- State Key Laboratory of Crop Stress Biology for Arid Areas and College of Life Sciences, Northwest A&F University, Yangling, China
In higher plants, the development of functional chloroplasts is essential for photosynthesis and many other physiological processes. With a long-term goal of elucidating the genetic regulation of chloroplast development, we identified two allelic leaf variegation mutants, variegated5-1 (var5-1) and var5-2. Both mutants showed a distinct leaf reticulation phenotype of yellow paraveinal regions and green interveinal regions, and the leaf reticulation phenotype correlated with photosynthetic defects. Through the identification of mutation sites in the two mutant alleles and the molecular complementation, we confirmed that VAR5 encodes a CorA family of Mg2+ transporters also known as AtMRS2-11/AtMGT10. Using protoplast transient expression and biochemical fractionation assays, we demonstrated that AtMRS2-11/AtMGT10/VAR5 likely localizes to the chloroplast envelope. Moreover, we established that AtMRS2-11/AtMGT10/VAR5 forms large molecular weight complexes in the chloroplast and the sizes of these complexes clearly exceed those of their bacterial counterparts, suggesting the compositions of CorA Mg2+ transporter complex is different between the chloroplast and bacteria. Our findings indicate that AtMRS2-11/AtMGT10/VAR5 plays an important role in the tissue specific regulation of chloroplast development.
Introduction
Magnesium ion (Mg2+) is one of the most abundant divalent cations in cells and is involved in myriads of essential metabolic processes (Maguire and Cowan, 2002). The electrically charged nature of Mg2+ necessitates the presence of membrane transporters to facilitate efficient Mg2+ trafficking across biological membranes. The functions and regulations of these transporters ensure the maintenance of Mg2+ homeostasis in different sub-cellular compartments in cells (Shaul, 2002).
Magnesium ion transport systems were most extensively studied in prokaryotes, and several types of putative Mg2+ transporters, including CorA, MgtE, and P-type ATPase Mgt, have been identified in bacteria (Smith and Maguire, 1995; Moomaw and Maguire, 2008). In eukaryotic cells, the CorA family Mg2+ transporter was identified in yeast mitochondria as MRS2 and is involved in group II intron splicing (Bui et al., 1999). Typical CorA Mg2+ transporters contain two C-terminal transmembrane domains and in between a conserved GMN motif as a Mg2+ binding site (Lunin et al., 2006; Pfoh et al., 2012). Genes coding for putative Mg2+ transporters are also present in plant genomes and the most extensively studied Mg2+ transporters in plants are the CorA family of transporters (Gardner, 2003). In the model system Arabidopsis thaliana, there are at least 10 genes encoding putative CorA Mg2+ transporters and they have been designated as encoding AtMRS2 or magnesium transport (AtMGT) (Schock et al., 2000; Li et al., 2001). Functionally, Arabidopsis CorA Mg2+ transporters were able to complement bacterial or yeast mutants that are defective in Mg2+ transport, suggesting that they are bona fide Mg2+ transporters (Schock et al., 2000; Li et al., 2001, 2008; Drummond et al., 2006; Mao et al., 2008; Chen et al., 2009; Gebert et al., 2009). Consistent with the important roles of Mg2+ in biological systems, AtMRS2/AtMGT genes have been shown to be important for several cellular processes. For example, AtMRS2-6/AtMGT5 and AtMRS2-2/AtMGT9 have been shown to be involved in pollen development (Li et al., 2008; Chen et al., 2009). Moreover, attempts to obtain insertional knock-out mutants of several AtMRS2/AtMGT genes have not yielded homozygous mutants for several genes including AtMRS2-2/AtMGT9, AtMRS2-6/AtMGT5, and AtMRS2-11/AtMGT10, suggesting these genes may be essential genes in Arabidopsis (Li et al., 2008; Chen et al., 2009; Gebert et al., 2009). Consistent with their membrane protein identities, these transporters are targeted to different sub-cellular compartments and are presumably responsible for Mg2+ homeostasis in various organelles. For example, AtMRS2-10/AtMGT1 and AtMRS2-4/AtMGT6 are plasma membrane Mg2+ transporters (Li et al., 2001; Mao et al., 2014). AtMRS2-6/AtMGT5 was shown to be targeted to mitochondria (Li et al., 2008). AtMRS2-7/AtMGT7 has been suggested to be located in the endoplasmic reticulum (Gebert et al., 2009). AtMRS2-1/AtMGT2 and AtMRS2-5/AtMGT3 are targeted to the tonoplast and involved in Mg2+ partition in leaf mesophyll cell vacuoles (Conn et al., 2011). In addition to the CorA Mg2+ transporters, other types of putative Mg2+ transporters have also been identified in plants including the AtMHX H+/Mg2+ transporter (Shaul et al., 1999;Shaul, 2002).
The chloroplast is the organelle for photosynthesis and many other essential processes in plants (Pogson and Albrecht, 2011). Mg2+ also plays critical roles in the chloroplast. For example, in the light reactions, Mg2+ is an integral part of photosynthetic pigment chlorophylls. In carbon reactions, it is known that Mg2+ can regulate the activities of ribulose 1,5-bisphosphate reductase/oxygenase (rubisco) and many other enzymes. Thus, it is not surprising that Mg2+ transporters are necessary in the chloroplast to facilitate the transport of the ion and to regulate chloroplast Mg2+ homeostasis. Of the AtMRS2/AtMGT family of Mg2+ transporters, AtMRS2-11/AtMGT10 is predicted to be targeted to the chloroplast based on the presence of a N-terminal chloroplast transit peptide (Drummond et al., 2006).
Plant variegation mutants generally refer to mutants that display differential leaf color in plant organs, especially in leaves (Yu et al., 2007). In variegation mutants defective in nuclear genes for chloroplast proteins, differential chlorophyll accumulation/chloroplast development can arise from a uniform mutant genetic background, and these mutants have long attracted attention because the mechanisms of variegation are instrumental in our understanding of chloroplast development. Among variegation mutants, one unique group of leaf variegation mutants is the reticulate mutants (Yu et al., 2007). These mutants usually show a differential chlorophyll accumulation/chloroplast development between paraveinal regions and interveinal regions, thus giving rise to a “net” like phenotype. Most of the reticulate mutants reported so far display green paraveinal regions and yellow interveinal regions and several reticulate mutants have been characterized at the molecular level (López-Juez et al., 1998; Yu et al., 2007). The mutation of CHLOROPHYLL A/B BINDING PROTEIN (CAB) GENE UNDEREXPRESSED 1 (CUE1) in Arabidopsis leads to a reticulate phenotype (Streatfield et al., 1999). CUE1 encodes a chloroplast envelope localized phosphoenolpyruvate (PEP)/phosphate translocator, which is responsible for transporting PEP into the chloroplast for the shikimate pathway and the synthesis of aromatic amino acids (Streatfield et al., 1999). The addition of aromatic amino acids can rescue the reticulate phenotype of cue1 mutants, suggesting a role for aromatic amino acids in the regulation of mesophyll chloroplast development (Streatfield et al., 1999). RETICULATA (RE) is another Arabidopsis locus that leads to a reticulate phenotype when mutated and RE encodes a novel protein of unknown functions that is also known as LOW CELL DENSITY 1 (LCD1) (Barth and Conklin, 2003; González-Bayón et al., 2006). The Arabidopsis differential development of vascular associated cells 1 (dov1) mutant defines another reticulate mutant with green paraveinal regions and yellow interveinal regions, and DOV1 encodes glutamine phosphoribosyl pyrophosphate aminotransferase 2 (ATase2), the first enzyme in the purine nucleotide biosynthesis pathway (Kinsman and Pyke, 1998; Rosar et al., 2012). Additional Arabidopsis reticulate mutants include venosa3 (ven3) and ven6 that are defective in carbamoyl phosphate synthetase and arginine biosynthesis (reviewed in Yu et al., 2007; Mollá-Morales et al., 2011; Lundquist et al., 2014). It is interesting to note that most of the genes that are associated with the reticulate mutant phenotypes code for proteins involved in primary metabolism (Lundquist et al., 2014). In addition, reticulate mutants with yellow paraveinal regions and yellow interveinal regions are rare. The intriguing feature of these mutants implies that there are distinct tissue specific regulatory schemes controlling the differentiation of mesophyll and vascular chloroplasts.
In this work, building on our long-term efforts to identify and characterize leaf variegation mutants, we isolated two allelic Arabidopsis thaliana variegation mutants, designated varigated5-1 (var5-1) and var5-2. Both mutant alleles showed a distinct reticulate leaf phenotype with yellow paraveinal regions and green interveinal regions. The molecular cloning of the VAR5 locus revealed that VAR5 encodes a putative magnesium transporter previously named as AtMRS2-11/AtMGT10. We confirmed that AtMRS2-11/AtMGT10/VAR5 is likely associated with the chloroplast envelope. In addition, blue native PAGE (BN-PAGE) analysis indicates that AtMRS2-11/AtMGT10/VAR5 forms large complexes in chloroplasts. Taken together, our data demonstrate that mutations in chloroplast Mg2+ transporter AtMRS2-11/AtMGT10/VAR5 can lead to a unique leaf reticulation phenotype and this intriguing tissue specific phenotype suggests that Mg2+ plays an important role in tissue specific regulation of chloroplast development.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana wild type, var5-1 and var5-2 mutants (all in Columbia-0 background) seeds were grown on commercial soil mix (Pindstrup, Denmark) or on Murashige and Skoog (MS) (Caisson Laboratories, Smithfield, UT, United States) containing 1% sucrose, and maintained under continuous light (∼100 μmol⋅m-2s-1) at 22°C in controlled growth rooms. The GABI T-DNA insertion line (TAIR stock number CS323329) was obtained from the TAIR.
