- 1Department of Plant Science, McGill University, Sainte Anne de Bellevue, QC, Canada
- 2Department of Crop and Soil Science, Washington State University, Pullman, WA, United States
- 3International Maize and Wheat Improvement Center (CIMMYT), El Batán, Texcoco, Mexico
Plant cell wall formation is a complex, coordinated and developmentally regulated process. Cellulose is the most dominant constituent of plant cell walls. Because of its paracrystalline structure, cellulose is the main determinant of mechanical strength of plant tissues. As the most abundant polysaccharide on earth, it is also the focus of cellulosic biofuel industry. To reduce culm lodging in wheat and for improved ethanol production, delineation of the variation for stem cellulose content could prove useful. We present results on the analysis of the stem cellulose content of 288 diverse wheat accessions and its genome-wide association study (GWAS). Cellulose concentration ranged from 35 to 52% (w/w). Cellulose content was normally distributed in the accessions around a mean and median of 45% (w/w). Genome-wide marker-trait association study using 21,073 SNPs helped identify nine SNPs that were associated (p < 1E-05) with cellulose content. Four strongly associated (p < 8.17E-05) SNP markers were linked to wheat unigenes, which included β-tubulin, Auxin-induced protein 5NG4, and a putative transmembrane protein of unknown function. These genes may be directly or indirectly involved in the formation of cellulose in wheat culms. GWAS results from this study have the potential for genetic manipulation of cellulose content in bread wheat and other small grain cereals to enhance culm strength and improve biofuel production.
Introduction
Increasing world population demands a sustainable increase in the production of food, feed and fuel crops (Scholey et al., 2016). Bread wheat (Triticum aestivum) occupies more agricultural area than any other food crop worldwide (http://www.wheatinitiative.org/). In addition to grain production, the annual worldwide production of wheat straw is around 3.5 × 108 tons, which is used as cattle fodder in developing countries and is a potential feedstock for cellulosic ethanol production (Singhania et al., 2014). Wheat straw, which is comprised of cellulose (~40%), hemicelluloses (~35%), and lignin (~25%), is one of the most abundant lignocellulosic raw materials in the world (Ruiz et al., 2013). Cellulose, a paracrystalline polysaccharide, is the main determinant of mechanical strength, which has implications in crop lodging, biotic and abiotic stresses. Cellulose amount in a unit length of the stem explains most of the variation in mechanical strength (Appenzeller et al., 2004; Dhugga, 2007). The proportion of cellulose in the cell wall also affects the total sugar release during the process of enzymatic hydrolysis (Fan et al., 2012; Lindedam et al., 2012). An understanding of the natural variability of cellulose in plants and its association with chromosomal regions could provide markers for enhancing grain and biomass yield (Ciesielski et al., 2014).
Cellulose consists of linear chains of β (1→4) linked glucan (polyglucose) known to be synthesized by the members of superfamily Glycosyltransferase 2 (GT2) called Cellulose synthase A (CesA; Fujii et al., 2010; Kumar et al., 2016). Twenty-two CesA genes have been reported in hexaploid wheat (Kaur et al., 2016). In addition to the CesA genes, the Glycosylhydrolase 9 (GH9) family genes are known to have an impact on the synthesis of cellulose in plants (Kotake et al., 2011). Based on the mutant analysis in Arabidopsis, a member of GH9 family called KORRIGAN1 (KOR1) has been reported to be involved in cellulose synthesis, cell expansion and intracellular trafficking of cellulose synthase complex (CSC; Szyjanowicz et al., 2004; Lei et al., 2014; Vain et al., 2014). Investigation of brittle culm 1 mutants in rice and brittle stalk 2 mutant in maize revealed the involvement of COBRA-like proteins in cellulose formation in secondary walls (Ching et al., 2006). Involvement of Sucrose synthase (SuSy) in channeling substrate to cellulose synthase has also been reported (Fujii et al., 2010). Similarly, several other proteins affect cellulose synthesis, including chitinase-like 1 (CSI1; Sánchez-Rodríguez et al., 2012), companion of cellulose synthase (CC; Endler et al., 2015), and tracheary element differentiation-related (TED) 6 and 7 (Rejab et al., 2015).
Variation for the proportion of cellulose in cell wall among wheat varieties is not yet known. This study was planned to identify the genomic regions affecting the variability of cellulose content among diverse spring wheat genotypes through GWAS.
