- 1National Institute of Agrobiological Sciences, Tsukuba, Japan
- 2Plant Breeding Institute, The University of Sydney, Cobbitty, NSW, Australia
- 3The Institute for Cereal Crops Improvement, Tel Aviv University, Tel Aviv, Israel
- 4Centre National de la Recherche Scientifique, Saint-André-de-Cruzières, France
- 5Plant Germplasm Institute, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
- 6Global Crop Diversity Trust, Bonn, Germany
- 7Institute of Crop Science, National Agriculture and Food Research Organization, Tsukuba, Japan
Einkorn and emmer wheat together with barley were among the first cereals domesticated by humans more than 10,000 years ago, long before durum or bread wheat originated. Domesticated einkorn wheat differs from its wild progenitor in basic morphological characters such as the grain dispersal system. This study identified the Non-brittle rachis 1 (btr1) and Non-brittle rachis 2 (btr2) in einkorn as homologous to barley. Re-sequencing of the Btr1 and Btr2 in a collection of 53 lines showed that a single non-synonymous amino acid substitution (alanine to threonine) at position 119 at btr1, is responsible for the non-brittle rachis trait in domesticated einkorn. Tracing this haplotype variation back to wild einkorn samples provides further evidence that the einkorn progenitor came from the Northern Levant. We show that the geographical origin of domesticated haplotype coincides with the non-brittle domesticated barley haplotypes, which suggest the non-brittle rachis phenotypes of einkorn and barley were fixed in same geographic area in today’s South-east Turkey.
Introduction
The diploid einkorn wheat Triticum monococcum L. subsp. monococcum (2n = 2x = 14, AmAm) (hereafter Tm) was among the first crops domesticated in the Fertile Crescent starting from its wild progenitor Triticum monococcum L. subsp. boeoticum (Boiss.) Á. Löve et D. Löve (2n = 2x = 14, AbAb) (hereafter Tb). The archeological record of einkorn domestication both in terms of date and place remains elusive maybe because it is a minor crop that usually is associated with emmer. However, using molecular markers, the present-day distribution of a progenitor of domestic einkorn has been located in south-eastern Turkey – between the volcanic mountain ranges of Kartal–Karadaǧ and Karacadaǧ (Heun et al., 1997; Kilian et al., 2007).
The geographical range and the long-time periods shown from the archeological evidence points to a protracted and geographically diffuse processes of domestication for most if not for all Near Eastern crops, rather than a single process (Lev-Yadun et al., 2000; Fuller et al., 2011, 2012; Willcox, 2013), however, some would disagree (Heun et al., 2012). This applies also to einkorn wheat, for which the ‘dispersed-specific model’ has been proposed (Kilian et al., 2007).
On Near Eastern archeological sites, emmer and barley were the staple cereals and those where einkorn dominate are the exception. One example is Tell Qaramel (Willcox and Herveux, 2013). Two-grained einkorn was identified at the late Pleistocene sites of Mureybet and Abu Hureyra. But there was a doubt with regard to the identification because of the confusion with wild Secale species. The term Triticum/Secale was adopted, however, we now know that this taxon is dominated by wild rye (Willcox, 2007, 2014). Einkorn on Near Eastern sites is represented predominantly by two-grained forms (Özkan et al., 2011) while on European sites the single-grained type dominates (Kreuz and Boenke, 2002). During the last millennia, hulled wheats were largely replaced by free-threshing tetraploid (durum wheat) and hexaploid wheats (bread wheat), which deliver higher yields. Today, einkorn is cultivated on a very small scale, i.e., in the Balkans, France, Italy, Morocco, Spain, and Turkey (Schiemann, 1956, 1957; Nesbitt and Samuel, 1996; Perrino et al., 1996; Zaharieva and Monneveux, 2014). This diploid wheat has been re-discovered as a source of genetic variation for wheat breeding, and it is increasingly used by the organic food industry in Europe due to its nutritional value and biodiversity that can be exploited for the improvement of the health-related traits of modern wheat (Arzani and Ashraf, 2017).
Multi-locus re-sequencing analysis of a comprehensive collection of 321 wild and 92 domesticated einkorn lines revealed, for instance, that wild einkorn consists of three distinct Tb races (i.e., alpha, beta, gamma) (Kilian et al., 2007). One of those races, the wild race beta, is genetically much more similar to domesticated einkorn, hence it is the presumed race that was the progenitor of all known present-day domestic einkorns. Today race beta occurs only in the Kartal–Karadaǧ and Karacadaǧ mountains. The diffusion of domesticated einkorn out of the Fertile Crescent was reconstructed based on archaeobotanical remains and genotyping data summarized by (Zaharieva and Monneveux, 2014; Brandolini et al., 2016).
For simplicity, we follow the classification system of J. MacKey (MacKey, 1988), although wild and domesticated einkorn are not separated by any significant crossing barrier (Harlan and Zohary, 1966). The rachis type is one of the most conspicuous features distinguishing Tb from Tm: The Tb forms have a fragile rachis, which promotes seed dispersal in the wild, while the latter’s rachis is non-fragile and breaks when threshed (Figure 1), a prominent component of the domestication syndrome (Hillman and Davies, 1990). The fracture point of rachis in einkorn wheat lies above the node to produce a wedge-shape rachis node (Sakuma et al., 2011). The wedge type is found in diploid and tetraploid wheats harboring the A genome (A, AB, AG) (Li and Gill, 2006) as well as for hexaploid (ABD) (Watanabe et al., 2002). Wild einkorn wheat (Tb) produces wedge-shape spikelets rather similar to those of wild barley (Hordeum vulgare ssp. spontaneum (C. Koch) Thell. In barley, the brittle rachis trait is conditioned by the allelic status at two physically linked genes Non-brittle rachis 1 (btr1) and Non-brittle rachis 2 (btr2): a loss-of-function in either gene results in the formation of a non-brittle rachis. Loss-of-function mutations in both genes are found in the domesticated barley genepool, implying at least three independent origins of the stiff rachis phenotype in barley (Pourkheirandish et al., 2015; Civan and Brown, 2017). The objective of this study was to identify the causative polymorphism responsible for the non-brittle rachis trait of domesticated einkorn and to find closest present-day representatives of the wild ancestors of cultivated einkorn that could indicate the possible location of einkorn domestication.

FIGURE 1. Wheat spike showing the brittle rachis of wild einkorn (Tb) accession KT1-1 (Left) and non-brittle rachis of domesticated einkorn (Tm) accession KT3-5 (Right).
Materials and Methods
Plant Materials
A recombinant inbred population of 115 lines (referred to as RILWA-1) bred from the cross Tm ‘KT3-5’ × Tb ‘KT1-1’ (Shindo et al., 2002) was obtained from Yokohama City University, Yokohama. Aneuploid stocks of bread wheat cv. Chinese Spring (CS) [nullisomic-tetrasomics (NTs); Sears, 1954, 1966 and ditelosomics (Dts); Sears and Sears, 1978)] were obtained from the National BioResource Project (NBRP)/KOMUGI, Kyoto University, as were a representative set of 33 accessions of Tb and 20 accessions of Tm (Supplementary Table S1). The full set of materials was fall-sown in October 2008 and 2009 in the field at Tsukuba, Japan.