Map-Based Cloning
To map the VAR5 locus, var5-1 was crossed with Ler to generate the F2 mapping population. Bulked segregant analysis using a pool of DNAs from 96 F2 mapping plants with the var5-1 phenotype and markers on all five Arabidopsis chromosomes placed VAR5 between two molecular markers T29J13#1 and nga139 on chromosome 5. For fine mapping, 1,140 individual mapping plants were used to place the VAR5 locus into a genomic region of 92 kb. In this region, genomic sequences of putative chloroplast genes were amplified by PCR and sequences and a point mutation in At5g22830 was detected. All primers used in this study are listed in Supplementary Table S1.
Chlorophyll Fluorescence Measurement and Starch Staining
The maximum quantum yield of PS II (FV/FM) was measured with the Open FluorCam FC800-O system (Photon Systems Instruments, Czechia) as described (Qi et al., 2016). To reveal the starch accumulation patterns in leaves, entire rosettes were boiled in 80% ethanol (v/v) until pigments were removed, then stained with fresh iodine solution (10 g/L KI and 1 g/L I2) for 5 min, and de-stained in water for 1 h.
Light Microscopy Observation of Leaf Vein
The middle main vein section from the third or fourth true leaves of wild type and var5-1 were hand cut and fixed in 4% (v/v) glutaraldehyde in 0.1 mM PBS buffer (pH 6.8) at 4°C overnight. After fixation, samples were dehydrated in a serial dilution of ethanol and embedded in Technovit 7100 resin (Kulzer, Wehrheim, Germany). Transverse semi-thin cross sections (3 μm) were prepared with Leica RM2265 microtome, placed on glass slides, stained with fresh iodine solution. Images of iodine stained leaf cross sections were obtained with a DM5000B microscope (Leica) equipped with a CCD camera.
Nucleic Acid Analysis and Arabidopsis Transformation
Arabidopsis DNA was isolated with the CTAB method (Yu et al., 2004). Total RNA was isolated with the TRIzol reagent according to the manufacturer’s manual (Invitrogen, Carlsbad, CA, United States). cDNAs were synthesized with the Transcriptor First Strand cDNA synthesis kit (Roche, Switzerland) with random primers. For the complementation of the var5-1 mutant, full-length At5g22830 cDNAs were amplified with TaKaRa PrimeSTAR polymerase using primers 22830F and 22830R and subsequently cloned into the binary vector pBI111L to generate P35S: At5g22830 (Yu et al., 2004). This construct was transformed in the var5-1 mutant using the floral dip method (Clough and Bent, 1998), and transgenic plants were screened on 1/2 MS plates containing 50 mg/L kanamycin.
Bioinformatic Analysis
AtMRS2-11/AtMGT10/VAR5 homologous proteins in different species were identified using the BLASTP program of the National Center for Biotechnology Information. Multiple sequence alignment and phylogenetic analysis were performed using the MEGA6 software (Tamura et al., 2013). Gene structures were constructed based on gene sequences retrieved from the Phytozome resource1 (Goodstein et al., 2012).
Protoplast Transient Expression Assay
To construct a transient expression vector P35S:VAR5-GFP, full-length coding sequences of AtMRS2-11/AtMGT10/VAR5 were amplified with primers 22830GFPF and 22830GFPR and cloned into pUC18-GFP vector with the GFP coding sequences fused at its 3′ terminus, and the expression of VAR5-GFP was under the CaMV 35S promoter. The 35S promoter in P35S:VAR5-GFP was replaced by a ∼1.9 kb promoter region of VAR5 to generate PV AR5:VAR5-GFP. These constructs, as well as the P35S:GFP control vector, were transformed into protoplasts isolated from 4-week-old wild-type plants as described (Yoo et al., 2007). Chlorophyll fluorescence and GFP signals were examined by a Nikon A1 confocal microscope (Nikon, Japan).
Chloroplast Isolation, Fractionation, and BN-PAGE
Intact chloroplasts were isolated from 4-week-old Arabidopsis plants that were dark treated for 16 h (Kunst, 1998). To fractionate sub-chloroplast compartments, intact chloroplasts were broken by passing through a 24-gauge syringe in hypotonic buffer (10 mM Hepes-KOH pH7.6, 5 mM MgCl2). The lysate was separated on sucrose gradients by ultracentrifugation with SW41 Ti rotor at 58,000 × g for 2 h (Beckman Coulter L100-XP), and fractions were collected as described (Qi et al., 2016). For BN-PAGE, chloroplast samples equivalent to 10 μg of total chlorophylls were solubilized in 25BTH20G buffer [25 mM Bis-Tris-HCl, pH 7.0, 20% (w/v) glycerol] containing 1.0% (w/v) of n-dodecyl-β-D-maltoside (β-DM) (Järvi et al., 2011). The solubilized chloroplast samples were resolved on 3–8% BN-PAGE with a constant voltage of 70 V at 4°C, and the first dimension gel strips were denatured and resolved on 10% SDS-PAGE. For immunoblot analysis, proteins separated by SDS-PAGE were transferred onto 0.2 μm PVDF membranes with the Trans-blot Semi-Dry Transfer Cell (Bio-Rad) following the manufacturer’s manual. PVDF membranes were incubated with specific primary antibodies (Agrisera), followed by horseradish peroxidase-conjugated secondary antibodies. Signals were detected with chemiluminescence solutions and ChemiDoc Imaging System (Bio-Rad).
Results
The Isolation of Two Reticulate Mutant Alleles var5-1 and var5-2
During the characterization of an Arabidopsis SALK T-DNA insertion line, one unique reticulate (‘net-like’) leaf variegation mutant was recovered (Figure 1A; early stage of this work was carried out in the lab of Dr. Steve Rodermel at Iowa State University). A second and similar reticulate leaf variegation mutant was identified in our genetic screens for chloroplast development mutants (Supplementary Figures S1A–D, Qi et al., 2016). Genetic tests revealed that these two mutants were allelic and we thus designated the two mutants as variegated5-1 (var5-1) and variegated5-2 (var5-2), respectively, following the variegation mutant naming sequence (Martínez-Zapater, 1993; Yu et al., 2007) (Supplementary Figures S1G,H). Both mutant alleles showed a similar reticulate leaf variegation phenotype (Figures 1A,B and Supplementary Figure S1A). This unique leaf variegation phenotype is more pronounced in adult leaves than juvenile leaves, as the seventh true leaf of var5-1 showed a clear reticulate phenotype, with overall yellow paraveinal regions and green interveinal regions (Figure 1B). Taken together, var5-1 and var5-2 represent two allelic mutants that display a unique leaf reticulation phenotype.
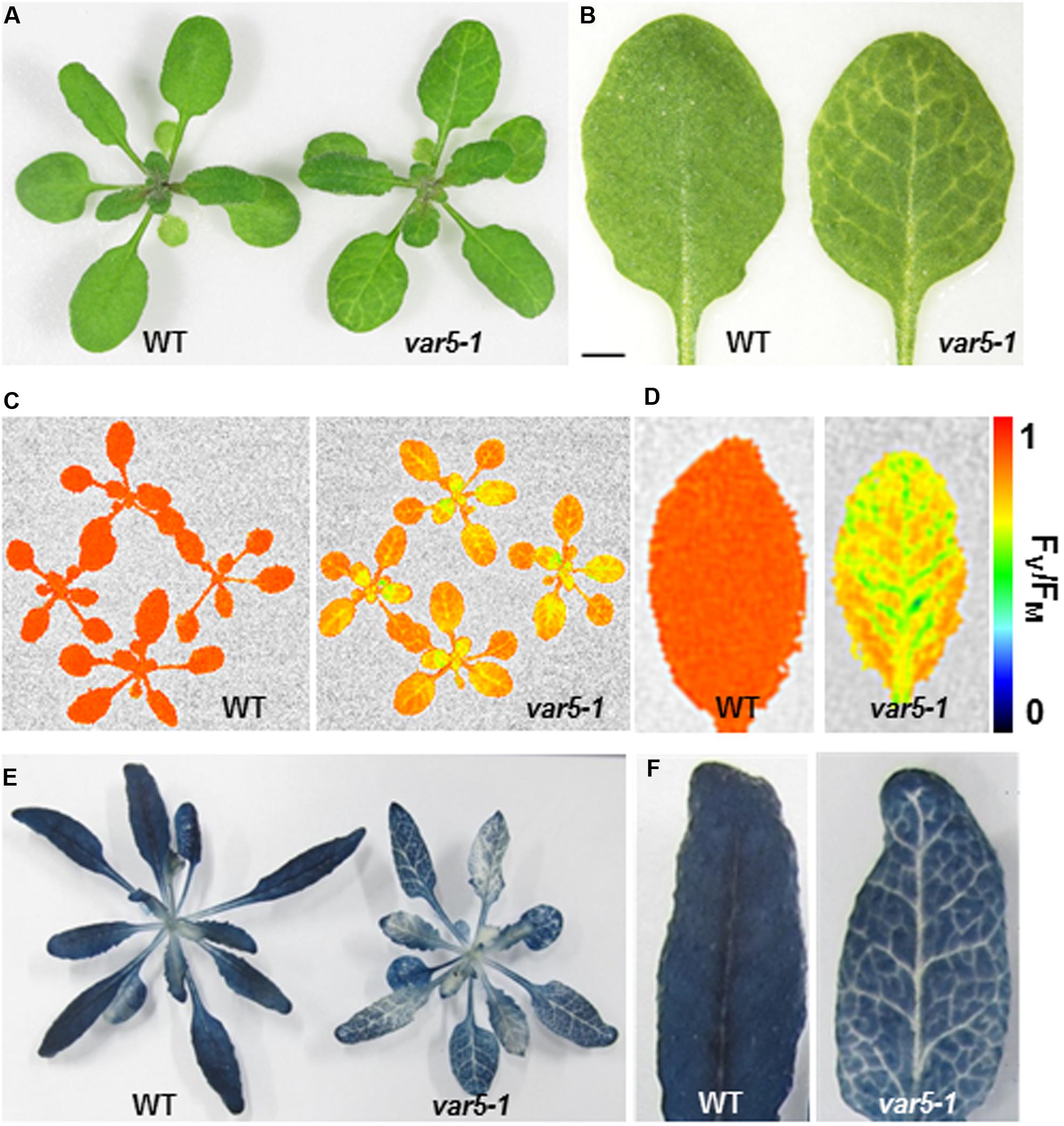
FIGURE 1. Phenotypes of the var5-1 mutant. (A). Representative 3-week-old seedlings of wild type (WT) and the var5-1 mutant. (B) Details of the third or fourth true leaf from 3-week-old WT and var5-1 mutant. (C,D) FV/FM measurement of WT and the var5-1 mutant (C for 3-week-old whole plants, and D for the detached seventh or eighth true leaves from 4-week-old plants). The color scale representing for the value of FV/FM is at the right of the figure. (E,F) Iodine staining with seedlings of 4-week-old WT and the var5-1 mutant.
