- Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, China
Drought, cold and salinity are the major environmental stresses that limit agricultural productivity. NAC transcription factors regulate the stress response in plants. Pumpkin (Cucurbita moschata) is an important cucurbit vegetable crop and it has strong resistance to abiotic stress; however, the biological functions of stress-related NAC genes in this crop are largely unknown. This study reports the function of CmNAC1, a stress-responsive pumpkin NAC domain protein. The CmNAC1-GFP fusion protein was transiently expressed in tobacco leaves for subcellular localization analysis, and we found that CmNAC1 is localized in the nucleus. Transactivation assay in yeast cells revealed that CmNAC1 functions as a transcription activator, and its transactivation domain is located in the C-terminus. CmNAC1 was ubiquitously expressed in different organs, and its transcript was induced by salinity, cold, dehydration, H2O2, and abscisic acid (ABA) treatment. Furthermore, the ectopic expression (EE) of CmNAC1 in Arabidopsis led to ABA hypersensitivity and enhanced tolerance to salinity, drought and cold stress. In addition, five ABA-responsive elements were enriched in CmNAC1 promoter. The CmNAC1-EE plants exhibited different root architecture, leaf morphology, and significantly high concentration of ABA compared with WT Arabidopsis under normal conditions. Our results indicated that CmNAC1 is a critical factor in ABA signaling pathways and it can be utilized in transgenic breeding to improve the abiotic stress tolerance of crops.
Introduction
Crops are affected by all kinds of abiotic stresses such as drought, cold, heat, and salinity stress that could lead toward extensive production losses worldwide. Therefore, understanding the stress response and improving the stress tolerance of crops is critical to boost agricultural productivity, ensure food security, and environmental sustainability (Mittler, 2006; Zhu, 2016). As a largest plant-specific transcription factor family, NAC domain proteins play an important role in plant development and regulation of abiotic stress tolerance. These proteins recently have received the attention as a major regulator in various stress signaling pathways and have been found to improve the abiotic stress tolerance of different crops through genetic engineering (Hu et al., 2006; Nakashima et al., 2007; Hong et al., 2016; Huang L. et al., 2016; Wang et al., 2017). As transcriptional factors, NAC domain proteins contain highly conserved DNA-binding domain in the N-terminal and diverse transcription activation or repression domain in the C-terminal (Olsen et al., 2005; Kim et al., 2007; Hao et al., 2010).
The NAC domain transcription factors in Arabidopsis function in plant development, senescence, and stress regulation. NAC1 and AtNAC2 regulate root development (Xie et al., 2000; He et al., 2005); CUC1, CUC2, and CUC3 control leaf serration and axillary bud development (Nikovics et al., 2006; Raman et al., 2008); SND1 and VND7 trigger the de novo xylem formation and regulate secondary wall synthesis in fibers (Zhong et al., 2006; Reusche et al., 2012). According to some recent reports, NAC domain proteins are also associated with senescence, such as ORE1, NAP, ATAF1, and JUNGBRUNNEN1 play an important role in promoting or delaying senescence (Balazadeh et al., 2010; Wu et al., 2012; Yang et al., 2014; Garapati et al., 2015; Qiu et al., 2015; Takasaki et al., 2015). NAC domain proteins also act as major regulators in abiotic stress response, for instance ATAF1, RD26, ANAC096, and ANAC013 are involved in the regulation of salinity, oxidative, and drought stress tolerance (Fujita et al., 2004; Wu et al., 2009; Clercq et al., 2013; Xu et al., 2013). In addition, NAC domain proteins have become a popular research topic for different plant species. The expression of OsNAC6 and ONAC022 genes is upregulated by various stresses, whereas transgenic rice plants enhance multiple stress tolerance (Nakashima et al., 2007; Hong et al., 2016). The overexpression of stress-responsive NAC domain protein, SNAC1, significantly improves the drought tolerance in the field while showing no yield penalty (Hu et al., 2006). GmNAC11 and GmNAC20 from soybean are induced by phytohormones and abiotic stress with different transcriptional activities. GmNAC11 is a transcriptional activator, whereas GmNAC20 is a mild transcriptional repressor. The EE of GmNAC20 improves the salt and freezing tolerance and promotes the lateral root formation in Arabidopsis. By contrast, the EE of GmNAC11 in Arabidopsis plants enhances only the salt tolerance (Hao et al., 2011). These findings revealed that NAC domain proteins are important regulators of plant development and stress response.
Cucurbits are one of the most important fruit vegetables cultivated worldwide. Most cucurbits, particularly watermelon, cucumber, and melon, are sensitive to abiotic stresses, such as salinity, cold, and drought (Xie et al., 2015). The use of stress tolerant pumpkin as rootstock was recently found to improve the salt tolerance of cucumbers and the low temperature, low magnesium and nitrogen stress tolerance of watermelons (Huang et al., 2010; Huang Y. et al., 2016; Xu et al., 2016; Nawaz et al., 2016, 2017; Niu et al., 2017). However, the possible molecular regulatory mechanism underlying pumpkin response to abiotic stress are not yet elucidated. Thus, an abiotic stress-related NAC transcription factor, CmNAC1, was characterized from the transcriptome data of salt-treated pumpkin. Expression patterns of CmNAC1 in response to dehydration, salinity, cold, ABA, GA, and H2O2 treatments were analyzed through real-time quantitative PCR (RT-qPCR). To evaluate its function in abiotic stress tolerance, we expressed CmNAC1 in Arabidopsis ectopically. Morphological assays revealed that the EE of CmNAC1 altered the plant phenotypes, such as relative dwarfism and large roots under normal growth conditions. Moreover, CmNAC1-EE plants showed good tolerance under abiotic stress treatment. In summary, our results suggest that CmNAC1 plays positive roles in plant stress response and can be a candidate gene for the improvement of crops stress tolerance in the future.
Materials and Methods
Plant Materials and Abiotic Stress Treatment
N15 an inbred line of pumpkin (Cucurbita moschata Duch.) developed at our laboratory was utilized as experimental material in this study. The seeds were germinated in distilled water and cultivated under a 12h light/12h dark cycle at 28°C/18°C in a growth chamber and photosynthetic photon flux density of 200 μmol m-2 s-1. The seedlings preparation method is described in our previous study (Xie et al., 2015). Fourteen-day-old pumpkin seedlings were treated with dehydration, 100 mM NaCl, 100 μM exogenous GA, 100 μM exogenous ABA, and 10 mM H2O2 at 4°C for 0, 0.5, 1, 4, 8, and 12 h. Different organs, including roots, young leaves, cotyledons, and hypocotyls (at the three-leaf stage); mature leaves and stems (at the six-leaf stage); and fruits (3 days after pollination) were used in measuring the organ-specific expression patterns of CmNAC1. All collected samples were immediately frozen in liquid nitrogen and stored at -80°C. Arabidopsis thaliana Columbia-0 plants cultured in substrate were used for the transgenic study of CmNAC1.
RNA Isolation and RT-qPCR
Total RNA was extracted using Tranzol (TransGen Biotech, Inc., Beijing, China), and reverse transcription was performed using 2 μg of RNA using HiScript II One Step RT-PCR Kit (Vazyme, Piscataway, NJ, United States) according to the manufacturer’s instructions. RT-qPCR was performed on Applied Biosystems QuantStudio system using an ABI 7500 real-time PCR machine (Applied Biosystems, Foster City, CA, United States) on it default PCR program with a reaction mixture volume of 10 μl. The melting curve was recorded after 40 cycles to verify the primer specificity by heating from 65°C to 95°C. One microliter of RNase-free H2O was also included in each plate as a control template. CmEF-1α, CmCAC (Obrero et al., 2011), and AtActin2 were used as the internal references. The 2-ΔΔct method was using to calculate relative gene expression values (Livak and Schmittgen, 2001). The Ct value of two reference genes was the square root of CtCmEF-1α × CtCmCAC. The primers are listed in Supplementary Table S2. All RT –qPCR reaction results were obtained from three independent replicates.
Cloning and Analysis of CmNAC1 and CmNAC1 Promoter
The CmNAC1 cDNA sequence was obtained from the transcriptome data of salt-treated pumpkin root in our laboratory. The NCBI accession number of the transcriptome is SRP066227. Primer pairs (Supplementary Table S2) were designed to amplify the coding sequence (CDS). The PCR products were cloned into pEASYT1-Blunt vector (TransGen Biotech, Inc., Beijing, China) and transfected into Escherichia coli Tran5αT1 competent cells (TransGen Biotech, Inc., Beijing, China). Finally, the target gene in positive clone strains was sequenced (Tsingke, Inc., Beijing, China). Multiple alignments of amino acid sequences of NAC domain proteins were performed using MEGA software (version 5.1) and DNAMAN (version 7). CmNAC1 promoter sequence was cloned from pumpkin genomic DNA using GenomeWalker Universal Kits (Takara, Shiga, Japan). The 1069 bp upstream from the start codon (ATG) of CmNAC1 was selected for further analysis. The online search tool PlantCARE was used to detect putative cis-acting regulatory elements1.
Subcellular Localization and Transactivation Assay Analysis
The full-length CDS of CmNAC1 was amplified by PCR using 2× High-Fidelity Master Mix (Tsingke, Inc., Beijing, China), and the fragments were inserted into the StuI site of the pH7LIC5.0-N-eGFP vector by using ClonExpress II One Step Cloning Kits (Vazyme, Piscataway, NJ, United States) to generate 35S::eGFP-CmNAC1 fusion protein under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (Zhang et al., 2015). The construct and negative control (pH7LIC5.0-N-eGFP) were transformed into Agrobacterium strain GV3101 and were infiltrated into tobacco (Nicotiana benthamiana) leaves via Agrobacterium-mediated transformation method (Sheludko et al., 2007). Laser scanning confocal microscope was used to detect the GFP fluorescence signal. The nucleus was stained with DNA dye 4,6-diamidino-2-phenylindole (DAPI).
