- 1Department of Plant Biotechnology, Chonnam National University, Gwangju, South Korea
- 2Department of Rural and Biosystems Engineering, Agricultural Robotics and Automation Research Center, Chonnam National University, Gwangju, South Korea
- 3Korea Atomic Energy Research Institute, Daejeon, South Korea
The modulation of glucose (Glc) homeostasis and signaling is crucial for plant growth and development. Nevertheless, the molecular signaling mechanism by which a plant senses a cellular Glc level and coordinates the expression of Glc-responsive genes is still incompletely understood. Previous studies have shown that Arabidopsis thaliana plasma membrane Glc-responsive regulator (AtPGR) is a component of the Glc-responsive pathway. Here, we demonstrated that a transcription factor bHLH34 binds to 5′-GAGA-3′ element of the promoter region of AtPGR in vitro, and activates beta-glucuronidase (GUS) activity upon Glc treatment in AtPGR promoter-GUS transgenic plants. Gain- and loss-of-function analyses suggested that the bHLH34 involved in the responses to not only Glc, but also abscisic acid (ABA) and salinity. These results suggest that bHLH34 functions as a transcription factor in the Glc-mediated stress responsive pathway as well as an activator of AtPGR transcription. Furthermore, genetic experiments revealed that in Glc response, the functions of bHLH34 are different from that of a bHLH104, a homolog of bHLH34. Collectively, our findings indicate that bHLH34 is a positive regulator of Glc, and may affect ABA or salinity response, whereas bHLH104 is a negative regulator and epistatic to bHLH34 in the Glc response.
Introduction
Sugar signaling plays pivotal roles in modulating many features of germination, metabolism, growth, and development throughout the whole plant life cycle. Glucose (Glc), one of the hexose hydrolytic products of sucrose, is a major sugar signaling metabolite. Three Glc signal transduction pathways in plants have been suggested (Xiao et al., 2000). These are Arabidopsis thaliana hexokinase 1 (AtHXK1)-dependent pathway, in which photosynthetic gene expression correlates with the AtHXK1-mediated signaling function. The second one is a glycolysis-dependent pathway that requires AtHXK1 catalytic activity and regulates the expression of the pathogenesis-related (PR) genes (Xiao et al., 2000). The third pathway is involved in the regulation of a restricted number of genes such as those coding for cell wall invertase and chalcone synthase, and is independent of increased AtHXK1 activity (Xiao et al., 2000). Genetic evidence also indicates that a HXK-independent Glc-sensing and signaling mechanism involved in a G protein-coupled receptor system exists in plants (Chen and Jones, 2004).
Multiple Glc signal transduction pathways are intimately linked to developmental stages, hormones, and environmental conditions (Xiao et al., 2000; Eckardt, 2002). Glc signaling mutants have revealed a relation between Glc and abscisic acid (ABA) signaling pathways (Laby et al., 2000; Price et al., 2003). The ABA signaling transcriptional regulator ABA insensitive 4 (ABI4) represses the promoter of the ribulose-1,5-bisphosphate carboxylase (RBCS) gene in response to Glc or ABA (Acevedo-Hernández et al., 2005). Moreover, ABI4 is a regulator of mannose-induced inhibition of seedling germination (Pego et al., 1999), indicating a general role of ABI4 in hexose signaling. Several groups also proposed that the sucrose non-fermenting 1 (SNF1)-related protein kinase 1 (SnRK1) recognizes stress-associated energy or sugar limitation and promotes plant stress tolerance (Polge and Thomas, 2006). Also, SnRK1 is inactivated by Type 2C protein phosphatases, known repressors of the ABA pathway, and provided with evidences for the molecular connection between sugar and ABA signaling (Rodrigues et al., 2013).
A number of cis-regulatory elements responsible for the sugar-mediated gene modulation have been identified (Rolland et al., 2006), including G-box (Giuliano et al., 1988), W-box (Pla et al., 1993), SP8 motif (Ishiguro and Nakamura, 1994), sugar-response element (SURE) (Grierson et al., 1994), GC-box (Lu et al., 1998), osamy element (Lu et al., 2002), and the GCCT element (Chung et al., 2016). Recently, we reported that the A. thaliana Storekeeper-like 1 and 2 (AtSTKL1 and AtSTKL2) bind to the GCCT element in the AtPGR promoter region and suppress the gene activation (Chung et al., 2016). Overexpression of AtPGR modulates the induction of Glc and 2-deoxyglucose insensitivity under stress. In contrast, cotyledon greening of mutant seeds with an atpgr RNAi knockdown shows increased sensitivity to Glc and 2-deoxyglucose (Chung et al., 2011). AtSTKL homologs are capable of modulating the Glc response through the alteration of AtPGR expression. AtSTKL-overexpressing transgenic plants enhance the sensitivity to Glc in comparison with the wild type (WT), whereas atstkls antisense plants show decreased sensitivity even with the high levels of Glc during cotyledon greening, indicating that AtSTKL play a significant role in Glc or Glc-mediated signaling pathways in Arabidopsis (Chung et al., 2016).
In the present study, we demonstrated that a transcription factor bHLH34 interacts with the 5′-GAGA-3′ cis-regulatory elements in vitro and activates the transcription of AtPGR in planta. bHLH34 was shown to act as a putative signaling protein in Glc, ABA, and high-salinity responses. The bHLH34-overexpressing transgenic plants showed enhanced resistance to Glc, ABA, and high salinity, whereas a decrease in bHLH34 expression reduced Glc, ABA, and salt stress resistance at the early seedling stage. In addition, the expression changes of the stress-associated genes in bHLH34 transgenic plants were consistent with the functions suggested by the physiological phenotype under abiotic stress conditions. Taken together, these results suggest that bHLH34 potentially functions as a factor linking Glc signaling and an abiotic stress response. Furthermore, genetic analyses suggested that bHLH34 has a function different from that of its highly homologous protein, bHLH104, in the Glc response.
Materials and Methods
Production of Recombinant bHLH34 Protein
For the DNA-binding analysis of the bHLH34 protein, full-length cDNA fragments were amplified using the following primers: for bHLH34, forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTATCCATCAATCGAAGACGA-3′ and reverse 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGCAACAGGAGGAAGATTTTTGA-3′. The DNA products were introduced into the pDONR/ZEO vector (Invitrogen, Carlsbad, CA, United States) for DNA sequence analysis. The DNA fragment was then cloned into a pET300/NT-DEST vector by the Gateway system according to the manufacturer’s instruction (Invitrogen). The recombinant protein was expressed in the Escherichia coli strain BL21 (DE3) codon+ (Stratagene, La Jolla, CA, United States), and purified by ion exchange chromatography using Ni-charged His-bind resin (Novagen, Darmstadt, Germany).
The Electrophoretic Mobility Shift Assay (EMSA)
Four DNA fragments derived from the 5′-upstream region of AtPGR [see Figure 1A; P1, between nucleotide (nt) positions -999 and -875 relative to the AtPGR start codon site; P2, -874 to -735 nt; P3, -734 to -595 nt; P4, -594 to -456 nt] were synthesized by PCR [initial denaturation at 94°C for 2 min, amplification for 30 cycles (denaturation at 94°C for 15 s, annealing 58°C for 15 s, and extension at 72°C for 20 s) and final extension at 72°C for 3 min] with 8 primers (Supplementary Table S1). Six DNA binding elements (DBEs), including DBE1 (-997 to -985 nt), DBE2 (-962 to -950 nt), DBE3 (-843 to -831 nt), DBE4 (-643 to -622 nt), DBE5 (-617 to -605 nt), and DBE6 (-500 to -488 nt), and various mutated DBE1 oligonucleotides were prepared using the two complementary sequences were synthesized as shown in Supplementary Table S1. The double-stranded complementary oligonucleotides used as probes were radiolabeled with γ-32P-ATP and T4 polynucleotide kinase (New England BioLabs).
