- 1State Key Laboratory of Hybrid Rice, College of Life Sciences, Wuhan University, Wuhan, China
- 2Hybrid Rice Research Center, Hubei Academy of Agricultural Sciences, Wuhan, China
It has been reported that the receptor-like cytoplasmic kinases (RLCKs) regulate many biological processes in plants, but only a few members have been functionally characterized. Here, we isolated a rice gene encoding AtRRK1 homology protein kinase, OsRRK1, which belongs to the RLCK VI subfamily. OsRRK1 transcript accumulated in many tissues at low to moderate levels and at high levels in leaves. Overexpression of OsRRK1 (OE-OsRRK1) caused adaxial rolling and erect morphology of rice leaves. In the rolled leaves of OE-OsRRK1 plants, both the number and the size of the bulliform cells are decreased compared to the wild-type (WT) plants. Moreover, the height, tiller number, and seed setting rate were reduced in OE-OsRRK1 plants. In addition, the brown planthopper (BPH), a devastating pest of rice, preferred to settle on WT plants than on the OE-OsRRK1 plants in a two-host choice test, indicating that OE-OsRRK1 conferred an antixenosis resistance to BPH. The analysis of transcriptome sequencing demonstrated that several receptor kinases and transcription factors were differentially expressed in OE-OsRRK1 plants and WT plants. These results indicated that OsRRK1 may play multiple roles in the development and defense of rice, which may facilitate the breeding of novel rice varieties.
Introduction
Increasing crop yield is a major challenge for modern agriculture (Ray et al., 2012). Regulating leaf development has been considered an effective way to achieve a breakthrough of potential yield for crops (Sinclair and Sheehy, 1999). Appropriate leaf shape is an important characteristic of the super-high-yield hybrid rice idiotype, in which the last three leaves from the top are long, erect, narrow, V-shaped (rolled), and thick (Yuan, 1997). Moderate leaf rolling in rice leads to erect leaf canopies, improves photosynthetic efficiency, accelerates dry-matter accumulation, and increases grain yield (Lang et al., 2004; Zhang et al., 2009; Zou et al., 2011). Isolation of genes controlling leaf rolling are expected to be beneficial for developing crops with the desired architecture (Zhang et al., 2009; Xu et al., 2014).
Leaf form was regulated by complicated developmental processes, including pattern formation, polarity establishment, and cell differentiation (Bowman et al., 2002; Micol and Hake, 2003; Lang et al., 2004). Bulliform cells are monocot-specific (with the exception of the Helobiae), large thin-walled, apparently empty, highly vacuolated and occur in groups between vascular bundles on the adaxial epidermis (Jane and Chiang, 1991; Itoh et al., 2005). In rice, two types of leaf rolling (adaxial rolling and abaxial rolling) have been related to the abnormal development of bulliform cells. Changes in the number, volume, and localization of bulliform cells can result in lack of osmotic pressure to support the normal form of the blade, and then, the leaf becomes rolled (Zhang et al., 2015). Generally, a leaf displays adaxial rolling when the number and size of bulliform cells are decreased in rice. For example, the mutant of a MYB transcription factor, sll1, has smaller and fewer bulliform cells than wild-type (WT), resulting in adaxial leaf rolling (Zhang et al., 2009). Conversely, a leaf displays abaxial rolling when the number and size of bulliform cells are increased. For example, enhanced expression of ACL1, encoding a protein with unknown conserved functional domains, causes increased bulliform cell number and abaxial leaf rolling (Li et al., 2010).
The receptor-like kinases (RLKs) play critical roles in plant development and response to stress stimuli (Gao and Xue, 2012). A typical RLK contains an extracellular receptor, a transmembrane domain and an intracellular kinase domain (Shiu and Bleecker, 2001). One of the RLK families lacking an extracellular domain has, therefore, been designated the receptor-like cytoplasmic kinases (RLCKs). Recent studies have shown that some RLCKs regulate both plant development and defense responses. In Arabidopsis, RLCKs are classed into 13 subfamilies according to the phylogenetic evaluation (Shiu et al., 2004). BIK1 (Botrytis-Induced Kinase 1), functions as an early-induced kinase in response to infection by Botrytis cinerea and is required for BRI1 (Brassinolide-Insensitive 1) mediated growth regulation through direct interaction with BRI1 (Veronese et al., 2006; Lin et al., 2013). PBL27 is an immediate downstream component of the chitin receptor CERK1 and contributes to the regulation of chitin-induced immunity in Arabidopsis (Shinya et al., 2014). It has been predicted that rice has a total of 379 RLCK genes (Vij et al., 2008). To date, few RLCK genes have been functionally characterized in rice. Of those that have, it has been reported that BSR1 (broad-spectrum resistance 1) positively regulates resistance against Xoo (Xanthomonas oryzae pv. oryzae) and M. oryzae (Magnaporthe grisea) both in Arabidopsis and rice (Dubouzet et al., 2011). OsRLCK185 and OsRLCK55 can interact with Xoo1488, which is a Xoo effector. OsRLCK185 also regulates PGN- and chitin-induced immunity (Yamaguchi et al., 2013). Five other RLCK genes, OsRLCK102, OsRLCK57, OsRLCLK107, OsRLCK118, and OsRLCLK176 are involved in innate immunity mediated by XA21 and in development by BR signaling in rice (Yamaguchi et al., 2013; Ao et al., 2014; Wang et al., 2016; Zhou et al., 2016). Most of these reported RLCK genes belong to the RLCK VII subfamily and are involved in disease resistance and growth development in Arabidopsis and rice. However, no other members of RLCK subfamilies in rice have been reported.
In this study, we characterized the RLCK gene OsRRK1, which was interacted with OsLecRK (a lectin RLK) that was involved in innate immune responses and seed germination (Cheng et al., 2013), belongs to the RLCK VI subfamily and encodes an AtRRK1 (Rop-interacting receptor-like kinase 1) homologous protein kinase. Overexpression of OsRRK1 caused rolling and erect leaves in rice plants. In addition, the degree of leaf rolling was positively correlated with the expression level of OsRRK1. OsRRK1 is also involved in rice development as well as in defense against BPH. The result of transcriptome sequencing indicated that OsRRK1 regulates plant development and defense to BPH mainly through receptor kinases, other RLCKs and transcription factors.
Materials and Methods
Plant Materials
In this study, the WT rice (Oryza sativa) cultivar used as controls in all the morphological and molecular comparisons was a japonica variety, Hejiang19. The template for gene amplification was derived from the cDNA of the rice indica variety Kasalath. All the transgenic lines including the OE and RNAi lines are in the background of Hejiang19. All experimental materials were transplanted in the experimental field at a spacing of 16.7 cm between plants within each row and 26.7 cm between rows at the Genetics Institute at Wuhan University (Wuhan, China). The plants were tended under the routine management regime.
Yeast Two-Hybrid Assay
The two-hybrid assay was performed using the GAL4-based transcription system. The OsRRK1 cDNA was cloned into a bait vector pGBKT7, while the OsLecRK cDNA was ligated into a prey vector pGADT7. Yeast strains AH109 (Clontech) were transformed with bait and prey cotransform constructs. Yeast diploids were selected on selection plates containing SD (Synthetic Dropout) medium lacking Leu, Trp, and His. The interactions between p53 and the SV40 large T-antigen (T), and between lamin (Lam) and the T served as positive and negative controls, respectively.
In Vivo Co-immunoprecipitation Assays
In vivo co-immunoprecipitation (Co-IP) assays were carried out by transient protein expression in rice protoplasts. HA- tagged OsRRK1 and MYC- tagged LecRK constructs were coexpressed in rice protoplasts, extracted in the buffer [50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% NP-40, 1 mM PMSF, plant protease inhibitor cocktail (Roche)], immunoprecipitated with anti-MYC antibody, then detected by the anti-HA (MBL, Catalog: M180) and anti-Myc (MBL, Catalog: M047), respectively.
Plasmid Constructs and Rice Transformation
To make an overexpression construct (OE-OsRRK1), a 1179-bp cDNA fragment encoding the full-length of OsRRK1 was PCR-amplified from the cDNA library of Kasalath using a pair of primers, OE-OsRRK1-F and OE-OsRRK1-R (Supplementary Table S1). The amplified DNA fragments were ligated with pCXUN-vector, which was digested with XcmI. The pCXUN-vector contains the maize ubiquitin promoter and a nos terminator.
The OsRRK1-RNAi constructs were generated by an overlapping PCR approach (Chen et al., 2009). Briefly, in the first-round PCR, two 521-bp fragments between +569 and +1089 (relative to the ATG at +1 bp) were amplified from the cDNA library of 9311 using the primers RNAi-F and RNAi-R1, RNAi-F and RNAi-R2. The PDK intron loop sequence fragment was amplified from pHAN using the primers PDK-F and PDK-R. Primers RNAi-R1 and PDK-F were used to introduce complementary adapters to the amplified fragments, as RNAi-R2 and PDK-R. The three amplified fragments were fused together as an inverted-repeat cassette in the second-round PCR by using a single RNAi-F primer. The resulting fragment was then directly cloned into the plant expression pCXUN-vector. All the primers used for vector construction are shown in Supplementary Table S1.
The constructs were introduced into Agrobacterium tumefaciens EHA105 via electroporation. The Agrobacterium-mediated transformations of rice (Hejiang19) were carried out as previously described (Chen et al., 2007).
Measurement of the Leaf Rolling Index and Leaf Erection Index
To determine the LRI, two measurements were taken, Lw (expand the leaf blade and determine the greatest width of the leaf blade) and Ln (measure the natural distance of the leaf blade margins at the same location on the leaf where Lw was measured). LRI was calculated as LRI (%) = (Lw -Ln)/Lw × 100%.
To determine the LEI, two measurements were taken, Lnl (the linear distance between the lamina joint and the tip of the leaf blade in the natural position) and Lsl (the length of the straightened leaf). LEI was calculated as LEI (%) = Lnl/Lsl × 100% (Shi et al., 2007).
Data were collected from the flag leaves of 20 individual plants at the heading stage.
Southern Blot Analysis
A probe (Supplementary Table S1) was labeled with [α-32P] dCTP using the Prime-a-Gene labeling system (Promega). Twenty microgram genomic DNA was digested with EcoRI restriction enzyme (Fermentas), then separated on a 1% agarose gel and blotted onto Hybond-N+ nylon membrane (Amersham Biosciences). The membranes were prehybridized for 3 h at 65°C and the hybridization buffer was refreshed with the labeled probe. The membranes were then incubated for 12 h at 65°C. Washing was conducted at 65°C for 15 min in 2 × SSC and 0.2% SDS, and subsequently for 15 min at 65°C in 1 × SSC and 0.1% SDS. The membranes were then exposed to storage phosphor screens (Amersham Biosciences), and the hybridization signals were detected using a Typhoon PhosphorImager (Amersham Biosciences).
