- 1Division of Vegetable Science, ICAR-Indian Agricultural Research Institute, New Delhi, India
- 2Crop Genetics and Informatics Group, School of Computational and Integrative Sciences, Jawaharlal Nehru University, New Delhi, India
In the present climate change scenario, controlling plant disease through exploitation of host plant resistance could contribute toward the sustainable crop production and global food security. In this respect, the identification of new sources of resistance and utilization of genetic diversity within the species may help in the generation of cultivars with improved disease resistance. Begomoviruses namely, Tomato yellow leaf curl virus (TYLCV) and Chilli leaf curl virus (ChLCV) are known to cause major yield losses in several economically important crop plants of the family Solanaceae. Though co-occurrence, association and synergistic interactions among these viruses in the host plants is reported, whether orthologous genetic loci in related host plants could be responsible for conferring resistance to these viruses has not been investigated yet. Several loci including Ty1, Ty2, Ty3, Ty4, and ty5 have been reported to confer resistance to leaf curl viruses in tomato. Here, we examined the pepper orthologous markers, corresponding to these QTL regions, for polymorphism between ChLCV susceptible and resistant genotypes of pepper. Further, to examine if the polymorphic markers are segregating with the disease resistance, Bulk Segregant Analysis (BSA) was performed on F2 population derived from crosses between resistant and susceptible lines. However, none of the markers showed polymorphism in BSA suggesting that the tested markers are not linked to genes/QTLs responsible for conferring resistance to ChLCV in the selected genotypes. In silico analysis was performed to study the synteny and collinearity of genes located within these QTL regions in tomato and pepper genomes, which revealed that more than 60% genes located in Ty2 and Ty4, 13.71% genes in Ty1, 23.07% in Ty3, and 44.77% genes located within ty5 QTL region in tomato are conserved in pepper genome. However, despite such a high conservation in gene content, the linkage relationship in these regions seems to be greatly affected by gross rearrangements in both the species.
Introduction
Hot pepper (Capsicum annuum L.) is extensively cultivated throughout the world, as an essential condiment and a cash crop (Bosland and Votava, 2000). In India, pepper is used as a vegetable, spice as well as for industrial purposes involving extraction of oleoresin and capsaicin (Kumar and Rai, 2005). Although pepper is infested by a large number of pathogens, viruses in-particular, cause heavy losses both in terms of quality and productivity of pepper. Whitefly (Bemisia tabaci Genn.) transmitted begomovirus ChLCV has been reported to cause havoc in the hot pepper growing areas of North India covering states of Delhi, Haryana, Rajasthan, Punjab, West Bengal, and Uttar Pradesh with up to 100% crop loss (Senanayake et al., 2007). Similar severity has also been reported in the major hot pepper growing districts of Andhra Pradesh in South India. The typical symptoms under field conditions include upward curling, puckering, smaller leaves, stunted stems, and lack of fruiting. Furthermore, several other begomoviruses including Chilli leaf curl India virus, Tomato leaf curl Joydebpur virus and Tomato leaf curl New Delhi virus (ToLCNDV) have been found to co-infect the pepper plants along with ChLCV under field conditions (Khan et al., 2006; Fortes et al., 2016; Srivastava et al., 2017). In fact, synergistic interactions between different begomoviruses infecting pepper have been reported to cause breakdown of natural resistance in the host plant (Singh et al., 2016; Al-Shihi et al., 2017).
At Division of Vegetable Science, ICAR-IARI, New Delhi, we had earlier tested 62 germplasm lines of hot pepper for resistance to the ChLCV under natural disease epiphytotic conditions in Trans-Gangetic plains of Northern India. In addition to field screening, the germplasm of hot pepper was subjected to virus indexing against ChLCV as well as ToLCNDV. The variables measured included disease incidence and severity. Scales for classifying the lines tested for leaf curl disease reactions were adopted from Kumar et al. (2006). Two genotypes of hot pepper namely, DLS-Sel-10 and WBC–Sel-5, were found resistant to ChLCV infection (Srivastava et al., 2017). Also, we observed, during these trials, that ToLCNDV infection was commonly prevalent along with ChLCV infection in the field conditions. However, resistant genotypes, DLS-Sel-10 and WBC-Sel-5, showed no incidence of any of the two viruses, indicating that these lines are likely resistant to ToLCNDV as well (Srivastava et al., 2015). The genetic basis of this resistance is yet to be examined.
Ty QTLs of tomato have been reported to confer resistance against both monopartite and bipartite tomato leaf curl viruses, prevalent in India (Ji et al., 2007b; Anbinder et al., 2009; Prasanna et al., 2015; Fortes et al., 2016). The Ty1 locus, mapped on chromosome 6 from Solanum chilense LA1969 (Zamir et al., 1994), was later reported to be linked to a major QTL Ty3 that has 60% contribution in symptom severity (Ji et al., 2007a). Ty2, first detected in S. habrochaites B6013, was mapped to chromosome 11 (Hanson et al., 2006; Ji et al., 2009a; Yang et al., 2014). Ty4, that accounts for only 16% of the variation, was located on the long arm of chromosome 3 (Ji et al., 2009b). A line derived from S. peruvianum was also reported to be the source of recessive resistance and the QTL responsible for resistance was named as ty5 (Friedmann et al., 1998; Anbinder et al., 2009). The ty5, mapped to chromosome 4, has been predicted to have originated from cultivar Tyking (Hutton et al., 2012). Hutton and Scott (2013) mapped another recessive resistance gene ty6 to chromosome 10, in Ty3-carrying S. chilense “LA2779”. Although initially these QTLs were identified as source of resistance to Tomato yellow leaf curl virus (TYLCV), recent studies reported that Ty3 is highly effective against monopartite Tomato leaf curl Bangalore virus (ToLCBV) as well as two bipartite viruses, ToLCNDV and Tomato leaf curl Palampur virus from India (Prasanna et al., 2015). Similarly, Ty1 gene has been shown to be effective against ToLCBV and ToLCNDV (Prasanna et al., 2015). Since the resistant lines selected in our study had shown resistant response to both ChLCV as well as ToLCNDV, we initiated this study to check if pepper genes orthologous to Ty loci of tomato, could be responsible for conferring resistance to ChLCV. We analyzed polymorphism in orthologous markers located within the QTL regions of Ty1, Ty2, Ty3, Ty4, and ty5 between a ChLCV-sensitive and -resistant genotype of hot pepper. Polymorphic markers between resistant and susceptible parents were then used for Bulk Segregant Analysis (BSA) on F2 population in order to find out if any of the markers is segregating with the resistance trait. Furthermore, in silico analysis was carried out to study the conservation and dispersion of genes within Ty QTL regions between tomato and pepper genomes.
