- 1Rice Research Institute of Sichuan Agricultural University, Chengdu, China
- 2Crop Research Institute, Sichuan Academy of Agricultural Sciences, Chengdu, China
- 3State Key Laboratory of Hybrid Rice, Sichuan Agricultural University, Chengdu, China
Anther and pollen development are crucial processes of plant male reproduction. Although a number of genes involved in these processes have been identified, the regulatory networks of pollen and anther development are still unclear. EMBRYONIC FLOWER 2b (OsEMF2b) is important for rice development. Its biological function in floral organ, flowering time and meristem determinacy have been well-studied, but its role, if only, on male reproduction is still unknown, because null mutants of OsEMF2b barely have anthers. In this study, we identified a weak allele of OsEMF2b, osemf2b-4. The T-DNA insertion was located in the promoter region of OsEMF2b, and OsEMF2b expression was significantly decreased in osemf2b-4. The osemf2b-4 mutant exhibited much more normal anthers than null mutants of OsEMF2b, and also showed defective floret development similar to null mutants. Cytological analysis showed various defects of anther wall and pollen development in osemf2b-4, such as slightly or extremely enlarged tapetum, irregular or normal morphology microspores, and partial or complete sterility. OsEMF2b was highly expressed in tapetum and microspores, and the protein was localized in the nucleus. The expression of 15 genes essential for anther and pollen development was investigated in both WT and osemf2b-4. Fourteen genes including GAMYB was up-regulated, and only PTC1 was down-regulated in osemf2b-4. This suggests that up-regulated GAMYB and down-regulated PTC1 might contribute to the defective anther and pollen development in osemf2b-4. Overall, our work suggests that OsEMF2b plays an essential role during post-meiotic anther and pollen development.
Introduction
Male reproductive development in plants is a complex biological process involving coordinated differentiation of anthers and the generation of gametophytes, then anther dehiscence and subsequently pollination. The anther usually consists of gametophytes at the center, surrounded by the tapetum and the anther wall with the middle cell layer, endothecium, and outer epidermis (Goldberg et al., 1993). During later anther development, major processes include programmed cell death (PCD) of the tapetum, and of the anther wall layers to release mature pollen. These highly coordinated processes require precise fine-tuning of gene regulatory networks such as transcriptional and epigenetic regulation. For example, histone H2B monoubiquitination mediated by HISTONE MONOUBIQUITINATION1 (OsHUB1) and OsHUB2 is involved in anther development by directly regulating tapetum degradation-related genes such as the transcription factor UNDEVELOPMENT TAPETUM1 (UDT1) (Cao et al., 2015). The chromatin protein MALE MEIOCYTE DEATH1 (MMD1) binds to H3K4me2 via the plant homeo domain (PHD) finger domain and this interaction is critical for function during Arabidopsis male meiosis (Andreuzza et al., 2015). SET DOMAIN GROUP 4 (SDG4) functions in the maintenance of methylated histone H3K4 and K36 levels in the mature pollen grain (Cartagena et al., 2008). SDG2 mediates H3K4me3 deposition at SPOROCYTELESS/NOZZLE (SPL/NZZ) and MALE STERILITY1 (MS1) for transcriptional activation, which are essential for differentiation of anther walls and pollen development in Arabidopsis (Berr et al., 2010). However, the molecular mechanisms regulating gene expression with histone methylations in gametophyte development remain elusive.
Polycomb group complexes (PcG) are conserved in molecular function and composition between Drosophila, mammals and plants, and regulate development through epigenetic control of gene expression via chromatin remodeling (Luo et al., 2009). One of the core polycomb repressive complexes, POLYCOMB REPRESSIVE COMPLEX2 (PRC2) is mainly responsible for tri-methylation of lysine 27 of histone H3, a hallmark of silenced genes in the euchromatin in Arabidopsis (Zhang et al., 2007). The EMF2-PRC2 complex composed of EMBRYONIC FLOWER2 (EMF2), FERTILIZATIONINDEPENDENT ENDOSPERM (FIE), MULTICOPY SUPPRESSOR OF IRA1 (MSI1), and CURLY LEAF/SWINGER (CLF/SWN) has been identified in Arabidopsis (Kohler and Villar, 2008). Loss of EMF2 causes direct flowering at the heading stage, passing vegetative shoot growth, but generates abnormal flower organs, and sterile florets (Moon et al., 2003). The EMF2 complex is not only involved in the transition from the vegetative to reproductive phase and flower organ development (Chanvivattana et al., 2004), but is also involved in seed development and stress response, via repression of the target genes such as ABSCISIC ACID INSENSITIVE3 (ABI3) (Kim et al., 2010).
Rice OsEMF2a and OsEMF2b are two paralogs of Arabidopsis EMF2, which are expressed in all tissues (Luo et al., 2009). The T-DNA insertions of OsEMF2b exhibited abnormal flowering time and defective flower organs (Luo et al., 2009; Conrad et al., 2014; Xie et al., 2014), although no flowering phenotype was observed in rice when the expression of OsEMF2a was enhanced (Li K. et al., 2006). OsEMF2b physically interacted with rice VERNALIZATION INSENSITIVE3-LIKE2 (OsVIL2) and controlled late flowering by increasing expression of LEAFY COTYEDON 2 AND FUSCA 3-LIKE1 (OsLFL1) and decreasing expression of EARLY HEADING DATE1 (Ehd1) (Yang et al., 2013). OsEMF2b also regulated meristem determinacy through modulating the expression of E-function homeotic genes such as OsMADS34 and the B-function gene OsMADS4, via H3K27me3 repression (Conrad et al., 2014; Xie et al., 2014). The null mutants of OsEMF2b exhibited very few anthers, therefore, it was difficult to investigate its molecular function during anther and pollen development. Here, we identify a weak function allele of OsEMF2b, osemf2b-4. The osemf2b-4 mutant exhibits much more normal anthers than the null mutants of OsEMF2b. Cytological analysis shows defective anther wall and pollen development. OsEMF2b is highly expressed in anthers, especially in the microspores, tapetum and connectives of anther. OsEMF2b is localized in nucleus, and required for fine-tuning the expression of 15 genes which are essential for anther and pollen development. Down-regulated PTC1 and up-regulated GAMYB possibly contribute to defective anther and pollen development in osemf2b-4. Our results suggest that OsEMF2b plays an essential role in the regulation of anther and pollen development gene through modulating the expression of genes involved in that process.
