- Department of Plant Science, Faculty of Agricultural and Environmental Sciences, McGill University, Sainte-Anne-de-Bellevue, QC, Canada
Salinity affects plant growth and is a major abiotic stress that limits crop productivity. It is well-understood that environmental adaptations and genetic traits regulate salinity tolerance in plants, but imparting the knowledge gained towards crop improvement remain arduous. Harnessing the potential of beneficial microorganisms present in the rhizosphere is an alternative strategy for improving plant stress tolerance. This review intends to elucidate the understanding of salinity tolerance mechanisms attributed by plant growth promoting rhizobacteria (PGPR). Recent advances in molecular studies have yielded insights into the signaling networks of plant–microbe interactions that contribute to salt tolerance. The beneficial effects of PGPR involve boosting key physiological processes, including water and nutrient uptake, photosynthesis, and source-sink relationships that promote growth and development. The regulation of osmotic balance and ion homeostasis by PGPR are conducted through modulation of phytohormone status, gene expression, protein function, and metabolite synthesis in plants. As a result, improved antioxidant activity, osmolyte accumulation, proton transport machinery, salt compartmentalization, and nutrient status reduce osmotic stress and ion toxicity. Furthermore, in addition to indole-3-acetic acid and 1-aminocyclopropane-1-carboxylic acid deaminase biosynthesis, other extracellular secretions of the rhizobacteria function as signaling molecules and elicit stress responsive pathways. Application of PGPR inoculants is a promising measure to combat salinity in agricultural fields, thereby increasing global food production.
Introduction
Climate change has exacerbated the severity of environmental stressors and affects crop production worldwide as part of the present Anthropocene Era. At the same time, there is a need to maintain food security for a growing global population through increases in crop production, while also forging agriculture more sustainable. Going forward, the quality of land and water will be critically pivotal for agriculture. Excess salt concentration in soil and water resources declines agricultural productivity, turns fertile fields to marginal lands, and leads to their abandonment. The Food and Agriculture Organization estimates that salinity has affected more than 6% of land area. Much of this land is not under cultivation but, a substantial proportion of the cultivated land, which constitutes 45 million ha of irrigated land (20% of total) and 32 million ha under dryland agriculture (about 2% of total) has been affected (Munns and Tester, 2008). The proportion of salinized land area might increase owing to climate change conditions conducive for salt accumulation (Othman et al., 2006).
Soluble salts deteriorate the fertility of soil by causing adverse effects on plant growth and development (Munns and Tester, 2008). Osmotic stress is the immediate impact of salinity (occurs within minutes) due to hypertonic conditions and ion toxicity (occurs over several hours to days and weeks) is the result of toxic ions (Na+ and Cl-) accumulating in the cells. Perturbed water balance and ion homeostasis affect hormonal status, transpiration, photosynthesis, translocation of nutrients, and other metabolic processes (Munns, 2002a). Beneficial soil microbiota enhance soil-water-plant relations through intricate mechanisms and subtle signaling cues that are not yet well-understood. A widely-proven notion is that the ability of soil microbes to manipulate phytohormonal signaling and trigger several other mechanisms to work in an integrated fashion contribute to enhanced stress tolerance in plants (Dodd and Perez-Alfocea, 2012). Inoculation of crop plants with beneficial microbes is gaining agronomic importance since they facilitate cultivation under saline-prone conditions by improving salt tolerance and hence, restoring yield (Lugtenberg et al., 2013). Bacteria isolated from extreme environments such as deserts and oceans have been shown to induce salt tolerance in crop plants. For example, a Pseudomonas fluorescens strain isolated from date-palm rhizosphere in Saharan region promoted root growth in maize (Zea mays) seedlings under salt stress (Zerrouk et al., 2016). Wheat plants (Triticum aestivum) inoculated with Serratia sp. Sl-12, a halophilic bacterium isolated from a salt lake showed improved salt tolerance and increased shoot biomass (Singh and Jha, 2016).
This review focuses on the evaluation of plant growth promoting rhizobacteria (PGPR) within the context of systems biology approaches for the alleviation of salinity stress with a brief overview of the causes for salinity and courses of plant tolerance. Recent advances in ‘omics’ technologies deliver a holistic understanding of the regulatory networks of stress responses modulated by the PGPR. Further, the reader may refer to comprehensive reviews on utilization of other beneficial microorganisms including arbuscular mycorrhizal fungi (AMF), endosymbionts, halotolerant, and phyllosphere bacteria to alleviate salinity stress (Yang et al., 2009; Dodd and Perez-Alfocea, 2012; Glick, 2012; Vorholt, 2012; Egamberdieva and Lugtenberg, 2014).
Salinity
Salinity is one of the major abiotic stressors that undermines plant growth and development (Pitman and Lauchli, 2002). Soil salinization is caused by natural or human activities that increase the concentration of dissolved salts, predominantly sodium chloride in the soil. Primary salinity is caused by natural processes, leading to significant salt accumulation in soil and groundwater over extended periods of time, which result in the formation of salt lakes, salt marshes, marine sediments, and salt scalds in the landscape. Sources of primary salinity may arise from weathering of rocks and minerals that releases soluble salts, precipitation that washes these salts downstream, wind-borne salts from oceans and sand dunes that are deposited inland, and influx of seawater followed by subsequent retreat (Pitman and Lauchli, 2002; Rengasamy, 2002).
Cultivation operations such as land clearing, excessive irrigation, and inadequate drainage are the reasons for secondary salinity. Native vegetation sustains the water table below the subsoil zone with deep roots in semi-arid and arid regions. Replacing perennial species with shallow rooted annual crops and long fallows increases water table leakage and groundwater recharge, which consecutively raises the water table level. Salt is deposited in the topsoil as the water evaporates, resulting in dryland salinity and may eventually form a salt scald. Salinity effects can be more detrimental when the groundwater table is high, as prominent in arid and coastal areas where only salt-tolerant plants (halophytes) grow (Doering and Sandoval, 1981; Rengasamy, 2002). Irrigated lands are more prone to salinity than drylands because irrigation water deposits salt behind, year after year. Secondary salinization has degenerated vast tracts of irrigated lands to the point that they are no longer economical for cultivation. Plants are often supplied with more water than they can utilize during evapotranspiration. For example, irrigation coupled with instances of heavy rainfall accelerates infiltration and groundwater recharge rates that raise the water table faster than it can drain. As the water table rises, it mobilizes dissolved salts from underground rocks close to the root zone. When the water table is within two meters of the soil surface in clay soils (less than a meter in sandy soils), there is a high probability of salt accumulation in the topsoil and salt stress to plants. Salt is also discharged and redistributed by surface runoff or leached down into soil profile by rainfall and then move laterally to watercourses (Sharma and Prihar, 1973; Pitman and Lauchli, 2002).
Poorly drained soils also suffer from waterlogging in irrigated areas. Clay soil (fine-textured) is less permeable than loam (medium-textured) and sandy soil (coarse-textured) and hence, it has high water holding capacity with low infiltration rate. Water can be stored and used by plants for a long time in clay soil but will not quickly transmit salt away from the root zone. The low porosity of clay soil acts as an impervious layer, causing inadequate drainage (Nassar and Horton, 1999). Inefficient irrigation and drainage systems lead to poor water distribution, resulting in over-irrigated waterlogged areas or under-irrigated water deficit areas, both causing salt accumulation. Waterlogging aggravates salinity stress by limiting aeration and nutrient supply to plants while proper grading and installation of drains to carry excess water and dissolved salts away from water stagnant areas may solve these problems. Groundwater mounds can develop in irrigated areas and force saline groundwater into waterways. Irrigation with salt-rich water increases salt being added to the soil and requires more water to leach out salts to prevent them from accumulating in the topsoil. Leaching reduces salinity levels when there is sufficient drainage and the groundwater table is deep. Conservation farming practices recommend appropriate methods to improve soil structure and irrigation efficiency (Shalhevet, 1994; Bauder and Brock, 2001).
The amount of salt stored in the soil also depends on soil type, with sandy soil having low and high capacity for clay loam minerals due to Na+ bound to negatively charged clay particles. Soil with ECe (electrical conductivity of saturated paste extract) of 4 dS m-1 is defined as saline by the USDA salinity laboratory. Most crop species are affected by ECe of less than 4 dS m-1 and thus, saline soil inhibits the yield of crops. Salinity caused by irrigation schemes has been recognized as a serious problem around the world since irrigated land is, on average, twice as productive as rain-fed land and produces about one-third of global food (Munns and Tester, 2008). Because salinity and water are inextricably linked, climate changes drive extreme consequences on agriculture when drought or flooding hit vulnerable regions. Salinization management has focused on improving irrigation water quality and soil drainage to strategically increase salt acclimation in crops (Pitman and Lauchli, 2002).
Salt Tolerance in Plants
Salinity tolerance in plants is dependent on its physiological mechanisms, duration of exposure to saline conditions, concentration of salt around roots, local soil–water relations, and microclimate conditions (temperature, humidity, etc.). Salt tolerance is usually quantified over a given period as survival, vegetative growth, or harvestable biomass at different physiological stages of the plant in saline versus non-saline conditions (Munns, 2002b). Crop yield decreases when salt concentration is above the threshold salinity level due to salt affecting the development of reproductive structures or translocation of nutrient reserves. There is a great diversity in salt tolerance between species and each species has a specific threshold salinity. Environmental adaptations and inherent genetic traits regulate salinity tolerance mechanisms in glycophytes and halophytes (Munns, 2002b). The majority of the plants are glycophytes (sensitive to salt) and tend to exclude the salts from roots, delaying salinity stress (Zhu, 2007). Halophytes grow in saline conditions and therefore, possess enhanced tolerance to high salt levels. They accumulate salts, carry through the xylem stream and precipitate them on leaves. Some species have evolved specialized cells called salt glands in shoots to excrete salt on its surface, which is then removed by water or wind. Few attempts have been made to introduce halophyte genes in crop plants and cultivate halophytes for food, forage, or fuel (Flowers et al., 1986; Flowers and Colmer, 2015).