Genes associated with cell wall have been previously explored through GWAS in miscanthus (Slavov et al., 2014), poplar (Porth et al., 2013), maize (Li et al., 2016), and barley (Houston et al., 2015). In barley, genes of Glycosyltransferase 2 and Glycosylhydrolase families were associated with culm cellulose variation. However, none of the genes found in maize through GWAS of stalk cellulose content was specifically involved in the cellulose biosynthetic pathway. In the present study, the stem internodes of 288 spring wheat varieties were analyzed for variation in cellulose content. Utilizing the 21,073 SNPs generated by DArT-seq GBS and cellulosic content, GWAS was performed using fixed and random model circulating probability unification (FarmCPU) method (Liu et al., 2016). Genes, which were not reported previously for their role in cellulose formation, were identified as associated with the culm cellulose content. Gene-trait associations identified in this study might be useful in altering the lignocellulose composition of wheat and other grasses.
Materials and Methods
Plant Material
A worldwide collection of 288 diverse spring wheat germplasm was used for the phenotypic and genotypic analysis. The collection included cultivars from different regions of United States, the International Maize and Wheat Improvement Centre (CIMMYT), Mexico, and historical lines dating back to 1871 (Mohan et al., 2013). The wide span of our collection was intended to capture the maximum variation possible while maintaining a manageable population size. This worldwide collection also represents the various market classes of wheat based on the kernel color, hardiness, and shape. The following types of genotypes were represented based on kernel type: soft white spring (SWS), soft red spring (SRS), hard red spring (HRS), hard white spring (HWS), and club wheat (Mohan et al., 2013). The plants were grown in the greenhouse of the Plant Growth Facilities, Washington State University, Pullman at 22°C/18°C temperature and 16/8 h day/night in 2014-15. Seeds were planted in a randomized design to accommodate the effect of light.
Phenotypic Analysis
The analysis on percent cellulose was performed for 288 diverse spring wheat genotypes, with three replicates per genotype. The first internode (from the base) of the main tiller of each of three mature plants was dried at 80°C. Measured amount of dried sample (45–55 mg) was placed into a pre-weighed 2 ml Eppendorf tubes with a screw cap. A mixture of acetic acid: water: nitric acid (8:2:1) was added to each tube (1.5 ml) and vortexed (Updegraff, 1969). All the tubes were transferred to a steel rack and placed in a boiling water bath for 4 h. The tubes were allowed to cool at room temperature and centrifuged in a swing-out rotor at 10,000 rpm for 10 min. The supernatant was aspirated off, the pellet washed with distilled water four times and finally washed with 90% ethanol. After each wash, the tubes were vortexed and centrifuged at 10,000 rpm for 10 min. The tubes were dried at 80°C followed by determination of percent cellulose on dry matter basis.
Population Structure and GWAS Analysis
Principal component analysis (PCA) was used to infer population structure through Genomic Association and Prediction Integrated Tool (GAPIT; Lipka et al., 2012; Ahmad et al., 2015; Tang et al., 2016). Twenty-one thousand and seventy-three SNP markers were obtained by analyzing the genomic DNA with a Genotyping-by-Sequencing (GBS) approach (Mohan et al. unpublished). In brief, genotyping was carried out at DArT Pyt Ltd in Canberra-Australia, using a combination of HiSeq 2000 (Illumina) next-generation sequencing with DArT-seq GBS technology (called DArTseq™). This method follows two-step complexity reductions by using two enzymes, PstI/HpaII and PstI/HhaI, along-with TaqI restriction enzyme to eliminate subsets of PstI -HpaII and PstI-HhaI fragments, respectively. The pooled barcoaded samples were run in a single lane on an Illumina Hiseq 2000 instrument for sequencing. A proprietary analytical pipeline developed by DArT Pyt Ltd was used to obtain the DArT score and SNP tables (http://www.diversityarrays.com/). Fixed and Random Model Circulating Probability Unification (FarmCPU; Liu et al., 2016) in R version 2.15.3 was used to calculate P-values for Manhattan Plot and Q-Q plots. A Manhattan plot was generated using the −log10(p) values for each SNP with 1% Bonferroni test threshold (Team, 2014). The significance of the genome-wide association between SNP marker and cellulose content was tested at FDR p < 0.001.
Gene Annotation
The sequences containing the SNPs were mapped against wheat unigenes downloaded from the NCBI database. Significant SNPs with associated unigenes were annotated using BLASTN with the International Wheat Genome Sequencing Consortium (IWGSC; Mayer et al., 2014) reference Sequence v1.0 (https://www.wheatgenome.org) posted on May 30, 2017. The functions of associated unigenes were also searched with BLASTN through identification of orthologs from other plant species.