Materials and Methods
Plant Growth Conditions and Genotyping
Lines PFG_K-03014.R of rice (Oryza sativa japonica cv. KitaaKe), which carries a T-DNA insertion in LOC_Os09g13630 (OsEMF2b), was obtained from RiceGE1. All plants were grown in rice paddies at Wenjiang, Sichuan and Lingshui, Hainan, where represent natural long-day and short-day conditions, respectively. Genotypes were determined by PCR using a pair of gene-specific primers listed in Supplementary Table S2.
Characterization of Anther and Pollen Phenotypes
For pollen viability, pollen was stained using I2-KI solution and observed under a Zeiss Imager A2 microscope. The panicle photograph was taken using the Canon EOS 60D digital camera and the spikelets were observed using a Zeiss Discovery V20 microscope. Anther development was classified into three stages as in Feng et al. (2000) and Zhang and Wilson (2009) and semi-thin sections were prepared as described (Qin et al., 2013). Briefly, spikelets at various developmental stages were fixed in 3% (v/v) glutaraldehyde in 0.2 M sodium phosphate buffer (pH 7.0), then dehydrated in a graded series of ethanol solutions, embedded in Technovit 7100 resin (Hereaus Kulzer, Germany), polymerized at 45°C, and sectioned at 2 μm thickness using a microtome (Leica RM2235, Germany). The sections were stained with 0.25% (w/v) toluidine blue (Sigma) and photographed using a Nikon E600 microscope.
RNA in Situ Hybridization
Wild type spikelets of different developmental stages were fixed in 5% acetic acid, 50% ethanol, and 3.7% formaldehyde in DEPC water for 16 h at 4°C, then dehydrated through an ethanol series, embedded in Paraplast Plus (Sigma), and sectioned at 8 μm using a microtome (Leica RM2235, Germany). A gene-specific region of OsEMF2b was amplified for probe preparation using OsEMF2b specific primers, and cloned into a pBluescript vector (TransGen Biotech, China), then digested with EcoRI or BamHI and transcribed with T7 RNA polymerase using the DIG RNA labeling kit (Roche), for both antisense and sense probes. RNA hybridization and probe detection were performed as described (Zhu et al., 2011).
RNA Isolation and Real-Time Quantitative Reverse Transcription PCR (RT-qPCR)
Total RNAs were extracted using TRIzol reagent (Invitrogen) from rice leaves and two stages of inflorescences, according to distance from the auricle of 0 or 1 cm, which are the standards for meiosis and early young microspore stage, respectively (Kerim et al., 2003). 1 μg total RNAs were used to synthesized first-strand cDNA using Primescript RT reagent Kit with gDNA Eraser following the manufacturer’s instructions [TaKaRa Biotechnology (Dalian), Co., Ltd, Japan]. Specific primers were designed according for individual genes (Supplementary Table S2). First-strand cDNA (2 μl) was amplified with 2 μl of specific primers in a total reaction volume of 20 μl, and each reaction was performed in triplicate using KAPA SYBR FAST qPCR Master Mix (KAPA BIOSYSTEMS) in a Thermal Cycler CFX96 (Bio-Rad, United States). The ubiquitin gene was used as an internal control for data normalization and changes in gene expression were calculated using the 2ΔΔCT algorithm (Liu et al., 2012). The means from three biological replicates were used for analysis.
Subcellular Localization Analysis
mCherry cDNA was amplified from pBIN20-ER-rk (Nelson et al., 2007) with the primer pairs mCherry F and mCherry R, and cloned into pC2300-35S-eGFP vector (Ma et al., 2017) to generate pC2300-mCherry with KpnI and BamHI restriction enzyme sites. The coding sequence of OsEMF2b was amplified using the primer pairs OsEMF2b-mCherry F and OsEMF2b-mCherry R listed in Supplementary Table S2. The amplified fragment and pC2300-mCherry backbone were digested with BamHI and XbaI, then ligated together to create pC2300-mCherry-OsEMF2b. The pC2300-mCherry and recombinant vector pC2300-mCherry-OsEMF2b were then transformed into rice protoplasts as described (Zhang et al., 2011). mCherry fluorescence was visualized using a ZEISS LSM 710 confocal microscope.
Complementation of the osemf2b-4 Mutant
The entire coding region of OsEMF2b was amplified with KOD DNA polymerase (TOYOBO, Japan) using the primer pairs OsEMF2b-comp F and OsEMF2b-comp R. The amplified fragment was digested with SpeI and PstI and then ligated to pCactN carrying the rice ACTIN promoter (Zhu et al., 2016) to generate pCactN-EMF2b, which was transformed into osemf2b-4 using Agrobacterium tumefaciens (Hiei et al., 1994). Positive transgenic plants were selected with G418 and identified using the primer pair for Neomycin phosphotransferase II gene (NPT II).
Statistical Analysis
Statistical significance of differences in the mean values was determined using the Student’s t-test function in Microsoft Excel software. Levels of significance were represented by asterisks as follows: ∗,∗∗ indicates significance at P ≤ 0.05 or 0.01, respectively.
Results
Identification and Morphological Characterization of osemf2b-4 Plants
Based on RiceGE database and sequencing results, the T-DNA insertion of osemf2b-4 was inserted -200 bp upstream of the OsEMF2b start codon (Supplementary Figure S1). RT-qPCR analysis revealed that the expression of OsEMF2b was significantly decreased in leaves of osemf2b-4 (Figure 1A), indicating that it was a weaker allele than the three null mutant alleles (Kim et al., 2010). The osemf2b-4 homozygous mutant was slightly shorter than wild type (Figure 1B). The most obvious phenotypes of osemf2b-4 were reduced grain number and seed set (Figure 1C). The grain number per panicle of wild type was 59 ± 3.6, whereas osemf2b-4 was 22.3 ± 2.1. Additionally, a variety of abnormal seeds were observed, for example, some seeds protruded from the hull, and some had a long sterile lemma (Figure 1D).
