- 1ICAR-National Research Centre on Plant Biotechnology, New Delhi, India
- 2Center for Bioinformatics and Computational Biology, University of Delaware, Newark, DE, United States
- 3Department of Biotechnology, Faculty of Science, Centre for Transgenic Plant Development, Jamia Hamdard University, New Delhi, India
- 4Department of Biotechnology, Indian Institute of Technology Madras, Chennai, India
- 5Division of Nematology, ICAR-Indian Agricultural Research Institute, New Delhi, India
The root-knot nematode (RKN), Meloidogyne incognita, is an obligate, sedentary endoparasite that infects a large number of crops and severely affects productivity. The commonly used nematode control strategies have their own limitations. Of late, RNA interference (RNAi) has become a popular approach for the development of nematode resistance in plants. Transgenic crops capable of expressing dsRNAs, specifically in roots for disrupting the parasitic process, offer an effective and efficient means of producing resistant crops. We identified nematode-responsive and root-specific (NRRS) promoters by using microarray data from the public domain and known conserved cis-elements. A set of 51 NRRS genes was identified which was narrowed down further on the basis of presence of cis-elements combined with minimal expression in the absence of nematode infection. The comparative analysis of promoters from the enriched NRRS set, along with earlier reported nematode-responsive genes, led to the identification of specific cis-elements. The promoters of two candidate genes were used to generate transgenic plants harboring promoter GUS constructs and tested in planta against nematodes. Both promoters showed preferential expression upon nematode infection, exclusively in the root in one and galls in the other. One of these NRRS promoters was used to drive the expression of splicing factor, a nematode-specific gene, for generating host-delivered RNAi-mediated nematode-resistant plants. Transgenic lines expressing dsRNA of splicing factor under the NRRS promoter exhibited upto a 32% reduction in number of galls compared to control plants.
Introduction
Meloidogyne incognita, the southern root-knot nematode (RKN), is an obligatory sedentary parasite that infects thousands of plant species. The life cycle of M. incognita (Kofoid and White, 1919) starts with the hatching of egg in the soil, maturing into pre-parasitic second-stage juvenile, which later penetrates the root tip, migrates along the vascular cylinder of the plant root, becomes sedentary and forms a feeding site. RKN infestation causes an estimated annual crop loss of hundreds of billions of dollars (Abad et al., 2008; Elling, 2013). The demonstration of host-delivered RNA interference (HD-RNAi) in plant-parasitic nematodes (PPNs) offers an effective strategy to control nematode infection in crop plants (Tamilarasan and Rajam, 2013; Dutta et al., 2015; Dong et al., 2016; Niu et al., 2016; Banerjee et al., 2017; Kumar et al., 2017). Two house-keeping genes (splicing factor and integrase) were successfully silenced in nematodes through host-mediated RNAi in tobacco plants (Yadav et al., 2006). Arabidopsis thaliana (L.) Heynth. transgenic line expressing dsRNA of the 16D10 gene of RKN show significant reduction in the number of galls (65–90%) compared to the control (Huang et al., 2006). HD-RNAi of the putative effector gene Mc16D10L in potato (Solanum tuberosum L.) and Arabidopsis show significant resistance to M. chitwoodi (Dinh et al., 2014).
RNAi based approaches for nematode control largely use the constitutive promoter CaMV35S for dsRNA production in host plants (Tamilarasan and Rajam, 2013). However, the utility of CaMV35S “constitutive” promoter in driving expression of RNAi constructs is highly debated due to possibility of off-targets effects (Goddijn et al., 1993; Urwin et al., 1997; Bertioli et al., 1999). Moreover, transgenic plants with a strong constitutive expression of stress-responsive genes often suffer from undesirable phenotypes. The stress-tolerant transgenic Arabidopsis expressing 35S::DREB1A displayed growth retardation and severe reduction in seed production (Liu et al., 1998; Yamaguchi-Shinozaki and Shinozaki, 2001). Similar observations were made in transgenic tomato (Solanum lycopersicum L.), rice (Oryza sativa L.), and tobacco (Nicotiana tabacum L.) plants.
Since expression of transgenes driven by constitutive promoters, especially for a HD-RNAi approach is less desired due its potential off-targets effects, it is imperative that stress-inducible and tissue-specific promoters are identified to minimize these effects. Several researchers have stressed the need for identification of tissue-specific nematode-responsive promoters (Rosso et al., 2009). Such conditional promoters responsive to M. incognita with preferential expression only in target tissues like roots are likely to be more effective in developing RNAi-based resistance. This strategy, therefore, can also reduce the cost for in vivo dsRNA expression (Bakhetia et al., 2005). Studies on nematode-responsive and root-specific (NRRS) promoters are very limited and largely based on promoter tagging and mutant line analysis. The first NRRS expression was reported in TobRB7, a tonoplast intrinsic protein (TIP) gene from tobacco that was selectively active in infested root tissue cells and was induced during the feeding cell development (Opperman et al., 1994). Similar to TOBRB7, a strawberry gene FARB7 showed near root-specific expression. Though, FARB7 shared regulatory elements with TOBRB7 (Vaughan et al., 2006), its response to nematode still needs to be validated. Also, three promoters of Arabidopsis, TUB-1, ARSK1, and RPL16A have been identified, which drive expression of cystatin mainly to the roots and eventually deliver significant level of resistance against M. incognita (Lilley et al., 2004).
To investigate the differential expression pattern of genes in response to nematodes, a number of microarray analyses have been performed in the last decade (Portillo et al., 2013). In a few such experiments, the whole root was considered (Hammes et al., 2005; Alkharouf et al., 2006; Ithal et al., 2007; Klink et al., 2007) and some analysis focused on nematode feeding sites enriched samples only (Xiao and Xue, 2001; Bar-Or et al., 2005; Jammes et al., 2005; Fuller et al., 2007; Schaff et al., 2007; Barcala et al., 2010). Recently, NEMATIC (NEMatode-Arabidopsis Transcriptomic Interaction Compendium) tool was launched, which uses transcriptome data for studying the interaction between Arabidopsis and plant-endoparasitic nematodes (Cabrera et al., 2014). Furthermore, in spite of several studies that examined differential promoter activity in nematode feeding cells (Opperman et al., 1994; Escobar et al., 1999), no specific regulatory elements responsible for low basal expression have been identified. Putative cis-elements present in nematode-responsive promoters such as ERE, Wun-Motif, EIRE (Sukno et al., 2006) and P-Box (Escobar et al., 1999) were identified by the comparative analysis with regulatory elements databases but their role during nematode interaction is yet to be confirmed.
The current work describes a computational approach to identify nematode-responsive root-specific (NRRS) promoters and demonstrates their utility in host-mediated resistance in Arabidopsis against M. incognita infection.
Materials and Methods
Propagation of Nematode
The southern root-knot nematode (RKN; M. incognita) culture was maintained on tomato and eggplant (Solanum melongena L.). Tomato and eggplant seeds were sterilized (soaked for 20 min in sterile distilled water, 5 min in 70% ethanol, and 15 min in 5% NaOCl and 0.1% Tween 20, and washed four times in sterile distilled water) and germinated on MS agar medium. After 3 weeks, the plant roots were infected with 500 second-stage juveniles (J2s) of RKN. Six weeks later, egg masses were hand-picked and hatched at 28°C in 10–15 ml of sterile water.
Plant Growth Conditions and Nematode Infection
Arabidopsis thaliana (Col-0) seeds were surface sterilized (by immersing for 2 min in 70% alcohol and 7 min in 0.1% Mercuric chloride and 0.1% SDS) and stratified for 72 h at 4°C before germination on Gamborg's B-5 medium. Plates were covered with parafilm M® and maintained at 21°C under a 16 h light/8 h dark photoperiod. Fourteen-day-old Arabidopsis seedlings were transferred from the Petri plates to trays containing a sand/vermicompost/cocopeat mixture (1:1:1 w/w). After 3 weeks, each plant was inoculated with 1,000 freshly hatched J2s of RKN using a 1 ml pipette. The trays were maintained in a growth chamber at 21°C and the M. incognita-infected galls and complete roots of A. thaliana were harvested at two different intervals viz., 10 and 21 days post inoculation (dpi). The uninfected root tissue, dissected from the plants at the same time points, served as control samples. At each time point, samples were harvested from both infected and uninfected plants and frozen in liquid nitrogen and stored at −80°C until further use.
