- 1Hubei Collaborative Innovation Centre for Grain Industry and Hubei Key Laboratory of Waterlogging Disaster and Agriculture Use of Wetland, College of Agriculture, Yangtze University, Jingzhou, China
- 2Western Barley Genetics Alliance, Murdoch University, Murdoch, WA, Australia
- 3Department of Genetics and Cell Biology, Yangtze University, Jingzhou, China
- 4Western Australian Agricultural Biotechnology Centre, Murdoch University, Murdoch, WA, Australia
- 5Institute of Nuclear Agricultural Science, Zhejiang University, Hangzhou, China
- 6Agriculture and Food, Department of Primary Industries and Regional Development, South Perth, WA, Australia
Leaf color is an important trait for not only controlling crop yield but also monitoring plant status under temperature stress. In this study, a thermo-inducible chlorophyll-deficient mutant, named V-V-Y, was identified from a gamma-radiated population of the barley variety Vlamingh. The leaves of the mutant were green under normal growing temperature but turned yellowish under high temperature in the glasshouse experiment. The ratio of chlorophyll a and chlorophyll b in the mutant declined much faster in the first 7–9 days under heat treatment. The leaves of V-V-Y turned yellowish but took longer to senesce under heat stress in the field experiment. Genetic analysis indicated that a single nuclear gene controlled the mutant trait. The mutant gene (vvy) was mapped to the long arm of chromosome 4H between SNP markers 1_0269 and 1_1531 with a genetic distance of 2.2 cM and a physical interval of 9.85 Mb. A QTL for grain yield was mapped to the same interval and explained 10.4% of the yield variation with a LOD score of 4. This QTL is coincident with the vvy gene interval that is responsible for the thermo-inducible chlorophyll-deficient trait. Fine mapping, based on the barley reference genome sequence, further narrowed the vvy gene to a physical interval of 0.428 Mb with 11 annotated genes. This is the first report of fine mapping a thermo-inducible chlorophyll-deficient gene in barley.
Introduction
Chlorophyll plays a unique and indispensable role for energy transfer in the reaction centers of photosynthesis (Tanaka and Tanaka, 2011; Lin et al., 2014). There are two types of chlorophyll in plants, chlorophyll a and chlorophyll b. Chlorophyll a is the principal photosynthetic pigment, harvesting energy directly from the sun, while chlorophyll b is an accessory photosynthetic pigment, transferring light energy to chlorophyll a. Chlorophyll content is a crucial trait for crop biomass and grain yield. Chlorophyll biosynthesis and biodegradation pathways are significant biochemical pathways and have received much attention from biochemists (Tanaka and Tanaka, 2007; Hörtensteiner and Kräutler, 2011).
Many types of chlorophyll-deficient mutants have been reported in plants and subdivided into four types based on the physiological mechanisms involved: total chlorophyll increase type, total chlorophyll deficiency type, chlorophyll a deficiency type and chlorophyll b deficiency type (Falbel and Staehelin, 1996). Thermo-sensitive chlorophyll mutants are a particular type of virescence, exhibiting normal or near-normal phenotypes at permissive temperatures, but mutational phenotypes at lower or higher temperatures.
Studies have shown that chlorophyll content is associated with plant heat stress (Havaux and Tardy, 1999; Mohammadi et al., 2009; Jespersen et al., 2016; Sun and Guo, 2016; Rossi et al., 2017), and chlorophyll fluorescence is a key indicator for monitoring the physiological status of plants under temperature stress (Jespersen et al., 2016; Oukarroum et al., 2016) and for screening heat-tolerant cultivars (Ristic et al., 2007; Kalaji et al., 2011; Sharma et al., 2012; Chen et al., 2016). Thus, thermo-sensitive mutants are of particular interest for studying chlorophyll biogenesis in response to temperature cues.
Mutants are key resources in gene cloning, functional analysis, and the elucidation of biochemical processes and pathways. In rice, more than 170 leaf color mutations have been described, and more than 160 leaf color genes have been identified across all 12 chromosomes, of which about 40 genes are directly involved in chlorophyll biosynthesis and/or catabolism (Deng et al., 2014; Zhu et al., 2016). In barley, a large number of mutant phenotypes with full or partial chlorophyll deficiency have been described (Gálová et al., 2000; Franckowiak and Lundqvist, 2012) but only a small number of genes were identified. Fructokinase-1-like (FLN) was identified as a candidate gene for barley stage green-revertible albino (HvSGRA) (Qin et al., 2015). A 95 bp insertion in proto chlorophyllide oxidoreductase (POR) resulted in chlorophyll deficiency in the NYB mutant barley (Liu Z.L. et al., 2008). Recently, genes controlling chlorophyll synthesis have been explored by RNA-seq in barley Alm gene near-isogenic lines (Shmakov et al., 2016). However, the genes responsible for chlorophyll biosynthesis and biodegradation and leaf color formation are largely unknown in barley.
Thermo-sensitive chlorophyll mutants have been reported in Arabidopsis thaliana (Markwell and Osterman, 1992), rice (Dong et al., 2001), maize (Pasini et al., 2005) and barley (Gálová et al., 2000). In rice, five genes participated in the temperature-induced abnormal leaf phenotype regulation have been cloned, including genes OsGluRS (Liu et al., 2007), tscd1 (Liu et al., 2015), tcd5 (LOC_Os05g34040) (Wang Y. et al., 2016) and NOA1 (Yang et al., 2011). In Arabidopsis thaliana, the genes CHLOROPHYLL SYNTHASE (chlg1-1) (Lin et al., 2014) and CHLOROPHYLL DEPHYTYLASE1 (cld1-1) (Lin et al., 2016) responsible for cotyledon-bleaching phenotype have been cloned. Barley chlorophyll mutants have displayed various temperature stress response patterns (Gálová et al., 2000). However, the genes and mechanisms of sense and response to temperature in barley chlorophyll biosynthesis are largely unknown.
Barley, as one of the most important sources for brewing and feeding in the world, is also a model plant for researchers in Triticale species with large genomes. The recent release of the barley genome sequence will accelerate study and cloning of genes relevant to agriculture (Mascher et al., 2017). In this study, a thermo-inducible chlorophyll-deficient mutant (V-V-Y) was identified from the cultivated barley Vlamingh using gamma-radiation treatment. The mutant displayed normal leaf color at permissive temperatures but turned yellowish when the temperature was above 30°C. We conducted genetic analysis of the chlorophyll-deficient mutant gene (vvy) and constructed a genetic map of the vvy gene. New markers were developed to fine mapping the vvy gene based on the barley reference genome sequence and whole-genome shotgun sequences of the two parents. Additionally, quantitative trait loci (QTL) analysis was performed to understand the effect of the mutant gene on yield.