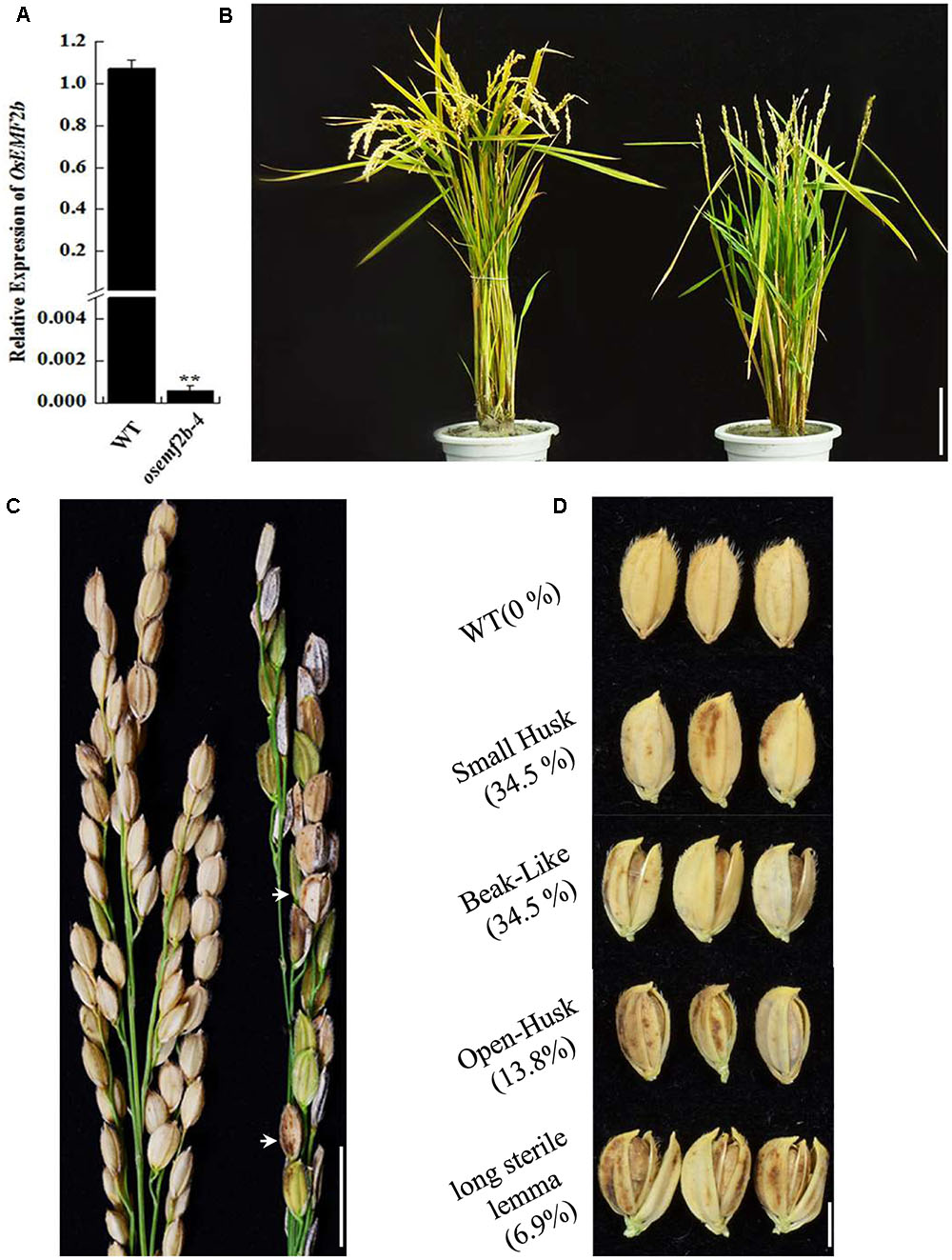
FIGURE 1. Phenotypic and RT-qPCR analyses of osemf2b-4. (A) Expression levels of OsEMF2b in the WT or osemf2b-4 leaves were detected by RT-qPCR. Error bars indicate SD. ∗∗ indicates significant difference by Student’s t-test (P ≤ 0.01). (B) Plant phenotypes of wild type (left) and osemf2b-4 (right) at maturity. Bars = 5 cm. (C) Panicles of wild type (left) and osemf2b-4 (right). Abnormal seeds are marked with white arrow in the osemf2b-4 panicle. Data are given as mean ± SEM (n = 3). Bars = 2 cm. (D) Seed phenotypes of wild type and osemf2b-4. The numbers to the left were calculated as the proportion of slightly defective types of abnormal seeds to spikelets for each plant at maturity. The 10.3% remaining were sterile spikelets. Bars = 5 mm.
To test whether the pleiotropic phenotype of osemf2b-4 was caused by the T-DNA insertion in its promoter region, we firstly carried out co-segregation analysis using T1 and T2 segregation populations (Supplementary Table S1). The coding sequence of OsEMF2b driven by rice ACTIN promoter was introduced into homogenous osemf2b-4, and 4 T0 transgenic plants were obtained (Supplementary Figure S2), the 3 with highly enhanced expression of OsEMF2b exhibited normal florets, anther and pollen, whereas the one with relatively low expression still exhibited defective floret, anther and pollen development (Supplementary Figures S2A–C, S3A–J). Together, these results suggested that the T-DNA insertion in OsEFM2b promoter region was responsible for the osemf2b-4 phenotypes.
Fertility Analysis of the osemf2b-4 Mutant
Previous studies on biological and molecular function of OsEMF2b were focused on flowering time, inflorescence and floret development using its null mutant (Luo et al., 2009; Conrad et al., 2014; Xie et al., 2014). Here, we used the weak allele osemf2b-4 to study its function during post-meiotic anther and pollen development. The spikelets of osemf2b-4 exhibited various morphologies of palea and/or lemma (Figure 2A). We divided these florets with stamen into two different types, slight defects (Figures 2B–E) and severe defects (Figures 2a–d). In osemf2b-4, there was a high percentage of slightly defective types including small husk, beak-like, open husk and long sterile lemma florets types, 34.5, 34.5, 13.8, and 6.9% (n = 142), respectively. The most severe cases, such as double lemma (3.4%, n = 142), coiled lemma and palea (3.4%, n = 142), missing lemma or palea (2%, n = 142), and leaf-like floret (1.4%, n = 142).