The CDS of CmNAC1 and the sequence encoding the N-terminus (1–450 bp) and C-terminus (450–879) were cloned. The PCR products were amplified with primers listed in Supplementary Table S2 and inserted into the BamHI site of pGBDKT7 vector and fused with the DNA-binding domain by using ClonExpress II One Step Cloning Kits to obtain pGBDKT7-CmNAC1-FL (1–292 aa), pGBDKT7-CmNAC1-N (1–150 aa) and pGBDKT7-CmNAC1-C (150–292 aa) constructs. These vectors and the pGBDKT7 (negative control) were separately transformed into the yeast strain Y2HGold. The transformed yeast cells were incubated on SD/-Trp, SD/-Trp/-His-Ade and SD/-Trp-His-x-gal plates. The transactivation activity was detected according to their growth status and α-galactosidase activity.
Generation of CmNAC1 Transgenic Arabidopsis Plants
The CDS of CmNAC1 was cloned into the pHellgate8 vector to generate the 35S::CmNAC1 construct by ClonExpress II One Step Cloning Kits. The construct was transformed into Agrobacterium tumefaciens strain GV3101 and then transferred into Arabidopsis thaliana ecotype Col-0 plants using the floral dip method (Clough and Bent, 1998). Transgenic Arabidopsis seeds were screened on MS medium suspended with kanamycin (50 mg/L). T3 homozygous lines were used for further experiments.
Abiotic Stress Tolerance Assays and ABA Sensitivity Analysis
The WT and CmNAC1-EE lines were used to evaluate the tolerance for various abiotic stresses. For the drought treatment, the water intake of 4-week-old potted Arabidopsis plants in water-saturated substrate was withheld for 2 weeks. Subsequently, the plants were re-watered for 7 days. Another batch of seedlings grown under normal conditions was subjected to -10°C cold treatment for 1 h to characterize the freezing tolerance of transgenic plants. These plants were incubated in 4°C growth chamber for 3 h before transferring to normal growth conditions (23 ± 1°C) for recovery. For the salt tolerance assay, 4-week-old potted Arabidopsis plants were subjected to 250 mM NaCl treatment for 2 weeks. The survival rate of transgenic and WT Arabidopsis lines were scored after each kind of stress treatment. The experiments were independently repeated three times and used approximately 40 plants each.
For ABA sensitivity analysis of transgenic plant, Arabidopsis seeds were cultivated on 1/2 MS medium supplemented with 0 and 2 μM ABA medium under 16 h light/8h dark cycle at 23°C/18°C in a growth chamber. The germination rate (seedlings with cotyledon) was scored from the 5th to 8th day after planting on the plates. The 7-day-old seedlings were transferred to vertical plates supplemented with 0 and 10 μM ABA for ABA response analysis. Images of roots and shoots were captured and scored using WinRHIZO Pro 2013 image analysis system.
Measurement of Electrolytic Leakage, Chlorophyll, ABA Content, Water Loss Rate, and Chlorophyll Fluorescence
Imaging PAM (MAXI; Heinz Walz, Effeltrich, Germany) was used to measure the chlorophyll fluorescence Fv/Fm value after the seedlings were stored in the dark for 30 min according to a previous method (Cheng F. et al., 2016). Fully expanded leaves from the 5-week-old seedlings of WT and EE plants were excised and weighed immediately to calculate the water loss rate. The leaves were dehydrated for 3 h on the dry filter paper (24°C–26°C) and weighed at designated time points. Images were captured at 0 and 3 h after treatment. The water loss rate was calculated based on the initial fresh weight of the leaves. Before and after cold treatment, the 4-week-old seedling leaves were collected and sampled for the measurement of electrolytic leakage according to a previous method (Hong et al., 2003). The relative chlorophyll content of 6-week-old seedling leaves from the control and salt-treated plants were measured as SPAD value using chlorophyll meter SPAD-502. For ABA content measurement, 0.10 g of 5-week-old seedling leaves were sampled and grinded on ice with cold extraction buffer (methanol:water:acetic acid, 80:19:1, v:v:v), shaken for 16 h at 4°C, centrifuged at 13,000 rpm for 15 min at 4°C. The supernatant was supplemented with internal standards, filtered using a syringe-facilitated 13-mm diameter nylon filter, dried by evaporation, and dissolved in 200 μl methanol. Simultaneous quantification of ABA by liquid chromatography was conducted using a UFLC with an autosampler according the method (Pan et al., 2008; Liu et al., 2012).
The experiment was repeated three times at least. All data were compared using Student’s t-test for statistical analysis.
Results
CmNAC1 Is a Stress-Responsive Gene
RT-qPCR was performed to elucidate the gene expression pattern in different tissues and the response of CmNAC1 to various abiotic stresses. Temporal and spatial expression results revealed that CmNAC1 has relative high expression levels in mature leaves, stem, roots, and the highest in hypocotyl. However, its expression was low in the new leaves and fruits (Figure 1A). Under H2O2 and NaCl stress conditions, CmNAC1 transcript was upregulated specifically at 0.5 and 8 h (Figures 1B,C). The expression of CmNAC1 was induced during 0.5–8 h under dehydration and ABA treatment, but only the transcript levels in ABA treatment reverted at 12 h (Figures 1F,G). However, GA consistently inhibited CmNAC1 expression during the treatment (Figure 1D). The cold stress repressed the CmNAC1 expression within 4 h, and then the transcript levels were induced until 12 h (Figure 1E). The qRT-PCR results suggest that similar to other stress-associated NAC domain proteins, CmNAC1 participates in plant stress response, and its expression is sensitive to ABA, H2O2, and GA signaling molecules.
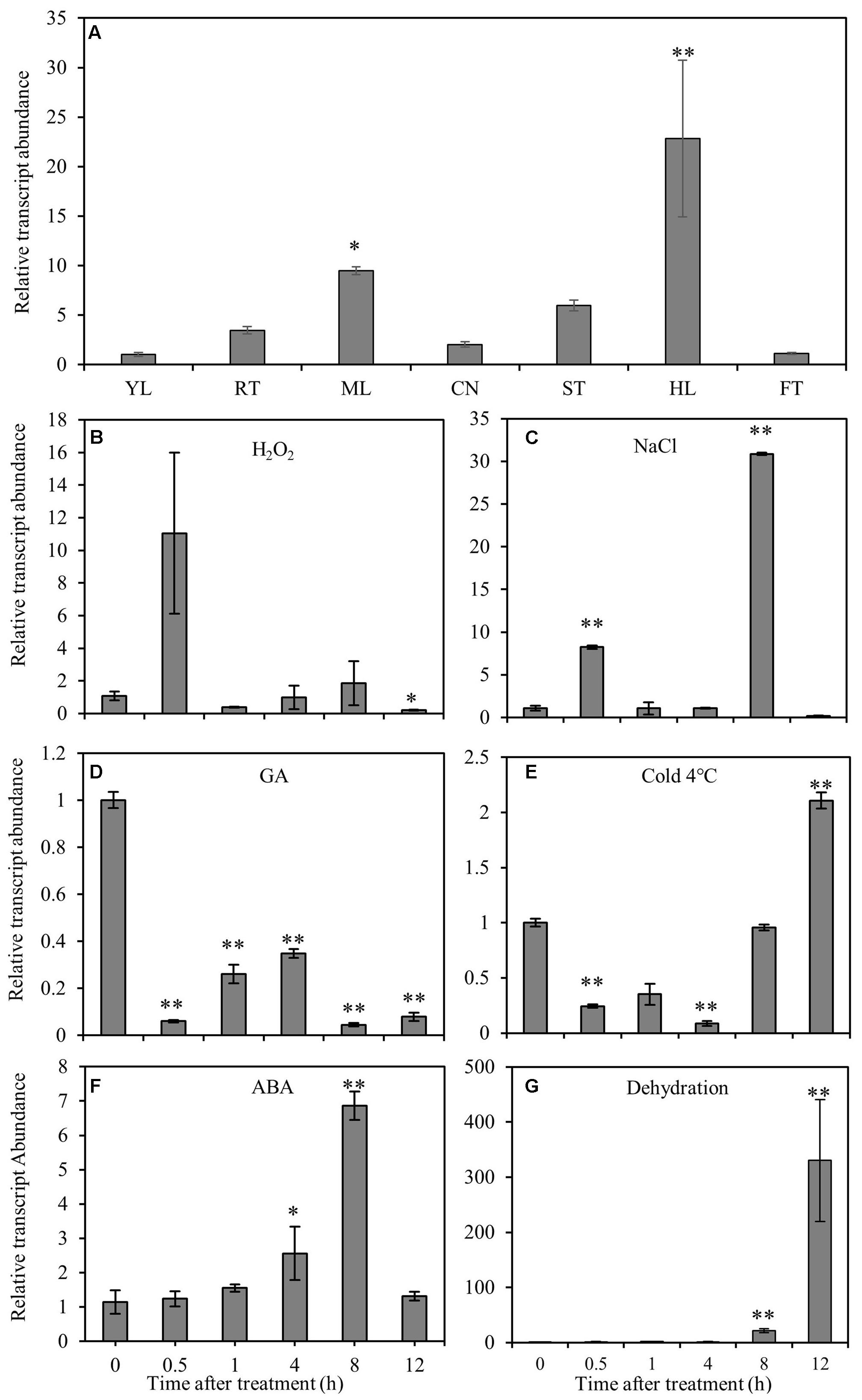
FIGURE 1. Expression patterns of CmNAC1 in pumpkin seedlings under different stress treatments or in different organs. (A) Expression patterns of CmNAC1 in various organs (root, RT; young leaves, YL; mature leaves, ML; cotyledon, CN; hypocotyl, HL; stem, ST; fruit, FT). Expression patterns of CmNAC1 in leaves after H2O2 (B), NaCl (C), GA (D), cold (E), ABA (F) and dehydration (G) treatments by qRT-PCR analysis. Zero represents leaf sample without any treatment; 0.5, 1, 4, 8, and 12 represent samples after 0.5, 1, 4, 8, and 12 h treatment, respectively. The 2-ΔΔCT method was used in qRT-PCR analysis. Transcript levels were normalized to CmEF1α and CmCAC. Values are means ± SD of three replicates. Three independent experiments were performed. Asterisks indicate a significant difference (∗P < 0.05; ∗∗P < 0.01) compared with the corresponding controls.