Genetic Analysis of the Non-brittle Rachis Trait
The rachis type of each of the RILWA-1 lines was characterized following the method described by Pourkheirandish et al. (2015). The awns emerging from opposing rows were pulled apart, and the proportion of brittle rachis nodes scored as 100× (the number of disarticulated rachis nodes/total number of rachis nodes). At least three mature spikes per plant of at least ten plants per line were tested. Genotypic scores for the lines, based on 211 EST markers, were obtained from Hori et al. (2007). A derived cleaved amplified polymorphic sequence (dCAPS) assay was developed to target a sequence within the Btr1 coding region to map the btr1 in diploid wheat: the relevant primer sequences were 5′-TCGCTCTGAGCAGGCTCGCGGCC/5′-GGTCCGGCTGTGAAGCATGAAC, and the resulting amplicons were digested with NotI. A linkage map (212 loci) was constructed from the genotypic data using MAPMAKER/EXP v3.0 software (Lander et al., 1987) and was used to perform a QTL analysis for rachis brittleness using Genstat v18 software1 (VSN International). The initial whole genome scan was performed applying a simple interval mapping (SIM) approach, which was followed up with composite interval mapping (CIM) once the appropriate co-factors had been selected.
Sequencing and Annotation of CS Bacterial Artificial Chromosomes (BACs)
The CS whole genome shotgun sequence (Brenchley et al., 2012) was queried using the barley Btr1 sequence (GenBank accession KR813335) in order to design a PCR assay for the wheat Btr1 coding region. The resulting primer sequences were 5′-CCGCAACGCTGCTGGGAGTT/5′-GGAGCGCGTCCTGGGCCT. The chromosomal origin of the amplicon was explored using DNA of the set of CS aneuploid lines (NTs and Dts) as template. A bacterial artificial chromosome (BAC) library constructed from CS genomic DNA (obtained from the John Innes Centre Genome Laboratory, Norwich, United Kingdom) was screened using the PCR assay designed above and shotgun sequencing of the BACs was carried out by the standard method (International Rice Genome Sequencing Project [IRGSP], 2005). Repetitive elements (retrotransposons and DNA transposons) were discarded with the aid of Repeat Masker software2. Gene-specific PCR and re-sequencing primers were designed for Btr1 and Btr2 from the contigs of wheat cv. CS BAC clone WCS1891K06 (Supplementary Table S2).
DNA Extraction and PCR Amplification
Genomic DNA was isolated from fresh single leaves as described elsewhere (Komatsuda et al., 1998) and used as template in a 10 μl PCR containing 0.25 U ExTaq polymerase (Takara, Tokyo, Japan), 0.3 μM of each primer, 200 μM dNTP, 2.0 mM MgCl2, 2.5% v/v DMSO, 25 mM TAPS buffer (pH 9.3), 50 mM KCl, 1 mM 2-mercaptoethanol and 20 ng template. The PCR regime comprised an initial denaturation of 94°C/5 min, followed by 30 cycles of 94°C/30 s, 60°C/30 s, 72°C/2 min, and was completed by a final extension of 72°C/10 min.
Amplicon Purification and Sequencing
The amplicons were purified using a QIAquick PCR purification kit (Qiagen, Germantown, MD, United States), then subjected to cycle sequencing on both strands using Big Dye Terminator v3.1 (Applied Biosystem, Foster City, CA, United States) technology. Each reaction comprised 25 cycles of 96°C/10 s, 50°C/5 s, 60°C/4 min. Salts, non-incorporated dNTPs and dye terminator were removed with an Agencourt CleanSeq system (Beckman Coulter, Fullerton, CA, United States), and the sequencing data was acquired using an ABI Prism 3130 genetic analyzer sequencer (Applied Biosystems, Foster City, CA, United States).
Phylogenetic Analysis
Sequencing reads were imported into Sequencher v5.4 software3 and low quality reads were removed manually. Sequence alignments were generated using ClustalW within MEGA 6 software (Tamura et al., 2013), and haplotypes defined using DnaSP 5.10.01 software (Librado and Rozas, 2009). Singleton SNPs and haplotypes (those detected in only one accession) were confirmed by manually inspecting the sequence chromatogram file.
A wild emmer wheat (T. turgidum ssp. dicoccoides (Körn.) Thell. ‘Zavitan’ (Avni et al., 2017) and a wild barley accession (Hordeum vulgare ssp. sponaneum, ‘OUH602’) were used as outgroups. The resulting phylogenetic trees were constructed using MEGA 6 software (Tamura et al., 2013) based on the neighbor-joining method, with uniform rates among sites, and applying the pairwise-deletion option to deal with gaps/missing data. A bootstrap analysis (1,000 replicates) was performed to provide confidence estimates for branch nodes.
Results
The Genetic Basis of the Non-brittle Rachis Trait in Einkorn Wheat
Phenotypic evaluation over two growing seasons showed that the Tb accession KT1-1 formed the wedge type brittle rachis in 67.0% rachis nodes, while the Tm accession KT3-5 formed the wedge type brittle rachis in only 1.5% rachis nodes (Figure 1). In the RILWA-1 population, the trait showed a continuous frequency distribution, but can be divided equally between those falling into the range 0–20% (non-brittle lines) and 50–90% (brittle lines), consistent with major locus effect that control over the trait (Supplementary Figure S1). QTL analysis revealed that rachis brittleness was determined by three loci, mapping to chromosomes 3A, 4A, and 7A (Figure 2 and Supplementary Table S3). The largest effect locus co-located with btr1 (dCAPS) at position 36.7 cM on chromosome 3A; the locus was associated with a LOD of 15.2 and explained 44.7% of the variance. The sequences of the two allelic forms of BTR1 revealed an alanine in wild einkorn (KT1-1) to threonine in domesticated einkorn (KT3-5) substitution at position 119. Whereas in BTR2 revealed an aspartic acid in wild einkorn (KT1-1) to glutamic acid in domesticated einkorn (KT3-5) substitution at position 10.
