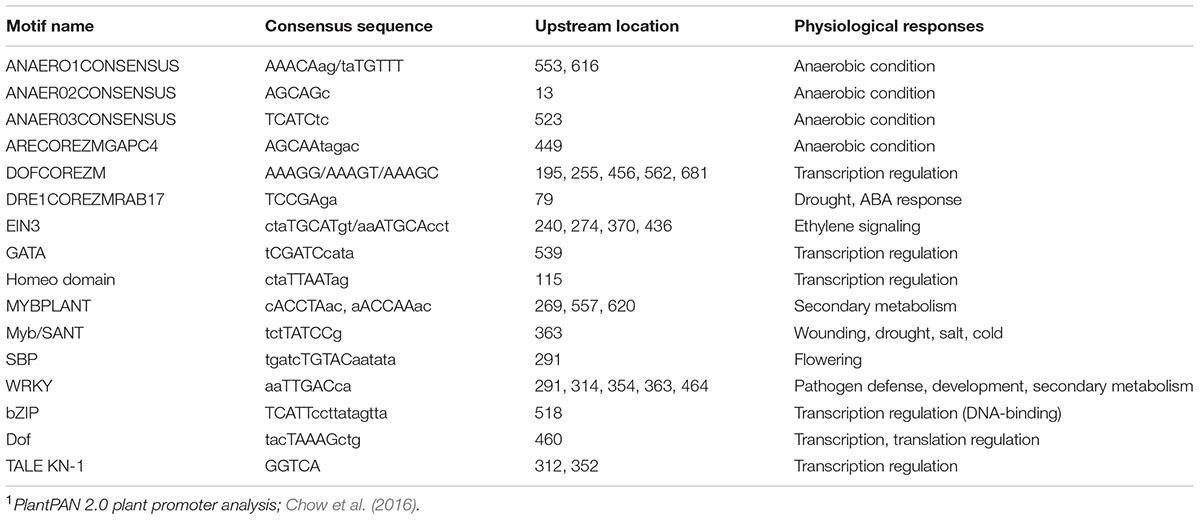
TABLE 2. Putative transcription factor–binding site motifs present in the 700 bp upstream promoter region of the maize PRms gene1.
Different approaches have been employed to develop resistance to A. flavus infection and aflatoxin contamination in maize. These include conventional breeding approaches that introgress resistance genes into agronomically important maize varieties. Natural sources of genetic resistance against A. flavus have been reported in maize but introgression of such resistance into elite germplasm is often hampered with long delays due to the quantitative nature of the trait as well as high phenotypic variability due to genotype–environment interactions (Zila et al., 2013; Warburton and Williams, 2014; Hawkins et al., 2015). As a result, there is a need to evaluate alternative approaches to provide durable resistance. Besides conventional breeding, modern genetic engineering tools have been employed to incorporate A. flavus resistance in susceptible maize varieties. These include transgenic expression of natural and synthetic resistance genes and host-induced RNAi-based gene silencing of A. flavus genes that are critical in fungal pathogenesis and aflatoxin production (reviewed by Bhatnagar-Mathur et al., 2015; Majumdar et al., 2017; Thakare et al., 2017).