Materials and Methods
Plant Materials
The mutant V-V-Y, which exhibits the thermo-inducible chlorophyll-deficient phenotype, was identified from a 200 gray gamma-radiation treatment of barley cultivar Vlamingh, a malting barley bred by the Department of Agriculture and Food Western Australia. The leaves of the mutant V-V-Y are green at normal temperature, but yellowish at temperatures above 30°C. A double haploid (DH) population with 203 lines was developed by anther culture from the F1 lines of V-V-Y × Buloke. Buloke is an Australian two-rowed malting barley with similar maturity, good agronomic traits, and green leaves.
Glasshouse Experiments
The 203 DH lines, mutant V-V-Y, and barley varieties Buloke and Vlamingh were planted in pots in a glasshouse at the Western Australian State Agricultural Biotechnology Centre. A high-temperature treatment of 30°C for 18 days was applied at seeding and tillering stages. After thermal treatment, the leaf color of each line was observed every 2 days. The DH lines with consistent phenotypes at both stages were used for mapping analysis.
Chlorophyll Content Measurements
To confirm the effect of temperature on chlorophyll content in the mutant V-V-Y, the chlorophyll a and chlorophyll b contents were determined with a spectrophotometer according to the method of Hiscox and Israelstam (1979). Approximately 0.5 g fresh leaves were cut into pieces and homogenized in 7 ml dimethyl sulfoxide (DMSO) and incubated at 65°C for 1 h. The extract was used to measure the absorbance value at 645 and 663 nm with a pure DMSO solution as a control. Dilute 5–10 times, if necessary. Each leaf sample was assayed with three replicates. Chlorophyll a and b contents were calculated as follows:
Chlorophyll a (g/L) = 0.0127A663–0.00269A645
Chlorophyll b (g/L) = 0.0029A663–0.00468A645
The unit of chlorophyll: mg/g fresh weight = (chlorophyll a or chlorophyll b × volume (L) × dilute times)/sample weight (g).
Genetic Linkage Map Construction
Genomic DNA was extracted using the SDS method (Edwards et al., 1991). The Fluidigm SNP genotyping system (Fluidigm, South San Francisco, CA, United States) was used to screen 446 previously designed SNP markers for polymorphism between parents (Close et al., 2009). The selected polymorphic markers between V-V-Y and Buloke were used to acquire genotype data of the DH population via the Fluidigm SNP genotyping system. The genotypic data was collected and inputted into JoinMap 4.1 software for marker linkage group analyses. The multipoint maximum likelihood mapping algorithm was used to calculate genetic distances with a LOD score > 3. The SNP marker linkage map was created using the integrated MapChart program in JoinMap (Voorrips, 2002). The final map was validated according to the barley reference genome sequence (Mascher et al., 2017).
Fine Mapping of vvy Gene and Candidate Gene Analysis
InDel markers were designed in the preliminary mapped region based on sequence alignments of Scope and Vlamingh with the Morex reference genome sequence. Scope was derived from Buloke with a single SNP variation for herbicide tolerance and thus used as the reference sequence for Buloke. The whole-genome shotgun sequences of Vlamingh and Scope were generated in-house and are available by request. The polymorphic InDel markers were used to analyze the genotype of the DH population (Table 1). Genotyping was performed by standard PCR. The PCR products were run on either 2.5% agarose gel or 6% sodium dodecyl sulfate-polyacrylamide gel. The phenotypic data and the genotypic date were combined for mapping of vvy gene according to the above method.

TABLE 1. The polymorphic InDel markers screened from the preliminary mapped region of the vvy gene used for fine mapping.
Information on the candidate genes in the target genetic region was extracted from the new barley genome annotation (Mascher et al., 2017). The amino acid sequence encoded by each candidate gene was searched for homologs using the UniProt database1. Moreover, the reviewed entry hits from UniProt were used as references for the biological functional prediction. Functional domain analyses were performed using the InterPro online tool2.
QTL Analysis for Yield
Vlamingh and its mutant V-V-Y were planted in 1 m long double rows with row spacing of 30 cm at Zhejiang University field stations. The materials were exposed to temperatures over 36°C for 2 days, at the later dough stage, under natural conditions. The yield trial was conducted under typical Mediterranean climate conditions with terminal drought and heat stresses to favor the trait expression. The DH population and parents were planted in 1 × 5 m plots in a randomized complete block design. Only the middle six rows (1 × 3 m) were harvested to evaluate yield. Parental and local barley varieties were used as grid controls for spatial analysis. The yield was adjusted according to the Best Linear Unbiased Prediction (BLUP) (Liu X.Q. et al., 2008). Individual yield data in the V-V-Y × Buloke double haploid population were used for QTL analysis. The software package MapQTL 5.0 (Van Ooijen, 2004) was used to conduct QTL analysis for barley yield after importing the files for genotypes, phenotypes, and genetic maps. Composite interval mapping (CIM) was used for QTL mapping. LOD threshold values declaring the presence of a QTL were estimated by performing whole-genome-wide permutation tests using 1,000 permutations. The QTL map was then generated using MapChart 2.2.
Results
Phenotypic Characterization of the Mutant V-V-Y
The leaves of both V-V-Y and Vlamingh are green at normal temperature (Figure 1A). In the glasshouse at high temperature (30°C), the leaves of V-V-Y started turning yellowish after 5 days and were fully yellow after 12 days treatment at both the seeding stage (Figure 1B) and the tillering stage (Figure 1C). In the field, when high temperatures occurred during the grain-filling stage, the leaves of V-V-Y turned yellowish (Figures 1D,F), while those of the parent Vlamingh turned yellow (Figures 1D,E). The leaves of V-V-Y took longer to senesce than the parent Vlamingh during the grain-filling stage at high temperature (Figures 1E,F).

FIGURE 1. Leaf colors in the mutant V-V-Y and the wild parent Vlamingh (VL) in the glasshouse and field. (A) leaf colors of VL and V-V-Y at the seeding stage at normal temperature, (B) leaf colors of VL and V-V-Y after 12 days of high-temperature treatment (30°C) at the seeding stage in the glasshouse, (C) leaf colors of VL and V-V-Y after 12 days of high-temperature treatment (30°C) at the tillering stage in the glasshouse, (D) plot performance of VL and V-V-Y at the grain-filling stage in the field under high temperature, (E) leaf color of VL at the grain-filling stage in the field under high temperature, (F) leaf color of V-V-Y at the grain-filling stage in the field under high temperature.
Chlorophyll Content in V-V-Y under High Temperature
Under the high-temperature treatment, chlorophyll a content in Vlamingh declined by 0.011 mg g-1 leaf per day (Figure 2A), which showed a good linear relationship with the treatment time (y = -0.0279x + 0.5468, R2 = 0.9869, “y” is the chlorophyll a content in Vlamingh, “x” is the treatment day). In V-V-Y, the chlorophyll a content declined by 0.017 mg g-1 leaf per day, on average; notably, the rate of decline in the first 7 days was five times higher than the last 11 days (Figure 2B). Under high-temperature treatment, the average rate of decline in chlorophyll b content in V-V-Y was about 1.6 times higher than in Vlamingh (Figure 2B) and markedly higher in the first 9 days (Figure 2B).