Materials and Methods
Extraction of Orthologous Markers in Pepper and Tomato Genomes
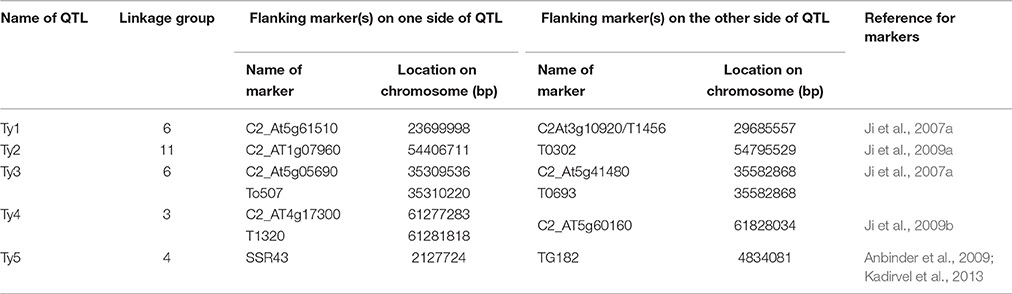
We used conserved ortholog set II (COSII) markers provided by Wu et al. (2009) for extracting the orthologous markers in and around Ty QTL regions between tomato and pepper genomes. Since the COSII markers mapped in this study provide a detail inference of syntenic regions between tomato and pepper genomes, we identified all the markers flanking Ty QTLs of tomato on genetic map of pepper and extracted all the markers located between and adjacent to these markers in pepper genome for further analysis. In case, the orthologs of flanking markers of Ty QTL of tomato were located on two different chromosomes of pepper, all the markers flanking these orthologous markers from both the chromosomes of pepper were extracted for the downstream analysis. The complete list of markers flanking Ty QTLs of tomato and list of orthologous markers extracted in pepper for further analysis are provided in Tables 1 and 2, respectively.
Analysis of Polymorphism between Resistant and Susceptible Parents and, F2 Population
Chilli leaf curl virus (ChLCV) susceptible genotypes, Phule Mukta (PM) and Anugraha, as well as resistant genotypes, DLS-Sel-10 and WBC-Sel 5, of hot pepper were used to study the polymorphism in selected markers. Genomic DNA was extracted from young leaf tissue following CTAB method (Murray and Thompson, 1980). DNA quality and quantity were assessed on a 1% (w/v) agarose gel stained with ethidium bromide (Sigma Aldrich Chemical Pvt. Ltd, Bangalore, India) and by using a NanoDrop®ND-1000 spectrophotometer, respectively.
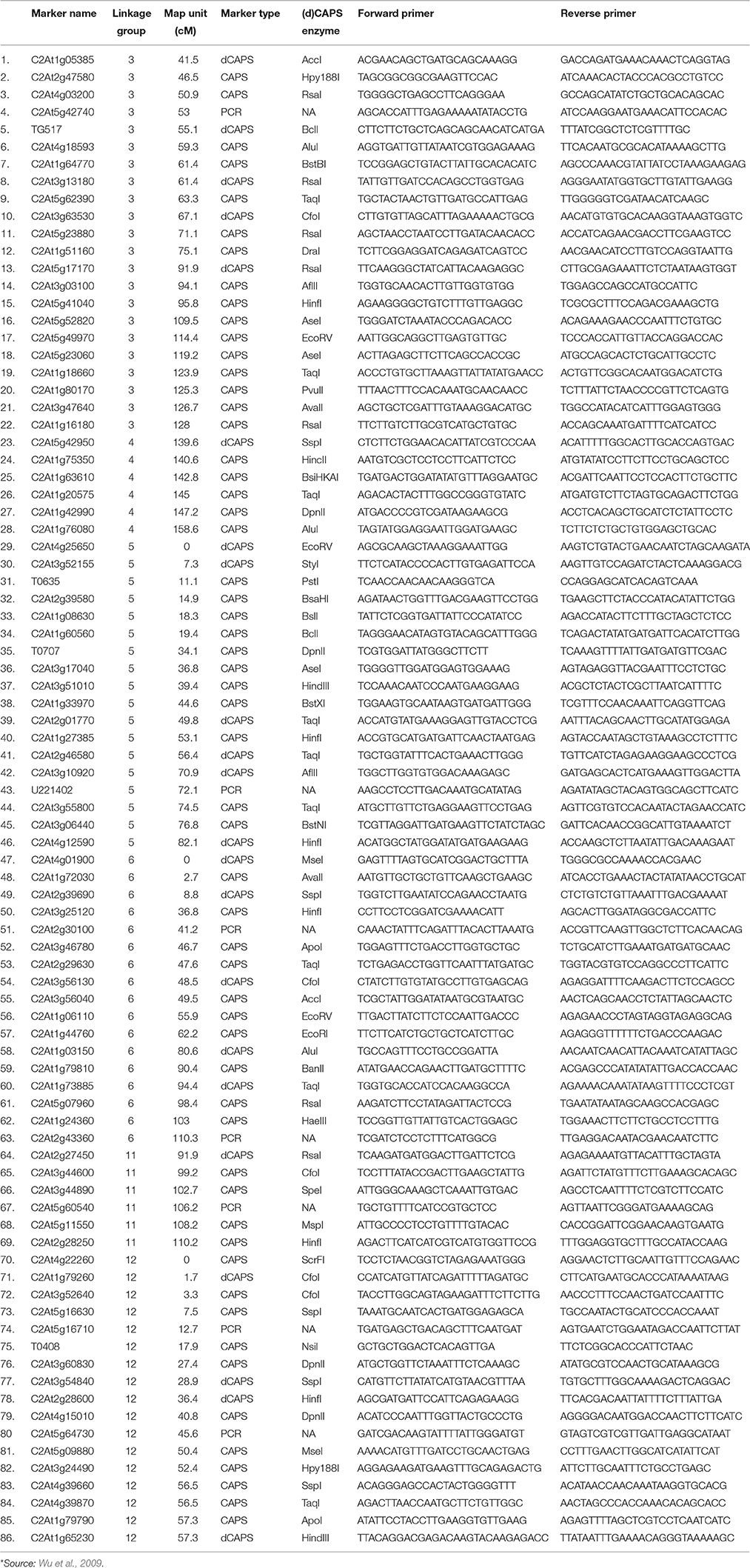
A total of 86 orthologous CAPS/dCAPS/PCR markers between tomato and pepper were selected for the polymorphism analysis (Table 2). Primers were custom synthesized (SBS Genetech Co. Ltd., Beijing, China). All the markers were amplified by PCR in 15 μl reaction volumes with 50 ng genomic DNA, 1.0 μM of each primer and 1.0 unit of Taq DNA polymerase (Hi media Laboratories, Mumbai, India). Amplification conditions involved: initial incubation at 94°C for 3 min followed by 30 cycles of 94°C for 0.5 min, 55–65°C (depending on annealing temperature of primers) for 1 min, and 72°C for 1 min; and a final incubation at 72°C for 5 min. Amplified products in case of CAPS and dCAPS markers were digested using specific restriction enzymes as given in the Table 2 as per manufacturer's instructions (Thermo Fisher Scientific Inc.). Amplified and digested products were resolved on 3.0% agarose gels with Tris/Acetate/EDTA (TAE), at a constant voltage of 60 V for 3 h using a horizontal gel electrophoresis system (BioRad, USA). The gels were visualized and photographed under UV light in a gel documentation unit (Alpha imager, Cell Biosciences, Santa Clara, CA).
The polymorphic markers between the parents were used for BSA in F2 population derived from crosses of PM X DLS-Sel-10 as well as Anugraha X WBC-Sel 5. Screening for ChLCV was done and symptom severity was scored on individual plants according to scale developed by Kumar et al. (2006) ranging from 0 to 5 with “0” indicating no symptoms and “5” indicating extreme susceptibility with more than 75% curling, deformed small leaves, stunted plant growth with small flowers and no or little fruit set. F2 plants showing extreme phenotypes of resistance and susceptibility under challenge inoculation were used for BSA. Ten F2 individuals showing resistant phenotype with a score of 0 and ten F2 individuals found susceptible with a score of 5 were separately used for the development of bulks. Equal quantities of DNA were bulked from susceptible individuals and resistant individuals to generate two DNA bulks, namely resistant bulk (RB) and susceptible bulk (SB). The susceptible and resistant bulks along with parents were screened with selected markers as described above.