Pathogenesis-related (PR) proteins are products of defense genes, which accumulate at the pathogen infection site and contribute to SAR (reviewed by Fu and Dong, 2013). Among different PR proteins reported in maize kernels, ZmPRms is believed to be involved in resistance to fungal pathogens. Murillo et al. (1999), using cellular and subcellular immunolocalization tools, demonstrated the accumulation of the ZmPRms protein in the aleurone and inner parenchyma cells of the scutellum post F. verticillioides infection. Early activation of the ZmPRms protein prior to onset of visual symptoms of A. flavus colonization clearly indicated a critical role of ZmPRms in kernel resistance to A. flavus (Shu et al., 2015). The work described in the present study functionally elucidates the contribution of ZmPRms toward A. flavus resistance and aflatoxin production during kernel infection. Higher levels of A. flavus growth (as determined by GFP fluorescence; Figure 3A) was primarily observed in the scutellar and adjacent endosperm tissue and correlated with the downregulation of ZmPRms production in PRms-RNAi kernels. This supports earlier observations on the spatial distribution of the ZmPRms protein in maize seeds during fungal infection (Murillo et al., 1999; Shu et al., 2015). The observed increase in aflatoxin content in the transgenic ZmPRms–RNAi maize kernels (Figure 6) correlated with increased fungal load in the kernels compared to a control line (Figure 3). Earlier work by Raventós et al. (1995) had demonstrated the role of ERE and enhancing element in the ZmPRms promoter in response to fungal (Fusarium sp.) elicitors. Presence of these cis elements in the ZmPRms promoter significantly induced the expression of the reporter gene (Tn9 chloramphenicol acetyltransferase; CAT) when exposed to fungal elicitors. Further in silico analysis of the ZmPRms promoter in the current study revealed the presence of several stress-related motifs (Table 2) associated with biotic/abiotic stress responses in plants (Chow et al., 2016). The role of these putative response elements in ZmPRms expression will be the subject of future studies. Besides ZmPRms, several other kernel-specific PR proteins are induced upon A. flavus infection but only a few are functionally characterized as to their contribution toward aflatoxin resistance (reviewed by Chen et al., 2015; Pechanova and Pechan, 2015). In future studies, it will be interesting to see whether the production of any of these PR proteins is affected due to the downregulation of ZmPRms in the kernels of ZmPRms–RNAi lines.
Defense response against fungal pathogens in maize is regulated by complex metabolic networks (Musungu et al., 2016). This involves typical transcriptional induction of genes associated with phytohormone-related defense signaling pathways associated with salicylic acid, ethylene, and jasmonic acid (JA) biosynthesis. Both A. flavus and F. verticillioides were shown to induce these phytohormone-related genes leading to the induction of PR proteins including ZmPRms (Murillo et al., 1999; Shu et al., 2015; Tang et al., 2015; Wang et al., 2016). Genome-wide association studies using aflatoxin-resistant inbred maize lines showed a strong correlation between high expression of genes associated with the JA biosynthetic pathway and reduced aflatoxin accumulation in the grains (Tang et al., 2015). Besides JA pathway genes, high expression of other genes, including a leucine-rich repeat protein kinase, expansin B3, reversion-to-ethylene sensitivity1, and an adaptor protein complex2 gene were also reported in the A. flavus resistant maize lines. Transcriptomic analysis has identified several candidate genes that are involved in peanut response to A. flavus (Wang et al., 2016). The involvement of specific metabolic pathway-related genes in aflatoxin resistance in certain crops/genotypes could be a genotype-related trait. Apparently, ZmPRms interacts with different categories of genes to orchestrate resistance against A. flavus. Several candidate genes (Table 1) identified from gene regulatory network analysis possibly regulated by ZmPRms include, genes associated with carbohydrate metabolism (anthocyanidin 3-O-glucosyltransferase), disease resistance (LRR-NB-ARC, RabGAP/TBC, Rab28), transcription factors [Myb, F-Box, and ereb44 (AP2-type)], and other metabolite-related (choline, inositol). Many of these putative ZmPRms-regulated genes are known to be induced by A. flavus and F. verticillioides in maize (Walley et al., 2013; Lanubile et al., 2014; Shu, 2014; Musungu et al., 2016). The majority of these genes were downregulated in the ZmPRms–RNAi lines except for a few that were upregulated in the ZmPRms silenced lines (Figure 5). The upregulated genes include mainly Rab28, ereb44, and microtubule (ATP-binding microtubule motor family protein). The significantly higher expression (∼6–10-fold) of the Rab28 gene in the ZmPRms–RNAi lines might indicate a compensatory role in A. flavus resistance due to reduced expression of ZmPRms. The involvement of many of these candidate genes in disease resistance and abiotic stress tolerance has been reported. Functional characterization (through overexpression in plants) of some of these genes such as Myb, F-Box, LRR, Rab28, and AP2 (ethylene responsive) has resulted in increased resistance against a wide variety of pathogens that include fungi, bacteria, and viruses besides contributing to abiotic stress tolerance in some cases (Cao et al., 2008; Yan et al., 2011; Amara et al., 2013; Jisha et al., 2015; Liu et al., 2016; Xing et al., submitted). Diverse sources of genetic background with substantial aflatoxin resistance have been described in maize (reviewed by Warburton and Williams, 2014; Brown et al., 2016), although, the exact mechanism of such resistance is not fully understood. It will be interesting to see whether any of these A. flavus resistant maize genotypes have a positive correlation between higher basal expression and/or rapid activation of ZmPRms (in response to A. flavus) and increased aflatoxin resistance.