Quantitative Real-time PCR (qRT-PCR)
Total RNA of various rice tissues including radicle and plumule in 48 h after emergence, root and leaf in the second tillering stage, flag leaf, second leaf from the top leaf, stem, leaf sheath, and young panicle at the heading stage were isolated using TRIzol reagent (TaKaRa) according to the manufacturer’s protocol. One microgram RNA was treated with DNase I (Fermentas) to remove genomic DNA and then used to synthesize cDNA with a RevertAidTM First Strand cDNA Synthesis Kit (Fermentas) following the manufacturer’s recommendations. The cDNA was then amplified by specific primers and SYBR Green PCR Master Mix (Applied Biosystems) in a CFX96 Real-Time System (Bio-Rad). The analysis of the results was performed as previously described (Wei et al., 2009). All primers used in this study are listed in Supplementary Table S1.
Histology and Microscopy Observation
To determine the detailed structure of leaf, mature flag leaves containing at least the bottom 1/2 of the tissues were used for a paraffin cross-section assay. Leaves were fixed in 70% formalin–acetic acid–alcohol solution (FAA) for 24 h at 4°C. After serial dehydration in various concentrations of ethanol, the samples were transferred into xylene and then embedded in paraffin. Sections (10 μm thick) were cut with a microtome (Leica RM2245) and mounted on microscope slides. Slices were spread on a platform at 40°C overnight and stained using 0.5% Toluidine Blue O at 37°C for 30 min. After dewaxing and rehydrating, the slices were examined and photographed using a LEICA CTR5000B microscope. Bulliform cell area was measured with Image J software1.
Analysis of Alignments and Phylogenesis
The BLASTP program2 was used to identify the homologous sequence of OsRRK1. Before phylogenetic analysis, multiple sequence alignments were generated using ClustalX Version 1.83 (Thompson et al., 1997). A phylogenetic tree was constructed by MEGA Version 5.1.0 using the neighbor-joining (NJ) method. Bootstrap analyses of 1,000 replicates were carried out.
BPH Host Choice Test
The host choice tests were conducted on 4-week-old rice plants in a cup (10 cm in diameter). Two WT plants and two OE-OsRRK1 plants were planted at opposite ends of roughly perpendicular diagonals. Thirty-fourth-instar BPH nymphs were then placed in the middle of the cup and the number of nymphs that settled on each plant was recorded each day after infestation. The experiment was repeated twice, each time with 20 biological replicates.
RNA-Sequencing and Data Analysis
The flag leaves of both Hejiang19 and OE-25 at the five-leaf stage were collected for RNA-sequencing analysis, with each sample containing a pool of 10 plants, each rice variety containing three biological replicates. Total RNAs were prepared using RNAiso Plus according to the manufacturer’s protocol (TaKaRa Code: D9108A). Half of RNA was performed to transcription expression analysis by Shanghai Biotechnology Corporation, and the rest was used to verify the RNA-sequencing results by qRT-PCR. All subsequent procedures, including mRNA purification, cDNA preparation, end repair of cDNA, adaptor ligation, and cDNA amplification were performed according to the manufacturer’s protocols accompanying the mRNA-Seq Sample Preparation Kit (Illumina). Each library had an insert size of 45 bp, and sequences of 50 bp on one end (1∗50 bp) were generated via Illumina HiSeq2000.
Cufflinks (version: 2.0.23) was used to calculate the FPKM value of every transcript. The P-values of different expressions were calculated using Fisher’s exact test. We used P < 0.05 and the absolute value of log2FC ≥ 0.585 as the threshold to judge the significance of each gene expression difference (Trapnell et al., 2010). Cluster analysis was performed and heat maps generated with the Genesis software based on a hierarchy (version: 1.7.64) (Sturn et al., 2002). For pathway analysis, we mapped all DEGs using the MapMan package with the Osa_MSU_v7 mapping file (Thimm et al., 2004).
Accession Numbers
Sequence data for OsRRK1 can be found in the GenBank database under accession number KY347802. The RNA-Seq raw data were submitted to Short Read Archive at NCBI with accession numbers of SAMN06448985, SAMN06448986, SAMN06448987, SAMN06448988, SAMN06448989, and SAMN06448990.
Results
Characterization of the OsRRK1 Gene
In the previous study, OsLecRK was used as a bait to screen a rice cDNA expression library established in a yeast two-hybrid system (Cheng et al., 2013). We isolated a full-length cDNA encoding a RLCK, which interacted with OsLecRK by a yeast two-hybrid assay (Supplementary Figure S1A) and co-immunoprecipitation (Supplementary Figure S1B). BLAST analysis in the NCBI database showed that this RLCK exhibits 57% amino acid sequence identity with AtRRK1 (Rop-interacting receptor-like kinase 1) from Arabidopsis, we named this gene OsRRK1 (LOC_Os06g47820). The full-length cDNA of OsRRK1 is 1567-bp long, and consists of a 237-bp 5’UTR, 1179-bp coding region and 151-bp 3’UTR (Supplementary Figure S2). The open reading frame (ORF) of OsRRK1 comprises six exons and five introns. It is predicted that it encodes a polypeptide of 392 amino acids with a serine-threonine/tyrosine-protein kinase catalytic (STYKc) domain (Supplementary Figures S3A,B).
The expression level of OsRRK1 in various rice organs and at different development stages was determined by quantitative real-time PCR (qRT-PCR). This demonstrated that the expression level of OsRRK1 was highest in leaves, especially in flag leaves, and then decreased gradually in leaf sheathes, young panicles, stems, plumules, radicles, and roots (Figure 1A).
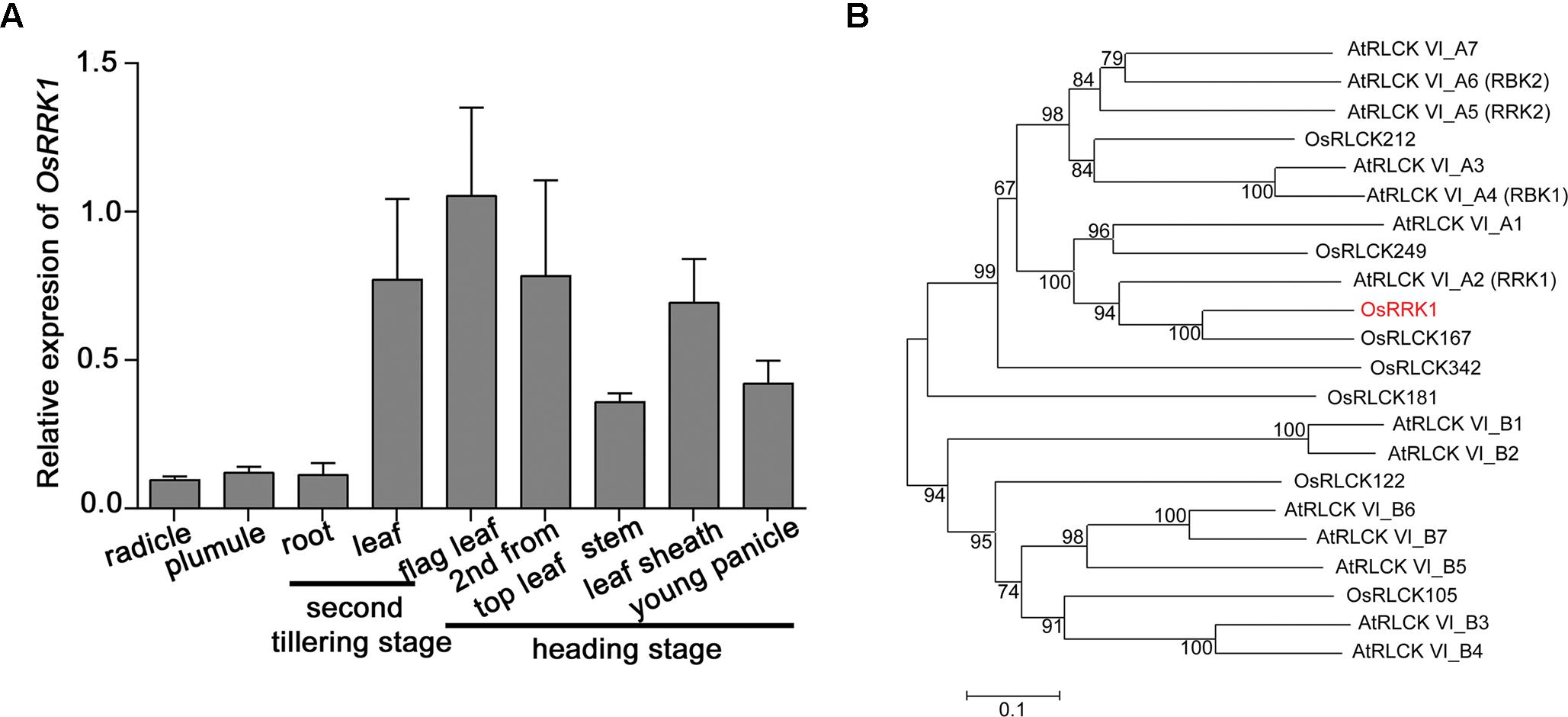
FIGURE 1. Characterization of the OsRRK1 gene. (A) Expression of OsRRK1 in various organs determined by qRT-PCR analysis. Rice ACTIN1 gene was used as an internal control. Error bars represent the SD of transcript levels determined from three independent replicates. (B) Phylogenetic analysis of OsRRK1 and its homologous protein with other RLCK VI sub-family members in Arabidopsis and rice by MEGA6.0 constructed using the Neighbor-Joining method. The OsRRK1 protein is shown in red. The GenBank accession numbers of OsRRK1 and its homologous proteins are as follows: OsRRK1: XP_015642177.1; OsRLCK212: XP_015642870.1; OsRLCK249: XP_015648938.1; OsRLCK167: XP_015634276.1; OsRLCK342: ABA95043.1; OsRLCK181: XP_015640273.1; OsRLCK122: XP_015632245.1; OsRLCK105: XP_015630796.1; AtRLCK VI_A1: NP_001078762.1; AtRLCK VI_A2: NP_179479.1; AtRLCK VI_A3: NP_201356.2; AtRLCK VI_A4: NP_568231.1; AtRLCK VI_A5: NP_198445.1; AtRLCK VI_A6: NP_001327889.1; AtRLCK VI_A7: NP_197392.2; AtRLCK VI_B1: NP_198595.1; AtRLCK VI_B2: NP_001321968.1; AtRLCK VI_B3: NP_001190918.1; AtRLCK VI_B4: NP_179266.2; AtRLCK VI_B5: NP_201199.1; AtRLCK VI_B6: NP_001319394.1; AtRLCK VI_B7: NP_173578.2.