The 1 kb upstream of ATG start codon CmNAC1 promoter sequence was cloned to detect the putative, stress-responsive cis-acting regulatory elements (Supplementary Figure S5). Stress response-related cis-acting elements, that include five ABA-responsive elements (ABREs), one low-temperature responsive motif, two GC motifs involved in anoxic specific inducibility, and two TGACG motifs involved in methyl jasmonate responsiveness (Table 1), were found in the promoter. These stress-related cis-acting elements could be necessary for the stress-regulated expression of CmNAC1. These findings indicate that CmNAC1 plays a critical role in early stress responsiveness.
CmNAC1 Belongs to ATAF Subfamily with Transcriptional Activity and Is Localized in the Nucleus
CmNAC1 cDNA had a length of 1200 bp and was predicted to contain an 879 bp CDS encoding 292 amino acid protein. The CmNAC1 gene accession number in NCBI GenBank database was MG199592. Amino acid sequence alignment shows that CmNAC1 shared an identity of 65.35% with AtATAF1 protein from Arabidopsis. CmNAC1 protein also contained a diverse activation domain in C-terminal and a conserved NAC domain in the N-terminal region including subdomains A to E (Supplementary Figure S1). To further reveal the divergence of CmNAC1 protein during evolution, we analyzed the phylogenetic relationship of CmNAC1 with SNACs orthologues, which were functionally characterized in Arabidopsis and crops. Phylogenetic analysis showed that CmNAC1 and AtATAF1 gene are in the same clade (Supplementary Figures S2, S7).
The eGFP-CmNAC1 fusion construct and the eGFP control in pH7LIC5.0-N-eGFP vector driven by CaMV35S promoter were transiently expressed in tobacco epidermal cells and visualized under a laser scanning confocal microscope to determine the subcellular localization of CmNAC1. The eGFP fluorescence signal was widely observed from the cytoplasm to nucleus, whereas the eGFP-CmNAC1 fusion protein fluorescence signal was mainly detected in the nucleus as confirmed by DAPI staining (Figure 2). Thus, above results demonstrated that CmNAC1 is a nuclear protein.
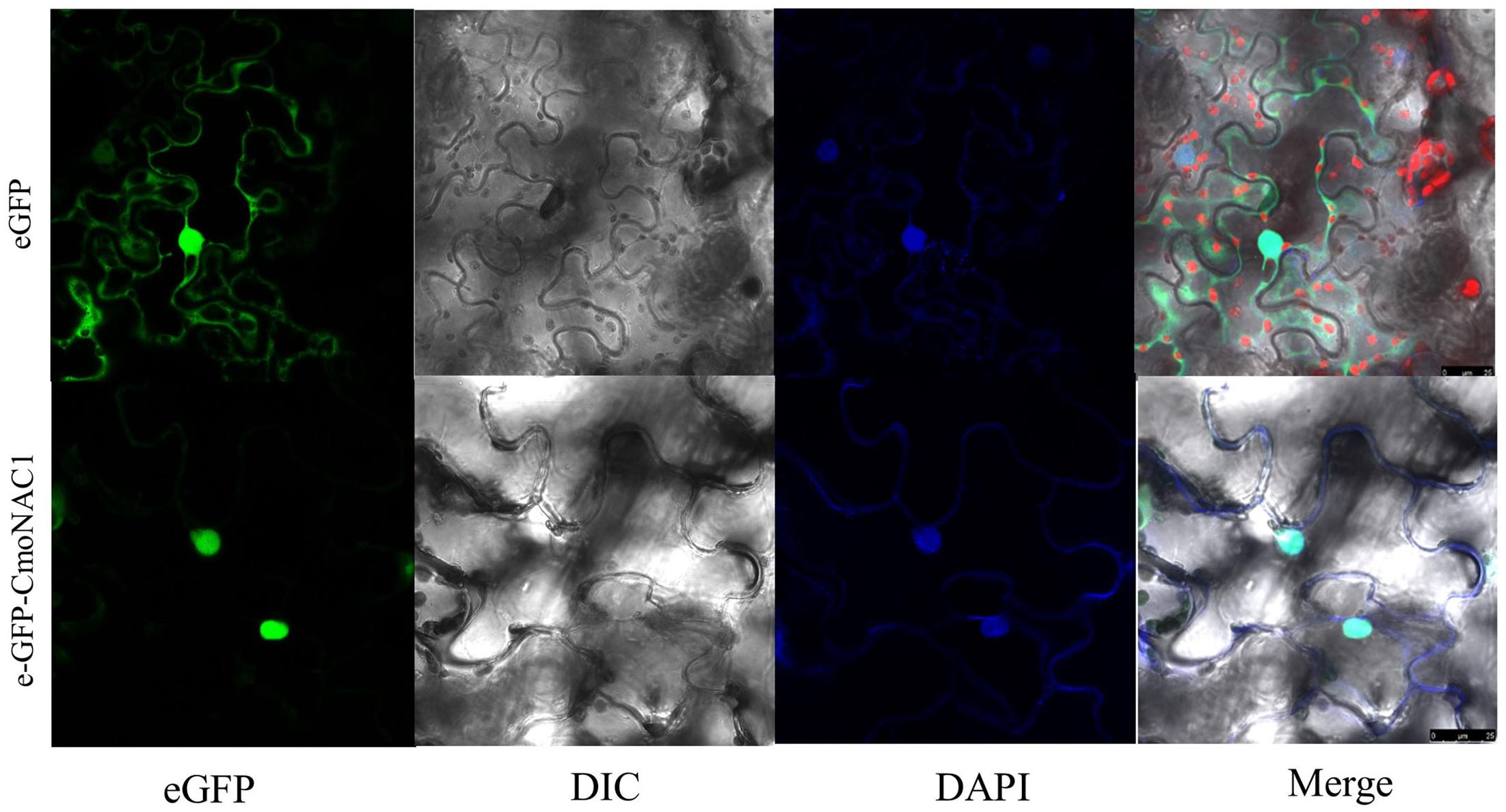
FIGURE 2. Nuclear localization of CmNAC1. e-GFP and e-GFP-CmNAC1 fusion proteins were transiently expressed in tobacco leaves under control of the CaMV 35S promoter and observed under a laser scanning confocal microscope. e-GFP images, DAPI stained images, differential interference contrast images (DIC), and merged images were taken.
The entire coding region, N-terminal and C-terminal domain coding sequence were inserted into the pGBDKT7 vector, which contains the GAL4 DNA-binding domain, to investigate the transcriptional activity of CmNAC1 protein (Figure 3A). These constructs and empty vector pGBDKT7 (negative control) were transformed into the yeast strain Y2HGold. The transactivation results show that all transformed yeast cells grew well on SD/-Trp medium. The yeast strain containing the full-length CmNAC1 (pGBDKT7-CmNAC1-A) and the C-terminus of CmNAC1 (pGBDKT7-CmNAC1-C) could grow well on the selected medium SD/-Trp/-His/-Ade, whereas the cells with the N-terminus of CmNAC1 (pGBDKT7-CmNAC1-N) and pGBDKT7 empty vector could not grow normally. Furthermore, the yeast cells that grew well on the SD/-Trp/-His-x-gal medium appeared blue in the presence of α-galactosidase, indicating the activation of the reporter gene LacZ (Figure 3B). These results indicate that CmNAC1 is a transcriptional activator, and its transactivation domain localizes in the C-terminus.
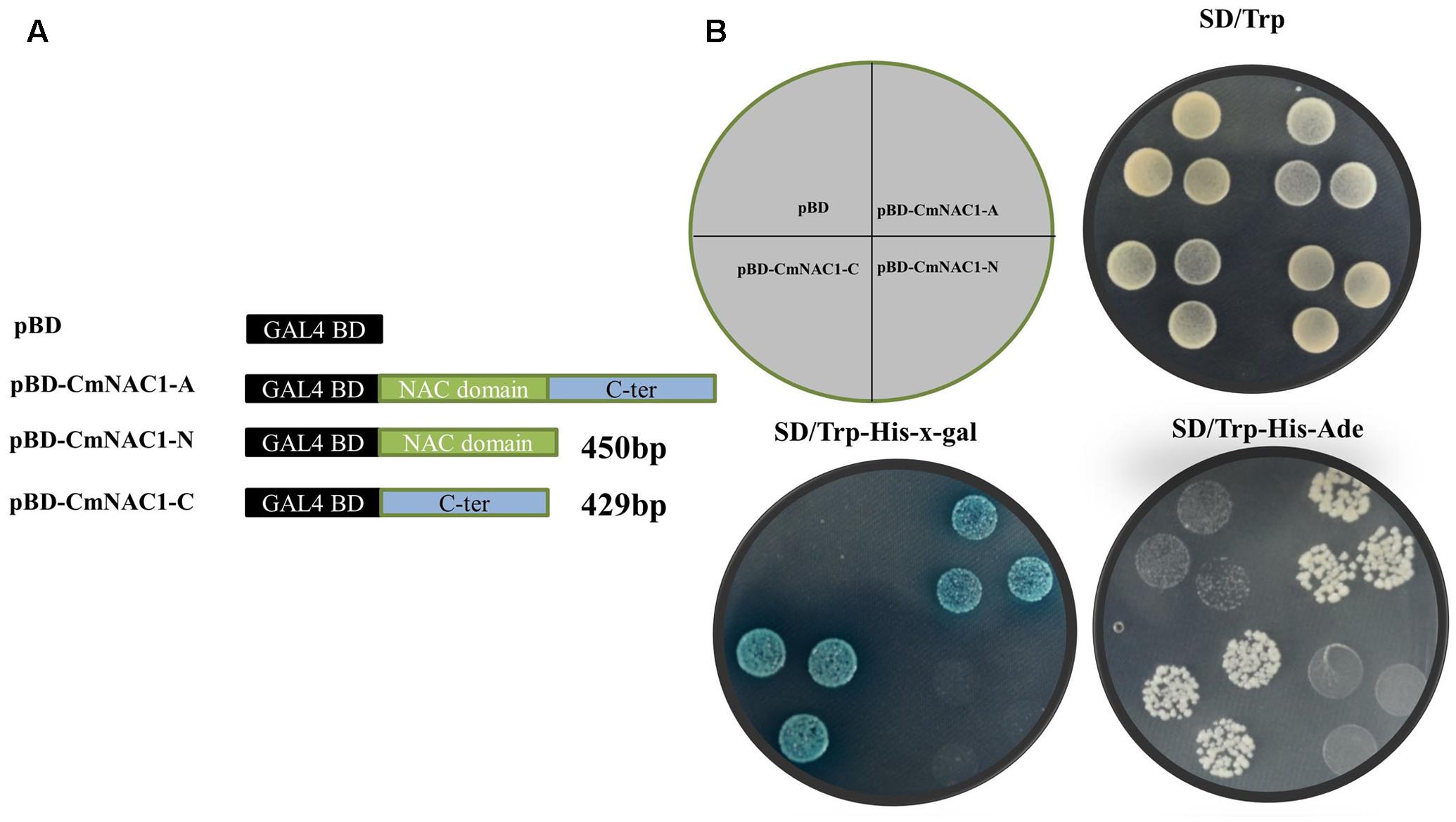
FIGURE 3. Transactivation assay of CmNAC1 in yeast cell Y2HGold. (A) Full-length protein (CmNAC1-A), N-terminal fragment (CmNAC1-N) and C-terminal fragment (CmNAC1-C) were fused with GAL4 DNA binding domain and expressed in yeast strain Y2HGold. The pGBDKT7 vector was used as a negative control. (B) Transformed yeasts were dripped on the SD/-Trp, SD/-Trp-His-Ade, and SD/-Trp-His-x-gal after being cultured for 3 days in the growth chamber.