Results
Cellulose Content
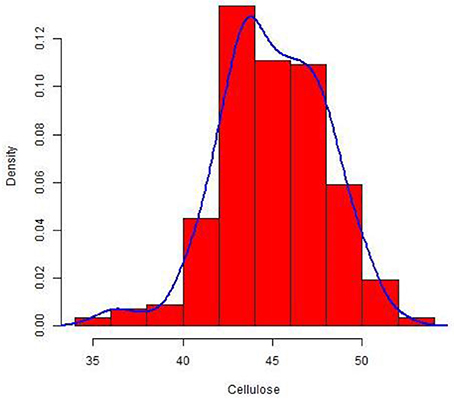
The culm cellulose content differed significantly in a set of 228 wheat lines with a range of 0.32–0.52 mg and an average of 0.45 mg cellulose/mg of dry weight (Table S2). The cellulose concentration was normally distributed around the mean in the set as depicted in the density plot (Figure 1). As expected from this plot, the calculated median was also similar to the overall mean across the population.
Principal Component Analysis and Marker-Trait Associations
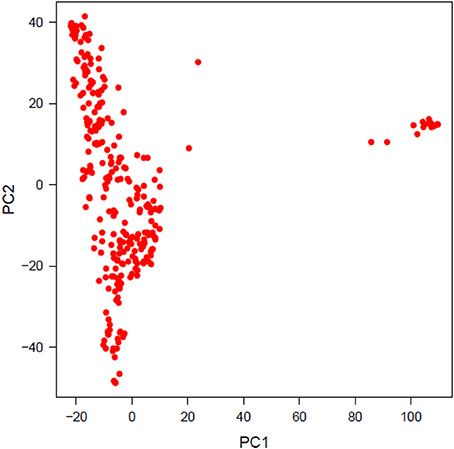
PCA was performed to investigate population structure. The first two PCs explained 8.13 and 4.90% variation in the population of lines. Plotting PC2 against PC1 revealed two distinct, a major and a minor, clusters. The minor cluster containing 20 genotypes was removed from the final analysis to account for population structure and the first PC was used as a covariate for GWAS analyses (Figure 2).
A total of 21,073 single nucleotide polymorphic (SNP) markers with a minor allele frequency (MAF) above 5% (Figure 3) and the cellulose content data from 268 lines were subjected to GWAS analysis, which revealed nine significant marker-trait associations with p-values of less than 1E-05 (Figure 4). The most significant SNP marker in our analysis corresponded to wheat chromosome 5AL with a p-value of 1.86E-07. The second most significant SNP was located on chromosome 1AL with a p-value of 2.24E-07. In addition, we found significant SNPs corresponding to chromosome 1AL, 6BS, 1DL, 2DS, 4DL, 5BL, and 3B with p-values of less than 1E-05 (Table 1). The quantile-quantile (QQ) plot drawn for calculated p-values was used to check spurious associations. The deviation of relatively a few markers from null expectations in the QQ plot supports the significant associations we have identified (Figure 5).
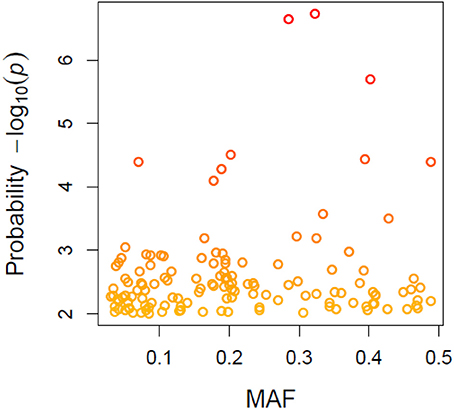
Figure 3. Minor allele frequency (MAF) patterns relative to allele calls for wheat genotypes based on 21073 SNPs.