In this study, we have demonstrated a significant role for ZmPRms in resistance to A. flavus infection of maize kernels through global regulation of genes associated with biotic and abiotic stress responses in plants. This includes genes associated with disease resistance, carbohydrate metabolism, and transcription factors that are known to be upregulated in plants under stress conditions. The observed increase in A. flavus growth and aflatoxin production in the ZmPRms–RNAi lines supports the RNA-seq interactome analysis that indicated ZmPRms might serve as a major network hub for regulation of downstream resistance-associated gene expression. ZmPRms–RNAi lines and their progenies were morphologically normal suggesting that it will be a good candidate host-resistance gene for overexpression in maize for increased resistance to A. flavus and possibly against other pathogens. The results presented here are promising and it might be possible to fine tune ZmPRms expression in a tissue-specific manner (using modern functional genomic tools) in future or use ZmPRms expression as a marker to screen for A. flavus resistant maize genotypes to reduce aflatoxin contamination in maize, and potentially in other economically important crop plants.
JC, KR, GP,DB, andRMconceived and designed the experiments. RM, CS, CC-W, and QW performed the experiments. BM, AF, MG, RM, CS, ML, and GP analyzed the data. RM, JC, KR, AF, and ML wrote the paper. All authors reviewed and approved the final manuscript.
This research was funded by the United States Department of Agriculture, Agricultural Research Service (No. 6054-42000-025-00D).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01758/full#supplementary-material
FIGURE S1 | Maize Interactome.
FIGURE S2 | Co-expression network.
FIGURE S3 | Transcriptional targets.
Amara, I., Capellades, M., Ludevid, M. D., Pagès, M., and Goday, A. (2013). Enhanced water stress tolerance of transgenic maize plants over-expressing LEA Rab28 gene. J. Plant Physiol. 170, 864–873. doi: 10.1016/j.jplph.2013.01.004
Bhatnagar-Mathur, P., Sunkara, S., Bhatnagar-Panwar, M., Waliyar, F., and Sharma, K. K. (2015). Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci. 234, 119–132. doi: 10.1016/j.plantsci.2015.02.009
Brown, R., Williams, W., Windham, G., Menkir, A., and Chen, Z.-Y. (2016). Evaluation of African-bred maize germplasm lines for resistance to aflatoxin accumulation. Agronomy 6:24.