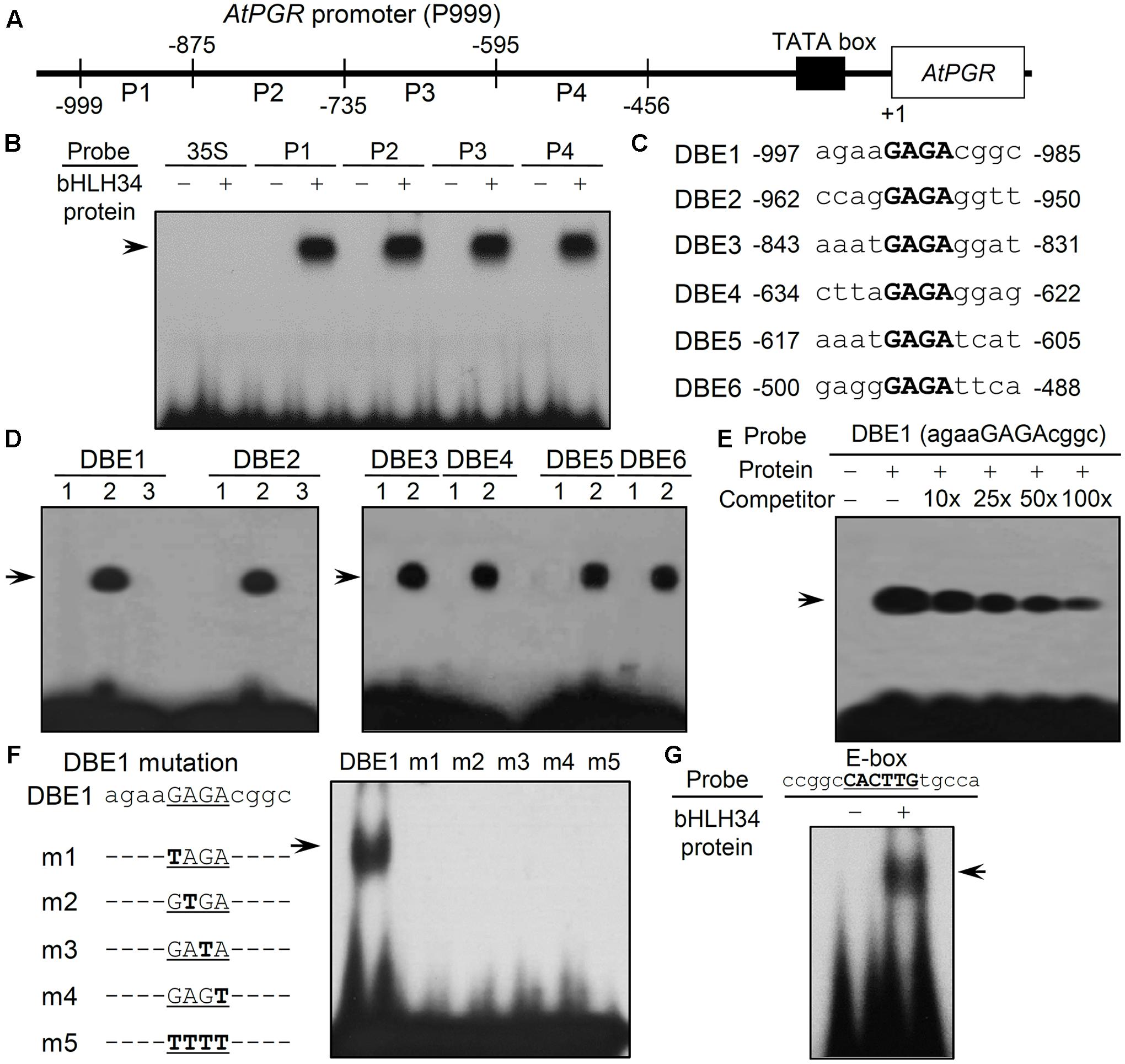
FIGURE 1. Electrophoretic mobility shift assay (EMSA) analyses of bHLH34. (A) A schematic diagram of the four (P1, P2, P3, and P4) fragments derived from the promoter 5′ upstream region (P999) of the AtPGR gene. The numbers indicate the nucleotide positions relative to the translation start site, ATG (A as +1). (B) Identification of the DNA-binding promoter fragment of the bHLH34 protein. Experiments were performed three times and similar results were obtained. The 32P-radiolabeled P1, P2, P3, and P4 DNA fragments incubated in the absence (–) or presence (+) of His-bHLH34 (arrow). 35S, 35S promoter without GAGA motif as a negative control. (C) Sequences of the six DNA binding elements (DBEs) in AtPGR promoter. GAGA motifs are shown in bold. (D) Identification of the DNA-binding promoter site in the bHLH34 protein. Experiments were carried out two times and similar results were obtained. Lane 1, the 32P-radiolabeled DBE1-6 oligonucleotides incubated in the absence of His-bHLH34; lane 2, the 32P-radiolabeled DBE1-6 oligonucleotides incubated in the presence of His-bHLH34 (arrow); and lane 3, the 32P-radiolabeled DBE1 or the 32P-radiolabeled DBE2 oligonucleotide incubated in the presence of MBP. (E) bHLH34 directly binds to the DBE1 oligonucleotide in the EMSA. The His-bHLH34 protein incubated with 32P-radiolabeled DBE1. An unlabeled DBE1 probe was used as the competitor (10-, 25-, 50- or 100-fold excess) to show binding specificity. The arrow indicates the shifted band. (F) bHLH34 specifically binds to the GAGA element. Mutations in GAGA element abolish bHLH34 binding to DBE1. A list of DBE1 and m1 to m4 point-mutated or four-nucleotide-mutated m5 probes. Mutated nucleotides are boldfaced. The binding sequence of bHLH34 was determined using various mutated sequences. The arrow indicates the position of the DBE1 probe-His-bHLH34 complex. (G) bHLH34 binds to E-box (ccggcCACTTGtgcca) in an EMSA. Lane 1, the 32P-radiolabeled- E-box (ccggcCACTTGtgcca) incubated in the absence of His-bHLH34; lane 2, the 32P-radiolabeled- E-box (ccggcCACTTGtgcca) incubated with His-bHLH34.
DNA–protein binding reactions were carried out at 25°C for 20 min in 20 μL volume containing 100 ng of each purified His-bHLH34 fusion protein, 1 μg of poly(dI-dC), 0.3 pmol of DNA fragments end-labeled with γ-32P-ATP, and DNA-binding buffer (10 mM Tris-HCl, pH 7.5, 2.5% glycerol, 50 mM KCl, 1 mM EDTA, and 1 mM DTT). The reaction mixture was subjected to 5% non-denaturing PAGE in TBE (44.5 mM Tris-hydroxymethyl aminomethane, 44.5 mM boric acid, and 1 mM EDTA) at 100 V for 2 h at 4°C. The gel with DNA–protein complexes was dried and X-ray film was exposed to this gel.
Plant Materials, Growth Conditions, and Stress Induction
Arabidopsis seedlings were grown in a growth room under intense light (110 μmol m-2 sec-1) at 22°C, 60% relative humidity, and 16 h/8 h light/dark conditions. The bHLH104 T-DNA insertion line SALK_005802 (bhlh104) was acquired from the Arabidopsis T-DNA insertion collection of the Salk Institute (Alonso et al., 2003). To select plants homozygous for the T-DNA insertion, the gene-specific primers 5′-CTCCAGAAAGCGGTGAGTTTTG-3′ and 5′-GAAGAATCACAAGTTTCTGGAG-3′ (forward and reverse, respectively) were utilized for the bhlh104 mutant. Plants yielding no PCR products with the gene-specific primers were subsequently tested for the presence of the T-DNA insertion using the gene-specific forward primer in combination with the T-DNA left border specific primer 5′-GCGTGGACCGCTGCACCT-3′.
The AtPGR promoter 999 (P999)-GUS construct was generated as described previously (Chung et al., 2016), and the overexpression of bHLH34 in the P999-GUS transgenic lines was achieved as described in Supplementary Materials and Methods.
For Glc treatment, 14-day-old Arabidopsis seedlings were submerged in sterilized water containing 6% Glc and sampled at 0, 6, 12, and 36 h under continuous light condition (110 μmol m-2 sec-1 light intensity, 24 h light condition) at 22°C, and 60% relative humidity conditions. For ABA or salt stress, 14-day-old Arabidopsis seedlings were submerged in the sterilized water containing 100 μM ABA or 150 mM NaCl and sampled at 0, 3, 6, and 12 h. For osmotic stress, 14-day-old Arabidopsis seedlings were submerged in the sterilized water containing 400 mM mannitol and sampled at 0, 6, 12, and 36 h. In each case, the collected seedlings were promptly frozen in liquid nitrogen and stored at -80°C.
Total RNA Extraction and qPCR Analysis
Total RNA was isolated from the frozen samples using the Plant RNeasy Extraction Kit (Qiagen, Valencia, CA, United States). To remove residual genomic DNA from the preparation, total RNA samples were treated with RNase-free DNase I in accordance with the manufacturer’s instructions (Qiagen). The concentration of RNA was exactly quantified via spectrophotometric measurements, and 3 μg of total RNA was separated on 1.2% formaldehyde agarose gels to verify its concentration and monitor its integrity. qPCR was performed on a Rotor-Gene 6000 quantitative PCR apparatus (Corbett Research, Mortlake, NSW, Australia), and each result was analyzed using the RG6000 1.7 software (Corbett Research). Total RNA samples were isolated from the variously treated 14-day-old seedlings using an RNeasy Plant Mini Kit (Qiagen). qPCR was conducted using the SensiMix One-Step Kit (Quantance, London, United Kingdom). Actin 1 (ACT1) served as an internal control, and the quantitative analyses were performed by the Delta Delta CT method (Livak and Schmittgen, 2001). Each sample was subjected to three independent experiments. RT-PCR was employed to measure the levels of bHLH34 expression in transgenic plants. The amount of RNA used in RT-PCR reactions was 300 ng. After 28 PCR cycles of amplification, 20 μL of each RT-PCR product was loaded onto a 1.2% (w/v) agarose gel in order to visualize the amplified DNA. The primers used for qPCR and RT-PCR reactions are shown in Supplementary Table S2.
Phenotypic Analysis and Stress Assays
For the Glc cotyledon greening test, seeds were sown on the Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with 5% or 6% Glc, and grown in a growth chamber under intense light (110 μmol m-2 sec-1) at 22°C, and 16 h/8 h light/dark conditions. Cotyledon greening was defined when a cotyledon was fully expanded and turned green. The green cotyledon was counted in triplicate (50 seeds/experiment).
For the ABA cotyledon greening or germination rate tests, seeds were sown on the MS medium supplemented with 0 or 1 μM ABA, and allowed to grow in the growth chamber under the same conditions as the Glc cotyledon greening assay. Germination was defined as an obvious protrusion of the root radicle through the seed coat. The germination rate of each line was measured after 1–8 days. Experiments were carried out in triplicate for each line (50 seeds each). Cotyledon greening of each seedling was measured at 14 days. The experiments were conducted in triplicate for each line (50 seeds each).
For the salt stress test, seeds were sown on the MS medium supplemented with 150 mM NaCl, grown in the growth chamber under the same conditions as the Glc cotyledon greening assay, and analyzed for the percentage of surviving seedlings after 3 weeks. The experiments were conducted in triplicate for each line (50 seeds each).
Measurement of Chlorophyll Content and Photochemical Efficiency (Fv/Fm)
Chlorophyll (Chl) content of leaves was determined by the spectrophotometric method (Lichtenthaler, 1987). Leaf powder was used to extract Chl pigments in 96% ethanol. Absorbance was analyzed at 648.6 and 664.2 nm. Chl fluorescence measurement was performed using a pulse-modulated fluorometer (Junior-PAM, Heinz Walz, Effeltrich, Germany). Photosystem (PS) II quantum yield (Fv/Fm) was calculated from the F0 and Fm values as described in the manufacturer’s instructions (Junior-PAM).
Accession Numbers
Arabidopsis Genome Initiative numbers for the sequences used in this study are as follows: bHLH34 (At3g23210), AtPGR (At5g19930), bHLH104 (At4g14410), bHLH105 (At5g54680), bHLH115 (At1g51070), bHLH28 (At5g46830), bHLH47 (At3g47640), bHLH11 (At4g36060), bHLH121 (At3g19860), AtHXK1 (At4g29130), RAB18 (At5g66400), RD29A (At5g52310), GIN6 (At2g40220), AtAPR2 (At1g62180), ABO3 (At1g66600), ABI1 (At4g26080), AtOZF2 (At4g29190), RD29B (At5g52300).