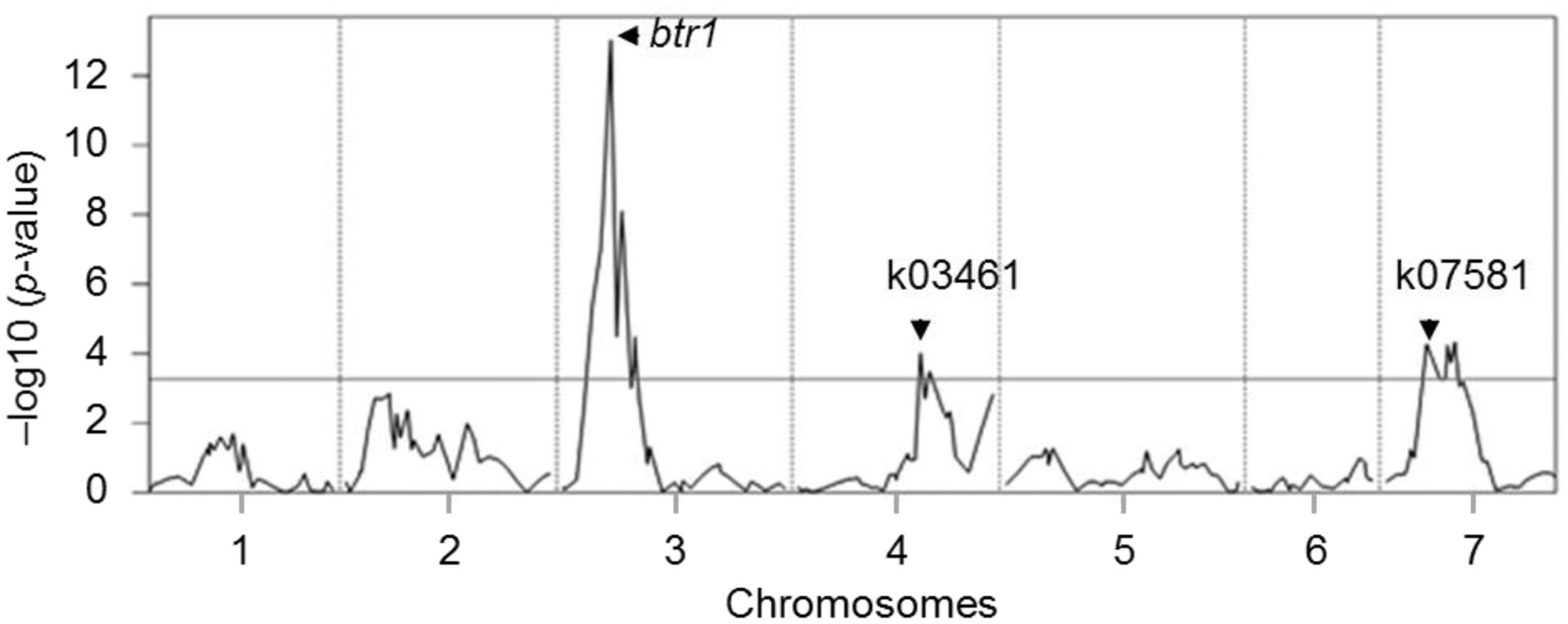
FIGURE 2. QTL mapping for rachis brittleness in einkorn wheat. The mean (across two seasons) rachis brittleness score was used to represent the trait in the analysis. The horizontal line represents the LOD threshold. The btr1 (dCAPS), k03461, and k07581 are molecular markers linked to the QTLs.
The Btr1 and Btr2 Homologs in Hexaploid Wheat
The primer pair 5′-CCGCAACGCTGCTGGGAGTT/5′-GGAGCGCGTCCTGGGCCT amplifies a 249 bp amplicon using a template of CS genomic DNA. An aneuploid analysis showed that the only templates, which failed to amplify this fragment were those which lacked the short arm of chromosome 3A (NT3A3B, NT3A3D and DT3AL, data not shown). The 126,452 bp sequence of CS BAC clone WCS1891K06 (GenBank accession MG324346), which tested positive for this amplicon, featured four Btr homologs, namely Btr1, Btr2, Btr1-like, and Btr2-like. Btr1 and Btr2 were orientated head-to-head, separated by 37,305 bp, while Btr1-like and Btr2-like were also orientated head-to-head, but were separated by just 440 bp (Figure 3).
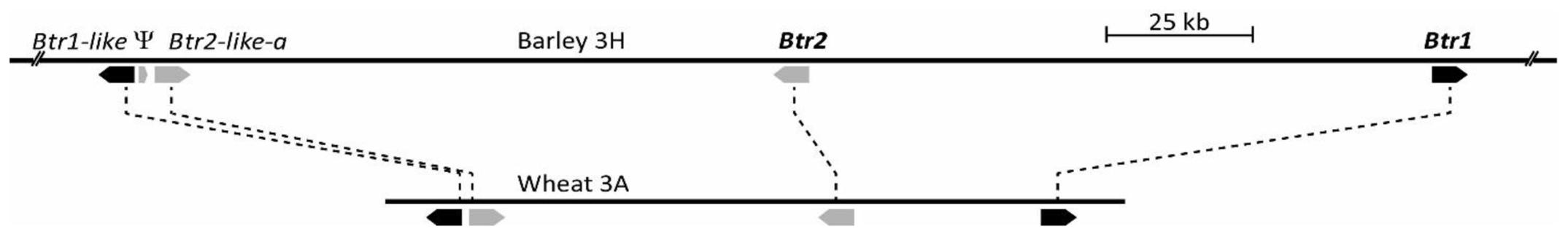
FIGURE 3. Comparison of orthologous Btr1/Btr2 loci from wild barley OUH602 (Upper, accession KR813335, Pourkheirandish et al., 2015) and bread wheat cultivar Chinese Spring (CS) A genome (Lower, accession MG324346, this study). Btr1 and Btr1-like are marked by black arrows, Btr2 and Btr2-like by gray arrows, while Ψ is a pseudogene sharing some homology with Btr2.
Sequence Variation at Btr1 and Btr2 Genes in Einkorn Wheat
The nucleotide sequence of CS (WCS1891K06) was exploited to design PCR primers suitable for re-sequencing einkorn Btr1 (TmB1) and Btr2 (TmB2) full genes (Supplementary Table S2). The einkorn Btr1 sequence was found to harbor a single 591 bp exon, predicted to encode a 196-residue protein (Figure 4), just as its wild barley ortholog does (Pourkheirandish et al., 2015). The Btr1 coding sequence present in KT1-1 was 91.7% identical to that present in wild barley, and the level of identity at the peptide level was 87.8%. The predicted transmembrane helices were highly conserved between barley and einkorn wheat.

FIGURE 4. BTR1 peptide alignment of wild diploid and tetraploid wheat and wild barley. The A119 residue is conserved across the species except for Tm. The two predicted transmembrane helices are indicated by boxes. Wheat 2X_BTR1-Tb stands for Tb accession KT1-1, Wheat 2X_BTR1-Tm stands for Tm accession KT3-5, Wheat 4X_BTR1-A and -B T. turgidum ssp. dicoccoides ‘Zavitan’, and Barley_BTR1-A is wild barley OUH602.
The einkorn Btr2 sequence was found to harbor a single 597 bp exon, predicted to encode a 198-residue protein, while the wild barley ortholog features a single exon of length 609 bp (202 residue product). The equivalent comparison between the KT1-1 and the wild barley Btr2 coding sequences revealed an 88.0% level of nucleotide identity and a level of identity at the peptide level of 82.0%.
When the 2,200 bp stretch spanning the einkorn Btr1 gene which includes 750 bp upstream and 900 bp downstream of the coding sequence was re-sequenced in a set of 20 Tm and 33 Tb accessions (Supplementary Table S1); eight Tb and three Tm haplotypes were found (Figure 5A). Two variants were identified in the coding region (Figure 5A), one at position 940 (C to G; S69 synonymous change), and the other at position 1088 (G to A; A119T non-synonymous change). All of the Tb accessions carried G at position 1088, and all of the Tm accessions carried A. The Btr1 and Btr1-like copies of both barley (Pourkheirandish et al., 2015) and emmer wheat (Avni et al., 2017) encoded alanine at the position indicating alanine is a well-conserved residue and threonine a mutation in the protein (Figure 4). The data supported the hypothesis that the A119T mutation is a functional change implying that the brittle/non-brittle rachis trait in einkorn wheat is controlled by the allelic status at Btr1. A phylogenetic analysis suggested a single origin for the mutation and that the latter (TmB1_Hap09, _Hap10 and _Hap11) were derived from haplotype TmB1_Hap01 (Figure 5B). The two accessions (KU-10873 and KU-10901) harboring TmB1_Hap01 were collected c. 50 km southeast of Maras in southeast Anatolia/Turkey close to the Kartal–Karadaǧ Mountains (Supplementary Table S1). The analysis also showed that TmB1_Hap09 was the haplotype of the founding domesticated genotype (Figure 5B), which subsequently became geographically highly dispersed. The further derived haplotypes TmB1_Hap10 and _Hap11 probably arose as a result of subsequent, natural mutations in the non-coding sequence and did not cause a functional change of Btr1 (Figure 5A).