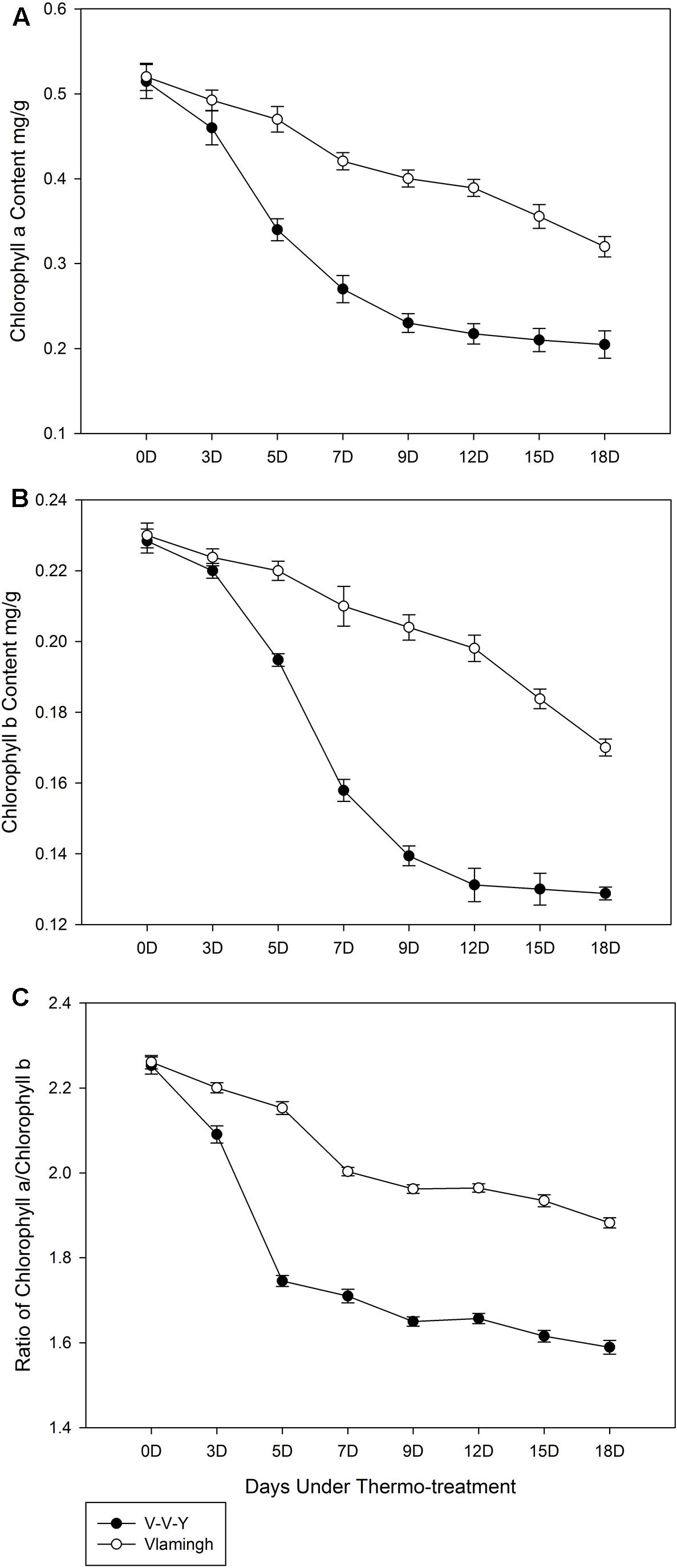
FIGURE 2. Chlorophyll contents of V-V-Y and Vlamingh at the tillering stage in the glasshouse high-temperature treatment (30°C). (A) chlorophyll a contents in V-V-Y and Vlamingh after 3, 5, 7, 9, 12, 15, and 18 days of high temperature, (B) chlorophyll b contents in V-V-Y and Vlamingh after 3, 5, 7, 9, 12, 15, and 18 days of high temperature, (C) the ratio of chlorophyll a/chlorophyll b in V-V-Y and Vlamingh after 3, 5, 7, 9, 12, 15, and 18 days of high temperature.
The ratio of chlorophyll a/chlorophyll b in Vlamingh remained relatively stable with a slight reduction throughout the high-temperature treatment, while that of V-V-Y declined more sharply in the first 5 days of treatment and remained relatively stable with a slight reduction over the next 13 days (Figure 2C). V-V-Y had a lower chlorophyll a/chlorophyll b ratio than Vlamingh throughout the high-temperature treatment, indicating that chlorophyll a declined faster than chlorophyll b in V-V-Y.
Inheritance and Preliminary Mapping of vvy Gene
In the V-V-Y × Buloke DH population, the high-temperature treatments in the glasshouse identified 100 individuals with the mutant phenotype of thermo-inducible yellowish leaf color and 103 individuals with the wild phenotype of green leaf color under high-temperature stress. Chi-square analysis showed that the ratio of mutant phenotype type: wild phenotype type fit the 1: 1 segregation ratio (χ2 = 0.0225, P > 0.90), revealing that the thermo-inducible yellowish leaf color phenotype is governed by a single mutant gene in V-V-Y.
A total of 446 pairs SNP markers were tested for polymorphism between V-V-Y and Buloke. Ninety-six polymorphic SNP markers were selected for genetic map construction. The total length of the genetic linkage map was 1033 cM with an average distance of 11.3 cM between two markers (Figure 3). The mutant gene (vvy) controlling the thermo-inducible yellowish leaf color phenotype in V-V-Y was mapped to the terminal region of the long arm of chromosome 4H between the SNP markers 1_0269 and 1_1531 with a genetic distance of 2.2 cM. These two molecular markers were located at 636.35 and 646.20 Mb of chromosome 4H in the new barley reference genome sequence (Mascher et al., 2017). The preliminary physical region of the vvy gene was mapped to a 9.85 Mb region.