Cao, Y., Yang, Y., Zhang, H., Li, D., Zheng, Z., and Song, F. (2008). Overexpression of a rice defense-related F-box protein gene OsDRF1 in tobacco improves disease resistance through potentiation of defense gene expression. Physiol. Plant. 134, 440–452. doi: 10.1111/j.1399-3054.2008.01149.x
Cary, J. W., Rajasekaran, K., Brown, R. L., Luo, M., Chen, Z.-Y., and Bhatnagar, D. (2011). Developing resistance to aflatoxin in maize and cottonseed. Toxins 3, 678–696. doi: 10.3390/toxins3060678
Cary, J. W., Uka, V., Han, Z., Buyst, D., Harris-Coward, P. Y., Ehrlich, K. C., et al. (2015). An Aspergillus flavus secondary metabolic gene cluster containing a hybrid PKS–NRPS is necessary for synthesis of the 2-pyridones, leporins. Fungal Genet. Biol. 81, 88–97. doi: 10.1016/j.fgb.2015.05.010
Casacuberta, J. M., Puigdomènech, P., and Segundo, B. S. (1991). A gene coding for a basic pathogenesis-related (PR-like) protein from Zea mays. Molecular cloning and induction by a fungus (Fusarium moniliforme) in germinating maize seeds. Plant Mol. Biol. 16, 527–536. doi: 10.1007/bf00023419
Casacuberta, J. M., Raventós, D., Puigdoménech, P., and San Segundo, B. (1992). Expression of the gene encoding the PR-like protein PRms in germinating maize embryos. Mol. Gen. Genet. 234, 97–104. doi: 10.1007/bf00272350
Chen, Z.-Y., Brown, R. L., Damann, K. E., and Cleveland, T. E. (2010). PR10 expression in maize and its effect on host resistance against Aspergillus flavus infection and aflatoxin production. Mol. Plant Pathol. 11, 69–81. doi: 10.1111/j.1364-3703.2009.00574.x
Chen, Z.-Y., Rajasekaran, K., Brown, R. L., Sayler, R. J., and Bhatnagar, D. (2015). Discovery and confirmation of genes/proteins associated with maize aflatoxin resistance. World Mycotoxin J. 8, 211–224. doi: 10.3920/wmj2014.1732
Chow, C.-N., Zheng, H.-Q., Wu, N.-Y., Chien, C.-H., Huang, H.-D., Lee, T.-Y., et al. (2016). PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44, D1154–D1160. doi: 10.1093/nar/gkv1035
Dolezal, A. L., Shu, X., Obrian, G. R., Nielsen, D. M., Woloshuk, C. P., Boston, R. S., et al. (2014). Aspergillus flavus infection induces transcriptional and physical changes in developing maize kernels. Front. Microbiol. 5:384. doi: 10.3389/fmicb.2014.00384
Fountain, J. C., Scully, B. T., Ni, X., Kemerait, R. C., Lee, R. D., Chen, Z.-Y., et al. (2014). Environmental influences on maize-Aspergillus flavus interactions and aflatoxin production. Front. Microbiol. 5:40. doi: 10.3389/fmicb.2014.00040
Frame, B., Main, M., Schick, R., and Wang, K. (2011). “Genetic transformation using maize immature zygotic embryos,” in Plant Embryo Culture: Methods and Protocols, eds T. A. Thorpe and E. C. Yeung (Totowa, NJ: Humana Press), 327–341.
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gómez-Ariza, J., Campo, S., Rufat, M., Estopà, M., Messeguer, J., Segundo, B. S., et al. (2007). Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related prms protein in rice plants. Mol. Plant Microbe Interact. 20, 832–842. doi: 10.1094/MPMI-20-7-0832
Hawkins, L. K., Mylroie, J. E., Oliveira, D. A., Smith, J. S., Ozkan, S., Windham, G. L., et al. (2015). Characterization of the maize chitinase genes and their effect on Aspergillus flavus and aflatoxin accumulation resistance. PLOS ONE 10:e0126185. doi: 10.1371/journal.pone.0126185
Ismaiel, A., and Papenbrock, J. (2015). Mycotoxins: producing fungi and mechanisms of phytotoxicity. Agriculture 5, 492–537.