In Silico Analysis of Conservation/Dispersion of Genes Located within Ty QTL Regions
In order to determine the physical location of genes present on selected Ty QTL regions and their synteny with hot pepper, following strategy was adopted:
First of all, the location of all the markers flanking the QTL of interest was checked on respective chromosomes in tomato EXPEN 2000 map in the Sol Genomics Network (http://www.sgn.cornell.edu) and the physical coordinates of these markers were recorded. Thereafter, the information regarding all the gene models between these coordinates was extracted from International Tomato Genome Sequencing project version 2.4 (https://solgenomics.net/organism/Solanum_lycopersicum/genome). The information regarding syntenic genes in hot pepper for all the tomato genes as well as the physical location of each gene on respective chromosome was downloaded from FTP site at pepper genome database (Pepper Institute, Zunyi Academy of Agricultural Sciences; http://peppersequence.genomics.cn/page/species/index.jsp). The information for selected genes was extracted from source file using Microsoft excel. Gene annotations for tomato and pepper genes were downloaded from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) and the pepper genome database, respectively (Tables S6–S10).
Results
Orthologous Markers between Pepper and Ty QTL Regions of Tomato
The detailed results obtained for orthologous markers in all five QTL regions are described below:
Orthologous Markers on Ty1 and Ty3 QTL Regions
Ji et al. (2007a) had mapped the begomovirus resistance locus Ty3 from S. chilense on chromosome 6 of tomato near TYLCV resistance locus Ty1. Ty1 locus was mapped to pericentromeric region, whereas, Ty3 to long arm of chromosome 6. The position of Ty3 locus in the map was shown between markers T0774 and T1079 and that of Ty1 locus between markers C2_At4g01900 and C2_At3g10920 (Table 1). We selected all the orthologous markers between C2_At4g01900 and T1079 so that all the markers located in the region harboring both Ty1 and Ty3 QTLs could be utilized to study the polymorphism in ChLCV resistant and susceptible pepper lines. The Ty1 flanking marker C2_At4g01900 was found on pepper chromosome 6 at 0 cM position, whereas, orthologous markers C2_At3g56130 and C2-At1g06110 flanked Ty3 locus on pepper chromosome 6. Therefore, all the markers between C2_At4g01900 (0 cM) and C2-At1g06110 (55.9c M) as well as few additional markers up to 110.3 cM in pepper CosII map of chromosome 6 were shortlisted for studying the polymorphism. In total, 17 orthologous CAPS markers were selected from this region (Table 2).
Orthologous Markers on Ty2 QTL Region
Ty2 QTL initially identified from S. habrochaites (B6013) was mapped to chromosome 11 in the 19 cM region on the long arm flanked by markers TG36 and cLET-24-J2A (Hanson et al., 2006; Ji et al., 2007b, 2009a). On comparison of Tomato EXPEN 2000 and pepper COSII maps, it was observed that markers between TG36 and CLET-24-J2A (85–95 cM) on chromosome 11 of tomato have dispersed in such a way that TG 36 which is at 84 cM on chromosome 11 of tomato occupied position at 115.5 cM on chromosome 11 of pepper. Whereas, orthologs of some of the markers located between 85 and 95 cM region in tomato mapped between 100 and 110 cM region in pepper. Some of the markers orthologous to chromosome 11 of tomato also mapped to chromosome 12 of pepper. We included all the orthologous markers located between 91.9 and 110.2 cM on pepper chromosome 11 and between 0 and 57.3 cM region on chromosome 12 of pepper of COSII map. In total, 23 orthologous CAPS markers were selected from this region (Table 2).
