- National Key Laboratory of Crop Genetic Improvement and National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan, China
Jasmonates (JAs) and abscisic acid (ABA) are phytohormones known play important roles in plant response and adaptation to various abiotic stresses including salinity, drought, wounding, and cold. JAZ (JASMONATE ZIM-domain) proteins have been reported to play negative roles in JA signaling. However, direct evidence is still lacking that JAZ proteins regulate drought resistance. In this study, OsJAZ1 was investigated for its role in drought resistance in rice. Expression of OsJAZ1 was strongly responsive to JA treatment, and it was slightly responsive to ABA, salicylic acid, and abiotic stresses including drought, salinity, and cold. The OsJAZ1-overexpression rice plants were more sensitive to drought stress treatment than the wild-type (WT) rice Zhonghua 11 (ZH11) at both the seedling and reproductive stages, while the jaz1 T-DNA insertion mutant plants showed increased drought tolerance compared to the WT plants. The OsJAZ1-overexpression plants were hyposensitive to MeJA and ABA, whereas the jaz1 mutant plants were hypersensitive to MeJA and ABA. In addition, there were significant differences in shoot and root length between the OsJAZ1 transgenic and WT plants under the MeJA and ABA treatments. A subcellular localization assay indicated that OsJAZ1 was localized in both the nucleus and cytoplasm. Transcriptome profiling analysis by RNA-seq revealed that the expression levels of many genes in the ABA and JA signaling pathways exhibited significant differences between the OsJAZ1-overexpression plants and WT ZH11 under drought stress treatment. Quantitative real-time PCR confirmed the expression profiles of some of the differentially expressed genes, including OsNCED4, OsLEA3, RAB21, OsbHLH006, OsbHLH148, OsDREB1A, OsDREB1B, SNAC1, and OsCCD1. These results together suggest that OsJAZ1 plays a role in regulating the drought resistance of rice partially via the ABA and JA pathways.
Introduction
Due to their sessile nature, plants have to face variable environmental stresses including drought, high salinity, cold, and heat during their growth and development. Plants respond and adapt to the adverse cues throughout their life cycle by coordinating an array of biochemical and physiological changes. Phytohormones, such as abscisic acid (ABA) and jasmonates (fatty acid-derived oxylipins, JAs), play important roles in promoting plant defense against abiotic stresses (Verma et al., 2016).
To date, key components of JA perception and signaling have been extensively studied in numerous plant species, particularly in Arabidopsis, tomato (Solanum lycopersicum), and rice (Oryza sativa) (Cheong and Choi, 2003; Goossens et al., 2016; Wasternack and Song, 2017). According to the updated model of JA perception and the regulation of JA-responsive genes (Xu, 2002; Chini et al., 2007; Sheard et al., 2010; Zhu et al., 2011), the COI1-JAZ complex acts as JA co-receptor and transcriptional repressor in JA signaling. Under normal or JA-absent conditions, JAZ proteins bind to transcription factors (TFs) and inhibit their activities by recruiting the TOPLESS (TPL) co-repressor or by directly recruiting histone-modifying proteins such as HDA6. These TFs can bind to the G-box of JA- and JA-Ile-responsive genes. Under any stress conditions or developmental processes in which the levels of endogenous JA-Ile (bioactive jasmonyl-isoleucine) are increased, the binding of JA-Ile to the COI1-JAZ co-receptor will lead to the degradation of JAZs via the 26S proteasome, and the JA signaling responses and downstream genes are activated.
JASMONATE ZIM-domain (JAZ) proteins, belonging to the group II of the TIFY family, have been identified as important regulators of JA signaling in numerous plants, including Arabidopsis thaliana, Solanum lycopersicum, and rice, etc. There are 20 and 18 members of the TIFY family in rice and Arabidopsis, respectively (Vanholme et al., 2007; Ye et al., 2009). In previous studies, overexpression of JAZ1 lacking the Jas domain in Arabidopsis showed an obvious JA-hyposensitive phenotype, and a similar phenotype was observed in a JAI3/JAZ mutant (Chini et al., 2007; Thines et al., 2007). Furthermore, JAZ proteins have been well characterized for their interaction with bHLH TFs to repress the transcriptional activities of bHLH, which are the core transcriptional activators of JA signaling mediated gene expression. For example, MYC2, MYC3, and MYC4, which are core bHLH factors in the regulation of JA signaling in rice and Arabidopsis, have been reported to interact with almost all the JAZ proteins (Cheng et al., 2011; Fernandez-Calvo et al., 2011; Sasaki-Sekimoto et al., 2014). JAZ proteins are also found to interact with the F-box protein COI1, leading to the degradation of the JAZ proteins (Song et al., 2011). The coi1 mutants in Arabidopsis were defective in the JA response. In addition, expression of the JA-induced genes AtVSP, Thi2.0, and PDF1.2, which encode defense-related proteins, were suppressed in the coi1 mutant (Penninckx et al., 1998; Xu et al., 2001), indicating that COI1 is a core player in the JA signaling pathway. JAZ proteins have been reported to be involved in the regulation of trichome formation, anthocyanin synthesis, and male fertility by interacting with various TFs and affecting their transcriptional function (Kirik et al., 2005; Gonzalez et al., 2008; Qi et al., 2011; Song et al., 2011). JAZ proteins directly interacted with a wide array of TFs (such as GL3, EGL3, TT8, and the MYB factor MYB75), repressed their transcriptional function, and then suppressed JA-induced anthocyanin biosynthesis and trichome initiation (Kirik et al., 2005; Gonzalez et al., 2008; Qi et al., 2011). In addition, JAZ proteins (JAZ1, JAZ8, and JAZ11) in Arabidopsis were reported to interact with two R2R3-MYB TFs MYB21 and MYB24, which inhibited the expression of downstream genes essential for JA-mediated stamen development (Song et al., 2011). JAZ proteins also have been reported to be involved in the regulation of abiotic stresses including cold, salinity, drought, wounding, and ozone (Savchenko et al., 2014; Wu et al., 2015). Hu et al. (2013) suggested that JAZ proteins (JAZ1 and JAZ4) physically interact with ICE1, resulting in a blockage in the ICE-CBF pathway in Arabidopsis. JAZ9, a transcriptional regulator in JA signaling, is known to modulate salt tolerance in rice (Wu et al., 2015).
Abscisic acid also plays an important role in regulating various stress responses and the expression of stress responsive genes (Stone et al., 2006; Nilson and Assmann, 2007; Sirichandra et al., 2009). A number of studies have shown that JA signaling can interact with ABA signaling (Lackman et al., 2011; Chen et al., 2012; Aleman et al., 2016). MED25 plays a positive role in JA signaling via OsMYC2, while it negatively regulates the protein abundance of ABI5, which plays a core role in ABA signaling (Chen et al., 2012). In addition, HDA6, an RPD3-type histone deacetylase, was found to be involved in the JA and ABA response (Wu et al., 2008; Chen and Wu, 2010). PYL4, an ABA receptor, is involved in the crosstalk between JA and the ABA pathway to regulate plant metabolism and growth (Lackman et al., 2011). The ABA-dependent signaling pathway is partially controlled by MYC/MYB TFs, which are target proteins of JAZ proteins. It has been demonstrated that ATMYC2 (JIN1) in Arabidopsis, an important player in JA signaling, can function as a positive activator in the expression of ABA and the drought inducible gene RD22 (Abe et al., 2003). Recently, several publications have demonstrated that JAs are involved in the regulation of numerous stress-responsive genes which are also regulated by ABA. The expression levels of SNAC1 and OsbHLH148, involved in the regulation of drought-responsive genes, were both increased by MeJA and ABA treatments (Takasaki et al., 2010; Seo et al., 2011). The crosstalk between JA and ABA signaling was also observed in guard cells. CPK6, an Arabidopsis calcium dependent protein kinase, was involved in both ABA and JA signaling in guard cells (Munemasa et al., 2011). Moreover, de Ollas et al. (2015) suggested that the ABA concentration was increased fourfold to sevenfold in an exogenous JA treatment, while a JA deficiency mutant jarl showed reduced ABA accumulation.
Although previous studies clearly suggest that JA is involved in drought responses, direct evidence for JAZ proteins in regulating drought resistance is lacking. In this study, we aimed to investigate whether and how a stress-responsive JAZ gene, OsJAZ1, regulates drought resistance in rice. The OsJAZ1-overexpression (OsJAZ1-OE) plants showed increased sensitivity to drought stress, while the jaz1 mutant plants were more hyposensitive to drought stress compared to the wild-type (WT) plants. Moreover, the jaz1 mutant plants were more hypersensitive to MeJA and ABA, indicating that JAZ1 may play a negative role in modulating JA and ABA signaling. Consistent with this, many genes in the JA and ABA signaling pathways were affected by OsJAZ1 overexpression under drought stress conditions. Our results suggest that OsJAZ1 plays a negative role in drought resistance, partially through regulation of the JA and ABA pathways.