Salinity impairs plant growth by causing osmotic imbalance and ion toxicity. The first osmotic phase occurs immediately when salt concentration increases above a threshold level around the roots. The osmotic stress induces water deficit in roots and shoot growth is arrested within minutes of exposure, but then recovers over several hours to a slow steady rate of growth. The second phase develops with time and is driven by the toxicity of excess Na+/Cl- ions that accumulate in the cytoplasm. When the salt concentration exceeds the rate of exclusion by roots or cellular ability to compartmentalize salts in the vacuoles, it builds up in the cytosol and disrupts cellular structures and functions (Munns, 2002b). Hence, all salinity tolerance in plants is directed towards maintaining osmotic balance and ion homeostasis. Even though the loss of cell turgor after the immediate osmotic shock is transient, reduction of cell elongation and cell division rates in root tips and young leaves over time lead to growth inhibition (Passioura and Munns, 2000). Osmotic stress affects shoot and reproductive development, for instance, younger leaves emerge slowly, lateral buds remain quiescent and flowering starts earlier. The growth regulating mechanisms are speculated to be long-distance signals of hormones and their precursors from roots to shoots. Phytohormone signaling is essential for regulation of cell division and differentiation, thereby controlling plant developmental morphogenesis (Santner and Estelle, 2009). The integrated signaling pathways are crucial in plant protection and adaptation mechanisms during abiotic and biotic stresses. In addition to five classical phytohormones, auxin, gibberellin, cytokinin, abscisic acid, and ethylene, other molecules including salicylic acid, jasmonic acid, nitric oxide, brassinosteroids, and strigolactones have been known to function as plant growth regulators. Phytohormone status is interdependent and both negative feedback and positive stimulation of synthesis have been reported. Many of the proteins including some transcription factors and protein kinases involved in plant hormone signaling have been elucidated. Phytohormone signaling cascades influence osmotic balance and other salt tolerance mechanisms (discussed below) and regulate plant acclimatization to salinity (reviewed in detail by Waśkiewicz et al., 2016). The plant roots encounter salinity first and root elongation rate recovers after initial exposure to salt but root architecture undergoes transition over time and high salt concentration represses formation of lateral roots. The aboveground symptoms of salinity induced osmotic stress overlap to that of drought stress, including leaf senescence and stunted growth (Munns, 2002a).
Osmotic stress affects stomatal conductance instantly due to perturbed water balance and abscisic acid (ABA) synthesis in guard cells, causing stomatal closure. Over the next several hours, transpiration rate is stabilized to a new reduced rate and ABA levels in situ are established (Fricke et al., 2006). Increased osmotic tolerance results in greater leaf expansion and stomatal conductance, which is beneficial only when there is sufficient soil water for transpiration losses (Munns and Tester, 2008). Photosynthesis rate decreases not only because of reduced leaf area and lesser gas exchange but also due to feedback inhibition of unused photosynthates, after exposure to salinity. The growth of sink tissues is constrained and carbohydrates accumulate in plant meristems and storage organs, which otherwise would be used in their proliferation and expansion. Modulating carbohydrate production in source leaves, phloem transport, and sink utilization downregulate the feedback photoinhibition and boost plant energy metabolism (Paul and Foyer, 2001; Perez-Alfocea et al., 2010). Reactive oxygen species (ROS) are constantly generated by cell organelles as a metabolic by-product and function as signaling molecules but their production is spiked under stressed environments. ROS including hydrogen peroxide, superoxide, and free oxygen radical are profoundly reactive with cellular components and induces programmed cell death. ROS cause chlorophyll degradation and lipid peroxidation that affects photosynthesis and membrane permeability, respectively (Apel and Hirt, 2004).
Plants have developed antioxidant mechanisms involving enzymes (superoxide dismutase, glutathione reductase, catalase, and peroxidases) and molecules (carotenoids, flavonoids, and other phenolics) that prevent tissues from oxidative damages by quenching and detoxifying ROS (Gill and Tuteja, 2010). Upregulation of antioxidant enzyme activity and metabolite synthesis is coordinated by gene networks in response to initial low levels of ROS and other signaling events (Mittler et al., 2004). Antioxidant production and osmolyte accumulation are considered as sensitive physiological markers of salt and other abiotic stresses (Munns, 2002a). A common metabolic change in response to salinity is the synthesis of low molecular weight organic compounds including polyols (sorbitol, mannitol, inositol, or glycerol), amino acids (proline or glutamate), and betaines (glycine betaine) that function as osmolytes. They are compatible solutes and accumulate in the cytosol to maintain osmotic balance both inside and outside the cell. Osmolytes also function as osmoprotectants by preventing desiccation of membranes and stabilize dehydrated enzymes rather playing role in osmoregulation. They facilitate stabilization of subcellular structures and free radical scavenging and protect plants from osmotic stress induced dehydration (Rhodes et al., 2002). Synthesis of osmolytes is an energy-demanding process yet enables the plant to recover from adverse effects of salt stress (Raven, 1985).
Effects of ionic stress are determinant under prolonged exposure to high salinity levels and predominant in salt-sensitive species. Sodium ions are toxic to many plants, so are high concentrations of chlorine, specifically those that are poor excluders of Na+ (ex: rice and beans) and sensitive to Cl- (ex: soybean and citrus). The influx of Na+ from roots is deposited in the xylem, carried through the transpiration stream and accumulated in the leaf blade rather than roots. Excluding Na+ is a daunting task because a relatively small proportion is recirculated through phloem and most of it remains in the shoot, causing toxicity (Munns, 2002a; Tester and Davenport, 2003). Hence, active efflux of Na+ from cells and retrieval of Na+ from xylem is required throughout the plant and achieved by regulatory networks of sodium/proton antiporters and high-affinity potassium transporters (Tester and Davenport, 2003; Davenport et al., 2005). A Na+/H+ antiporter SOS1 (salt overly sensitive) localized on the plasma membrane is involved in the transport of Na+ out of the cell and its activity is dependent on substrate (Na+) concentration (Qiu et al., 2002). Excess Na+ ion concentration affects low-affinity potassium uptake system because of the similar chemical nature of Na+ and K+ ions thereby, inhibiting K+ uptake by the roots. Plants activate high-affinity K+ transporters (HKT) to increase the uptake of K+ ions over Na+ ions and K+ concentration relative to Na+ in cytoplasm increases salinity tolerance (Rodriguez-Navarro and Rubio, 2006). Salt accumulation in intracellular spaces restrain enzymes involved in photosynthesis and respiration and interfere with vesicular trafficking (Baral et al., 2015; Jacoby et al., 2016). Cytosolic activities are inhibited under a high Na+/K+ ratio and cells need to effectively compartmentalize sodium into vacuoles, which further improves osmotic adjustments. Intracellular compartmentation of Na+ is regulated by Na+/H+ antiporters and Na+/H+ exchangers (NHX) on the tonoplast, which are driven by a proton gradient (Halfter et al., 2000).
Plants with adequate calcium supply have demonstrated enhanced salt tolerance and supplemental Ca2+ stimulates rapid leaf elongation rate (Cramer, 1992). Calcium mediated signaling is important in maintaining Na+/K+ ratios by sustaining potassium transporters and suppressing non-selective cation channels and a rise in cytosolic Ca2+ levels is the first detectable response to sodium stress (Epstein, 1998). Membrane depolarization activates Ca2+ channels in cellular membranes that regulate Ca2+ oscillations in the cytosol and generate Ca2+ signals under salt stress. The calcium signal sensor, calcineurin B-like protein (CBL4, previously identified as SOS3) forms a complex with a CBL-interacting protein kinase (CIPK24, identified as SOS2) to phosphorylate SOS1, thus enabling its activation (Halfter et al., 2000; Zhu, 2002). Other sensor proteins are calcium dependent protein kinases (CDPKs), SOS3-like calcium binding proteins (SCaBPs), and calmodulins (CaMs) (Chinnusamy et al., 2006). Progressive accumulation of Cl- is toxic to chloroplasts and mitochondria, and tolerance of high Cl- concentrations requires compartmentalization and exclusion. The active influx of Cl- is catalyzed by a Cl-/2H+ symporter but passive uptake also occurs under saline conditions and efflux takes place through Cl- permeable channels (Yamashita et al., 1994). Transport of Cl- to shoots is limited by reduced xylem loading of Cl- via anion channels (downregulated by ABA) and Cl- is actively retrieved from the xylem stream (Gilliham and Tester, 2005).
Biochemical analysis, gene expression and mutant studies conducted to investigate molecular functions of plants in response to salinity revealed that complex signal transduction pathways and gene regulatory networks exist to alleviate stress (Hasegawa et al., 2000). Breeding of salt-tolerant genotypes to improve crop production has been persevered by plant scientists but in spite of the advances, relatively few determinant genetic traits for salt tolerance in crop species have been identified to date (Munns and Tester, 2008). However, the acquired knowledge will lead to the development of tolerant cultivars and implementation of sustainable crop protection measures that are environmentally safe. Conventional breeding practices and genetic engineering techniques could be the most relevant but often time-consuming and cost-intensive strategies. Meanwhile, application of beneficial microbes to increase salt tolerance in plants is a feasible alternative to reclaim salinity prone lands under cultivation (Berg, 2009). A plant, together with its associated microbial community, the phytomicrobiome function as a holobiont. The physiology and metabolism of the host plant are influenced by the phytomicrobiome, facilitating its adaptation to the habitat. Members of the phytomicrobiome, which include PGPR, AMF and other facultative endosymbionts are inoculated as microbial consortia and this strategy has gained interest lately to enhance crop productivity in stressed environments (Smith et al., 2015b).
Salt Tolerance Mediated by Plant Growth Promoting Rhizobacteria
During the past century, research has continuously demonstrated numerous beneficial associations between plants and microbes, beginning with the classic legume–rhizobia symbiosis. The plant rhizosphere is enriched with nutrient sources excreted from roots that support the higher abundance of microbial population than the surrounding bulk soil (Lugtenberg and Kamilova, 2009). Free-living beneficial bacteria dwelling in the rhizosphere that exert beneficial activities are known as plant growth promoting rhizobacteria (PGPR). Some of them are facultative endophytes that further invade intercellular spaces of host tissues and thrive as endophytes to establish a mutually beneficial association. PGPR living outside the plant cell are differently associated with plant roots and directly relate to the underlying mechanisms of plant–microbe interactions. The majority of the PGPR colonize the root surface and thrive in spaces between root hairs and rhizodermal layers whereas, some are not physically in contact with the roots (Gray and Smith, 2005). Root exudates are an integral part of rhizosphere signaling events and regulate communication in beneficial plant–microbe interactions. Phenols, flavonoids, and organic acids secreted by roots have been known to act as chemical signals for bacterial chemotaxis, secretion of exopolysaccharides, quorum sensing and biofilm formation during rhizosphere colonization (Bauer and Mathesius, 2004; Badri et al., 2009; Narula et al., 2009). Isolated from rhizosphere soils, PGPR are screened in vitro for plant growth promoting characteristics and tested for beneficial effects in greenhouse and field trials prior to commercialization. PGPR promote plant growth and development through diverse mechanisms such as enhanced nutrient assimilation (biofertilizers) by biological nitrogen fixation, phosphorous solubilisation or iron acquisition (Rodriguez and Fraga, 1999; Steenhoudt and Vanderleyden, 2006; Sharma et al., 2013; Jin et al., 2014; Kuan et al., 2016), control pathogens by antagonism and competition (biocontrol agents) (Compant et al., 2005; Beneduzi et al., 2012; Chowdhury et al., 2015), degrade organic pollutants and reduce metal toxicity of contaminated soils (bioremediation), and facilitate phytoremediation (Divya and Kumar, 2011; Nie et al., 2011; Janssen et al., 2015; Weyens et al., 2015).