OsRRK1 is OsRLCK216 and belongs to the RLCK VI subfamily (Vij et al., 2008; Gao and Xue, 2012). In order to analyze the genetic relationship between OsRRK1 and other RLCKs in rice and Arabidopsis, we constructed an unrooted phylogenetic tree for OsRRK1 and found that OsRRK1 exhibits relatively higher homology with OsRLCK167 and AtRLCK VI_A2 (AtRRK1) (Dorjgotov et al., 2009) (Figure 1B).
Overexpression of OsRRK1 Results in a Leaf Rolling Phenotype
In order to elucidate the function of OsRRK1 in rice, an OsRRK1 overexpression construct driven by the ubiquitin promoter was produced and introduced into WT plants via A. tumefaciens-mediated transformation. Seventeen positive transgenic lines (OE-OsRRK1) were generated and confirmed by Southern blot analysis (Supplementary Figure S4). Eight of them (OE-5, 7, 16, 19, 21, 22, 24, 25) had a single copy of OsRRK1 and the others had multiple copies. We chose three single copy lines (OE-22, 24, and 25) for further analysis.
Compared to WT plants, all of the OE-OsRRK1 plants displayed the leaf rolling phenotype. During the heading stage, we were able to observe that leaves of WT plants were flat, whereas the flag leaves of OE-OsRRK1 plants were rolled to different degrees in the field (Figure 2A) and the laboratory (Figure 2B). We transected the middle part of these leaves and found that the cross-section revealed rolling to varying degrees (Figure 2C).

FIGURE 2. The leaf phenotype of the WT and OE-OsRKK1 plants. (A) Morphology of WT plants and the OE-OsRRK1 plants at the heading stage in the paddy field. Red arrow indicated the leaf morphology. Leaves of WT plants were flat, whereas the flag leaves of OE-OsRRK1 plants were rolled to different degrees in the field. (B) Mature leaves of the WT and OE-OsRRK1 plants at the heading stage in a bucket (Bar = 10 cm). The corresponding close-up images of the flag leaf architecture details are shown besides each bucket image (Bar = 5 cm). (C) Cross-section of the middle part of the flag leaf in the WT and the OE-OsRRK1 plants at the heading stage. ad, adaxial; ab, abaxial. Bars = 2 mm.
To evaluate the degree of leaf rolling accurately, we calculated the LRI, for which a higher value indicates a higher rolling degree (Shi et al., 2007). At the heading stage, we measured the maximum leaf width (Lw) and natural leaf width (Ln). The maximum width of flag leaves was not significantly different in WT plants and the OE-OsRRK1 plants (Figure 3A). However, the natural width of flag leaves in OE-OsRRK1 plants was significantly reduced compared to WT (Figure 3B). In WT plants, the natural width of flag leaves was almost the same as the maximum width, while in OE-OsRRK1 plants the natural width of flag leaves was much smaller than the maximum width. The LRI of the WT plants was 0, while the LRIs of the OE-OsRRK1 plants were at least 20% higher than that of the WT plants (Figure 3C). These results indicate that the leaf rolling of the OE-OsRRK1 plants could be attributed to changes in the natural width of the leaf rather than the maximum width.
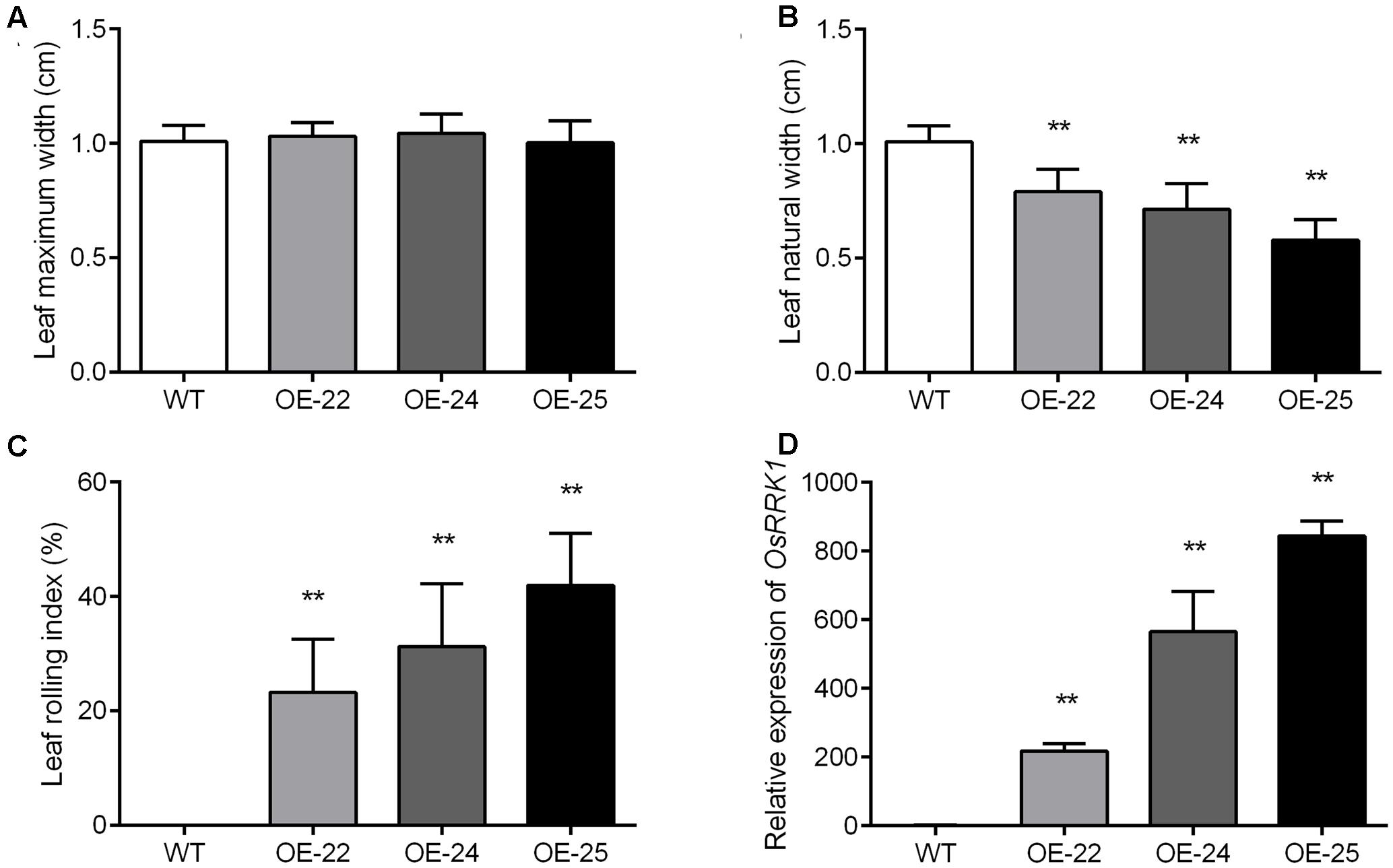
FIGURE 3. Evaluation of the degree of leaf rolling in WT and OE-OsRRK1 plants. (A,B) Leaf maximum width (A) and leaf natural width (B) of the WT and OE-OsRRK1 plants. Twenty plants of each line were measured at the heading stage. (C) LRI of the flag leaves of WT and the OE-OsRRK1 plants based on the leaf maximum and natural width. Error bars indicate SD (n = 20). (D) The relative expression levels of OsRRK1 in the WT and OE-OsRRK1 plants. Flag leaves at the heading stage were used in this experiment. Values are the means ± SD of three biological replicates. ∗Student’s t-test, P < 0.05; ∗∗Student’s t-test, P < 0.01.
In general, leaf rolling leads to changes in the erectness of leaves (Shi et al., 2007). Thus, we measured the LEI, which quantifies how upright the leaves are. At the heading stage, we measured the linear distance between the lamina joint and the tip of the leaf blade (Lnl) and the length of the straightened leaf in its natural position (Lsl). The LEIs in the flag leaves of OE-OsRRK1 plants were clearly different from those of the WT plants. The LEI calculations yielded values for the flag leaves of OE-OsRRK1 plants all close to 100%, while the LEIs of the WT plants averaged 95.1 ± 3.08% (Supplementary Figure S5).
To explain the different degrees of rolling in the three OE-OsRRK1 plants, we tested the expression level of OsRRK1 in these transgenic plants by qRT-PCR analyses. The results demonstrated that the expression of OsRRK1 in these transformants was upregulated to varying degrees. They were 200 times, 500 times, and 800 times upregulated in OE-22, OE-24, and OE-25, respectively (Figure 3D). The results revealed that with the increase of the OsRRK1 relative expression level, the natural width of the flag leaves becomes smaller, so they may be negatively correlated. While with the increase of the OsRRK1 relative expression level, the LRI also increase, so they may be positively correlated. However, LEI did not show significant correlation with the relative expression of OsRRK1.
To investigate the role of OsRRK1 in rice leaf development further, RNAi analysis was also conducted. Unexpectedly, although the expression of OsRRK1 was notably downregulated, no obvious change in leaf development was detected (Supplementary Figures S6A–C). The LRIs of all the RNAi plants were 0 in the flag leaves, which is similar to the WT plants. In addition, the LEIs of all the RNAi plants exhibited no obvious difference from the WT plants (Supplementary Figure S6D). OsRLCK167 is the closest homolog of OsRRK1. We compared the expression abundance of OsRRK1 and OsRLCK167 in WT rice leaves and found they showed a similar expression level in flag leaf at heading stage (Supplementary Figure S7A). And we found that the expression of OsRLCK167 in RNAi plants showed no difference from the WT plants (Supplementary Figure S7B). Therefore, we suggested that functional redundancy might explain no changes in phenotypes by RNAi approaches. In addition, the transcript level of the RNAi plants was not inhibited completely, which was very possible these RNAi lines still have more than sufficient expression of OsRRK1 for normal function. Thus, we concluded that overexpression of OsRRK1 results in a rolled-leaf phenotype, while moderate downregulation of OsRRK1 was associated with no abnormality in leaf development.