Microarray Data Mining and Statistical Analysis
Publicly available microarray data for Arabidopsis was analyzed using packages for R-statistical language and using an online microarray resource, Genevestigator (v 3.0) (Zimmermann et al., 2004). Microarray datasets from studies done by different labs and using platforms were difficult to process through our in-house scripts, primarily due to the batch-effects, and differences in numbers and identifiers of probe-sets corresponding to different platforms. Such datasets were analyzed through Genevestigator tool (Hruz et al., 2008); this study is referred to as “Genevestigator” analysis from here on. Independent to the Genevestigator analysis, the comprehensive microarray datasets from “AtGenExpress” study were processed and analyzed through in-house R-language scripts; this is referred to as “stand-alone” analysis from here on. Please see Supplementary Text Presentation 1 for more information on both the Genevestigator and stand-alone analysis.
Genes that preferentially up-regulate upon nematode infection were identified using Genevestigator; stand-alone analysis to identify such nematode-responsive genes could not be performed because the microarray datasets were not available in public domain at that time this study was performed. Genevestigator (v3) classified the microarray data from nematode infection assays into two stages—“early” and “late” based on days post infection. Probe sets that showed an up-regulation (>1.5-fold-change, log2-scale) in both “early” and “late” stages were downloaded in tab-separated file format and used for downstream analysis. On other hand, genes that show root-specific expression were identified using both the Genevestigator and stand-alone analysis. In stand-alone analysis, probe sets that showed a consistent up-regulation (>2-fold-change, log2) across all three contrasts, including with vegetative stem, inflorescence and leaf samples, at each stage were recorded (see Supplementary Text Presentation 1); this process was repeated for three time points (7, 17, and 21 days). Since, “AtGenExpress” data represented select developmental stages, we collected probe sets that are preferentially expressed (>1.5-fold-change, log2-scale) in root samples (six root zones) compared to other tissues or plant samples from all datasets (except AtGenExpress) available in Genevestigator (v3).
Motif Discovery Using MEME
Multiple EM for Motif Elicitation (MEME) was used for the discovery of novel putative motifs. A third-order Markov chain background model was prepared from the upstream sequence of all the Arabidopsis genes. The upstream sequences for 27,144 genes were extracted using RSAT (Thomas-Chollier et al., 2008). Repetitive DNA elements were masked from the input sequences using Repeat Masker program (Smyth, 2004). Two different modes, Any Number of Repetitions (ANR) and Zero or One Occurrence per Sequence (ZOOPS) were used with the following parameters:
-nmotifs 30 -minsites 4 -maxsites 12 -minw 6 -maxw 12 -revcomp -dna -mod anr–bfile
Where, number of -minsites and -maxsites is given based on number of input sequences and the third order background model is provided for–bfile.
-nmotifs 30 -minw 6 -maxw 8 -revcomp -dna -mod zoops–bfile
Where, -minw 6–10 and -maxw 8–12 and the third order background model is provided for –bfile.
For ANR mode, we considered 30 motifs ranked by their significance levels of length between 6 and 12 nt, while for ZOOPS mode we had three runs, each predicting 10 statistically ranked motifs corresponding to motif length of 6–8 nt, 8–10 nt, and 10–12 nt, respectively. The statistically-significant motifs were screened for similarity with known transcription factor binding motifs using POXO (Kankainen et al., 2006) (http://ekhidna.biocenter.helsinki.fi/poxo) and STAMP (Mahony and Benos, 2007).
Identification of Known, Conserved Cis-Regulatory Elements from NRRS Gene Promoters
Promoters of NRRS genes identified along with the previously reported genes as positive control (TobRB7, Atcel1, Hahsp17, and Lemmi9) were subjected to comparative analysis for identification of conserved cis-elements. The upstream region from the start codon was extracted to include only the intergenic region (~1.5 kb) using Regulatory Sequence Analysis Tool (RSAT) (http://www.rsat.eu/) (Thomas-Chollier et al., 2008). The known cis-elements present in each of these promoters were collected using two different public resources, PLACE (Higo et al., 1999) and ATcisDB (Davuluri et al., 2003). The set of reported motifs were further used to elucidate conserved cis-elements using a Python script.
Expression Analysis of Putative NRRS Genes
Total RNA was isolated from M. incognita-infested galls and whole roots of Arabidopsis using RNeasy mini kit (Qiagen, Germany) following the manufacturer's protocol. The RNA integrity was checked by formaldehyde gel electrophoresis with ethidium bromide dye. Nanodrop spectrophotometer 8000 (Thermo Scientific, USA) was used to calculate purity ratios and quantify total RNA. cDNA was prepared using Protoscript M-MuLV first strand synthesis kit (NEB, USA) using oligo-d (T)23VN primers. cDNA was normalized and re-quantified before qRT-PCR. RT Primers (Supplementary Table 1) were designed from the cDNA sequence of selected genes using “Primer3” portal (http://frodo.wi.mit.edu/primer3/) (Rozen and Skaletsky, 2000).
The cDNA from infected and control samples was quantified and used as template with three biological replicates along with three technical replicates for each biological replicate. To ensure purity of the master mix and reaction mix setup, a non-template control reaction was included in every plate. A 20 μl reaction volume consisting of SYBR FAST qRT-PCR Master Mix (2x) Universal (KAPA Biosystems) and 10 pmol of each primer was used in all qRT-PCR reactions. The qRT-PCR reactions were performed on a StepOnePlus™ Real-Time PCR system with the following cyclic conditions: initial denaturation temperature of 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 61°C for 45 s. The PCR products were exposed to melting curve analysis; the conditions were incubation at 60–95°C with a temperature increment of 0.3°C s−1 (Applied Biosystems®). The threshold cycle values were normalized by plant UBQ10 (Supplementary Table 1) as endogenous control and fold changes of the target gene were calculated by 2−ΔΔCt method (Livak and Schmittgen, 2001).
Preparation of Promoter::GUS Construct
The promoter regions (1.5 kb upstream of the start codon) of NRRS genes, At1g74770 and At2g18140, were amplified from the Arabidopsis genomic DNA using the gene specific oligonucleotides (Supplementary Table 1), with flanking restriction sites, BamHI and SalI. The amplified products were eluted from gel using Pure Link Gel Extraction kit (Invitrogen, USA), quantified and digested with BamHI and SalI. The 1.5 kb BamHI/SalI promoter fragment was cloned upstream of the GUS gene, using T4 DNA ligase (NEB, USA), with linearized BamHI/SalI digested pORE-R2 (Coutu et al., 2007) vector and transformed to DH5-α strain of E. coli (NEB, Massachusetts, USA). The prm::GUS fusion constructs were validated by nucleotide sequencing and introduced into Agrobacterium tumefaciens (Smith and Townsend, 1907) strain GV3101. Arabidopsis plants were transformed using the floral dip method (Clough and Bent, 1998). The primary transformants were selected on medium containing kanamycin (50 μg/ml) and further grown to develop T3 seeds. For each promoter, five independent Arabidopsis transgenic lines were tested for their response to nematode infection.
GUS Assay
Histochemical localization of GUS activity was performed with the substrate 5-bromo-4-chloro-3-indolyl-β-D glucuronide (X-Gluc) (Jefferson, 1989). The infected plants were uprooted and washed with water until roots were free of soil. The soil-free plants were immersed in freshly prepared GUS assay buffer [0.5 mM X-Gluc, 0.1 M NaHPO4 pH 8.0, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.01 M EDTA pH 8.0, 20% methanol, and 0.1% Triton X-100] and vacuum infiltrated for 5–10 min in a desiccator. The tubes were then incubated overnight in dark at 37°C. The tissues were cleared by replacing the buffer with 70% ethanol and then imaged with a stereomicroscope (Nikon®, Japan) with an external fiber optic light source. The plants were monitored for GUS activity at 10 and 21 days after nematode infection.
Cloning of At1g74770 Promoter in pBC-06 RNAi Vector
A 349-bp splicing factor sequence (AW828516) was amplified from M. incognita and cloned in pBC06 RNAi vector in sense and anti-sense directions (Yadav et al., 2006). CaMV35 promoter was removed from above vector and replaced with At1g74770 root-specific promoter using Sbf I and BamHI restriction enzymes. All cloning steps were performed according to the protocols described by Sambrook et al. (1989), and the constructs were confirmed by restriction fragment analysis and sequencing. The binary vectors were transferred to A. tumefaciens strain GV3101 by freeze and thaw method (Weigel and Glazebrook, 2006). A. thaliana (Col-0) plants were transformed with pAt1g74770::splicing factor and an empty pBC-06 vector through floral dip method (Jefferson, 1989). The transformed plants were selected on kanamycin (50 μg/ml). T2 transgenic plants were raised and confirmed through PCR analysis (data not given). Five independent transgenic lines were developed and two transgenic lines were evaluated against M. incognita infection. Nematode infection assays were performed in T3 plants and the numbers of galls were calculated.