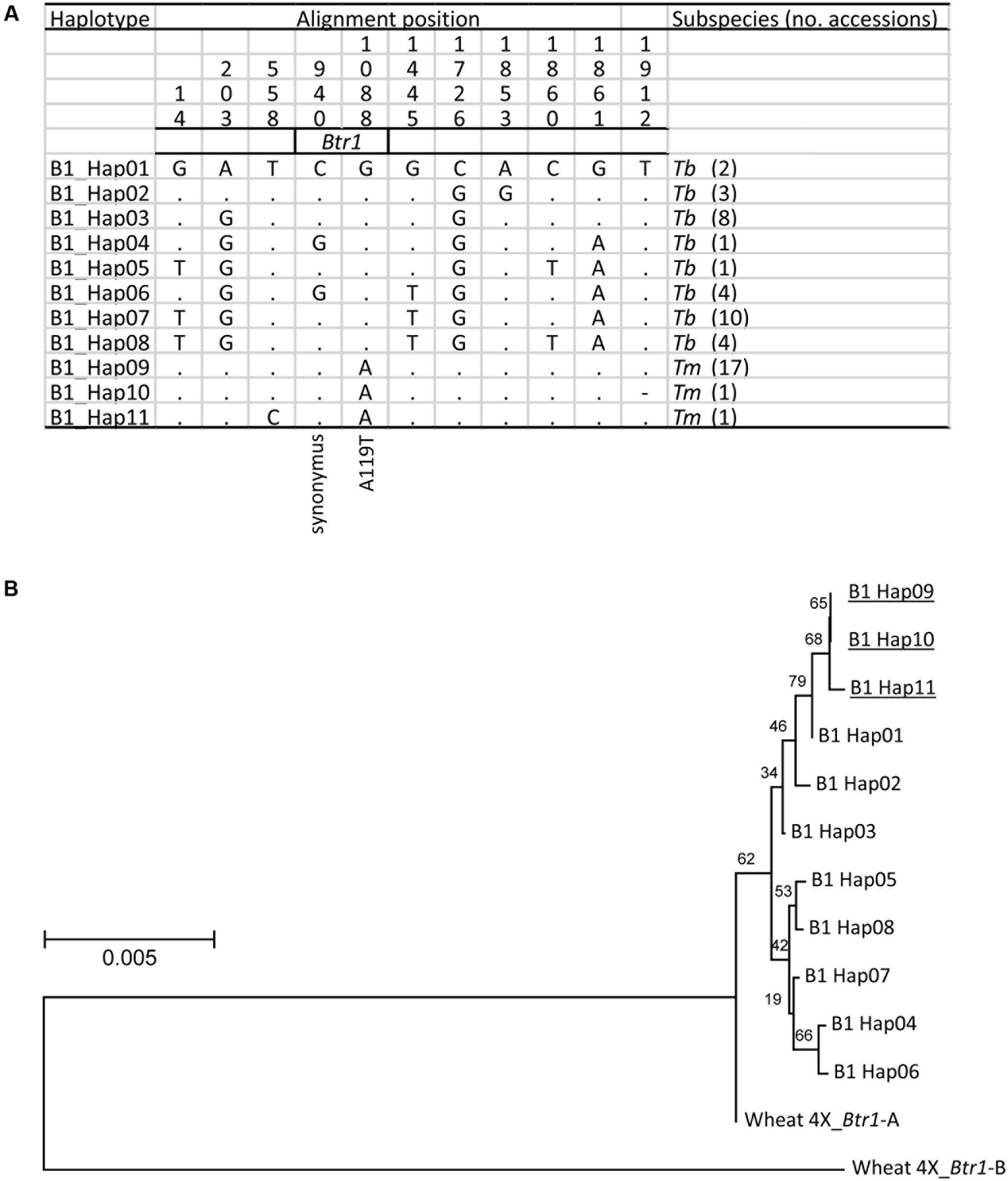
FIGURE 5. The origin of the btr1 allele found in domesticated einkorn wheat. (A) The eleven recognized Btr1 haplotypes found in Tm (three haplotypes)and Tb (eight haplotypes). Identical nucleotides are shown in the first line as dots. The A119T amino acid substitution caused non-brittle phenotype in Tm. (B) A phylogenetic analysis of Btr1 based on nucleotide sequence spanning the Btr1 coding sequence plus 750 bp upstream and 900 bp downstream of the coding sequence. Wheat 4X_Btr1-A and -B T. turgidum ssp. dicoccoides ‘Zavitan’ (A and B genome) formed the paraphyletic outgroup. Domesticated haplotypes are indicated by underline. Local bootstrap values after 1000 replicates are indicated near the branches.
Similarly, when a 1,600 bp stretch of the Btr2 sequence which includes 350 bp upstream and 650 bp downstream of the coding sequence was re-sequenced among the Tm and Tb accessions, a total of 14 haplotypes was revealed in wild einkorn and three in domesticated einkorn wheat (Figure 6A); these included two non-synonymous variants in the Btr2 coding sequence. The first of these (position 422, G to T) resulted in a predicted E10D substitution; as this polymorphism occurred within the Tb genepool, it cannot be responsible for the brittle rachis trait. The second non-synonymous polymorphism lay at position 618 (C to T), causing a R76C substitution. The R76C variant in Btr2 was only found in one domesticated einkorn accession (KU-11047) originating from northern Turkey carrying haplotype B2_Hap16. It is unknown whether R76C mutation causes a functional change of Btr2, but this polymorphism is less likely to be responsible for the initial occurrence of non-brittle rachis in Tm because of its rare appearance. These findings suggest that the A119T at Btr1 is the critical polymorphism, which causes the non-brittle rachis phenotype of domesticated einkorn.