Bulliform Cell Number and Size Are Decreased in OE-OsRRK1 Plants
Bulliform cells are located between the vascular veins of the leaf in rice. Previous studies have demonstrated that changes in the number and area of the bulliform cells result in leaf rolling (Zhang et al., 2009, 2015; Li et al., 2010). Therefore, we performed transverse sectioning so that we could examine the leaves of OE-OsRRK1 and WT plants. The bulliform cells were smaller and fewer in OE-OsRRK1 leaves than in the WT leaves, whereas no significant changes were found for other cell types and their arrangements between OE-OsRRK1 and the WT leaves (Figures 4A,B). The bulliform cells of WT plants were typically arranged in groups of 4.64 ± 0.59 cells, whereas the bulliform cells in OE-OsRRK1 leaves were only in groups of 4.05 ± 1.23, 3.68 ± 0.83, and 3.41 ± 0.59 cells in OE-22, OE-24, and OE-25, respectively (Figure 4C). In addition, the bulliform cell area in OE-OsRRK1 leaves was reduced compared to WT leaves (Figure 4D). We also found that with the increase of the OsRRK1 expression level, the number of bulliform cells and the average cell area were all decreased, suggesting they may be negatively correlated. These results suggest that overexpression of OsRRK1 decreased bulliform cell number and size, which resulted in leaf rolling.
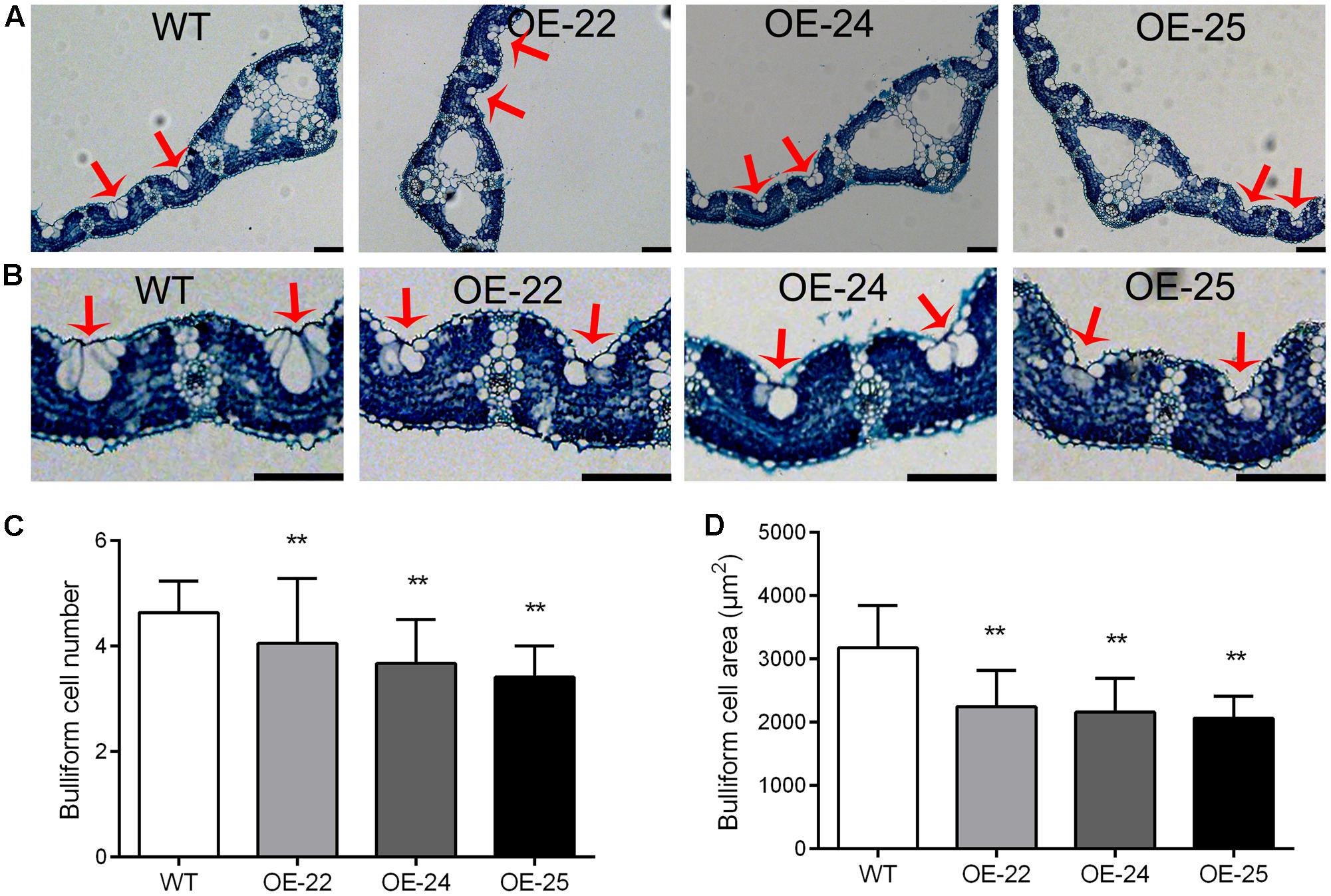
FIGURE 4. Bulliform cell number and size in the leaves of OE-OsRRK1 plants. (A) Cross sections of flag leaves in WT and OE-OsRRK1 plants. (B) Enlarged view of cell structure of flag leaves in WT and OE-OsRRK1 plants. Bulliform cells are indicated by red arrows. Bars = 100 μm. (C,D) The bulliform cell number (C) and bulliform cell area (D) in the leaves of WT and OE-OsRRK1 plants. Ten samples were investigated. ∗Student’s t-test, P < 0.05; ∗∗Student’s t-test, P < 0.01.
Overexpression of OsRRK1 in Rice Conferred an Antixenosis Effect to BPH
OsRRK1 was identified as an interactor of OsLecRK, which was involved in defense to BPH (Cheng et al., 2013). We compared the performance of BPH on OE-OsRRK1 and WT plants to determine whether OsRRK1 is involved in defense to this herbivorous insect of rice. Host preference was assessed on the basis of the number of BPHs that settled on WT and OE-OsRRK1 plants after releasing BPHs into the center of containers where BPHs could choose between WT and OE-OsRRK1 plants. Comparing the OE-22 and WT plants, there was no significant difference in the number of BPHs on each from the first day to the sixth day (Figure 5A). However, more insects choose to settle on the WT plants than on the OE-24 and OE-25 plants. From the fourth day, the number of BPHs settled on each plant type was significantly different between the OE-24 and WT plants (Figure 5B). For the OE-25 and WT plants, a significant difference of BPH choice was observed from the first day to the sixth day (Figure 5C). These results indicated that OsRRK1 confers defense to BPH attacks via antixenosis, which is known to be an important mechanism of plant defense against insect attacks (Walling and Thompson, 2012). As the OsRRK1 overexpression level can be ranked OE-25 > OE-24 > OE-22, our results suggest that the level of defense is determined by the accumulation level of OsRRK1 transcripts.
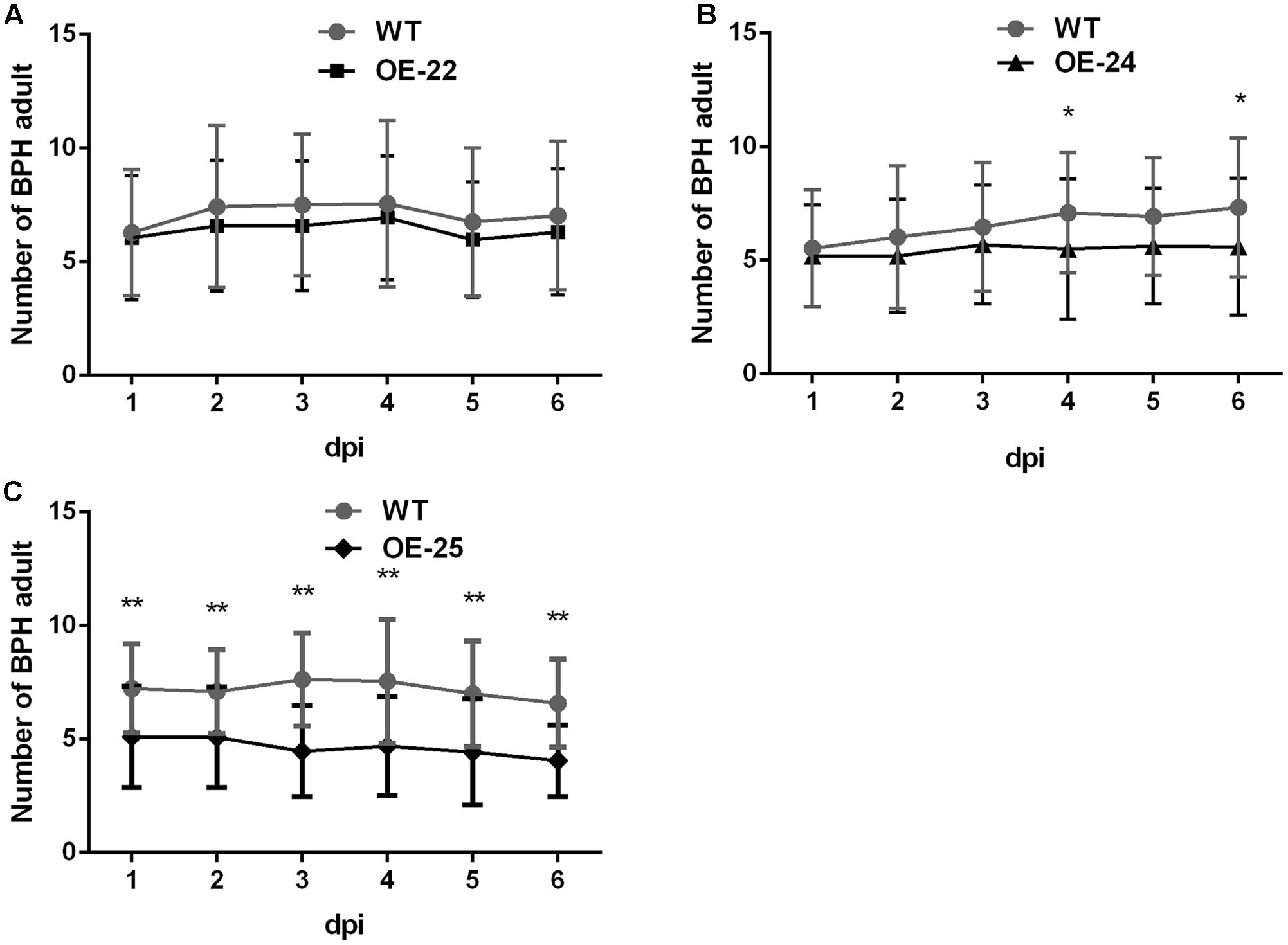
FIGURE 5. Two-host choice test for BPHs on WT and OE-OsRRK1 plants. (A) No significant difference between BPHs settling on WT plants and OE-22 plants. (B) Significant difference between BPHs settling on WT plants and OE-24 plants was only observed on the fourth day and the sixth day. (C) BPHs congregated on the WT plants to a greater extent than on OE-25 plants and there was a significant difference from the first day to the sixth day. ∗Student’s t-test, P < 0.05; ∗∗Student’s t-test, P < 0.01.