Tissue Specific Starch Accumulation Defects in the var5-1 Mutant
The reticulate leaf color phenotype suggests that photosynthetic capacities are compromised in var5-1 mutants. To correlate the visible leaf color phenotype with the photosynthetic status of the mutant, we measured parameters that indicate photosynthetic performance. For the light reactions, we measured the maximum quantum yield of PS II, also known as FV/FM, using a whole plant chlorophyll fluorescence imaging system. The average FV/FM of 15 min dark adapted wild-type whole plants was 0.82, while that of var5-1 was 0.66, suggesting that photosynthesis is compromised in the mutant (Figure 1C). In addition, while a single leaf of wild-type plant showed a uniform distribution of FV/FM values, the FV/FM from var5-1 also showed a ‘net-like’ pattern, with paraveinal regions showing higher value while interveinal regions having lower FV/FM (Figure 1D). We next examined the accumulation of starch in the wild type and var5-1 plants as one indicator of photosynthetic productivities. Interestingly, the starch accumulation in the var5-1 mutant shows clearly a reticulate pattern, clearly matching both the leaf reticulation and FV/FM patterns (Figures 1E,F). In addition, iodine stained leaf cross sections showed that chloroplasts in the palisade mesophyll cells on the top of vascular bundles in the var5-1 mutant, were significantly less stained compared to that of wild type (Supplementary Figures S2A–D), suggesting the development of chloroplasts in the yellow paraveinal regions are compromised in the var5-1 mutant. These data indicate that the mutation in var5-1 leads to a unique expression of leaf reticulation and a pronounced reduction of photosynthetic capacities as indicated by PSII activities and starch accumulation in paraveinal regions than in interveinal regions.
The Molecular Cloning of VAR5
Genetic analysis revealed that the reticulate phenotype of var5-1 was likely caused by a single nuclear recessive mutation, and map-based cloning procedures were used to clone the VAR5 locus (Figure 2). Initial mapping with molecular markers on all five chromosomes and subsequent fine mapping using 1,140 mapping plants placed VAR5 in a ∼92 kb genomic region between markers MDJ22#2 and MRN17#1 on chromosome 5 (Figure 2A). Based on the leaf color phenotype, genomic DNA sequences of nuclear genes for chloroplast proteins in this interval were examined. A single G to A transition mutation was identified in the first nucleotide of intron 9 in At5g22830 (Figure 2A). At the mRNA level, amplification products spanning the mutation site appeared longer in the mutant than in wild type, suggesting pre-mRNA splicing might be affected in var5-1 (Figure 2B). Sequencing of the full-length At5g22830 cDNA in var5-1 revealed the nature of pre-mRNA splicing defects in var5-1. The mutation in the intron 9 led to the retention of the entire intron 9 in the At5g22830 transcript (Figure 2C). Surprisingly, a partial segment of intron 6 (the first 17 bp of the 328 bp wild-type intron 6) was also retained (Figure 2C). The translation of this aberrant mRNA would in theory give rise to a protein with the first 262 amino acids of the annotated 459 amino acids of wild-type protein plus four additional amino acids before a premature stop codon (Figure 2D). In theory, this early stop codon would abolish the C-terminal transmembrane domains and the conserved GMN motif (Figure 2E). We also determined the mutation site in var5-2 and identified a G to A mutation at the last nucleotide of intron 4, which also caused abnormal At5g22830 transcript in var5-2 (Supplementary Figures S1E,F). This mutation led to the retention of the entire intron 4 (80 bp) in the At5g22830 transcript and a putative translation product with the first 208 amino acids of wild-type protein before a premature stop codon (Supplementary Figures S1E,F).
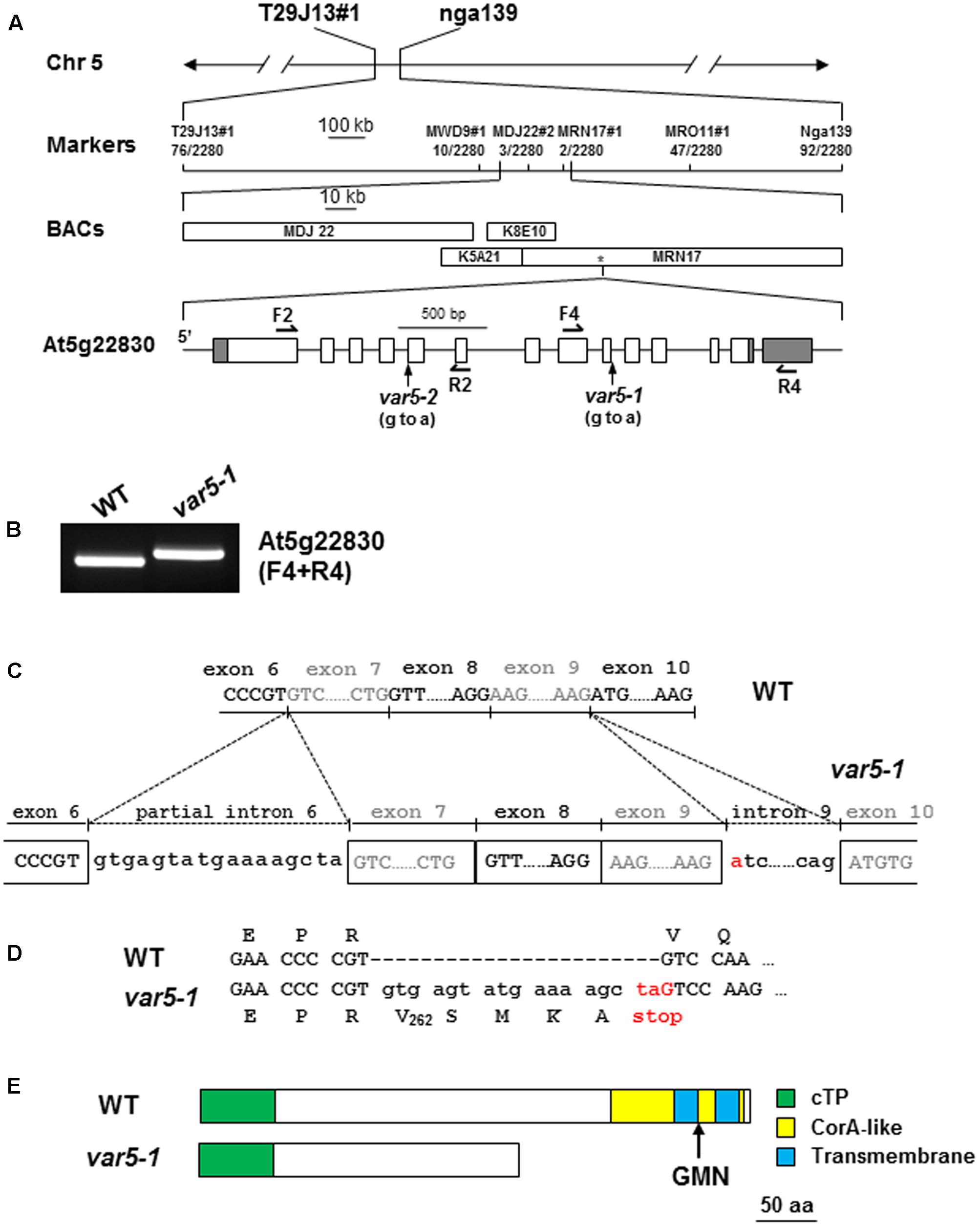
FIGURE 2. The molecular cloning of VAR5. (A) The map based cloning of VAR5. The mutation of var5-1 was linked to markers T29J13#1 and nga139 on chromosome 5. A total of 1,140 F2 individual plants (2,280 chromosomes) were used in fine mapping, and the numbers of recombinants are listed under each marker. The asterisk indicates the position of At5g22830. In At5g22830 gene model, gray boxes and white boxes represent UTR and exons, respectively, and solid lines indicate introns. The position of the G to A mutation in var5-1 was shown. (B) RT-PCR analysis of At5g22830 transcripts in WT and var5-1. Primers used in the amplifications were indicated in the parentheses. (C) The molecular nature of mutations in var5-1. The G to A transition mutation was shown red in the first nucleotide of intron 9 in At5g22830. Intron 9 and partial intron 6 remain in the At5g22830 transcripts in var5-1. (D) The var5-1 mutation leads to a premature stop codon at the end of partial intron 6. (E) Based on the sequences of abnormal mRNAs of At5g22830 in var5-1, the putative translation products of mutant mRNA of At5g22830 was indicated. Domains are indicated with colored boxes, and cTP for chloroplast transit peptide.