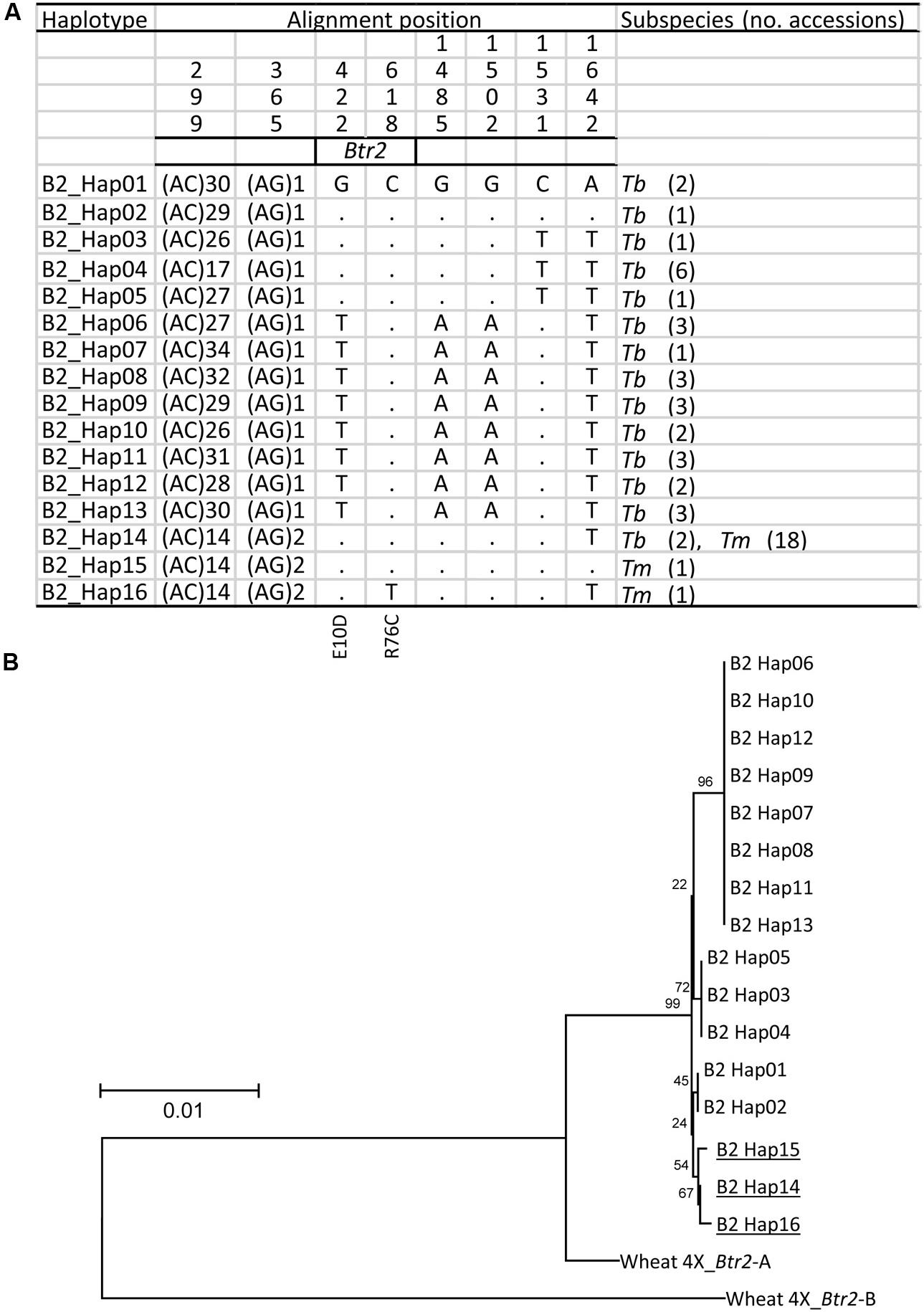
FIGURE 6. The origin of the Btr2 allele found in domesticated einkorn wheat. (A) The 16distinct Btr2 haplotypes found in Tm (three haplotypes) and Tb (14 haplotypes), of which one (B2_Hap14) is represented in both genepools. Identical nucleotides are shown in the first line as dots. (B) A phylogenetic analysis of Btr2 based on nucleotide sequence spanning the Btr2 coding sequence plus 350 bp upstream and 650 bp downstream of the coding sequence. Wheat 4X_Btr1-A and -B T. turgidum ssp. dicoccoides ‘Zavitan’ (A and B genome) formed the paraphyletic outgroup. Cultivated haplotypes are indicated by underline. Local bootstrap values after 1000 replicates are indicated near the branches.
The Tm haplotypes (TmB2_Hap14, _Hap15 and _Hap16) all appear to have evolved from the Tb haplotype TmB2_Hap14 (Figure 6B). As TmB2_Hap14 is the only haplotype in common between Tb and Tm, the inference is that this was the haplotype of the progenitor of the current Tm lineage. As expected, the two Tb accessions collected close to the Kartal–Karadaǧ Mountains (KU-10873 and KU-10901) were found to harbor both TmB1_Hap01 and TmB2_Hap14, a combination, which is identical to the make-up of the domesticated einkorn accessions (apart from the BTR1 A119T substitution).
Discussion
In barley, Btr1 and Btr2 act as complementary genes, with loss-of-function mutations in either of them generating the non-brittle rachis phenotype (Pourkheirandish et al., 2015). In einkorn wheat, a critical A119T substitution at Btr1 was found to discriminate between wild and domesticated einkorn wheat. A119T lies outside either of the two predicted transmembrane helices formed by BTR1, but it nevertheless appears to represent a component of the active site of the einkorn wheat BTR1 protein. The replacement of a threonine by an alanine residue has been suggested to facilitate receptor internalization in a rat opiod receptor (Wolf et al., 1999), and has also been shown to affect the activity of a histone protein (Jacob et al., 2014). Whether the A119T substitution in some way impairs the receptor-ligand structure, as suggested by Pourkheirandish et al. (2015), remains to be established. These findings suggest that the A119T at Btr1 is the critical polymorphism, which causes the non-brittle (a stiffer) rachis phenotype of domesticated einkorn. In this study, we did not find any einkorn wheat carrying functional allele of Btr1 in combination with loss-of-functional allele of Btr2. Whether Btr1 and Btr2 are functionally linked and act complementary genes as for barley has still to be investigated.
Today the most extensive natural stands of wild einkorn wheat are found in the northern part of the Fertile Crescent (Harlan and Zohary, 1966). However, we do not know the extent of natural stands at the end of the Pleistocene and the beginning of the Holocene. Archeological finds provide evidence for wild einkorn cultivation in northern Syria and Eastern Turkey between 11500 and 10500 cal BP. Signs of morphological domestication only appear after this long period of pre-domestication cultivation (Tanno and Willcox, 2012). Multi-locus genotyping (incl. re-sequencing) studies have concluded that the most likely site of the progenitor of einkorn wheat lies in south-eastern Turkey (Heun et al., 1997; Kilian et al., 2007). In the previous study by Kilian et al. (2007), the closest wild to domesticated einkorn were collected within the Kartal–Karadaǧ and Karacadaǧ mountain ranges on basaltic soils (Kilian et al., 2007).
Here, a haplotype analysis of Btr1 and Btr2 sequences showed that two wild einkorn accessions, which resembled the immediate progenitor – at Btr1 and Btr2 – of all domesticated einkorn wheats included in this study, both were collected between Kahramanmaraş and Gaziantep. This location lies immediately east of the Kartal–Karadaǧ mountain range and is the region where the progenitor of einkorn survives until today (Kilian et al., 2007). Furthermore, it has to be seen, if the A119T mutation led to the foundation of all known ‘eco-geographical’ or ‘geographical provenance’ groups/ lineages of domesticated einkorn (Flaksberger, 1935; Dorofeev et al., 1979; Szabó and Hammer, 1996; Zaharieva and Monneveux, 2014; Brandolini et al., 2016). Re-sequencing of Btr1 and Btr2 in a comprehensive collection of wild and domesticated accessions from diverse regions will answer if the non-brittle einkorn is selected only once (A119T) or multiple times.