Jisha, V., Dampanaboina, L., Vadassery, J., Mithöfer, A., Kappara, S., and Ramanan, R. (2015). Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLOS ONE 10:e0127831. doi: 10.1371/journal.pone.0127831
Kebede, H., Abbas, H., Fisher, D., and Bellaloui, N. (2012). Relationship between aflatoxin contamination and physiological responses of corn plants under drought and heat stress. Toxins 4, 1385–1403. doi: 10.3390/toxins4111385
Kumar, P., Mahato, D. K., Kamle, M., Mohanta, T. K., and Kang, S. G. (2016). Aflatoxins: a global concern for food safety, human health and their management. Front. Microbiol. 7:2170. doi: 10.3389/fmicb.2016.02170
Lanubile, A., Ferrarini, A., Maschietto, V., Delledonne, M., Marocco, A., and Bellin, D. (2014). Functional genomic analysis of constitutive and inducible defense responses to Fusarium verticillioides infection in maize genotypes with contrasting ear rot resistance. BMC Genomics 15:710. doi: 10.1186/1471-2164-15-710
Liu, Z., Luan, Y., Li, J., and Yin, Y. (2016). Expression of a tomato MYB gene in transgenic tobacco increases resistance to Fusarium oxysporum and Botrytis cinerea. Eur. J. Plant Pathol. 144, 607–617. doi: 10.1007/s10658-015-0799-0
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Majumdar, R., Rajasekaran, K., and Cary, J. W. (2017). RNA interference (RNAi) as a potential tool for control of mycotoxin contamination in crop plants: concepts and considerations. Front. Plant Sci. 8:200. doi: 10.3389/fpls.2017.00200
McGinnis, K., Chandler, V., Cone, K., Kaeppler, H., Kaeppler, S., Kerschen, A., et al. (2005). Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol. 392, 1–24. doi: 10.1016/S0076-6879(04)92001-0
Mitchell, N. J., Bowers, E., Hurburgh, C., and Wu, F. (2016). Potential economic losses to the USA corn industry from aflatoxin contamination. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 33, 540–550. doi: 10.1080/19440049.2016.1138545
Murillo, I., Cavallarin, L., and Segundo, B. S. (1999). Cytology of infection of maize seedlings by Fusarium moniliforme and immunolocalization of the pathogenesis-related PRms protein. Phytopathology 89, 737–747. doi: 10.1094/PHYTO.1999.89.9.737
Musungu, B., Bhatnagar, D., Brown, R. L., Fakhoury, A. M., and Geisler, M. (2015). A predicted protein interactome identifies conserved global networks and disease resistance subnetworks in maize. Front. Genet. 6:201. doi: 10.3389/fgene.2015.00201
Musungu, B. M., Bhatnagar, D., Brown, R. L., Payne, G. A., Obrian, G., Fakhoury, A. M., et al. (2016). A network approach of gene co-expression in the Zea mays/Aspergillus flavus pathosystem to map host/pathogen interaction pathways. Front. Genet. 7:206. doi: 10.3389/fgene.2016.00206
Pechanova, O., and Pechan, T. (2015). Maize-pathogen interactions: an ongoing combat from a proteomics perspective. Int. J. Mol. Sci. 16, 28429–28448. doi: 10.3390/ijms161226106
Rajasekaran, K., Cary, J. W., Cotty, P. J., and Cleveland, T. E. (2008). Development of a GFP-expressing Aspergillus flavus strain to study fungal invasion, colonization, and resistance in cottonseed. Mycopathologia 165, 89–97. doi: 10.1007/s11046-007-9085-9
Rajasekaran, K., Majumdar, R., Sickler, C., Wei, Q., Cary, J., and Bhatnagar, D. (2017). Fidelity of a simple liberty leaf-painting assay to validate transgenic maize plants expressing the selectable marker gene, bar. J. Crop Improv. 31, 628–636. doi: 10.1080/15427528.2017.1327913
Rajasekaran, K., Sickler, C. M., Brown, R. L., Cary, J. W., and Bhatnagar, D. (2013). Evaluation of resistance to aflatoxin contamination in kernels of maize genotypes using a GFP-expressing Aspergillus flavus strain. World Mycotoxin J. 6, 151–158. doi: 10.3920/wmj2012.1497
Raventós, D., Jensen, A. B., Rask, M.-B., Casacuberta, J. M., Mundy, J., and San Segundo, B. (1995). A 20 bp cis-acting element is both necessary and sufficient to mediate elicitor response of a maize PRms gene. Plant J. 7, 147–155. doi: 10.1046/j.1365-313X.1995.07010147.x
Ryals, J. A., Neuenschwander, U. H., Willits, M. G., Molina, A., Steiner, H. Y., and Hunt, M. D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819.