Orthologous Markers on Ty4 QTL Region
The Ty4 locus was mapped to chromosome 3 of tomato in some resistant breeding lines derived from LA1932 of S. Chilense in the 2.3 cM interval between C2_At4g17300 and C2_At5g60160. Although these markers were not found on COS II map, markers C2_At5g62390 and C2_At5g52820, which flank Ty4 locus on both sides (Ji et al., 2009b) could be located at 63.3 and 109.5 cM, respectively on the long arm of chromosome 3 of pepper. We selected all the 22 orthologous markers located between 41.5 and 128 cM region of chromosome 3 of pepper for the current study (Table 2).
Orthologous Markers on ty5 QTL Region
Tomato yellow leaf curl virus (TYLCV) resistance gene ty5 has been reported to account for 39.7–46.6% of the variation in symptom severity among segregating plants. Anbinder and coworkers (2009) mapped this gene to chromosome 4 near the marker SlNAC1 which was at 13.5 cM from marker J04-1 and at 17.1 cM from TG182. Later, Kadirvel and co-workers (2013) reported that SSR43 flank SINAC1 on one side. We tried to locate these markers on chromosome 4 of tomato and compared it with the map of chromosome 4 of pepper in COSII. However, there were no orthologous markers in this region. Whereas, when chromosome 4 of Tomato EXPEN 2000 map was compared with chromosome 5 of pepper COSII map, some orthologous markers could be identified in this region. Therefore, we selected orthologous markers located between 139.6 and 158.6 cM on chromosome 4 (6 markers) as well as between 0 and 82.1 cM on chromosome 5 (18 markers) of pepper for our study (Table 2).
Survey of Polymorphism between Resistant and Susceptible Parents and, F2 Population
A total of 86 orthologous markers were evaluated in four pepper genotypes: the susceptible genotypes, PM and Anugraha, as well as resistant genotypes, DLS-Sel-10 and WBC-Sel -5. Four markers, namely C2At5g11550 (located on chromosome 11 and selected for testing Ty2 QTL synteny), C2At5g23060 (located on chromosome 3 and selected for Ty4 QTL synteny), C2At3g55800 (located on chromosome 5 and selected for ty5 QTL synteny) and C2At5g17170 (located on chromosome 3 and selected for Ty4 QTL synteny) were found to be polymorphic between the resistant and susceptible parents (Figure S1). These markers were then tested using BSA on F2 population derived from the cross between PM X DLS-Sel-10 and Anugraha X WBC-Sel. None of the markers showed polymorphism in BSA thereby, indicating lack of linkage between the tested markers and resistance trait.
Analysis of Synteny and Micro-Collinearity between Pepper and Tomato Genomes at Ty QTL Regions
Since, we did not observe the linkage in polymorphic markers tested in this study, we further examined the syntenic relationship and order of the genes in the selected QTL regions between tomato and pepper, so as to unravel the pattern of conservation/dispersion of homologous segments within these QTL regions. The markers flanking Ty QTLs in tomato and their physical location on respective chromosomes is shown in Table 1. The details of tomato genes located within each QTL region, their orthologs and physical location have been provided in Tables S1–S5. The pattern of conservation and dispersion of genes located within each QTL region between tomato and pepper has been displayed in Figures 1A–E.
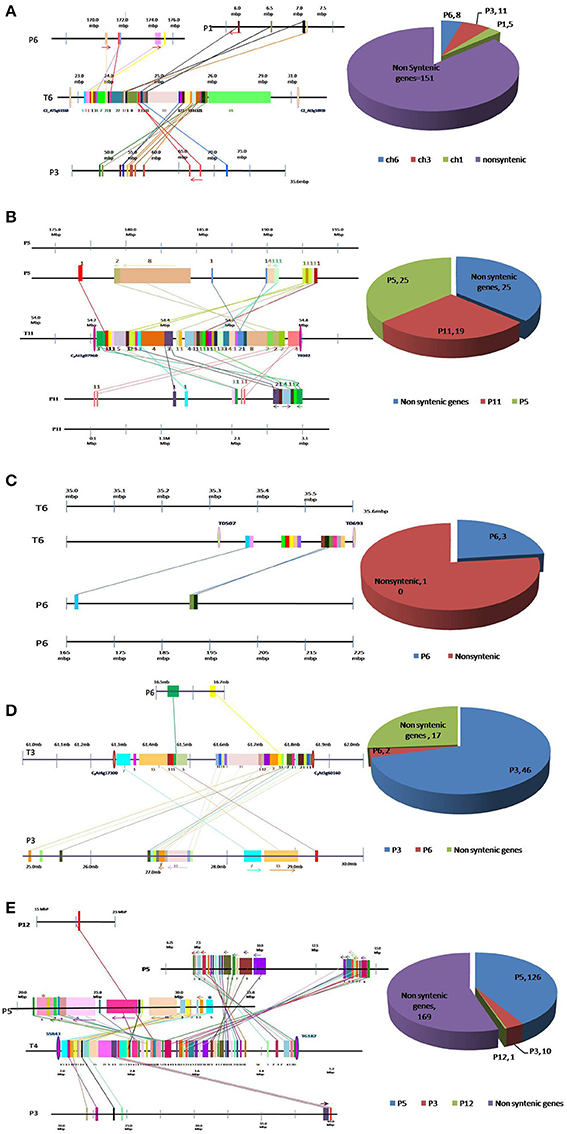
Figure 1. Conservation and dispersion of genes located in (A) Ty1, (B) Ty2, (C) Ty3, (D) Ty4, and (E) ty5 QTL regions of tomato and orthologous regions from pepper. The region on the left represents synteny and micro-collinearity between tomato Ty QTL region and orthologous regions in pepper. The chromosomal regions are represented by horizontal lines, whereas, vertical bars represent genes. The tomato chromosome number is indicated with “T” followed by number, whereas pepper chromosome number is indicated by “P” followed by number. Orthologous genes are shown by same color bar and are joined through a line. The arrows indicate change in orientation of genes in pepper genome. The region on the right presents distribution of syntenic and non-syntenic orthologs identified in pepper. Numbers of non-syntenic genes and syntenic genes found on each chromosome are given.