FIGURE 2. Floret and pollen fertility comparisons of the wild type and osemf2b-4. (A) A wild type spikelet; (B–E) four types of slight floret defects with stamens including small husk (B), beak-like (C), open husk (D), and long sterile lemma (E) spikelets in osemf2b-4 plants, bars = 2 mm; (F–J) spikelets after removing the lemma and palea, bars = 2 mm; (K–R) I2-KI pollen staining, (L–O) pollen from yellow anthers, (P–R) pollen from three white anthers (white arrowheads), respectively. Bars = 200 μm. (a–d) Four types of severe floret defects with stamens such as double lemma (a), coiled lemma and palea (b), missing lemma or palea (c), and leaf-like (d) spikelets in osemf2b-4 mutant, Bars = 2 mm; (e–h) spikelets after removing the lemma and palea, bars = 2 mm; (i–l) Pollen fertility using I2-KI pollen staining. Numbers indicate the ratio of different types of floral organ defects with stamens per plant. Bars = 200 μm.
The palea and lemma were removed for clearly visualizing anthers (Figures 2F–J,e–h). In osemf2b-4, normal anthers were observed in most florets, but the number of anthers in each floret were variable. In the three null mutants anthers were rarely produced but did not contain any pollen grains (Luo et al., 2009; Kim et al., 2010). I2-KI staining showed that the pollen fertility of osemf2b-4 anthers was variable, from semi-sterility to complete sterility (Figures 2L–O,i–l), when compared with the fertility of WT anthers (Figure 2K). Abortive pollen grains were observed in the transparent and white stamens (Figures 2P–R). These results suggested that the partial spikelet sterility of the osemf2b-4 mutant was likely caused by reduced pollen viability, and that osemf2b-4 was an excellent allele for studying the function of OsEMF2b during post-meiotic anther and pollen development.
Cytological Characterization of Post-meiotic Anther and Pollen Development in osemf2b-4
To more precisely determine the cytological reason for the post-meiosis anther and pollen defects in osemf2b-4, we performed semi-thin sections for at three stages of anther development (Feng et al., 2000; Zhang and Wilson, 2009). According to the anther defects in different locules, osemf2b-4 anthers were grouped into three different categories from young microspore stage to mature pollen stage, respectively (Figures 3A–L). At the young microspore stage, the tapetal cells was deeply stained, and exine was deposited on the round microspores in wild type anthers (Figure 3A). However, three different phenotypes were observed in osemf2b-4 anthers: type I (74%, n = 45) with likely normal tapetum and irregular shape of microspores (Figure 3B); type II (20%, n = 45) with lightly stained and weakly expanded tapetum and absent middle layer (Figure 3C); type III (6%, n = 45) with extremely enlarged and disordered tapetum and no exine on microspores (Figure 3D). At the vacuolated stage in wild type anthers, the tapetal cells had become more degenerated and the microspores appeared vacuolated (Figure 3E). The mutant anthers exhibited three different types, including type I (16%, n = 146) with likely normal locules (Figure 3F), type II (17%, n = 146) with shrunken microspores (Figure 3G), and type III (67%, n = 146) with delayed degradation of tapetum and folded microspores (Figure 3H).
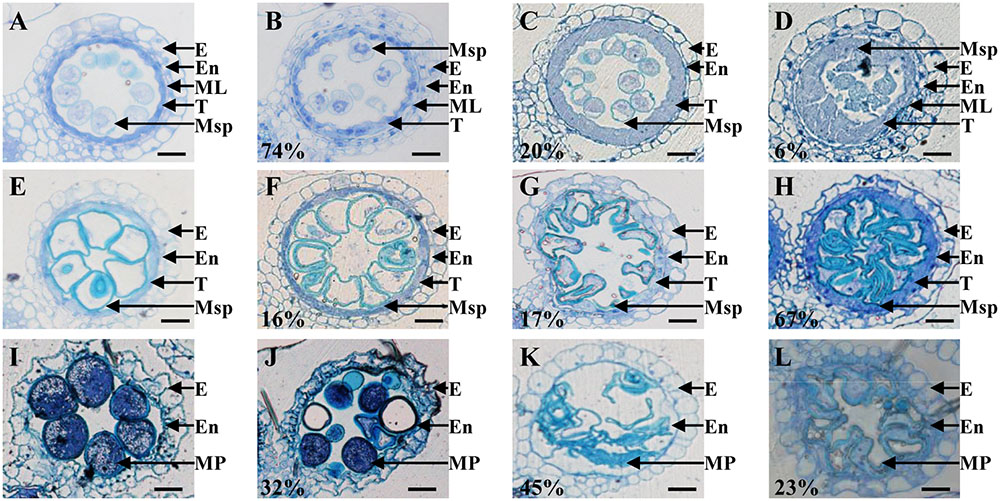
FIGURE 3. Histological comparison of anther development in the wild type and osemf2b-4. Three stages of anther development in the wild type and osemf2b-4 are compared. The images are of cross sections through a single locule. Wild type sections are shown in (A,E,I), and others show osemf2b-4 sections. (A–D) Young microspore stage; (E–H) vacuolated pollen stage; (I–L) mature pollen stage. Numbers indicate the percentage of different anther defects at one stage. E, epidermis; En, endothecium; ML, middle layer; T, tapetum; Ms, microsporocyte; Msp, microspore; MP, mature pollen. Bars = 15 μm.
By the mature pollen stage, the tapetum and middle layer had completely degenerated and the endothecial cell layer eventually was disrupted to release pollen grains in wild type (Figure 3I). In osemf2b-4, 32% of anther locules (n = 38) displayed linear shaped surface, similar to wild type anthers. Some pollen grains were stained weakly with toluidine blue, while some grains were irregularly shaped and collapsed in type I (Figure 3J). Another two types of anther defects, type II (45%, n = 38) and type III (23%, n = 38) exhibited glossy and smooth anther surface and shrunken pollen, as well as delayed degradation of the endothecial cell layer (Figures 3K,L). Together, these observations suggested that osemf2b-4 had developmental defects in post-meiotic anther wall layers and microspores, and that OsEMF2b plays an essential role in post-meiotic anther and pollen development.
Expression Pattern of OsEMF2b in Anther and Subcellular Localization of OsEMF2b
Given the anther defects observed in osemf2b-4, we examined the expression pattern of OsEMF2b at two developmental stages using in situ hybridization (Figures 4A–C). OsEMF2b transcripts were detected in microsporocyte, tapetum and vascular bundle of anther connective at the meiosis stage (Figure 4A). OsEMF2b had high expression in tapetum and weak expression in microspores and vascular bundle at the young microspore stage (Figure 4B). Only background level of signal was detected with the sense probe (Figure 4C).