For gene expression study, RNA isolated from dsRNA expressing transgenic plant and control plant used as template, with setup of three biological replicates along with three technical replicates for each sample. To ensure purity of the master mix and reaction mix setup, a non-template control was included in every plate. A 20 μl reaction volume consisting of SYBR FAST qRT-PCR Master Mix (2x) Universal (KAPA Biosystems) and 10 pmol of each primer was used in all qRT-PCR reactions. The qRT-PCR reactions were performed on a StepOnePlus™ Real-Time PCR system with the following cyclic conditions: initial heating temperature of 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 61°C for 45 s. The PCR products were exposed to melting curve analysis; the conditions were incubation at 60–95°C with a temperature increment of 0.3°C s−1 (Applied Biosystems®). The threshold cycle values were normalized by nematode 18S ribosomal RNA (Supplementary Table 1) as endogenous control and fold changes of the target gene were calculated by 2−ΔΔCt method (Livak and Schmittgen, 2001).
Results
Identification and Selection of Nematode-Responsive and Root Specific (NRRS) Candidate Genes
Candidate NRRS genes were identified in three steps: (a) an analyses of microarray data was performed to find genes that were induced by nematode infection and genes with root-specific expression patterns, (b) these root-specific and nematode-responsive gene sets were collated to find genes with desired NRRS gene expression pattern, and (c) finally, a cis-element based screening of candidate NRRS gene promoters was performed to select final candidates for downstream analyses.
The Genevestigator analysis i.e., meta-analysis of microarray datasets identified 1,374 probe sets that showed statistically significant and consistent up-regulation (>1.5-fold-change, log2 scale) in different (n = 6) root zones (Figure 1). On other hand, the stand-alone analysis, comparing root samples with vegetative shoot, inflorescence and leaf samples identified a total of 672 probe sets from three developmental time points (7, 17, and 21 days). In process, we also identified, 79 probe sets that were consistently enriched in root across all three time points investigated in stand-alone analysis (Figure 2A, Supplementary Table 2) and 336 probe sets that were common between Genevestigator and stand-alone analysis (Figure 2B). The low overlap between probes sets from Genevestigator and stand-alone analysis could be because of the additional data processing and summarization steps that Genevestigator performs to reduce batch effects and to provide robust expression estimates from different microarray platforms. In addition to root-specific probe sets, the Genevestigator analysis for nematode-induced genes (>1.5-fold-change, log2 scale) from “early” and “late” stages of nematode infection yielded 780 probe sets, with 42 probe sets showing a constant up-regulation in both stages (Figure 2C, Supplementary Table 3).
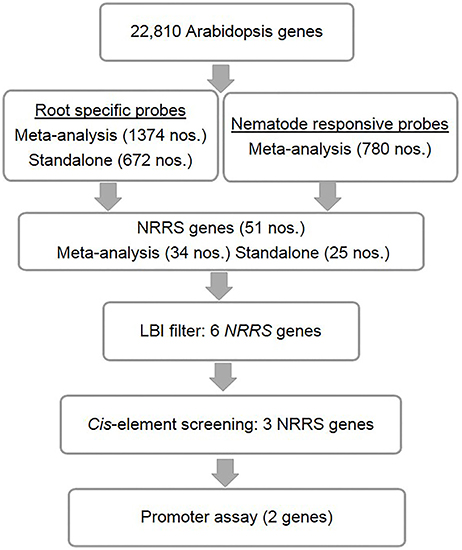
Figure 1. Identification of nematode-responsive root-specific (NRRS) genes. The top level consists of A. thaliana root-specific probes identified through both meta-analysis and stand-alone analysis. However, M. incognita nematode-responsive probes were identified through meta-analysis alone. Below that are the genes that were the result of cross-comparison with nematode-responsive genes and root-specific genes sets. After identifying 51 NRRS genes, an LBI filter was imposed which reduced the number of genes to six with low basal expression under control conditions. Further screening, based on cis-elements, led to identification of three NRRS genes. The lowest level includes the final two LBI genes that have been identified based on the results of screening by experimentally validated cis-elements, identified in earlier studies.
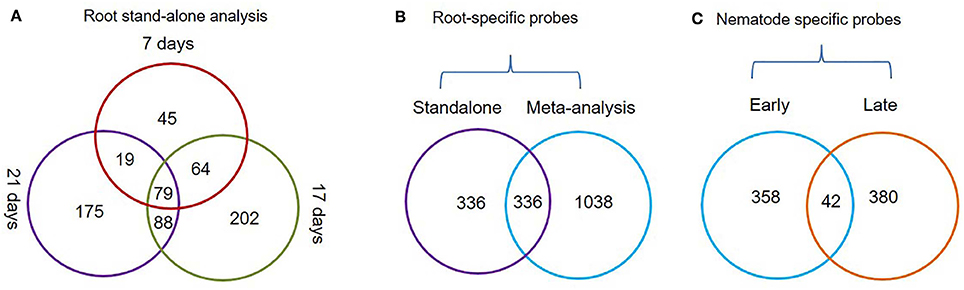
Figure 2. Venn diagrams depicting the status of root-specific and nematode-responsive probes. (A) Probes differentially expressed in the root at three different time points (7, 17, and 21 days) and a subset of common probes (79 nos.) constantly up-regulated in roots using the stand-alone studies. (B) Probes that were identified as root-specific by both meta-analysis and stand-alone analysis. (C) Probes that were constantly up-regulated during early and late stages of M. incognita nematode infection.
These probe sets were converted to Arabidopsis Gene Identifiers (AGIs) and collated to identify candidate NRRS genes. If multiple probes represented a single gene then the probe with highest median expression across samples was selected to represent the gene, and if a single probe represented multiple genes then all of these were included in final set. This conversion resulted in 700 root-specific AGIs from stand-alone analysis, 1,452 root-specific AGIs from meta-analysis and 850 nematode-responsive AGIs (Table 1). The nematode-responsive (NR) AGIs (n = 700) were then compared with root-specific (RS) AGIs from both the Genevestigator analysis (n = 1,452) and the stand-alone analysis (n = 700) to identify genes with nematode responsive and root-specific expression (n = 51). We refer to these 51 genes as candidate NRRS genes; these included 25 and 34 genes from NR vs. RS comparisons, where RS genes corresponded to stand-alone (Supplementary Table 4) and Genevestigator analysis (Supplementary Table 5) respectively. Both these categories NR vs. RS from stand-alone and NR vs. RS from Genevestigator analysis has eight common genes (Supplementary Table 6). Finally, to select genes that have a low expression under normal developmental stages, which is a desired characteristic for gene promoters that will drive expression of RNAi construct, we filtered genes based on average expression values in root tissues (see Supplementary Text Presentation 1). Six genes passed the low basal intensity (LBI) filter and were considered for next step i.e., a cis-element based screening (Table 2).
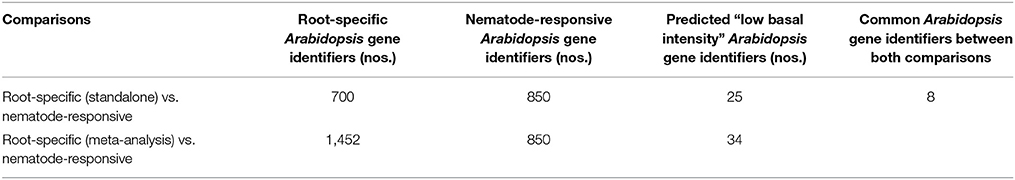
Table 1. Low Basal Intensity genes (in absence of nematode) from meta-analysis and stand-alone analysis.
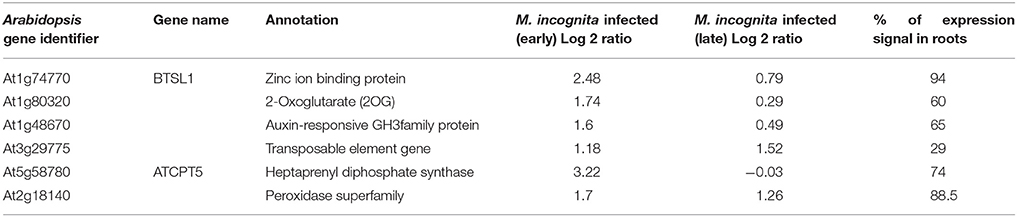
Table 2. Summary of the genes (n = 6) qualifying the Low Basal Intensity filter along with other details.