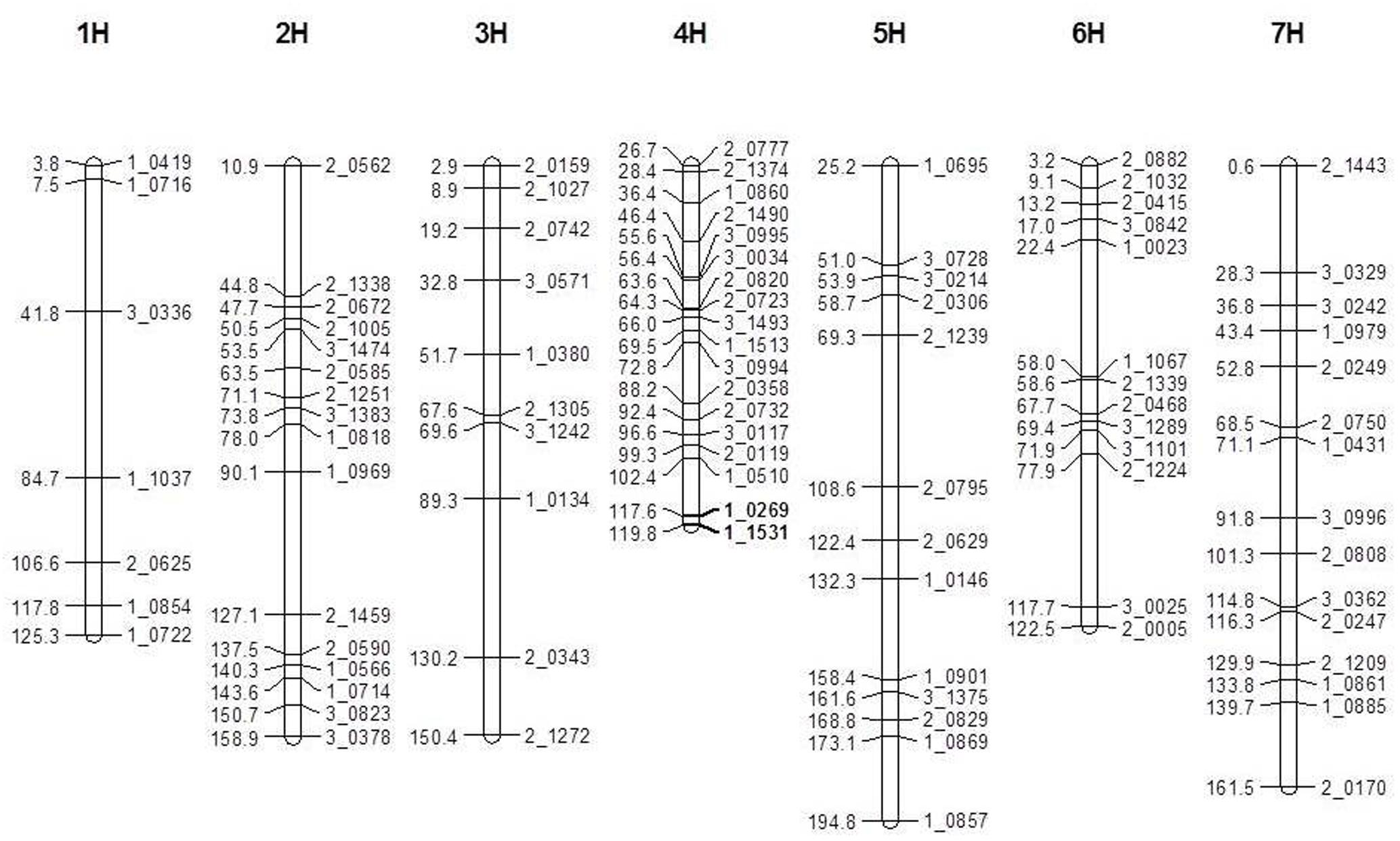
FIGURE 3. Linkage map of the V-V-Y × Buloke DH population on the barley 7 chromosomes and preliminary mapping interval of vvy gene. Marker locus names are to the right of each linkage group and distances (Kosambi cM) are to the left of each marker interval. Solid black bars indicate the preliminary mapping interval of vvy gene.
Fine Mapping of vvy Gene and Candidate Gene Analysis
To fine map the vvy gene, 117 InDel markers were designed within the preliminary mapped region. Polymorphism was screened between the parent lines, which yielded 12 polymorphic markers (Table 1). These markers were used to genotype the V-V-Y × Buloke DH population. The linkage and fine mapping results showed that the vvy gene was located between marker WR12 (640.660333 Mb) and marker WR25 (641.088341 Mb), with a physical interval of 0.428 Mb (Figures 4A,B).
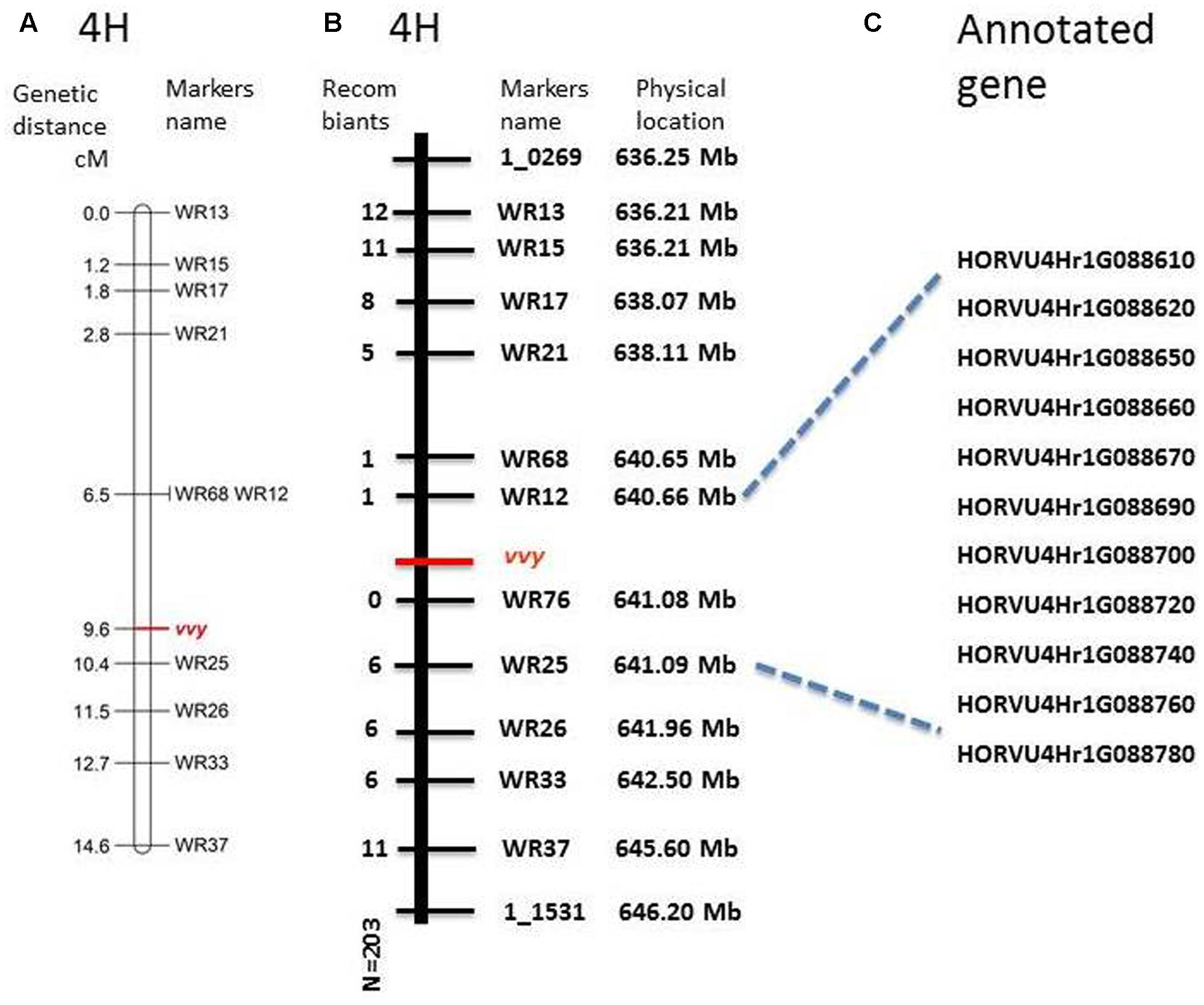
FIGURE 4. Fine mapping of vvy gene on barley chromosome 4H and the candidate genes. (A) Linkage map of vvy gene, marker locus names are to the right of each linkage group and distances (Kosambi cM) are to the left of each marker interval, (B) physical mapping, marker names and the physical location on the right, the number of detected recombinant individuals on the left, (C) predicated candidate genes.