Materials and Methods
Plant Materials and Growth Conditions
The OsJAZ1 mutant line 4A-00845 in the background of the rice variety Dongjin (Oryza sativa L. ssp. japonica) was obtained from the POSTECH RISD1. Homozygous mutant lines (mut-1 and mut-2) were segregated from the heterozygous mutant 4A-00845. Genotyping was performed using the OsJAZ1 genomic primers and the T-DNA left-border primer (Supplementary Table 1). The full coding sequence of OsJAZ1 (LOC_Os04g55920.1) was amplified from the total cDNA of rice leaves. The amplified fragment of OsJAZ1 was cloned into pCAMBIA1301H, which was driven by the OsLEA3 promoter (Xiao et al., 2007). The plasmid was then introduced into the japonica rice cultivar Zhonghua11 (ZH11) via Agrobacterium-mediated transformation (Hiei and Komari, 2008). The rice plants used for drought resistance testing were grown in pots or paddy field under the natural conditions at Wuhan (114.36°E, 30.48°N), China. The seedlings for phytohormone treatments were grown in a phytotron (PPFD 75 μmol/m2.s) with a 14 h light/10 h dark cycle for 7 days with a temperature at 28°C and 25°C, for the light and dark conditions, respectively.
Drought Resistance Testing
To investigate the drought stress resistance of OsJAZ1-OE plants and jaz1 mutant plants at the seedling stage, WT and transgenic seeds were germinated on 1/2 strength MS medium for 3 days in the dark and 4 days in a greenhouse. WT and transgenic lines were transplanted into barrels (30 cm in diameter and 25 cm in height) filled with a mixture of sand and soil (1:1). Each barrel was planted with about 10 transgenic seedlings and 10 WT seedlings in a half-and-half manner. Seedlings at the four-leaf stage were subjected to drought stress treatment for 4 days with the water content of the soil maintained at 5–10%. After recovery by watering for 7 days, the plants were photographed and the survival rates were recorded. Each of the OE or mutant lines were tested with three biological repeats for drought resistance. Drought stress treatment was also performed in the barrels at the reproductive stage. In this testing, each barrel was planted with a single plant. A rice automatic phenotyping (RAP) platform (Yang et al., 2014) was used to extract phenotypic traits of the plants in the barrels. At the booting stage, the WT and transgenic plants were subjected to drought stress treatment for 7 days with the soil moisture content maintained at 15%. All the materials in this had three repeats under both normal growth and drought stress conditions. To reflect the morphological changes of plants in response to drought stress treatment, we adopted an image index called PAR, which was defined as the plant perimeter divided by the projected plant area. Since rice leaves exhibit rolling under moderate and severe drought stress conditions, the projected area will decrease, but the plant perimeter will not change too much. Therefore, the change in PAR will largely reflect the degree of leaf rolling and plant architecture. Drought stress treatment at the reproductive stage was performed in a refined paddy field facilitated with a movable rain-off shelter. Each material was planted in a plot of 10 plants with three repeats. The transgenic plants and WT plants at the booting stage were subjected to drought stress treatment for about 3 weeks with the soil moisture content maintained at 15% for the final 5 days. Then the field was fully irrigated for recovery. The relative water content (RWC) in plant leaves were determined as follows: RWC % = 100% × (fresh weight – dry weight)/(turgid weight – dry weight). The dry weight was determined by placing the samples at 80°C for 24 h. Turgid weight was obtained after placing the samples in ddH2O at room temperature for 3 h.
Phytohormone Treatment
To test the phytohormones (ABA and JA) sensitivity of the OsJAZ1-OE transgenic plants and T-DNA mutant plants at the seedling stage, WT plants and transgenic lines were germinated on 1/2 strength MS medium for 3 days in the dark. After germination, seedlings with similar shoot length and root length were transplanted to transparent plastic plates (13 cm × 13 cm, seven plants half by half, at least three repeats) with 1/2 strength MS medium containing phytohormones (3 μM for ABA, 10 μM for MeJA) or water as a control. Finally, the phenotypes were recorded and shoot length and root length of these seedling were measured. To check the expression level of OsJAZ1 under phytohormone treatments, the seedlings at four-leaf stage were sprayed with ABA (100 μmol/L), MeJA (100 μmol/L), and SA (100 μmol/L), respectively, each with three replicates. Then seedling leaves were sampled in a designed time course indicated in Figure 1. The concentration for testing MeJA and ABA sensitivity of OsJAZ1-OE and T-DNA mutant plants were selected by referring to Cai et al. (2014) and Tang et al. (2016).
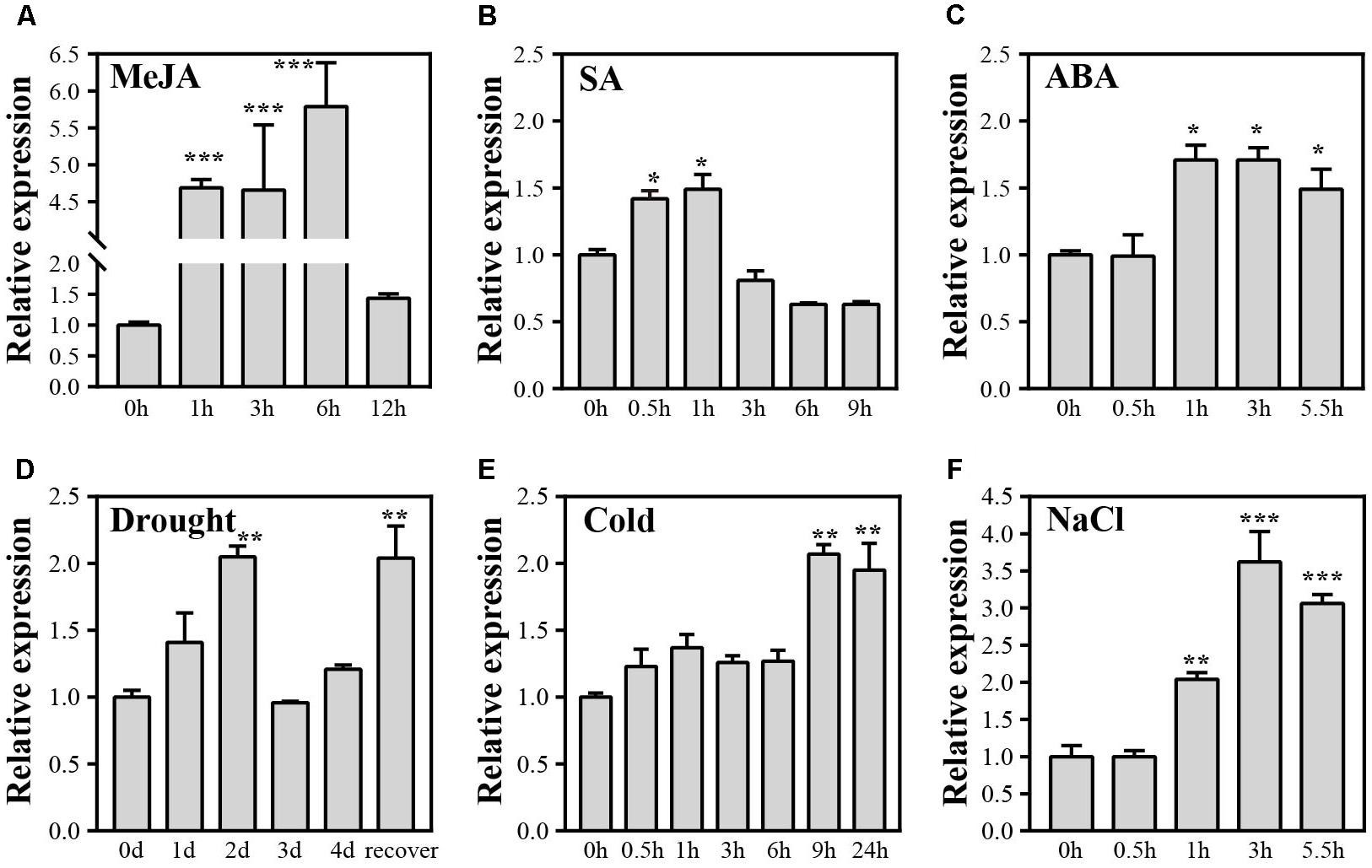
FIGURE 1. Expression levels of OsJAZ1 under various abiotic stresses and phytohormone treatments. (A–F) Seedlings at the four-leaf stage were subjected to treatment with drought stress, NaCl (200 mmol/L), cold (4°C), ABA (100 μmol/L), MeJA (100 μmol/L), and SA (100 μmol/L), respectively. Error bars represent the SE of three replicates. Significance was determined by Student’s t-test (∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05).