Conserved Cis-Regulatory Elements in the Promoters of Nematode-Responsive and NRRS Gene
The six candidate NRRS genes were screened for the presence of cis-regulatory elements (Table 3) that have been experimentally validated to play a role in root-specificity i.e., AS1, Sorlip1, and FaRB7, and typical for promoters of genes that are upregulated in nematode feeding site (NFS) upon infection i.e., TobRB7 box A/B etc. The purpose of this regulatory element based screening was to further reduce the number of genes selected for downstream molecular cloning and in planta validation. Out of six NRRS genes, only three genes—At1g74770, At2g18140, and At1g48670 (Figure 3) included regulatory elements from both the nematode responsive and root specific (NRRS) categories in their promoter regions. ATCEL2, earlier implicated in nematode-responsiveness as well as root specificity used as positive control (Wieczorek et al., 2008). Gene At2g39230, coding for a LOJ protein (Saha et al., 2007) served as negative control and had no nematode-responsive as well as root-specific elements except for single FaRB7 element. Two genes, At5g58780 and At3g29775 lacked root-specific elements whereas the At1g80320 gene promoter contained only the E-box motif out of all the studied elements (n = 12). Surprisingly, except for the E-Box motif, all other putative motifs described by Sukno et al. (2006) were found to be distributed in all three categories of genes, TobRB7, Atcel1, Hahsp17, and Lemmi9 that show elevated expression upon nematode infection in the NFS (Opperman et al., 1994; Escobar et al., 1999, 2003; Sukno et al., 2006), genes that are downregulated in NFS—AtPAl1, AtTIP, and AtANT1 (Hammes et al., 2005) and UBP22/At5g10790 that does not show any significant difference in expression pattern in response to nematode infection (Favery et al., 2004). Therefore, these putative motifs (W-Box, ElRE, ERE, and P-Box) proposed by were not included in this step to filter NRRS gene based on presence of cis-regulatory elements (Table 3).
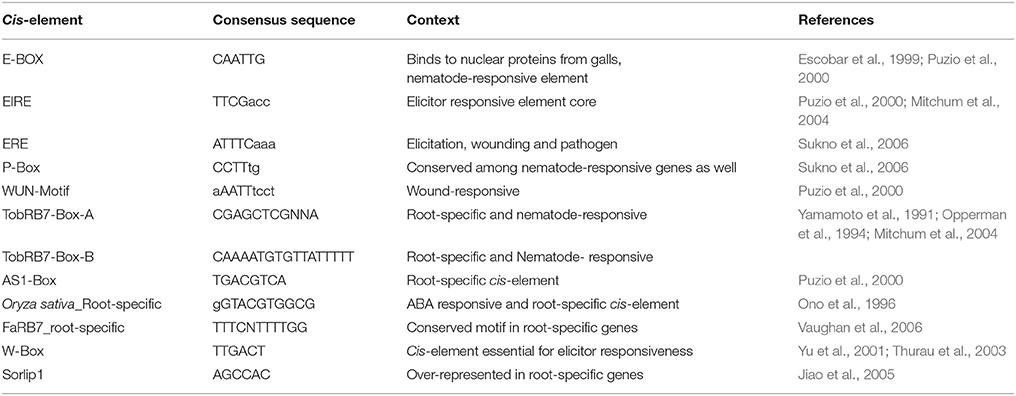
Table 3. Nematode-responsive (NR) and Root-specific (RS) cis-elements used for screening of 6 NRRS genes passing Low Basal Intensity filter.
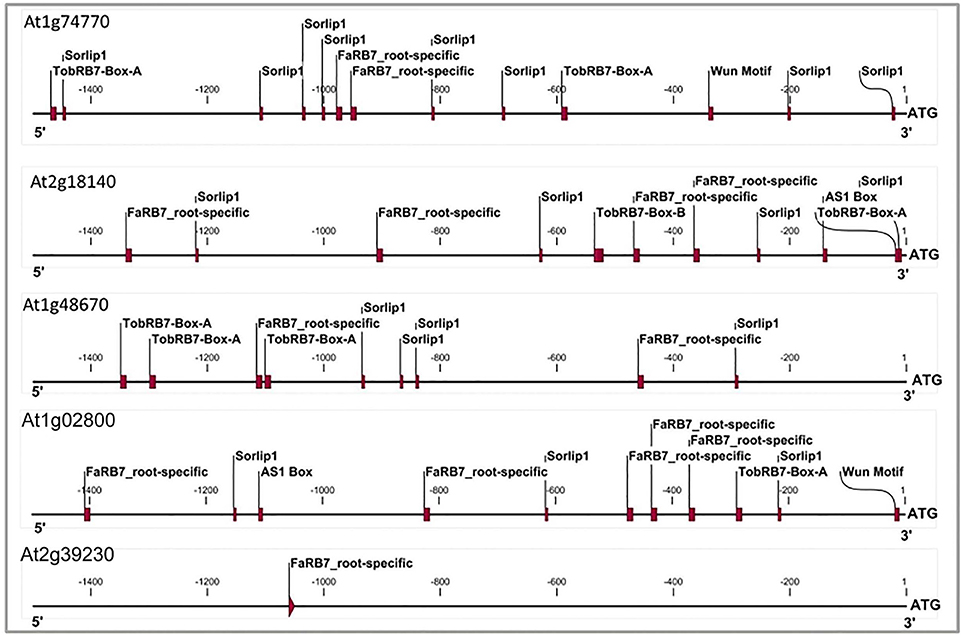
Figure 3. Cis-elements profile of promoters of three genes which qualified NRRS genes screening along with At1g02800 gene (AtCel2 positive) and At2g39230 gene (LOJ gene- lateral organ Junction gene and considered as negative gene unlikely to have any role in root development and nematode responsiveness). The TobRB7-Box-A/Box-B elements are nematode-responsive and Sorlip1, AS1 box and FaRB7 are root-specific elements. Please note overabundance of NRRS elements in NRRS genes and AtCel2 gene as compared to gene unlikely to be involved in nematode responsiveness and root development. The 1.5 kb region of each promoter was used for the profiling.
To find new candidates that may play role in upregulation of nematode-responsive genes in NFS, we used promoters from two different set of genes. First set consisted of genes that are well-studied for role in establishing compatible plant nematode interaction and show elevated expression upon infection (Lemmi9, TobRB7, and Hahsp17) along with the six NRRS genes predicted in this study (At1g74770, At1g48670, At2g18140, At3g29775, At5g58780, and At1g80320). The second set consisted just the six NRRS gene promoters from this study. We identified four motifs which were common between promoters of well-studied nematode-responsive genes and our NRRS genes (Figure 4A). In addition, we found three new motifs that are present only in NRRS genes (Figure 4B). Later on, MEME analysis has been carried out for the promoter region of all 51 genes to identify NRRS motif (Supplementary Figure Presentation 1; Supplementary Figure 1). To identify the transcription factors that might bind to these sites, we used POXO (Kankainen et al., 2006) and STAMP (Mahony and Benos, 2007) but no statistically significant similarity to known motifs was found. Therefore, further characterization of these novel motifs is required to establish their functional roles in NRRS activity.
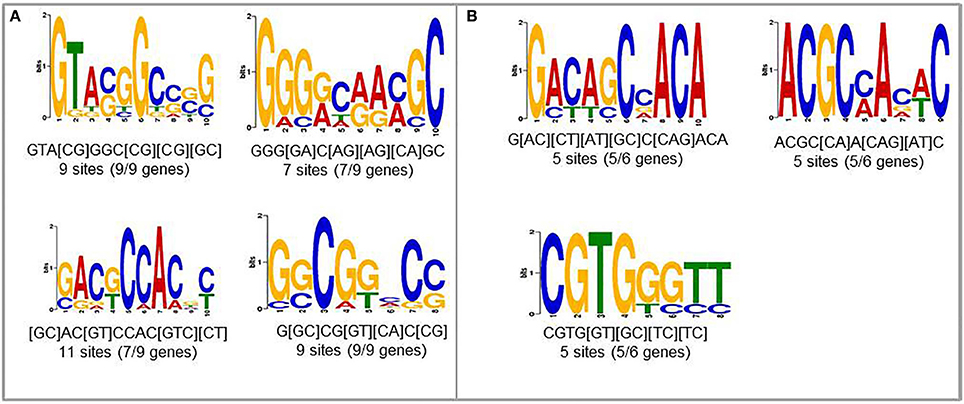
Figure 4. Novel cis-elements predicted using MEME tool. (A) Input set containing both experimentally validated (earlier published) and predicted (present study) NRRS promoters. (B) Novel NRRS cis-elements predicted from set of NRRS promoters identified in this study.
Confirmation of Nematode-Responsive Expression Pattern of NRRS Genes
Genes that passed the cis-regulatory element based criteria were evaluated for expression patterns in response to nematode infection before proceeding for molecular cloning their promoter regions. First, the expression patterns of six genes, UBP7, OXA1, Actin, RPN7, ATPase (Jammes et al., 2005), and UBQ10 (accession number DQ793132.1) was investigated in response to nematode infection for selection of appropriate internal controls. UBQ10 gene showed a consistent expression in both wild-type and nematode infected plants and was used as an internal controls for quantitative real time (qRT) PCR assay. The expression of six NRRS genes—At1g74770, At2g18140, At1g80320, At1g48670, At5g58780, and At3g29775 was studied at two different time points (10 and 21 dpi) along with At5g26530 gene, earlier implicated in nematode-responsiveness, which served as positive control (Kumar et al., 2016). The relative expression of At2g18140 and At1g74770 was maximum at 10 and 21 dpi, respectively as compared to control (Figure 5).