Ectopic Expression of CmNAC1 Changes the Phenotype of Arabidopsis
When grown in the substrate, the CmNAC1-EE plants showed two different phenotypes. One was similar to WT, and the other was dwarf and light green in color (Supplementary Figure S3B). Eight transgenic positive lines with sufficient seeds and stable phenotype were selected for further gene expression experiment. RT-PCR results show that three EE lines had the same phenotype that is different from that of WT, and PCR analysis using vector specific primer indicated that all transgenic lines are positive (Supplementary Figure S3A). Therefore, CmNAC1-EE lines EE19 and EE26 with high CmNAC1 expression levels were selected for further analysis. EE19 and EE26 lines showed sterility and dwarfism phenotype with light green and round-shaped leaves (Figures 4B,C). These plants also showed accelerated chlorophyll degradation during senescence (shown as white color) as compared with wild type plants with red senescence leaves (Figure 4A). When grown in the 1/2 MS dish, both EE line seedlings showed downward cotyledon and light color (Figures 4D,E). Also, both EE lines had larger roots with larger total root surface area, longer total root length, and more root tips than the WT plants (Figure 5). These findings suggest that CmNAC1 is an important regulator of plant phenotype development.

FIGURE 4. Phenotypic analysis of CmNAC1-EE lines. (A) Seven-week-old WT and CmNAC1-EE plants were grown in substrate under long-day (LD) conditions. (B) Five-week old WT and CmNAC1-EE plants grown in substrate under LD conditions. The leaves of CmNAC1-EE lines were light green. (C) The plant height of WT and CmNAC1-EE plants grown under LD conditions after 6 weeks. (D) Transgenic plants grown in the 1/2 MS medium for 8 days. (E) Transgenic plants grown in the 1/2 MS medium for 16 days. EE lines show curly cotyledons and leaves.
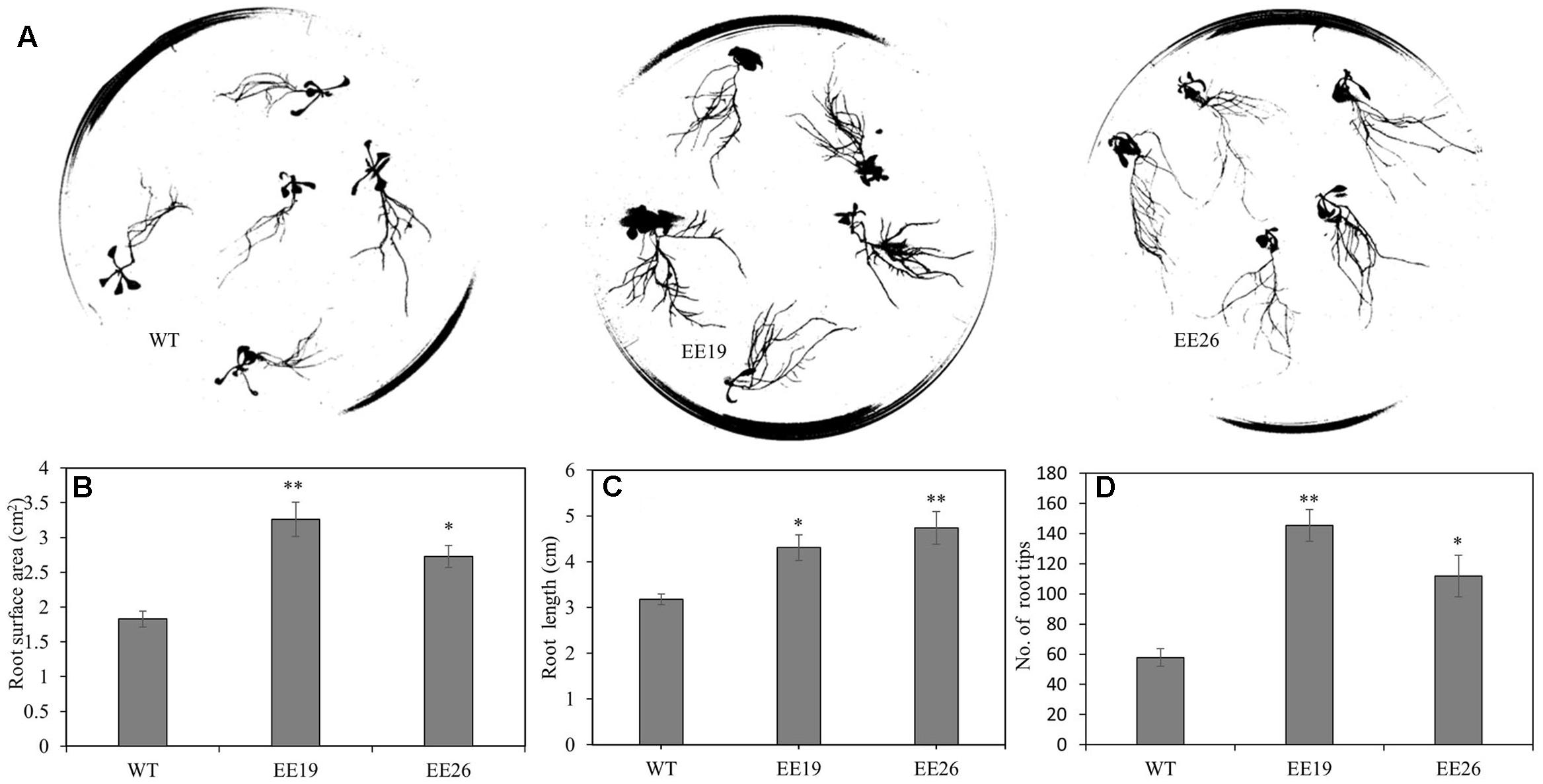
FIGURE 5. CmNAC1-EE lines have enhanced root growth. (A) Root scanning picture of WT, EE19 and EE26 lines. (B) Quantification of total root surface area. (C) Quantification of total root length. (D) Quantification of total root tips. Three independent experiments were performed. Asterisks indicate statistically significant differences compared with WT (∗P < 0.05; ∗∗P < 0.01).
Ectopic Expression of CmNAC1 Improves Drought Tolerance and Increases ABA Content in Arabidopsis
The 4-week-old transgenic lines EE19, EE26 and WT Arabidopsis seedlings were transferred to water-saturated substrate to determine the function of CmNAC1 to drought stress tolerance in plants. Water was suspended to gradually reduce water availability in the substrate. After 2 weeks of treatment, most WT plants wilted because of the extreme water deficit. By contrast, the CmNAC1-EE Arabidopsis plants showed little wilting and a slight leaf senescence phenotype. After re-watering, most of the CmNAC1-EE plants remained vigorous and survived, whereas only few of the WT plants recovered (Figure 6A). Furthermore, the survival ratio of the two CmNAC1 transgenic lines (100%) was higher than those of the WT plants (19.5%) under drought stress (Figure 6B). The CmNAC1- EE plants showed lower water loss rate and better water retaining phenotype than WT during the 3 h dehydration stress (Figures 6C,D). The EE plants also had significantly high expression levels of key ABA biosynthetic gene AtNCED3 in accordance to CmNAC1 expression levels and ABA content (Figures 6E,F,G). These results suggest that CmNAC1-EE leads to high ABA content and efficient water use in Arabidopsis.