Subcellular Localization
For investigating the subcellular localization of the OsJAZ1 protein, the coding region of OsJAZ1 was cloned into the PM999-35 vector with C-terminal YFP tag by a one-step in vitro recombination method (Gibson et al., 2009). The resulting construct (35S: JAZ1-YFP) and the nuclear marker 35S: CFP-GHD7 (Xue et al., 2008) were used for transient transformation of protoplasts. The protocol for isolation and transformation of rice protoplasts was described previously (Xie and Yang, 2013). The fluorescence signal was observed with a confocal laser-scanning microscope (FV1200) at 16–18 h after transformation. To confirm the nuclear signal of JAZ1, the protoplasts were treated with 100 μM MeJA for 3 h in the dark before collection for microscope observation.
RNA Extraction and Quantification of Gene Expression
All the harvested leaf samples were frozen in liquid nitrogen, and RNA was extracted using TRIzol® reagent (AmbionTM, Lot No.15596018) according to the manufacturer’s instructions. RNA was reverse transcribed using the EasyScript One-Step gDNA Removal and cDNA Synthesis Kit (TRANS) according to the manufacturer’s protocol. The quality of cDNA product was checked by amplifying OsActin (LOC_Os03g50885), which was used as an internal control and then the cDNA sample was 10-fold diluted with ddH2O for qRT-PCR. Real-time PCR was performed using a Quant Studio 6 Flex Real-time system with a SYBR Green Master Mix kit (Applied BiosystemsTM). Three repeated reactions were routinely performed for each sample. The PCR reactions were performed as follows: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 1 s, 60°C for 30 s. All primers used in this study for qPCR are shown in Supplementary Table 1. The quantification of the relative expression level followed the reported 2-ΔΔCT method (Livak and Schmittgen, 2001).
RNA-Seq and Transcriptome Profiling Analysis
Total RNA was extracted from WT Zhonghua 11 (ZH11) and the OsJAZ1-OE plants at the four-leaf stage under normal and drought stress conditions with three biological replicates. The RNA samples were sequenced by Novogene (Tianjin, China) with Hiseq-PE150 and the raw data was analyzed using the customized RNA-seq data processing platform at the BMKCloud cloud server2. According to the results of Pearson’s correlation coefficient (Schulze et al., 2012), the third biological replicate of WT ZH11 and OsJAZ1-OE under drought stress was excluded from further analysis because of a low correlation coefficient. The expression levels of the genes were quantified by FPKM (fragments per kilobase of transcript per million fragments mapped) (Florea et al., 2013). The fold change of the expression level of the stressed sample over the corresponding non-stressed sample was calculated, and the genes with an absolute value of |log2(fold change)|≥ 1 and FDR < 0.05 were selected as differentially expressed genes (DEGs) (Anders and Huber, 2010). The Gene Ontology Consortium and Kyoto Encyclopedia of Genes and Genomes were referenced by GO enrichment analysis and the functional associated pathways, respectively, for the DEGs.
The RNA-Seq data were deposited in the Gene Expression Omnibus under accession number GSE107425.
Statistical Analysis
The Student’s t-test was used for statistical analysis. Statistical significance was determined at P ∗< 0.05, P ∗∗< 0.01, and P ∗∗∗< 0.001.
Results
Expression Analysis of OsJAZ1
To investigate if OsJAZ1 is involved in stress response, we applied quantitative real-time PCR (qPCR) to check the expression profiles of OsJAZ1 under various phytohormone treatments and abiotic stresses at the four-leaf stage of rice. The results showed that OsJAZ1 was strongly induced by MeJA (Figure 1A), suggesting that JAZ1 may be involved in JA signaling. OsJAZ1 was slightly up-regulated by exogenous SA and ABA (Figures 1B,C). OsJAZ1 was also induced by multiple abiotic stresses including drought, cold, and salinity (Figures 1D–F). Considering the biochemical function of JAZ proteins, these results suggest that OsJAZ1 may participate in the regulation of the responses to abiotic stresses and multiple phytohormones.
OsJAZ1 Negatively Regulates Drought Resistance at the Seedling Stage
To answer whether OsJAZ1 is involved in drought resistance regulation, we requested and obtained a heterozygous mutant 4A-00845 (Dongjin rice background) from the POSTECH RISD3. As shown in Figure 2A, the T-DNA was inserted in the promoter of OsJAZ1, and homozygous mutant plants (mut-1 and mut-2) were identified by PCR analysis using the primers Z1-mutF, Z1-mutR, and PGAR (Supplementary Table 1). The expression level of OsJAZ1 in the mutant was dramatically repressed (Figure 2B). The primers JAZ1rtF and JAZ1rtR (Supplementary Table 1) were used for OsJAZ1 expression analysis. For testing drought resistance at the seedling stage, the jaz1 mutant plants and the WT Dongjin plants of 3–4 cm length were transplanted to a blue barrel which was filled with a mixture of sand and soil (1:1). The mutant lines and WT seedlings were subjected to drought stress at the four-leaf stage. The water content of the soil was maintained at 5–10% for 3–4 days, and then watered for recovery. All the jaz1 mutant lines (mut-1 and mut-2) showed increased tolerance to drought stress treatment (Figure 2D). After recovery for 7 days, about 80% of the jaz1 mutant plants survived, while only 10–20% of the WT plants survived (Figure 2F). This result suggested that JAZ1 may have a negative role in drought resistance.
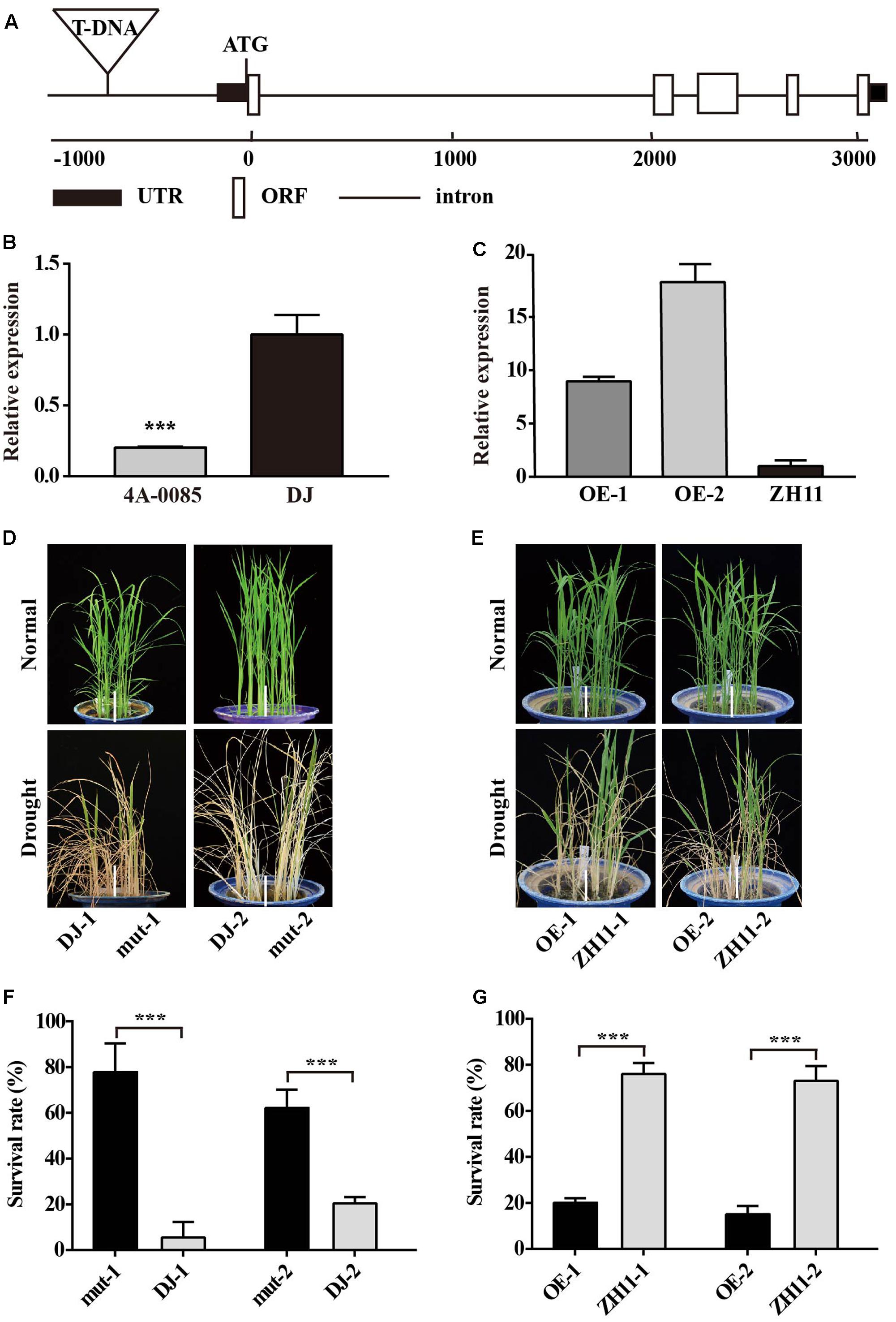
FIGURE 2. Phenotypes of the OsJAZ1-OE plants and the jaz1 T-DNA insertion mutant plants under drought stress treatment at the seedling stage. (A) Schematic diagram of the OsJAZ1 gene and 4A-00845 T-DNA insertion mutant. UTR, ORF, and introns are indicated in the black box, white box, and line, respectively. (B) Expression levels of OsJAZ1 in the 4A-00845 homozygous line and Dongjin. (C) The expression levels of OsJAZ1 in the OE lines and WT ZH11. (D,E) Drought tolerance testing of jaz1 T-DNA mutant plants and over-expression plants. (D) The jaz1 mutant lines showed increased drought resistance. (E) The OsJAZ1-OE plants were more sensitive to drought stress treatment compared to WT ZH11. (F,G) The survival rates of the transgenic and WT plants after recovery from drought stress treatment for 7 days. Mut-1 and mut-2 were heterozygous mutants segregated from 4A-00845. Error bars represent the SE of three replicates (∗∗∗P < 0.005, Student’s t-test).