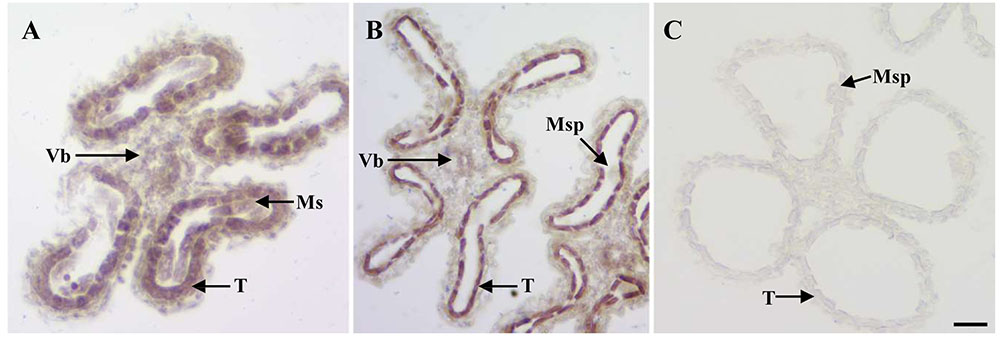
FIGURE 4. RNA in situ analysis of OsEMF2b. (A) A wild type anther at the meiosis stage showing OsEMF2b expression in microsporocytes, tapetum and connectives of anther. (B) A wild type anther at the young microspore stage showing stronger OsEMF2b expression in tapetal cells, and weaker expression in microspores and connectives of anther. (C) A wild type anther at young microspore stage with the sense probe. Ms, microsporocyte; T, tapetum; Msp, microspore; Vb, vascular bundle of anther connective. Bars = 15 μm.
Even though OsEMF2b was predicted to be localized in the nucleus (Yoshida et al., 2001; Luo et al., 2009), due to two putative nuclear localization signals, a single C2H2-type zinc finger motif and a VRN2-EMF2-FIS2-SUZ12 (VEFS) domain (Figure 5A), no experimental data showed its protein localization. In order to test the subcellular localization of OsEMF2b, a 35S::mCherry-OsEMF2b fusion construct was introduced into rice leaf protoplasts. As expected, the mCherry-OsEMF2b fusion protein was observed in the nucleus (Figures 5Bd–f), whereas the empty vector with 35S::mCherry was observed in cytoplasm (Figures 5Ba–c).
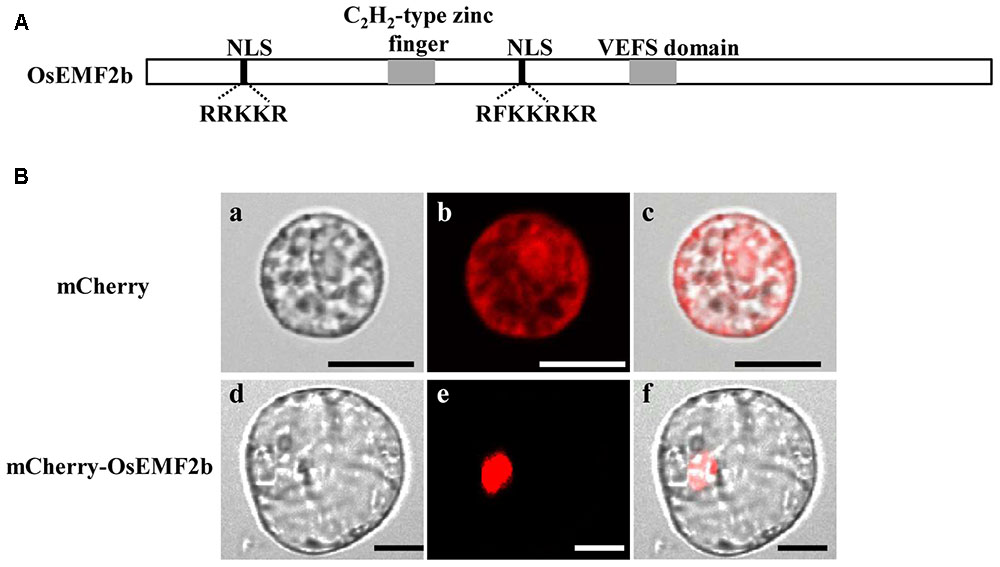
FIGURE 5. Structure and nuclear localization of OsEMF2b. Schematic representation of OsEMF2b. The black boxes indicate two putative nuclear localization signals (NLS) with a basic segment (Lys-Lys-Arg-Lys-Arg), and the gray boxes indicate a C2H2-type zinc finger motif and the VRN2-EMF2-FIS2-SUZ12 (VEFS) domain, respectively. (A) Nuclear localization of OsEMF2b in rice protoplasts. (B) (a–c) Images of mCherry. (d–f) Images of mCherry-OsEMF2b fusion. (a,d) Bright-field images. (b,e) Red fluorescence images. (c,f) Merged images of (a,b,d,e), respectively. Bars = 2 μm in (a–c), 4 μm in (d–f).
Expression of Genes Involved in Anther and Pollen Development Was Affected in osemf2b-4
Defective anther development was observed in osemf2b-4, probably by affecting the gene expressions involved in post-meiotic anther and pollen development. We used the auricle distance between the penultimate leaf and flag leaf for assessing anther development stage; 0 and 1 cm respectively represented meiosis and young microspore stage (Kerim et al., 2003). Then we analyzed the expression of several nuclear genes involved in anther and pollen development at these two stages. Most anther and pollen-related genes had tissue-specific expression patterns. TAPETUM DEGENERATION RETARDATION (TDR) (Li N. et al., 2006) and ETERNAL TAPETUM1 (EAT1) (Niu et al., 2013) were preferentially expressed in the tapetum, as HISTONE MONOUBIQUITINATION1/2 (OsHUB1/2) (Cao et al., 2015), GAMYB (Aya et al., 2009), APPOTOSIS INHIBITOR5 (OsAPI5), OsAIP1/2 (Li X. et al., 2011), CARBON STARVED ANTHER (CSA) (Zhu et al., 2015), MYB IMPORTANT FOR DROUGHT RESPONSE1 (MID1) (Guo et al., 2016) and PERSISTENT TAPETAL CELL1 (PTC1) (Li H. et al., 2011) were expressed in the tapetum and microspores. OsRH2/34 (Huang et al., 2016), OsMYB106 and OsSPX1 (Zhang et al., 2016) were also essential for anther and/or pollen development. PTC1 expression was significantly decreased in osemf2b-4, and the other 14 genes were significantly up-regulated at stage 2 (Figure 6), suggesting that OsEMF2b was required for fine-tuning the expression of these genes involved in the anther and pollen development.