To confirm that the reticulate phenotype of var5-1 was indeed caused by the mutation identified in At5g22830, a binary vector (P35S: At5g22830) containing the wild-type At5g22830 cDNA under the control of the CaMV 35S promoter was constructed and transformed into var5-1. Multiple transgenic lines that showed completely green leaf color and the presence of wild-type cDNA amplification products were identified, indicating that the reticulate phenotype of var5-1 can be rescued by the wild-type At5g22830 (Figures 3A–D). At the mRNA level, the complementation lines showed the presence of wild-type At5g22830 cDNA (Figure 3E). At the protein level, we generated polyclonal antibodies against amino acid residues from 61 to 263 of At5g22830 and Western blotting analyses detected the accumulation of At5g22830 protein in the wild type, but not in the var5-1 mutant (Figure 3F). In contrast, At5g22830 protein accumulation was greatly enhanced in the var5-1 P35S:At5g22830 complementation line (Figure 3F). Similar results were obtained with var5-2 mutant (Supplementary Figure S1G). The identifications of mutations sites in var5-1 and var5-2, the molecular complementation and the western data show that At5g22830 is VAR5.
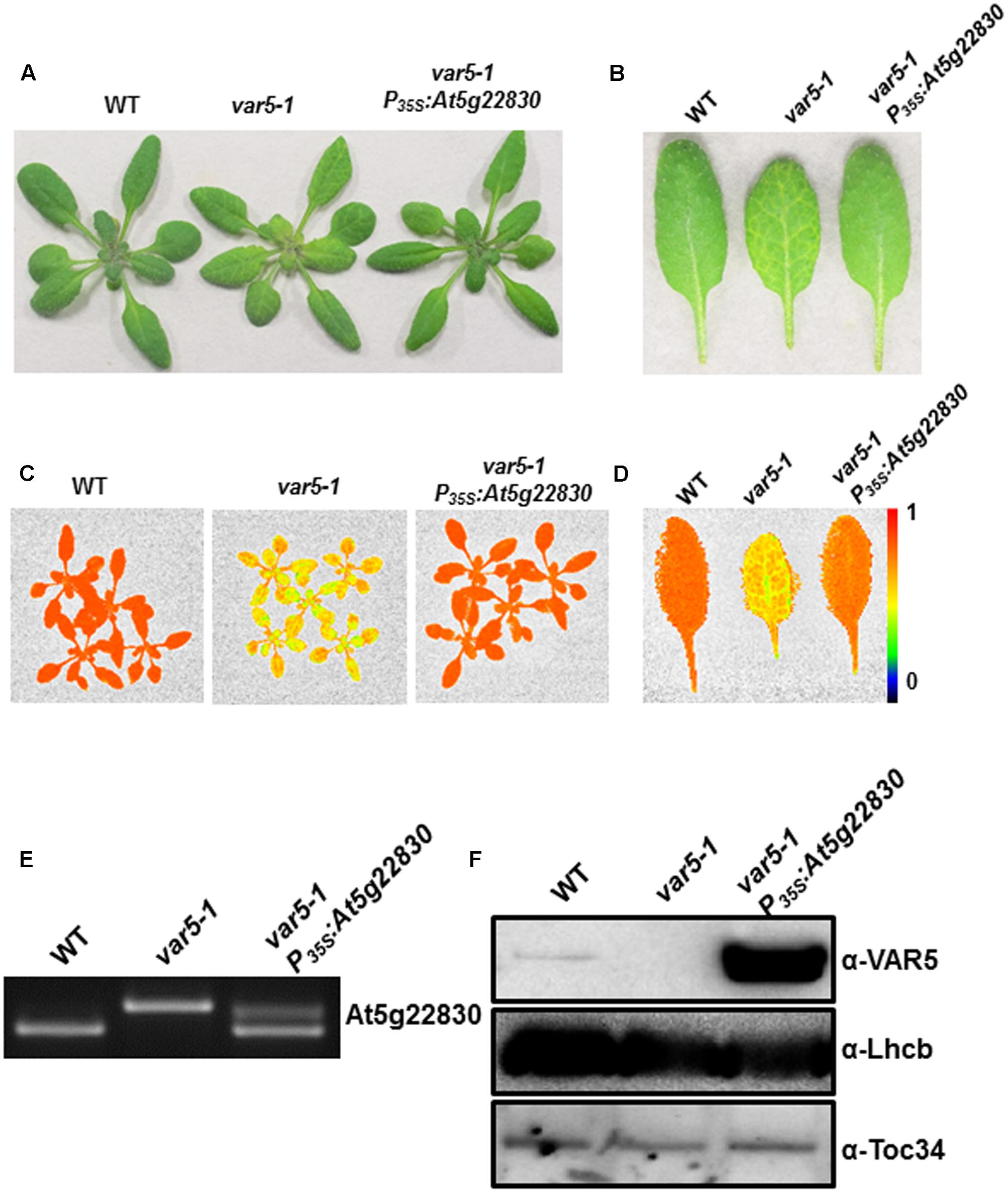
FIGURE 3. Complementation of var5-1. (A) Representative 3-week-old WT, var5-1, and the var5-1 complementation line (var5-1 P35S:At5g22830). (B) Representative individual leaves of WT, var5-1, and var5-1 P35S:At5g22830. (C,D) FV/FM measurement of WT, var5-1 and var5-1 P35S:At5g22830 (C for 18-day-old whole plants, and D for the detached third or fourth true leaves from 18-day-old plants). The color scale representing for the value of FV/FM is at the right of the figure. (E) At5g22830 transcript accumulations in WT, var5-1, and var5-1 P35S:At5g22830. Total leaf RNA was extracted, and subjected to reverse transcription PCR with primers F4 and R4. (F) At5g22830 protein accumulations in WT, var5-1, and var5-1 P35S:At5g22830. Membrane fractions were isolated from leaf tissues and proteins were separated with 10% SDS-PAGE. Immunoblots were probed with antibodies against At5g22830, Lhcb and Toc34.
VAR5/At5g22830 Encodes a Putative CorA Family Magnesium Transporter, AtMRS2-11/AtMGT10, in Arabidopsis
VAR5/At5g22830 was previously reported to encode a putative CorA family Mg2+ transporter that is also known as AtMRS2-11/AtMGT10 (Schock et al., 2000; Li et al., 2001; Drummond et al., 2006). At the protein level, AtMRS2-11/AtMGT10/VAR5 contains the characteristic features of CorA Mg2+ transporters, including the two C-terminal transmembrane domains and in between the Mg2+-binding GMN motif (Lunin et al., 2006; Pfoh et al., 2012). There are at least 10 members of CorA family Mg2+ transporters in Arabidopsis thaliana, and some of them are predicted to be targeted into organelles including the chloroplast and the mitochondrion (Gebert et al., 2009). We analyzed the relationship between AtMRS2-11/AtMGT10/VAR5 and its putative chloroplast-localized homologs from various eukaryotic photosynthetic organisms, as well as CorA homologs from bacteria (Figure 4A). Overall, AtMRS2-11/AtMGT10/VAR5 homologs from different organisms are highly conserved (Figure 4A). In addition, these proteins can be grouped into major clades including eudicot, monocot, algae and prokaryotic organisms (Figure 4A). The evolutionary relationship between these genes were also obvious at the DNA level as putative higher plant AtMRS2-11/AtMGT10/VAR5 related genes shared identical gene structures, suggesting these genes emerged before the divergence of eudicot and monocot plants (Figure 4B). In contrast, AtMRS2-11/AtMGT10/VAR5 homologs from green algae did not share conserved gene structures with their plant counterparts (Figure 4B). These data suggest that the CorA family Mg2+ transporter is prevalent and highly conserved, consistent with the critical roles Mg2+ plays in prokaryotic and eukaryotic cells.