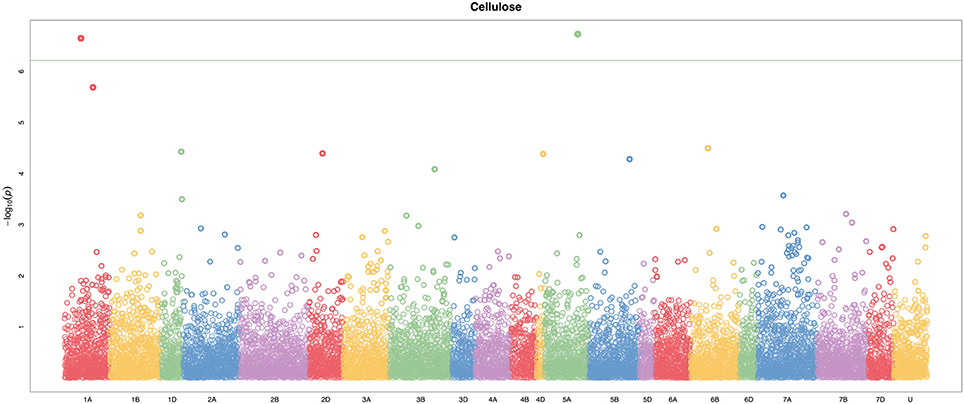
Figure 4. Manhattan plot of genome-wide association study (GWAS) on stem cellulose content (mg cellulose/mg dry weight) by using the FarmCPU. The −log10(p–values) from GWAS are plotted against the position on each of the 21 bread wheat chromosomes. U represents unassigned chromosome scaffolds. Two loci on chromosomes 1A and 5A were identified above the Bonferroni threshold correcting for genome-wide multiple tests at type I error of 0.001 (green line).
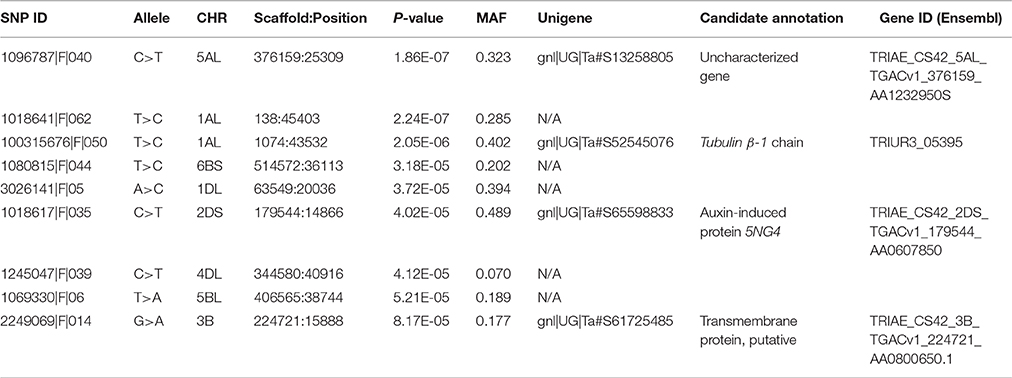
Table 1. Regions of wheat genome showing significant associations with stem cellulose content variation based on GWAS.
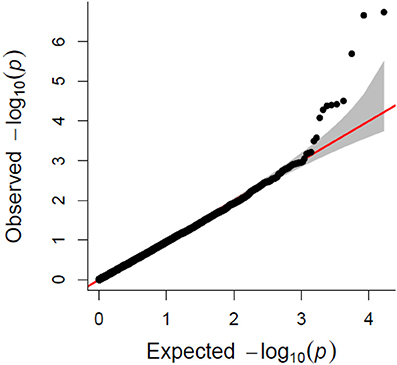
Figure 5. Quantile-quantile (QQ) plot showing the deviation from null hypothesis for associated SNP makers.
Gene Identification
Significant SNP markers resulting from GWAS were mapped to the wheat unigene database. The identified unigenes were annotated based on the sequence comparisons using NCBI BLAST and EnsemblPlant databases. The first and second most significant SNP markers were associated with the unigenes TRIAE_CS42_5AL_TGACv1_376159_AA1232950 and gnl|UG|Ta#S52545076, respectively. The third and fourth SNPs respectively were associated with the genes TRIAE_CS42_2DS_TGACv1_179544_AA0607850 and TRIAE_CS42_3B_TGACv1_224721_AA0800650.1. The gene TRIAE_CS42_5AL_TGACv1_376159_AA1232950 is uncharacterized as judged from a lack of its functional annotation in wheat and other plant species. The unigene gnl|UG|Ta#S52545076 was 60% identical at the amino acid level with a gene in the Tubulin superfamily, Tubulin β-1 chain, of Triticum urartu. TRIAE_CS42_2DS_TGACv1_179544_AA0607850 showed 82% identity over 97% amino acid coverage with the Auxin-induced protein 5NG4 of Aegilops tauschii, whereas TRIAE_CS42_3B_TGACv1_224721_AA0800650.1 was annotated based on 51% amino acid identity and 97% coverage with a putative transmembrane protein of Medicago truncatula (Table 1).