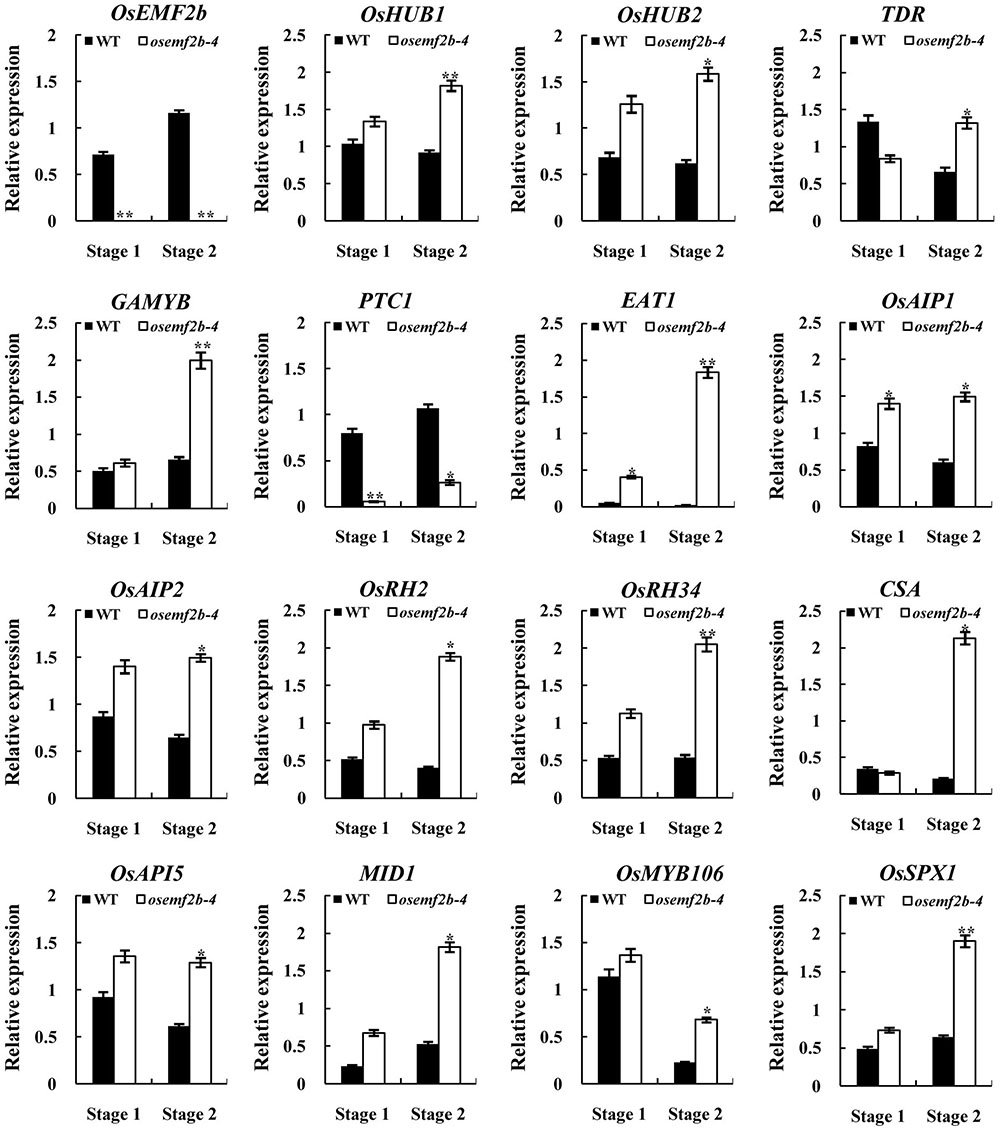
FIGURE 6. RT-qPCR expression analysis of OsEMF2b and genes involved in rice anther and pollen development in the inflorescence of the wild type and osemf2b-4 plants. Stage 1 and stage 2 represent meiosis stage and young microspore stage, respectively. The ubiquitin gene was used as an internal control. Each data point is the average of three biological repeats. Error bars indicate SD. ∗,∗∗ indicate significant difference by Student’s t-test (P ≤ 0.05 and 0.01).
Discussion
In this study, we identified a weak OsEMF2b mutant, osemf2b-4, which exhibited defective floret development and seed set. Cytological analysis showed various defects during anther and pollen development in osemf2b-4, such as delayed tapetum degradation. RNA in situ showed that OsEMF2b was expressed in the tapetum and microspores. These results suggested that OsEMF2b plays an important role in anther and pollen development.
EMF2 played an important role in repression of genes by promoting H3K27me3 at target genes in Arabidopsis (Conrad et al., 2014). We therefore investigated the expression of 15 genes whose encoded proteins are nuclearly localized and are essential for anther and pollen development. Among these genes, the expression of PTC1 was significantly decreased in osemf2b-4. PTC1 plays an essential role in rice anther and pollen development, as a knockout (Li H. et al., 2011) or knock-down (results not published) of PTC1 resulted in developmental defects in anthers and pollen. Therefore, the down-regulated expression of PTC1 in osemf2b-4 possibly played an important contribution to the defective anther and pollen development observed in osemf2b-4. On the other hand, OsEMF2b physically interacts with VERNALIZATION INSENSITIVE3-LIKE2 (OsVIL2), a PHD-finger domain protein showing homologies to the Arabidopsis VIN3-group proteins, to promote H3K27me3 deposition in rice (Yang et al., 2013). The Arabidopsis VIN3-group proteins with PHD-finger domain are known to work together with the PRC2 core complex containing EMF2 for constituting the so-called PHD-PRC2 complexes (Ietswaart et al., 2012). PTC1 encodes a PHD-finger protein and its ortholog protein in Arabidopsis, MALE STERILITY1 (MS1), contains an aromatic cage required for H3K4me2/3 binding (Lee et al., 2009). Therefore, OsEMF2b and PTC1 might function together through histone methylation modification in anther and microspore development. PTC1 is expressed in tapetum and microspores at the young microspore stage (Li H. et al., 2011), and the overlapped spatiotemporal expression pattern with OsEMF2b (Figure 4) are consistent with the idea that OsEMF2b and PTC1 function together.