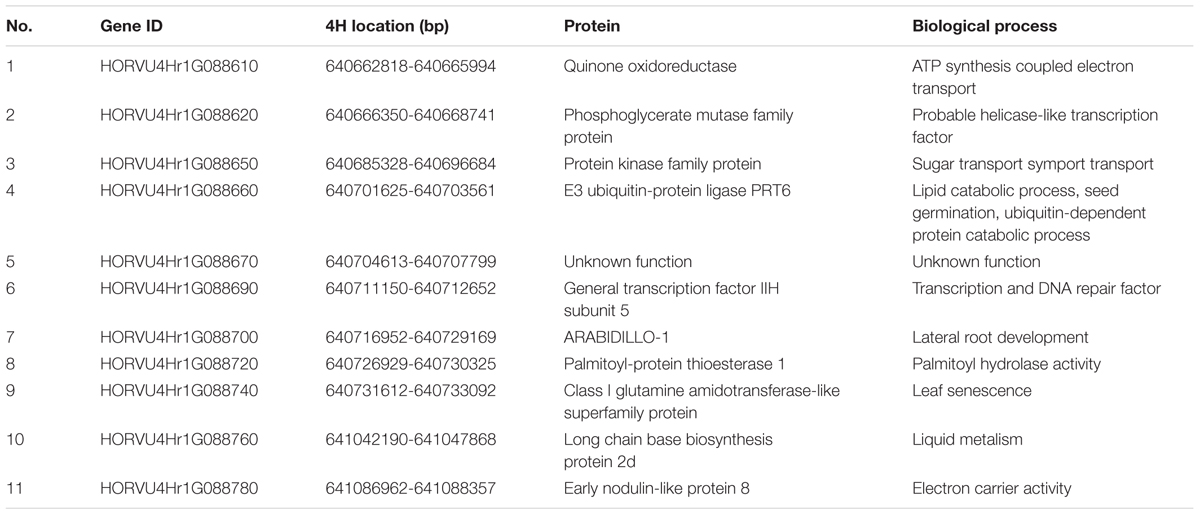
According to the most recent barley genome annotation (Mascher et al., 2017), 11 candidate genes are located in this region (Figure 4C and Table 2). The predicted protein function and biological process for each candidate gene are listed in Table 2. Of these 11 genes, HORVU4Hr1G88650 is the kinase family protein, HORVU4Hr1G88690 is the transcription factor, HORVU4Hr1G88670 has unknown function, HORVU4Hr1G88700 controls lateral root development, and the other seven participate in several biochemical metabolism processes (Table 2). It should be noted that HORVU4Hr1G88610 was predicted to target the chloroplast3.
QTL Analysis for Yield
Individual yield data from the V-V-Y × Buloke DH population and the parental lines were collected. The average yield in the mutant V-V-Y and Vlamingh was 3755 ± 246.7 and 4194 ± 249.6 kg ha-1. The average yield of Buloke was 4340 ± 254 kg ha-1. The yield of the DH lines varied from 470 to 4953 kg ha-1. QTL analysis was performed to explore the genetic loci that control yield. A QTL located on chromosome 4H linked with marker WR33 explained 10.4% of the yield variation with LOD = 4 (Figure 5). This QTL is coincident with the vvy gene interval responsible for the thermo-inducible chlorophyll-deficient trait.
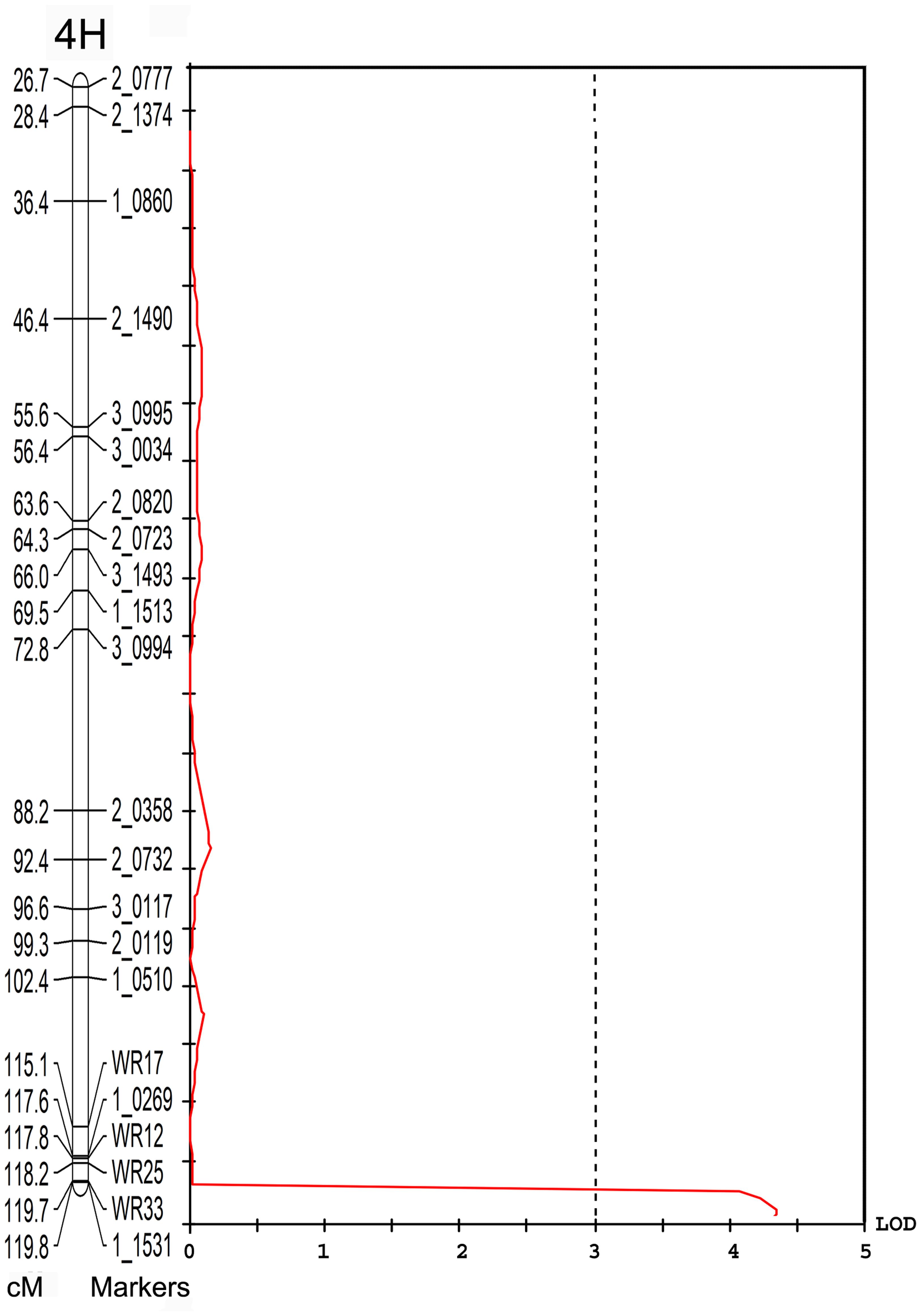
FIGURE 5. Quantitative trait loci associated with the 4H marker WR33 (LOD = 4) for grain yield in the field in the V-V-Y × Buloke DH population. The X-axis is the LOD score and the Y-axis is the markers and genetic distance (Kosambi cM) of chromosome 4H.