One of the few early Neolithic sites where einkorn is the dominant crop, Tell Qaramel (Willcox and Herveux, 2013), lies less than 100 km to the south of where the two Tb accessions were collected (Figure 7). This adds weight to the hypothesis that it was in this general region that einkorn was first taken into cultivation. Willcox and Herveux (2013) suggested on the basis of an association with arable weeds that morphologically wild einkorn was cultivated at Tell Qaramel by approximately 11500 cal BP. Could this be the earliest example of cultivated wild einkorn, which later diffused as a minor component of emmer fields over a wider area of northern Levant and into Cyprus by 10600 cal BP? More archaeobotanical evidences are needed to answer this question but we can speculate that at some point the btr1 allele appeared, fixed and dispersed to agrarian societies far away from the Karadaǧ mountain range. Einkorn is diploid and mutation in Btr1, rather than accumulated mutations in multiple loci or multiple genomes, was sufficient to produce non-brittle rachis. Therefore, producing non-brittle rachis seems to be achieved in rather a shorter time than domestic emmer wheat which required fixation of two non-functional mutations in the homoeologous Btr1 genes in the A and B sub-genomes (Avni et al., 2017). The possible unintentional fixation of non-brittle rachis in diploid wheat or barley may led to an intentional selection of two mutations in tetraploid wheat genome A and B. It is true that QTLs other than the btr1 were identified in wheat and barley but with a milder phenotypic effect (Komatsuda et al., 2004; Jiang et al., 2014).
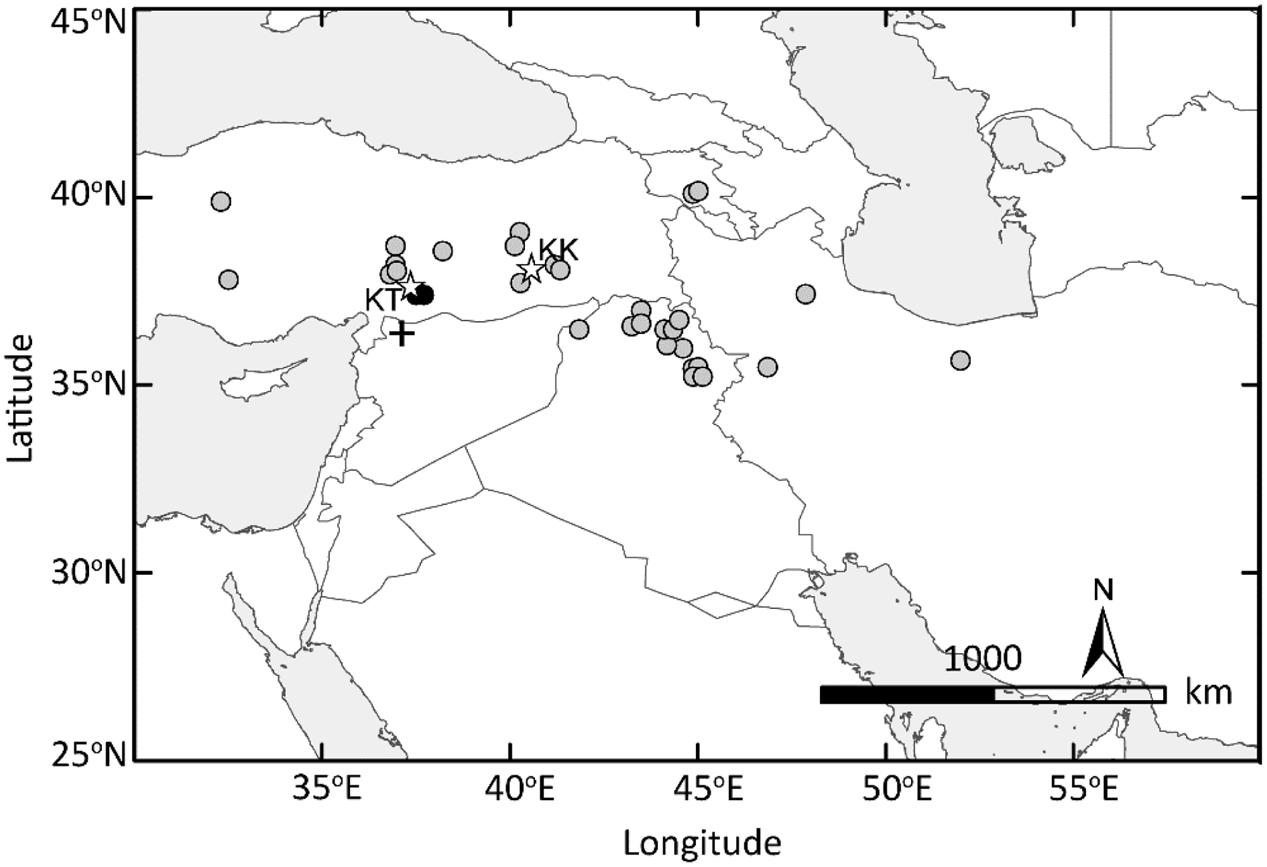
FIGURE 7. Domesticated einkorn originated in the Northern Levant. The GIS-based map of the Fertile Crescent indicates the collection sites of wild einkorn accessions analyzed in this study. Two wild einkorn lines equally closest to domesticated einkorn types at Btr1 and Btr2 are indicated by black dots. KT, Kartal–Karadaǧ; KK, Karacadaǧ mountains are indicated with stars. The archeological site of Tell Qaramel is indicated with plus sign.
Archaeobotanical finds indicate that it probably took over one millennium between the start of cultivation and the establishment of populations dominated by semi-brittle types (Tanno and Willcox, 2012). These finds provide evidence for multiple “false starts” and “dead ends” in the long history of plant and animal domestication, i.e., early domesticated lineages went extinct or diffused out of their original domestication area – and became slowly adapted to other environments (Fuller et al., 2011, 2012). In other words, we do not know how many domesticates were lost and we do not know to what extend the modern crop genepool (incl. the crop wild relatives) represents the situation in the past. However, Kilian et al. (2007) presented evidence that einkorn, in contrast to more intensely bred crops, possibly underwent little reduction of diversity during domestication.
The wild ancestors of domesticated einkorn btr1 (A119T) have been found in the same geographical area as the immediate wild ancestors of the btr2 (Pourkheirandish et al., 2015) and btr1b (Civan and Brown, 2017) lineages of domesticated barley. More precisely, the closest wild relatives to these non-brittle rachis phenotypes were all found northwest and southwest of Gaziantep and one of which (wild barley accession FT730) was collected in the Kartal–Karadaǧ mountains. This coincidence supports the ‘Cradle of Agriculture’ theory of Lev-Yadun et al. (2000) and in this case similar results should be found for the other founder crops of the Near East (Emmer wheat, pea, chickpea, lentil, and bitter vetch).