To further confirm the negative role of JAZ1 in drought resistance, we constructed an overexpression vector of OsJAZ1 which was driven by the drought-inducible OsLEA3-1 promoter (Xiao et al., 2007). We selected two independent OsJAZ1-OE transgenic lines (ZH11 background) with the expression levels of OsJAZ1 in the transgenic plants confirmed by quantitative real-time PCR (Figure 2C). The OsJAZ1-OE plants and WT ZH11 were treated with drought stress at the four-leaf stage. During the drought stress treatment, the OsJAZ1-OE lines were more sensitive to the stress compared to the WT ZH11 (Figure 2E). After recovery, the survival rate of the OsJAZ1-OE lines (20%) was significantly lower than that of ZH11 (80%) (Figure 2G). These results together further supported that JAZ1 may act as a negative regulator in drought resistance.
OsJAZ1 Negatively Affects Drought Resistance at the Reproductive Stage
Our results described above indicated that OsJAZ1 has a negative effect on drought resistance at the seedling stage. Therefore, we wondered whether JAZ1 affects drought resistance at the reproductive stage. We conducted a drought resistance phenotyping of the OsJAZ1-OE lines and T-DNA mutant lines along with their WT controls (ZH11 and Dongjin) at the panicle development reproductive stage by using the high-throughput rice phenotyping facility (HRPF) (Yang et al., 2014). The dynamic changes of the plants to drought stress treatment were recorded by RGB digital cameras (Figures 3A,D) installed in the facility. As shown in Figure 3B, an image index named PAR (plant perimeter divided by projected area), which mainly reflects the degree of leaf-rolling (see description in Materials and Methods), was significantly greater in the OsJAZ1-OE plants than the WT ZH11 under drought stress conditions. This result indicated that the OE plants were more sensitive to drought stress treatment. The RWC is a commonly used indicator for the evaluation of the degree of cell and tissue hydration. As shown in Figure 3C, the RWC values of the OsJAZ1-OE plants (OE-1 and OE-2) were significantly lower (59.36% and 65.21%) than the WT ZH11 (82.15%) under drought stress conditions. This result further supported the negative effect of OsJAZ1 in drought resistance. For the jaz1 mutant lines, the PAR was significantly lower compared to the WT Dongjin under drought stress conditions, while the RWC values of the jaz1 mutant lines (mut-1 and mut-2) were significantly greater (83.38% and 79.44%) than the WT (61.88%) under drought stress conditions (Figures 3E,F). At the same time, we performed drought testing in a paddy field facilitated with a movable rain-off shelter. We found that the green leaves of the OsJAZ1-OE plants were significantly fewer than that of ZH11 after severe drought stress treatment. However, we did not observe a significant difference in the drought sensitivity phenotype between the jaz1 mutant and the WT in the field at the reproductive stage (Supplementary Figure 1). However, there were no differences in yield-related traits between the transgenic and WT plants under drought stress conditions, which may be mainly due to the fact that OsJAZ1/EG2 has been reported with a role in the regulation of spikelet development (Cai et al., 2014), and so the ectopic expression or mutation of this gene may lead to abnormal fertility.
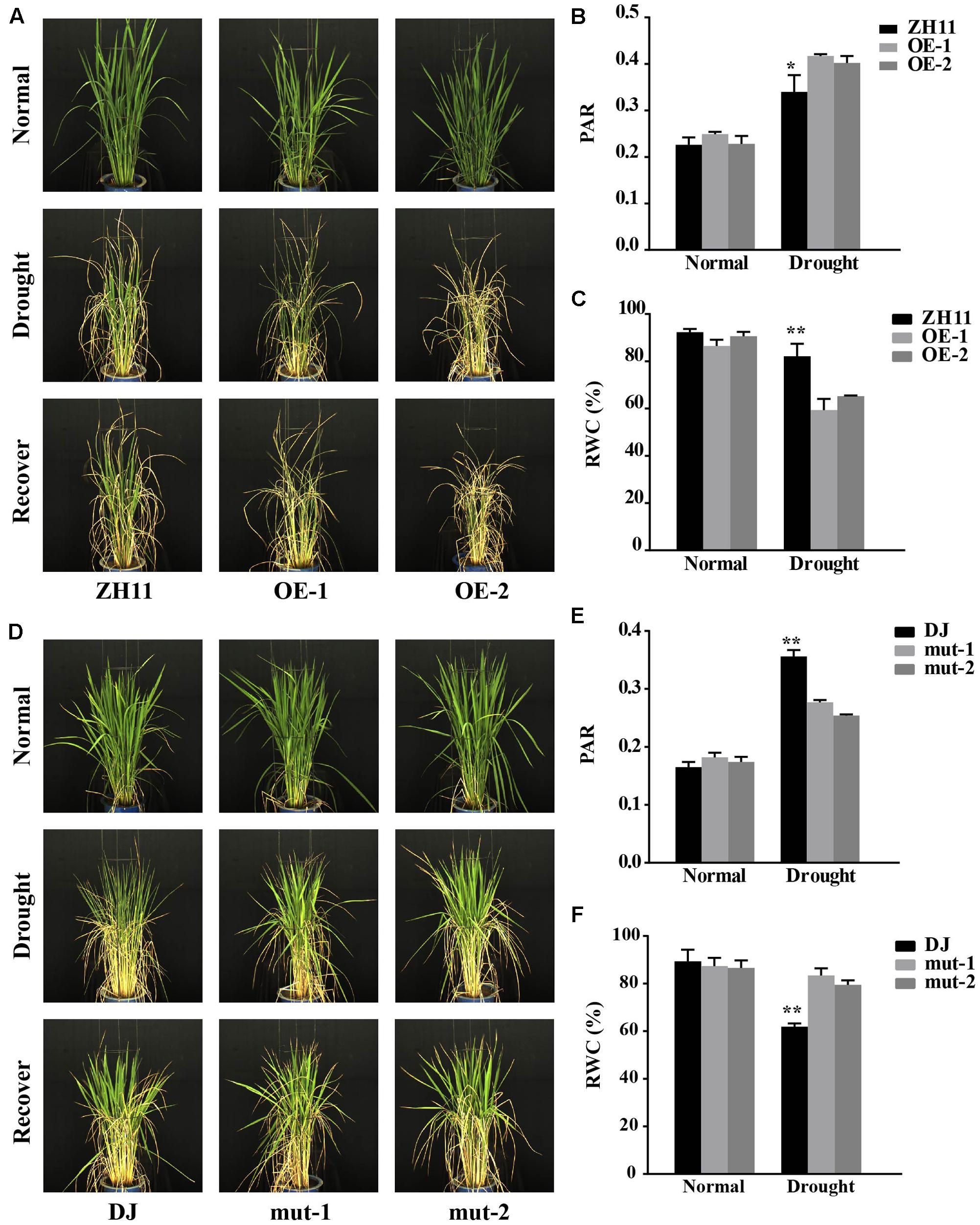
FIGURE 3. Drought resistance testing of the OsJAZ1-OE and mutant lines at the reproductive stage. (A) The OsJAZ1-OE lines and the WT plants grown in barrels at the booting stage were subjected to drought stress treatment for 7 days with the soil moisture content maintained at 15%. The degree of leaf rolling is indicated by the PAR (B) and the relative water content (RWC) (C) of the OsJAZ1-OE lines under normal and drought stress conditions. PAR = plant perimeter/plant projected area. (D) The jaz1 mutant lines showed increased drought resistance. PAR (E) and RWC (F) of the jaz1 mutant lines and Dongjin under normal and drought stress conditions. Error bars represent the SE of three replicates (∗∗P < 0.01, ∗P < 0.05, Student’s t-test).