Discussion
Various leaf color mutants have been described in barley (Gálová et al., 2000) but only a few genes controlling leaf color have been well characterized (Liu Z.L. et al., 2008; Qin et al., 2015; Shmakov et al., 2016). In this study, we showed that V-V-Y is a thermo-inducible chlorophyll-deficient mutant at the seeding and tillering stages. The genetic analysis showed that the thermo-inducible chlorophyll-deficient trait is controlled by a single mutant gene (vvy). Preliminary mapping showed that the vvy gene is located on the distal region of the long arm of chromosome 4H, which was further narrowed to a physical distance of 0.428 Mb by fine mapping, covering 11 annotated genes (Mascher et al., 2017). This is the first report of fine mapping a thermo-inducible chlorophyll-deficient gene in barley.
Several temperature-sensitive chlorophyll synthesis and regulation genes have been cloned. The tcd5 (LOC_Os05g34040) gene, encoding a monooxygenase, is responsible for albino leaves at low temperatures in rice (Wang Y. et al., 2016). The NOA1 gene (LOC_Os02g0104700) encodes a circularly permuted GTPase (cGTPase) that can regulate chlorophyll synthesis in a temperature-dependent manner (Yang et al., 2011). The OsGluRS gene on chromosome 2, encoding glutamyl-Trna synthetase, is responsible for high-temperature-induced yellow leaves in rice (Liu et al., 2007). Recently, the thermo-sensitive chlorophyll deficit 1 (tscd1) gene was mapped in a 34.95 kb region on the long arm of chromosome 2, which controls low-temperature-induced yellow leaves at the seeding and tillering stages in rice (Liu et al., 2015). In Arabidopsis thaliana, missense mutations in CHLOROPHYLL SYNTHASE (chlg1-1) (Lin et al., 2014) and CHLOROPHYLL DEPHYTYLASE1 (cld1-1) (Lin et al., 2016) were identified as responsible for the light-dependent, heat-induced cotyledon-bleaching phenotype. However, of the 11 candidate genes in this study, none was an orthologue of the cloned temperature-dependent chlorophyll biosynthesis and regulation gene.
The chlorophyll biosynthetic deficiency is one of the main reasons for low chlorophyll content in many plants (Zhou Y. et al., 2013). Yellow–green leaf is a type of mutant in rice; ygl1 (Wu et al., 2007), ygl2 (Chen et al., 2013), ygl3 (Tian et al., 2013), ygl6 (Shi et al., 2015), ygl7 (allele of the OsChlD gene) (Deng et al., 2014), ygl8 (Zhu et al., 2016), ygl9 (Wang Z.-W. et al., 2016), and ygl138 (Zhang et al., 2013) have been cloned, and most of these genes are involved in the chlorophyll biosynthesis process. However, none of the candidate genes of vvy are orthologous of the cloned rice yellow–green leaf genes.
The chlorophyll biosynthesis pathways have been well characterized in higher plants (Tanaka and Tanaka, 2007; Hörtensteiner and Kräutler, 2011). At least 27 genes covering 15 steps are involved in chlorophyll biosynthesis from glutamyl-Trna to chlorophyll a and chlorophyll b (Beale, 2005; Nagata et al., 2005). However, none of the candidate genes identified in this study was homologous to these 27 genes. Low chlorophyll contents might also result from deficient signal transduction between the nucleus and chloroplast (Zhou K. et al., 2013), restrained heme feedback (Terry and Kendrick, 1999), impaired synthesis and importation of chloroplast proteins (Gothandam et al., 2005; Miura et al., 2007) or harmful photooxidation (Richter et al., 2013). However, no known structural or transcription factor gene involved in the chlorophyll pathway was found in the target region. It is reasonable to speculate that a new gene for chlorophyll biosynthesis and regulation is involved in V-V-Y. Expression analysis of the annotated genes in V-V-Y and Vlamingh may provide more information for identification of the candidate gene, especially how these genes respond to high temperature treatment.
Heat stress during the reproductive and grain-filling phases is a severe threat to barley and wheat production. Chloroplasts, as the sensors to heat stress, are the major target of thermal damage in plants (Sun and Guo, 2016). Studies in barley (Havaux and Tardy, 1999), wheat (Mohammadi et al., 2009) and bentgrass (Jespersen et al., 2016; Rossi et al., 2017) indicated that chlorophyll losses with limited reductions in photosynthesis might be an adaptive response to high light and heat stress. The rice tscd1 mutant can mature normally without any significant effects on key related-yield traits (Liu et al., 2015). In this study, the vvy gene interval overlapped with a QTL for barley yield under typical Mediterranean climate conditions with terminal drought and heat stresses. Heat stress can severely damage photosystem II (PSII), the most heat-sensitive photosynthetic apparatus within chloroplast thylakoid membrane protein complexes involved in photosynthetic electron transfer and ATP (Sun and Guo, 2016). Thus, we propose that the autonomic deficient chlorophyll biosynthesis under temperature stress might be a protective mechanism for crops to develop an adaptive response mechanism to temperature stress.
In most cases, leaf color mutants will retard plant growth (Wu et al., 2007; Jiang et al., 2012; Qin et al., 2015). However, some mutants can mature normally without any significant effect on yield-related traits (Chen et al., 2007; Liu et al., 2015). In the present study, the mutant V-V-Y leaves took longer to senesce than its wild parent under short period (2 days) of extreme high temperature (over 36°C) and thus maintained higher grain plumpness. The thermo-sensitive mutant may provide an ideal system for exploring why the reduction in chlorophyll content is an adaptive response to heat stress. It may also provide an opportunity to develop dynamic varieties that are temperature-sensitive to chlorophyll content with the potential to improve crop heat tolerance. However, the mutant gene could not provide yield advantage under a typical Mediterranean climate conditions with constant terminal drought and heat stresses. Thus, further research is required to understand the impact of the mutant gene on other yield components under both normal and heat stress conditions.