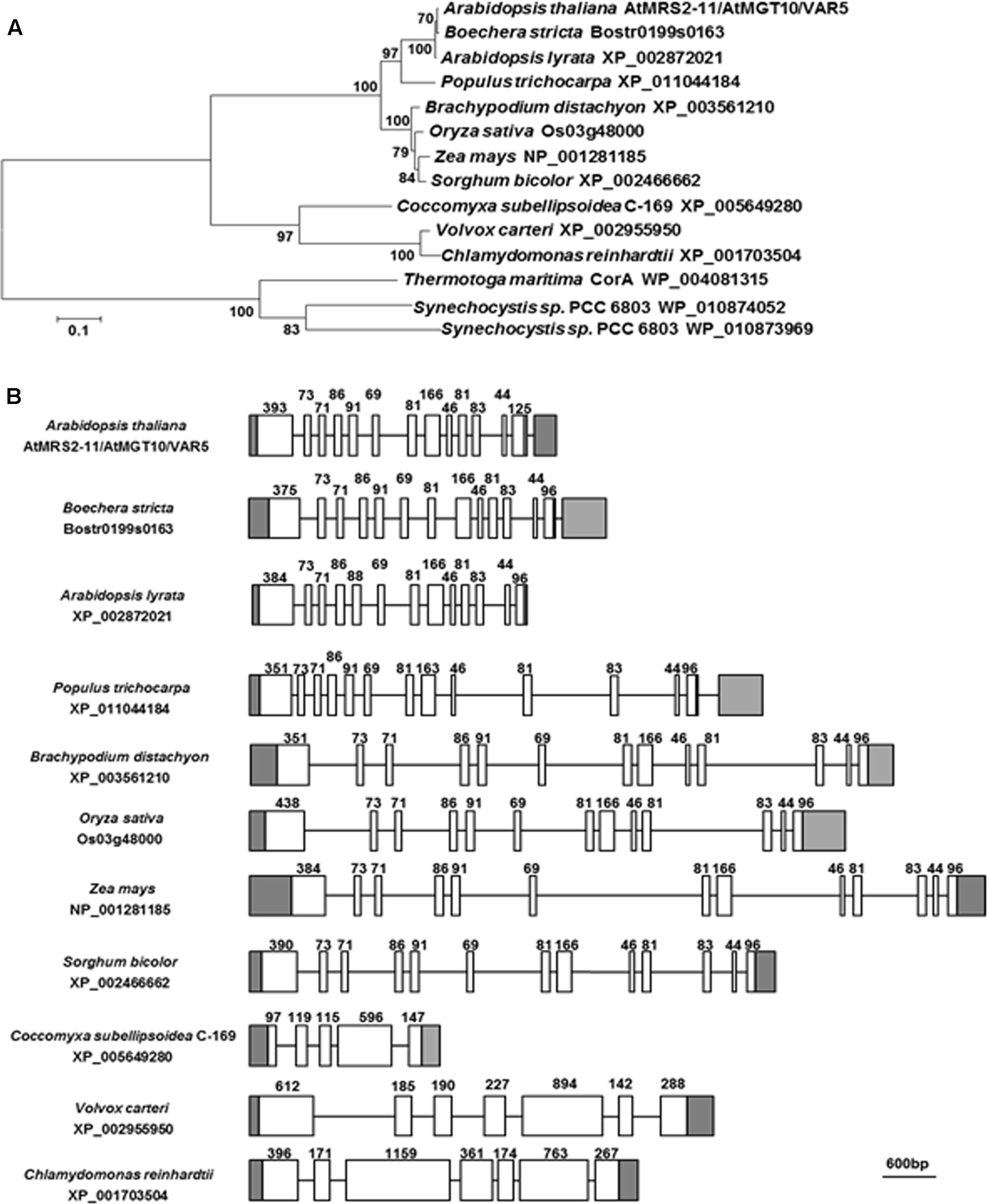
FIGURE 4. The AtMRS2-11/AtMGT10/VAR5 gene family. (A) Phylogenetic analysis of the AtMRS2-11/AtMGT10/VAR5 protein family. Full-length protein sequences of AtMRS2-11/AtMGT10/VAR5 homologous proteins from dicots Arabidopsis thaliana, Boechera stricta, Arabidopsis lyrata, Populus trichocarpa, monocots Brachypodium distachyon, Oryza sativa, Zea mays, Sorghum bicolor, green algae Coccomyxa subellipsoidea, Volvox carteri, Chlamydomonas reinhardtii, and prokaryotes Thermotoga maritime and Synechocystis sp. PCC6803 were obtained from NCBI. The phylogenetic tree was constructed with MEGA6 (Tamura et al., 2013). The accession numbers were included. (B) Gene Structures of AtMRS2-11/AtMGT10/VAR5 homologous genes. Genomic DNA sequences were retrieved from the Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). In gene structures, white boxes and lines indicate exons and introns, respectively. Gray boxes indicate UTRs. Lengths of coding exons sequences were shown.
AtMRS2-11/AtMGT10/VAR5 Localizes to the Chloroplast Envelope
AtMRS2-11/AtMGT10/VAR5 is predicted by the ChloroP program to contain a putative chloroplast transit peptide (Emanuelsson et al., 1999). Experimentally, it was shown to be localized on the periphery of chloroplasts (Drummond et al., 2006). We took two approaches to independently confirm the sub-cellular localization of AtMRS2-11/AtMGT10/VAR5. First, we used the protoplast transient expression assay to examine the localization of VAR5-GFP under the control of the 35S promoter (P35S:VAR5-GFP) and the endogenous VAR5 promoter (PV AR5:VAR5-GFP), respectively (Figure 5A). Control vector (P35S:GFP) gave GFP signals mainly in the cytosol and the nucleus (Figure 5A). In contrast, both P35S:VAR5-GFP and PV AR5:VAR5-GFP gave GFP signals that outlined the periphery of the chloroplast, as well as some puncta inside the chloroplast, suggesting both promoters can direct VAR5 to sub-chloroplast locations that likely correspond to the chloroplast envelope (Figure 5A). Next, we determined the AtMRS2-11/AtMGT10/VAR5 sub-chloroplast localization biochemically (Figure 5B). Intact chloroplasts were fractionated into the thylakoids (Thy), the stroma (Str), the envelope fraction 1 (Env1) and 2 (Env2) with sucrose gradient centrifugations (Qi et al., 2016). Using antibodies against chloroplast proteins with known sub-chloroplast localizations, we showed that RbcL (Rubisco large subunit) was enriched in the stroma fraction, and Lhcb (PSII light harvesting complex protein) was enriched in the thylakoids fraction as expected. The enrichment of chloroplast envelope membranes in the Env2 fraction was indicated by chloroplast envelope proteins Tic40 and Toc34 (components of the Translocon at the Inner/Outer Envelope Membrane of Chloroplasts, TIC/TOC) (Figure 5B; Nakai, 2015). Using VAR5 antibodies that we had generated, we found that the majority of AtMRS2-11/AtMGT10/VAR5 was present in the Env2 fraction, similar to the distribution patterns of TIC/TOC components, suggesting that it is likely a chloroplast envelope membrane protein (Figure 5B).
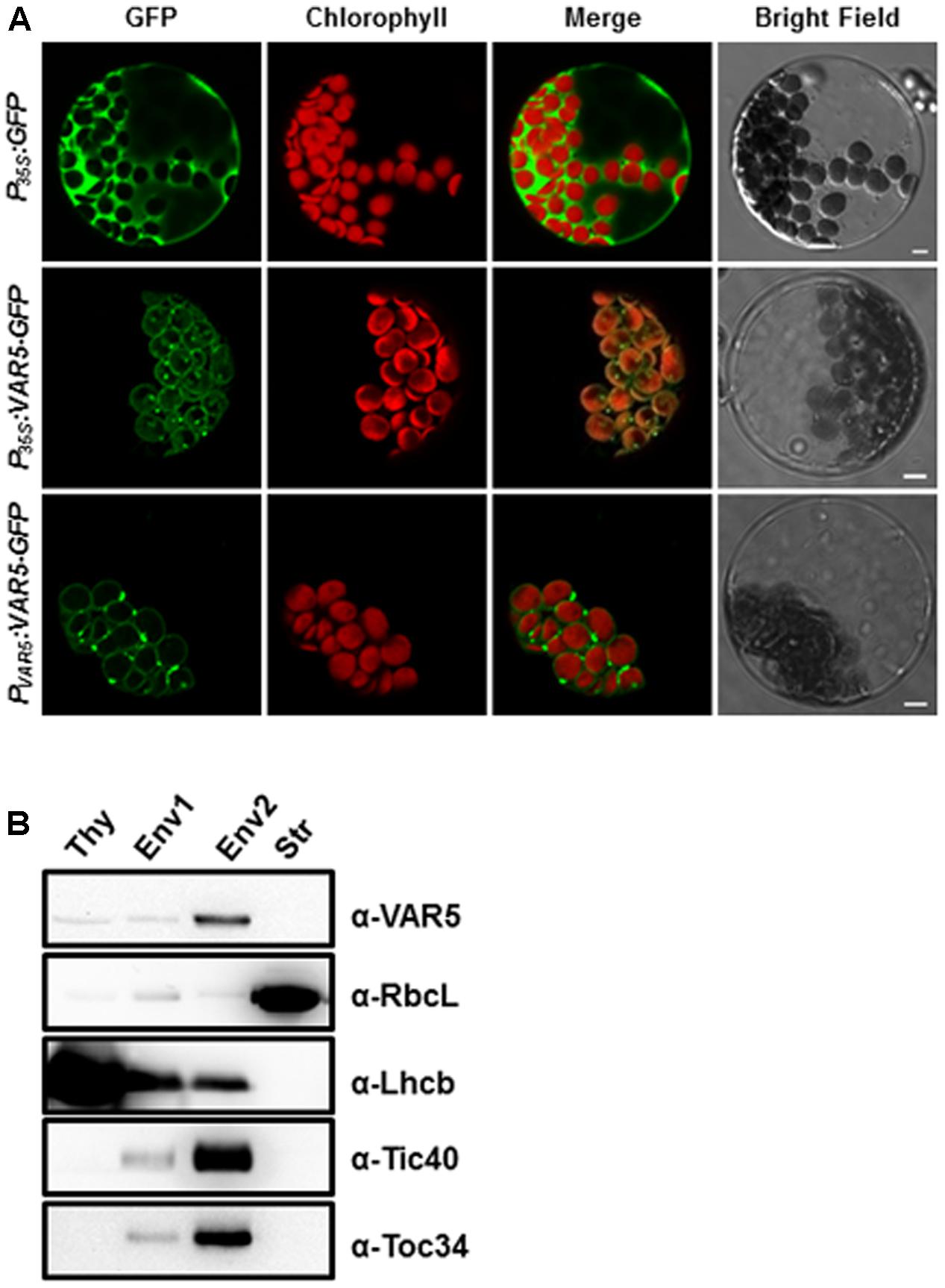
FIGURE 5. Subcellular localization of AtMRS2-11/AtMGT10/VAR5. (A) Protoplasts from 3-week-old WT plants were transformed with vectors containing the full-length VAR5 (P35S:VAR5-GFP and PV AR5:VAR5-GFP) and the GFP control (P35S:GFP) and examined via confocal microcopy. Representative protoplasts with GFP fluorescence were shown. Chlorophyll is for chlorophyll fluorescence, and bars are 5 μm. (B) Fractions of thylakoid (Thy), Env1 (mixed envelope and thylakoids), Env2 (mainly envelope) and stroma (Str) were isolated from WT chloroplasts and subjected to immunoblot. Antibodies against RbcL (stroma control), Lhcb (thylakoid control), Tic40 and Toc34 (envelope control) were used to indicate different fractions.