Discussion
From a larger set of 288 diverse bread wheat lines, we used 268 well-structured accessions to study the genetic association of cellulose content in wheat. The most appropriate model was selected to obtain a higher level of confidence in the association results. GWAS was conducted using Fixed and Random Model Circulating Probability Unification (FarmCPU); a new and more efficient recently developed method, which accounts for fixed and random effects to control false positives (Liu et al., 2016). Most of the GWAS mapping studies in wheat thus far have been conducted to identify genes or QTLs related to agronomic performance (Lopes et al., 2015; Jaiswal et al., 2016), grain yield (Sukumaran et al., 2015), or disease resistance (Kollers et al., 2013; Gurung et al., 2014). Cellulose is a major component of cell walls and a key determinant of mechanical strength of plant tissues (Appenzeller et al., 2004; Ching et al., 2006). The involvement of the CesA genes in cellulose synthesis is well-documented, and recently 22 CesA genes were reported in wheat (Kaur et al., 2016). These genes are differentially expressed in primary and secondary cell wall forming cells. Although, we identified 9 SNP markers in this study to be associated [−log10(p) = 7 to −log10(p) = 5] with cellulose content (Table S1), we were able to map only four of these to the wheat unigene database. A high marker density and population size in our study increased the confidence about these SNP associations (Wang et al., 2012). The genes associated with the cellulose content may contribute to its natural variation in wheat lines. The involvement of many genes other than CesAs in controlling cellulose synthesis supports our suggestion (Kotake et al., 2011).
Only a few studies have explored the genes other than CesA involved in the cellulose biosynthetic pathway (Porth et al., 2013; Slavov et al., 2014; Houston et al., 2015; Li et al., 2016). Recently in a GWA study in barley, a species syntenic to wheat, the association of several genes from the Glycosyltransferases and Glycosylhydrolases families was shown with the culm cellulose content (Houston et al., 2015). Similar to barley GWAS associations, our results also pointed to the involvement of the GT gene family in cellulose formation. We also identified some unique associations not reported in the barley study.
Our results pointed to the involvement of β-tubulin in the regulation of cellulose content. β-tubulins proteins form heterodimers with α-tubulins to form microtubules, which have long been known to guide the deposition of cellulose microfibrils in the cell wall in a helical pattern (Rao et al., 2016). Functional association of cortical microtubules with cellulose synthase complexes is well-documented (Paredez et al., 2006; Chan et al., 2007, 2010; Wightman and Turner, 2008; Crowell et al., 2009; Gutierrez et al., 2009).
Another important association in our study is for the Auxin-induced protein, 5NG4. This gene is a member of the plant drug/metabolite exporter (P-DME; TC 2.A.7.4) family, also called WALLS ARE THIN1 (WAT1)-related proteins. Mutant studies in Arabidopsis helped demonstrate its involvement in secondary cell wall formation. Comparative transcriptomics and metabolomics demonstrated synchronized downregulation of the secondary cell wall CesAs (CesA8, CesA7, and CesA4) and auxin metabolism genes (auxin-responsive genes and auxin influx transporter genes) in wat1 mutants (Ranocha et al., 2010). Higher expression of PIN-like auxin efflux carrier and auxin-induced protein 5NG4 genes in relation to both cell division and cell expansion was found in an expression profiling study of Chinese fir (Cunninghamia lanceolata; Qiu et al., 2013). With regard to the association of a putative transmembrane protein of unknown function with cellulose content in our study, several membrane proteins other than CesA are known to be involved in cellulose formation. We could not, however, assign a putative function to this protein. The additional genes we have reported in this study to be associated with cellulose content in the wheat culm are potential candidates for improving culm strength and the potential of wheat stover for biofuels by increasing the cellulose content.
Conclusion
Cellulose content in wheat culms of 288 diverse cultivars varied widely. Genome-wide association analysis helped identify four genetic associations for cellulose content, which have the potential as molecular markers to manipulate cellulose content in wheat with the goal of improving culm strength and cellulosic biofuel production.