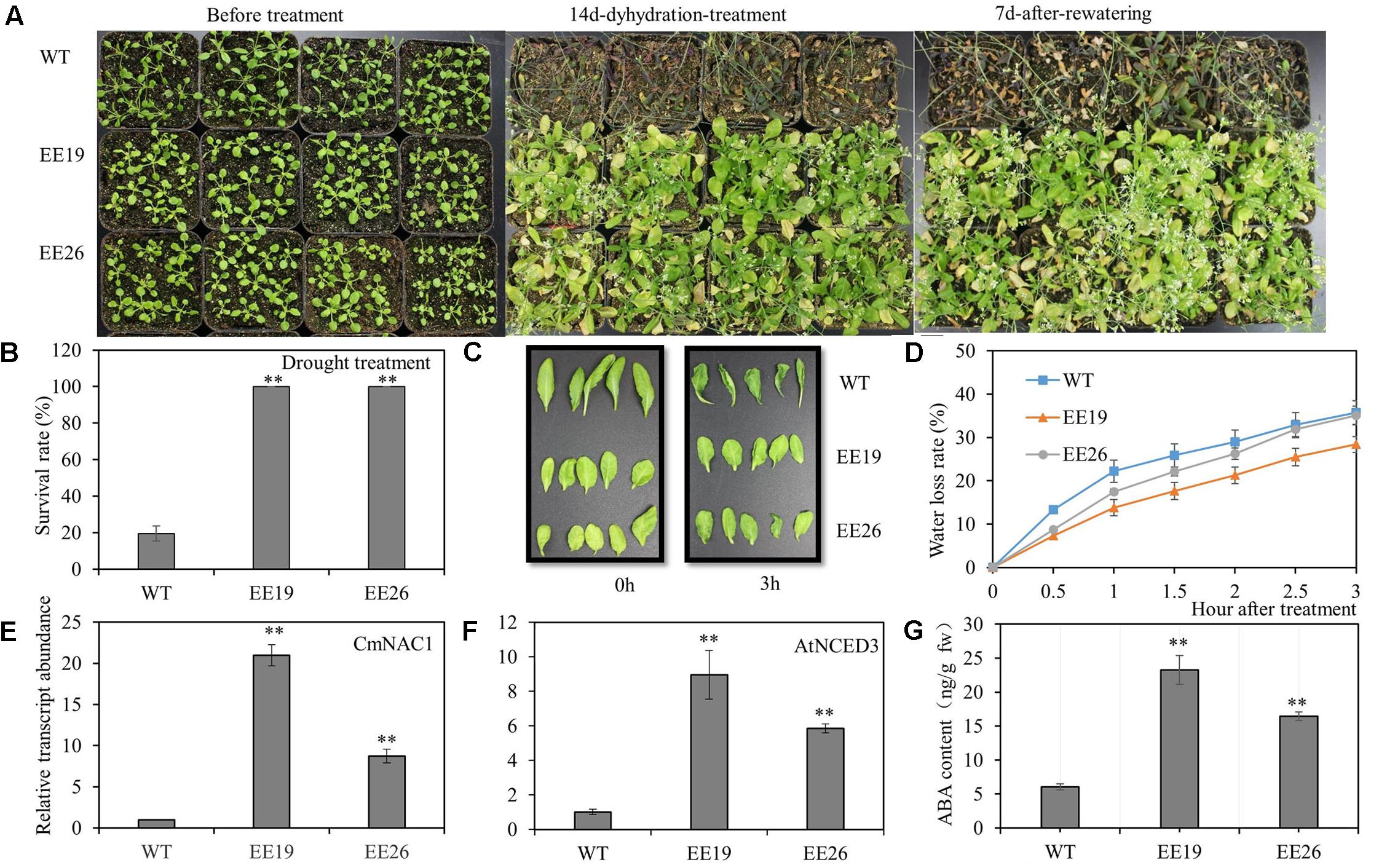
FIGURE 6. CmNAC1-EE lines have enhanced drought stress tolerance. Four-week-old seedlings of WT and EE plants were grown in substrate for treatment. (A) Phenotypes of WT and EE plants during drought stress. (B) Quantitative analysis of survival rate after drought stress. (C) Fully expanded leaves of Arabidopsis plants (5 weeks old) were excised and exposed to dehydration stress for 3 h on dry filter paper at room temperature. Pictures were taken at 0 and 3 h after treatment. (D) Water loss assay from detached leaves of 5-week-old seedlings. (E) CmNAC1 expression levels in transgenic Arabidopsis EE19, EE26 lines and WT plants. (F) Key ABA biosynthesis gene AtNCED3 expression levels in transgenic Arabidopsis EE19, EE26 lines and WT plants. (G) Concentration of ABA in transgenic Arabidopsis EE19, EE26 lines and WT plants. Three independent experiments were performed. Asterisks indicate statistically significant differences compared with WT plants (∗∗P < 0.01).
CmNAC1 Ectopic Expressing Plants Are Tolerant to Salt and Cold stress
Salt tolerance of CmNAC1-EE plants were examined. Four-week-old seedlings cultivated in substrate were irrigated with 250 mM NaCl solution. After 30 days of salt treatment, CmNAC1-EE lines showed better growth status than the WT plants, thereby indicating serious chlorosis (Figure 7A). As shown in Figure 7C, the Fv/Fm value decreased significantly starting at the 10th day of treatment, this indicated that the leaves were damaged starting at the 10th day of salt treatment, and the EE lines consistently performed better than WT Arabidopsis. Moreover, the Fv/Fm images also revealed that salt damage at the 7th day of treatment was not visible. However, at the 14th day of treatment, the WT plants were significantly damaged compared with the EE plants (Figure 7B). Consistent with the Fv/Fm value changes, the transgenic plants had lighter green leaves (lower SPAD value) without salinity. However, after 2 weeks of salt stress treatment, their SPAD value was higher than that of the WT plants (Figure 7E). All EE lines also had high survival rate (Figure 7D). In conclusion, EE lines are more tolerant to salinity stress.
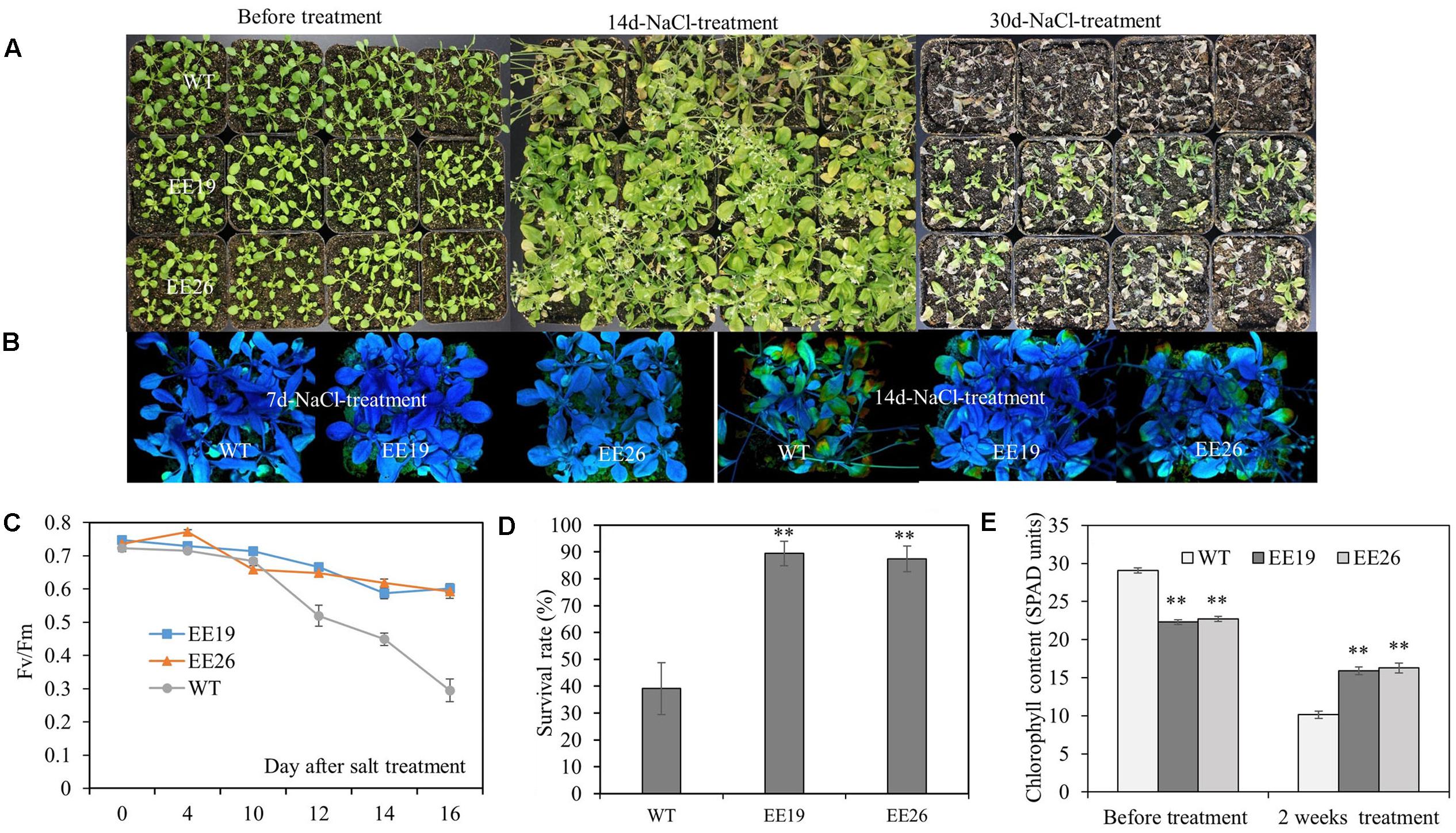
FIGURE 7. CmNAC1-EE lines have enhanced salt stress tolerance. Four-week-old seedlings of WT and EE plants were grown in substrate for NaCl (250 mM) treatment. (A) Phenotypes of WT and EE plants before salt stress and after 14 and 30 days of salt treatment. (B) Fv/Fm images during seven and 14 days after salt treatment. (C) Fv/Fm value was measured 0, 4, 10, 12, 14, and 16 days after salt treatment. (D) Survival rate after salt stress. (E) Chlorophyll content of CmNAC1-EE lines and WT plants was measured by SPAD before and after 14 days of salt treatment. Three independent experiments were performed. Asterisks indicate statistically significant differences compared with WT plants (∗P < 0.05; ∗∗P < 0.01).