AtMRS2-11/AtMGT10/VAR5 Forms Large Protein Complexes in the Chloroplast
The crystal structure of the thermophilic bacterium Thermotoga maritima CorA Mg2+ transporter indicates that it forms a homopentamer (Lunin et al., 2006). To determine whether AtMRS2-11/AtMGT10/VAR5 forms complexes in Arabidopsis, two-dimensional BN-PAGE was carried out (Järvi et al., 2011). BN-PAGE can effectively separate chloroplast membrane protein complexes, and the estimated sizes of the major photosynthetic protein complexes are known (Figure 6). For instance, the sizes of the four large PSII supercomplexes were estimated to be 880–1,400 kDa (Caffarri et al., 2009; Hou et al., 2015), while the PSI monomer and the rubisco holoenzyme migrate at approximately 600 and 540 kDa, respectively (Berghöfer and Klösgen, 1999; Järvi et al., 2011). Western blot analysis with second dimension gels using antibodies against AtpA (the α subunit of the chloroplast ATP synthase), Lhcb, PetC (a subunit of the cytochrome b6f complex) and PsaF (a subunit of the PSI) clearly identified various photosynthetic complexes at expected sizes (Figure 6). Interestingly, AtMRS2-11/AtMGT10/VAR5 antibodies detected a streak of signals in the second dimension gels (Figure 6). Based on our results, it is clear that different forms of complexes containing AtMRS2-11/AtMGT10/VAR5 exist in the chloroplast, and the sizes of these complexes are clearly larger than the PSI monomer (∼600 kDa) (Figure 6). The distribution of AtMRS2-11/AtMGT10/VAR5 complexes overlapped partially with the PSII supercomplexes, which suggest the complexes could reach over 800 kDa (Figure 6). This is in stark contrast to the CorA complexes in Thermotoga maritime, which have molecular weights around 210 kDa as homopentamers (Lunin et al., 2006; Matthies et al., 2016). We did not detect AtMRS2-11/AtMGT10/VAR5 signals in var5-1 plants, agreeing with our one dimension SDS-PAGE results (Figure 2). These data suggest that AtMRS2-11/AtMGT10/VAR5 forms large complexes and the complex compositions are different in Arabidopsis from those in bacteria.
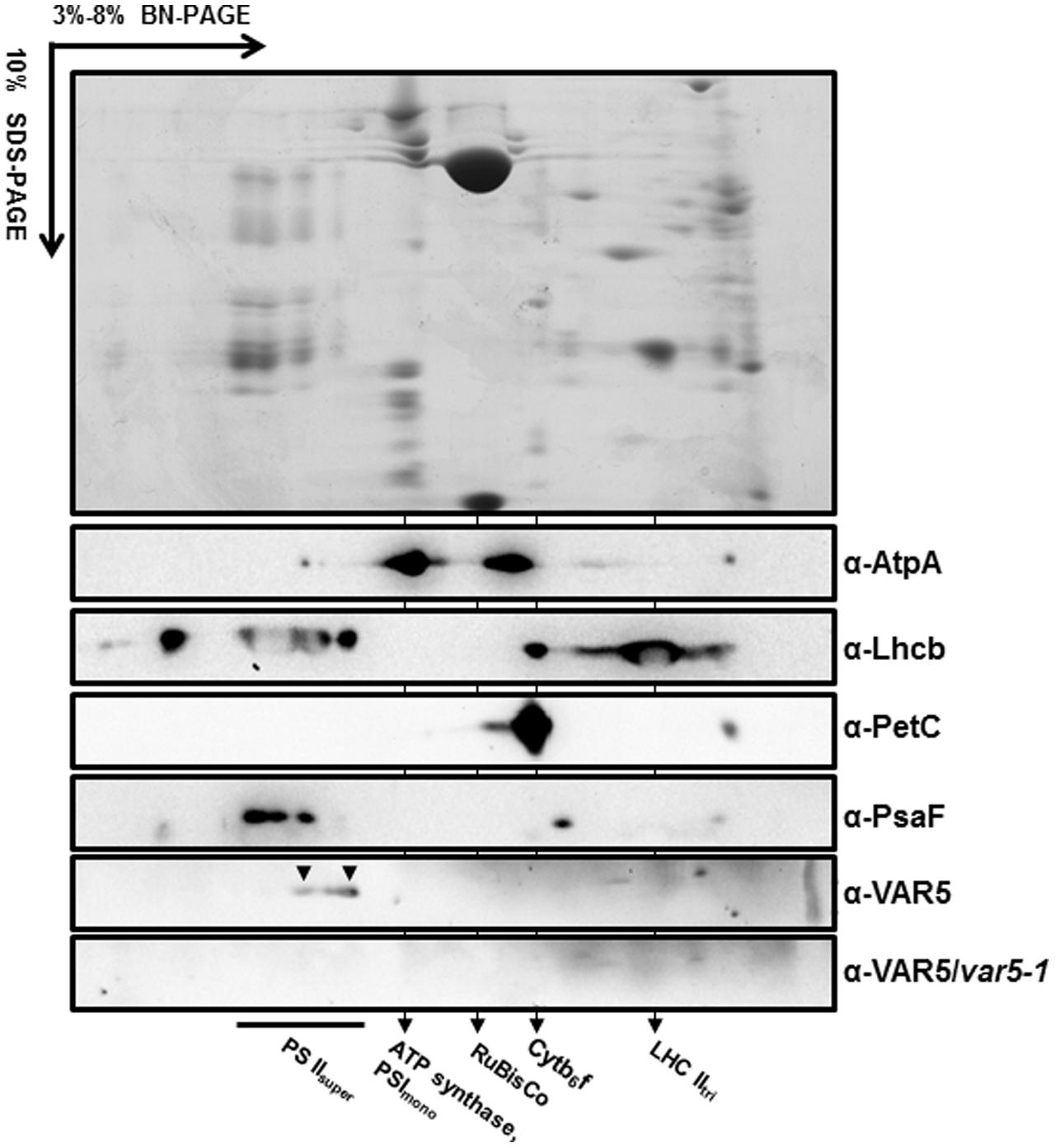
FIGURE 6. AtMRS2-11/AtMGT10/VAR5 forms large molecular weight complexes. Intact chloroplasts were solubilized with 1.0% β-DM, and subjected to first dimension 3–8% blue native PAGE (BN-PAGE). The first dimension gel strips were cut, denatured and resolved in 10% SDS-PAGE for the second dimension. Antibodies against subunits of different photosynthetic complexes were used for immunoblotting. The immunoblotting of second dimension BN-PAGE of var5-1 sample was used for a negative control.
Discussion
We are interested in the genetic regulation of chloroplast development and have characterized Arabidopsis leaf variegation mutants that are results of mutations in nuclear genes (Yu et al., 2007; Liu et al., 2010; Putarjunan et al., 2013). These variegation mutants are intriguing because non-uniform leaf color phenotypes can arises from uniform genetic mutant backgrounds. Plant variegation mutants can be categorized based on their variegation patterns. The first type of variegation mutants shows apparently random distributions of chlorophyll accumulation and/or chloroplast development. Mutants in this category include immutans (im), yellow variegated (var2), and thylakoid formation (thf) (reviewed in Yu et al., 2007). The second type, the virescent mutant, displays differential chlorophyll accumulation and/or chloroplast development along the leaf proximal-distal axis, usually with the leaf base regions containing less chlorophylls and tip regions containing higher levels of chlorophylls (López-Juez et al., 1998). The third type displays differential chlorophyll accumulation and/or chloroplast development between paraveinal regions and interveinal regions, giving rise to a reticulate pattern. A number of reticulate mutants including chlorophyll a/b binding protein (CAB) gene underexpressed 1 (cue1), reticulata (re; also known as lower cell density 1, lcd1), and differential development of vascular associated cells 1(dov1) have been reported and characterized (Kinsman and Pyke, 1998; Streatfield et al., 1999; Barth and Conklin, 2003; González-Bayón et al., 2006).
In this report, we identified two new allelic leaf variegation mutants in Arabidopsis thaliana that display a striking reticulate phenotype and designated these mutants as var5-1 and var5-2. We cloned VAR5 via map-based cloning procedures and identified mutations in At5g22830 in var5-1 and var5-2, respectively. Both mutants contain mutations in the conserved sequences that form the plant intron borders and these mutations lead to the mis-splicing of pre-mRNA of At5g22830 (Figure 2; Lorković et al., 2000). It is worthy of noting that the mutation in intron 9 caused mis-splicing of intron 6 in var5-1, suggesting that downstream intron sequences can influence the splicing of upstream introns (Nesic and Maquat, 1994). Our data from two mutant alleles, the molecular cloning of VAR5 and molecular complementation results clearly establish that the reticulate leaf phenotype of var5-1 and var5-2 is the result of mutations in At5g22830, which is also known as AtMRS2-11/AtMGT10.