Results
Identification of the bHLH34 Candidate Related to AtPGR Expression
Recently, we demonstrated that the promoter sequences between nucleotide (nt) positions -999 and -456 of the AtPGR gene modulate transcription expression of the gene during Glc treatment (Chung et al., 2016). To identify transcription factors involved in the regulation of AtPGR, we performed yeast one-hybrid screening of a cDNA library of Arabidopsis (Chung et al., 2016). The yeast one-hybrid method involved in the X-galactosidase (Gal) filter assay was used to isolate transcription factor candidates that bind to P999-binding site (between positions -999 and -456 nt) in the upstream region of AtPGR (Figure 1A and Supplementary Figure S1A). Sequence analysis of the clone isolated from the P999 binding site (-999 to -456 nt) revealed that the gene encoded a basic helix-loop-helix (bHLH) family protein (At3g23210), which have been designated as a bHLH34 in Arabidopsis (Supplementary Figure S1B). bHLH34 cDNA is 963 bp long and encodes a protein of 320 amino acid residues with a calculated molecular weight of 35.6 kDa, which shows 30–56% identity and 52–67% similarity with Arabidopsis proteins bHLH115, bHLH105, bHLH104, bHLH28, bHLH47, bHLH11, and bHLH121. A phylogenetic tree representing the distance groups of sequences was built using a cluster algorithm (Supplementary Figure S1C). bHLH34 harbors a conserved single DNA-binding domain within a basic helix-loop-helix motif in its central region that is 88, 76, and 69% identical to the corresponding region of Arabidopsis bHLH104, bHLH115, and bHLH105 proteins, respectively (Supplementary Figure S1D). Recently, Li et al. (2016) demonstrated that bHLH34 and bHLH104 modulate iron homeostasis in Arabidopsis. Protein interaction assays indicate that both heterodimers and homodimers can form among bHLH34, bHLH104, and bHLH105. The bHLH105, also known as IAA-LEUCINE RESISTANT 3 (ILR3), modulate metal homeostasis, which influences IAA-conjugate hydrolysis (Rampey et al., 2006).
bHLH34 Interacts Specifically with the GAGA Cis-Element of AtPGR Promoter in Vitro
We attempted to express and purify the bHLH34 protein in E. coli to undertake electrophoretic mobility shift assay (EMSA) with the promoter in the region -999 to -456 of AtPGR. An EMSA was carried out to test whether the candidate protein bHLH34 can interact with each of the four DNA fragments (P1: -999 to -875 nt, P2: -874 to -735 nt, P3: -734 to -595 nt, and P4: -594 to -456; Figure 1A). Purified bHLH34 protein bound to all four fragments: P1, P2, P3, and P4 (Figure 1B).
To further investigate bHLH34-binding sites, we aligned the sequences of the P1-P4 fragments, and found that there were conserved cis-elements (5′-GAGA-3′) at six regions in the alignment (Figure 1C). The six DNA binding elements (DBEs), named as DBE1, DBE2, DBE3, DBE4, DBE5, or DBE6, were employed as a probe in an EMSA (Figure 1C). When we used the maltose-binding protein (MBP) as a negative control, there was no DNA–protein complex in the binding assay with DBE1 and DBE2 (Figure 1D, lane 3). Purified bHLH34 protein bound strongly to all six oligonucleotides DBE1–DBE6 (Figure 1D, lane 2). These results indicated that bHLH34 is a DNA-binding protein. To investigate that bHLH34 recognizes these oligonucleotides, excess unlabeled DBE1 was added. An addition of a molar excess of the unlabeled DBE1 oligonucleotide probe gradually diminished the shifted bands of bHLH34 protein in a dose-dependent manner (Figure 1E). These findings indicate that bHLH34 recognizes the six DBE fragments containing the consensus GAGA sequence.
To verify the specificity of binding between bHLH34 and the GAGA element, an EMSA was performed using various mutated DBE1 oligonucleotide fragments (Figure 1F). Mutations introduced in the GAGA sequence abrogated the binding of bHLH34 (Figure 1F, lanes m1, m2, m3, m4, and m5). From these results, we concluded that bHLH34 has sequence-specific binding affinity for the GAGA element in vitro.
On the other hand, consensus DNA sequence recognized by the bHLH proteins is known well as the E-box (5′-CANNTG-3′) (Robinson et al., 2000) and, single copy E-box (between positions -788 and -783 nt) exists in P999 site of the AtPGR gene. To determine whether bHLH34 protein could recognize the E-box, we conducted the EMSA assay with probe containing the E-box sequence (5′-CACTTG–3′; Supplementary Table S1), showing that bHLH34 could bind to E-box sequence (Figure 1G). This result indicate that bHLH34 binds to not only GAGA cis-element but also E-box sequence.
bHLH34 Activates the Expression of the AtPGR
To determine the transcriptional activity of AtPGR by the bHLH34, we examined the AtPGR expression level in P999-GUS transgenic plants, when the bHLH34 was constitutively expressed under the control of Cauliflower Mosaic Virus (CaMV) 35S promoter (OX1-1/P999-GUS, OX2-5/P999-GUS). Two independent lines showing high levels of bHLH34 transgene expression (Figure 2A) were selected for transcription activity analyses. As shown in Figures 2B,C, under normal conditions, OX1-1/P999-GUS and OX2-5/P999-GUS lines increased GUS expression to approximately double that of P999-GUS alone (1.73- and 1.66-fold, respectively). When 6% Glc was applied to seedlings for 12 h, GUS activity in the P999-GUS transgenic plant was elevated in the leaves and roots (Figure 2B). In addition, a significant increase in GUS activity levels (1.44- to 1.47-fold) was observed in P999-GUS transgenic lines overexpressing bHLH34 constructs in comparison with the P999-GUS transgenic plant (Figures 2B,C). Furthermore, to investigate the level and tissue specificities of the AtPGR expression, we measured GUS activity and the transcript level of AtPGR on root and leaf tissues of the P999-GUS transgenic plants treated with or without Glc. As shown in Supplementary Figure S2, GUS and AtPGR expressions analyses indicated that the P999-GUS transgenic lines were expressed more strongly in leaves than in roots under the same condition. Constitutive CaMV35S promoter (35S pro)-GUS served as a positive control for analysis of GUS activity (Figures 2B,D). Taken together, these data indicate that bHLH34 is a functional transcription factor that regulates the expression of the AtPGR gene. Moreover, it appears that bHLH34 functions as a transcriptional activator.
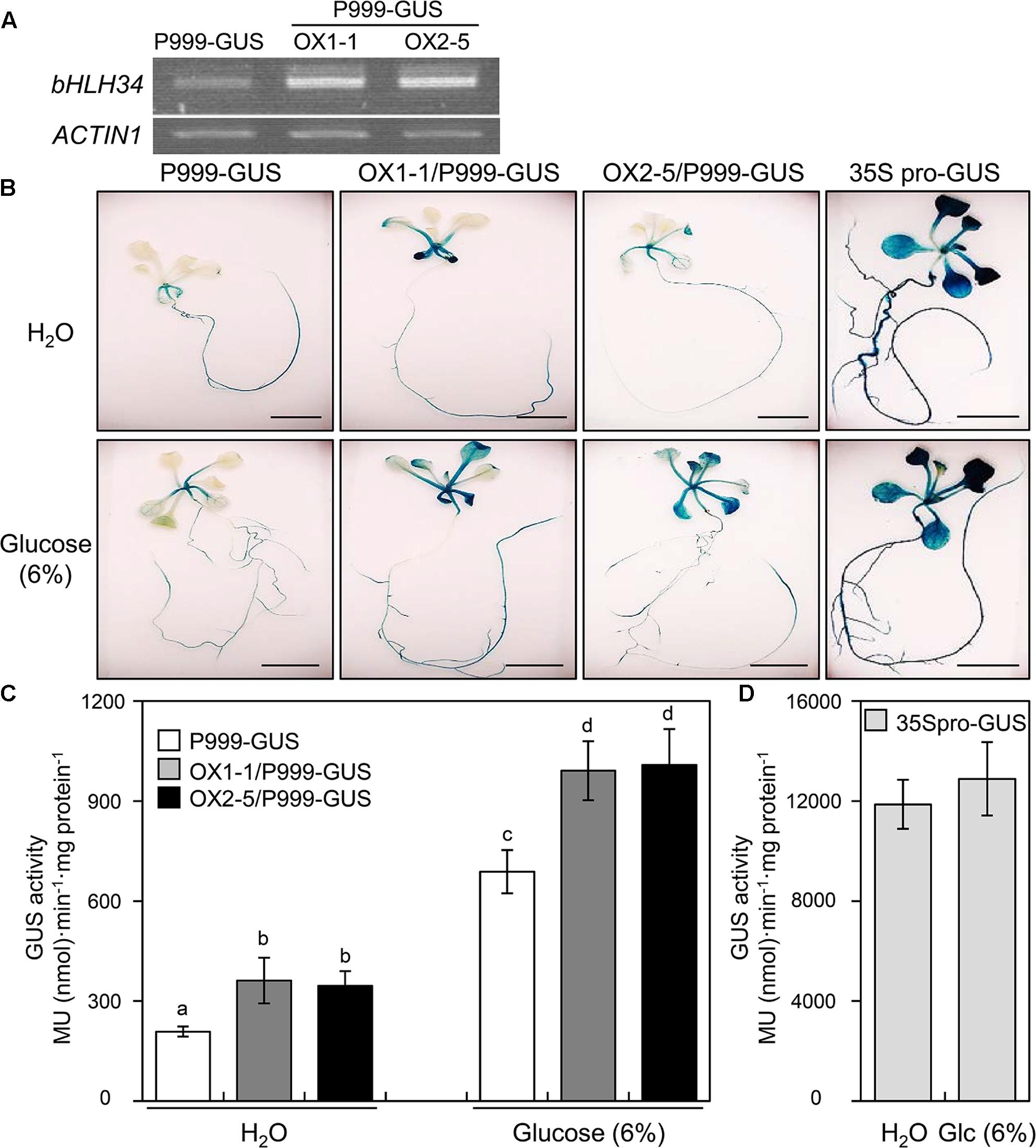
FIGURE 2. bHLH34 activates AtPGR expressions. (A) Expression levels of bHLH34 in P999-GUS and two independent P999-GUS/bHLH34-overexpressing (OX1-1/P999-GUS, OX2-5/P999-GUS) transgenic plants were confirmed by reverse transcription (RT)-PCR using RNA extracted from 14-day-old seedlings. Actin 1 served as an internal RT-PCR control. (B) The activities of the 999-bp regulatory promoter region (P999) of the AtPGR gene were examined by means of GUS as a reporter in transgenic plants (P999-GUS). These transgenic plants overexpressing bHLH34 (OX1-1/P999-GUS, OX2-5/P999-GUS) were analyzed histochemically by treatment with H2O or 6% glucose for 12 h. 35S pro-GUS served as a positive control for analysis of GUS activity. Scale bars = 5 mm. (C,D) P999-GUS, OX1-1/P999-GUS, OX2-5/P999-GUS (C), and 35S pro-GUS (D) seedlings grown on the MS medium for 12 days were carefully taken out and treated with H2O or 6% Glc for 12 h. Subsequently, seedlings were subjected to GUS staining, and GUS activity was measured. The values of GUS activities are averages of three independent enzymatic assays. Each assay was performed with extracts obtained from three individual seedlings of each transgenic plant. Error bars indicate standard deviations, and different letters above bars indicate a statistical difference (ANOVA, P < 0.05).