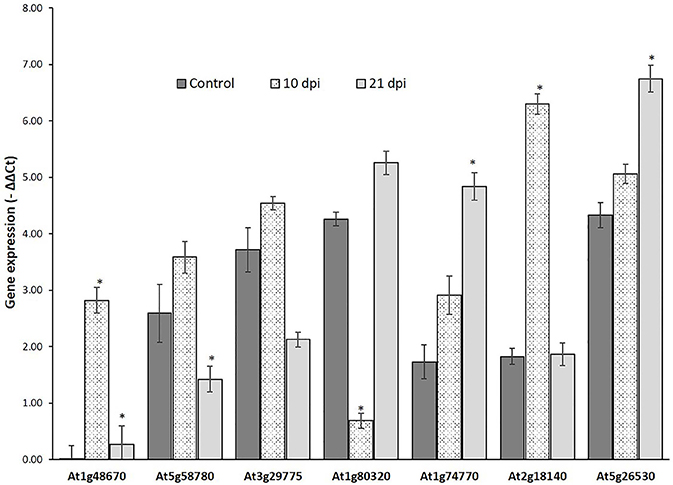
Figure 5. Expression analysis of nematode-responsive root-specific genes using Quantitative Real time PCR. Relative expression of six NRRS genes (At1g48670, At5g58780, At3g29775, At1g80320, At1g74770, and At2g18140) along with At5g26530, a nematode-responsive root-specific gene used as positive control (Kumar et al. 2016) in control and nematode infected plant root samples at 10- and 21- dpi. The transcripts levels were normalized to the expression of a plant UBQ10 gene. The data are shown as −ΔΔCt and each bar represents the mean ± SE (n = 3). An asterisk indicates statistical significance difference in a one-way ANOVA and Tukey test (p ≤ 0.05).
In Vivo Validation of NRRS Promoters
To experimentally confirm the root-specific and nematode-responsive behavior of the shortlisted genes, we generated promoter::GUS constructs for both the putative NRRS genes. Five independent transgenic lines for each promoter construct and 15 plants from each independent line were evaluated. In transgenic plants harboring pAt1g74770::GUS constructs, the nematode infected plants (T3-P6) showed strong GUS expression in roots at 21 dpi (Figure 6A complete plant right side and Figure 6C) as compared to uninfected (control) plants (Figure 6A complete plant left side and Figure 6B). On examination of pAt1g74770::GUS lines under microscope, large numbers of galls were visible along with blue staining throughout the root system (Figure 6D). In transgenic plants harboring pAt2g18140::GUS constructs, the nematode infected transgenic line (T3-P2) revealed maximum GUS activity only in galls during early stages of infection at 10 dpi (Figures 6F,G) as compared to uninfected (control) plants (Figures 6E,H). The microscopic examination revealed strong GUS activity in the gall (Figure 6I). Thus, the histochemical GUS assay of promoter-reporter gene constructs upon nematode infection confirmed the root-specificity and nematode-responsiveness of both the promoters. GUS activity was not detected in control (uninfected) transgenic plants for either of the promoters used.
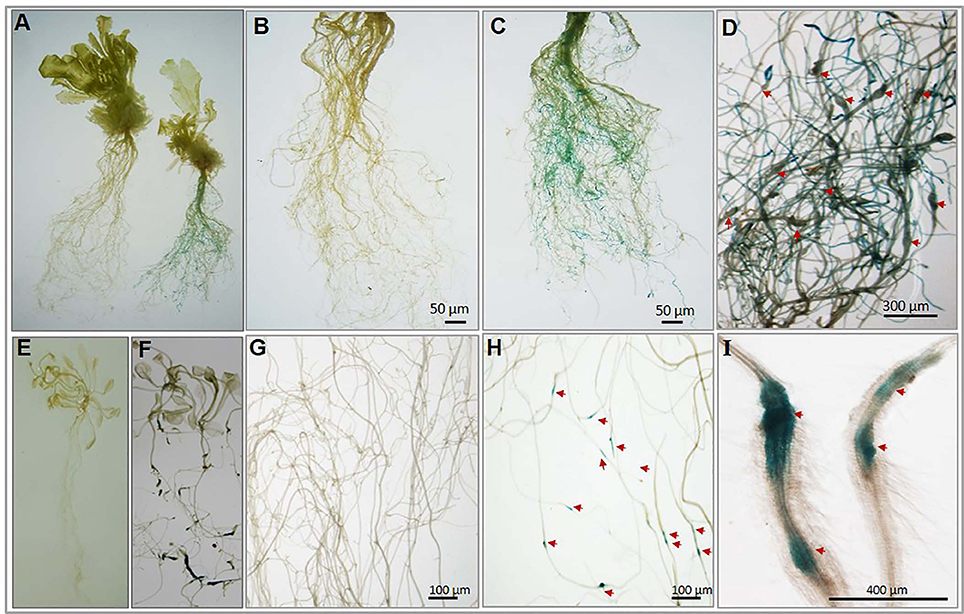
Figure 6. Histochemical GUS analysis of transgenic Arabidopsis lines harboring nematode-responsive root-specific promoters. (A) Control plant (w/o nematode infection) plant (left) and nematode infected plant (right) at 21 dpi. (B) Control (w/o nematode infection) root of transgenic (pAt1g74770::GUS) plant. (C) Nematode infected root of transgenic (pAt1g74770::GUS) plant exhibiting strong GUS activity throughout the root at 21 dpi. (D) Enlarged microscopic view of nematode infected root of transgenic (pAt1g74770::GUS) plant exhibiting strong GUS activity and multiple galls at 21 dpi. (E) Control (w/o nematode infection) transgenic plant (pAt2g18140::GUS). (F) Nematode infected transgenic plant (pAt2g18140::GUS) showing GUS activity at 10 dpi. (G) Control (w/o nematode infection) root of transgenic (pAt2g18140::GUS) plant. (H) Nematode infected root of transgenic (pAt2g18140::GUS) plant exhibiting GUS activity only in the nematode induced galls at 10 dpi. (I) Enlarged microscopic view of galls from nematode infected roots of transgenic (pAt2g18140::GUS) plant showing GUS activity at 10 dpi. The red arrow in (D,H) and (I) point to the nematode galls.
Screening of pAt1g74770::Splicing Factor RNAi Transgenic Lines for Nematode Resistance
NRRS promoter (At1g74770) was used for expressing dsRNA of a nematode gene, splicing factor, to evaluate the efficacy of NRRS promoters in inducing HD-RNAi mediated resistance in Arabidopsis. Of the five independent transgenic lines containing promoter At1g74770::splicing factor, two transgenic lines (At1g74770::SF E1 and At1g74770::SF E2) were tested against nematode. For each transgenic line 15 plants were evaluated. Transgenic lines of the splicing factor gene exhibited 20–32% reduction in number of galls compared to control plants (Figure 7). To study the effect of transgenic lines on gene expression in nematodes, females were isolated from transgenic lines expressing splicing factor dsRNA and control plants at 42 dpi. Quantitative Real-Time PCR (qRT-PCR) analysis revealed slightly reduced expression of splicing factor gene in the females isolated from transgenic plants compared to control, indicating that the splicing factor gene, driven by the NRRS promoter, has not been silenced significantly (Figure 8).
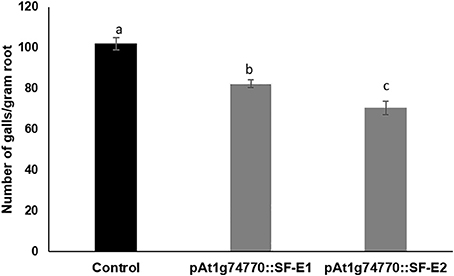
Figure 7. Root-knot nematode infection assay in control (empty vector) and transgenic plants expressing dsRNA of splicing factor gene using At1g74770 promoter. Fifteen plants each of control and transgenic lines (two independent events, E1 and E2) were evaluated and the values shown are average number of knots per plant. Each bar denotes the mean ± SE (n = 15), and bars with different letters (a–c) indicate statistical significance difference in a one-way ANOVA (p ≤ 0.05).