Sensitivity of OsJAZ1-Overexpression and Mutant Plants to ABA and MeJA Treatment
To investigate the possible roles of OsJAZ1 in ABA and JA signaling, the OsJAZ1-OE plants were treated with 10 μM MeJA and 3 μM ABA. As shown in Figure 4A, the OsJAZ1-OE lines exhibited hyposensitivity to MeJA and ABA. This result was consistent with a previous study that showed Arabidopsis JAZ9-OE plants were insensitive to MeJA (Yang et al., 2012). Moreover, the shoot and root length of the OsJAZ1-OE lines were significantly longer than that of WT ZH11 under MeJA and ABA treatments (Figures 4C,D), but there was no significant difference under normal conditions. Therefore, we proposed that JAZ1 might repress ABA and JA signaling. To further confirm this, the jaz1 mutant seeds were germinated for MeJA and ABA sensitivity analysis. We found that the germination of jaz1 mutant seeds were more sensitive to MeJA and ABA compared to the WT Dongjin (Figure 4B). In addition, the shoot and root length of the jaz1 mutant lines were much shorter than that of the WT Dongjin under the MeJA and ABA treatments (Figures 4E,F). Nevertheless, there was no significant difference in these phenotypes under normal germination conditions. Taken together, our results suggest that JAZ1 may negatively modulate ABA and JA signaling.
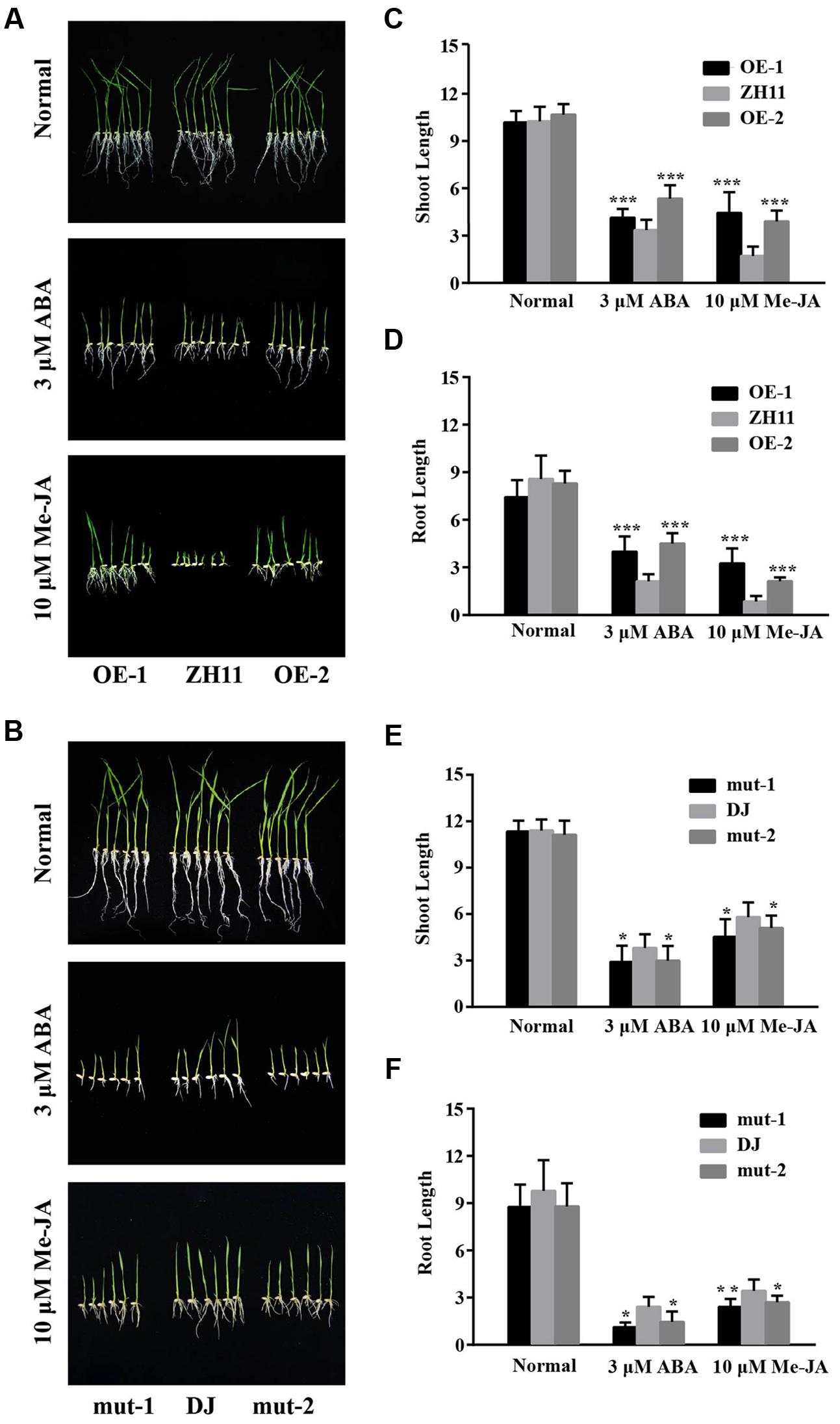
FIGURE 4. ABA and MeJA sensitivity assay of the OsJAZ1-OE and T-DNA mutant plants. (A,B) The OE lines and the mutant lines were treated with 3 μM ABA and 10 μM MeJA, under normal conditions (no addition of ABA or MeJA) as a control. (C–F) Shoot and root length of the seedlings were measured to estimate the sensitivity of the WT and transgenic plants. Error bars represent the SE of three replicates (∗∗∗P < 0.005, ∗∗P < 0.01, ∗P < 0.05, Student’s t-test).
Subcellular Localization of OsJAZ1
In previous studies, JAZ proteins, such as JAZ9 and JAZ10, were reported to locate in nucleus (Chung and Howe, 2009; Ye et al., 2009; Withers et al., 2012). Therefore, we wondered whether OsJAZ1 is also a nuclear protein. To investigate the subcellular localization of the OsJAZ1, the coding region of OsJAZ1 was cloned into PM999-35 vector with C-terminal YFP tag. The tag-fused construct (35S: JAZ1-YFP) and the nuclear marker 35S: CFP-GHD7 (Xue et al., 2008) were used for transient transformation of protoplast in rice (Xie and Yang, 2013). To our surprise, OsJAZ1-YFP was found to be localized in cytoplasm (Figure 5A), and the YFP signal of OsJAZ1 was completely separated from the nuclear signal. In addition, YFP alone was observed in both nucleus and cytoplasm in a diffuse manner (Figure 5C). As we know, MYC2, a bHLH TFs, interacts with JAZ1 in nucleus (Cai et al., 2014). To further examine whether OsJAZ1 is also localized in nucleus in presence JA signaling, the protoplasts contained the construct 35S: OsJAZ1-YFP were treated with 100 μM MeJA for 3 h. As shown in Figure 5B, OsJAZ1-YFP was obviously observed in the nucleus and was co-localized with Ghd7 which has been reported as nuclear protein in rice. These results indicate that OsJAZ1 can function as a nuclear protein in the presence JA signaling.