Pleiotropic Roles of OsRRK1 in Multiple Plant Developmental Processes
In addition to rolled and erect leaves, other phenotype changes in OE-OsRRK1 plants were also observed. Compared with WT plants, the plant height, tiller numbers and seed setting rate of OE-OsRRK1 plants were significantly reduced. The height of WT plants was 60.28 ± 2.45 cm, whereas the height was 55.77 ± 3.65 cm, 58.51 ± 2.15 cm, and 57.79 ± 2.38 cm in OE-22, OE-24, and OE-25, respectively (Figure 6A). There were about 9.75 ± 1.45 tillers in WT plants, but only 7.35 ± 1.14, 7.65 ± 1.90, and 7.55 ± 0.89 tillers in OE-22, OE-24, and OE-25, respectively (Figure 6B). In WT plants, the seed setting rate was about 88.61% ± 2.09%, but was 61.2% ± 5.48%, 65.96% ± 8.38%, and 67.67% ± 6% in OE-22, OE-24, and OE-25, respectively (Figure 6C). These results indicate that overexpression of OsRRK1 changed multiple plant developmental processes.
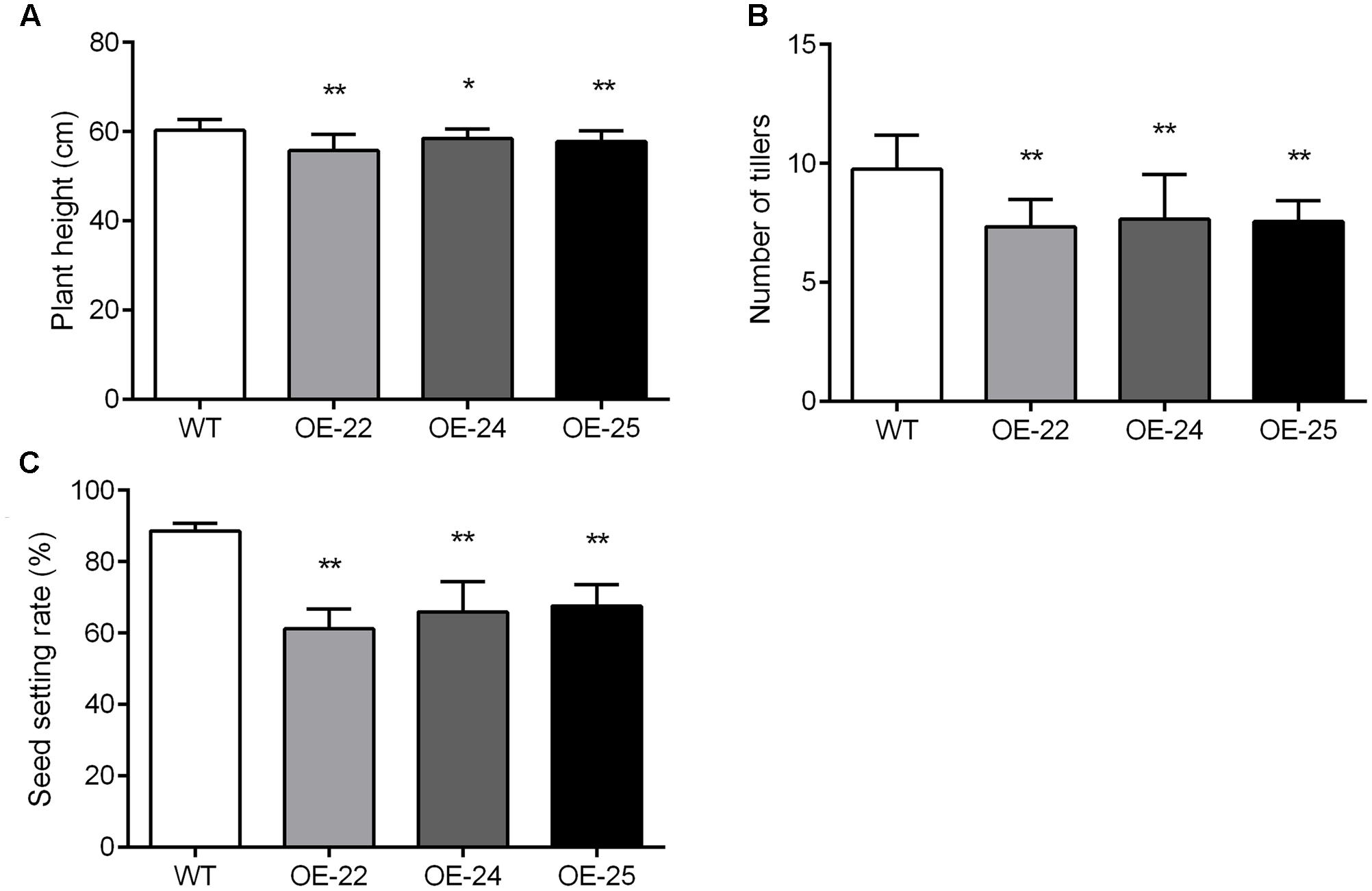
FIGURE 6. Pleiotropic effects of the OsRRK1 gene. (A,B) Plant height (A) and tiller number (B) in WT plants and OE-OsRRK1 plants at the heading stage. Data are averages for 20 plants. (C) The seed setting rate of WT plants and OE-OsRRK1 plants. Seeds from 20 plants were measured after harvest. The seed setting rate was determined as: (filled grain number per panicle)/(grain number per panicle)∗100%. Asterisks indicate a significant difference between WT plants and OE-OsRRK1 plants according to a t-test, ∗Student’s t-test, P < 0.05; ∗∗Student’s t-test, P < 0.01.
Transcript Profiles Are Distinct in WT Plants and OE-OsRRK1 Plants
To understand the molecular mechanism of rice development and defense to BPH mediated by OsRRK1, the expression profiles of WT plants Hejiang19 (H) and a line of OE-OsRRK1 plants (OE-25, abbreviated as OE hereafter) were determined using deep RNA-sequencing. As a result, 116.4 million paired-end sequence reads of 50-bp in length were generated from the six samples. After removing low-quality reads, a total of 113.8 million high quality clean reads were retained, of which more than 97% were aligned to the reference genome using TopHat (Supplementary Table S2).
One fundamental use of transcriptome sequencing is the analysis of differentially expressed genes (DEGs) between groups (Wang et al., 2009). In our study, we defined DEGs as the transcripts showing at least a 1.5-fold change in the FPKM (fragments per kilobase of exon per million fragments mapped) (log2FC ≥ 0.585 or log2FC ≤ –0.585) and a P-value < 0.05. In total, 625 DEGs were detected among the paired comparisons (H vs. OE), including 366 upregulated and 259 downregulated genes. DEGs in H and OE of the three biological replicates were hierarchically clustered in the heat map (Supplementary Figure S8). The majority of DEGs had similar expression patterns among three biological replicates, showing consistent upregulation or downregulation.
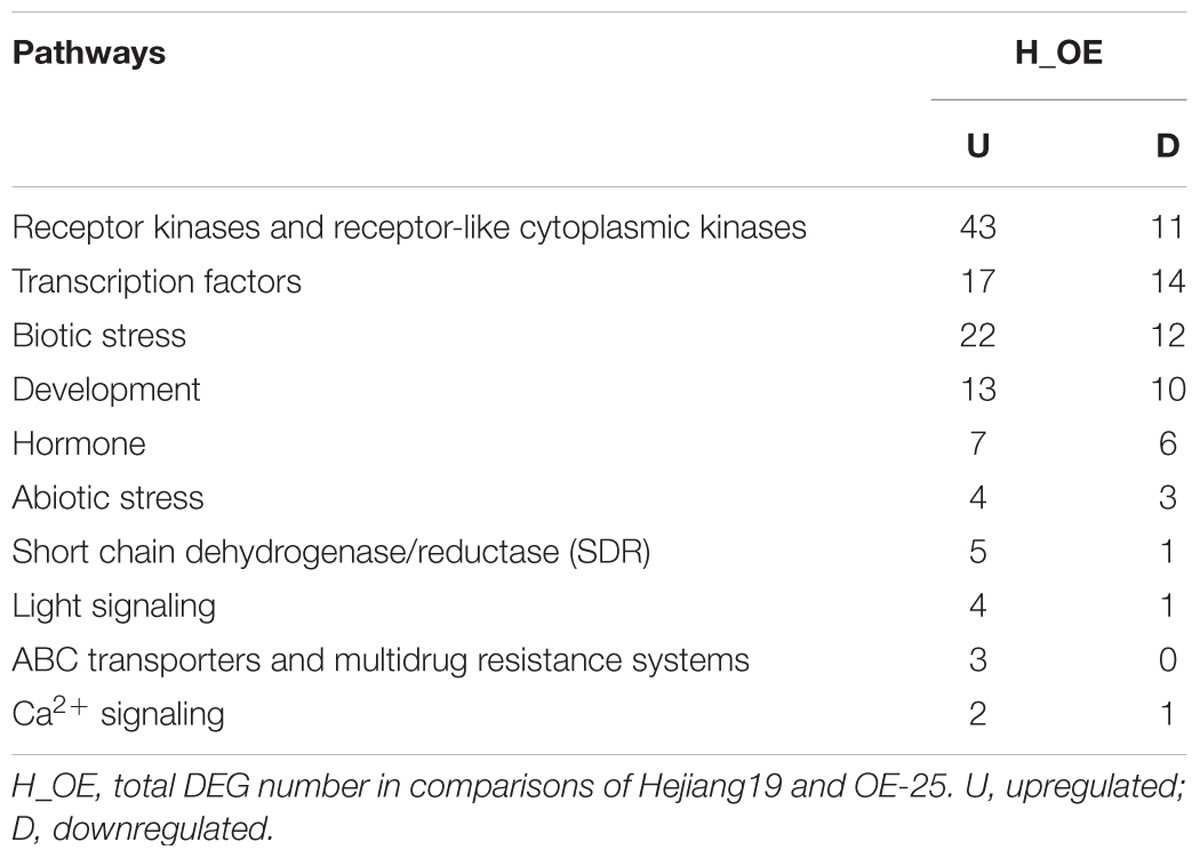
To enhance our understanding of the biological function of DEGs, those assigned to MapMan pathways and important classifications are provided in Table 1. Receptor kinases and receptor-like cytoplasmatic kinase VII-related genes are important components of signal transduction and play roles in plant development. In this study, 43 genes encoding receptor kinases and receptor-like cytoplasmatic kinase VII were upregulated, 11 genes encoding receptor kinases were downregulated (Table 1). Among them, legume lectins beta domain containing protein (LOC_Os07g04110) and receptor-like protein kinase 2 precursor (LOC_Os08g14950) are involved in plant resistance; brassinosteroid insensitive 1-associated receptor kinase 1 (LOC_Os11g31540) and receptor-like protein kinase 2 (LOC_Os11g40480) are involved in plant development. To verify the RNA-sequencing results, the expressions of the aforementioned four genes were analyzed by qRT-PCR with gene specific primers. The qRT-PCR results were consistent with RNA-sequencing data (Supplementary Table S3). Compared to WT, all of them were upregulated in all three OE-OsRRK1 plants (Figures 7A–D).