Inoculation with PGPR has been known to modulate abiotic stress regulation via direct and indirect mechanisms that induce systemic tolerance (Yang et al., 2009). Many PGPR have been investigated for their role in improving plant-water relations, ion homeostasis and photosynthetic efficiency in plants under salt stress (Figure 1); their amelioration mechanisms are intricate and often not well-understood. These mechanisms are regulated by a complex network of signaling events occurring during the plant–microbe interaction and consequently ensuing stress alleviation (Smith et al., 2017). Accumulating evidence using high-throughput techniques implies that understanding the dynamic function of PGPR in relation to stomatal conductance, ion transport, water and nutrient uptake, phytohormonal status, signal transduction proteins, antioxidant enzymes, and carbohydrate metabolism in plants is important for determining the induced systemic tolerance (Figure 2).
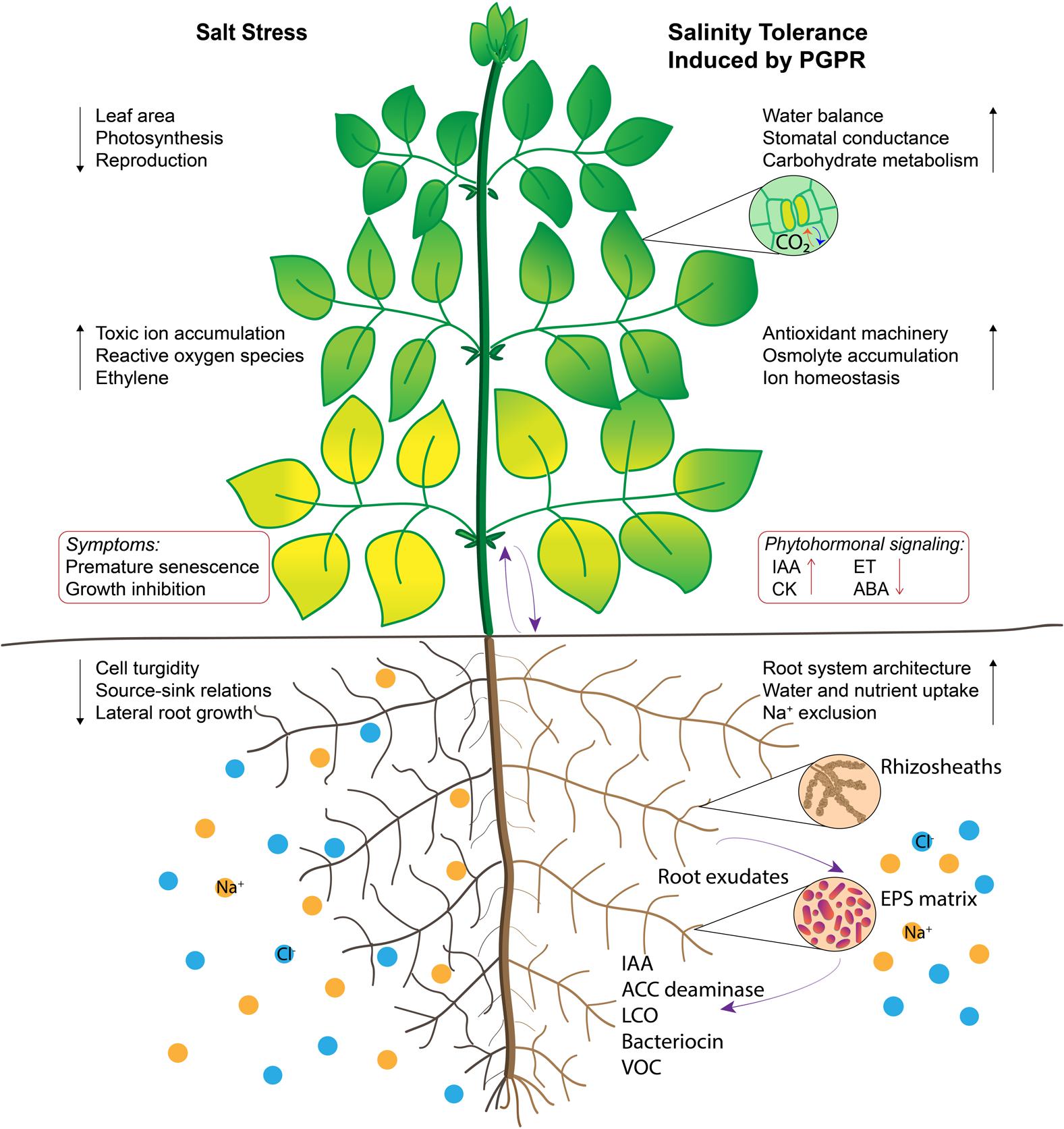
FIGURE 1. Illustration of salt tolerance mechanisms induced by plant growth promoting rhizobacteria (PGPR). Root surfaces are colonized by PGPR and extracellular polysaccharide matrix acts as a protective barrier against salt stress. Some extracellular molecules function as signaling cues that manipulate phytohormonal status in plants. Enhanced root-to-shoot communication improves water and nutritional balance, source-sink relations and stomatal conductance. Stimulating osmolyte accumulation, carbohydrate metabolism and antioxidant activity delay leaf senescence, which inturn contribute to photosynthesis. Regulation of physiological processes are indicated by black arrows and signaling pathways are indicated by purple arrows.
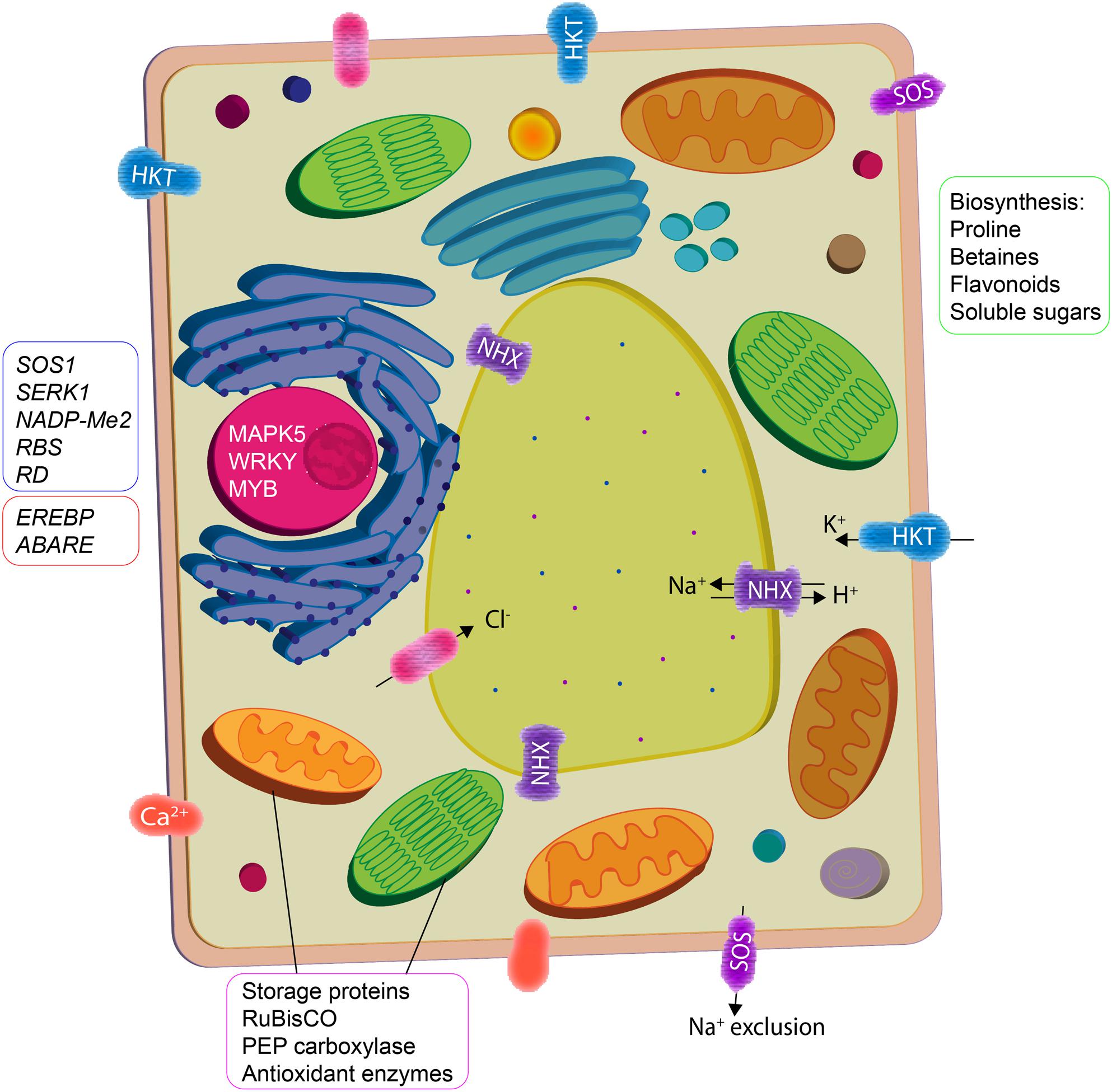
FIGURE 2. Plant growth promoting rhizobacteria interaction mediate cellular activity in plants to ameliorate salinity stress. Osmotic imbalance and oxidative damage are reduced by enhanced biosynthesis of compatible solutes and antioxidants. Ion homeostasis is maintained by increase in activity of K+ transporters (HKT) and H+ exchangers (NHX) that facilitate salt compartmentalization/exclusion. PGPR also upregulate the expression of stress responsive genes (phytohormone signaling) and proteins (vegetative storage, photosynthesis, and antioxidant enzymes).