Considering these 14 genes with up-regulated expression in osemf2b-4, all mutants of them exhibited anther and/or pollen developmental defects (Cao et al., 2015; Shi et al., 2015; Huang et al., 2016; Zhang et al., 2016). However, it was unknown whether the over-expression of these 14 genes caused defects in anther and pollen development process in rice. But in barley, over-expressing HvGAMYB resulted in partial or complete sterility (Murray et al., 2003). As the molecular pathway of anther development is conserved between rice and barley (Fernández Gómez and Wilson, 2014), it is not surprising that over-expressing GAMYB in rice led to anther and pollen developmental defects, and possibly contributes to the phenotype of osemf2b-4. As similar with GAMYB, both knockout and over-expressed MS1 exhibited defective anther and pollen in Arabidopsis (Yang et al., 2007), indicating that the expression of genes involved in anther and pollen development was tightly controlled. Based on that, these 14 genes with up-regulated expression in osemf2b-4 should be also tightly controlled in wild type, in order to keep normal expression level to function normally. This tight control in anther and pollen could be done directly or indirectly by OsEMF2b. Overall, our studies suggested that OsEMF2b played an essential role in anther and pollen development, and possibly by fine-tuning the expression of genes expressed in tapetum and microspores, such as PTC1 and GAMYB.
Author Contributions
LD performed most of the experiments and wrote the manuscript. SZ and ML performed RT-qPCR analyses and complementation test. GW performed the semi-thin sections. SF, WC, and JT finished the subcellular localization analysis. BT provided technical assistance. PQ supervised and complemented the writing. YW, BM, and SL supervised the research.
Funding
This work was supported by funds from the National Natural Science Foundation of China (31471475; 31571634) and State Key Laboratory of Hybrid Rice.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Zhixue Zhao for assistance with the confocal microscope assistance. We thank Prof. Ming Luo and Dr. Fang Huang (CSIRO, Australia) for their helpful discussions, and Dr. Sheila McCormick for editing the English.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01998/full#supplementary-material
FIGURE S1 | Schematic characterization of four independent mutant alleles of OsEMF2b. Three mutants are T-DNA insertions including osemf2b-1 in intron 19, osemf2b-2 in intron 10 and osemf2b-4 in this study. The osemf2b-3 mutant is generated by targeting induced local lesions in genomes (TILLIING), and is a guanine to thymine substitution.
FIGURE S2 | Phenotype (A,B) and corresponding relative expression levels (C) of osemf2b-4 and complementation plants at heading stage in T0 progenies. (A) Plant phenotypic comparison. Bars = 10 cm. (B) Observation of inflorescence. Bars = 2 cm. Comp, complementation plants. The white arrows indicated abnormal spikelets. ∗∗ indicates significant difference by Student’s t-test (P ≤ 0.01).
FIGURE S3 | Spikelets after removing the lemma and palea (A–E) and I2-KI pollen staining (F–J) of osemf2b-4 and complementation plants. The numbers are percentage of stained pollen grains. Bars = 2 mm and 200 μm, respectively.
TABLE S1 | Segregation of the osemf2b-4 lines in T1 and T2 progenies.
TABLE S2 | Primers used in this study.
Footnotes
References
Andreuzza, S., Nishal, B., Singh, A., and Siddiqi, I. (2015). The chromatin protein DUET/MMD1 controls expression of the meiotic gene TDM1 during male meiosis in Arabidopsis. PLOS Genet. 11:e1005396. doi: 10.1371/journal.pgen.1005396
Aya, K., Ueguchi-Tanaka, M., Kondo, M., Hamada, K., Yano, K., Nishimura, M., et al. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21, 1453–1472. doi: 10.1105/tpc.108.062935
Berr, A., McCallum, E. J., Menard, R., Meyer, D., Fuchs, J., Dong, A., et al. (2010). Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22, 3232–3248. doi: 10.1105/tpc.110.079962
Cao, H., Li, X., Wang, Z., Ding, M., Sun, Y., Dong, F., et al. (2015). Histone H2B monoubiquitination mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 Is involved in anther development by regulating tapetum degradation-related genes in rice. Plant Physiol. 168, 1389–1405. doi: 10.1104/pp.114.256578
Cartagena, J. A., Matsunaga, S., Seki, M., Kurihara, D., Yokoyama, M., Shinozaki, K., et al. (2008). The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen. Dev. Biol. 315, 355–368. doi: 10.1016/j.ydbio.2007.12.016
Chanvivattana, Y., Bishopp, A., Schubert, D., Stock, C., Moon, Y. H., Sung, Z. R., et al. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276. doi: 10.1242/dev.01400
Conrad, L. J., Khanday, I., Johnson, C., Guiderdoni, E., An, G., Vijayraghavan, U., et al. (2014). The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice. Plant J. 80, 883–894. doi: 10.1111/tpj.12688
Feng, J., Lu, Y., Liu, X., and Xu, X. (2000). Pollen development and its stages in rice (Oryza sativa L.). Zhongguo Shuidao Kexue 15, 21–28.