Author Contributions
SK performed all the experiments in the greenhouse and laboratory set up and conducted genetic, genomic, bioinformatics analyses, and wrote the manuscript. AM helped in writing and editing the manuscript. JS helped in troubleshooting, provided constructive comments, suggestions and financial support to conduct the experiments. KD provided the protocols for cellulose content analysis and thoroughly edited the manuscript. PV and SS provided the genotyping data, XZ, AM, and HD helped in the creating the SNP data and GWAS analysis. ZZ and KG provided their expert advice and suggestions to interpret the results.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors acknowledge financial support received from BioFuelNet and NSERC discovery program. Genotyping was made possible by the kind support of the Mexican Secretariat of Agriculture, Livestock, Rural Development, Fisheries, and Food (SAGARPA) through the project “Seeds of Discovery” Sustainable Modernization of Traditional Agriculture project (MasAgro).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01913/full#supplementary-material
References
Ahmad, S., Kaur, S., Lamb-Palmer, N. D., Lefsrud, M., and Singh, J. (2015). Genetic diversity and population structure of Pisum sativum accessions for marker-trait association of lipid content. Crop J. 3, 238–245. doi: 10.1016/j.cj.2015.03.005
Appenzeller, L., Doblin, M., Barreiro, R., Wang, H., Niu, X., Kollipara, K., et al. (2004). Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11, 287–299. doi: 10.1023/B:CELL.0000046417.84715.27
Chan, J., Calder, G., Fox, S., and Lloyd, C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9, 171–175. doi: 10.1038/ncb1533
Chan, J., Crowell, E., Eder, M., Calder, G., Bunnewell, S., Findlay, K., et al. (2010). The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J. Cell Sci. 123, 3490–3495. doi: 10.1242/jcs.074641
Ching, A., Dhugga, K. S., Appenzeller, L., Meeley, R., Bourett, T. M., Howard, R. J., et al. (2006). Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224, 1174–1184. doi: 10.1007/s00425-006-0299-8
Ciesielski, P. N., Resch, M. G., Hewetson, B., Killgore, J. P., Curtin, A., Anderson, N., et al. (2014). Engineering plant cell walls: tuning lignin monomer composition for deconstructable biofuel feedstocks or resilient biomaterials. Green Chem. 16:2627. doi: 10.1039/c3gc42422g
Crowell, E. F., Bischoff, V., Desprez, T., Rolland, A., Stierhof, Y.-D., Schumacher, K., et al. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21, 1141–1154. doi: 10.1105/tpc.108.065334
Dhugga, K. S. (2007). Maize biomass yield and composition for biofuels. Crop Sci. 47, 2211–2227. doi: 10.2135/cropsci2007.05.0299
Endler, A., Kesten, C., Schneider, R., Zhang, Y., Ivakov, A., Froehlich, A., et al. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell 162, 1353–1364. doi: 10.1016/j.cell.2015.08.028
Fan, W.-X., Hou, Y.-X., Feng, S.-W., Zhu, F.-K., and Ru, Z.-G. (2012). Study on cellulose and lodging resistance of wheat straw. J. Henan Agric. Sci. 9. Available online at: http://en.cnki.com.cn/Article_en/CJFDTotal-HNNY201209010.htm
Fujii, S., Hayashi, T., and Mizuno, K. (2010). Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant Cell Physiol. 51, 294–301. doi: 10.1093/pcp/pcp190
Gurung, S., Mamidi, S., Bonman, J. M., Xiong, M., Brown-Guedira, G., and Adhikari, T. B. (2014). Genome-wide association study reveals novel quantitative trait Loci associated with resistance to multiple leaf spot diseases of spring wheat. PLoS ONE 9:e108179. doi: 10.1371/journal.pone.0108179
Gutierrez, R., Lindeboom, J. J., Paredez, A. R., Emons, A. M. C., and Ehrhardt, D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806. doi: 10.1038/ncb1886
Houston, K., Burton, R. A., Sznajder, B., Rafalski, A. J., Dhugga, K. S., Mather, D. E., et al. (2015). A genome-wide association study for culm cellulose content in barley reveals candidate genes co-expressed with members of the cellulose synthase a gene family. PLoS ONE 10:e0130890. doi: 10.1371/journal.pone.0130890