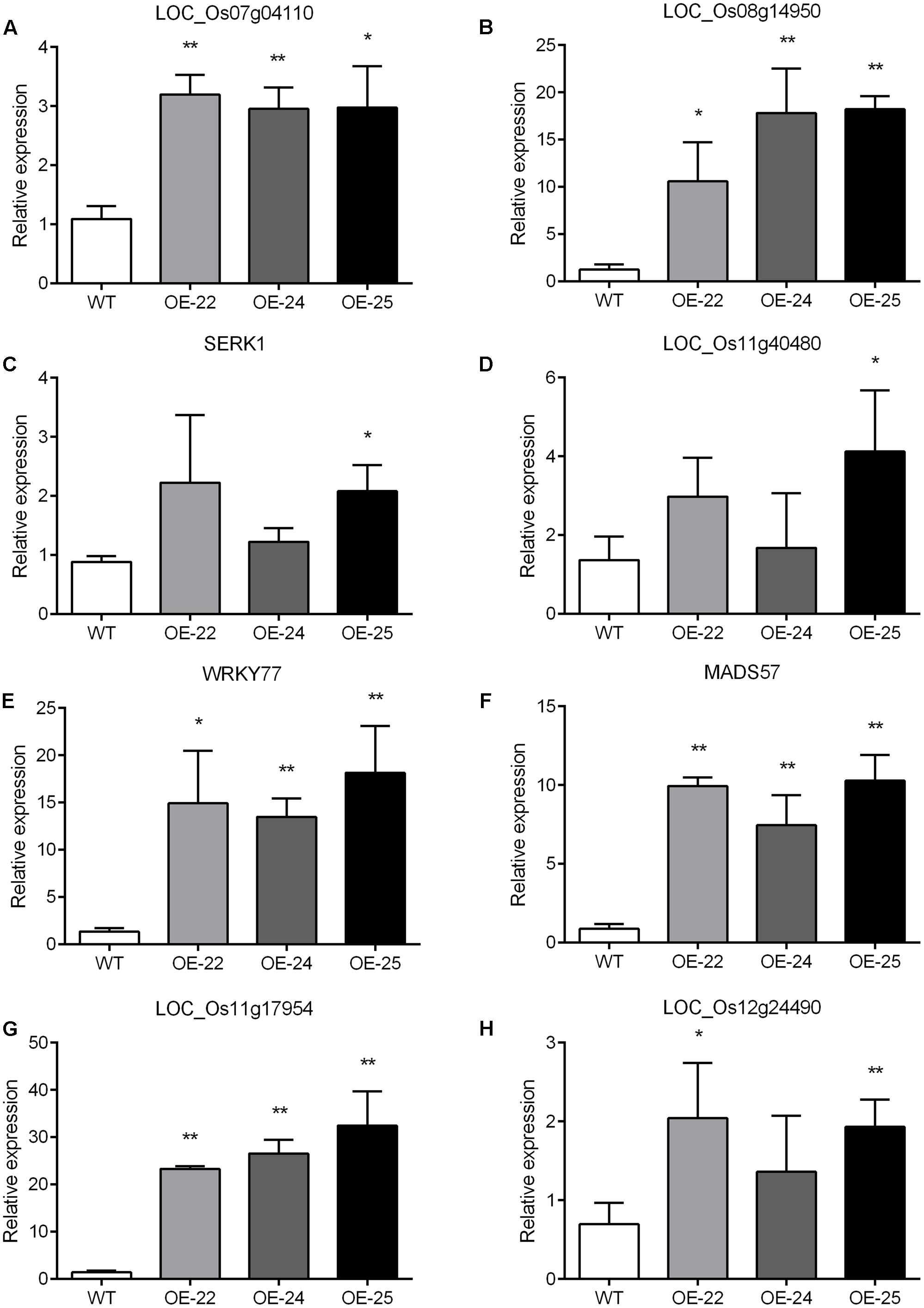
FIGURE 7. (A–H) qRT-PCR analysis of differentially expressed genes detected by the transcriptomic analysis. SERK1: LOC_Os11g31540; WRKY77: LOC_Os01g40260; MADS57: LOC_Os02g49840. Significant differences are indicated by ∗P < 0.05, ∗∗P < 0.01 (Student’s t-test).
Transcription factors (TFs) also play important roles in defense responses and plant development (Wang et al., 2012). There are 31 TF-related DEGs between the WT and OE plants, including 17 upregulated genes and 14 downregulated genes (Table 1). In previous studies, overexpression of OsWRKY77 (LOC_Os01g40260) repressed growth of a pathogen by enhancing expression of defense-related PR1, PR2, and PR5 genes, and overexpression of OsMADS57 (LOC_Os02g49840) resulted in increased tillers (Wang et al., 2008; Guo et al., 2013; Lan et al., 2013). Moreover, MYB domain TFs and ZF domain TFs also participate in plant development and resistance (Sugimoto et al., 2000; Shin et al., 2002; Wang et al., 2008). We also confirmed these four TF gene expressions in WT and OE plants by qRT-PCR. The qRT-PCR results showed that all of them were consistent with the transcriptome data (Supplementary Table S3) and were upregulated in all three OE-OsRRK1 plants (Figures 7E–H). OsRRK1 may operate by means of transducing signals activated by receptor kinases and then regulating related TFs, finally resulting in leaf rolling and a BPH-defense phenotype.
In rice, at least 35 leaf rolling mutants have been reported, 13 genes of which were related to the bulliform cells; these include RL14, ACL1, SRL1 and SLL1. However, none of these genes were found differentially expressed between WT and OE-25 plants in the transcriptome data (Supplementary Table S3). These results suggest that overexpression of the OsRRK1 gene resulting in leaf rolling is independent of the previously reported leaf rolling-related genes.
Discussion
The RLCKs represent a large gene family in plants with diverse biological roles, which include development and stress responses (Vij et al., 2008; Liu et al., 2016). They are divided into 13 subfamilies in Arabidopsis and 17 subfamilies in rice based on phylogenetic clades of amino acid sequences (Shiu et al., 2004). Most well studied RLCK genes belong to the RLCK VII subfamily, while there have been few studies of the RLCK VI subfamily. In Arabidopsis, only four RLCK VI subfamily genes have been studied. Rop binding protein kinases 1 and 2 (RBK1 and 2) directly bind to AtRop4 GTPase, which has emerged as a central regulator of diverse signaling pathways in plant growth and pathogen defense (Molendijk et al., 2008). Rop-interacting receptor-like kinase 1 and 2 (AtRRK1 and 2) kinases could be specifically activated by GTP-bound Rop GTPases in vitro (Dorjgotov et al., 2009). However, the characterization of RLCK VI subfamily genes in rice has not been reported. In this study, we identified a RLCK VI subfamily gene OsRRK1 as a potential novel molecular interactor of OsLecRK (Supplementary Figure S7). The overexpression of OsRRK1 caused leaf rolling, agriculturally relevant traits and defense to BPH in rice, indicating that the RLCK VI subfamily also may play roles in both rice development and immunity.
To date, many studies have been performed to characterize genes controlling leaf rolling by analysis of mutants in rice. More than 35 leaf rolling mutants have been reported, of which 13 genes have been cloned and affected leaf rolling through the regulation of bulliform cells. For example, RL14 encodes a 2OG-Fe (II) oxygenase. The rl14 mutant reduced bulliform cell size and caused inward leaf rolling (Fang et al., 2012). ACL1, encoding a protein with unknown conserved functional domains, positively regulated bulliform cell development since the enhanced expression of ACL1 increased bulliform cell number and resulted in outward leaf rolling (Li et al., 2010). SRL1, encoding a putative glycosylphosphatidylinositol-anchored protein, negatively regulated the bulliform cell number since its loss-of-function mutant increased bulliform cell number and led to inward leaf rolling (Xiang et al., 2012). SLL1 encodes a transcriptional factor of the MYB family. The sll1 mutant also displays inward leaf rolling due to defective development of abaxial sclerenchymatous cells and formation of bulliform cells on the abaxial epidermis (Zhang et al., 2009). In this study, OsRRK1 negatively regulates the bulliform cell number and size. The degree of rolling was positively correlated with the expression level of OsRRK1. Through analysis of transcriptome and qRT-PCR data between the WT and OE-25 plants, none of the 13 reported genes associated with leaf rolling were found differentially expressed (Supplementary Table S3 and Figures 8A–D). These results suggest that leaf rolling resulting from overexpression of OsRRK1 was activated by a novel pathway, which was independent of these previously reported genes related to leaf rolling.