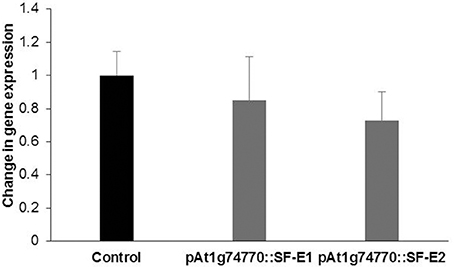
Figure 8. Quantitative Real-time PCR expression analysis of splicing factor gene. Expression levels of splicing factor gene in nematode females, developed in the dsRNA expressing Arabidopsis transgenic lines (pAt1g74770::SF-E1 and pAt1g74770::SF-E1) compared to control. The transcripts levels were normalized to the expression of a nematode 18S rRNA gene. The data are shown as fold change and each bar represents the mean ± SE (n = 3). There was no statistical difference in data sets.
Discussion
Plant parasitic nematodes (PPNs) are one of the major threat to crops across the globe, and RKN (Meloidogyne spp.) account for a major proportion of damage caused by nematodes (Elling, 2013). Among different biotechnological approaches to control nematode infestation, silencing of nematode genes crucial for primary infection and reproduction through RNAi has emerged an effective method (Dutta et al., 2015). However, this controlled targeting of nematode genes is contingent upon expression of RNAi constructs in response to nematode infection, therefore depends upon the identification of NRRS genes. In this study, we identify such gene using in silico analysis of available microarray data and demonstrate the proof-of-concept for host-delivered, RNAi-mediated nematode resistance in Arabidopsis.
Nematode-Responsive Root-Specific (NRRS) Genes
The NRRS gene set identified in this study comprises of several putative nematode responsive and root specific genes such as those from family of expansins, peroxidases, and PINFORMED auxin transporter, that have well-established roles in plant–nematode interactions. Expansins facilitate nematode penetration into the roots (Gal et al., 2006; Fudali et al., 2008); up-regulation of expansins and expansin-like genes are reported in developing galls in Arabidopsis (Jammes et al., 2005), tomato (Gal et al., 2006) and in soybean syncytia induced by soybean cyst nematodes (Ithal et al., 2007). Peroxidases play an essential role in cell wall strengthening and are also reported to be up-regulated in nematode-infected roots (Ithal et al., 2007). The up-regulation of both expansins as well as peroxidases is proposed to be part of mechanism that maintains a balance between cell wall loosening and cell wall strengthening during feeding site development of nematodes (Gheysen and Mitchum, 2009). PIN are efflux facilitators that mediate auxin transport; these express in specific regions of roots and show elevated expression upon nematode infection (Grunewald et al., 2009). The presence of these gene members of families that have established roles in mediating successful plant nematode interactions such as ATEXPA14, ATEXP6, ATEXP4, ATEXPB3, and ATAXP4 in nematode-responsive gene set along with ATCYP86 (a peroxidase), ATRFNR2 in NRRS gene set shows the effectiveness of such strategy to identify tissue-specific and pathogen responsive genes. In addition to, the presence of cis-regulatory elements, other than those used for filtering NRRS genes, such as those that play role wounding, pathogen response, plant defense signaling, disease, and pathogen responses and most importantly, the elements involved in organ specificity (Table 3) further supports that validity of approach used for selection of NRRS genes. Thereby, demonstrating that expression-based surveys for identifying condition-specific gene promoters may have a potential utility in developing plants with host-mediated response to biotic and abiotic response.
Nematode-Responsive Root-Specific Promoter Driven Expression upon Nematode Infection
In planta prm::GUS constructs of both NRRS genes tested in this study (At1g74770 and At2g18140) demonstrates highly restricted expression in roots in response to nematode infection. The At2g18140, expression is highly restricted to galls that encapsulates NFS with no trace in aerial portion of plant. At2g18140 encodes for a peroxidase protein and plant peroxidases have earlier been implicated in host-plant parasitic nematode interaction (Vercauteren et al., 2001; Jammes et al., 2005; Severino et al., 2012), including Coffea canephora sp. Thereby, confirming important and likely conserved roles of peroxidases in response to PPNs. The At1g74770 gene promoter displays GUS expression throughout the root, strong and constitutively present across all cell layers. Thereby, it seems like an ideal candidate to develop nematode-resistant plants.
Nematode Infection Assay of pAt1g74770::Splicing Factor Transgenic Plant
The number of galls in Arabidopsis transgenic plants expressing pAt1g74770::splicing factor gene was lower (20–32%) compared to control plants. However, transgenic tobacco plants expressing dsRNA of splicing factor gene through CaMV35S promoter exhibited about 95% reduction in gall formation as well as in number of nematode females (Yadav et al., 2006). Similarly, the splicing factor gene driven by 35S promoter in Arabidopsis transgenic lines exhibited up to 71% reduction in gall number (Kumar et al., 2017). In the last few years, several promoters have been identified in various crops including tobacco (Opperman et al., 1994), LEMMI9 in tomato (Escobar et al., 1999), Hahsp17.7G4 in Helianthus annuus (Escobar et al., 2003), AtCel-1 in Arabidopsis (Sukno et al., 2006), AtWRKY23 (Grunewald et al., 2008), and ZmRCP-1 in maize, banana, and plantains (Onyango et al., 2016). None of the promoters identified have been utilized for driving the production of dsRNA of nematode genes for HD-RNAi silencing. Only TobRB7, a gall specific promoter was used to drive M. javanica gene, MjTis11, in tobacco but no sign of gall reduction was observed in nematode infected plants (Fairbairn et al., 2007). The lack of silencing observed in the TobRB7 promoter lines was attributed to the weakness of the TobRB7 promoter. However, we observed up to 32% reduction in nematode infection by using our NRRS promoter as compared to 71% reduction with use of CaMV35S promoter. The CaMV35S promoter, being constitutive in nature is likely to produce more dsRNA as compared to the conditional NRRS promoter used in the present study. However, the use of 35S promoter has to be exercised with caution since dsRNA is produced in all tissues all the time and can lead to undesirable effects in transgenic plants. There is a need to identify and evaluate large number of tissue-specific promoters and use the best ones for developing nematode resistant plants using HD-RNAi approach.
Conclusion
Plant-parasitic nematodes (PPNs) are primary biotic factors that limit crop production. RNA interference (RNAi) presents a practical approach for silencing of multiple nematode parasitism and developmental genes via the host-mediated response. This study presents a computational approach to identify NRRS gene promoters and demonstrates its practical utility for host-induced RNAi-mediated control of nematode infestation in Arabidopsis as a proof-of-concept. Although further work is required to improve the efficiency of nematode control by testing other nematode gene targets, the study provides a general framework that addresses concerns regarding the use of constitutive promoters that may lead to off-targets effects and represent one more step toward the development of crops with a built-in defense mechanism against invading pathogens.
Author Contributions
AtK, AnK, AS, KS, RS, and PKJ planned the experiments. AtK designed and performed in silico analysis for NRRS promoters, cis-element identification. AtK performed molecular cloning of promoters and expression based validation. AnK generated nematode infected root samples, carried out gene expression studies through qRT-PCR, developed transgenic lines, carried out GUS analysis and RNAi construct preparation. AtK and AnK took the photographs. AtK, AnK, VS, AS, MA, RB, KS, RS, and PKJ analyzed the results. AtK, AnK, and PKJ wrote and finalized the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge the financial support from Indian Council of Agricultural Research (ICAR) through National Agricultural Innovative Project (NAIP/C4/C1092) and National Agricultural Science Fund (NFBSFARA/RNA-3022/2012-13). The authors thank the staff of National Phytotron Facility (NPF), IARI, New Delhi, India, for providing space in green house and growth chambers.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02049/full#supplementary-material
Supplementary Table 1. Primers used for qRT-PCR analysis and amplification of target promoter sequences from the Arabidopsis (Col 0) genomic DNA.
Supplementary Table 2. Probes preferentially expressed (FC ≥ 4 and corrected p-value ≤ 0.01) in roots among all three studied time points (7, 17 and 21 days).
Supplementary Table 3. Genes that were found to be up-regulated, in Arabidopsis thaliana on Meloidogyne incognita infection at the early and late stages, through meta-analysis.
Supplementary Table 4. NRRS genes identified using standalone analysis.
Supplementary Table 5. NRRS genes identified using meta-analysis.
Supplementary Table 6. NRRS genes (8 nos.) common between the standalone and meta-analysis.
Supplementary Text Presentation 1. Details of meta-analysis and stand-alone analysis.
Supplementary Figure Presentation 1. MEME analysis using promoter region of 51 nematode-responsive root-specific genes.
Supplementary Figure 1. MEME output for 51 NRRS genes for motif AAAxAAAA.
Abbreviations
HD-RNAi, Host-delivered RNA interference; NRRS, Nematode-responsive root-specific; RKNs, Root-Knot Nematodes; PPNs, Plant Parasitic Nematodes; J2s, Second-stage Juveniles; DPI, Days Post Inoculation; LBI, Low Basal Intensity; G-MAI, Global-Mean of Average Intensities.