Four-week-old potted Arabidopsis seedlings were subjected to freezing at -10°C for 1 h to characterize the function of CmNAC1 in cold tolerance. After that, incubate the plants in a cold growth chamber (4°C) for 3 h, followed by transferring the plants to normal growth conditions (23 ± 1°C) for recovery. EE lines had less cold injury and better recovery than the WT plants after cold treatment (Supplementary Figure S4A). Moreover, the survival ratio of the two transgenic lines were significantly higher than WT plants (Supplementary Figure S4B). During cold treatment, the two CmNAC1 transgenic lines also had lower electrolyte leakage than WT plants (Supplementary Figure S4C). These results indicate that EE of CmNAC1 also improves the tolerance of plants to low-temperature stress.
CmNAC1 Ectopic Expressing Arabidopsis Lines Are ABA Hypersensitive
Abscisic acid is a critical stress regulator in plants, and EE lines had high ABA content. Therefore, the ABA sensitivity of EE plants was assessed. ABA significantly inhibited Arabidopsis germination when the seeds were cultivated on 1/2 MS medium supplemented with 0 and 2 μM ABA (Figure 8A). However, the emergence ratio of WT seeds was higher than that of EE lines seeds in 1/2 MS medium containing 2 μM ABA (Figure 8C). The seedlings of 7-day-old WT and EE lines were transplanted to vertical 1/2 MS medium containing 0 and 10 μM ABA for ABA sensitivity analysis. Both EE lines showed yellow and senescent shoot phenotypes compared with the control (Figure 8B). Additionally, when the seedlings were cultivated in 1/2 MS medium supplemented with 2 μM ABA for 2 weeks, both transgenic lines showed yellow seedling cotyledon leaves compared with the light green seedling cotyledon leaves of WT plants (Figure 8D). Therefore, we suggested that CmNAC1 transgenic line shoots are more hypersensitive to ABA than WT plants.
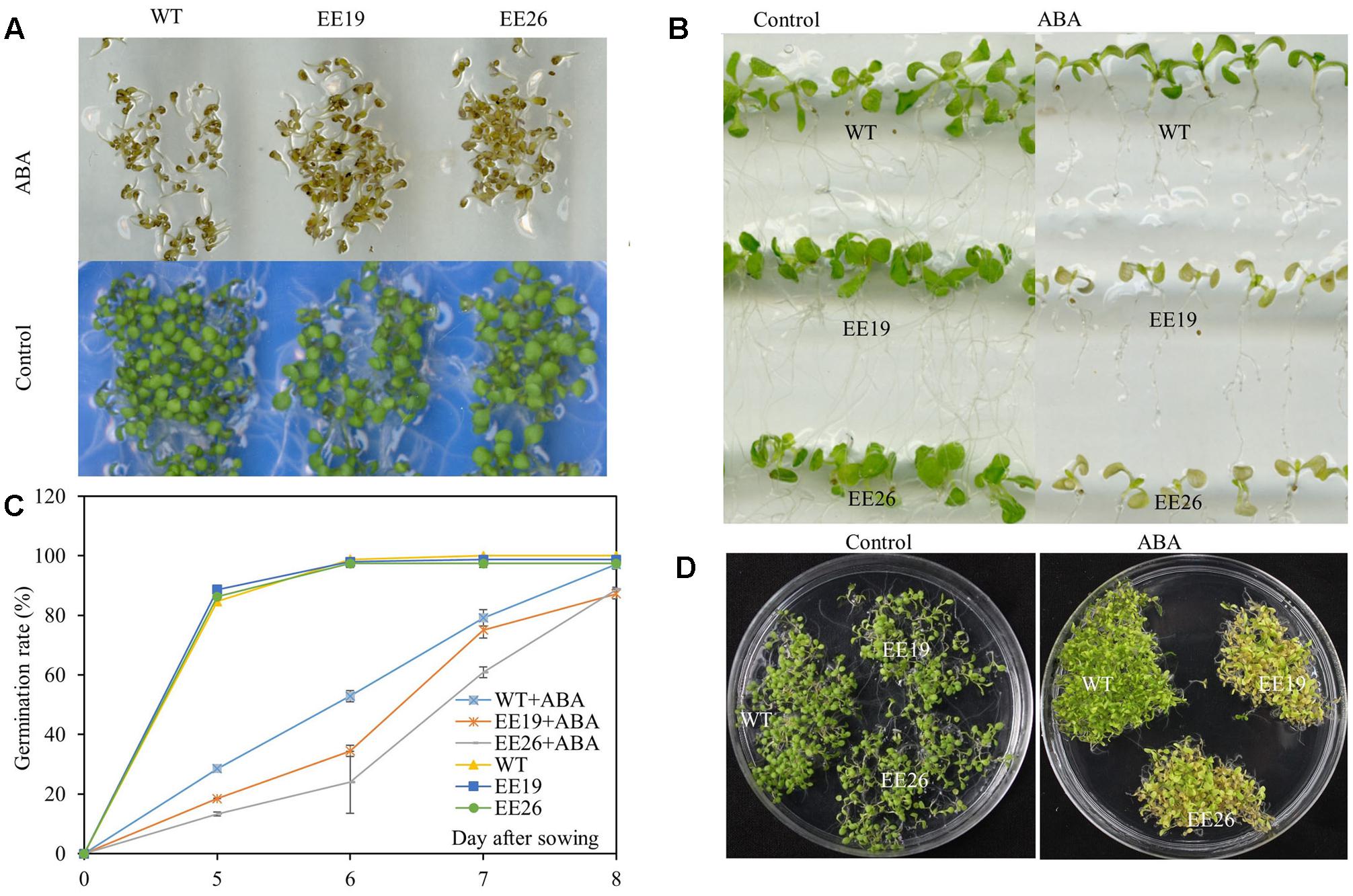
FIGURE 8. Hypersensitivity of CmNAC1-EE lines to ABA. Phenotypes of WT and EE plants grown in 1/2 MS medium supplemented with ABA for sensitivity quantitation. (A) Pictures of germination phenotype with 0 and 2 μM ABA were taken after 8 days of treatment. (B) Phenotype of WT and EE lines in vertical 1/2 MS medium with 0 and 10 μM ABA. (C) Germination rate were scored at five, six, seven, and 8 days after planting in the 1/2 MS plates supplemented with 0 and 2 μM ABA. (D) Phenotype of WT and EE lines grown in 1/2 MS medium with 0 and 2 μM ABA for 2 weeks and EE lines showing yellow phenotype compared with WT green phenotype.
Discussion
CmNAC1 Is a Nucleus Localized Stress-Responsive NAC Transcriptional Activator
NAC proteins contain highly conserved DNA-binding domain; but, they play different roles in various stress signaling pathway and several developmental programs across different plants (Puranik et al., 2012). However, the function of NAC transcriptional factors in pumpkin is still unknown. In this study 76 NAC domain proteins were identified from the transcriptome data (Supplementary Figure S6), CmNAC1 is the greatest induced NAC genes by salinity. CmNAC1 is also the first identified stress-related NAC transcription factor in pumpkin containing diverse C-terminal activation domain and conserved N-terminal NAC domain. Amino acid sequence alignment shows that CmNAC1 has high sequence identity with AtATAF1 of stress-responsive NAC domain proteins from Arabidopsis (Supplementary Figure S1). Recently plenty of NAC domain proteins were identified to be stress-responsive and those proteins have important function in different stress signaling pathways (Hao et al., 2011; Xu et al., 2013; Yang et al., 2015; Hong et al., 2016; Zhang et al., 2016). The expression of CmNAC1 is induced by drought, salt, ABA and H2O2 (Figures 1B,C,F,G), and several stress-related cis-elements are present in promoter of CmNAC1 (Table 1), suggesting it is a stress-responsive NAC domain protein. Most of transcription factors are localized in nucleus and have transactivation activity, CmNAC1 protein is also localized in nucleus (Figure 2) and both the C-terminal and full-length CmNAC1 have high transactivation ability in yeast (Figure 3B). However previous studies reported that the conserved hydrophobic LVFY motif in N-terminal of NARD would lead to abolished transactivation (Hao et al., 2010; Wang et al., 2017). CmNAC1 also contains NARD, but we have not observed significant difference of transactivation ability between the C-terminal and full-length CmNAC1. This might be due to its high transcriptional activation motif in C-terminal, but the mechanisms need to be further investigated. These results suggest that CmNAC1 is a nucleus localized stress-responsive transcriptional activator.
CmNAC1 Is Involved in Developmental Processes
Growth retardation with a dwarf phenotype has been observed in Arabidopsis EE of NTM1, AtATAF1, OsNAC2, MlNAC5, and ANAC036 (Kim et al., 2006; Wu et al., 2009; Kato et al., 2010; Chen et al., 2015; Yang et al., 2015). CmNAC1-EE also leads to a dwarf phenotype (Figure 4C), indicating that CmNAC1 regulates shoot development. Plants can change their phenotypes to alleviate the harmful impacts of environmental stresses (Pierik and Testerink, 2014). AtATAF1 are reported to play an important role in promoting senescence and chlorophyll degradation (Garapati et al., 2015). CmNAC1-EE also showed light color and accelerated chlorophyll degradation during senescence with white leaves (Figure 4A). The results suggested the function of CmNAC1 are conserved in some aspects. Interestingly, we also found different root phenotype in EE lines. Several studies have proved that rice, Arabidopsis, and tobacco plants with optimized root systems have good tolerance to drought and limited nutrient supply (Yu et al., 2008; Werner et al., 2010; Uga et al., 2013). These findings are in agreement with our results that EE plants have large roots and good stress tolerance (Figure 5). NAC1 and AtNAC2 from Arabidopsis can promote later root development (Xie et al., 2000; He et al., 2005), and NAC domain proteins function as homo- or heterodimers (Olsen et al., 2005). However, the mechanisms of CmNAC1 in regulating root development and its cooperation with other NAC transcriptional factors related with auxin and root architecture development are still unknown.