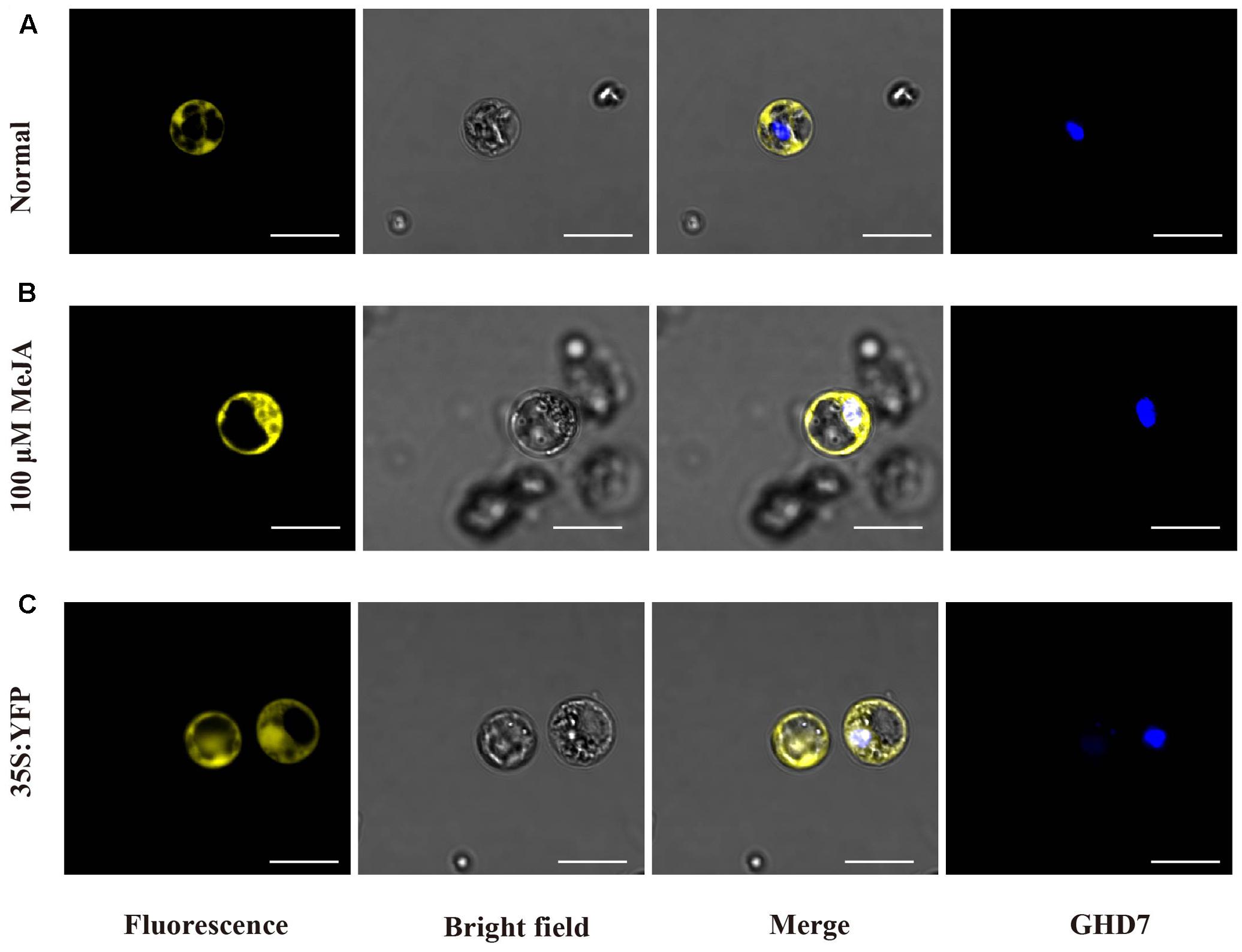
FIGURE 5. OsJAZ1 subcellular localization. (A,B) Confocal microscopy of OsJAZ1-YFP localization in rice protoplasts under normal conditions and 100 μM MeJA treatment, respectively. (C) 35S: YFP (PM999-YFP) was transformed into rice protoplasts as a control. GHD7 was used as a nucleus marker. Bars = 20 μm.
Transcriptome Analysis of the OsJAZ1-Overexpressed Plants
To investigate the transcriptomic changes that may explain the drought-hypersensitive phenotype of the OsJAZ1-OE plants, the whole genome expression profiles of OsJAZ1-OE and WT ZH11 plants under normal and drought stress conditions were analyzed by RNA-seq. Each sample was collected with three biological replicates. We set the threshold as FDR < 0.05 between sample replicates, and |log2(fold change)|≥ 1 relative to the respective control. As a result, 2,206 and 2,519 DEGs were detected in the OsJAZ1 OE and WT ZH11 plants, respectively, under drought stress conditions compared to normal conditions (Supplementary Files 2, 3). As shown in Figure 6A, the heat map revealed the different expression patterns of the DEGs (Supplementary File 1) in the OsJAZ1-OE and ZH11 plants. There were 899 and 946 up-regulated DEGs, and 1,307 and 1,573 down-regulated DEGs in the OsJAZ1-OE and WT ZH11 plants, respectively. Gene ontology (GO) enrichment analysis was conducted to identify the major functional gene groups of the DEGs. We found that most GO enrichments of the DEGs belonged to the ‘biological function’ super-group, both in the OsJAZ1-OE and WT ZH11 plants (Supplementary Files 2, 3). As shown in Figure 6B, there were 484 (Group II) and 797 (Group I) DEGs (Supplementary Files 4, 5) specifically in OsJAZ1-OE and WT ZH11, respectively, and the expression levels of most genes (1,722) were changed both in OsJAZ1-OE and WT ZH11. Among the OsJAZ1-OE or WT ZH11-specific DEGs, 197 and 244 DEGs were up-regulated, and 287 and 553 DEGs were down-regulated in the OsJAZ1-OE and WT ZH11 plants, respectively. It is noticeable that the number of up-regulated and down-regulated DEGs in the OsJAZ1-OE plants were much less than that in the WT ZH11, indicating that JAZ1 may repress some of the expression levels of drought-responsive genes.
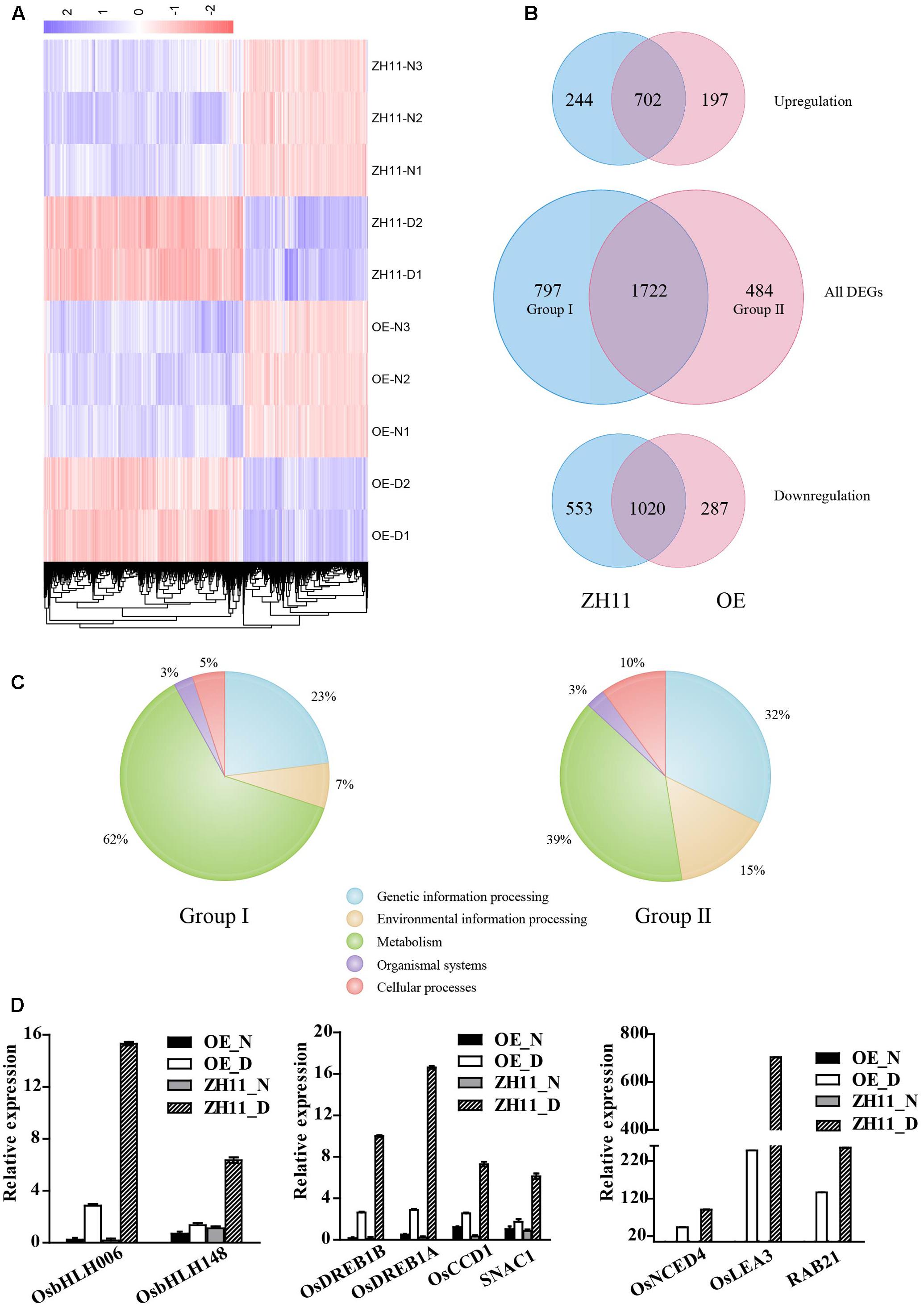
FIGURE 6. Transcriptome profiling of OsJAZ1-OE plants. (A) The expression patterns of the DEGs under normal (–N) and drought stress conditions (–D). Color scale is in log2 of the fold change. (B) Number of specific and common drought-responsive DEGs in the WT and OE plants. (C) KEGG classification of the specific drought-responsive DEGs (Group I and Group II) in the WT and OsJAZ1-OE plants. (D) Confirmation of several genes involved in the ABA and JA pathways and stress resistance showing lower drought-induced levels in the OsJAZ1-OE plants than in WT ZH11. Error bars represent the SE of three replicates.