bHLH34 Expression in Various Organs and Different Developmental Stages
To examine the tissue expression pattern of the bHLH34 and AtPGR, we analyzed the expression patterns of the bHLH34 and AtPGR in various organs and at different developmental stages of Arabidopsis seedlings by quantitative real-time PCR (qPCR). The maximal expression levels of the bHLH34 and AtPGR were detected in leaves, while moderate expression levels were detected in roots and flowers; low expression levels were detected in stem (Figure 3A). The expression patterns of bHLH34 and AtPGR during leaf development from 1 week after germination (WAG) to 5 WAG are also examined (Figure 3B). The expression patterns of bHLH34 and AtPGR were similar at the 1 and 2 WAG stages. In contrast, different expression patterns were observed for bHLH34 and AtPGR at the 3, 4 and 5 WAG stages, indicating that the expression level of the AtPGR is likely to be involved other unidentified regulatory factors or be independent of bHLH34 at different developmental stage.
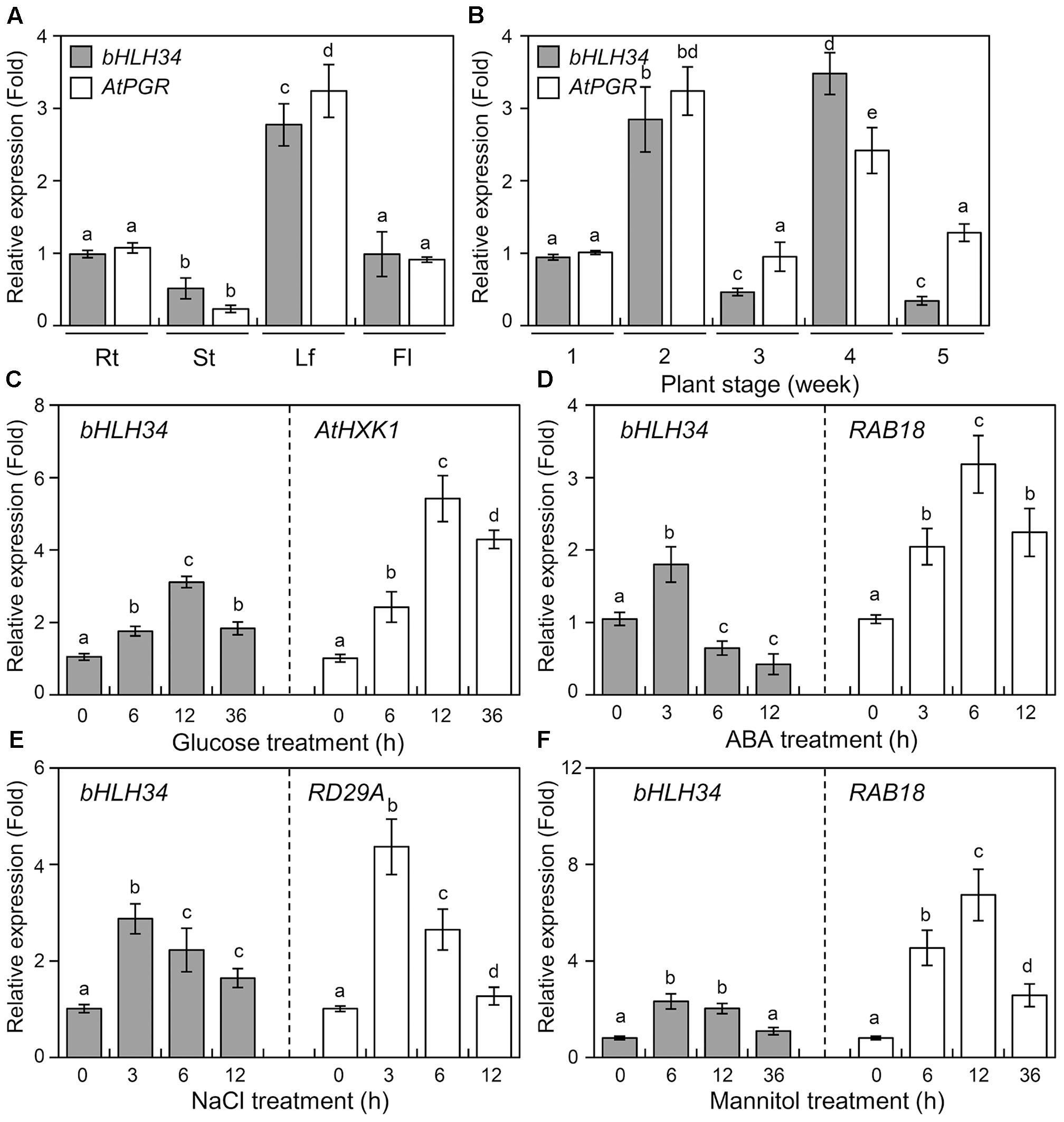
FIGURE 3. Expression of bHLH34 in Arabidopsis. (A,B) Expression profiles for AtPGR gene in various organs and at different developmental stages. Results are relative to the bHLH34 expression and represent the average of three independent biological replicates (n = 5–25, mean values ± SD, ANOVA, P < 0.05). Different letters above bars indicate a statistical difference. (A) RNA levels were confirmed by qPCR using total RNA isolated from roots (Rt), stems (St), leaf (Lf), and flowers (Fl). (B) RNA levels were determined by qPCR using total RNA isolated at the indicated plant sampling stages. (C–F) Expression of the bHLH34 in Arabidopsis under glucose, ABA, NaCl, and mannitol stress conditions. qPCR analyses of the expression of bHLH34 involved in glucose (C), ABA (D), NaCl (E), or mannitol (F) responses. Total RNA samples were obtained from 14-day-old seedlings treated with 6% glucose, 100 μM ABA, 150 mM NaCl, or 400 mM mannitol at the indicated time points. Error bars indicate standard deviations of three independent experiments, and different letters above bars indicate a statistical difference (ANOVA, P < 0.05). Each experiment was performed with total RNA of each sample obtained from 20 seedlings. Arabidopsis Actin 1 was used as the internal control. HXK1 (C), RAB18 (D), RD29A (E), or RAB18 (F) gene served as a control for the glucose, ABA, salt, or mannitol stress treatment, respectively.
The bHLH34 Gene Is Regulated by Abiotic Stress
To determine the in vivo functions of bHLH34, the accumulation of bHLH34 mRNA was assessed in Arabidopsis using qPCR during Glc, ABA, salt, and mannitol treatment. bHLH34 was induced in Arabidopsis seedlings after Glc treatment, and the bHLH34 transcript reached its maximum amount at 12 h followed by a slight decline (Figure 3C). In contrast, only slight upregulation of bHLH34 was observed after 3 h of ABA treatment (Figure 3D). In particular, transcript level of bHLH34 reached a peak within 3 and 6 h after the salt and mannitol treatment, respectively, and decreased for the remainder of the salt and mannitol experiments (Figures 3E,F). The stress-inducible A. thaliana Hexokinase 1 (AtHXK1), Responsive to ABA 18 (RAB18), and Responsive to Desiccation 29A (RD29A) genes served as references for the abiotic stress treatment (Figures 3C–F). These results indicate that bHLH34 is modulated by Glc, ABA, salinity, and osmotic stress.
Abiotic Stress Responses in bHLH34 Transgenic Plants
To investigate bHLH34 function in vivo, bHLH34 was overexpressed in Arabidopsis transgenic plants under the control of the 35S promoter. Twelve homozygous lines (T3 generation) were obtained, and two independent lines (OX2-1 and OX3-5) showing high levels of transgene expression (Supplementary Figure S3A) were selected for phenotypic characterization. To further evaluate the functional consequences of the loss of bHLH34, bhlh34 RNA interference (RNAi) lines were generated using the exon 1 to exon 2 cDNA sequence. bHLH34 expression was assessed by qPCR in two independent bhlh34 RNAi (ri2-2 and ri5-1) lines. bHLH34 expression was found to be knocked down in the RNAi lines (Supplementary Figure S3A). In addition, to determine whether the bHLH34 RNAi construct influences on the expression levels of the bHLH34 homologs, bHLH104, bHLH105 and bHLH115, in bhlh34 RNAi lines, we performed qPCR assay. The assay revealed that expression of the bhlh34 RNAi construct do not inhibit the expression bHLH104, bHLH105 and bHLH115, indicating that bhlh34 RNAi construct is highly specific to its own target transcripts (Supplementary Figures S3B–D). No morphological differences were observed among WT, bHLH34-overexpressing and bhlh34 RNAi plants, when grown on the Murashige and Skoog (MS) medium containing 1% sucrose (Murashige and Skoog, 1962) (Supplementary Figures S3E–G).