CmNAC1 Improves Tolerance to Multiple Abiotic Stress
Recently, numerous NAC domain proteins from different crops were reported to play a positive role in stress responsiveness and regulation of abiotic stress tolerance (Hong et al., 2016; Huang L. et al., 2016; Zhang et al., 2016; Wang et al., 2017). The observations that CmNAC1 significantly improves the tolerance of Arabidopsis to salt, drought and cold stress and EE lines shows better growth performance and higher survival ratio under stress conditions (Figures 6A,B, 7A,D and Supplementary Figures S4A,B) indicates that CmNAC1 also functions as a positive stress-responsive transcription factor of salt, drought and cold stress tolerance in pumpkin. In the represent study, several physiological changes in transgenic Arabidopsis seem involved in the mechanisms of abiotic stress tolerance. First, CmNAC1 significantly improves the tolerance of Arabidopsis to salt stress, and reducing the accumulation of Na+ in leaves is one of plant salinity tolerance mechanisms (Roy et al., 2014). The leaves of CmNAC1-EE plants got less damaged when grown under salt stress (Figures 7B,E), it is might be due to less Na+ content in EE leaves. Secondly, as one of the most important phytohormones, ABA regulates plant development, closing of stomata, and stress tolerance (Raghavendra et al., 2010; Yoshida et al., 2014). It is observed that CmNAC1-EE Arabidopsis lead to higher ABA content and ABA hypersensitive (Figures 6G, 8), this could lead to increased closing of stomata under drought, and then control water loss ratio and enhance drought tolerance (Figures 6A,C,D). Thus increased ABA content and sensitivity in EE plants may partially account for the enhanced drought tolerance, which is consistent with previous findings that overexpression of MlNAC5, AtATAF1, ONAC022, TaNAC47 in plants led toward improved drought stress (Wu et al., 2009; Yang et al., 2015; Hong et al., 2016; Zhang et al., 2016). Finally, cold-responsive genes in plants were also partially regulated by ABA, ABA-mimicking ligand application could improve cold stress tolerance (Cheng Z.M. et al., 2016), and we also found that CmNAC1 could improve cold tolerance (Supplementary Figures S4A,B). Our result are in accordance with the previous studies that ectopic expression of MlNAC5, GmNAC20 and TaNAC47 conferred ABA hypersensitivity and enhanced cold tolerance in Arabidopsis (Hao et al., 2011; Yang et al., 2015; Zhang et al., 2016), indicating that CmNAC1 may function as a positive regulator of abiotic stress tolerance in ABA-dependent pathway.
CmNAC1 Plays a Critical Role in ABA-Dependent Signaling
Stress induced ABA biosynthesis in leaves was mainly controlled by AtNCED3 (Tan et al., 2003). AtATAF1 is a rapid stress response transcription factor that controls ABA biosynthesis by binding to AtNCED3 promoters and enhancing its expression (Jensen et al., 2013; Garapati et al., 2015). CmNAC1 has a close relationship with the AtATAF1 in Arabidopsis (Supplementary Figure S1), CmNAC1-EE lines have high AtNCED3 expression levels and ABA content (Figures 6F,G). Therefore we speculate that CmNAC1 could bind to the promoter of AtNCED3 to regulate its expression, and this might be a conserved pathway in regulating ABA synthesis in plants. In addition, the expression of CmNAC1 is significantly induced by dehydration and ABA (Figures 1F,G). These results indicate that CmNAC1 play dual role in ABA signaling responsiveness and ABA synthesis regulation. ABREs are recognized by a group of transcription factors, ABRE-binding factors (ABRE/ABFs) (Yoshida et al., 2014). In Arabidopsis AREB1, AREB2, and ABF3 collectively regulate ABRE-dependent ABA signaling involved in drought stress tolerance (Yoshida et al., 2010). Several conserved ABREs were enriched in almost the exact position from promoter of Arabidopsis AtATAF1, cucumber CsATAF1 (Supplementary Table S1), and rice OsNAC6 (Nakashima et al., 2007). Similarly five ABREs are found in the promoter of CmNAC1 (Table 1); suggesting that ATAF1 homologous genes from different plant species have pivotal functions in ABRE-dependent ABA signaling. Therefore, CmNAC1 might be a downstream gene of AREB/ABFs transcription factors and cooperatively regulate the ABRE-dependent gene expression. In summary, we can speculate that once the plants are exposed to stress, ABA quickly promotes CmNAC1 expression through the AREB/ABF pathway. CmNAC1 expression subsequently produces additional NCED3 transcripts to finally obtain increased ABA in a positive feedback loop.
Conclusion
The function of CmNAC1 as a new stress-response transcription factor in pumpkin was evaluated in Arabidopsis plants. CmNAC1 plays a positive role in promoting root growth and adapting to salt, cold, and drought stress. The EE plants also had significantly high ABA level, indicating that the ABA signaling pathway is involved in CmNAC1-mediated abiotic stress tolerance. CmNAC1 is a functional transcription factor in pumpkin and a candidate gene for stress tolerance regulation and genetic modification of crops.
Author Contributions
ZB, QK, YH, and FC designed the research. HC, LW, MLN, JS, and JX did experiment and analyzed the data. HC and MAN wrote the manuscript. The manuscript is revised by HC and ZB. All authors read and approved the manuscript.
Funding
This research work was supported by National Natural Science Foundation of China (31172000, 31372110), China Agriculture Research System (CARS-25), and the International Science and Technology Cooperation Program of China (2015DFG32310).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02052/full#supplementary-material
FIGURE S1 | Multiple alignment of CmNAC1 amino acid sequences and other NAC domain proteins from selected plant species. Alignment was performed using DNAMAN program. Four conserved NAC subdomains were indicated: GhNAC2 from Gossypium hirsutum, GmNAC2 from Glycine max, and CUC1 and AtATAF1 from Arabidopsis thaliana.
FIGURE S2 | Phylogenetic relationship of CmNAC1 with Arabidopsis SNACs gene. CmNAC1 from pumpkin and ATAF1, ATAF2, RD26, ANAC102, ANAC055, SHYG, AtNAP, ORE1, ANAC056, ANAC016, ANAC018, ANAC019, ANAC025, and ANAC053 from Arabidopsis thaliana.
FIGURE S3 | Expression of CmNAC1 in transgenic Arabidopsis lines. (A) PCR identification and RT-PCR results of CmNAC1 expression levels in different transgenic lines. AtActin was used as the internal control gene. (B) Phenotypes of T0 generation plants grown for 5 weeks in substrate.
FIGURE S4 | Four-week-old seedlings of WT, EE19 and EE26 plants grown in substrate for cold treatment. (A) Phenotypes of EE lines and WT plants before and after cold stress treatment. (B) Survival rate quantitation. (C) Electrolyte leakage was measured during cold treatment. Asterisks indicate statistically significant differences compared with control (∗P < 0.05; ∗∗P < 0.01). Three independent experiments were performed.
FIGURE S5 |CmNAC1 promoter sequence of pumpkin. The 1069 bp upstream from the start codon of CmNAC1 was cloned from pumpkin using GenomeWalker Universal Kits.
FIGURE S6 | Multiple alignment of NAC domain proteins from pumpkin transcriptome. Alignment was performed using DNAMAN program. Seventy six NAC domain proteins were analyzed.
FIGURE S7 | Phylogenetic relationship of CmNAC1 with SNACs proteins from different crops. SNAC1, SNAC3, OSNAC48 from Oryza Sativa, SlNAC4 from Solanum lycopersicum, GhNAC2 from Gossypium hirsutum, GmNAC2 from Glycine max, and AtATAF1 from Arabidopsis thaliana.
Acknowledgments
The authors are thankful to Prof. Junhong Zhang and Prof. Botao Song for providing plant expression vector Phellsgate8.
Abbreviations
ABA, abscisic acid; ABRE, ABA response elements; DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast images; GA, gibberellin; NARD, NAC repression domain; SNACs, stress-responsive NAC transcription factors.