Jaiswal, V., Gahlaut, V., Meher, P. K., Mir, R. R., Jaiswal, J. P., Rao, A. R., et al. (2016). Genome wide single locus single trait, multi-locus and multi-trait association mapping for some important agronomic traits in common wheat (T. aestivum L.). PLoS ONE 11:e0159343. doi: 10.1371/journal.pone.0159343
Kaur, S., Dhugga, K. S., Gill, K., and Singh, J. (2016). Novel structural and functional motifs in cellulose synthase (CesA) genes of bread wheat (Triticum aestivum, L.). PLoS ONE 11:e0147046. doi: 10.1371/journal.pone.0147046
Kollers, S., Rodemann, B., Ling, J., Korzun, V., Ebmeyer, E., Argillier, O., et al. (2013). Genetic architecture of resistance to Septoria tritici blotch (Mycosphaerella graminicola) in European winter wheat. Mol. Breed. 32, 411–423. doi: 10.1007/s11032-013-9880-6
Kotake, T., Aohara, T., Hirano, K., Sato, A., Kaneko, Y., Tsumuraya, Y., et al. (2011). Rice Brittle culm 6 encodes a dominant-negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls. J. Exp. Bot. 62, 2053–2062. doi: 10.1093/jxb/erq395
Kumar, M., Wightman, R., Atanassov, I., Gupta, A., Hurst, C. H., Hemsley, P. A., et al. (2016). S-Acylation of the cellulose synthase complex is essential for its plasma membrane localization. Science 353, 166–169. doi: 10.1126/science.aaf4009
Lei, L., Zhang, T., Strasser, R., Lee, C. M., Gonneau, M., Mach, L., et al. (2014). The jiaoyao1 mutant is an allele of korrigan1 that abolishes endoglucanase activity and affects the organization of both cellulose microfibrils and microtubules in Arabidopsis. Plant Cell 26, 2601–2616. doi: 10.1105/tpc.114.126193
Li, K., Wang, H., Hu, X., Liu, Z., Wu, Y., and Huang, C. (2016). Genome-wide association study reveals the genetic basis of stalk cell wall components in maize. PLoS ONE 11:e0158906. doi: 10.1371/journal.pone.0158906
Lindedam, J., Andersen, S. B., DeMartini, J., Bruun, S., Jørgensen, H., Felby, C., et al. (2012). Cultivar variation and selection potential relevant to the production of cellulosic ethanol from wheat straw. Biomass Bioenergy 37, 221–228. doi: 10.1016/j.biombioe.2011.12.009
Lipka, A. E., Tian, F., Wang, Q., Peiffer, J., Li, M., Bradbury, P. J., et al. (2012). GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399. doi: 10.1093/bioinformatics/bts444
Liu, X., Huang, M., Fan, B., Buckler, E. S., and Zhang, Z. (2016). Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 12:e1005767. doi: 10.1371/journal.pgen.1005767
Lopes, M., Dreisigacker, S., Peña, R., Sukumaran, S., and Reynolds, M. (2015). Genetic characterization of the wheat association mapping initiative (WAMI) panel for dissection of complex traits in spring wheat. Theor. Appl. Genet. 128, 453–464. doi: 10.1007/s00122-014-2444-2
Mayer, K. F., Rogers, J., DoleŽel, J., Pozniak, C., Eversole, K., Feuillet, C., et al. (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788. doi: 10.1126/science.1251788
Mohan, A., Schillinger, W. F., and Gill, K. S. (2013). Wheat seedling emergence from deep planting depths and its relationship with coleoptile length. PLoS ONE 8:e73314. doi: 10.1371/journal.pone.0073314
Paredez, A. R., Somerville, C. R., and Ehrhardt, D. W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495. doi: 10.1126/science.1126551
Porth, I., Klapšte, J., Skyba, O., Hannemann, J., McKown, A. D., Guy, R. D., et al. (2013). Genome-wide association mapping for wood characteristics in populus identifies an array of candidate single nucleotide polymorphisms. New Phytol. 200, 710–726. doi: 10.1111/nph.12422
Qiu, Z., Wan, L., Chen, T., Wan, Y., He, X., Lu, S., et al. (2013). The regulation of cambial activity in Chinese fir (Cunninghamia lanceolata) involves extensive transcriptome remodeling. New Phytol. 199, 708–719. doi: 10.1111/nph.12301
Ranocha, P., Denancé, N., Vanholme, R., Freydier, A., Martinez, Y., Hoffmann, L., et al. (2010). Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 63, 469–483. doi: 10.1111/j.1365-313X.2010.04256.x
Rao, G., Zeng, Y., He, C., and Zhang, J. (2016). Characterization and putative post-translational regulation of α-and β-tubulin gene families in Salix arbutifolia. Sci. Rep. 6:19258. doi: 10.1038/srep19258