Chung et al. (2011) demonstrated that AtPGR is required for the modulation of Arabidopsis insensitivity to Glc exposure. To determine whether bHLH34 is also related to the Glc response, Glc sensitivity was evaluated by measuring the rate of cotyledon greening. WT, bHLH34-overexpressing (OX2-1, OX3-5), and bhlh34 (ri2-2, ri5-1) seeds were grown on the MS medium containing 5% Glc for 10 days. The cotyledon greening rate of WT seedlings was 33%, and this rate was approximately 25.6% (range 24.3–26.9%) for bhlh34 RNAi lines (Figures 4A,B). In contrast, approximately 75% (range 67.6–82.4%) of the bHLH34-overexpressing cotyledons expanded and turned green (Figures 4A,B). To assess the effects of bHLH34 expression on the ABA response, the seeds of the WT, bhlh34, and bHLH34-overexpressing plants were germinated on the MS medium containing 1% sucrose and 1 μM ABA. The germination percentage of bhlh34 lines was much more affected than that of WT and bHLH34-overexpressing plants by treatment with 1 μM ABA. The germination rate of bHLH34-overexpressing plants was higher than that of the WT (Figure 4C). These results show that the bHLH34 is required for ABA-modulated seed germination in Arabidopsis.
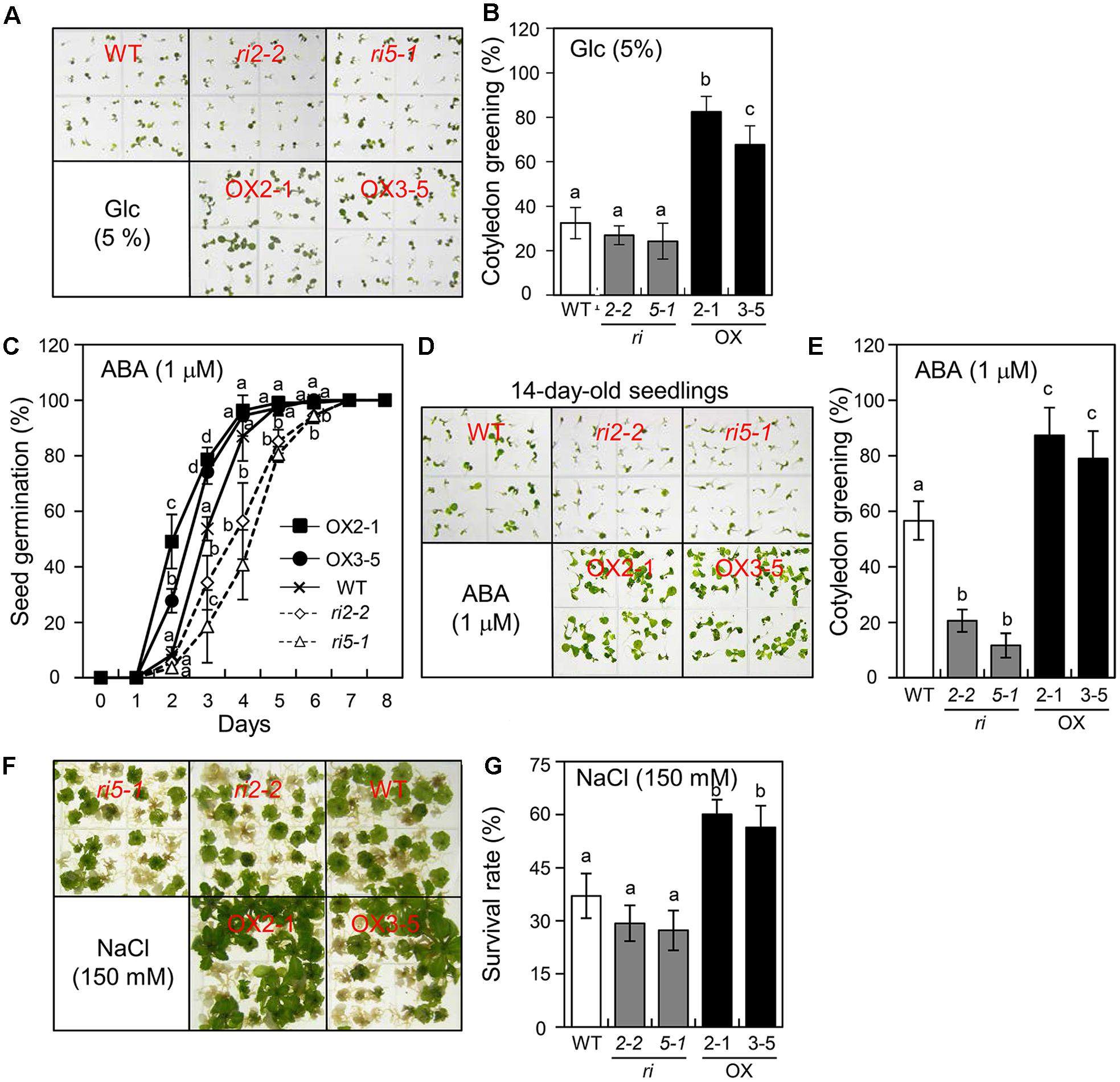
FIGURE 4. The influence of bHLH34 transgenic plants on glucose (Glc), ABA, and salt stress insensitivity. (A,B) Effects of Glc treatment on cotyledon greening. Seeds of the samples were sown on MS agar plates supplement with 5% Glc and allowed to grow for 10 days. The photograph shows that bHLH34-overexpressing lines (OX2-1 and OX3-5) were better development and greener than the WT and bhlh34 RNAi (ri2-2 and ri5-1) plants under Glc stress condition (A). Seedlings with green cotyledons were counted (triplicates, n = 50 each). Error bars indicate standard deviations, and different letters above bars indicate a statistical difference (ANOVA, P < 0.05) (B). (C) Sensitivity of germination to ABA. Seeds were sown on MS agar plates supplement with 1 μM ABA and allowed to grow for indicated days, and germination was counted (triplicates, n = 50 each). Error bars indicate standard deviations for three independent experiments, and different letters above bars indicate a statistical difference (ANOVA, P < 0.05). (D) Seeds of the samples were sown on the MS medium containing 1 μM ABA and permitted to grow for 14 days. The photograph shows that bHLH34-overexpressing lines were greener than the WT and bhlh34 RNAi plants under ABA condition. (E) Sensitivity of cotyledon greening to ABA. Seeds were sown on the MS medium containing 1 μM ABA and permitted to grow for 14 days, and seedlings with green cotyledons were counted (triplicates, n = 50 each). Error bars indicate standard deviations for three independent experiments, and different letters above bars indicate a statistical difference (ANOVA, P < 0.05). (F) Effects of salt stress on plant growth. Seeds were sown on MS medium containing 150 mM NaCl and permitted to grow for 21 days. The photograph shows that bHLH34-overexpressing lines were better development than the WT and bhlh34 RNAi plants under salt stress condition. (G) Effects of salt treatment on cotyledon greening. Seeds were sown on MS medium supplement with 150 mM NaCl and allowed to grow for 21 days, and surviving plants was counted (triplicates, n = 35 each). Error bars indicate standard deviations, and different letters above bars indicate a statistical difference (ANOVA, P < 0.05).
The ABA-induced effect was also evaluated by measuring the cotyledon greening rate. The relative reduction in cotyledon greening of the bhlh34 lines in response to ABA was more profound than that of the WT and bHLH34-overexpressing plants at 14 days after germination. In the WT, the cotyledon greening efficiency was 56.7%, whereas the cotyledon greening efficiency of bHLH34-overexpressing plants was 78.9–87.4% during treatment with ABA. In contrast, the cotyledon greening efficiency of bhlh34 RNAi lines was 11.7–20.6% (Figures 4D,E). These results indicated that the bhlh34 RNAi lines were more likely to be sensitive to Glc and ABA than the WT plant was. On the other hand, the bHLH34-overexpressing plants were less sensitive to exogenous Glc and ABA than were the WT and bhlh34 RNAi plants.
To characterize the effects of salt stress on the bHLH34-overexpression plants, we evaluated the response to treatment with 150 mM NaCl. The survival rate of the WT was slightly above 35% at 3 weeks after germination. Fewer than 30% of plants of the bhlh34 RNAi line (ri2-2) remained alive, and 27% of the bhlh34 leaves (ri5-1) turned green, as compared with 56–60% in OX2-1 and OX3-5 (Figures 4F,G). These results are consistent with the idea that bHLH34 is necessary for the regulation of developmental growth under abiotic stress.
AtPGR Expression in bHLH34 Transgenic Plants upon Glc Treatment
As shown in Figures 2B,C, the induction of GUS activity caused by bHLH34 upon exposure to exogenous Glc may indicate that AtPGR participates in the Glc response through a bHLH34-mediated signaling pathway. To quantify AtPGR expression in the bHLH34 transgenic plants during Glc treatment, a second set of experiments using qPCR was conducted. Figure 5A shows that AtPGR mRNA expression in bHLH34-overexpressing transgenic plants without Glc treatment appears to be higher than that in WT and bhlh34 RNAi lines. By contrast, the transcript abundance of Glc-inducible AtPGR showed greater reduction in bhlh34 RNAi (ri2-2 and ri5-1) plants as compared with WT after Glc treatment. As expected, AtPGR expression was more strongly induced by Glc treatment in the bHLH34-overexpressing transgenic (OX2-1 and OX3-5) lines than in the WT and bhlh34 RNAi plants. Basically, these results are consistent with those obtained in the GUS activity assay after Glc treatment (Figure 2).