Footnotes
References
Balazadeh, S., Siddiqui, H., Allu, A. D., Matallana-Ramirez, L. P., Caldana, C., Mehrnia, M., et al. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62, 250–264. doi: 10.1111/j.1365-313X.2010.04151.x
Chen, X., Lu, S. C., Wang, Y. F., Zhang, X., Lv, B., Luo, L. Q., et al. (2015). OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J. 82, 302–314. doi: 10.1111/tpj.12819
Cheng, F., Lu, J., Gao, M., Shi, K., Kong, Q., Huang, Y., et al. (2016). Redox signaling and CBF-responsive pathway are involved in salicylic acid-improved photosynthesis and growth under chilling stress in watermelon. Front. Plant Sci. 7:519. doi: 10.3389/fpls.2016.01519
Cheng, Z. M., Jin, R., Cao, M. J., Liu, X. D., and Chan, Z. L. (2016). Exogenous application of ABA mimic 1 (AM1) improves cold stress tolerance in bermudagrass (Cynodon dactylon). Plant Cell Tissue Organ Cult. 125, 231–240. doi: 10.1007/s11240-016-0941-5
Clercq, I. D., Vermeirssen, V., Aken, O. V., Vandepoele, K., Murcha, M. W., Law, S. R., et al. (2013). The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25, 3472–3490. doi: 10.1105/tpc.113.117168
Clough, S., and Bent, A. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Garapati, P., Xue, G. P., Munne-Bosch, S., and Balazadeh, S. (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168, 1122–1139. doi: 10.1104/pp.15.00567
Hao, Y. J., Song, Q. X., Chen, H. W., Zou, H. F., Wei, W., Kang, X. S., et al. (2010). Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232, 1033–1043. doi: 10.1007/s00425-010-1238-2
Hao, Y. J., Wei, W., Song, Q. X., Chen, H. W., Zhang, Y. Q., Wang, F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68, 302–313. doi: 10.1111/j.1365-313X.2011.04687.x
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., and Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. doi: 10.1111/j.1365-313X.2005.02575.x
Hong, S. W., Lee, U., and Vierling, E. (2003). Arabidopsis hot mutants define multiple functions required for acclimation to high temperatures. Plant Physiol. 132, 757–767. doi: 10.1104/pp.102.017145
Hong, Y., Zhang, H., Huang, L., Li, D., and Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7:4. doi: 10.3389/fpls.2016.00004
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Huang, L., Hong, Y. B., Zhang, H. J., Li, D. Y., and Song, F. M. (2016). Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 16:203. doi: 10.1186/s12870-016-0897-y
Huang, Y., Bie, Z. L., He, S. P., Hua, B., Zhen, A., and Liu, Z. X. (2010). Improving cucumber tolerance to major nutrients induced salinity by grafting onto Cucurbita ficifolia. Environ. Exp. Bot. 69, 32–38. doi: 10.1016/j.envexpbot.2010.02.002
Huang, Y., Jiao, Y., Nawaz, M. A., Chen, C., Liu, L., Lu, Z., et al. (2016). Improving magnesium uptake, photosynthesis and antioxidant enzyme activities of watermelon by grafting onto pumpkin rootstock under low magnesium. Plant Soil 409, 229–246. doi: 10.1007/s11104-016-2965-3
Jensen, M. K., Lindemose, S., de Masi, F., Reimer, J. J., Nielsen, M., Perera, V., et al. (2013). ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio. 3, 321–327. doi: 10.1016/j.fob.2013.07.006
Kato, H., Motomura, T., Komeda, Y., Saito, T., and Kato, A. (2010). Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 167, 571–577. doi: 10.1016/j.jplph.2009.11.004
Kim, H. S., Park, B. O., Yoo, J. H., Jung, M. S., Lee, S. M., Han, H. J., et al. (2007). Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J. Biol. Chem. 282, 36292–36302. doi: 10.1074/jbc.M705217200
Kim, Y.-S., Kim, S.-G., Park, J.-E., Park, H.-Y., Lim, M.-H., Chua, N.-H., et al. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144. doi: 10.1105/tpc.106.043018
Liu, H., Li, X., Xiao, J., and Wang, S. (2012). A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8:2. doi: 10.1186/1746-4811-8-2
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-DELTADELTACT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. doi: 10.1016/j.tplants.2005.11.002
Nakashima, K., Tran, L. S. P., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Nawaz, M. A., Imtiaz, M., Kong, Q., Cheng, F., Ahmed, W., Huang, Y., et al. (2016). Grafting: a technique to modify ion accumulation in horticultural crops. Front. Plant Sci. 7:1457. doi: 10.3389/fpls.2016.01457
Nawaz, M. A., Wang, L., Jiao, Y., Chen, C., Zhao, L., Mei, M., et al. (2017). Pumpkin rootstock improves nitrogen use efficiency of watermelon scion by enhancing nutrient uptake, cytokinin content, and expression of nitrate reductase gene. Plant Growth Regul. 82, 233–246. doi: 10.1007/s10725-017-0254-7
Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945. doi: 10.1105/tpc.106.045617
Niu, M., Junjun, X., Sun, J., Huang, Y., Kong, Q., Nawaz, M. A., et al. (2017). A shoot based Na+ tolerance mechanism observed in pumpkin-an important consideration for screening salt tolerant rootstocks. Sci. Hort. 218, 38–47. doi: 10.1016/j.scienta.2017.02.020
Obrero, A., Die, J. V., Roman, B., Gomez, P., Nadal, S., and Gonzalez-Verdejo, C. I. (2011). Selection of reference genes for gene expression studies in zucchini (Cucurbita pepo) using qPCR. J. Agric. Food Chem. 59, 5402–5411. doi: 10.1021/jf200689r
Olsen, A. N., Ernst, H. A., Lo Leggio, L., and Skriver, K. (2005). DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 169, 785–797. doi: 10.1016/j.plantsci.2005.05.035
Pan, X., Welti, R., and Wang, X. (2008). Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69, 1773–1781. doi: 10.1016/j.phytochem.2008.02.008
Pierik, R., and Testerink, C. (2014). The art of being flexible: how to escape from shade, salt, and drought. Plant Physiol. 166, 5–22. doi: 10.1104/pp.114.239160
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Qiu, K., Li, Z. P., Yang, Z., Chen, J. Y., Wu, S. X., Zhu, X. Y., et al. (2015). EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLOS Genet. 11:e1005399. doi: 10.1371/journal.pgen.1005399
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395. doi: 10.1016/j.tplants.2010.04.006
Raman, S., Greb, T., Peaucelle, A., Blein, T., Laufs, P., and Theres, K. (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55, 65–76. doi: 10.1111/j.1365-313X.2008.03483.x
Reusche, M., Thole, K., Janz, D., Truskina, J., Rindfleisch, S., Drubert, C., et al. (2012). Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. Plant Cell 24, 3823–3837. doi: 10.1105/tpc.112.103374
Roy, S. J., Negrao, S., and Tester, M. (2014). Salt resistant crop plants. Curr. Opin. Plant Biol. 26, 115–124. doi: 10.1016/j.copbio.2013.12.004
Sheludko, Y. V., Sindarovska, Y. R., Gerasymenko, I. M., Bannikova, M. A., and Kuchuk, N. V. (2007). Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression. Biotechnol. Bieng. 96, 608–614. doi: 10.1002/bit.21075
Takasaki, H., Maruyama, K., Takahashi, F., Fujita, M., Yoshida, T., Nakashima, K., et al. (2015). SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 84, 1114–1123. doi: 10.1111/tpj.13067
Tan, B. C., Joseph, L. M., Deng, W. T., Liu, L. J., Li, Q. B., Cline, K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35, 44–56. doi: 10.1046/j.1365-313X.2003.01786.x
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Wang, L. Q., Li, Z., Lu, M. Z., and Wang, Y. C. (2017). ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic tamarix and Arabidopsis. Front. Plant Sci. 8:635. doi: 10.3389/fpls.2017.00635
Werner, T., Nehnevajova, E., Kollmer, I., Novak, O., Strnad, M., Kramer, U., et al. (2010). Root-Specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22, 3905–3920. doi: 10.1105/tpc.109.072694
Wu, A. H., Allu, A. D., Garapati, P., Siddiqui, H., Dortay, H., Zanor, M. I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24, 482–506. doi: 10.1105/tpc.111.090894
Wu, Y. R., Deng, Z. Y., Lai, J. B., Zhang, Y. Y., Yang, C. P., Yin, B. J., et al. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290. doi: 10.1038/cr.2009.108
Xie, J. J., Lei, B., Niu, M. L., Huang, Y., Kong, Q. S., and Bie, Z. L. (2015). High throughput sequencing of small RNAs in the two cucurbita germplasm with different sodium accumulation patterns identifies novel microRNAs involved in salt stress response. PLOS ONE 10e:0127412. doi: 10.1371/journal.pone.0127412
Xie, Q., Frugis, G., Colgan, D., and Chua, N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. doi: 10.1101/gad.852200
Xu, J. H., Zhang, M., Liu, G., Yang, X. P., and Hou, X. L. (2016). Comparative transcriptome profiling of chilling stress responsiveness in grafted watermelon seedlings. Plant Physiol. Biochem. 109, 561–570. doi: 10.1016/j.plaphy.2016.11.002.70
Xu, Z. Y., Kim, S. Y., Hyeon do, Y., Kim, D. H., Dong, T., Park, Y., et al. (2013). The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724. doi: 10.1105/tpc.113.119099
Yang, J. D., Worley, E., and Udvardi, M. (2014). A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26, 4862–4874. doi: 10.1105/tpc.114.133769
Yang, X. W., Wang, X. Y., Ji, L., Yi, Z. L., Fu, C. X., Ran, J. C., et al. (2015). Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 34, 943–958. doi: 10.1007/s00299-015-1756-2
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Yu, H., Chen, X., Hong, Y. Y., Wang, Y., Xu, P., Ke, S. D., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20, 1134–1151. doi: 10.1105/tpc.108.058263
Zhang, B., Liu, C., Wang, Y., Yao, X., Wang, F., Wu, J., et al. (2015). Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 206, 1513–1526. doi: 10.1111/nph.13335
Zhang, L. N., Zhang, L. C., Xia, C., Zhao, G. Y., Jia, J. Z., and Kong, X. Y. (2016). The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front. Plant Sci. 6:174. doi: 10.3389/fpls.2015.01174
Zhong, R. Q., Demura, T., and Ye, Z. H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. doi: 10.1105/tpc.106.047399
Keywords: pumpkin, ABA, NAC domain protein, ABRE, abiotic stress tolerance
Citation: Cao H, Wang L, Nawaz MA, Niu M, Sun J, Xie J, Kong Q, Huang Y, Cheng F and Bie Z (2017) Ectopic Expression of Pumpkin NAC Transcription Factor CmNAC1 Improves Multiple Abiotic Stress Tolerance in Arabidopsis. Front. Plant Sci. 8:2052. doi: 10.3389/fpls.2017.02052
Received: 11 August 2017; Accepted: 16 November 2017;
Published: 28 November 2017.
Edited by:
Andy Pereira, University of Arkansas, United StatesReviewed by:
Ratna Karan, University of Florida, United StatesCharu Lata, National Botanical Research Institute (CSIR), India
Copyright © 2017 Cao, Wang, Nawaz, Niu, Sun, Xie, Kong, Huang, Cheng and Bie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhilong Bie, biezhilong@hotmail.com
 Haishun Cao
Haishun Cao Li Wang
Li Wang Muhammad A. Nawaz
Muhammad A. Nawaz Mengliang Niu
Mengliang Niu Jingyu Sun
Jingyu Sun Qiusheng Kong
Qiusheng Kong Yuan Huang
Yuan Huang Fei Cheng
Fei Cheng Zhilong Bie
Zhilong Bie