Out of 175 tomato genes located within Ty1 QTL region, only 24 were found to be conserved in pepper genome (Figure 1A). Moreover, these 24 genes were dispersed in three pepper chromosomes viz. chromosome 1 (5 genes), chromosome 3 (11 genes) and chromosome 6 (8 genes). Clearly, orthologous genes in this QTL region were not located on big syntenic blocks but instead seem to have dispersed to different chromosomes of pepper genome (Figure 1A; Table S1).
Tomato chromosome 11 harbors 69 genes within Ty2 QTL region, out of which 44 were found to be conserved in the pepper genome (Figure 1B). Among these, 19 were located on chromosome 11, whereas, 25 genes were found on chromosome 5 of pepper. Though this region exhibited longer syntenic regions, the order of genes was not always same as in tomato genome (Figure 1B; Table S2). For example, a block of eight tomato genes comprising Solyc11g070100.1, Solyc11g070110.1, Solyc11g070120.1, Solyc11g070130.1, Solyc11g070140.1, Solyc11g070150.1, Solyc11g070160.1, and Solyc11g070170.1 was located between 56.63 and 54.69 Mb region of tomato chromosome 11. The orthologous genes to this region were identified on a single chromosome (chromosome 5) in pepper but the order of genes had reversed (between 179.74 and 184.53 Mb).
In LA1932-derived advanced lines, Ty3 locus has been mapped within a short distance between markers T0507 and T0693 on chromosome 6 (Ji et al., 2007a). Thirteen genes were located between these markers in tomato. However, orthologs of only three of them could be located in pepper genome (Figure 1C, Table S3).
Sixty-five genes were found in a 0.54 Mb region (from 61.28 to 61.82 Mb) harboring Ty4 QTL on tomato chromosome 3. Orthologs for 48 of them were identified in pepper genome (2 on chromosome 6 and 46 on chromosome 3; Figure 1D) Among these, 46 genes were dispersed in 4.15 Mb region on pepper chromosome 3 (from 25.19 to 29.34 Mb). Syntenic blocks in this region comprised 7–15 genes, however, gene order was altered in most of the segments (indicated by forward and backward arrows in Figure 1D). Overall, in spite of high level of synteny, gross changes in position of genes were observed in this region of pepper and tomato genomes (Table S4).
Due to non-availability of closely-linked markers, a 2.7 Mb region (from 2.13 to 4.83 Mb) of tomato chromosome 4 harboring ty5 QTL was used to study the synteny between tomato and pepper genomes in this region. A total of 306 genes were found in this region in tomato genome, however orthologs of only 137 genes could be identified in pepper genome (Figure 1E; Table S4). Among these, 126 genes were found on pepper chromosome 5, concentrated in two regions, one region from 7.58 to 14.68 Mb harboring 68 genes and another region from 21.46 to 32.03 Mb harboring 58 genes. The gene order in different segments which had dispersed to either different chromosomes or different regions of the same chromosome was conserved in some cases, reversed in others and reshuffled in a few cases.
Discussion
Whitefly-transmitted begomoviruses are responsible for heavy losses in the yields of several vegetable and staple food crops, worldwide. With the changing climatic conditions and agricultural practices, the prevalence and distribution of these viruses has significantly increased causing a global concern. TYLCV, a monopartite virus, is one of the examples from this category where novel strains have been reported from diverse hosts and habitats, not detected earlier (Pratap et al., 2011; Fortes et al., 2016). Considerable work has been carried out in identification and utilization of QTLs/genes responsible for conferring resistance to the TYLCV in tomato. Leaf curl begomoviruses are known to infect a range of host species within family Solanaceae and often mixed infection with multiple viruses have been reported which lead to increase in severity of symptoms (Srivastava et al., 2015; Singh et al., 2016). Although, the role of Ty genes in conferring resistance to ChLCV, another monopartite virus (George et al., 2014) remains to be tested, recent studies show that Ty genes can confer resistance to both monopartite and bipartite begomoviruses (Prasanna et al., 2015; Fortes et al., 2016).
Structural and functional conservation in R genes has been reported among several related hosts species (Yu et al., 1996; Dijan-Caporalino et al., 1998; Grube et al., 2000). Several studies in the past, using common genetic markers, revealed conservation of large tracts of collinear markers in solanaceous genomes (Bonierbale et al., 1988; Tanksley et al., 1992; Doganlar et al., 2002). Since, there is high level of synteny and collinearity between tomato and pepper genomes, analysis of structural and functional conservation of Ty QTL regions in tomato and pepper genomes has promising implications in deciphering possible role of Ty orthologs in conferring resistance to ChCLV in pepper (Grube et al., 2000).
Pepper (C. annuum L.) is one of the few plant species in which pioneering work of comparative genetic mapping using DNA-based markers was done (Tanksley et al., 1988; Prince et al., 1993). Thereafter, several genetic maps were developed in other solanaceous crops based on different DNA marker systems, further improving the genome coverage, marker density and insights into synteny (Livingstone et al., 1999; Kang et al., 2001; Lefebvre et al., 2002; Lee et al., 2004; Paran et al., 2004; Ben-Chaim et al., 2006; Minamiyama et al., 2006). Wu et al. (2009) developed a linkage map of pepper comprising 299 conserved orthologous markers (COSII) and inferred the probable positions of additional 288 COSII markers utilizing synteny between pepper and tomato genomes. Overall, 587 orthologous markers were reported by them in pepper genome. Based on the results, 35 conserved syntenic segments with well-preserved order of genes were reported in pepper and tomato genomes (Wu et al., 2009). In this study, we leveraged this resource to identify the orthologous markers in pepper corresponding to Ty QTL regions of tomato.