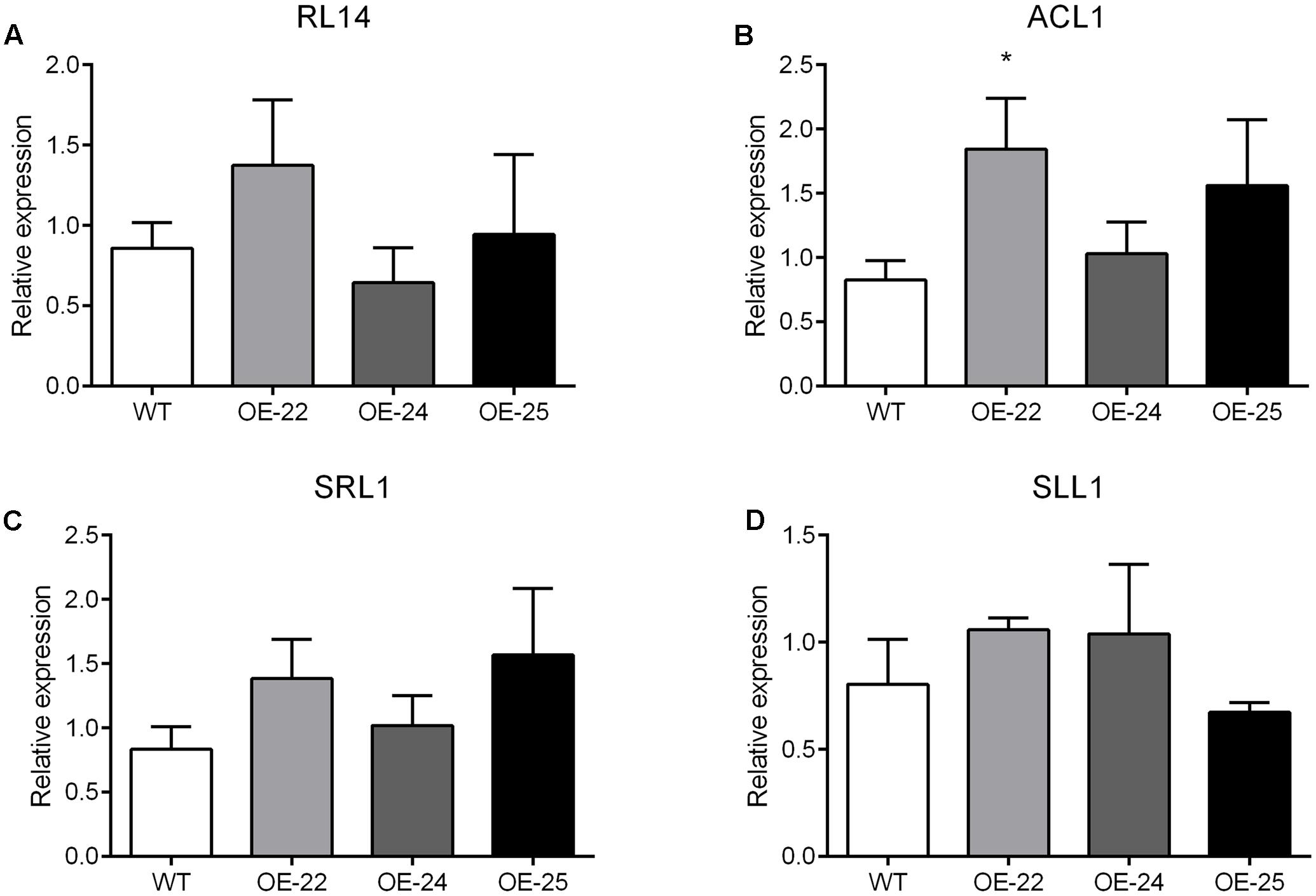
FIGURE 8. (A–D) qRT-PCR analysis of the reported genes related to leaf rolling. Significant differences are indicated by ∗P < 0.05, (Student’s t-test).
OsRRK1 encodes an intracellular protein with a STYKc domain (Supplementary Figure S3) and interacts with OsLecRK to confer defense to BPH attacks via antixenosis (Figures 5B,C), which is known to be one of the major mechanisms of plant defense against insect attacks (Cheng et al., 2013). Previous studies have provided evidence that RLCKs can be phosphorylated by receptor kinases. For example, OsCERK1 (chitin elicitor receptor kinase 1), a plasma membrane protein, phosphorylated OsRLCK185 with a STKc domain. OsRLCK185 is a possible transmitter which links the PRR OsCERK1 with MAP kinase cascades and regulates chitin induced immune responses (Yamaguchi et al., 2013). Other RLCKs, such as OsRLCK57, OsRLCK107, and OsRLCK176 with the STYKc domain, interact with OsBRI1, a rice brassinosteroid receptor, and they also regulate immune responses by the immune receptor XA21 (Zhou et al., 2016). OsRRK1 is an interactor of OsLecRK. The BPH resistance protein BPH3 is also composed of three lectin receptor kinases (OsLeckRK1-3). Both of these lectin receptor kinases confer rice resistance to BPH. Thus, we hypothesize that OsRRK1 may be similar to other RLCKs. It can be phosphorylated by OsLecRK, which could recognize HAMPs (herbivore-associated molecular patterns), and subsequently trigger immune responses to BPH. However, how OsRRK1 accepts signals from OsLecRK and what proteins are activated by OsRRK1 remain to be determined.
To understand the molecular mechanism of rice development and defense to BPH mediated by OsRRK1, we determined the expression profiles of WT plants and OE-OsRRK1 plants by deep RNA sequencing. We found that many DEGs were involved in receptor kinases and transcription factors. Receptor kinases are pivotal components of signal transduction and play roles in plant development. In this study, we selected four genes, LOC_Os07g04110, LOC_Os08g14950, LOC_Os11g31540, and LOC_Os11g40480 for qRT-PCR confirmation (Figures 7A–D). In rice, the functions of these four genes are unknown. By analysis of homologous genes in Arabidopsis, we found that they act roles either in the process of plant development or plant defense. Perception of pathogen (or microbe)-associated molecular patterns (PAMPs/MAMPs) by pattern recognition receptors (PRRs) is a key component of plant innate immunity. The Arabidopsis EF-Tu receptor (EFR) recognizes the bacterial elongation factor Tu (EF-Tu) and confers broad-spectrum bacterial disease resistance (Schoonbeek et al., 2015). SERK1 is essential to the early events of BR signaling, which can regulate plant growth and development (Gou et al., 2012). Besides OsLecRK, OsRRK1 may interact with other receptor kinases like the aforementioned ones to influence rice growth and development.
Transcription factors also play key roles in defense response and plant development (Wang et al., 2012). In this study, we also selected four genes, LOC_Os01g40260, LOC_Os02g49840, LOC_Os11g17954, and LOC_Os12g24490 for qRT-PCR analysis (Figures 7E–H). LOC_Os01g40260 (OsWRKY77) is a positive regulator of PR gene expression and basal resistance to the bacterial pathogen PstDC3000 (Lan et al., 2013). In OsRRK1 overexpression lines, the expression of OsWRKY77 was upregulated that could improve defense to BPH attacks. Overexpression of LOC_Os02g49840 (OsMADS57) can increase rice tillers by interacting with other proteins (Guo et al., 2013), and it was upregulated in OsRRK1 overexpression lines. However, OsRRK1 overexpression lines had reduced number of tillers compared to WT plants. We thought that there are a number of factors that affect the numbers of tillers, OsMADS57 maybe just one of them. Even though the expression of OsMADS57 was upregulated, some other genes may also affect the numbers of tillers in OsRRK1 overexpression lines. LOC_Os11g17954 is a MYB domain TFs. LOC_Os12g24490 is a ZF domain TFs. Both MYB and ZF domain TFs participate in plant development and defense. For example, AtMYB21 confers some features of constitutive photomorphogenesis even in the dark, and causes seedling lethality (Shin et al., 2002). NtMYB2 is involved in the stress response of the retrotransposon and defense-related genes (Sugimoto et al., 2000). Many members of the ZF subfamily are regulated by abiotic or biotic stresses, suggesting that they could have an effective role in stress tolerance (Wang et al., 2008). OsRRK1 could activate these TF genes to control rice leaf rolling and defense to BPH.
In addition to receptor kinases and TFs, 34 DEGs were involved in biotic stress (Table 1). Among them, 16 genes encode PR-proteins, which include NBS-LRR disease resistance protein. Plant NBS-LRR disease resistance protein is the largest protein family involved in disease resistance, pathogen sensing and host defense (Moffett et al., 2002; DeYoung and Innes, 2006). Previous studies have shown that BPH resistance genes can activate PR-proteins (Du et al., 2009). OsRRK1 conferred antixenosis to BPH and may also be related to the PR-protein. In addition, 33 DEGs were involved in development (Table 1). Although their functions in rice remain to be elucidated, the expression changes still reflect their role in OE-OsRRK1 development. Thirteen DEGs are related to hormones (Table 1). Hormone signaling pathways play pivotal roles in plant development and defense. We found traditional defense hormones-related genes including three JA-related genes and two SA-related DEGs. It has been reported that JA and SA are related to defense against BPH (Du et al., 2009; Zhao et al., 2016). Three auxin-related genes were also found amongst the DEGs. Auxin biosynthesis affects the development of plants.
In summary, we characterized a gene that regulates not only development but also plant immunity. Moreover, the phenotypes of leaf rolling and BPH defense were positively correlated to the expression level of OsRRK1. This feature has the potential to facilitate artificial control of these phenotypes and may eventually contribute to the development of desired rice variety.
Author Contributions
YM, BD, and GH designed the research; YM, YZh, XS, SS, YZe, YW, RC, AY, and LZ performed research; YM, BD, and GH analyzed data and wrote the manuscript. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by grants from National Program on the Development of Basic Research of China (2013CB126904, 2013CBA01403), the National Natural Science Foundation of China (grant numbers 31470102, 31071059).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01783/full#supplementary-material
Abbreviations
BPH, brown planthopper; LEI, leaf erect index; LRI, leaf rolling index; OE, overexpression; RNAi, RNA interference.