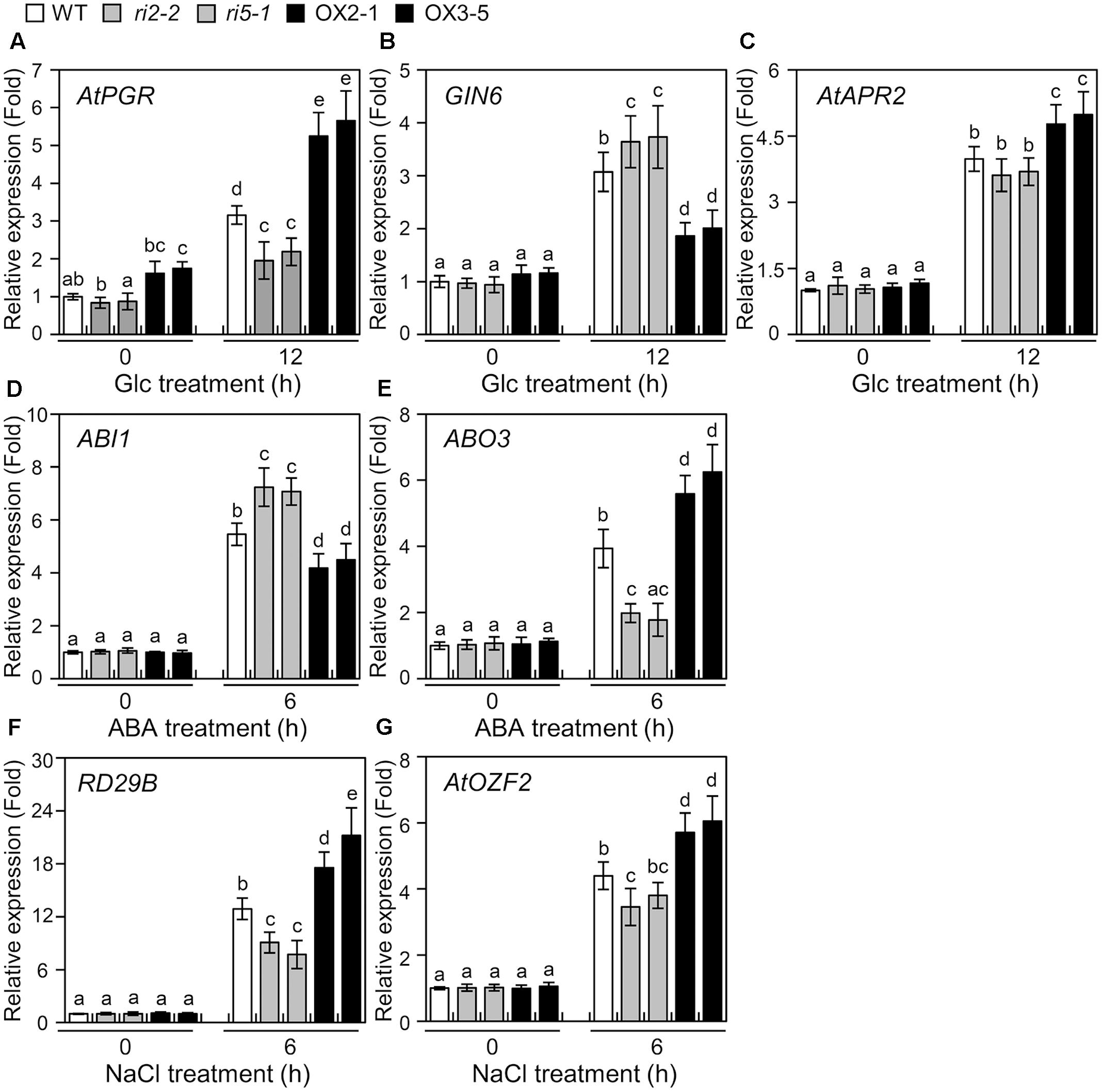
FIGURE 5. Expression of abiotic stress-regulated genes in bHLH34 transgenic plants. (A–G) mRNA levels of AtPGR (A), GIN6 (B), AtAPR2 (C), ABI1 (D), ABO3 (E), RD29B (F), or AtOZF2 (G) were measured by qPCR using total RNA from 14-day-old WT, two independent bhlh34 RNAi (ri2-2, ri5-1), and two independent bHLH34-overexpressing (OX2-1, OX3-5) seedlings, which were treated with 6% Glc, 100 μM ABA, or 150 mM NaCl with gentle shaking for the indicated period. The mean value of three technical replicates was normalized to the level of Actin 1 mRNA, an internal control. Error bars indicate standard deviations, and different letters above bars indicate a statistical difference (n = 20 each, ANOVA, P < 0.05).
Expression of Stress-Regulated Genes in bHLH34 Transgenic Plants
To investigate the transcriptional regulatory roles of bHLH34, we screened genes involved in Glc, ABA, and salt stress response and harboring the promoter including GAGA cis-element and E-box sequences. Evaluated were six genes, Glucose-Insensitive 6 (GIN6), Arabidopsis thaliana Adenosine 5′-Phosphosulfate Reductase 2 (AtAPR2), Abscisic acid Insensitive 1 (ABI1), ABA Overly Sensitive 3 (ABO3), Responsive to Desiccation 29B (RD29B), and A. thaliana Oxidation-related Zinc Finger 2 (AtOZF2), which are induced by various abiotic stresses (Leung et al., 1997; Arenas-Huertero et al., 2000; Ren et al., 2010; Huang et al., 2012; Chung et al., 2015).
As shown in Figures 5B–F, Glc- or ABA-induced expression of GIN6 or ABI1 was significantly reduced in bHLH34-overexpressing lines (OX2-1, OX3-5), in comparison with WT and bhlh34 RNAi (ri2-2, ri5-1) plants (Figures 5B,D). However, the induction of transcript levels of AtAPR2 and ABO3 were more pronounced in bHLH34-overexpressing lines than in WT and bhlh34 RNAi plants during Glc or ABA conditions (Figures 5C,E). The induction of transcript levels of dehydration-inducible RD29B and AtOZF2 genes were more pronounced in bHLH34-overexpressing lines than in WT and bhlh34 RNAi lines, but less pronounced in the bhlh34 RNAi lines than in WT following salt treatment (Figures 5F,G). These results indicated that bHLH34 may act positively or negatively depending on the types of abiotic stress-related genes harboring the promoter including GAGA cis-element and E-box sequences.
Genetic Interaction between bHLH34 and bHLH104 in Glc Response
bHLH34 and bHLH104 sequences are highly homologous, with a significant 56% identity at the amino acid level (Supplementary Figure S4). Furthermore, bHLH34 and bHLH104 harbor a conserved DNA-binding domain within the basic helix-loop-helix motif in their central region that is 88% identical between these proteins (Supplementary Figure S1D). To determine whether the bHLH104 (At4g14410) gene is related to Glc, we obtained the At4g14410-tagged T-DNA insertion mutant SALK_005802. The absence of bHLH104 transcript was verified using qPCR (Figure 6B). The mutant was designated as bhlh104.
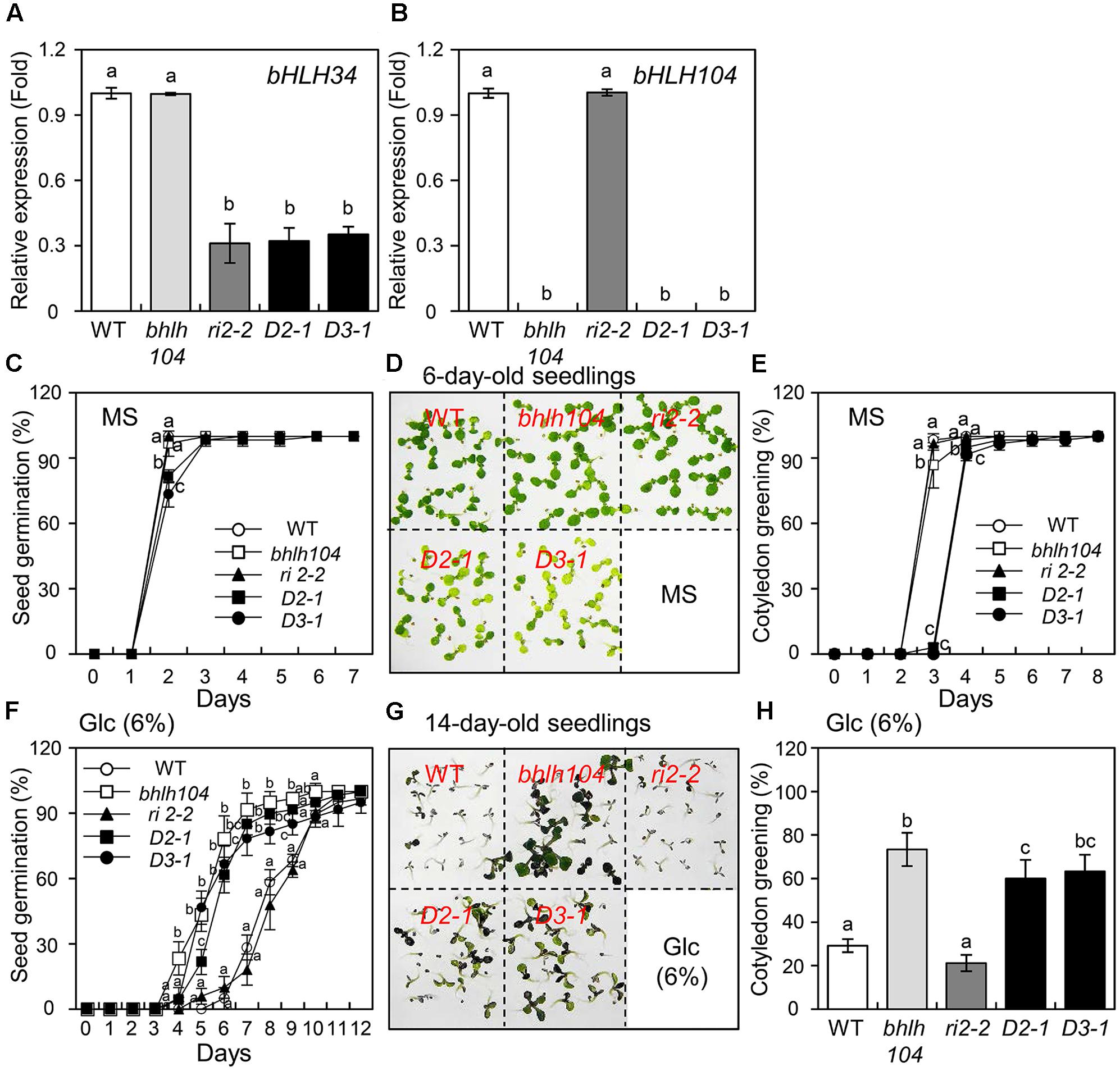
FIGURE 6. Genetic analysis of bhlh104, bhlh34 RNAi (ri2-2) and bhlh34/bhlh104 double mutants (D2-1 and D3-1) under normal and glucose (Glc) conditions. (A,B) bHLH34 (A) and bHLH104 (B) expression levels in WT, bhlh104, bhlh34, and two independent bhlh34/bhlh104 mutant lines (D2-1 and D3-1) were confirmed by qPCR using RNA extracted from 10-day-old seedlings. The mean value of three technical replicates was normalized to the level of Actin 1 mRNA, an internal control. Error bars indicate standard deviations, and different letters above bars indicate a statistical difference (n = 20 each, ANOVA, P < 0.05). (C) Seed germination assays in the normal condition. Seeds of WT, bhlh104, bhlh34 RNAi (ri2-2) and bhlh34/bhlh104 mutants (D2-1 and D3-1) were sown on the sterile MS medium and permitted to grow for indicated days, germination was scored (triplicates, n = 50 each). Error bars indicate standard deviations for three independent experiments, and different letters above bars indicate a statistical difference (50 seeds per point, ANOVA, P < 0.05). (D) Seeds of the samples were sown on the MS medium and permitted to grow for 6 days. The photograph indicates that bhlh104 showed development and green cotyledons similar to those of WT and bhlh34, while bhlh34/bhlh104 lines had paler green leaves. (E) Cotyledon greening assays in the normal condition. Seeds were sown on the MS medium and permitted to grow for indicated days, green cotyledons were counted (triplicates, n = 50 each). Error bars indicate standard deviations for three independent experiments, and different letters above bars indicate a statistical difference (50 seeds per point, ANOVA, P < 0.05). (F) Effects of Glc treatment on seed germination. Seeds were sown on the MS medium containing 6% Glc and permitted to grow for indicated days, germination was scored (triplicates, n = 50 each). Error bars indicate standard deviations for three independent experiments, and different letters above bars indicate a statistical difference (50 seeds per point, ANOVA, P < 0.05). (G) Seeds of the samples were sown on the MS medium and permitted to grow for 14 days. The photograph shows that bhlh104 and bhlh34/bhlh104 mutant lines show better development and are greener than the WT and bhlh34 at high Glc concentration. (H) Effects of Glc treatment on cotyledon greening. Seeds were sown on the MS medium and permitted to grow for 14 days, green cotyledons were counted (triplicates, n = 50 each). Error bars indicate standard deviations for three independent experiments, and different letters above bars indicate a statistical difference (ANOVA P < 0.05).