Osmotic Balance
Plant growth promoting rhizobacteria regulate water potential and stomatal opening by affecting hydraulic conductivity and transpiration rate. Maize plants inoculated with Bacillus megaterium showed increased root hydraulic conductivity compared to uninoculated plants when exposed to salinity (2.59 dS m-1) and this was correlated with increased expression of two ZmPIP (plasma membrane aquaporin protein) isoforms (Marulanda et al., 2010). PGPR induce osmolyte accumulation and phytohormone signaling that facilitate plants to overcome initial osmotic shock after salinization. Enhanced proline synthesis in transgenic Arabidopsis thaliana with proBA genes derived from Bacillus subtilis conferred salt tolerance to the plants (Chen et al., 2007). Inoculation of salt tolerant Bacillus amyloliquefaciens SN13 onto rice (Oryza sativa) plants exposed to salinity (200 mM NaCl) in hydroponic and soil conditions increased plant salt tolerance and affected expression of 14 genes, of which, four (SOS1, ethylene responsive element binding proteins EREBP, somatic embryogenesis receptor-like kinase SERK1 and NADP-malic enzyme NADP-Me2) were upregulated and two [glucose insensitive growth GIG and (SNF1) serine-threonine protein kinase SAPK4] were downregulated under hydroponic conditions whereas, only MAPK5 (Mitogen activated protein kinase 5) was upregulated under greenhouse conditions. Genes involved in osmotic and ionic stress response mechanisms were modulated by SN13 inoculation (Nautiyal et al., 2013).
Beneficial microorganisms can stimulate carbohydrate metabolism and transport, which directly implicate source-sink relations, photosynthesis, growth rate and biomass reallocation. Seed inoculated B. aquimaris strains increased total soluble sugars and reducing sugars in wheat under saline (ECe = 5.2 dS m-1) field conditions and resulted in higher shoot biomass, NPK accumulation, and Na reduction in leaves (Upadhyay and Singh, 2015). Higher plant dry matter accumulation after 36 days in pepper (Capsicum annuum) plants inoculated with Azospirillum brasilense and Pantoea dispersa under salinity was related to enhanced stomatal conductance and photosynthesis, but neither chlorophyll concentration nor photochemical efficiency of photosystem II was affected (del Amor and Cuadra-Crespo, 2012). Microbes exposed to osmolality fluctuations in their surrounding environment accumulate large quantities of osmoprotectants in their cytosol (Kempf and Bremer, 1998). Under such circumstances, biosynthesis of osmolytes including proline, trehalose, and glycine betaines by PGPR is most likely to be quicker than their associated host plants. The compatible solutes absorbed through plant roots aid in maintaining osmotic balance and preventing cellular oxidative damage under saline conditions. Co-inoculation of bean (Phaseolus vulgaris) with Rhizobium tropici and Paenibacillus polymyxa strain modified to overexpress trehalose 6-phosphate gene resulted in increased nodulation, N content and plant growth. A microarray analysis of nodules revealed upregulation of stress tolerance genes suggesting that extracellular trehalose, which functions as an osmoprotectant can induce salinity tolerance (Figueiredo et al., 2008).
Ion Homeostasis
Bacteria limit plant salt uptake by trapping cations in the exopolysaccharide matrix, altering root structure with extensive rhizosheaths, and regulating expression of ion affinity transporters. PGPR have been known to increase the mineral nutrient exchange of both macro and micronutrients and alleviate nutrient imbalance caused by the high influx of Na+ and Cl- ions. Microbial induced nutrient cycling (mineralization), rhizosphere pH changes (organic acids), and metal chelation (siderophores) increase plant nutrient availability (Dodd and Perez-Alfocea, 2012; Lugtenberg et al., 2013). PGPR help maintaining ion homeostasis and high K+/Na+ ratios in shoots by reducing Na+ and Cl- accumulation in leaves, increasing Na+ exclusion via roots, and boosting the activity of high-affinity K+ transporters. Inoculation of Azotobacter strains C5 (auxin producing) and C9 in maize plants under salt stress improved K+ uptake and Na+ exclusion. Chlorophyll, proline and polyphenol contents in leaves increased and PGPR inoculation enhanced plant stress responses (Rojas-Tapias et al., 2012). In a study conducted with Arabidopsis thaliana and Burkholderia phytofirmans PsJN to understand the spatiotemporal regulation of short and long-term salt stress, colonized plants exhibited higher tolerance to sustained salt stress. The expressional patterns of genes involved in ion homeostasis (KT1, HKT1, NHX2, and SOS1) were altered after stress and rapid molecular changes induced by PsJN may be linked to the observed salt tolerance (Pinedo et al., 2015). A halophyte grass, Puccinellia tenuiflora inoculated with B. subtilis GB03 showed less Na+ accumulation and validated by upregulation of PtHKT1 and PtSOS1 genes but PtHKT2 was downregulated in roots under high salt concentrations (200 mM NaCl) (Niu et al., 2016).
Phytohormone Signaling
Soil bacteria modulate plant hormone status by releasing exogenous hormones, metabolites, and enzymes that may contribute to increased salt tolerance. Besides, phytohormones and metabolites are synthesized de novo in the plants in response signaling events of plant–microbe interactions during stress (Dodd et al., 2010).
Auxin
Auxin biosynthesis occurs via multiple pathways in rhizobacteria and one is the utilization of tryptophan present in root exudates and its conversion into indole-3-acetic acid (IAA), which is absorbed by the plant roots. Together with the plant’s endogenous IAA pool, an auxin signaling pathway is triggered and results in stimulation of cell growth and proliferation. IAA produced by PGPR is one of the most common and widely studied bacterial signaling molecules in plant–microbe interactions. The function of exogenous IAA is dependent on the endogenous IAA levels in plants. At optimal IAA concentration, acquisition of bacterial IAA may result in neutral, promotion or inhibition of plant growth (Dodd et al., 2010; Spaepen and Vanderleyden, 2011).
Bacillus amyloliquefaciens SQR9 enhanced salt stress tolerance (100 mM NaCl) of maize seedlings in vitro and bacterial inoculation increased chlorophyll and total soluble sugar contents, improved peroxidase and catalase activity, enhanced glutathione content, and K+/Na+ ratio. In addition, salinity induced ABA level was counteracted by SQR9 inoculation, which maintained it at the normal level. These physiological mechanisms to relieve salt stress were confirmed by the upregulation of genes RBCS, RBCL (encoding RuBisCo subunits), H(+)-Ppase (encoding H+ pumping pyrophosphatase), HKT1, NHX1, NHX2 and NHX3, and also the downregulation of NCED expression (encoding 9-cis-epoxycarotenoid dioxygenase) in inoculated seedlings (Chen et al., 2016). Enterobacter sp. EJ01 isolated from a halophyte plant, sea china pink (Dianthus japonicus thunb) improved plant growth and salt stress tolerance (200 mM) in Arabidopsis and tomato (Solanum lycopersicum) plants. Short-term treatment (6 h) with EJ01 increased expression of genes involved in salt stress response such as DRE-binding proteins DREB2b, Relative to Desiccation (RD29A, RD29B), late embryogenesis abundant (LEA) genes (RAB18), proline biosynthesis (P5CS1 and P5CS2), and stress-inducible priming processes (MPK3 and MPK6) in Arabidopsis seedlings. GFP-tagged EJ01 displayed colonization of the bacteria in the rhizosphere and endosphere of Arabidopsis roots. In addition, ROS scavenging activities including antioxidant enzyme, ascorbate peroxidase were enhanced in inoculated tomato plants under salt stress (Kim et al., 2014).
The role of bacterial cytokinins in salt stress tolerance is largely unknown yet with relatively fewer studies. Pseudomonas strains (P. aurantiaca TSAU22, P. extremorientalis TSAU6 and P. extremorientalis TSAU20) enhanced growth up to 52%, compared to control plants and alleviated salinity (100 mM NaCl) induced dormancy of wheat seeds (Egamberdieva, 2009). Cytokinin producing B. subtilis inoculated onto lettuce seedlings under water deficit conditions increased accumulation of shoot biomass and shortened roots with only small effect on root biomass. Despite increased shoot cytokinins, the possible role in root-to-shoot signaling was latent seemingly hindered by shoot ABA (Arkhipova et al., 2007).
Ethylene
Synthesis of ethylene in response to stress may increase plant tolerance or expedite senescence (Morgan and Drew, 1997). Ethylene regulates plant adaptation to stress at the expense of growth and development. As ethylene levels increase under stress, transcription of auxin response factors is inhibited and it constraints plant growth. PGPR that secrete 1-aminocyclopropane-1-carboxylase (ACC) deaminase restrict ethylene biosynthesis in plants. The enzyme converts ACC, the precursor of ethylene to ammonia and α-ketobutyrate. Many studies have shown enhanced stress tolerance and growth promotion in plants conferred by soil bacteria producing ACC deaminase (Glick et al., 2007). The following examples illustrate some of the salt tolerance mechanisms induced by PGPR producing ACC deaminase.
Pseudomonas putida UW4 inoculated tomato (Solanum lycopersicum) seedlings showed increased shoot growth after 6 weeks in saline conditions up to 90 mM NaCl. The expression of Toc GTPase, a gene of the chloroplast protein import apparatus was upregulated, which may facilitate import of proteins involved as a part of stress response (Yan et al., 2014). A nutrient flow study of pea (Pisum sativum cv. Alderman) inoculated with Variovorax paradoxus 5C-2 under salt stress of 70 and 130 mM NaCl showed increased root to shoot K+ flow and Na+ deposition in roots, thereby increasing K+/Na+ ratio in shoots. Inoculation with PGPR also increased the photosynthesis rate and electron transport, while decreased stomatal resistance and xylem balancing pressure; overall improved the plant biomass (Wang et al., 2016). Enterobacter sp. UPMR18 inoculated okra (Abelmoschus esculentus) plants exhibited increase in antioxidant enzyme activities and transcription of ROS pathway genes when grown in 75 mM NaCl and showed enhanced salt tolerance (Habib et al., 2016). ACC deaminase producing strains of Pseudomonas fluorescens and Enterobacter spp. significantly improved maize yield in salt-affected fields. Higher K+/Na+ ratios and NPK uptake were also recorded in inoculated plants under salt stress (Nadeem et al., 2009).