Schubert, M., Houdelet, M., Kogel, K.-H., Fischer, R., Schillberg, S., and Nölke, G. (2015). Thanatin confers partial resistance against aflatoxigenic fungi in maize (Zea mays). Transgenic Res. 24, 885–895. doi: 10.1007/s11248-015-9888-2
Shepherd, C. T., and Scott, M. P. (2009). Construction and evaluation of a maize (Zea mays) chimaeric promoter with activity in kernel endosperm and embryo. Biotechnol. Appl. Biochem. 52, 233–243. doi: 10.1042/BA20070269
Shu, X. (2014). Pathogenesis and Host Response during Infection of Maize Kernels by Aspergillus flavus and Fusarium verticillioides. Raleigh, NC: North Carolina State University.
Shu, X., Livingston, D. P., Franks, R. G., Boston, R. S., Woloshuk, C. P., and Payne, G. A. (2015). Tissue-specific gene expression in maize seeds during colonization by Aspergillus flavus and Fusarium verticillioides. Mol. Plant Pathol. 16, 662–674. doi: 10.1111/mpp.12224
Tang, J. D., Perkins, A., Williams, W. P., and Warburton, M. L. (2015). Using genome-wide associations to identify metabolic pathways involved in maize aflatoxin accumulation resistance. BMC Genomics 16:673. doi: 10.1186/s12864-015-1874-9
Thakare, D., Zhang, J., Wing, R. A., Cotty, P. J., and Schmidt, M. A. (2017). Aflatoxin-free transgenic maize using host-induced gene silencing. Sci. Adv. 3:e1602382. doi: 10.1126/sciadv.1602382
Umesha, S., Manukumar, H. M. G., Chandrasekhar, B., Shivakumara, P., Shiva Kumar, J., Raghava, S., et al. (2017). Aflatoxins and food pathogens: impact of biologically active aflatoxins and their control strategies. J. Sci. Food Agric. 97, 1698–1707. doi: 10.1002/jsfa.8144
Walley, J. W., Shen, Z., Sartor, R., Wu, K. J., Osborn, J., Smith, L. G., et al. (2013). Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc. Natl. Acad. Sci. U.S.A. 110, E4808–E4817. doi: 10.1073/pnas.1319113110
Wang, H., Lei, Y., Yan, L., Wan, L., Ren, X., Chen, S., et al. (2016). Functional genomic analysis of Aspergillus flavus interacting with resistant and susceptible peanut. Toxins 8:46. doi: 10.3390/toxins8020046
Warburton, M. L., and Williams, W. P. (2014). Aflatoxin resistance in maize: what have we learned lately? Adv. Bot. 2014:352831. doi: 10.1155/2014/352831
Yan, Y.-S., Chen, X.-Y., Yang, K., Sun, Z.-X., Fu, Y.-P., Zhang, Y.-M., et al. (2011). Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant 4, 190–197. doi: 10.1093/mp/ssq066
Keywords: PRms, RNAi, Aspergillus flavus, maize, aflatoxin resistance, gene network analysis
Citation: Majumdar R, Rajasekaran K, Sickler C, Lebar M, Musungu BM, Fakhoury AM, Payne GA, Geisler M, Carter-Wientjes C, Wei Q, Bhatnagar D and Cary JW (2017) The Pathogenesis-Related Maize Seed (PRms) Gene Plays a Role in Resistance to Aspergillus flavus Infection and Aflatoxin Contamination. Front. Plant Sci. 8:1758. doi: 10.3389/fpls.2017.01758
Received: 22 June 2017; Accepted: 25 September 2017;
Published: 17 October 2017.
Edited by:
Essaid Ait Barka, University of Reims Champagne-Ardenne, FranceReviewed by:
Zonghua Wang, Fujian Agriculture and Forestry University, ChinaCopyright © 2017 Majumdar, Rajasekaran, Sickler, Lebar, Musungu, Fakhoury, Payne, Geisler, Carter-Wientjes, Wei, Bhatnagar and Cary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeffrey W. Cary, jeff.cary@ars.usda.gov
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.