References
Abad, P., Gouzy, J., Aury, J. M., Castagnone-Sereno, P., Danchin, E. G., Deleury, E., et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915. doi: 10.1038/nbt.1482
Alkharouf, N. W., Klink, V. P., Chouikha, I. B., Beard, H. S., MacDonald, M. H., Meyer, S., et al. (2006). Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224, 838–852. doi: 10.1007/s00425-006-0270-8
Bakhetia, M., Charlton, W., Atkinson, H. J., and McPherson, M. J. (2005). RNA interference of dual oxidase in the plant nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 18, 1099–1106. doi: 10.1094/MPMI-18-1099
Banerjee, S., Banerjee, A., Gill, S. S., Gupta, O. P., Dahuja, A., Jain, P. K., et al. (2017). RNA Interference: a novel source of resistance to combat plant parasitic nematodes. Front. Plant Sci. 8:834. doi: 10.3389/fpls.2017.00834
Barcala, M., García, A., Cabrera, J., Casson, S., Lindsey, K., Favery, B., et al. (2010). Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 61, 698–712. doi: 10.1111/j.1365-313X.2009.04098.x
Bar-Or, C., Kapulnik, Y., and Koltai, H. (2005). A broad characterization of the transcriptional profile of the compatible tomato response to the plant parasitic root-knot nematode Meloidogyne javanica. Eur. J. Plant Pathol. 111, 181–192. doi: 10.1007/s10658-004-2134-z
Bertioli, D. J., Smoker, M., and Burrows, P. R. (1999). Nematode-responsive activity of the cauliflower mosaic virus 35S promoter and its subdomains. Mol. Plant Microbe Interact. 12, 189–196. doi: 10.1094/MPMI.1999.12.3.189
Cabrera, J., Bustos, R., Favery, B., Fenoll, C., and Escobar, C. (2014). Technical advance NEMATIC : a simple and versatile tool for the in silico analysis of plant – nematode interactions. Mol. Plant Pathol. 15, 627–636. doi: 10.1111/mpp.12114
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coutu, C., Brandle, J., Brown, D., Miki, B., and Simmonds, J. (2007). PORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res. 16, 771–781. doi: 10.1007/s11248-007-9066-2
Davuluri, R. V., Sun, H., Palaniswamy, S. K., Matthews, N., Molina, C., Kurtz, M., et al. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4:25. doi: 10.1186/1471-2105-4-25
Dinh, P. T. Y., Brown, C. R., and Elling, A. A. (2014). RNA Interference of effector gene Mc16D10L confers resistance against Meloidogyne chitwoodi in Arabidopsis and Potato. Phytopathology 104, 1098–1106. doi: 10.1094/PHYTO-03-14-0063-R
Dong, L., Xu, J., Chen, S., Li, X., and Zuo, Y. (2016). Mi-flp-18 and Mi-mpk-1 genes are potential targets for Meloidogyne incognita control. J. Parasitol. 102, 20–213. doi: 10.1645/15-768
Dutta, T. K., Banakar, P., and Rao, U. (2015). The status of RNAi- based transgenic research in plant nematology. Front. Micrbiol. 5:760. doi: 10.3389/fmicb.2014.00760
Elling, A. A. (2013). Major emerging problems with minor Meloidogyne species. Phytopathology103, 1092–1102. doi: 10.1094/PHYTO-01-13-0019-RVW
Escobar, C., Barcala, M., Portillo, M., Almoguera, C., Jordano, J., and Fenoll, C. (2003). Induction of the Hahsp17.7G4 promoter by root-knot nematodes: involvement of heat-shock elements in promoter activity in giant cells. Mol. Plant Microbe Interact. 16, 1062–1068. doi: 10.1094/MPMI.2003.16.12.1062
Escobar, C., De Meutter, J., Aristizábal, F. A., Sanz-Alférez, S., del Campo, F. F., Barthels, N., et al. (1999). Isolation of the LEMMI9 gene and promoter analysis during a compatible plant-nematode interaction. Mol. Plant Microbe Interact. 12, 440–449. doi: 10.1094/MPMI.1999.12.5.440
Fairbairn, D. J., Cavallaro, A. S., Bernard, M., Mahalinga-Iyer, J., Graham, M. W., and Botella, J. R. (2007). Host-delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta 226, 1525–1533. doi: 10.1007/s00425-007-0588-x
Favery, B., Chelysheva, L. A., Lebris, M., Jammes, F., Marmagne, A., De Almeida-Engler, J., et al. (2004). Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16, 2529–2540. doi: 10.1105/tpc.104.024372
Fudali, S., Janakowski, S., Sobczak, M., Griesser, M., Grundler, F. M., Golinowski, W., et al. (2008). Two tomato alpha-expansins show distinct spatial and temporal expression patterns during development of nematode-induced syncytia. Physiol. Plant 132, 370–383. doi: 10.1111/j.1399-3054.2007.01017.x
Fuller, V. L., Lilley, C. J., Atkinson, H. J., and Urwin, P. E. (2007). Differential gene expression in Arabidopsis following infection by plant-parasitic nematodes Meloidogyne incognita and Heterodera schachtii. Mol. Plant Pathol. 8, 595–609. doi: 10.1111/j.1364-3703.2007.00416.x
Gal, T. Z., Aussenberg, E. R., Burdman, S., Kapulnik, Y., and Koltai, H. (2006). Expression of a plant expansin is involved in the establishment of root-knot nematode parasitism in tomato. Planta 224, 155–162. doi: 10.1007/s00425-005-0204-x
Gheysen, G., and Mitchum, M. (2009). “Molecular insights in the susceptible plant response to nematode infection,” in Cell Biology of Plant Nematode Parasitism, eds R. H. Berg and C. Taylor (Berlin: Springer), 45–81.
Goddijn, O. J., Lindsey, K., van der Lee, F. M., Klap, J. C., and Sijmons, P. C. (1993). Differential gene expression in nematode-induced feeding structures of transgenic plants harbouring promoter-gusA fusion constructs. Plant J. 4, 863–873. doi: 10.1046/j.1365-313X.1993.04050863.x
Grunewald, W., Cannoot, B., Friml, J., and Gheysen, G. (2009). Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog. 5:e1000266. doi: 10.1371/journal.ppat.1000266
Grunewald, W., Karimi, M., Wieczorek, K., Van de Cappelle, E., Wischnitzki, E., Grundler, F., et al. (2008). A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 148, 358–368. doi: 10.1104/pp.108.119131
Hammes, U. Z., Schachtman, D. P., Berg, R. H., Nielsen, E., Koch, W., McIntyre, L. M., et al. (2005). Nematode-induced changes of transporter gene expression in Arabidopsis roots. Mol. Plant Microbe Interact. 18, 1247–1257. doi: 10.1094/MPMI-18-1247
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., et al. (2008). Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008:420747. doi: 10.1155/2008/420747
Huang, G., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2006). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. U.S.A. 103, 14302–14306. doi: 10.1073/pnas.0604698103
Ithal, N., Recknor, J., Nettleton, D., Maier, T., Baum, T. J., and Mitchum, M. G. (2007). Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant Microbe Interact. 20, 510–525. doi: 10.1094/MPMI-20-5-0510
Jammes, F., Lecomte, P., de Almeida-Engler, J., Bitton, F., Martin-Magniette, M. L., Renou, J. P., et al. (2005). Genome-wide expression profiling of the host-response to root-knot nematode infection in Arabidopsis. Plant J. 44, 447–458. doi: 10.1111/j.1365-313X.2005.02532.x
Jiao, Y., Ma, L., Strickland, E., and Deng, X. W. (2005). Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17, 3239–3256. doi: 10.1105/tpc.105.035840
Kankainen, M., Pehkonen, P., Rosenstöm, P., Törönen, P., Wong, G., and Holm, L. (2006). POXO: A web-enabled tool series to discover transcription factor binding sites. Nucleic Acids Res. 34, 534–540. doi: 10.1093/nar/gkl296
Klink, V. P., Overall, C. C., Alkharouf, N. W., MacDonald, M. H., and Matthews, B. F. (2007). A time-course comparative microarray analysis of an incompatible and compatible response by Glycine max (soybean) to Heterodera glycines (soybean cyst nematode) infection. Planta 226, 1423–1447. doi: 10.1007/s00425-007-0581-4
Kumar, A., Joshi, I., Kohli, D., Satheesh, V., Abdin, M. Z., Sirohi, A., et al. (2016). Characterization of root-knot nematode responsive and root-specific promoter containing PIN domain from Arabidopsis thaliana (L.) Heynh. Indian J. Genet. Plant Breed. 76, 75–83. doi: 10.5958/0975-6906.2016.00011.0