To gain insights into the putative functions of the specific DEGs, GO enrichment and KEGG analyses were conducted for the DEGs in Group I and Group II. According to the GO term enrichment, the top three terms in Group I were ‘response to oxygen-containing compound’ (GO:1901700), ‘response to nitrate’ (GO:0010167), and ‘response to oxidative stress’ (GO:0006979), while the top three categories in Group II were ‘cell proliferation’ (GO:0008283), ‘regulation of DNA replication’ (GO:0006275), and ‘histone H3-K9 methylation’ (GO:0051567) (Supplementary Files 4, 5). In addition, there were 49 DEGs and 32 DEGs, respectively, in the WT ZH11 and OsJAZ1-OE plants belonging to the GO of ‘water deprivation’ (GO:0009414). According to the results of the KEGG analysis (Supplementary Files 4, 5), 58 and 99 DEGs were identified, respectively, in Group II and Group I. According to the KEGG analysis, 62% and 7% of the DEGs in Group I belong to ‘metabolism’ and ‘environmental information processing’ (Plant hormone signal transduction), while 39% and 15% of the DEGs in Group II belong to these two classifications, respectively (Figure 6C). These results suggested that the DEGs in the plant metabolism and hormone signaling pathways may mainly contribute to the hypersensitive phenotype of the OsJAZ1-OE plants. To further confirm this speculation, we performed quantitative real-time PCR to examine the expression profiles of some DEGs belonging to plant hormone metabolism and signaling pathways. As shown in Figure 6D, we found that the expressions levels of OsNCED4 (ABA biosynthesis), RAB21 and OsLEA3 (typical downstream genes of ABA signaling), OsbHLH006 and OsbHLH148 (related to JA signaling), and OsDREB1A, OsDREB1B, SNAC1, and OsCCD1 (well-known drought responsive genes) were significantly lower in the OsJAZ1-OE plants than in the WT ZH11 under drought stress conditions. These results indicate that the JA and ABA signaling pathways might be attenuated in the OsJAZ1-OE plants under drought stress conditions, and the expression levels of some drought resistance-related genes were also repressed to some extent. Together with the hyposensitive and hypersenstive phenotypes for the OsJAZ1-OE and the jaz1 mutant, respectively, under MeJA and ABA treatments, these results suggest that JAZ1 may affect drought resistance partially in an ABA and JA-dependent manner.
Discussion
Phytohormones play important roles in promoting the adaptation of plants to various abiotic stresses. The roles of JA and ABA under abiotic stress have been intensively studied. However, JAZ proteins act as a repressor of JA signaling, and there are only a few reports about the roles of JAZ proteins under abiotic stress treatment in rice (Hu et al., 2013; Wu et al., 2015; Lv et al., 2017). Here, we found that the jaz1 mutant plants showed increased drought resistance at the seedling and reproductive stages, while the OsJAZ1-OE plants were more sensitive to drought stress treatment compared to the WT plants.
OsJAZ1 Negatively Regulates JA and ABA Signaling
According to previous studies, JAZ proteins play a negative role for JA perception and the regulation of JA responsive genes. JA-insensitive and hypersensitive mutants contributed to confirming the negative role of JAZ proteins in JA signaling. MeJA treatment was performed to examine the JA responsiveness of OsJAZ1 (Figure 1A). The changed expression level of OsJAZ1 showed that OsJAZ1 was responsive to JA. A MeJA sensitivity test for the OsJAZ1-OE lines and jaz1 mutant lines suggests that the OsJAZ1-OE plants were hyposensitive to MeJA (Figure 4A), whereas the jaz1 mutant plants were hypersensitive to MeJA (Figure 4B). The result is consistent with previous results that the JAZ overexpression plants showed a hyposensitive phenotype under MeJA treatment (Chini et al., 2007; Thines et al., 2007). The expression level of OsJAZ1 was much greater in the overexpression plants than that in WT ZH11, resulting in stronger repression on the activation of JA-responsive TFs. Therefore, we propose that OsJAZ1 acts as a repressor in the JA signaling pathway.
Previous studies have reported that the ABA-dependent signaling pathway and the JA dependent pathway have common TFs (MYB/MYC), such as OsMYC2 (Abe et al., 2003; Adie et al., 2007). However, crosstalk between the JA and ABA signaling pathways is still ambiguous, since both synergistic and antagonistic interactions have been reported. In our study, we found that OsJAZ1 is also significantly induced by ABA (Figure 1C), and a hyposensitive phenotype is present in the OsJAZ1-OE plants under ABA treatment (Figure 4A). What’s more, the jaz1 mutant lines were hypersensitive to ABA (Figure 4B), indicating that OsJAZ1 plays a negative role in the ABA signaling pathway. Our finding that OsJAZ1 negatively regulates both the JA and ABA signaling pathways may contribute to the study of crosstalk between JA and ABA signaling. Nevertheless, the phenotypic changes of the jaz1 mutant lines under the MeJA and ABA treatments were less obvious than that of the OsJAZ1-OE plants, which may be due to potential functional redundancy of OsJAZ1 homologs in rice.
OsJAZ1 Negatively Modulates Drought Resistance
Plant adaptation to variable environmental stresses relies on arrays of signaling networks. JAs play important roles in plant adaptation and mediate several biotic responses, plant growth and developmental processes, and recently they have been reported to regulate tolerance to abiotic stresses. The JAZ proteins, belonging to the TIFY plant-specific family, are key components of the JA signaling pathway. A previous study indicated that JAZ9 repressed the expression of OsbHLH062 and OsMYB30, resulting in increased salt and cold tolerance (Wu et al., 2015; Lv et al., 2017). However, there is no direct evidence that JAZ proteins can affect drought resistance. In this study, we found that the jaz1 mutant lines showed increased drought resistance compared to the Dongjin control rice at both the seedling and reproductive stages (Figures 2D, 3D). As expected, the OsJAZ1-OE plants exhibited decreased drought resistance at both the seedling and reproductive stages (Figures 2E, 3A). In addition, we also tested the drought resistance of the OsJAZ1 transgenic rice in the field. The results showed that the OsJAZ1-OE lines were more sensitive to drought stress treatment in terms of the green leaves which remained after drought recovery, which is consistent with the increased drought sensitivity of the OsJAZ1-OE lines observed at the seedling stage. However, the jaz1 mutant lines in the field did not show an obvious hyposensitive phenotype as we expected (Supplementary Figure 1). One of the possible reasons may be that the suppression of OsJAZ1 only has a relatively small genetic effect in changing drought resistance, especially under the severe drought stress treatment we applied in this experiment. Nevertheless, it cannot be excluded that other homologs of OsJAZ1 may have similar functions in regulating drought resistance. In addition, we suspect that the overexpression or mutation of OsJAZ1 may cause fertility abnormalities, since this gene has been reported with a role in the control of spikelet development (Cai et al., 2014). Therefore, the relative yield or spikelet fertility are not suitable traits to evaluate the effect of this gene on drought resistance at the reproductive stage.
We conducted transcriptome profiling to find clues to the mechanisms responsible for the drought-hypersensitive phenotype of the OsJAZ1-OE plants. According to the profiling results, we speculate that the hypersensitive phenotype of the OsJAZ1-OE plants under drought stress treatment may be partially attributed to the significantly decreased expression levels of abiotic stress and ABA and JA signaling and stress-responsive genes. These include OsNCED4, OsLEA3, RAB21, OsbHLH006, OsbHLH148, OsDREB1A, OsDREB1B, SNAC1, and OsCCD1 which were confirmed by qRT-PCR analysis. OsNCED4, OsLEA3, and RAB21 are well known ABA and abiotic stress responsive genes (Ganguly et al., 2012). Furthermore, OsNCED4 is a key gene in ABA biosynthesis and involved in the regulation of drought resistance (Zong et al., 2016). OsLEA3 encodes a group 3 late-embryogenesis abundant protein and plays an important role in drought and salt stress responses (Hu, 2008). The suppressed expression of OsNCED4, OsLEA3, and RAB21 may result in an attenuation of the ABA pathway and subsequently lead to the drought-hypersensitive phenotype of the OsJAZ1-OE plants. Furthermore, no DEGs in Group II were enriched in the GO terms related to ABA signaling (Supplementary File 4), such as ‘response to ABA’ (GO:0009737) and ‘regulation of ABA biosynthetic process’ (GO:0010115). These results further suggest that ABA signaling was suppressed to some extent in the process of OsJAZ1-regulated drought resistance.