Author Contributions
Project concept and design: CL, YX, WZ, and FY. Mutant identification: DW and CL. Population development: SB, SW, and X-QZ. Field trial: SB, SW, and X-QZ. Genotyping: RW, FY, X-QZ, CT, and CL. Wrote the MS: RW, YX, CL, and WZ.
Funding
The project funds were provided by the National Key R&D Program of China (2016YFD0102101), Australian Grain Research and Development Corporation (UMU00049), and National Natural Science Foundation of China (31501309, 31201212).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Beale, S. I. (2005). Green genes gleaned. Trends Plant Sci. 10, 309–312. doi: 10.1016/j.tplants.2005.05.005
Chen, H., Cheng, Z., Ma, X., Wu, H., Liu, Y., Zhou, K., et al. (2013). A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice. Plant Cell Rep. 32, 1855–1867. doi: 10.1007/s00299-013-1498-y
Chen, S., Yang, J., Zhang, M., Strasser, R. J., and Qiang, S. (2016). Classification and characteristics of heat tolerance in Ageratina adenophora populations using fast chlorophyll a fluorescence rise O-J-I-P. Environ. Exp. Bot. 122, 126–140. doi: 10.1016/j.envexpbot.2015.09.011
Chen, T., Zhang, Y., Zhao, L., Zhu, Z., Lin, J., Zhang, S., et al. (2007). Physiological character and gene mapping in a new green-revertible albino mutant in rice. J. Genet. Genomics 34, 331–338. doi: 10.1016/S1673-8527(07)60035-6
Close, T. J., Bhat, P. R., Lonardi, S., Wu, Y., Rostoks, N., Ramsay, L., et al. (2009). Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10:582. doi: 10.1186/1471-2164-10-582
Deng, X.-J., Zhang, H.-Q., Wang, Y., He, F., Liu, J.-L., Xiao, X., et al. (2014). Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLOS ONE 9:e99564. doi: 10.1371/journal.pone.0099564
Dong, Y., Dong, W., Shi, S., and Jin, Q. (2001). Identification and genetic analysis of a thermo-sensitive seedling-colour mutant in rice (Oryza sativa L.). Breed. Sci. 51, 1–4. doi: 10.1270/jsbbs.51.1
Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19:1349. doi: 10.1093/nar/19.6.1349
Falbel, T. G., and Staehelin, L. A. (1996). Partial blocks in the early steps of the chlorophyll synthesis pathway: a common feature of chlorophyll b-deficient mutants. Physiol. Plant. 97, 311–320. doi: 10.1034/j.1399-3054.1996.970214.x
Franckowiak, J. D., and Lundqvist, U. (2012). Descriptions of barley genetic stocks for 2012. Barley Genet. Newsl. 42, 36–173.
Gálová, E., Böhmová, B., and Ševčovičová, A. (2000). Analysis of some barley chlorophyll mutants and their response to temperature stress. Photosynthetica 38, 29–35. doi: 10.1023/A:1026735605804
Gothandam, K. M., Kim, E.-S., Cho, H., and Chung, Y.-Y. (2005). OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol. Biol. 58, 421–433. doi: 10.1007/s11103-005-5702-5
Havaux, M., and Tardy, F. (1999). Loss of chlorophyll with limited reduction of photosynthesis as an adaptive response of Syrian barley landraces to high-light and heat stress. Funct. Plant Biol. 26, 569–578. doi: 10.1071/PP99046
Hiscox, J. T., and Israelstam, G. (1979). A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 57, 1332–1334. doi: 10.1139/b79-163
Hörtensteiner, S., and Kräutler, B. (2011). Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 1807, 977–988. doi: 10.1016/j.bbabio.2010.12.007
Jespersen, D., Zhang, J., and Huang, B. (2016). Chlorophyll loss associated with heat-induced senescence in bentgrass. Plant Sci. 249, 1–12. doi: 10.1016/j.plantsci.2016.04.016
Jiang, S., Zhang, X., Zhang, F., Xu, Z., Chen, W., and Li, Y. (2012). Identification and fine mapping of qCTH4, a quantitative trait loci controlling the chlorophyll content from tillering to heading in rice (Oryza sativa L.). J. Hered. 103, 720–726. doi: 10.1093/jhered/ess041
Kalaji, H. M., Bosa, K., Kościelniak, J., and Hossain, Z. (2011). Chlorophyll a fluorescence—a useful tool for the early detection of temperature stress in spring barley (Hordeum vulgare L.). OMICS 15, 925–934. doi: 10.1089/omi.2011.0070
Lin, Y. P., Lee, T. Y., Tanaka, A., and Charng, Y. Y. (2014). Analysis of an Arabidopsis heat-sensitive mutant reveals that chlorophyll synthase is involved in reutilization of chlorophyllide during chlorophyll turnover. Plant J. 80, 14–26. doi: 10.1111/tpj.12611
Lin, Y. P., Wu, M. C., and Charng, Y. Y. (2016). Identification of a chlorophyll dephytylase involved in chlorophyll turnover in Arabidopsis. Plant Cell 28, 2974–2990. doi: 10.1105/tpc.16.00478
Liu, J., Wang, J., Yao, X., Zhang, Y., Li, J., Wang, X., et al. (2015). Characterization and fine mapping of thermo-sensitive chlorophyll deficit mutant1 in rice (Oryza sativa L.). Breed. Sci. 65, 161–169. doi: 10.1270/jsbbs.65.161
Liu, W., Fu, Y., Hu, G., Si, H., Zhu, L., Wu, C., et al. (2007). Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.). Planta 226, 785–795. doi: 10.1007/s00425-007-0525-z
Liu, X.-Q., Rong, J.-Y., and Liu, X.-Y. (2008). Best linear unbiased prediction for linear combinations in general mixed linear models. J. Multiv. Anal. 99, 1503–1517. doi: 10.1016/j.jmva.2008.01.004
Liu, Z.-L., Yuan, S., Liu, W.-J., Du, J.-B., Tian, W.-J., Luo, M.-H., et al. (2008). Mutation mechanism of chlorophyll-less barley mutant NYB. Photosynthetica 46, 73–78. doi: 10.1007/s11099-008-0013-0
Markwell, J., and Osterman, J. C. (1992). Occurrence of temperature-sensitive phenotypic plasticity in chlorophyll-deficient mutants of Arabidopsis thaliana. Plant Physiol. 98, 392–394. doi: 10.1104/pp.98.1.392
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Miura, E., Kato, Y., Matsushima, R., Albrecht, V., Laalami, S., and Sakamoto, W. (2007). The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19, 1313–1328. doi: 10.1105/tpc.106.049270
Mohammadi, M., Karimizadeh, R. A., and Naghavi, M. R. (2009). Selection of bread wheat genotypes against heat and drought tolerance based on chlorophyll content and stem reserves. J. Agric. Soc. Sci. 5, 119–122.