To investigate the genetic relation between bHLH34 and bHLH104, a bHLH34 RNAi construct was transformed into the bhlh104 mutant. Ten homozygous lines (T3 generation) were obtained, and bHLH34 expression was assessed by qPCR in two independent bhlh34/bhlh104 lines (D2-1 and D3-1). bHLH34 expression was knocked down successfully in the double mutant lines (Figure 6A), and the effects of Glc on seed germination or cotyledon greening efficiency in the WT, bhlh34 RNAi (ri2-2), bhlh104, and bhlh34/bhlh104 (D2-1 and D3-1) lines were assessed. Under normal condition, the germination percentage and cotyledon greening rate were similar between WT, bhlh34, and bhlh104 mutant on the MS medium (Figures 6C–E). In contrast, the germination percentage or cotyledon greening rate of bhlh34/bhlh104 lines was much more affected than that of WT, bhlh34, and bhlh104 at 2 days after germination (Figure 6C). In addition, 6-day-old bhlh34/bhlh104 lines displayed paler green leaves (Figures 6D,E), more strongly decreased chlorophyll content, and lower photosynthetic efficiency (Fv/Fm) in comparison with the WT, bhlh34, and bhlh104 mutant, implying that the bhlh34/bhlh104 double mutant had a chlorotic leaf phenotype (Supplementary Figure S5). These results suggest that both bHLH34 and bHLH104 are most likely involved in seed germination and leaf chlorophyll synthesis in Arabidopsis.
To characterize the correlation of bHLH34 and bHLH104 functions in the Glc response, seeds of WT, bhlh34 RNAi (ri2-2),bhlh104, and bhlh34/bhlh104 lines (D2-1 and D3-1) were germinated on the MS medium containing Glc. At 6% Glc, 28.3% of WT and 18.1% of bhlh34 seeds germinated at 7 days, and this rate increased to 91.7% and approximately 81.7% (range 78.3–85%) for bhlh104 and bhlh34/bhlh104 seeds, respectively (Figure 6F). When the bhlh34/bhlh104 lines were allowed to grow for 14 days prior to the assessment of cotyledon greening rates in response to Glc, 23.3% of WT and 21.2% of bhlh34 leaves expanded and turned green, as compared to 73.3% in the bhlh104 mutant. Furthermore, approximately 62.7% (range 60–63.3%) of two bhlh34/bhlh104 lines survived at 14 days (Figures 6G,H). These results show that bhlh104 and bhlh34/bhlh104 mutants are more likely to be insensitive to Glc than the WT and bhlh34 RNAi. This finding indicated that the bhlh104 mutant is epistatic to the bhlh34 under high Glc condition, and bHLH104 may modulate the sensitivity of seed germination and cotyledon greening phenotype more tightly than bHLH34 does in the Glc response as a negative regulator.
Discussion
Glucose signaling has been implicated in regulation of plant growth and development. To date, no common or conserved cis-element to regulate the Glc-responsive genes has been reported (Rolland et al., 2006). These findings are key to the identification of specific transcription factors in Glc signaling. In addition, Glc signaling is associated with ABA biosynthesis and signaling (Finkelstein and Gibson, 2001; Rook et al., 2006), and other processes affected by ABA, such as osmotic stress (Rolland et al., 2002).
In this study, a transcription factor involved in the Glc response, bHLH34, was isolated by yeast one-hybrid screening (Supplementary Figure S1A) and was found to be capable of modulating the Glc response through the regulation of AtPGR expression. As shown in Figure 2, bHLH34-overexpressing constructs in P999-GUS transgenic lines increased GUS activity more strongly during Glc treatment than those without Glc exposure. In addition, the level of GUS activities in leaves of both P999-GUS and P999-GUS/bHLH34-overexpressing plants was stronger than those in roots regardless of Glc treatment (Supplementary Figure S2A). The expression levels of Glc-inducible AtPGR showed enhanced induction in bHLH34-overexpressing transgenic plants relative to the WT and bhlh34 RNAi plants during Glc treatment. Nevertheless, AtPGR expression was decreased more strongly by Glc treatment in bhlh34 RNAi transgenic lines than in WT plants (Figure 5A). These results suggest that bHLH34 acts as an activator of the AtPGR gene in Glc signaling. EMSA results showed that bHLH34 binds to the GAGA and E-box sequences in the upstream promoter region of the AtPGR gene (Figure 1). Previously, it was reported that AtMYC2 (bHLH family protein) can bind to both G-box (CACGTG) and Abscisic acid-responsive element (ABRE, ACGTGG/TC) (Abe et al., 2003). Thus, a coupling cis-element is required to regulate some ABA-responsive genes. Therefore, it can be implied that bHLH34 binds to the GAGA cis-element and E-box sequence as a coupling element to regulate the AtPGR gene.
Several sugar-responsive promoters were shown to contain the carbohydrate metabolite signal-responsive element (CMSRE)-1 (TGGACGGCC), TATCCA box, translation elongation factor (TEF) element (CATAAT), and SUC-6 element (GAANGAGANGA) (Lu et al., 2002; Morikami et al., 2005; Geisler et al., 2006; Li et al., 2006). No conserved cis-acting element has been reported, indicating perhaps the complex nature of sugar signaling pathways. Moreover, the SUC-6 box has a sequence similar to that of the GAGA box (bHLH34-binding site), indicating that the SUC-6 cis-element may be required for the activation of gene expression through competition for the binding site with bHLH34 or other bHLH domain transcription factors. Although the gene-regulatory mechanism is still unknown, we propose that bHLH34 may affect downstream genes involved in sugar signaling.
At present, it is known that plant bHLHs are involved in developmental processes such as development of an embryo, root hair, stomata, carpel margin, petals, and anther (Pires and Dolan, 2010). Furthermore, numerous bHLHs play roles in the regulation of light (Castelain et al., 2012) and hormonal signaling, such as ABA, brassinosteroid, jasmonate, ethylene, and gibberellin signaling pathways (Friedrichsen et al., 2002; Abe et al., 2003; Song et al., 2014). Recent research revealed that overexpression of some bHLH genes elevates tolerance to freezing, cold, pathogen, salinity, oxidative and osmotic stresses (Chinnusamy et al., 2003; Babitha et al., 2013; Fan et al., 2014; Liu et al., 2015), indicating that bHLHs are closely associated with the responses to biotic and abiotic stress. Here, we demonstrated that bHLH34-overexpressing transgenic lines were more resistant to Glc, ABA, and salinity stress than were the WT and bhlh34 RNAi plants, whereas bhlh34 RNAi lines showed increased sensitivity to Glc, ABA, and salinity stress during cotyledon greening (Figure 4). These data imply that bHLH34 is a positive regulator of the Glc and abiotic stress responses.
It is widely known that Glc signaling can link ABA and a salinity response (Quesada et al., 2000; Rolland et al., 2002). The transcript of bHLH34 is regulated not only by Glc but also by ABA, salt and osmotic stress (Figure 3), implying that bHLH34 possibly regulates a plant abiotic stress response in a Glc-dependent manner. To investigate this possibility, the expression of several stress-related genes (GIN6, AtAPR2, ABI1, ABO3, AtOZF2, and RD29B) was monitored in WT and bHLH34 transgenic plants with and without Glc, ABA, or NaCl treatment. Figure 5 showed that bHLH34 positively regulates AtAPR2, ABO3, AtOZF2, and RD29B but negatively regulates genes GIN6 and ABI1. This result may be the reason for the increased resistance to abiotic stresses in bHLH34-overexpressing transgenic plants. Therefore, it is possible that bHLH34 is an important regulator in Glc dependent-mediated stress signaling responses or that it indirectly exerts the effect on the ABA and salt stress responses.