Kumar, A., Kakrana, A., Sirohi, A., Subramaniam, K., Srinivasan, R., Abdin, M. Z., et al. (2017). Host-delivered RNAi-mediated root-knot nematode resistance in Arabidopsis by targeting splicing factor and integrase genes. J. Gen. Plant Pathol. 83, 91–97. doi: 10.1007/s10327-017-0701-3
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lilley, C. J., Urvin, P. E., Johnston, K. A., and Atkinson, H. J. (2004). Preferential expression of a plant cystatin at nematode feeding sites confers resistance to Meloidogyne incognita and Globodera pallida. Plant Biotechnol. J. 2, 3–12. doi: 10.1046/j.1467-7652.2003.00037.x
Mahony, S., and Benos, P. V. (2007). STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35, 253–258. doi: 10.1093/nar/gkm272
Mitchum, M., Sukno, S., Shani, Z., Shoseyov, O., and Davis, E. L. (2004). The promoter of the Arabidopsis thaliana cel1 endo-1,4-β-glucanase gene is differentially expressed in plant feeding cells induced by root-knot and cyst nematodes. Mol. Plant Pathol. 5, 175–181. doi: 10.1111/j.1364-3703.2004.00216.x
Niu, J., Liu, P., Liu, Q., Chen, C., Guo, Q., Yin, J., et al. (2016). Msp40 effector of root-knot nematode manipulates plant immunity to facilitate parasitism. Sci. Rep. 6:19443. doi: 10.1038/srep19443
Ono, A., Izawa, T., Chua, N. H., and Shimamoto, K. (1996). The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol. 112, 483–491. doi: 10.1104/pp.112.2.483
Onyango, S. O., Roderick, H., Tripathi, J. N., Collins, R., Atkinson, H. J., Oduor, R. O., et al. (2016). The ZmRCP-1 promoter of maize provides root tip specific expression of transgenes in plantain. J. Biol. Res. Thessaloniki 23:4. doi: 10.1186/s40709-016-0041-z
Opperman, C. H., Taylor, C. G., and Conkling, M. A. (1994). Root-knot nematode-directed expression of a plant root-specific gene. Science 263, 221–223. doi: 10.1126/science.263.5144.221
Portillo, M., Cabrera, J., Lindsey, K., Topping, J., Andrés, M. F., Emiliozzi, M., et al. (2013). Distinct and conserved transcriptomic changes during nematode-induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol. 197, 1276–1290. doi: 10.1111/nph.12121
Puzio, P., Lausen, J., Heinen, P., and Grundler, F. (2000). Promoter analysis of pyk20, a gene from Arabidopsis thaliana. Plant Sci. 157, 245–255. doi: 10.1016/S0168-9452(00)00287-9
Rosso, M. N., Jones, J. T., and Abad, P. (2009). RNAi and functional genomics in plant parasitic nematodes. Annu. Rev. Phytopathol. 47, 207–232. doi: 10.1146/annurev.phyto.112408.132605
Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Saha, D., Kumar, V., Bhat, S. R., and Srinivasan, R. (2007). Characterization of upstream sequences of the LOJ gene leads to identification of a novel enhancer element conferring lateral organ junction-specific expression in Arabidopsis thaliana. Plant Mol. Biol. Rep. 29, 265–277. doi: 10.1007/s11105-010-0229-6
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Schaff, J. E., Nielsen, D. M., Smith, C. P., Scholl, E. H., and Bird, D. M. (2007). Comprehensive transcriptome profiling in tomato reveals a role for glycosyltransferase in Mi-mediated nematode resistance. Plant Physiol. 144, 1079–1092. doi: 10.1104/pp.106.090241
Severino, F. E., Brandalise, M., Costa, C. S., et al. (2012). CaPrx, a Coffea arabica gene encoding a putative class III peroxidase induced by root-knot nematode infection. Plant Sci. 191–192, 35–42. doi: 10.1016/j.plantsci.2012.04.012
Smith, E. F., and Townsend, C. O. (1907). A plant-tumor of Bacterial origin. Science 25, 671–673. doi: 10.1126/science.25.643.671
Smyth, G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:3. doi: 10.2202/1544-6115.1027
Sukno, S., Shimerling, O., McCuiston, J., Tsabary, G., Shani, Z., Shoseyov, O., et al. (2006). Expression and regulation of the Arabidopsis thaliana cel1 endo 1,4 beta glucanase gene during compatible plant-nematode interactions. J. Nematol. 38, 354–361.
Tamilarasan, S., and Rajam, M. V. (2013). Engineering crop plants for nematode resistance through host-derived RNA interference. Cell Dev. Biol. 2:114. doi: 10.4172/2168-9296.1000114
Thomas-Chollier, M., Sand, O., Turatsinze, J. V., Janky, R., Defrance, M., Vervisch, E., et al. (2008). RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 36, 119–127. doi: 10.1093/nar/gkn304
Thurau, T., Kifle, S., Jung, C., and Cai, D. (2003). The promoter of the nematode resistance gene Hs1pro-1 activates a nematode-responsive and feeding site-specific gene expression in sugar beet (Beta vulgaris L.) and Arabidopsis thaliana. Plant Mol. Biol. 52, 643–660.
Urwin, P. E., Lilley, C. J., McPherson, M. J., and Atkinson, H. J. (1997). Resistance to both cyst and root-knot nematodes conferred by transgenic Arabidopsis expressing a modified plant cystatin. Plant J. 12, 455–461. doi: 10.1046/j.1365-313X.1997.12020455.x
Vaughan, S. P., James, D. J., Lindsey, K., and Massiah, A. J. (2006). Characterization of FaRB7, a near root-specific gene from strawberry (Fragaria x ananassa Duch.) and promoter activity analysis in homologous and heterologous hosts. J. Exp. Bot. 57, 3901–3910. doi: 10.1093/jxb/erl185
Vercauteren, I., Van Der Schueren, E., Van Montagu, M., and Gheysen, G. (2001). Arabidopsis thaliana genes expressed in the early compatible interaction with root-knot nematodes. Mol. Plant Microbe Interact. 14, 288–299. doi: 10.1094/MPMI.2001.14.3.288
Weigel, D., and Glazebrook, J. (2006). Transformation of Agrobacterium using the freeze–thaw method. Cold Spring Harb. Protoc. 7, 1031–1036. doi: 10.1101/pdb.prot4666
Wieczorek, K., Hofmann, J., Blöchl, A., Szakasits, D., Bohlmann, H., and Grundler, F. M. W. (2008). Arabidopsis endo-1,4-β-glucanases are involved in the formation of root syncytia induced by Heterodera schachtii. Plant J. 53, 336–351. doi: 10.1111/j.1365-313X.2007.03340.x
Xiao, F. H., and Xue, G. P. (2001). Analysis of the promoter activity of late embryogenesis abundant protein genes in barley seedlings under conditions of water deficit. Plant Cell Rep. 20, 667–673. doi: 10.1007/s002990100384
Yadav, B. C., Veluthambi, K., and Subramaniam, K. (2006). Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 148, 219–222. doi: 10.1016/j.molbiopara.2006.03.013
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2001). Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Novartis Found Symp. 236, 176–186. doi: 10.1002/9780470515778.ch13
Yamamoto, Y. T., Taylor, C. G., Acedo, G. N., Cheng, C. L., and Conkling, M. A. (1991). Characterization of cis-acting sequences regulating root-specific gene expression in tobacco. Plant Cell 3, 371–382.
Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1540. doi: 10.1105/tpc.13.7.1527
Keywords: Arabidopsis, HD-RNAi, in silico analysis, nematode-responsive genes, promoter analysis, root-specific genes
Citation: Kakrana A, Kumar A, Satheesh V, Abdin MZ, Subramaniam K, Bhattacharya RC, Srinivasan R, Sirohi A and Jain PK (2017) Identification, Validation and Utilization of Novel Nematode-Responsive Root-Specific Promoters in Arabidopsis for Inducing Host-Delivered RNAi Mediated Root-Knot Nematode Resistance. Front. Plant Sci. 8:2049. doi: 10.3389/fpls.2017.02049
Received: 15 June 2017; Accepted: 15 November 2017;
Published: 12 December 2017.
Edited by:
Peng Zhang, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Charles Melnyk, Swedish University of Agricultural Sciences, SwedenSailendra Nath Sarkar, University of Calcutta, India
Copyright © 2017 Kakrana, Kumar, Satheesh, Abdin, Subramaniam, Bhattacharya, Srinivasan, Sirohi and Jain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pradeep K. Jain, jainpmb@gmail.com
†These authors have contributed equally to this work.
 Atul Kakrana1,2†
Atul Kakrana1,2† Anil Kumar
Anil Kumar Viswanathan Satheesh
Viswanathan Satheesh Ramamurthy Srinivasan
Ramamurthy Srinivasan Anil Sirohi
Anil Sirohi Pradeep K. Jain
Pradeep K. Jain