Plant growth promoting rhizobacteria that produce both IAA and ACC deaminase can effectively protect plants from a range of stresses. IAA accumulation induces transcription of ACC synthase genes, which increases ACC concentration, leading to the production of ethylene. PGPR containing ACC deaminase may break down some of the excess ACC and lower plant ethylene levels during an advent of environmental stress and simultaneously allow IAA to promote plant growth (Glick, 2012). Endophytic bacteria (Arthrobacter sp. and Bacillus sp.) producing ACC deaminase and IAA increased proline content in sweet pepper (Capsicum annuum). The inoculated plants manifested downregulation of stress-inducible genes CaACCO (ACC oxidase) and CaLTPI (Lipid transfer protein) under mild osmotic stress (Sziderics et al., 2007). Pantoea dispersa PSB3 is a native bacterium in chickpea (Cicer arietinum) and produces IAA and ACC deaminase. Upon inoculation to chickpea cv. GPF2, it significantly improved plant biomass, pod number, pod weight, seed number, and seed weight in salt (150 mM NaCl) affected plants. The improved salt tolerance was associated with significant reduction of Na+ uptake and electrolyte leakage and increase of relative leaf water content, chlorophyll content, and K+ uptake (Panwar et al., 2016).
Abscisic Acid
There are relatively few studies on determining the role of exogenous ABA in plant–microbe interactions and whether bacterial ABA influences ABA status of plants under salt stress. However, PGPR modulate ABA biosynthesis and ABA-mediated signaling pathways that may contribute to the enhanced growth of salt-stressed plants. Halotolerant Dietzia natronolimnaea STR1 induced salinity (150 mM NaCl) tolerance mechanisms in wheat plants via modulation of an ABA-signaling cascade, validated by the upregulation of TaABARE (ABA-responsive gene) and TaOPR1 (12-oxophytodienoate reductase 1) leading to TaMYB and TaWRKY stimulation, followed by expression of stress response genes including upregulation of TaST (a salt stress-induced gene). Expression of SOS pathway related genes and tissue-specific responses of ion transporters were modulated. Gene expression of various antioxidant enzymes and proline content were increased, contributing to enhanced protection against salt stress in PGPR inoculated plants (Bharti et al., 2016). Cucumber (Cucumis sativus) plants inoculated with Burkholderia cepacia SE4, Promicromonospora sp. SE188 and Acinetobacter calcoaceticus SE370 had significantly higher biomass under salinity stress (120 mM NaCl). PGPR increased water potential and decreased electrolyte leakage. The inoculated plants showed down-regulation of ABA compared with control plants, while salicylic acid and gibberellin GA4 contents were increased (Kang et al., 2014a). Seed inoculation of cotton (Gossypium hirsutum) with Pseudomonas putida Rs-198 reduced ABA accumulation and increased plant biomass in salinized soil but the induced salt tolerance can also be attributed to regulated ionic balance and improved endogenous IAA content (Yao et al., 2010). Wheat plants inoculated with PGPR strains Arthrobacter protophormiae SA3 and B. subtilis LDR2 built up IAA while conflicted the increase of ABA and ACC content under salt stress conditions (100 mM NaCl). The amelioration effect was further validated by the upregulation of TaCTR1 (Serine/Threonine protein kinase – ethylene responsive) and TaDRE2 (drought-responsive element) genes (Barnawal et al., 2017).
Extracellular Molecules
The extracellular secretions of PGPR including proteins, hormones, volatiles, polyamines, and other compounds have been determined to manipulate signaling pathways and regulatory functions that positively impact plant defense and development by stimulating growth, inducing disease resistance and eliciting stress tolerance (Barnawal et al., 2013; Kang et al., 2014b; Bhattacharyya et al., 2015; Smith et al., 2015a; Zhou et al., 2016).
Exopolysaccharides
Bacteria secrete exopolysaccharides (EPS) which are responsible for attachment, often along with other bacteria, to soil particles and root surfaces. EPS bind soil particles to aggregates, stabilizing soil structures, and increasing water holding capacity and cation exchange capacity (Upadhyay et al., 2011). EPS usually form an enclosed matrix of microcolonies, which confer protection against environmental fluctuations, water and nutrient retention, and epiphytic colonization (Balsanelli et al., 2014). They are also indispensable for mature biofilm formation and functional nodules in legume–rhizobia symbiosis (Stoodley et al., 2002; Skorupska et al., 2006). Inoculation of EPS producing Pseudomonas mendocina with an arbuscular mycorrhizal fungus, Glomus intraradices onto lettuce (Lactuca sativa) resulted in stabilization of soil aggregates under field conditions (Kohler et al., 2006). Inoculation with salt-tolerant Halomonas variabilis HT1 and Planococcus rifietoensis RT4 increased the growth of chickpea (Cicer arietinum var. CM-98) and soil aggregation with roots under high salt concentrations (up to 200 mM NaCl) (Qurashi and Sabri, 2012). Quinoa (Chenopodium quinoa) seeds inoculated with Enterobacter sp. MN17 and Bacillus sp. MN54 improved plant-water relations under saline irrigation conditions of 400 mM NaCl (Yang et al., 2016). EPS production and composition improve bacterial resistance to abiotic stress (Sandhya and Ali, 2015) but the role of EPS in plant salinity tolerance deserves further investigation.
Lipo-chitooligosaccharides
Legume–rhizobia symbiosis is affected by salt stress and high levels of salinity inhibit nodule formation and nitrogen fixation (Tu, 1981; Zahran, 1999). Lipo-chitooligosaccharides (LCOs) are secreted by rhizobia as Nod-factors (NFs) in response to flavonoids present in root exudates and initiate nodule formation. LCOs are conserved at the core but diverge in the N-Acetyl chain length, degree of saturation, and substitutions (glycosylation or sulfation), which are crucial in host specificity (Oldroyd, 2013). Nod-factors also act as stress response signals in legumes and NF synthesis is modulated by other PGPR and abiotic stresses. High salinity (100–200 mM NaCl) inhibited root hair deformation responses to increase in NF concentrations in Soybean (Glycine max) – Bradyrhizobium japonicum symbiosis (Duzan et al., 2004). Inoculation of IAA producing Azospirillum brasilense Cd into the Rhizobium-Bean (Phaseolus vulgaris cv. Negro Jamapa) symbiosis increased root branching and flavonoid synthesis under 50 mM NaCl. The co-inoculation also promoted Nod-genes expression in R. tropici CIAT899 and R. etli ISP42 grown in the presence of root exudates (Dardanelli et al., 2008). Free-living rhizobia are more resistant to salt stress than inside their legume hosts. R. tropici CIAT899 is highly tolerant to stress and high salt concentrations enhance Nod-gene expression, Nod-factor synthesis and diversity; 46 different NFs were identified compared to 29 NFs under control with only 15 NFs common to both (Estevez et al., 2009). Inoculation of B. japonicum 532C grown in genistein (a flavonoid) induced media significantly enhanced nodulation and growth of soybean under salinity levels (36 and 61 mM NaCl) and such positive effects become more evident with time (Miransari and Smith, 2009) and increased yield up to 21% under salinized field conditions in an earlier study.
Bacteriocins
Bacteriocins are small peptides secreted by rhizobacteria that are bactericidal or bacteriostatic against relative bacteria, thus providing a competitive advantage to the producer strain but might also promote microbial diversity in an ecologic niche (Kirkup and Riley, 2004). Application of thuricin 17, isolated from a soybean endosymbiont Bacillus thuriengenesis NEB 17 differentially altered the proteome of salt-stressed (250 mM NaCl) Arabidopsis plants. Expression of proteins involved in carbon and energy metabolism pathways were modulated by the bacterial signals. Proteins involved in photosynthesis including PEP carboxylase, RuBisCo-oxygenase large subunit, pyruvate kinase and proteins of photosystems I and II were upregulated along with other stress related proteins (Subramanian et al., 2016b). These bacterial signal compounds also induced similar changes in the proteome of soybean seeds at 48 h under 100 mM NaCl. In addition, isocitrate lyase and antioxidant glutathione-S-transferase were increased. These findings by shotgun proteomics suggested that thuricin 17 positively manipulate plant proteome profile and enhance physiological tolerance to salinity (Subramanian et al., 2016a).
Polyamines
Polyamines (PAs) are low molecular weight aliphatic amines with pronounced antioxidant activity that are ubiquitous in all living organisms and modulate ROS homeostasis by scavenging free radicals and stimulating antioxidant enzymes. The most abundant polyamines, spermidine, spermine, and putrescine are implicated in various developmental processes and stress responses in plants (Gupta et al., 2013). Application of exogenous polyamines increase abiotic stress tolerance but PGPR secretion of polyamines is largely unexplored. Spermidine from Bacillus megaterium BOFC15 increased cellular polyamine accumulation in Arabidopsis, thereby activating PA-mediated signaling pathways contributing to the osmotic stress tolerance of plants. The bacterial inoculation resulted in greater biomass, elevated photosynthetic capacity and higher antioxidant enzyme activity. Other tolerance mechanisms involved robust root system architecture and ABA dependent stress responses, which maintained water balance and stomatal conductance (Zhou et al., 2016).
Volatile Compounds
Volatile organic compounds (VOC) released from PGPR are known to stimulate plant growth, resulting in increased shoot biomass, and modulated stress responses. Perception of volatiles by plants and subsequently induced mechanisms require further research (Bailly and Weisskopf, 2012). B. subtilis GB03 VOCs mediated tissue specific regulations of Na+ homeostasis in salt-stressed plants. Arabidopsis under 100 mM NaCl treated with VOCs decreased Na+ accumulation by concurrently downregulating expression of HKT1 in roots but upregulating it in shoots. Presumably, the induction of HKT1 dependent shoot-to-root recirculation resulted in reduced Na+ accumulation up to ∼50% throughout the plant. Treatment with VOCs increased leaf surface area, root mass, and total K+ content when compared with controls whereas, inoculated athkt1 mutants showed stunted growth. Exposure to VOCs reduced the total Na+ level by 18% and enhanced shoot and root growth of sos3 mutants in 30 mM NaCl (Zhang et al., 2008). A putative VOCs blend released from Pseudomonas simiae AU induced salt-tolerance in soybean (Glycine max) under 100 mM NaCl by decreasing root Na+ accumulation and increasing proline and chlorophyll content. Protein expression analysis confirmed upregulation of vegetative storage proteins (Na+ homeostasis), RuBisCO large chain proteins (photosynthesis) in exposed soybean seedlings (Vaishnav et al., 2015).