Considering the high similarity between bHLH34 and bHLH104 (Supplementary Figure S4), it was expected bHLH34 and bHLH104 to perform a similar function in the Glc response. In the present study, the comparison of phenotypes of bhlh34/bhlh104 double mutants with WT, bhlh34, and bhlh104 mutant revealed delayed seed germination, delayed cotyledon greening, paler green leaves, decreased total chlorophyll contents, and reduced photosynthetic efficiency when grown under normal condition (Figure 6 and Supplementary Figure S5), suggesting that the bHLH34-bHLH104 complex may be involved in photosynthesis and development processes. As shown in Figure 6, the germination or cotyledon greening percentage of bhlh104 and bhlh34/bhlh104 double mutant lines was higher than that of the WT and bhlh34 RNAi upon the treatment with high Glc concentration, indicating that bhlh34/bhlh104 double mutant phenocopied Glc insensitivity of the bhlh104 mutant. Thus, bHLH104 is epistatic to bHLH34. Therefore, it is possible that bHLH104 regulates the Glc response more tightly than bHLH34 does. Recently, Li et al. (2016) reported that bHLH34 and bHLH104 positively regulate iron homeostasis in Arabidopsis as a homo- and heterodimer. However, here, bHLH34 and bHLH104 displayed different function in Glc signaling, implying that bHLH34 and bHLH104 can be possible to bind to different cis-element for Glc signaling. Therefore, we need to further study cis-element of bHLH104 for Glc signaling. Overall, our results suggest that the bHLH34-bHLH104 complex plays a significant role in modulating Glc-dependent early seedling development and the Glc signaling.
Author Contributions
J-HM and CK designed the experiments and interpreted the results. SL provided technical assistance with the EMSA analysis. J-HM, H-WJ, DY, and K-HL carried out the experiments and interpreted the results. CK supervised the project and complemented the writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by a grant to CK from the Next-Generation BioGreen21 Program (SSAC, PJ011160) funded by the Rural Development Administration, and by the Basic Science Research Program funded by the Ministry of Education, Science and Technology of Korea (2015R1D1A1A09059125).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02100/full#supplementary-material
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Acevedo-Hernández, G. J., León, P., and Herrera-Estrella, L. R. (2005). Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 43, 506–519. doi: 10.1111/j.1365-313X.2005.02468.x
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and León, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14, 2085–2096. doi: 10.1101/gad.14.16.2085
Babitha, K. C., Ramu, S. V., Pruthvi, V., Mahesh, P., Nataraja, K. N., and Udayakumar, M. (2013). Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 22, 327–341. doi: 10.1007/s11248-012-9645-8
Castelain, M., Hir, R. L., and Bellini, C. (2012). The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiol. Plant. 145, 450–460. doi: 10.1111/j.1399-3054.2012.01600.x
Chen, J. G., and Jones, A. M. (2004). AtRGS1 function in Arabidopsis thaliana. Methods Enzymol. 389, 338–350. doi: 10.1016/S0076-6879(04)89020-7
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Chung, J. S., Lee, H. N., Leustek, T., Knaff, D. B., and Kim, C. S. (2015). The Arabidopsis thaliana adenosine 5’-phosphosulfate reductase 2 (AtAPR2) participates in flowering time and glucose response. J. Plant Biol. 58, 128–136. doi: 10.1007/s12374-014-0514-2
Chung, M. S., Huang, P., Ha, C. M., Jun, J. H., Ahn, S. J., Zhang, F. C., et al. (2011). Molecular identification and physiological characterization of a putative novel plasma membrane protein from Arabidopsis involved in glucose response. J. Plant Biol. 54, 57–64. doi: 10.1007/s12374-011-9145-z
Chung, M. S., Lee, S., Min, J. H., Huang, P., Ju, H. W., and Kim, C. S. (2016). Regulation of Arabidopsis thaliana plasma membrane glucose-responsive regulator (AtPGR) expression by A. thaliana storekeeper-like transcription factor, AtSTKL, modulates glucose response in Arabidopsis. Plant Physiol. Biochem. 104, 155–164. doi: 10.1016/j.plaphy.2016.03.029
Eckardt, N. A. (2002). Abscisic acid biosynthesis gene underscores the complexity of sugar, stress, and hormone interactions. Plant Cell 14, 2645–2649. doi: 10.1105/tpc.141110
Fan, M., Bai, M. Y., Kim, J. G., Wang, T., and Oh, E. (2014). The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern–triggered immunity in Arabidopsis. Plant Cell 26, 828–841. dio: 10.1105/tpc.113.121111
Finkelstein, R. R., and Gibson, S. I. (2001). ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr. Opin. Plant Biol. 5, 26–32. doi: 10.1016/S1369-5266(01)00225-4
Friedrichsen, D. M., Nemhauser, J., Muramitsu, T., Maloof, J. N., Alonso, J., Ecker, J. R., et al. (2002). Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162, 1445–1456.
Geisler, M., Kleczkowski, L. A., and Karpinski, S. (2006). A universal algorithm for genome-wide in silicio identification of biologically significant gene promoter putative cis-regulatory-elements; identification of new elements for reactive oxygen species and sucrose signaling in Arabidopsis. Plant J. 45, 384–398. doi: 10.1111/j.1365-313X.2005.02634.x
Giuliano, G., Pichersky, E., Malik, V. S., Timko, M. P., Scolnik, P. A., and Cashmore, A. R. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. U.S.A. 85, 7089–7093. doi: 10.1073/pnas.85.19.7089
Grierson, C., Du, J. S., Zabala, M. D. T., Beggs, K., Smith, C., Holdsworth, M., et al. (1994). Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. Plant J. 5, 815–826. doi: 10.1046/j.1365-313x.1994.5060815.x
Huang, P., Ju, H. W., Min, J. H., Zhang, X., Chung, J. S., Cheong, H. S., et al. (2012). Molecular and physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 2, a plasma membrane protein involved in ABA and salt stress response through the ABI2-mediated signaling pathway. Plant Cell Physiol. 53, 193–203. doi: 10.1093/pcp/pcr162
Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/bf00282746
Laby, R. J., Kincaid, M. S., Kim, D., and Gibson, S. I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23, 587–596. doi: 10.1046/j.1365-313x.2000.00833.x
Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9, 759–771. doi: 10.2307/3870430
Li, X., Zhang, H., Ai, Q., Liang, G., and Yu, D. (2016). Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 170, 2478–2493. doi: 10.1104/pp.15.01827
Li, Y., Lee, K. K., Walsh, S., Smith, C., Hadingham, S., Sorefan, K., et al. (2006). Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a relevance vector machine. Genome Res. 14, 414–427. doi: 10.1101/gr.4237406
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Liu, Y., Ji, X., Nie, X., Qu, M., Zheng, L., Tan, Z., et al. (2015). Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 207, 692–709. doi: 10.1111/nph.13387
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, C. A., Ho, T. H. D., Ho, S. L., and Yu, S. M. (2002). Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14, 1963–1980. doi: 10.1105/tpc.001735
Lu, C. A., Lim, E. K., and Yu, S. M. (1998). Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 273, 10120–10131. doi: 10.1074/jbc.273.17.10120
Morikami, A., Matsunaga, R., Tanaka, Y., Suzuki, S., Mano, S., and Nakamura, K. (2005). Two cis-acting regulatory elements are involved in the sucrose-inducible expression of the sporamin gene promoter from sweet potato in transgenic tobacco. Mol. Genet. Genomics 272, 690–699. doi: 10.1007/s00438-004-1100-y
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Pego, J. V., Weisbeek, P. J., and Smeekens, S. C. M. (1999). Mannose inhibits Arabidopsis germination via a hexokinase-mediated step. Plant Physiol. 199, 1017–1023. doi: 10.1104/pp.119.3.1017
Pires, N., and Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Pla, M., Vilardell, J., Guiltinan, M. J., Marcotte, W. R., Niogret, M. F., Quatrano, R. S., et al. (1993). The cis-regulatory element CCACGTGG is involved in ABA and water-stress response of the maize gene rab18. Plant Mol. Biol. 21, 259–266. doi: 10.1007/bf00019942
Polge, C., and Thomas, M. (2006). SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12, 20–28. doi: 10.1016/j.tplants.2006.11.005
Price, J., Li, T. C., Kang, S. G., Na, J. K., and Jang, J. C. (2003). Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol. 132, 1424–1438. doi: 10.1104/pp.103.020347
Quesada, V., Ponce, M. R., and Micol, J. L. (2000). Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154, 421–436.
Rampey, R. A., Woodward, A. W., Hobbs, B. N., Tierney, M. P., Lahner, B., Salt, D. E., et al. (2006). An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174, 1841–1857. doi: 10.1534/genetics.106.061044
Ren, X., Chen, Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313x.2010.04248.x
Robinson, K. A., Koepke, J. I., Kharodawala, M., and Lopes, J. M. (2000). A network of yeast basic helix-loop-helix interactions. Nucleic Acids Res. 28, 4460–4466. doi: 10.1093/nar/28.22.4460
Rodrigues, A., Adamo, M., Crozet, P., Margalha, L., and Confraria, A. (2013). ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 25, 3871–3884. doi: 10.1105/tpc.113.114066
Rolland, F., Baena-Gonzales, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14, S185–S205.
Rook, F., Hadingham, S. A., Li, Y., and Bevan, M. W. (2006). Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ. 29, 426–434. doi: 10.1111/j.1365-3040.2005.01477.x
Song, S., Huang, H., Gao, H., Wang, J., and Wu, D. (2014). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26, 263–279. doi: 10.1105/tpc.113.120394
Keywords: activator, AtPGR, bHLH34, glucose-responsive element, transcription factor
Citation: Min J-H, Ju H-W, Yoon D, Lee K-H, Lee S and Kim CS (2017) Arabidopsis Basic Helix-Loop-Helix 34 (bHLH34) Is Involved in Glucose Signaling through Binding to a GAGA Cis-Element. Front. Plant Sci. 8:2100. doi: 10.3389/fpls.2017.02100
Received: 25 June 2017; Accepted: 27 November 2017;
Published: 11 December 2017.
Edited by:
Bernd Mueller-Roeber, University of Potsdam, GermanyReviewed by:
Charanpreet Kaur, Jawaharlal Nehru University, IndiaLászló Szabados, Biological Research Centre (MTA), Hungary
Copyright © 2017 Min, Ju, Yoon, Lee, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheol S. Kim, cskim626@jnu.ac.kr;, cskim626@chonnam.ac.kr
 Ji-Hee Min
Ji-Hee Min Hyun-Woo Ju
Hyun-Woo Ju Dayoung Yoon
Dayoung Yoon Kyeong-Hwan Lee
Kyeong-Hwan Lee Sungbeom Lee
Sungbeom Lee Cheol S. Kim
Cheol S. Kim