Fernández Gómez, J., and Wilson, Z. A. (2014). A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnol. J. 12, 765–777. doi: 10.1111/pbi.12181
Goldberg, R. B., Beals, T. P., and Sanders, P. M. (1993). Anther development: basic principles and practical applications. Plant Cell 5, 1217–1229. doi: 10.1105/tpc.5.10.1217
Guo, C., Yao, L., You, C., Wang, S., Cui, J., Ge, X., et al. (2016). MID1 plays an important role in response to drought stress during reproductive development. Plant J. 88, 280–293. doi: 10.1111/tpj.13250
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Huang, C. K., Sie, Y. S., Chen, Y. F., Huang, T. S., and Lu, C. A. (2016). Two highly similar DEAD box proteins, OsRH2 and OsRH34, homologous to eukaryotic initiation factor 4AIII, play roles of the exon junction complex in regulating growth and development in rice. BMC Plant Biol. 16:84. doi: 10.1186/s12870-016-0769-5
Ietswaart, R., Wu, Z., and Dean, C. (2012). Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 28, 445–453. doi: 10.1016/j.tig.2012.06.002
Kerim, T., Imin, N., Weinman, J. J., and Rolfe, B. G. (2003). Proteome analysis of male gametophyte development in rice anthers. Proteomics 3, 738–751. doi: 10.1002/pmic.200300424
Kim, S. Y., Zhu, T., and Sung, Z. R. (2010). Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiol. 152, 516–528. doi: 10.1104/pp.109.143495
Kohler, C., and Villar, C. B. (2008). Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 18, 236–243. doi: 10.1016/j.tcb.2008.02.005
Lee, W. Y., Lee, D., Chung, W. I., and Kwon, C. S. (2009). Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J. 58, 511–524. doi: 10.1111/j.1365-313X.2009.03795.x
Li, H., Yuan, Z., Vizcay-Barrena, G., Yang, C., Liang, W., Zong, J., et al. (2011). PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156, 615–630. doi: 10.1104/pp.111.175760
Li, K., Yang, J., Liu, J., Du, X., Wei, C., Su, W., et al. (2006). Cloning, characterization and tissue-specific expression of a cDNA encoding a novel EMBRYONIC FLOWER 2 gene (OsEMF2) in Oryza sativa. DNA Seq. 17, 74–78. doi: 10.1080/10425170500151961
Li, N., Zhang, D. S., Liu, H. S., Yin, C. S., Li, X. X., Liang, W. Q., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014. doi: 10.1105/tpc.106.044107
Li, X., Gao, X., Wei, Y., Deng, L., Ouyang, Y., Chen, G., et al. (2011). Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-Box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell 23, 1416–1434. doi: 10.1105/tpc.110.082636
Liu, M. S., Chen, L. F., Lin, C. H., Lai, Y. M., Huang, J. Y., and Sung, Z. R. (2012). Molecular and functional characterization of broccoli EMBRYONIC FLOWER 2 genes. Plant Cell Physiol. 53, 1217–1231. doi: 10.1093/pcp/pcs063
Luo, M., Platten, D., Chaudhury, A., Peacock, W. J., and Dennis, E. S. (2009). Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol. Plant 2, 711–723. doi: 10.1093/mp/ssp036
Ma, X., Sun, X., Li, C., Huan, R., Sun, C., Wang, Y., et al. (2017). Map-based cloning and characterization of the novel yellow-green leaf gene ys83 in rice (Oryza sativa). Plant Physiol. Biochem. 111, 1–9. doi: 10.1016/j.plaphy.2016.11.007
Moon, Y. H., Chen, L., Pan, R. L., Chang, H. S., Zhu, T., Maffeo, D. M., et al. (2003). EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15, 681–693. doi: 10.1105/tpc.007831
Murray, F., Kalla, R., Jacobsen, J., and Gubler, F. (2003). A role for HvGAMYB in anther development. Plant J. 33, 481–491. doi: 10.1046/j.1365-313X.2003.01641.x
Nelson, B., Cai, X., and Nebenfuhr, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Niu, N., Liang, W., Yang, X., Jin, W., Wilson, Z. A., Hu, J., et al. (2013). EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4:1445. doi: 10.1038/ncomms2396
Qin, P., Tu, B., Wang, Y., Deng, L., Quilichini, T. D., Li, T., et al. (2013). ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol. 54, 138–154. doi: 10.1093/pcp/pcs162
Shi, J., Cui, M., Yang, L., Kim, Y. J., and Zhang, D. (2015). Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 20, 741–753. doi: 10.1016/j.tplants.2015.07.010
Xie, S., Chen, M., Pei, R., Ouyang, Y., and Yao, J. (2014). OsEMF2b acts as a regulator of flowering transition and floral organ identity by mediating H3K27me3 deposition at OsLFL1 and OsMADS4 in rice. Plant Mol. Biol. Rep. 33, 121–132. doi: 10.1007/s11105-014-0733-1
Yang, C., Vizcay-Barrena, G., Conner, K., and Wilson, Z. A. (2007). MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19, 3530–3548. doi: 10.1105/tpc.107.054981
Yang, J., Lee, S., Hang, R., Kim, S. R., Lee, Y. S., Cao, X., et al. (2013). OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 73, 566–578. doi: 10.1111/tpj.12057
Yoshida, N., Yanai, Y., Chen, L., Kato, Y., Hiratsuka, J., Miwa, T., et al. (2001). EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13, 2471–2481. doi: 10.1105/tpc.13.11.2471
Zhang, D., and Wilson, Z. A. (2009). Stamen specification and anther development in rice. Chin. Sci. Bull. 54, 2342–2353. doi: 10.1007/s11434-009-0348-3
Zhang, K., Song, Q., Wei, Q., Wang, C., Zhang, L., Xu, W., et al. (2016). Down-regulation ofOsSPX1caused semi-male sterility, resulting in reduction of grain yield in rice. Plant Biotechnol. J. 14, 1661–1672. doi: 10.1111/pbi.12527
Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLOS Biol. 5:e129. doi: 10.1371/journal.pbio.0050129
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 1–14. doi: 10.1186/1746-4811-7-30
Zhu, J., Lou, Y., Xu, X., and Yang, Z. N. (2011). A genetic pathway for tapetum development and function in Arabidopsis. J. Integr. Plant Biol. 53, 892–900. doi: 10.1111/j.1744-7909.2011.01078.x
Zhu, X., Liang, W., Cui, X., Chen, M., Yin, C., Luo, Z., et al. (2015). Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J. Cell Mol. Biol. 82, 570–581. doi: 10.1111/tpj.12820
Keywords: rice, OsEMF2b, semi-sterility, anther development, male reproductive development
Citation: Deng L, Zhang S, Wang G, Fan S, Li M, Chen W, Tu B, Tan J, Wang Y, Ma B, Li S and Qin P (2017) Down-Regulation of OsEMF2b Caused Semi-sterility Due to Anther and Pollen Development Defects in Rice. Front. Plant Sci. 8:1998. doi: 10.3389/fpls.2017.01998
Received: 15 August 2017; Accepted: 08 November 2017;
Published: 22 November 2017.
Edited by:
Jin-Gui Chen, Oak Ridge National Laboratory (DOE), United StatesReviewed by:
Dongfang Wang, Spelman College, United StatesMartin Fellner, Palacký University Olomouc, Czechia
Copyright © 2017 Deng, Zhang, Wang, Fan, Li, Chen, Tu, Tan, Wang, Ma, Li and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Qin, qinpeng626@gmail.com Shigui Li, lishigui@sicau.edu.cn
†These authors have contributed equally to this work.
 Luchang Deng
Luchang Deng Siwei Zhang1†
Siwei Zhang1† Yuping Wang
Yuping Wang