The genomic organization of Btr1/Btr2 and Btr1-like/Btr2-like gene pairs on chromosome 3A of hexaploid bread wheat cultivar CS was exactly the same as that on barley chromosome 3H (Pourkheirandish et al., 2015). This demonstrates that the configuration must have been present in the common ancestor of wheat and barley, which diverged some eight million years ago (Middleton et al., 2014). The implied duplication event, which formed the Btr1/Btr2 and the Btr1-like/Btr2-like pairs must therefore have occurred prior to the separation of the wheat and barley lineages. The sequence of the einkorn Btr genes is highly homologous to that of their barley orthologs. Their predicted products are of the same size and its BTR1 protein forms the same two transmembrane helices seen in both barley and emmer wheat BTR1 (Pourkheirandish et al., 2015; Avni et al., 2017). As for BTR2, neither the ‘CAR’ nor the ‘PIP’ motifs found in barley were reproduced in the wheat protein of different ploidy levels. The wild forms of both einkorn and emmer wheat, just as does that of barley produce spikelets with a wedge type rachis internode, promoting spike disarticulation above each rachis node (Zohary et al., 2013). The implication is that the products of the wild type allele at each of the Btr orthologs all act similarly to form a disarticulation layer above the rachis node to produce a wedge-shaped rachis internode. This form of brittle rachis is produced by species belonging to about half of the Triticeae genera (specifically, the genera Hordeum, Triticum, Secale, Psathyrostachys, Heteranthelium, Crithopsis, Taeniatherum, Thinopyrum, Dasypyrum, Eremopyrum, and Sitanion, along with a few Aegilops species; Sakuma et al., 2011). The extrapolation that allelic variation at the Btr1 and/or Btr2 orthologs is universally responsible for the brittle rachis in these taxa will need to be verified by further research.
Conclusion
The non-brittle einkorn wheat originated by a single nucleotide change at Btr1 that caused an amino acid substitution. How the single amino acid substitution (A119T) can change the specificity of BTR1 protein is an open question. This mutation was selected as a result of cultivation by early farmers. Today the nearest wild relatives of non-brittle einkorn have been found growing only in the northern Levant suggesting the latter may have originated there too along with non-brittle barley. The work described here provides a foundation for future studies on spike disarticulation of wheat and other members of Triticeae tribe.
Author Contributions
TKa selected and provided the Tb and Tm materials. MP and SS assessed rachis type and performed QTL analysis. MP constructed the physical contig, which was sequenced by HK, TM, and annotated by MP. FD and MP carried out haplotype analysis. AD provided the tetraploid sequence of btr1 and btr2 orthologs. GW provided the archaeobotanical data. MP and TKo designed the experiments. MP, AD, BK, GW, and TKo wrote the manuscript. All authors have reviewed and commented on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. A. M. Alqudah (IPK, Germany) for help with QTL analysis. We are very grateful to Dr. A. Bogaard (Oxford, United Kingdom) and Dr. H. Özkan (Adana, Turkey) for valuable comments on an earlier version of this MS. This work was partially supported by Japanese Society for Promotion of Science to FD, MP, and TKo.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02031/full#supplementary-material
Footnotes
- ^ http://www.vsni.co.uk/software/genstat/
- ^ www.repeatmasker.org/cgi-bin/WEBRepeatMasker
- ^ www.genecodes.com/
References
Arzani, A., and Ashraf, M. (2017). Cultivated ancient wheats (Triticum spp.): a potential source of health-beneficial food products. Comp. Rev. Food Sci. Food Saf. 16, 477–488. doi: 10.1111/1541-4337.12262
Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. doi: 10.1126/science.aan0032
Brandolini, A., Volante, A., and Heun, M. (2016). Geographic differentiation of domesticated einkorn wheat and possible Neolithic migration routes. Heredity 117, 135–141. doi: 10.1038/hdy.2016.32
Brenchley, R., Spannagl, M., Pfeifer, M., Barker, G. L., D’amore, R., Allen, A. M., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710. doi: 10.1038/nature11650
Civan, P., and Brown, T. A. (2017). A novel mutation conferring the nonbrittle phenotype of cultivated barley. New Phytol. 214, 468–472. doi: 10.1111/nph.14377
Dorofeev, V., Filatenko, A., Migushova, E., Udachin, R., and Yakubtsiner, M. (1979). “Flora of cultivated plants,” in Wheat, Vol. 1, eds V. Dorofeev and O. Korovina (Leningrad: Springer).
Flaksberger, C. (1935). “Flora of cultivated plants,” in Cereals, Wheat, Vol. 1, ed. E. V. Wulf (Moscow: State Agricultural Publishing Company).
Fuller, D. Q., Willcox, G., and Allaby, R. G. (2011). Cultivation and domestication had multiple origins: arguments against the core area hypothesis for the origins of agriculture in the Near East. World Archaeol. 43, 628–652. doi: 10.1093/jxb/err307
Fuller, D. Q., Willcox, G., and Allaby, R. G. (2012). Early agricultural pathways: moving outside the ‘core area’ hypothesis in Southwest Asia. J. Exp. Bot. 63, 617–633. doi: 10.1093/jxb/err307
Harlan, J. R., and Zohary, D. (1966). Distribution of Wild Wheats and Barley. Science 153, 1074–1080. doi: 10.1126/science.153.3740.1074
Heun, M., Abbo, S., Lev-Yadun, S., and Gopher, A. (2012). A critical review of the protracted domestication model for Near-Eastern founder crops: linear regression, long-distance gene flow, archaeological, and archaeobotanical evidence. J. Exp. Bot. 63, 4333–4341. doi: 10.1093/jxb/ers162
Heun, M., Schaferpregl, R., Klawan, D., Castagna, R., Accerbi, M., Borghi, B., et al. (1997). Site of einkorn wheat domestication identified by DNA fingerprinting. Science 278, 1312–1314. doi: 10.1126/science.278.5341.1312
Hillman, G. C., and Davies, M. S. (1990). Measured domestication rates in wild wheats and barley under primitive cultivation, and their archaeological implications. J. World Prehist. 4, 157–222. doi: 10.1007/BF00974763
Hori, K., Takehara, S., Nankaku, N., Sato, K., Sasakuma, T., and Takeda, K. (2007). Barley EST markers enhance map saturation and QTL mapping in diploid wheat. Breeding Sci. 57, 39–45. doi: 10.1270/jsbbs.57.39
International Rice Genome Sequencing Project [IRGSP] (2005). The map-based sequence of the rice genome. Nature 436, 793–800.
Jacob, Y., Bergamin, E., Donoghue, M. T. A., Mongeon, V., Leblanc, C., Voigt, P., et al. (2014). Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343, 1249–1253. doi: 10.1126/science.1248357
Jiang, Y.-F., Lan, X.-J., Luo, W., Kong, X.-C., Qi, P.-F., Wang, J.-R., et al. (2014). Genome-wide quantitative trait locus mapping identifies multiple major loci for brittle rachis and threshability in Tibetan semi-wild wheat (Triticum aestivum ssp. tibetanum Shao). PLOS ONE 9:e114066. doi: 10.1371/journal.pone.0114066
Kilian, B., Ozkan, H., Walther, A., Kohl, J., Dagan, T., Salamini, F., et al. (2007). Molecular diversity at 18 loci in 321 wild and 92 domesticate lines reveal no reduction of nucleotide diversity during Triticum monococcum (Einkorn) domestication: implications for the origin of agriculture. Mol. Biol. Evol. 24, 2657–2668. doi: 10.1093/molbev/msm192
Komatsuda, T., Maxim, P., Senthil, N., and Mano, Y. (2004). High-density AFLP map of nonbrittle rachis 1 (btr1) and 2 (btr2) genes in barley (Hordeum vulgare L.). Theor. Appl. Genet. 109, 986–995. doi: 10.1007/s00122-004-1710-0
Komatsuda, T., Nakamura, I., Takaiwa, F., and Oka, S. (1998). Development of STS markers closely linked to the vrs1 locus in barley, Hordeum vulgare. Genome 41, 680–685. doi: 10.1139/g98-069
Kreuz, A., and Boenke, N. (2002). The presence of two-grained einkorn at the time of the Bandkeramik culture. Veget. Hist. Archaeobot. 11, 233–240. doi: 10.1007/s003340200026
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E., et al. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Lev-Yadun, S., Gopher, A., and Abbo, S. (2000). Archaeology. The cradle of agriculture. Science 288, 1602–1603. doi: 10.1126/science.288.5471.1602
Li, W., and Gill, B. S. (2006). Multiple genetic pathways for seed shattering in the grasses. Funct. Integr. Genomics 6, 300–309. doi: 10.1007/s10142-005-0015-y
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
MacKey, J. (1988). A plant breeder’s perspective on taxonomy of cultivated plants. Biol. Zentralbl. 107, 369–379.