We shortlisted 86 orthologous markers, corresponding to different Ty loci of tomato, in pepper and tested them for polymorphism in ChCLV-susceptible and resistant genotypes of pepper. Four markers, polymorphic between the resistant and susceptible parents, were subsequently used for BSA in F2 population. The fundamental principle of BSA is that if there is a molecular marker that shows polymorphism between the parents of a population and is closely linked to major QTL/gene controlling a particular trait, it should co-segregate with the QTL (Quarrie et al., 1999). Therefore, two DNA pools developed from F2 plants showing contrasting trait (extreme resistance and extreme susceptibility to ChLCV) were evaluated to identify polymorphic markers between them so as to confirm the linkage of the marker to the loci determining the trait. However, since in the current study, none of the markers showed polymorphism in BSA, it indicates that these polymorphic markers were not segregating with the trait controlling resistance/susceptibility to ChLCV and, therefore, are not linked to the trait of our interest. This can be explained by gross rearrangements in plant genome during evolution. The decay of R-gene collinearity among plant species has been especially attributed to tandem and segmental duplications that eventually lead to copy number and presence-absence variations (Zhang et al., 2014).
To further examine the level of synteny and micro-collinearity among genes lying in Ty QTLs of tomato and the orthologous regions in pepper, we leveraged the information available for syntenic regions in tomato and pepper genomes at pepper genome database. The concept of synteny which pertains to the “preserved co-location of homologous genes on chromosomes between species, irrespective of genetic linkage and gene order” was introduced in 1971 (Ehrlich et al., 1997; Passarge et al., 1999; Peters et al., 2012). Organization of genome, diversification of genes and evolutionary relationships between various solanaceous crops has been investigated by different workers using synteny and conserved linkage (Ku et al., 2000; Fulton, 2002; Wu et al., 2006; Wang et al., 2008). Conservation in the order and sequence of orthologs was reported in most of these cases, barring a few small-scale differences and positive gene selections (Doganlar et al., 2002; Wang et al., 2008). Two recent publications also suggested strong collinearity of the pepper genome with that of tomato (Kim et al., 2014; Qin, 2014).
In the present study, we observed that 13.71% genes located in the Ty1 QTL region, 63.76% in Ty2 QTL region, 23.07% in Ty3 QTL region, 73.84% located in Ty4 QTL region and 44.77% of genes located in ty5 QTL region in tomato were found to be conserved in pepper genome. However, despite such high conservation in some QTLs, the linkage relationship between different genes was greatly affected due to gross rearrangements with respect to order and position of genes in these species. The results appear to be in agreement with the earlier reports showing that the linear order of the genes in tomato and pepper chromosomes has been greatly modified due to rearrangements (Tanksley et al., 1988; Prince et al., 1993; Livingstone et al., 1999). Chromosomal rearrangements within solanaceous genomes (5 between potato and tomato and 30 rearrangements between pepper and tomato) have been reported in several other studies as well (Bonierbale et al., 1988; Tanksley et al., 1988; Prince et al., 1993; Livingstone et al., 1999; Wang et al., 2008). Significant rearrangements involving inversions and segmental translocations have been reported in the euchromatin regions of tomato, potato and pepper (Peters et al., 2012). We also noticed several gene rearrangements both within and between chromosomes as well as local gene rearrangements between tomato and pepper genomes in the Ty QTL regions investigated in this study. A closer look at the annotations of orthologous Ty regions in tomato and pepper genomes (Tables S6–S10) revealed that a large number of genes coding for proteins belonging to the nucleotide binding site-leucine-rich repeat (NBS-LRR) family present in tomato Ty QTLs are not conserved in pepper which might be partially responsible for the lack of linkage between the markers used in the study. Conversely, several disease resistance-related genes and transcription factors in the selected QTLs were found to be conserved in both the genomes. Further, functional characterization of these genes has potential to provide a way forward for begomovirus resistance breeding in pepper.
Conclusion
The present study provides useful information regarding conservation and dispersion of genes located within five quantitative trait loci conferring resistance to TYLCV of tomato between pepper and tomato genomes. The study revealed significant synteny in Ty QTLs in both the genomes. The micro-collinearity however, seems to be greatly affected by genomic rearrangements including inversions, deletions and reshuffling of gene order. These rearrangements are largely responsible for the lack of linkage relationship between orthologous markers in these species. The study provides an important insight into structural changes that may lead to variability in disease resistance in related plant species.
Author Contributions
MM performed the experiments, carried out in silico analysis and wrote the MS. AS developed mapping populations and looked after the breeding aspect of study. RS helped in the bioinformatics analysis and revision of the manuscript. PK supervised the work and contributed in the revision of manuscript.
Funding
The research was carried out under the project “Identification and molecular tagging of gene(s) controlling resistance to Chilli leaf curl virus infection in hot pepper (C. annuum L.)” funded by National Agricultural Science Fund, Indian Council of Agricultural Research, New Delhi.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01803/full#supplementary-material
References
Al-Shihi, A. A., Ammara, U. E., Amin, I., Deadman, M., and Al-Sadi, A. M. (2017). Association of tomato yellow leaf curl virus and chili leaf curl virus with leaf curl disease of radish and the synergistic interaction on Nicotiana benthamiana. Int. J. Agric. Biol. 19, 266–272. doi: 10.17957/IJAB/15.0273
Anbinder, I., Reuveni, M., Azari, R., Paran, I., Nahon, S., Shlomo, H., et al. (2009). Molecular dissection of tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 119, 519–530. doi: 10.1007/s00122-009-1060-z
Ben-Chaim, A., Borovsky, Y., Falise, M., Mazourek, M., Kang, B.-C., Paran, I., et al. (2006). QTL analysis for capsaicinoid content in capsicum. Theor. Appl. Genet. 113, 1481–1490. doi: 10.1007/s00122-006-0395-y
Bonierbale, M. W., Plaisted Rl Fau-Tanksley, S. D., and Tanksley, S. D. (1988). RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120, 1095–1103.
Bosland, P. W., and Votava, E. J. (2000). “Peppers: vegetable and spice capsicums. crop production science in horticulture,” in Peppers: Vegetable And Spice Capsicums (Wallingford, CT: CAB International Publishing), 204.