Paraburkholderia phytofirmans PsJN VOCs stimulate plant growth and induce salinity tolerance that have been demonstrated both in vitro (150 mM NaCl/15 mM CaCl2) and in soil (200 mM NaCl/20 mM CaCl2). Growth parameters of Arabidopsis plants measured as rosette area, fresh weight, and primary root length were higher than the control plants and exposure to VOCs showed parallel growth promoting effects of direct bacterial inoculation. The emitted compounds were analyzed and the plants were exposed to a blend of 2-undecanone, 7-hexanol, 3-methylbutanol molecules, which mimicked the effect of VOCs (Ledger et al., 2016). Genome wide mapping association of Arabidopsis accession lines revealed 10 genetic loci associated with growth stimulation in response to the presence of P. simiae WCS417r in vitro, which is partly caused by VOC produced by the bacterium. Even though the study was conducted to select lines for breeding strategies, it is interesting to note that the genotype variation of host plants has different interactions with the associated root microbiome (Wintermans et al., 2016).
Conclusion
Application of PGPR inoculants as biofertilizers and biocontrol agents is an integral component in organic farming practices (Babalola, 2010). With rising emphasis on sustainable agriculture, environmental protection, and food security, exploitation of beneficial soil microbiota is imperative. Abiotic stresses constraint yield and turn agriculture production systems fragile; in addition, persisting climate change intensify the frequency, degree, and resultant damage of stressful conditions. Plants have evolved complex mechanisms to tolerate abiotic stresses caused by various environmental factors, including salinity. Plant associated bacteria in soil mitigate the adverse effects of these stresses in a more time-sensitive and cost-effective manner, where the development of tolerant cultivars has been somewhat overwhelmed. Research directed towards the application of PGPR in salt-affected fields encourages commercialization of inoculants for salinity tolerance. The systems biology of plant–microbe interactions in response to environmental stimuli such as salinity, opens up new prospects of understanding the regulatory networks of plant salt tolerance modulated by rhizosphere bacteria (Table 1). While the induced salt tolerance may be contributed by the release of extracellular compounds that function as chemical signals to the plant, improved soil properties that reduce the impact of salinity is another important benefit yet to be explored. Stress adaptation of plants are induced by associated microbiota and cutting-edge research as discussed above may be successfully applied to improve crop yield in saline prone regions. The potential application of PGPR to help plants deal with stress in agricultural fields seems vastly large, yet much is left to be utilized.
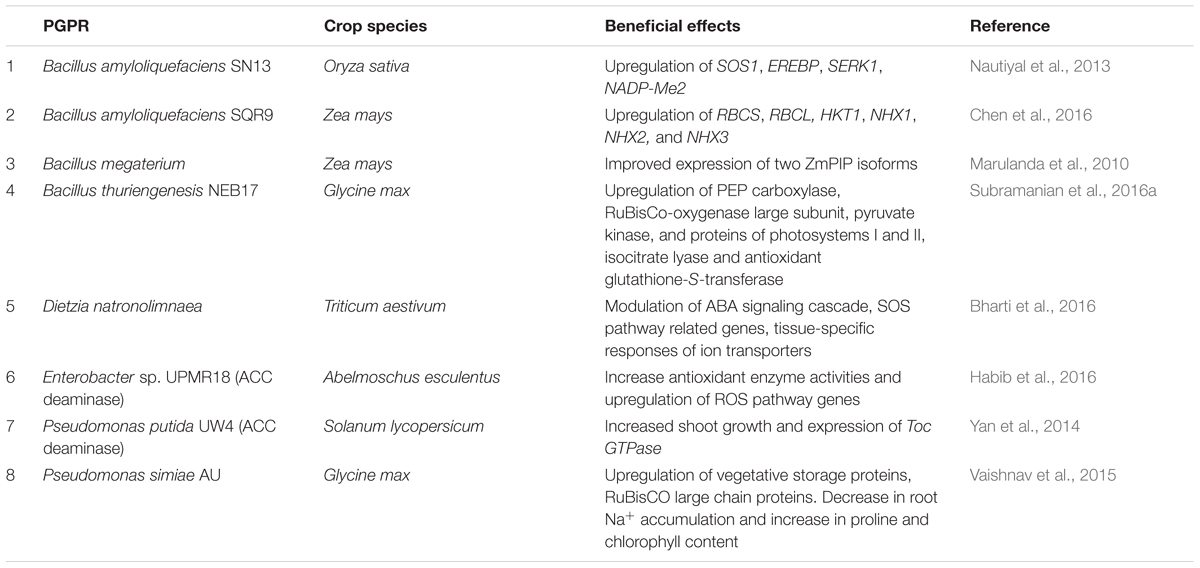
TABLE 1. Summary of PGPR interaction effects in crop plants under salinity stress from recent studies using systems biology approaches.
Author Contributions
GI gathered literature and prepared the manuscript. DS provided feedback and oversaw progression of the manuscript.
Funding
The submitted work is part of MITACS Accelerate funded Ph.D. program in partnership with Inocucor, Montreal, Canada.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors sincerely thank the members of Donald Smith laboratory, Department of Plant Science, Faculty of Agricultural and Environmental Sciences, McGill University and Inocucor, Montreal who offered guidance and help.
References
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Arkhipova, T. N., Prinsen, E., Veselov, S. U., Martinenko, E. V., Melentiev, A. I., and Kudoyarova, G. R. (2007). Cytokinin producing bacteria enhance plant growth in drying soil. Plant Soil 292, 305–315. doi: 10.1007/s11104-007-9233-5
Babalola, O. O. (2010). Beneficial bacteria of agricultural importance. Biotechnol. Lett. 32, 1559–1570. doi: 10.1007/s10529-010-0347-0
Badri, D. V., Weir, T. L., van der Lelie, D., and Vivanco, J. M. (2009). Rhizosphere chemical dialogues: plant–microbe interactions. Curr. Opin. Biotechnol. 20, 642–650. doi: 10.1016/j.copbio.2009.09.014
Bailly, A., and Weisskopf, L. (2012). The modulating effect of bacterial volatiles on plant growth: current knowledge and future challenges. Plant Signal. Behav. 7, 79–85. doi: 10.4161/psb.7.1.18418
Balsanelli, E., de Baura, V. A., Pedrosa, F. D., de Souza, E. M., and Monteiro, R. A. (2014). Exopolysaccharide biosynthesis enables mature biofilm formation on abiotic surfaces by Herbaspirillum seropedicae. PLOS ONE 9:e110392. doi: 10.1371/journal.pone.0110392
Baral, A., Shruthi, K. S., and Mathew, M. K. (2015). Vesicular trafficking and salinity responses in plants. IUBMB Life 67, 677–686. doi: 10.1002/iub.1425
Barnawal, D., Bharti, N., Pandey, S. S., Pandey, A., Chanotiya, C. S., and Kalra, A. (2017). Plant growth promoting rhizobacteria enhances wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression. Physiol. Plant. doi: 10.1111/ppl.12614 [Epub ahead of print].
Barnawal, D., Maji, D., Bharti, N., Chanotiya, C. S., and Kalra, A. (2013). ACC deaminase-containing Bacillus subtilis reduces stress ethylene-induced damage and improves mycorrhizal colonization and rhizobial nodulation in Trigonella foenum-graecum under drought stress. J. Plant Growth Regul. 32, 809–822. doi: 10.1007/s00344-013-9347-3
Bauder, J. W., and Brock, T. A. (2001). Irrigation water quality, soil amendment, and crop effects on sodium leaching. Arid Land Res. Manag. 15, 101–113. doi: 10.1080/15324980151062724
Bauer, W. D., and Mathesius, U. (2004). Plant responses to bacterial quorum sensing signals. Curr. Opin. Plant Biol. 7, 429–433. doi: 10.1016/j.pbi.2004.05.008
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. P. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051.
Berg, G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Bharti, N., Pandey, S. S., Barnawal, D., Patel, V. K., and Kalra, A. (2016). Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 6:34768. doi: 10.1038/srep34768
Bhattacharyya, D., Garladinne, M., and Lee, Y. H. (2015). Volatile indole produced by rhizobacterium Proteus vulgaris JBLS202 stimulates growth of Arabidopsis thaliana through auxin, cytokinin, and brassinosteroid pathways. J. Plant Growth Regul. 34, 158–168. doi: 10.1007/s00344-014-9453-x
Chen, L., Liu, Y., Wu, G., Veronican Njeri, K., Shen, Q., Zhang, N., et al. (2016). Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol. Plant. 158, 34–44. doi: 10.1111/ppl.12441
Chen, M., Wei, H., Cao, J., Liu, R., Wang, Y., and Zheng, C. (2007). Expression of Bacillus subtilis proBA genes and reduction of feedback inhibition of proline synthesis increases proline production and confers osmotolerance in transgenic Arabidopsis. J. Biochem. Mol. Biol. 40, 396–403.
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2006). Salt stress signaling and mechanisms of plant salt tolerance. Genet. Eng. (N Y) 27, 141–177.
Chowdhury, S. P., Hartmann, A., Gao, X. W., and Borriss, R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42-a review. Front. Microbiol. 6:780. doi: 10.3389/fmicb.2015.00780
Compant, S., Duffy, B., Nowak, J., Clement, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/Aem.71.9.4951-4959.2005
Cramer, G. R. (1992). Kinetics of maize leaf elongation.2. Responses of a Na-excluding cultivar and a Na-including cultivar to varying Na/Ca salinities. J. Exp. Bot. 43, 857–864. doi: 10.1093/jxb/43.6.857
Dardanelli, M. S., Fernández de Córdoba, F. J., Espuny, M. R., Rodríguez Carvajal, M. A., Soria Díaz, M. E., Gil Serrano, A. M., et al. (2008). Effect of Azospirillum brasilense coinoculated with Rhizobium on Phaseolus vulgaris flavonoids and Nod factor production under salt stress. Soil Biol. Biochem. 40, 2713–2721.
Davenport, R., James, R. A., Zakrisson-Plogander, A., Tester, M., and Munns, R. (2005). Control of sodium transport in durum wheat. Plant Physiol. 137, 807–818. doi: 10.1104/pp.104.057307
del Amor, F. M., and Cuadra-Crespo, P. (2012). Plant growth-promoting bacteria as a tool to improve salinity tolerance in sweet pepper. Funct. Plant Biol. 39, 82–90. doi: 10.1071/Fp11173
Divya, B., and Kumar, M. D. (2011). Plant -microbe interaction with enhanced bioremediation. Res. J. Biotechnol. 6, 72–79.