AtMRS2-11/AtMGT10/VAR5 encodes a CorA family of Mg2+ transporter in Arabidopsis. In bacteria, the canonical CorA is situated at the plasma membrane and regulates Mg2+ homeostasis inside the cell (Maguire, 2006). CorA family Mg2+ transporters are also present in higher plants (Schock et al., 2000; Li et al., 2001). In Arabidopsis thaliana, the 10 members of the CorA family of Mg2+ transporters are shown to be targeted into different sub-cellular compartments, with AtMRS2-11/AtMGT10/VAR5 as the sole member that is predicted to contain a chloroplast transit peptide (Gebert et al., 2009). Given the many processes Mg2+ is involved in the chloroplast, it is reasonable to assume that Mg2+ transporters need to be present in the chloroplast envelope to facilitate the transport of Mg2+. Indeed, a previous report has shown that AtMRS2-11/AtMGT10/VAR5 is possibly located at the periphery of the chloroplast through fluorescent microscopy observations (Drummond et al., 2006). In this work, we demonstrated that AtMRS2-11/AtMGT10/VAR5 is likely located to the chloroplast envelope using two approaches. First, through protoplast transient expression and confocal microscopy, we showed that VAR5-GFP can be localized to sub-chloroplast compartments that likely correspond to the chloroplast envelope (Figure 5). We also confirmed the chloroplast envelope localization of VAR5 biochemically and found that VAR5 showed similar localization patterns with components of the TIC/TOC complexes at the chloroplast enveloped (Figure 5; Nakai, 2015). To our surprise, we did observe VAR5-GFP signals inside the chloroplast with both the 35S and the endogenous VAR5 promoter driven VAR5-GFP, and we do not yet know the nature of this phenomenon. Consistent with the chloroplast localization of AtMRS2-11/AtMGT10/VAR5, the two VAR5 mutant alleles we identified show clear defects of chloroplast development (Figure 1). The reticulate phenotype of both mutants indicates a tissue specific expression of mutant phenotype. There are several possible explanations for this intriguing phenotype. It is possible that the essential roles of vascular tissues for transporting may necessitate higher levels of AtMRS2-11/AtMGT10/VAR5 activities and the lack of the transporter in var5-1 mutants would lead to more detrimental effects in these tissues. Alternatively, other types of Mg2+ channels, such as fast activating chloroplast cation channels (FACC), may also be present in the chloroplast envelope to transport Mg2+ (Pottosin and Dobrovinskaya, 2015). It is thus possible that they may compensate for the lack of AtMRS2-11/AtMGT10/VAR5 in the interveinal regions. It was shown that AtMRS2-11/AtMGT10/VAR5 displays tissue specific expression patterns (Drummond et al., 2006; Gebert et al., 2009). However, the tissue expression pattern does not correlate with the reticulate mutant phenotype. Because both alleles we identified contain point mutations that lead to mis-regulated splicings of VAR5 pre-mRNA, we also sought to identify a T-DNA insertion line for VAR5. In a previous report, no homozygous mutant plant was recovered in a GABI T-DNA insertion line (TAIR stock number CS323329), which has a T-DNA insertion in the sixth intron, suggesting that the complete loss of AtMRS2-11/AtMGT10/VAR5 might be lethal (Gebert et al., 2009). Characterizing the same T-DNA line, we identified the precise insertion site of the T-DNA in VAR5 but also failed to obtain homozygous insertion plants (data not shown). It is possible that the null allele of VAR5 is lethal at early stage of embryo development. However, we could not rule out the possibility that additional secondary mutations may contribute to this lethality, as reported for other cases (Gebert et al., 2009; Bergelson et al., 2016).
In this report, we clearly demonstrated that AtMRS2-11/AtMGT10/VAR5 forms large complexes and the estimates of the molecular weights of these complexes are exceeding 800 kDa (Figure 6). The formation of complexes for the CorA family of Mg2+ transporters is not unexpected as it has been shown in bacteria that CorA forms homopentamers with a molecular weight of ∼200 kDa (Lunin et al., 2006; Matthies et al., 2016). However, the sizes we observed for AtMRS2-11/AtMGT10/VAR5 complexes are surprising and well beyond its bacterial counterparts. Although we don’t know the precise nature of these AtMRS2-11/AtMGT10/VAR5-containing complexes, it does suggest that the formation and regulation of CorA transporter complexes are different in plants, at least in the chloroplast. It is possible that in Arabidopsis chloroplasts, AtMRS2-11/AtMGT10/VAR5 forms homo-oligomers that contain more than five subunits. Alternatively, AtMRS2-11/AtMGT10/VAR5 complexes in the chloroplast may contain additional components. Considering the importance roles Mg2+ plays in the chloroplast, it is reasonable to assume that addition subunits may be necessary to serve structural or regulatory functions. The future determination of the components of these complexes containing AtMRS2-11/AtMGT10/VAR5 would shine more light on the functions of CorA family Mg2+ transporters in the chloroplast and the roles it plays in chloroplast development and biomass production, especially under low-magnesium or plus-magnesium conditions.
Author Contributions
FY conceived and coordinated the study. SL and YQ designed, performed, and analyzed the experiments shown in Figures 1–3 and supplemental data. YQ and JZ designed, performed, and analyzed the experiments shown in Figures 4, 6. YL and RW designed, performed, and analyzed the experiments shown in Figure 5. JS, LA, and XL provided technical assistance and contributed to the preparation of the figures. FY and YQ wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China Grants 31400216 (to JZ), 31300988 (to YQ), and 31470290 (to LA), by Natural Science Foundation of Shaanxi Province Grants 2015JM3081 (to FY), and by Fundamental Research Funds for the Central Universities Grants QN2013034 (to JS), 2014YB036 (to LA), and 2452015215 (to XL).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02007/full#supplementary-material
FIGURE S1 | The molecular nature of var5-2. (A) Representative 3-week-old seedlings of wild type (WT) and the var5-2 mutant. (B) Details of the third or fourth true leaf from 3-week-old WT and var5-2 mutant. (C,D) FV/FM measurement of WT and the var5-2 mutant (C for 3-week-old whole plants, and D for the detached third or fourth true leaves from 3-week-old plants). The color scale representing for the value of FV/FM is at the right of the figure. (E) The molecular nature of mutations in var5-2. Intron 4 remains in the At5g22830 transcripts in var5-2. (F) The var5-2 mutation leads to a premature stop codon at the beginning of intron 4. Based on the sequences of abnormal mRNAs of At5g22830 in var5-2, the putative translation products of mutant mRNA of At5g22830 was indicated. Domains are indicated with colored boxes. H. The representative first or second true leaf from 3-week-old WT, var5-1, var5-2, and var5-1 var5-2 F1 generation. (G) The accumulation of VAR5 in WT, var5-1, var5-2 and var5-1 var5-2 F1. TIC40 was used for the envelope protein control.
FIGURE S2 | Light microscopy of leaf sections. The middle main vein section from the third or fourth true leaves of 2-week-old WT and var5-1 were fixed and observed with iodine staining. (A) WT; (B) var5-1. (C,D) The dash line areas from A and B were enlarged.
Footnotes
Abbreviations
AtMGT, Arabidopsis thaliana Mg2+ transporters; AtMRS2, Arabidopsis thaliana mitochondrial RNA splicing2; CaMV, Cauliflower Mosaic Virus; CorA, Co2+ resistance A; GFP, green fluorescence protein; GMN, glycine–methionine–asparagine motif; Ler, Landsberg erecta; PSI, photosystem I; PSII, photosystem II; UTR: untranslated region; VAR5, Variegated5.
References
Barth, C., and Conklin, P. L. (2003). The lower cell density of leaf parenchyma in the Arabidopsis thaliana mutant lcd1-1 is associated with increased sensitivity to ozone and virulent Pseudomonas syringae. Plant J. 35, 206–218. doi: 10.1046/j.1365-313X.2003.01795.x
Bergelson, J., Buckler, E. S., Ecker, J. R., Nordborg, M., and Weigel, D. (2016). A proposal regarding best practices for validating the identity of genetic stocks and the effects of genetic variants. Plant Cell 28, 606–609. doi: 10.1105/tpc.15.00502
Berghöfer, J., and Klösgen, R. B. (1999). Two distinct translocation intermediates can be distinguished during protein transport by the TAT (Δph) pathway across the thylakoid membrane. FEBS Lett. 460, 328–332. doi: 10.1016/S0014-5793(99)01365-4
Bui, D. M., Gregan, J., Jarosch, E., Ragnini, A., and Schweyen, R. J. (1999). The bacterial magnesium transporter CorA can functionally substitute for its putative homologue Mrs2p in the yeast inner mitochondrial membrane. J. Biol. Chem. 274, 20438–20443. doi: 10.1074/jbc.274.29.20438
Caffarri, S., Kouril, R., Kereïche, S., Boekema, E. J., and Croce, R. (2009). Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3063. doi: 10.1038/emboj.2009.232
Chen, J., Li, L., Liu, Z., Yuan, Y., Guo, L., Mao, D., et al. (2009). Magnesium transporter AtMGT9 is essential for pollen development in Arabidopsis. Cell Res. 19, 887–898. doi: 10.1038/cr.2009.58
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Conn, S. J., Conn, V., Tyerman, S. D., Kaier, B. N., Leigh, R. A., and Gilliham, M. (2011). Magnesium transporters, MGT2/MRS2-1 and MGT2/MRS2-5, are important for magnesium partitioning within Arabidopsis thaliana mesophyll vacuoles. New Phytol. 190, 583–594. doi: 10.1111/j.1469-8137.2010.03619.x
Drummond, R. S. M., Tutone, A., Li, Y., and Gardner, R. C. (2006). A putative magnesium transporter AtMRS2-11 is localized to the plant chloroplast envelope membrane system. Plant Sci. 170, 78–89. doi: 10.1016/j.plantsci.2005.08.018
Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Gardner, R. C. (2003). Genes for magnesium transport. Curr. Opin. Plant Biol. 6, 263–267. doi: 10.1016/S1369-5266(03)00032-3
Gebert, M., Meschenmoser, K., Svidová, S., Weghuber, J., Schweyen, R., Eifler, K., et al. (2009). A root-expressed magnesium transporter of the MRS2/MGT gene family in Arabidopsis thaliana allows for growth in low-Mg2+ environments. Plant Cell 21, 4018–4030. doi: 10.1105/tpc.109.070557
González-Bayón, R., Kinsman, E. A., Quesada, V., Vera, A., Robles, P., Ponce, M. R., et al. (2006). Mutations in the RETICULATA gene dramatically alter internal architecture but have little effect on overall organ shape in Arabidopsis leaves. J. Exp. Bot. 57, 3019–3031. doi: 10.1093/jxb/erl063
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Hou, X., Fu, A., Garcia, V. J., Buchanan, B. B., and Luan, S. (2015). PSB27: a thylakoid protein enabling Arabidopsis to adapt to changing light intensity. Proc. Natl. Acad. Sci. U.S.A. 112, 1613–1618. doi: 10.1073/pnas.1424040112
Järvi, S., Suorsa, M., Paakkarinen, V., and Aro, E. M. (2011). Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem. J. 439, 207–214. doi: 10.1042/BJ20102155
Kinsman, E. A., and Pyke, K. A. (1998). Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development 125, 1815–1822.