Middleton, C. P., Senerchia, N., Stein, N., Akhunov, E. D., Keller, B., Wicker, T., et al. (2014). Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe. PLOS ONE 9:e85761. doi: 10.1371/journal.pone.0085761
Nesbitt, M., and Samuel, D. (1996). From staple crop to extinction? The archaeology and history of the hulled wheats. Hulled Wheats 4, 41–100.
Özkan, H., Willcox, G., Graner, A., Salamini, F., and Kilian, B. (2011). Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genet. Resour. Crop Evol. 58, 11–53. doi: 10.1007/s10722-010-9581-5
Perrino, P., Laghetti, G., D’antuono, L., Al Ajlouni, M., Kanbertay, M., Szabó, A., et al. (1996). “Ecogeographical distribution of hulled wheat species,” in Hulled Wheats, eds S. Padulosi, K. Hammer, and J. Heller (Rome: International Plant Genetic Resources Institute), 102–118.
Pourkheirandish, M., Hensel, G., Kilian, B., Senthil, N., Chen, G., Sameri, M., et al. (2015). Evolution of the grain dispersal system in barley. Cell 162, 527–539. doi: 10.1016/j.cell.2015.07.002
Sakuma, S., Salomon, B., and Komatsuda, T. (2011). The domestication syndrome genes responsible for the major changes in plant form in the Triticeae crops. Plant Cell Physiol. 52, 738–749. doi: 10.1093/pcp/pcr025
Schiemann, E. (1956). New dates for recent cultivation of Triticum monococcum and Triticum dicoccum in Jugoslavie. Wheat Inform. Serv. 3, 1–3.
Schiemann, E. (1957). Once more: “new dates for recent cultivation of einkorn and emmer in Jugoslavia”. Wheat Inform. Serv. 5, 3.
Sears, E. R. (1954). The aneuploids of common wheat. Missouri Agric. Exp. Stn. Res. Bull. 572, 1–58.
Sears, E. R. (1966). “Nullisomic-tetrasomic combinations in hexaploid wheat,” in Chromosome Manipulations and Plant Genetics, eds R. Riley and K. R. Lewis (Edinburgh: Springer), 29–47. doi: 10.1007/978-1-4899-6561-5_4
Sears, E. R., and Sears, L. M. (1978). “The telocentric chromosomes of common wheat,” in Proceedings of the 5th International Wheat Genetics Symposium: Indian Society for Genetics and Plant Breeding, New Delhi, 389–407.
Shindo, C., Sasakuma, T., Watanabe, N., and Noda, K. (2002). Two-gene systems of vernalization requirement and narrow-sense earliness in einkorn wheat. Genome 45, 563–569. doi: 10.1139/g02-015
Szabó, A., and Hammer, K. (1996). “Notes on the taxonomy of farro: Triticum monococcum, T. dicoccon and T. spelta.” in Proceedings of the 1st International Workshop on Hulled Wheats, held at Castelvecchio Pascoli, Tuscany, 2–40.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanno, K., and Willcox, G. (2012). Distinguishing wild and domestic wheat and barley spikelets from early Holocene sites in the Near East. Veget. Hist. Archaeobot. 21, 107–115. doi: 10.1007/s00334-011-0316-0
Watanabe, N., Sugiyama, K., Yamagishi, Y., and Sakata, Y. (2002). Comparative telosomic mapping of homoeologous genes for brittle rachis in tetraploid and hexaploid wheats. Hereditas 137, 180–185. doi: 10.1034/j.1601-5223.2002.01609.x
Willcox, G. (2007). “The adoption of farming and the beginnings of the Neolithic in the Euphrates valley: cereal exploitation between the 12th and 8th millennia cal BC,” in The Origins and Spread of Domestic Plants in Southwest Asia and Europe, eds S. Colledge and J. Conolly (Walnut Creek, CA: Left Coast Press), 21–36.
Willcox, G. (2013). Anthropology. The roots of cultivation in southwestern Asia. Science 341, 39–40. doi: 10.1126/science.1240496
Willcox, G. (2014). “Les premiers indices de la culture des céréales au Proche-Orient,” in La transition néolithique en Méditerranée, eds C. Manen, T. Perrin, and J. Guilaine (Paris: Errance Arles), 46–58.
Willcox, G., and Herveux, L. (2013). “Late Pleistocene/early Holocene charred plant remains: preliminary report,” in Tell Qaramel 1999-2007. Protoneolithic and Early Pre-pottery Neolithic Settlement in Northern Syria, eds R. Mazurowski and Y. Kanjou (Warsaw: PCMA), 120–130.
Wolf, R., Koch, T., Schulz, S., Klutzny, M., Schroder, H., Raulf, E., et al. (1999). Replacement of threonine 394 by alanine facilitates internalization and resensitization of the rat mu opioid receptor. Mol. Pharmacol. 55, 263–268.
Zaharieva, M., and Monneveux, P. (2014). Cultivated einkorn wheat (Triticum monococcum L. subsp. monococcum): the long life of a founder crop of agriculture. Genet. Resour. Crop Evol. 61, 677–706. doi: 10.1007/s10722-014-0084-7
Keywords: agricultural origins, einkorn, wheat, non-brittle rachis, domestication
Citation: Pourkheirandish M, Dai F, Sakuma S, Kanamori H, Distelfeld A, Willcox G, Kawahara T, Matsumoto T, Kilian B and Komatsuda T (2018) On the Origin of the Non-brittle Rachis Trait of Domesticated Einkorn Wheat. Front. Plant Sci. 8:2031. doi: 10.3389/fpls.2017.02031
Received: 18 September 2017; Accepted: 14 November 2017;
Published: 04 January 2018.
Edited by:
Paul Gepts, University of California, Davis, United StatesReviewed by:
Ahmad Arzani, Isfahan University of Technology, IranGuoxiong Chen, Chinese Academy of Sciences, China
Copyright © 2018 Pourkheirandish, Dai, Sakuma, Kanamori, Distelfeld, Willcox, Kawahara, Matsumoto, Kilian and Komatsuda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takao Komatsuda, takao@affrc.go.jp Mohammad Pourkheirandish, mohammad.pourkheirandish@sydney.edu.au
†Present address: Shun Sakuma, Faculty of Agriculture, Tottori University, Tottori, Japan
 Mohammad Pourkheirandish
Mohammad Pourkheirandish Fei Dai
Fei Dai Shun Sakuma
Shun Sakuma Hiroyuki Kanamori1
Hiroyuki Kanamori1 Assaf Distelfeld
Assaf Distelfeld George Willcox
George Willcox Taihachi Kawahara
Taihachi Kawahara Benjamin Kilian
Benjamin Kilian Takao Komatsuda
Takao Komatsuda