Rejab, N. A., Nakano, Y., Yoneda, A., Ohtani, M., and Demura, T. (2015). Possible contribution of TED6 and TED7, secondary cell wall-related membrane proteins, to evolution of tracheary element in angiosperm lineage. Plant Biotechnol. 32, 343–347. doi: 10.5511/plantbiotechnology.15.0826a
Ruiz, H. A., Cerqueira, M. A., Silva, H. D., Rodriguez-Jasso, R. M., Vicente, A. A., and Teixeira, J. A. (2013). Biorefinery valorization of autohydrolysis wheat straw hemicellulose to be applied in a polymer-blend film. Carbohydr. Polym. 92, 2154–2162. doi: 10.1016/j.carbpol.2012.11.054
Sánchez-Rodríguez, C., Bauer, S., Hématy, K., Saxe, F., Ibáñez, A. B., Vodermaier, V., et al. (2012). Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24, 589–607. doi: 10.1105/tpc.111.094672
Scholey, D., Burton, E., and Williams, P. (2016). The bio refinery; producing feed and fuel from grain. Food Chem. 197, 937–942. doi: 10.1016/j.foodchem.2015.11.063
Singhania, R. R., Saini, J. K., Saini, R., Adsul, M., Mathur, A., Gupta, R., et al. (2014). Bioethanol production from wheat straw via enzymatic route employing Penicillium janthinellum cellulases. Bioresour. Technol. 169, 490–495. doi: 10.1016/j.biortech.2014.07.011
Slavov, G. T., Nipper, R., Robson, P., Farrar, K., Allison, G. G., Bosch, M., et al. (2014). Genome-wide association studies and prediction of 17 traits related to phenology, biomass and cell wall composition in the energy grass Miscanthus sinensis. New Phytol. 201, 1227–1239. doi: 10.1111/nph.12621
Sukumaran, S., Dreisigacker, S., Lopes, M., Chavez, P., and Reynolds, M. P. (2015). Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor. Appl. Genet. 128, 353–363. doi: 10.1007/s00122-014-2435-3
Szyjanowicz, P. M., McKinnon, I., Taylor, N. G., Gardiner, J., Jarvis, M. C., and Turner, S. R. (2004). The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J. 37, 730–740. doi: 10.1111/j.1365-313X.2003.02000.x
Tang, Y., Liu, X., Wang, J., Li, M., Wang, Q., Tian, F., et al. (2016). GAPIT Version 2: an enhanced integrated tool for genomic association and prediction. Plant Genome 9. doi: 10.3835/plantgenome2015.11.0120
Team, R. C. (2014). R: A Language and Environment for Statistical Computing. Vienna :R Foundation for Statistical Computing.
Updegraff, D. M. (1969). Semimicro determination of cellulose content in biological materials. Anal. Biochem. 42, 120–124.
Vain, T., Crowell, E. F., Timpano, H., Biot, E., Desprez, T., Mansoori, N., et al. (2014). The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol. 165, 1521–1532. doi: 10.1104/pp.114.241216
Wang, H., Smith, K. P., Combs, E., Blake, T., Horsley, R. D., and Muehlbauer, G. J. (2012). Effect of population size and unbalanced data sets on QTL detection using genome-wide association mapping in barley breeding germplasm. Theor. Appl. Genet. 124, 111–124. doi: 10.1007/s00122-011-1691-8
Keywords: GWAS, cellulose, wheat, cereals, grasses, SNPs, bioenergy, cell wall
Citation: Kaur S, Zhang X, Mohan A, Dong H, Vikram P, Singh S, Zhang Z, Gill KS, Dhugga KS and Singh J (2017) Genome-Wide Association Study Reveals Novel Genes Associated with Culm Cellulose Content in Bread Wheat (Triticum aestivum, L.). Front. Plant Sci. 8:1913. doi: 10.3389/fpls.2017.01913
Received: 24 April 2017; Accepted: 23 October 2017;
Published: 06 November 2017.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Liezhao Liu, Southwest University, ChinaHarsh Raman, New South Wales Department of Primary Industries, Australia
Rogelio Santiago, University of Vigo, Spain
Copyright © 2017 Kaur, Zhang, Mohan, Dong, Vikram, Singh, Zhang, Gill, Dhugga and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kanwarpal S. Dhugga, k.dhugga@cgiar.org
Jaswinder Singh, jaswinder.singh@mcgill.ca
 Simerjeet Kaur1
Simerjeet Kaur1 Amita Mohan
Amita Mohan Prashant Vikram
Prashant Vikram Sukhwinder Singh
Sukhwinder Singh Kulvinder S. Gill
Kulvinder S. Gill Kanwarpal S. Dhugga
Kanwarpal S. Dhugga Jaswinder Singh
Jaswinder Singh