Kunst, L. (1998). Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol. Biol. 82, 43–48. doi: 10.1385/0-89603-391-0:43
Li, L., Sokolov, L. N., Yang, Y., Li, D., Ting, J., Pandy, G. K., et al. (2008). A mitochondrial magnesium transporter functions in Arabidopsis pollen development. Mol. Plant 1, 675–685. doi: 10.1093/mp/ssn031
Li, L., Tutone, A. F., Drummond, R. S. M., Gardner, R. C., and Luan, S. (2001). A novel family of magnesium transport genes in Arabidopsis. Plant Cell 13, 2761–2775. doi: 10.1105/tpc.010352
Liu, X., Yu, F., and Rodermel, S. (2010). Arabidopsis chloroplast ftsH, var2 and suppressors of var2 leaf variegation: a review. J. Integr. Plant Biol. 52, 750–761. doi: 10.1111/j.1744-7909.2010.00980.x
López-Juez, E., Jarvis, R. P., Takeuchi, A., Page, A. M., and Chory, J. (1998). New Arabidopsis cue mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol. 118, 803–815. doi: 10.1104/pp.118.3.803
Lorković, Z. J., Wieczorek-Kirk, D. A., Lambermon, M. H., and Filipowicz, W. (2000). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5, 160–167. doi: 10.1016/S1360-1385(00)01595-8
Lundquist, P. K., Rosar, C., Bräutigam, A., and Weber, A. P. M. (2014). Plastid signals and the bundle sheath: mesophyll development in reticulate mutants. Mol. Plant 7, 14–29. doi: 10.1093/mp/sst133
Lunin, V. V., Dobrovetsky, E., Khutoreskaya, G., Zhang, R., Joachimiak, A., Doyle, D. A., et al. (2006). Crystal structure of the CorA Mg2+ transporter. Nature 404, 833–837. doi: 10.1038/nature04642
Mao, D., Chen, J., Tian, L., Liu, Z., Yang, L., Tang, R., et al. (2014). Arabidopsis transporter MGT6 mediates magnesium uptake and is required for growth under magnesium limitation. Plant Cell 26, 2234–2248. doi: 10.1105/tpc.114.124628
Mao, D., Tian, L., Li, L., Chen, J., Deng, P., Li, D., et al. (2008). AtMGT7: an Arabidopsis gene encoding a low-affinity magnesium transporter. J. Integr. Plant Biol. 50, 1530–1538. doi: 10.1111/j.1744-7909.2008.00770.x
Maguire, M. E. (2006). The structure of CorA: a Mg2+-selective channel. Curr. Opin. Struct. Biol. 16, 432–438. doi: 10.1016/j.sbi.2006.06.006
Maguire, M. E., and Cowan, J. A. (2002). Magnesium chemistry and biochemistry. Biometals 15, 203–210. doi: 10.1023/A:1016058229972
Martínez-Zapater, J. M. (1993) Genetic analysis of variegated mutants in Arabidopsis. J. Hered. 84, 138–140. doi: 10.1093/oxfordjournals.jhered.a111298
Matthies, D., Dalmas, O., Borgnia, M. J., Dominik, P. K., Merk, A., Rao, P., et al. (2016). Cryo-EM structures of the magnesium channel CorA reveal symmetry break upon gating. Cell 164, 747–756. doi: 10.1016/j.cell.2015.12.055
Mollá-Morales, A., Sarmiento-Mañús, R., Robles, P., Quesada, V., Pérez-Pérez, J. M., González-Bayón, R., et al. (2011). Analysis of ven3 and ven6 reticulate mutants reveals the importance of arginine biosynthesis in Arabidopsis leaf development. Plant J. 65, 335–345. doi: 10.1111/j.1365-313X.2010.04425.x
Moomaw, A. S., and Maguire, M. E. (2008). The unique nature of Mg2+ channels. Physiology 23, 275–285. doi: 10.1152/physiol.00019.2008
Nakai, M. (2015). The TIC complex uncovered: the alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim. Biophys. Acta 1847, 957–967. doi: 10.1016/j.bbabio.2015.02.011
Nesic, D., and Maquat, L. E. (1994). Upstream introns influence the efficiency of final intron removal and RNA 3’-end formation. Genes Dev. 8, 363–375. doi: 10.1101/gad.8.3.363
Pfoh, R., Li, A., Chakrabarti, N., Payandeh, J., Pomes, R., and Pai, E. F. (2012). Structural asymmetry in the magnesium channel CorA points to sequential allosteric regulation. Proc. Natl. Acad. Sci. U.S.A. 109, 18809–18814. doi: 10.1073/pnas.1209018109
Pogson, B. J., and Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155, 1545–1551. doi: 10.1104/pp.110.170365
Pottosin, I., and Dobrovinskaya, O. (2015). Ion channels in native chloroplast membranes: challenges and potential for direct patch-clamp studies. Front. Physiol. 6:396. doi: 10.3389/fphys.2015.00396
Putarjunan, A., Liu, X., Nolan, T., Yu, F., and Rodermel, S. (2013). Understanding chloroplast biogenesis using second-site suppressors of immutans and var2. Photosynth. Res. 116, 437–453. doi: 10.1007/s11120-013-9855-9
Qi, Y., Liu, X., Liang, S., Wang, R., Li, Y., Zhao, J., et al. (2016). A putative chloroplast thylakoid metalloprotease VIRESCENT3 regulates chloroplast development in Arabidopsis thaliana. J. Biol. Chem. 291, 3319–3332. doi: 10.1074/jbc.M115.681601
Rosar, C., Kanonenberg, K., Nanda, A. M., Mielewczik, M., Bräutigam, A., Novák, O., et al. (2012). The leaf reticulate mutant dov1 is impaired in the first step of purine metabolism. Mol. Plant 5, 1227–1241. doi: 10.1093/mp/sss045
Schock, I., Gregan, J., Steinhauser, S., Schweyen, R., Brennicke, A., and Knoop, V. (2000). A member of a novel Arabidopsis thaliana gene family candidate Mg2+ ion transporters complements a yeast mitochondrial group II intron-splicing mutant. Plant J. 24, 489–501. doi: 10.1111/j.1365-313X.2000.00895.x
Shaul, O. (2002). Magnesium transport and function in plants: the tip of the iceberg. Biometals 15, 309–323. doi: 10.1023/A:1016091118585
Shaul, O., Hilgemann, D. W., de-Almeida-Engler, J., Van Montagu, M., Inzé, D., and Galili, G. (1999). Cloning and characterization of a novel Mg2+/H+ exchanger. EMBO J. 18, 3973–3980. doi: 10.1093/emboj/18.14.3973
Smith, R. L., and Maguire, M. E. (1995). Distribution of the CorA Mg2+ transport system in gram-negative bacteria. J. Bacteriol. 177, 1638–1640. doi: 10.1128/jb.177.6.1638-1640.1995
Streatfield, S. J., Weber, A., Kinsman, E. A., Häusler, R. E., Li, J., Post-Beittenmiller, D., et al. (1999). The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell 11, 1609–1621. doi: 10.1105/tpc.11.9.1609
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yu, F., Fu, A., Aluru, M., Park, S., Xu, Y., Liu, H., et al. (2007). Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 30, 350–365. doi: 10.1111/j.1365-3040.2006.01630.x
Keywords: AtMRS2-11/AtMGT10/VAR5, chloroplast development, leaf variegation, Mg2+ transporters, chloroplast envelope
Citation: Liang S, Qi Y, Zhao J, Li Y, Wang R, Shao J, Liu X, An L and Yu F (2017) Mutations in the Arabidopsis AtMRS2-11/AtMGT10/VAR5 Gene Cause Leaf Reticulation. Front. Plant Sci. 8:2007. doi: 10.3389/fpls.2017.02007
Received: 12 July 2017; Accepted: 10 November 2017;
Published: 27 November 2017.
Edited by:
Julian Eaton-Rye, University of Otago, New ZealandReviewed by:
Sheng Luan, University of California, Berkeley, United StatesJinpeng Gao, Washington State University, United States
Copyright © 2017 Liang, Qi, Zhao, Li, Wang, Shao, Liu, An and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Yu, flyfeiyu@gmail.com
†These authors have contributed equally to this work.
 Shuang Liang†
Shuang Liang† Yafei Qi
Yafei Qi Jun Zhao
Jun Zhao Fei Yu
Fei Yu