Dijan-Caporalino, C. L., Pijarowski, A., Januel, V., Lefebvre, C., and Al, C. E. (1998). “Characterising and fine mapping of the Me3 gene conferring heat-stable resistance to root-knot nematodes (Meloidogyne spp.) in pepper (Capsicum annuum L.) using AFLPs,” in Xth Eucarpia Meeting on Genetics and Breeding of Capsicum and Eggplant. Institut National de la Recherce Agro-nomique, (Avignon), 125–128.
Doganlar, S., Frary, A., Daunay, M. C., Lester, R. N., and Tanksley, S. D. (2002). Conservation of gene function in the solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 161, 1713–1726.
Ehrlich, J., Sankoff, D., and Nadeau, J. H. (1997). Synteny conservation and chromosome rearrangements during mammalian evolution. Genetics 147, 289–296.
Fortes, I. M., Sanchez-Campos, S., Fiallo-Olive, E., Diaz-Pendon, J. A., Navas-Castillo, J., and Moriones, E. (2016). A novel strain of tomato leaf curl New Delhi virus has spread to the Mediterranean basin. Viruses 8:307. doi: 10.3390/v8110307
Friedmann, M., Lapidot, M., and Cohen, S, and, M., P. (1998). A novel source of resistance to tomato yellow leaf curl virus exhibiting a symptomless reaction to viral infection. J. Am. Soc. Hortic. Sci. 123, 1004–1007.
Fulton, T. M. (2002). Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell Online 14, 1457–1467. doi: 10.1105/tpc.010479
George, B., Vinoth Kumar, R., and Chakraborty, S. (2014). Molecular characterization of chilli leaf curl virus and satellite molecules associated with leaf curl disease of Amaranthus spp. Virus Genes 48, 397–401. doi: 10.1007/s11262-013-1027-7
Grube, R. C., Radwanski, E. R., and Jahn, M. (2000). Comparative genetics of disease resistance within the solanaceae. Genetics 155, 873–887.
Hanson, P., Green, S. K., and Kuo, G. (2006). Ty-2, a gene on chromosome 11 conditioning geminivirus resistance in tomato. Tomato Genet. Coop. 56, 17–18.
Hutton, S. F., and Scott, J. W. (2013). “Fine-mapping and cloning of Ty-1 and Ty-3; and mapping of a new TYLCV resistance locus, “Ty-6”,” in Tomato Breeders Round Table Proceedings 2013, Chiang Mai.
Hutton, S. F., Scott, J. W., and Schuster, D. J. (2012). Recessive resistance to tomato yellow leaf curl virus from the tomato cultivar Tyking is located in same region as Ty-5 on chromosome 4. J. Am. Soc. Hortic. Sci. 47, 324–327.
Ji, Y., Schuster, D. J., and Scott, J. W. (2007a). Ty-3, a begomovirus resistance locus near the tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breed. 20, 271–284. doi: 10.1007/s11032-007-9089-7
Ji, Y., Scott, J. W., Hanson, P., Graham, E., and Maxwell, D. P. (2007b). “Sources of resistance, inheritance, and location of genetic loci conferring resistance to members of the tomato-infecting begomoviruses,” in Tomato Yellow Leaf Curl Virus Disease, ed H. Czosnek (Netherlands: Springer), 343–362.
Ji, Y., Scott, J. W., and Schuster, D. J. (2009a). Toward fine mapping of the Tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. Hortic. Sci. 44, 614–618.
Ji, Y., Scott, J. W., Schuster, D. J., and Maxwell, D. P. (2009b). Molecular mapping of Ty-4, a tomato yellow leaf curl virus resistance locus on chromosome 3 of tomato. J. Am. Soc. Hortic. Sci. 134, 281–288.
Kadirvel, P., De La Pana, R., Schaffeitner, R., Huang, S., Geethanjali, S., Kenyon, L., et al. (2013). Mapping of QTLs in tomato line FLA456 associated with resistance to a virus causing yellow leaf curl disease. Euphytica 190, 297–308. doi: 10.1007/s10681-012-0848-0
Kang, B. C., Nahm, S. H., Huh, J. H., Yoo, H. S., Yu, J. W., Lee, M. H., et al. (2001). An interspecific (Capsicum annuum × C. chinese) F 2 linkage map in pepper using RFLP and AFLP markers. TAG Theor. Appl. Genet. 102, 531–539. doi: 10.1007/s001220051678
Khan, M. S., Raj, S. K., and Singh, R. (2006). First report of tomato leaf curl New Delhi virus infecting chilli in India. Plant Pathology 55, 289–289. doi: 10.1111/j.1365-3059.2006.01324.x
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Ku, H. M., Vision, T., Liu, J., and Tanksley, S. D. (2000). Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. U.S.A. 97, 9121–9126. doi: 10.1073/pnas.160271297
Kumar, S., Kumar, S., Singh, M., Singh, A. K., and Rai, M. (2006). Identification of host plant resistance to pepper leaf curl virus in chilli (Capsicum species). Sci. Hortic. 110, 359–361. doi: 10.1016/j.scienta.2006.07.030
Lee, J. M., Nahm, S. H., Kim, Y. M., and Kim, B. D. (2004). Characterization and molecular genetic mapping of microsatellite loci in pepper. TAG Theor. Appl. Genet. 108, 619–627. doi: 10.1007/s00122-003-1467-x
Lefebvre, V., Pflieger, S., Thabuis, A., Caranta, C., Blattes, A., Chauvet, J. C., et al. (2002). Towards the saturation of the pepper linkage map by alignment of three intraspecific maps including known-function genes. Genome 45, 839–854. doi: 10.1139/g02-053
Livingstone, K. D., Lackney, V. K., Blauth, J. R., Van Wijk, R., and Jahn, M. K. (1999). Genome mapping in capsicum and the evolution of genome structure in the solanaceae. Genetics 152, 1183–1202.