Nagata, N., Tanaka, R., Satoh, S., and Tanaka, A. (2005). Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17, 233–240. doi: 10.1105/tpc.104.027276
Oukarroum, A., El Madidi, S., and Strasser, R. J. (2016). Differential heat sensitivity index in barley cultivars (Hordeum vulgare L.) monitored by chlorophyll a fluorescence OKJIP. Plant Physiol. Biochem. 105, 102–108. doi: 10.1016/j.plaphy.2016.04.015
Pasini, L., Bruschini, S., Bertoli, A., Mazza, R., Fracheboud, Y., and Marocco, A. (2005). Photosynthetic performance of cold-sensitive mutants of maize at low temperature. Physiol. Plant. 124, 362–370. doi: 10.1111/j.1399-3054.2005.00522.x
Qin, D., Dong, J., Xu, F., Guo, G., Ge, S., Xu, Q., et al. (2015). Characterization and fine mapping of a novel barley stage green-revertible albino gene (HvSGRA) by bulked segregant analysis based on SSR assay and specific length amplified fragment sequencing. BMC Genomics 16:838. doi: 10.1186/s12864-015-2015-1
Richter, A. S., Peter, E., Rothbart, M., Schlicke, H., Toivola, J., Rintamäki, E., et al. (2013). Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant Physiol. 162, 63–73. doi: 10.1104/pp.113.217141
Ristic, Z., Bukovnik, U., and Prasad, P. V. (2007). Correlation between heat stability of thylakoid membranes and loss of chlorophyll in winter wheat under heat stress. Crop Sci. 47, 2067–2073. doi: 10.2135/cropsci2006.10.0674
Rossi, S., Burgess, P., Jespersen, D., and Huang, B. (2017). Heat-induced leaf senescence associated with Chlorophyll metabolism in Bentgrass lines differing in heat tolerance. Crop Sci. 57, 169–178. doi: 10.2135/cropsci2016.06.0542
Sharma, D. K., Andersen, S. B., Ottosen, C.-O., and Rosenqvist, E. (2012). Phenotyping of wheat cultivars for heat tolerance using chlorophyll a fluorescence. Funct. Plant Biol. 39, 936–947. doi: 10.1071/FP12100
Shi, J., Wang, Y., Guo, S., Ma, L., Wang, Z., Zhu, X., et al. (2015). Molecular mapping and candidate gene analysis of a yellow-green leaf 6(ygl6) mutant in rice. Crop Sci. 55, 669–680. doi: 10.2135/cropsci2014.07.0483
Shmakov, N. A., Vasiliev, G. V., Shatskaya, N. V., Doroshkov, A. V., Gordeeva, E. I., Afonnikov, D. A., et al. (2016). Identification of nuclear genes controlling chlorophyll synthesis in barley by RNA-seq. BMC Plant Biol. 16:119. doi: 10.1186/s12870-016-0926-x
Sun, A. Z., and Guo, F. Q. (2016). Chloroplast retrograde regulation of heat stress responses in plants. Front. Plant Sci. 7:398. doi: 10.3389/fpls.2016.00398
Tanaka, R., and Tanaka, A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346. doi: 10.1146/annurev.arplant.57.032905.105448
Tanaka, R., and Tanaka, A. (2011). Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim. Biophys. Acta. 1807, 968–976. doi: 10.1016/j.bbabio.2011.01.002
Terry, M. J., and Kendrick, R. E. (1999). Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 119, 143–152. doi: 10.1104/pp.119.1.143
Tian, X., Ling, Y., Fang, L., Du, P., Sang, X., Zhao, F., et al. (2013). Gene cloning and functional analysis of yellow green leaf3 (ygl3) gene during the whole-plant growth stage in rice. Genes Genom. 35, 87–93. doi: 10.1007/s13258-013-0069-5
Van Ooijen, J. (2004). MapQTL® 5. Software for the Mapping of Quantitative Trait Loci in Experimental Populations. Wageningen: Kyazma BV.
Voorrips, R. E. (2002). MapChart software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, Y., Zhang, J., Shi, X., Peng, Y., Li, P., Lin, D., et al. (2016). Temperature-sensitive albino gene TCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. J. Exp. Bot. 67, 5187–5202. doi: 10.1093/jxb/erw287
Wang, Z.-W., Zhang, T.-Q., Xing, Y.-D., Zeng, X.-Q., Wang, L., Liu, Z.-X., et al. (2016). YGL9, encoding the putative chloroplast signal recognition particle 43 kDa protein in rice, is involved in chloroplast development. J. Integr. Agric. 15, 944–953. doi: 10.1016/S2095-3119(15)61310-7
Wu, Z., Zhang, X., He, B., Diao, L., Sheng, S., Wang, J., et al. (2007). A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 145, 29–40. doi: 10.1104/pp.107.100321
Yang, Q., He, H., Li, H., Tian, H., Zhang, J., Zhai, L., et al. (2011). NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and rubisco formation in rice. PLOS ONE 6:e20015. doi: 10.1371/journal.pone.0020015
Zhang, F., Luo, X., Hu, B., Wan, Y., and Xie, J. (2013). YGL138 (t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice 6:7. doi: 10.1186/1939-8433-6-7
Zhou, K., Ren, Y., Lv, J., Wang, Y., Liu, F., Zhou, F., et al. (2013). Young leaf chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta 237, 279–292. doi: 10.1007/s00425-012-1756-1
Zhou, Y., Gong, Z., Yang, Z., Yuan, Y., Zhu, J., Wang, M., et al. (2013). Mutation of the light-induced yellow leaf 1 gene, which encodes a geranylgeranyl reductase, affects chlorophyll biosynthesis and light sensitivity in rice. PLOS ONE 8:e75299. doi: 10.1371/journal.pone.0075299
Keywords: barley, thermo-inducible chlorophyll-deficient mutant, fine mapping, QTL, vvy gene
Citation: Wang R, Yang F, Zhang X-Q, Wu D, Tan C, Westcott S, Broughton S, Li C, Zhang W and Xu Y (2017) Characterization of a Thermo-Inducible Chlorophyll-Deficient Mutant in Barley. Front. Plant Sci. 8:1936. doi: 10.3389/fpls.2017.01936
Received: 06 October 2017; Accepted: 27 October 2017;
Published: 14 November 2017.
Edited by:
Guijun Yan, University of Western Australia, AustraliaReviewed by:
Xuechen Zhang, New South Wales Department of Primary Industries, AustraliaMeixue Zhou, University of Tasmania, Australia
Copyright © 2017 Wang, Yang, Zhang, Wu, Tan, Westcott, Broughton, Li, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhao Xu, xyh09@yangtzeu.edu.cn Wenying Zhang, wyzhang@yangtzeu.edu.cn Chengdao Li, c.li@murdoch.edu.au
†These authors have contributed equally to this work.
 Rong Wang1,2†
Rong Wang1,2† Sue Broughton
Sue Broughton Chengdao Li
Chengdao Li Wenying Zhang
Wenying Zhang Yanhao Xu
Yanhao Xu