Dodd, I. C., and Perez-Alfocea, F. (2012). Microbial amelioration of crop salinity stress. J. Exp. Bot. 63, 3415–3428. doi: 10.1093/jxb/ers033
Dodd, I. C., Zinovkina, N. Y., Safronova, V. I., and Belimov, A. A. (2010). Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 157, 361–379. doi: 10.1111/j.1744-7348.2010.00439.x
Doering, E. J., and Sandoval, F. M. (1981). Chemistry of seep drainage in southwestern north-dakota. Soil Sci. 132, 142–149. doi: 10.1097/00010694-198108000-00003
Duzan, H. M., Zhou, X., Souleimanov, A., and Smith, D. L. (2004). Perception of Bradyrhizobium japonicum Nod factor by soybean [Glycine max (L.) Merr.] root hairs under abiotic stress conditions. J. Exp. Bot. 55, 2641–2646. doi: 10.1093/jxb/erh265
Egamberdieva, D. (2009). Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Physiol Plant 31, 861–864. doi: 10.1007/s11738-009-0297-0
Egamberdieva, D., and Lugtenberg, B. (2014). “Use of plant growth-promoting rhizobacteria to alleviate salinity stress in plants,” in Use of Microbes for the Alleviation of Soil Stresses, Vol. 1, ed. M. Miransari (New York, NY: Springer), 73–96.
Estevez, J., Soria-Diaz, M. E., de Cordoba, F. F., Moron, B., Manyani, H., Gil, A., et al. (2009). Different and new Nod factors produced by Rhizobium tropici CIAT899 following Na+ stress. FEMS Microbiol. Lett. 293, 220–231. doi: 10.1111/j.1574-6968.2009.01540.x
Figueiredo, M. V. B., Burity, H. A., Martinez, C. R., and Chanway, C. P. (2008). Alleviation of drought stress in the common bean (Phaseolus vulgaris L.) by co-inoculation with Paenibacillus polymyxa and Rhizobium tropici. Appl. Soil Ecol. 40, 182–188. doi: 10.1016/j.apsoil.2008.04.005
Flowers, T. J., and Colmer, T. D. (2015). Plant salt tolerance: adaptations in halophytes. Ann. Bot. 115, 327–331. doi: 10.1093/aob/mcu267
Flowers, T. J., Hajibagheri, M. A., and Clipson, N. J. W. (1986). Halophytes. Q. Rev. Biol. 61, 313–337. doi: 10.1086/415032
Fricke, W., Akhiyarova, G., Wei, W. X., Alexandersson, E., Miller, A., Kjellbom, P. O., et al. (2006). The short-term growth response to salt of the developing barley leaf. J. Exp. Bot. 57, 1079–1095. doi: 10.1093/jxb/erj095
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gilliham, M., and Tester, M. (2005). The regulation of anion loading to the maize root xylem. Plant Physiol. 137, 819–828. doi: 10.1104/pp.104.054056
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica (Cairo) 2012, 963401. doi: 10.6064/2012/963401
Glick, B. R., Cheng, Z., Czarny, J., and Duan, J. (2007). Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur. J. Plant Pathol. 119, 329–339. doi: 10.1007/s10658-007-9162-4
Gray, E. J., and Smith, D. L. (2005). Intracellular and extracellular PGPR: commonalities and distinctions in the plant-bacterium signaling processes. Soil Biol. Biochem. 37, 395–412. doi: 10.1016/j.soilbio.2004.08.030
Gupta, K., Dey, A., and Gupta, B. (2013). Plant polyamines in abiotic stress responses. Acta Physiol. Plant. 35, 2015–2036. doi: 10.1007/s11738-013-1239-4
Habib, S. H., Kausar, H., and Saud, H. M. (2016). Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ROS-scavenging enzymes. Biomed. Res. Int. 2016:6284547. doi: 10.1155/2016/6284547
Halfter, U., Ishitani, M., and Zhu, J. K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. U.S.A. 97, 3735–3740. doi: 10.1073/pnas.040577697
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., and Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/annurev.arplant.51.1.463
Jacoby, R. P., Che-Othman, M. H., Millar, A. H., and Taylor, N. L. (2016). Analysis of the sodium chloride-dependent respiratory kinetics of wheat mitochondria reveals differential effects on phosphorylating and non-phosphorylating electron transport pathways. Plant Cell Environ. 39, 823–833. doi: 10.1111/pce.12653
Janssen, J., Weyens, N., Croes, S., Beckers, B., Meiresonne, L., Van Peteghem, P., et al. (2015). Phytoremediation of metal contaminated soil using willow: exploiting plant-associated bacteria to improve biomass production and metal uptake. Int. J. Phytoremediation 17, 1123–1136. doi: 10.1080/15226514.2015.1045129
Jin, C. W., Ye, Y. Q., and Zheng, S. J. (2014). An underground tale: contribution of microbial activity to plant iron acquisition via ecological processes. Ann. Bot. 113, 7–18. doi: 10.1093/aob/mct249
Kang, S.-M., Khan, A. L., Waqas, M., You, Y.-H., Kim, J.-H., Kim, J.-G., et al. (2014a). Plant growth-promoting rhizobacteria reduce adverse effects of salinity and osmotic stress by regulating phytohormones and antioxidants in Cucumis sativus. J. Plant Interact. 9, 673–682. doi: 10.1080/17429145.2014.894587
Kang, S. M., Radhakrishnan, R., Khan, A. L., Kim, M. J., Park, J. M., Kim, B. R., et al. (2014b). Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal and stress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol. Biochem. 84, 115–124. doi: 10.1016/j.plaphy.2014.09.001
Kempf, B., and Bremer, E. (1998). Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170, 319–330. doi: 10.1007/s002030050649
Kim, K., Jang, Y.-J., Lee, S.-M., Oh, B.-T., Chae, J.-C., and Lee, K.-J. (2014). Alleviation of salt stress by Enterobacter sp. EJ01 in tomato and Arabidopsis is accompanied by up-regulation of conserved salinity responsive factors in plants. Mol. Cells 37, 109–117. doi: 10.14348/molcells.2014.2239
Kirkup, B. C., and Riley, M. A. (2004). Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428, 412–414. doi: 10.1038/nature02429
Kohler, J., Caravaca, F., Carrasco, L., and Roldan, A. (2006). Contribution of Pseudomonas mendocina and Glomus intraradices to aggregate stabilization and promotion of biological fertility in rhizosphere soil of lettuce plants under field conditions. Soil Use Manage. 22, 298–304. doi: 10.1111/j.1475-2743.2006.00041.x
Kuan, K. B., Othman, R., Abdul Rahim, K., and Shamsuddin, Z. H. (2016). Plant growth-promoting rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilisation of maize under greenhouse conditions. PLOS ONE 11:e0152478. doi: 10.1371/journal.pone.0152478
Ledger, T., Rojas, S., Timmermann, T., Pinedo, I., Poupin, M. J., Garrido, T., et al. (2016). Volatile-mediated effects predominate in Paraburkholderia phytofirmans growth promotion and salt stress tolerance of Arabidopsis thaliana. Front. Microbiol. 7:1838. doi: 10.3389/fmicb.2016.01838
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Lugtenberg, B. J., Malfanova, N., Kamilova, F., and Berg, G. (2013). Plant growth promotion by microbes. Mol. Microb. Ecol. Rhizosphere 1-2, 559–573.
Marulanda, A., Azcon, R., Chaumont, F., Ruiz-Lozano, J. M., and Aroca, R. (2010). Regulation of plasma membrane aquaporins by inoculation with a Bacillus megaterium strain in maize (Zea mays L.) plants under unstressed and salt-stressed conditions. Planta 232, 533–543. doi: 10.1007/s00425-010-1196-8
Miransari, M., and Smith, D. L. (2009). Alleviating salt stress on soybean (Glycine max (L.) Merr.) - Bradyrhizobium japonicum symbiosis, using signal molecule genistein. Eur. J. Soil Biol. 45, 146–152. doi: 10.1016/j.ejsobi.2008.11.002
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Morgan, P. W., and Drew, M. C. (1997). Ethylene and plant responses to stress. Physiol. Plant. 100, 620–630. doi: 10.1034/j.1399-3054.1997.1000325.x
Munns, R. (2002a). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nadeem, S. M., Zahir, Z. A., Naveed, M., and Arshad, M. (2009). Rhizobacteria containing ACC-deaminase confer salt tolerance in maize grown on salt-affected fields. Can. J. Microbiol. 55, 1302–1309. doi: 10.1139/W09-092
Narula, N., Kothe, E., and Behl, R. K. (2009). Role of root exudates in plant-microbe interactions. J. Appl. Bot. Food Qual. Angewandte Botanik 82, 122–130.
Nassar, I. N., and Horton, R. (1999). Salinity and compaction effects on soil water evaporation and water and solute distributions. Soil Sci. Soc. Am. J. 63, 752–758.
Nautiyal, C. S., Srivastava, S., Chauhan, P. S., Seem, K., Mishra, A., and Sopory, S. K. (2013). Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol. Biochem. 66, 1–9. doi: 10.1016/j.plaphy.2013.01.020
Nie, M., Wang, Y. J., Yu, J. Y., Xiao, M., Jiang, L. F., Yang, J., et al. (2011). Understanding plant-microbe interactions for phytoremediation of petroleum-polluted soil. PLOS ONE 6:e17961. doi: 10.1371/journal.pone.0017961
Niu, S. Q., Li, H. R., Pare, P. W., Aziz, M., Wang, S. M., Shi, H. Z., et al. (2016). Induced growth promotion and higher salt tolerance in the halophyte grass Puccinellia tenuiflora by beneficial rhizobacteria. Plant Soil 407, 217–230. doi: 10.1007/s11104-015-2767-z
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Othman, Y., Al-Karaki, G., Al-Tawaha, A., and Al-Horani, A. (2006). Variation in germination and ion uptake in barley genotypes under salinity conditions. World J. Agric. Sci. 2, 11–15.
Panwar, M., Tewari, R., Gulati, A., and Nayyar, H. (2016). Indigenous salt-tolerant rhizobacterium Pantoea dispersa (PSB3) reduces sodium uptake and mitigates the effects of salt stress on growth and yield of chickpea. Acta Physiol. Plant. 38:278. doi: 10.1007/s11738-016-2284-6
Passioura, J. B., and Munns, R. (2000). Rapid environmental changes that affect leaf water status induce transient surges or pauses in leaf expansion rate. Aust. J. Plant Physiol. 27, 941–948.