Footnotes
- ^ http://rsbweb.nih.gov/ij/
- ^ https://blast.ncbi.nlm.nih.gov
- ^ http://cufflinks.cbcb.umd.edu
- ^ http://genome.tugraz.at
References
Ao, Y., Li, Z., Feng, D., Xiong, F., Liu, J., Li, J. F., et al. (2014). OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 80, 1072–1084. doi: 10.1111/tpj.12710
Bowman, J. L., Eshed, Y., and Baum, S. F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18, 134–141. doi: 10.1016/S0168-9525(01)02601-4
Chen, R., Zhao, X., Shao, Z., Wei, Z., Wang, Y., Zhu, L., et al. (2007). Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19, 847–861. doi: 10.1105/tpc.106.044123
Chen, S., Songkumarn, P., Liu, J., and Wang, G. L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121. doi: 10.1104/pp.109.137125
Cheng, X., Wu, Y., Guo, J., Du, B., Chen, R., Zhu, L., et al. (2013). A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 76, 687–698. doi: 10.1111/tpj.12328
DeYoung, B. J., and Innes, R. W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249. doi: 10.1038/ni1410
Dorjgotov, D., Jurca, M. E., Fodor-Dunai, C., Szucs, A., Otvos, K., Klement, E., et al. (2009). Plant Rho-type (Rop) GTPase-dependent activation of receptor-like cytoplasmic kinases in vitro. FEBS Lett. 583, 1175–1182. doi: 10.1016/j.febslet.2009.02.047
Du, B., Zhang, W., Liu, B., Hu, J., Wei, Z., Shi, Z., et al. (2009). Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. U.S.A. 106, 22163–22168. doi: 10.1073/pnas.0912139106
Dubouzet, J. G., Maeda, S., Sugano, S., Ohtake, M., Hayashi, N., Ichikawa, T., et al. (2011). Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice. Plant Biotechnol. J. 9, 466–485. doi: 10.1111/j.1467-7652.2010.00568.x
Fang, L., Zhao, F., Cong, Y., Sang, X., Du, Q., Wang, D., et al. (2012). Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 10, 524–532. doi: 10.1111/j.1467-7652.2012.00679.x
Gao, L., and Xue, H. (2012). Global analysis of expression profiles of rice receptor-like kinase genes. Mol. Plant 5, 143–153. doi: 10.1093/mp/ssr062
Gou, X., Yin, H., He, K., Du, J., Yi, J., Xu, S., et al. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLOS Genet. 8:e1002452. doi: 10.1371/journal.pgen.1002452
Guo, S., Xu, Y., Liu, H., Mao, Z., Zhang, C., Ma, Y., et al. (2013). The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 4, 1566. doi: 10.1038/ncomms2542
Itoh, J., Nonomura, K., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., et al. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46, 23–47. doi: 10.1093/pcp/pci501
Jane, W. N., and Chiang, S. H. T. (1991). Morphology and development of bulliform cells in Arundo formosana Hack. Taiwania Int. J. Life Sci. 36, 85–96.
Lan, A., Huang, J., Zhao, W., Peng, Y., Chen, Z., and Kang, D. (2013). A salicylic acid-induced rice (Oryza sativa L.) transcription factor OsWRKY77 is involved in disease resistance of Arabidopsis thaliana. Plant Biol. 15, 452–461. doi: 10.1111/j.1438-8677.2012.00664.x
Lang, Y., Zhang, Z., Gu, X., Yang, J., and Zhu, Q. (2004). Physiological and ecological effects of crimpy leaf character in rice (Oryza sativa L.) I. Leaf orientation, canopy structure and light distribution. Acta Agron. Sin. 30, 806–810.
Li, L., Shi, Z., Li, L., Shen, G., Wang, X., An, L., et al. (2010). Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol. Plant. 3, 807–817. doi: 10.1093/mp/ssq022
Lin, Z., Zhan, X., Cheng, S., and Cao, L. (2013). Progress on mapping and cloning genes for leaf architecture in rice (Oryza sativa. L). J. Nucl. Agric. Sci. 27, 1662–1669.
Liu, X., Li, M., Liu, K., Tang, D., Sun, M., Li, Y., et al. (2016). Semi-Rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J. Exp. Bot. 67, 2139–2150. doi: 10.1093/jxb/erw029
Micol, J. L., and Hake, S. (2003). The development of plant leaves. Plant Physiol. 131, 389–394. doi: 10.1104/pp.015347
Moffett, P., Farnham, G., Peart, J., and Baulcombe, D. C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519. doi: 10.1093/emboj/cdf453
Molendijk, A. J., Ruperti, B., Singh, M. K., Dovzhenko, A., Ditengou, F. A., Milia, M., et al. (2008). A cysteine-rich receptor-like kinase NCRK and a pathogen-induced protein kinase RBK1 are Rop GTPase interactors. Plant J. 53, 909–923. doi: 10.1111/j.1365-313X.2007.03384.x
Ray, D. K., Ramankutty, N., Mueller, N. D., West, P. C., and Foley, J. A. (2012). Recent patterns of crop yield growth and stagnation. Nat Commun. 3, 1293. doi: 10.1038/ncomms2296
Schoonbeek, H., Wang, H., Stefanato, F. L., Craze, M., Bowden, S., Wallington, E., et al. (2015). Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 206, 606–613. doi: 10.1111/nph.13356
Shi, Z., Wang, J., Wan, X., Shen, G., Wang, X., and Zhang, J. (2007). Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226, 99–108. doi: 10.1007/s00425-006-0472-0
Shin, B., Choi, G., Yi, H. K., Yang, S. C., Cho, I. S., Kim, J., et al. (2002). AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J. 30, 23–32. doi: 10.1046/j.1365-313X.2002.01264.x
Shinya, T., Yamaguchi, K., Desaki, Y., Yamada, K., Narisawa, T., Kobayashi, Y., et al. (2014). Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 79, 56–66. doi: 10.1111/tpj.12535
Shiu, S. H., and Bleecker, A. B. (2001). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001:re22. doi: 10.1126/stke.2001.113.re22
Shiu, S. H., Karlowski, W. M., Pan, R., Tzeng, Y. H., Mayer, K. F., and Li, W. H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
Sinclair, T. R., and Sheehy, J. E. (1999). Erect leaves and photosynthesis in rice. Science 283, 1456–1457. doi: 10.1126/science.283.5407.1455c
Sturn, A., Quackenbush, J., and Trajanoski, Z. (2002). Genesis: cluster analysis of microarray data. Bioinformatics 18, 207–208. doi: 10.1093/bioinformatics/18.1.207
Sugimoto, K., Takeda, S., and Hirochika, H. (2000). MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 12, 2511–2527. doi: 10.1105/tpc.12.12.2511
Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., et al. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273. doi: 10.1105/tpc.105.035576
Vij, S., Giri, J., Dansana, P. K., Kapoor, S., and Tyagi, A. K. (2008). The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 1, 732–750. doi: 10.1093/mp/ssn047
Walling, L. L., and Thompson, G. A. (2012). “Behavioral and molecular-genetic basis of resistance against phloem-feeding insects,” in Phloem: Molecular Cell Biology, Systemic Communication, Biotic Interactions, eds G. A. Thompson and A. J. E. van Bel (Oxford: Wiley-Blackwell), 328–351. doi: 10.1002/9781118382806.ch16
Wang, D., Guo, Y., Wu, C., Yang, G., Li, Y., and Zheng, C. (2008). Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9:44. doi: 10.1186/1471-2164-9-44
Wang, J., Wu, G., Peng, C., Zhou, X., Li, W., He, M., et al. (2016). The receptor-like cytoplasmic kinase OsRLCK102 regulates XA21-mediated immunity and plant development in rice. Plant Mol. Biol. Rep. 34, 628–637. doi: 10.1007/s11105-015-0951-1
Wang, Y., Guo, H., Li, H., Zhang, H., and Miao, X. (2012). Identification of transcription factors potential related to brown planthopper resistance in rice via microarray expression profiling. BMC Genomics 13:687. doi: 10.1186/1471-2164-13-687
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wei, Z., Hu, W., Lin, Q., Cheng, X., Tong, M., Zhu, L., et al. (2009). Understanding rice plant resistance to the brown planthopper (Nilaparvata lugens): a proteomic approach. Proteomics 9, 2798–2808. doi: 10.1002/pmic.200800840
Xiang, J., Zhang, G., Qian, Q., and Xue, H. (2012). SEMI-ROLLED LEAF1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 159, 1488–1500. doi: 10.1104/pp.112.199968
Xu, Y., Wang, Y., Long, Q., Huang, J., Wang, Y., Zhou, K., et al. (2014). Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 239, 803–816. doi: 10.1007/s00425-013-2009-7
Yamaguchi, K., Yamada, K., Ishikawa, K., Yoshimura, S., Hayashi, N., Uchihashi, K., et al. (2013). A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe. 13, 347–357. doi: 10.1016/j.chom.2013.02.007
Zhang, G., Xu, Q., Zhu, X., Qian, Q., and Xue, H. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21, 719–735. doi: 10.1105/tpc.108.061457
Zhang, J., Wu, S., Jiang, L., Wang, J., Zhang, X., Guo, X., et al. (2015). A detailed analysis of the leaf rolling mutant sll2 reveals complex nature in regulation of bulliform cell development in rice (Oryza sativa L.). Plant Biol. 17, 437–448. doi: 10.1111/plb.12255
Zhao, Y., Huang, J., Wang, Z., Jing, S., Wang, Y., Ouyang, Y., et al. (2016). Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc. Natl. Acad. Sci. U.S.A. 113, 12850–12855. doi: 10.1073/pnas.1614862113
Zhou, X., Wang, J., Peng, C., Zhu, X., Yin, J., Li, W., et al. (2016). Four receptor-like cytoplasmic kinases regulate development and immunity in rice. Plant Cell Environ. 39, 1381–1392. doi: 10.1111/pce.12696
Keywords: rice, leaf rolling, bulliform cell, RLCK, defense
Citation: Ma Y, Zhao Y, Shangguan X, Shi S, Zeng Y, Wu Y, Chen R, You A, Zhu L, Du B and He G (2017) Overexpression of OsRRK1 Changes Leaf Morphology and Defense to Insect in Rice. Front. Plant Sci. 8:1783. doi: 10.3389/fpls.2017.01783
Received: 06 June 2017; Accepted: 02 October 2017;
Published: 24 October 2017.
Edited by:
Madelaine Elisabeth Bartlett, University of Massachusetts Amherst, United StatesReviewed by:
Yuguo Xiao, Brigham Young University, United StatesMichael Gerard Muszynski, University of Hawaii at Manoa, United States
Copyright © 2017 Ma, Zhao, Shangguan, Shi, Zeng, Wu, Chen, You, Zhu, Du and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangcun He, gche@whu.edu.cn Bo Du, bodu@whu.edu.cn
 Yinhua Ma
Yinhua Ma Yan Zhao1
Yan Zhao1 Ya Zeng
Ya Zeng Bo Du
Bo Du Guangcun He
Guangcun He