Minamiyama, Y., Tsuro, M., and Hirai, M. (2006). An SSR-based linkage map of Capsicum annuum. Mol. Breed. 18, 157–169. doi: 10.1007/s11032-006-9024-3
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Paran, I., Van Der Voort, J. R., Lefebvre, V., Jahn, M., Landry, L., Van Schriek, M., et al. (2004). An integrated genetic linkage map of pepper (Capsicum spp.). Mol. Breed. 13, 251–261. doi: 10.1023/B:MOLB.0000022526.30914.31
Passarge, E., Horsthemke, B., and Farber, R. A. (1999). Incorrect use of the term synteny. Nat. Genet. 23, 387–387. doi: 10.1038/70486
Peters, S. A., Bargsten, J. W., Szinay, D., Van De Belt, J., Visser, R. G. F., Bai, Y., et al. (2012). Structural homology in the Solanaceae: analysis of genomic regions in support of synteny studies in tomato, potato and pepper. Plant J. 71, 602–614. doi: 10.1111/j.1365-313X.2012.05012.x
Prasanna, H. C., Sinha, D. P., Rai, G. K., Krishna, R., Kashyap, S. P., Singh, N. K., et al. (2015). Pyramiding Ty-2 and Ty-3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Pathol. 64, 256–264. doi: 10.1111/ppa.12267
Pratap, D., Kashikar, A. R., and Mukherjee, S. K. (2011). Molecular characterization and infectivity of a tomato leaf curl New Delhi virus variant associated with newly emerging yellow mosaic disease of eggplant in India. Virol. J. 8:305. doi: 10.1186/1743-422X-8-305
Prince, J. P., Pochard, E., and Tanksley, S. D. (1993). Construction of a molecular linkage map of pepper and a comparison of synteny with tomato. Genome 36, 404–417. doi: 10.1139/g93-056
Qin, C. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Agrotechnology 2:4. doi: 10.4172/2168-9881.S1.013
Quarrie, S., Lazi-Jani, V., Kovaevi, D., Steed, A., and Peki, S. (1999). Bulk segregant analysis with molecular markers and its use for improving drought resistance in maize. J. Exp. Bot. 337, 1299–1306. doi: 10.1093/jxb/50.337.1299
Senanayake, D. M. J. B., Mandal, B., Lodha, S., and Varma, A. (2007). First report of Chilli leaf curl virus affecting chilli in India. Plant Pathol. 56, 343–343. doi: 10.1111/j.1365-3059.2007.01513.x
Singh, A. K., Kushwaha, N., and Chakraborty, S. (2016). Synergistic interaction among begomoviruses leads to the suppression of host defense-related gene expression and breakdown of resistance in chilli. Appl. Microbiol. Biotechnol. 100, 4035–4049. doi: 10.1007/s00253-015-7279-5
Srivastava, A., Mangal, M., Saritha, R. K., Jat, S. L., Gosavy, G. U., and Kalia, P. (2015). Natural epiphytotic screening of chilli germplasm lines against leaf curl virus complex. Int. J. Trop. Agric. 33, 3581–3586.
Srivastava, A., Mangal, M., Saritha, R. K., and Kalia, P. (2017). Screening of chilli pepper (Capsicum spp.) lines for resistance to the begomoviruses causing chilli leaf curl disease in India. Crop Prot. 100, 177–185. doi: 10.1016/j.cropro.2017.06.015
Tanksley, S. D., Bernatzky, R., Lapitan, N. L., and Prince, J. P. (1988). Conservation of gene repertoire but not gene order in pepper and tomato. Proc. Natl. Acad. Sci. U.S.A. 85, 6419–6423. doi: 10.1073/pnas.85.17.6419
Tanksley, S. D., Ganal, M. W., Prince, J. P., De Vicente, M. C., Bonierbale, M. W., Broun, P., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160.
Wang, Y., Diehl, A., Wu, F., Vrebalov, J., Giovannoni, J., Siepel, A., et al. (2008). Sequencing and comparative analysis of a conserved syntenic segment in the Solanaceae. Genetics 180, 391–408. doi: 10.1534/genetics.108.087981
Wu, F., Eannetta, N. T., Xu, Y., Durrett, R., Mazourek, M., Jahn, M. M., et al. (2009). A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor. Appl. Genet. 118, 1279–1293. doi: 10.1007/s00122-009-0980-y
Wu, F., Mueller, L. A., Crouzillat, D., Pétiard, V., and Tanksley, S. D. (2006). Combining bioinformatics and phylogenetics to identify large sets of single-copy Orthologous Genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174, 1407–1420. doi: 10.1534/genetics.106.062455
Yang, X., Caro, M., Hutton, S. F., Scott, J. W., Guo, Y., Wang, X., et al. (2014). Fine mapping of the tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. Mol. Breed. 34, 749–760. doi: 10.1007/s11032-014-0072-9
Yu, G. X., Bush, A. L., and Wise, R. P. (1996). Comparative mapping of homoeologous group 1 regions and genes for resistance to obligate biotrophs in Avena, Hordeum, and Zea mays. Genome 39, 155–164. doi: 10.1139/g96-021
Zamir, D., Ekstein-Michelson, I., Zakay, Y., Navot, N., Zeidan, M., Sarfatti, M., et al. (1994). Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor. Appl. Genet. 88:141. doi: 10.1007/BF00225889
Keywords: begomoviruses, ChLCV, pepper, tomato, TYLCV, genome conservation
Citation: Mangal M, Srivastava A, Sharma R and Kalia P (2017) Conservation and Dispersion of Genes Conferring Resistance to Tomato Begomoviruses between Tomato and Pepper Genomes. Front. Plant Sci. 8:1803. doi: 10.3389/fpls.2017.01803
Received: 29 July 2017; Accepted: 04 October 2017;
Published: 07 November 2017.
Edited by:
Manoj Prasad, National Institute of Plant Genome Research (NIPGR), IndiaReviewed by:
Pranav Pankaj Sahu, Max Planck Institute for Plant Breeding Research (MPG), GermanyEmanuela Noris, Consiglio Nazionale Delle Ricerche (CNR), Italy
Copyright © 2017 Mangal, Srivastava, Sharma and Kalia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manisha Mangal, manishamangal@rediffmail.com
 Manisha Mangal
Manisha Mangal Arpita Srivastava
Arpita Srivastava Rita Sharma
Rita Sharma Pritam Kalia
Pritam Kalia