Paul, M. J., and Foyer, C. H. (2001). Sink regulation of photosynthesis. J. Exp. Bot. 52, 1383–1400. doi: 10.1093/jexbot/52.360.1383
Perez-Alfocea, F., Albacete, A., Ghanem, M. E., and Dodd, I. C. (2010). Hormonal regulation of source-sink relations to maintain crop productivity under salinity: a case study of root-to-shoot signalling in tomato. Funct. Plant Biol. 37, 592–603. doi: 10.1071/Fp10012
Pinedo, I., Ledger, T., Greve, M., and Poupin, M. J. (2015). Burkholderia phytofirmans PsJN induces long-term metabolic and transcriptional changes involved in Arabidopsis thaliana salt tolerance. Front. Plant Sci. 6:466. doi: 10.3389/fpls.2015.00466
Pitman, M. G., and Lauchli, A. (2002). “Global impact of salinity and Agricultural ecosystems,” in Salinity: Environment - Plants - Molecules, eds A. Lauchli and U. Luttage (Amsterdam: Kluwer Academic Publishers), 3–20.
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Qurashi, A. W., and Sabri, A. N. (2012). Bacterial exopolysaccharide and biofilm formation stimulate chickpea growth and soil aggregation under salt stress. Braz. J. Microbiol. 43, 1183–1191. doi: 10.1590/S1517-838220120003000046
Raven, J. A. (1985). Regulation of Ph and generation of osmolarity in vascular plants - a cost-benefit analysis in relation to efficiency of use of energy, nitrogen and water. New Phytol. 101, 25–77. doi: 10.1111/j.1469-8137.1985.tb02816.x
Rengasamy, P. (2002). Transient salinity and subsoil constraints to dryland farming in Australian sodic soils: an overview. Aust. J. Exp. Agric. 42, 351–361.
Rhodes, D., Nadolska-Orczyk, A., and Rich, P. J. (2002). “Salinity, osmolytes and compatible solutes,” in Salinity: Environment - Plants - Molecules, eds A. Läuchli and U. Lüttge (Dordrecht: Springer), 181–204.
Rodriguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Rodriguez-Navarro, A., and Rubio, F. (2006). High-affinity potassium and sodium transport systems in plants. J. Exp. Bot. 57, 1149–1160. doi: 10.1093/jxb/erj068
Rojas-Tapias, D., Moreno-Galván, A., Pardo-Díaz, S., Obando, M., Rivera, D., and Bonilla, R. (2012). Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl. Soil Ecol. 61, 264–272.
Sandhya, V., and Ali, S. Z. (2015). The production of exopolysaccharide by Pseudomonas putida GAP-P45 under various abiotic stress conditions and its role in soil aggregation. Microbiology 84, 512–519. doi: 10.1134/S0026261715040153
Santner, A., and Estelle, M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459, 1071–1078. doi: 10.1038/nature08122
Shalhevet, J. (1994). Using water of marginal quality for crop production - major issues. Agric. Water Manage. 25, 233–269. doi: 10.1016/0378-3774(94)90063-9
Sharma, D. R., and Prihar, S. S. (1973). Effect of depth and salinity of groundwater on evaporation and soil salinization. Indian J. Agric. Sci. 43, 582–586.
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2:587. doi: 10.1186/2193-1801-2-587
Singh, R. P., and Jha, P. N. (2016). Alleviation of salinity-induced damage on wheat plant by an ACC deaminase-producing halophilic bacterium Serratia sp SL-12 isolated from a salt lake. Symbiosis 69, 101–111. doi: 10.1007/s13199-016-0387-x
Skorupska, A., Janczarek, M., Marczak, M., Mazur, A., and Król, J. (2006). Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb. Cell Fact. 5:7. doi: 10.1186/1475-2859-5-7
Smith, D. L., Gravel, V., and Yergeau, E. (2017). Editorial: signaling in the phytomicrobiome. Front. Plant Sci. 8:611. doi: 10.3389/fpls.2017.00611
Smith, D. L., Praslickova, D., and Ilangumaran, G. (2015a). Inter-organismal signaling and management of the phytomicrobiome. Front. Plant Sci. 6:722. doi: 10.3389/fpls.2015.00722
Smith, D. L., Subramanian, S., Lamont, J. R., and Bywater-Ekegard, M. (2015b). Signaling in the phytomicrobiome: breadth and potential. Front. Plant Sci. 6:709. doi: 10.3389/fpls.2015.00709
Spaepen, S., and Vanderleyden, J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a001438. doi: 10.1101/cshperspect.a001438
Steenhoudt, O., and Vanderleyden, J. (2006). Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 24, 487–506. doi: 10.1111/j.1574-6976.2000.tb00552.x
Stoodley, P., Sauer, K., Davies, D. G., and Costerton, J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209. doi: 10.1146/annurev.micro.56.012302.160705
Subramanian, S., Ricci, E., Souleimanov, A., and Smith, D. L. (2016a). A proteomic approach to lipo-chitooligosaccharide and thuricin 17 effects on soybean germinationunstressed and salt stress. PLOS ONE 11:e0160660. doi: 10.1371/journal.pone.0160660
Subramanian, S., Souleimanov, A., and Smith, D. L. (2016b). Proteomic studies on the effects of lipo-chitooligosaccharide and thuricin 17 under unstressed and salt stressed conditions in Arabidopsis thaliana. Front. Plant Sci. 7:1314. doi: 10.3389/fpls.2016.01314
Sziderics, A. H., Rasche, F., Trognitz, F., Sessitsch, A., and Wilhelm, E. (2007). Bacterial endophytes contribute to abiotic stress adaptation in pepper plants (Capsicum annuum L.). Can. J. Microbiol. 53, 1195–1202. doi: 10.1139/W07-082
Tester, M., and Davenport, R. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 91, 503–527. doi: 10.1093/aob/mcg058
Tu, J. C. (1981). Effect of salinity on rhizobium-root-hair interaction, nodulation and growth of soybean. Can. J. Plant Sci. 61, 231–239.
Upadhyay, S. K., and Singh, D. P. (2015). Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 17, 288–293. doi: 10.1111/plb.12173
Upadhyay, S. K., Singh, J. S., and Singh, D. P. (2011). Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 21, 214–222. doi: 10.1016/S1002-0160(11)60120-3
Vaishnav, A., Kumari, S., Jain, S., Varma, A., and Choudhary, D. K. (2015). Putative bacterial volatile-mediated growth in soybean (Glycine max L. Merrill) and expression of induced proteins under salt stress. J. Appl. Microbiol. 119, 539–551. doi: 10.1111/jam.12866
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Wang, Q. Y., Dodd, I. C., Belimov, A. A., and Jiang, F. (2016). Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase growth and photosynthesis of pea plants under salt stress by limiting Na+ accumulation. Funct. Plant Biol. 43, 161–172. doi: 10.1071/Fp15200
Waśkiewicz, A., Gładysz, O., and Goliñski, P. (2016). “Participation of phytohormones in adaptation to salt stress,” in Plant Hormones Under Challenging Environmental Factors, eds G. J. Ahammed and J.-Q. Yu (Dordrecht: Springer), 75–115.
Weyens, N., Beckers, B., Schellingen, K., Ceulemans, R., Van der Lelie, D., Newman, L., et al. (2015). The potential of the Ni-resistant TCE-degrading Pseudomonas putida W619-TCE to reduce phytotoxicity and improve phytoremediation efficiency of poplar cuttings on A Ni-TCE Co-contamination. Int. J. Phytoremediation 17, 40–48. doi: 10.1080/15226514.2013.828016
Wintermans, P. C., Bakker, P. A., and Pieterse, C. M. (2016). Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol. Biol. 90, 623–634. doi: 10.1007/s11103-016-0442-2
Yamashita, K., Kasai, M., Yamamoto, Y., and Matsumoto, H. (1994). Stimulation of plasma-membrane H+-transport activity in barley roots by salt stress - possible role of increase in chloride permeability. Soil Sci. Plant Nutr. 40, 555–563.
Yan, J. M., Smith, M. D., Glick, B. R., and Liang, Y. (2014). Effects of ACC deaminase containing rhizobacteria on plant growth and expression of Toc GTPases in tomato (Solanum lycopersicum) under salt stress. Botany 92, 775–781. doi: 10.1139/cjb-2014-0038
Yang, A. Z., Akhtar, S. S., Iqbal, S., Amjad, M., Naveed, M., Zahir, Z. A., et al. (2016). Enhancing salt tolerance in quinoa by halotolerant bacterial inoculation. Funct. Plant Biol. 43, 632–642. doi: 10.1071/Fp15265
Yang, J., Kloepper, J. W., and Ryu, C. M. (2009). Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14, 1–4. doi: 10.1016/j.tplants.2008.10.004
Yao, L. X., Wu, Z. S., Zheng, Y. Y., Kaleem, I., and Li, C. (2010). Growth promotion and protection against salt stress by Pseudomonas putida Rs-198 on cotton. Eur. J. Soil Biol. 46, 49–54. doi: 10.1016/j.ejsobi.2009.11.002
Zahran, H. H. (1999). Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 63, 968–989.
Zerrouk, I. Z., Benchabane, M., Khelifi, L., Yokawa, K., Ludwig-Muller, J., and Baluska, F. (2016). A Pseudomonas strain isolated from date-palm rhizospheres improves root growth and promotes root formation in maize exposed to salt and aluminum stress. J. Plant Physiol. 191, 111–119. doi: 10.1016/j.jplph.2015.12.009
Zhang, H., Kim, M. S., Sun, Y., Dowd, S. E., Shi, H., and Paré, P. W. (2008). Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol. Plant Microbe Interact. 21, 737–744. doi: 10.1094/MPMI-21-6-0737
Zhou, C., Ma, Z., Zhu, L., Xiao, X., Xie, Y., Zhu, J., et al. (2016). Rhizobacterial strain Bacillus megaterium BOFC15 induces cellular polyamine changes that improve plant growth and drought resistance. Int. J. Mol. Sci. 17:976. doi: 10.3390/ijms17060976
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: salinity stress, plant tolerance, rhizobacteria, phytohormones, signaling
Citation: Ilangumaran G and Smith DL (2017) Plant Growth Promoting Rhizobacteria in Amelioration of Salinity Stress: A Systems Biology Perspective. Front. Plant Sci. 8:1768. doi: 10.3389/fpls.2017.01768
Received: 26 April 2017; Accepted: 27 September 2017;
Published: 23 October 2017.
Edited by:
Helena Freitas, Universidade de Coimbra, PortugalReviewed by:
Lianghui Ji, Temasek Life Sciences Laboratory, SingaporeAdele Muscolo, Mediterranea University of Reggio Calabria, Italy
Copyright © 2017 Ilangumaran and Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donald L. Smith, donald.smith@mcgill.ca
 Gayathri Ilangumaran
Gayathri Ilangumaran Donald L. Smith
Donald L. Smith