Previous studies have shown that JA also plays an important role in abiotic stress resistance. In this study, the expression levels of OsbHLH006 and OsbHLH148 were repressed in the OsJAZ1-OE plants under drought stress treatment. OsbHLH006 and OsbHLH148 are both basic helix-loop-helix proteins that regulate drought resistance via the JA-dependent pathway (Seo et al., 2011; Miyamoto et al., 2013). Previous studies have reported that OsDREB1A, OsDREB1B, SNAC1, and OsCCD1 positively regulate drought resistance in rice (Dubouzet et al., 2003; Figueiredo et al., 2012; Redillas et al., 2012; Jing et al., 2016). OsDREB1A and OsDREB1B regulate the expression of abiotic stress-related genes via an ABA-independent pathway. Here, we showed that the expression levels of OsDREB1A, OsDREB1B, SNAC1, and OsCCD1 were suppressed in the OsJAZ1-OE plants under drought stress conditions, which further supported the hypersensitive phenotype in the OsJAZ1-OE plants under drought stress treatment. Besides these reported genes related to the JA and ABA pathways or drought resistance, there are many DEGs in Group I enriched in several GO terms related to other stress responses, such as ‘response to oxidative stress’ (GO:0006979), ‘response to water deprivation’ (GO:0009414), and ‘response to salt stress’ (GO:0009651), while there were none or fewer DEGs in Group II enriched in these GO terms.
Conclusion
We have demonstrated that OsJAZ1 functions as a negative regulator in the drought resistance of rice, partially in an ABA-dependent and JA-dependent manner.
Author Contributions
JF designed and performed the experiments and wrote the manuscript. HW generated the transgenic materials and performed the experiments. SM, DX, and RL provided assistance with subcellular localization and data analysis. LX designed the experiments and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Program of China for Transgenic Research (2016ZX08009003-002 and 2016ZX08001-003), National Program on High Technology Development (2016YFD0100604), and the Fundamental Research Funds for the Central Universities to LX.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02108/full#supplementary-material.
Footnotes
- ^ http://www.postech.ac.kr/life/pfg/risd/
- ^ http://www.biocloud.net/
- ^ http://www.postech.ac.kr/life/pfg/risd/
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Adie, B. A., Perez-Perez, J., Perez-Perez, M. M., Godoy, M., Sanchez-Serrano, J. J., Schmelz, E. A., et al. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19, 1665–1681. doi: 10.1105/tpc.106.048041
Aleman, F., Yazaki, J., Lee, M., Takahashi, Y., Kim, A. Y., Li, Z., et al. (2016). An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: a putative link of ABA and JA signaling. Sci. Rep. 6:28941. doi: 10.1038/srep28941
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Cai, Q., Yuan, Z., Chen, M., Yin, C., Luo, Z., Zhao, X., et al. (2014). Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5:3476. doi: 10.1038/ncomms4476
Chen, L. T., and Wu, K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5, 1318–1320. doi: 10.4161/psb.5.10.13168
Chen, R., Jiang, H., Li, L., Zhai, Q., Qi, L., Zhou, W., et al. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24, 2898–2916. doi: 10.1105/tpc.112.098277
Cheng, Z., Sun, L., Qi, T., Zhang, B., Peng, W., Liu, Y., et al. (2011). The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4, 279–288. doi: 10.1093/mp/ssq073
Cheong, J.-J., and Choi, Y. D. (2003). Methyl jasmonate as a vital substance in plants. Trends Genet. 19, 409–413. doi: 10.1016/s0168-9525(03)00138-0
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Chung, H. S., and Howe, G. A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21, 131–145. doi: 10.1105/tpc.108.064097
de Ollas, C., Arbona, V., and Gomez-Cadenas, A. (2015). Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant Cell Environ. 38, 2157–2170. doi: 10.1111/pce.12536
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Fernandez-Calvo, P., Chini, A., Fernandez-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Figueiredo, D. D., Barros, P. M., Cordeiro, A. M., Serra, T. S., Lourenco, T., Chander, S., et al. (2012). Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J. Exp. Bot. 63, 3643–3656. doi: 10.1093/jxb/ers035
Florea, L., Song, L., and Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res. 2:188. doi: 10.12688/f1000research.2-188.v2
Ganguly, M., Datta, K., Roychoudhury, A., Gayen, D., Sengupta, D. N., and Datta, S. K. (2012). Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance. Plant Signal. Behav. 7, 502–509. doi: 10.4161/psb.19646
Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A. III, and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/NMETH.1318
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Goossens, J., Fernandez-Calvo, P., Schweizer, F., and Goossens, A. (2016). Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91, 673–689. doi: 10.1007/s11103-016-0480-9
Hiei, Y., and Komari, T. (2008). Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3, 824–834. doi: 10.1038/nprot.2008.46
Hu, T. Z. (2008). OsLEA3, a late embryogenesis abundant protein gene from rice, confers tolerance to water deficit and salt stress to transgenic rice. Russ. J. Plant Physiol. 55, 530–537. doi: 10.1134/s1021443708040158
Hu, Y., Jiang, L., Wang, F., and Yu, D. (2013). Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
Jing, P., Zou, J., Kong, L., Hu, S., Wang, B., Yang, J., et al. (2016). OsCCD1, a novel small calcium-binding protein with one EF-hand motif, positively regulates osmotic and salt tolerance in rice. Plant Sci. 247, 104–114. doi: 10.1016/j.plantsci.2016.03.011
Kirik, V., Lee, M. M., Wester, K., Herrmann, U., Zheng, Z., Oppenheimer, D., et al. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132, 1477–1485. doi: 10.1242/dev.01708
Lackman, P., Gonzalez-Guzman, M., Tilleman, S., Carqueijeiro, I., Perez, A. C., Moses, T., et al. (2011). Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc. Natl. Acad. Sci. U.S.A. 108, 5891–5896. doi: 10.1073/pnas.1103010108
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, Y., Yang, M., Hu, D., Yang, Z., Ma, S., Li, X., et al. (2017). The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing beta-amylase expression. Plant Physiol. 173, 1475–1491. doi: 10.1104/pp.16.01725
Miyamoto, K., Shimizu, T., Mochizuki, S., Nishizawa, Y., Minami, E., Nojiri, H., et al. (2013). Stress-induced expression of the transcription factor RERJ1 is tightly regulated in response to jasmonic acid accumulation in rice. Protoplasma 250, 241–249. doi: 10.1007/s00709-012-0400-z
Munemasa, S., Hossain, M. A., Nakamura, Y., Mori, I. C., and Murata, Y. (2011). The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol. 155, 553–561. doi: 10.1104/pp.110.162750
Nilson, S. E., and Assmann, S. M. (2007). The control of transpiration. Insights from Arabidopsis. Plant Physiol. 143, 19–27. doi: 10.1104/pp.106.093161
Penninckx, I. A., Thomma, B. P., Buchala, A., Metraux, J. P., and Broekaert, W. F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113.
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., et al. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814. doi: 10.1105/tpc.111.083261
Redillas, M. C., Jeong, J. S., Kim, Y. S., Jung, H., Bang, S. W., Choi, Y. D., et al. (2012). The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. doi: 10.1111/j.1467-7652.2012.00697.x
Sasaki-Sekimoto, Y., Saito, H., Masuda, S., Shirasu, K., and Ohta, H. (2014). Comprehensive analysis of protein interactions between JAZ proteins and bHLH transcription factors that negatively regulate jasmonate signaling. Plant Signal. Behav. 9:e27639. doi: 10.4161/psb.27639
Savchenko, T., Kolla, V. A., Wang, C. Q., Nasafi, Z., Hicks, D. R., Phadungchob, B., et al. (2014). Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 164, 1151–1160. doi: 10.1104/pp.113.234310
Schulze, S. K., Kanwar, R., Golzenleuchter, M., Therneau, T. M., and Beutler, A. S. (2012). SERE: single-parameter quality control and sample comparison for RNA-Seq. BMC Genomics 13:524. doi: 10.1186/1471-2164-13-524
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65, 907–921. doi: 10.1111/j.1365-313X.2010.04477.x
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Sirichandra, C., Wasilewska, A., Vlad, F., Valon, C., and Leung, J. (2009). The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J. Exp. Bot. 60, 1439–1463. doi: 10.1093/jxb/ern340
Song, S., Qi, T., Huang, H., Ren, Q., Wu, D., Chang, C., et al. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000–1013. doi: 10.1105/tpc.111.083089
Stone, S. L., Williams, L. A., Farmer, L. M., Vierstra, R. D., and Callis, J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428. doi: 10.1105/tpc.106.046532
Takasaki, H., Maruyama, K., Kidokoro, S., Ito, Y., Fujita, Y., Shinozaki, K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 284, 173–183. doi: 10.1007/s00438-010-0557-0
Tang, N., Ma, S., Zong, W., Yang, N., Lv, Y., Yan, C., et al. (2016). MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28, 2161–2177. doi: 10.1105/tpc.16.00171
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12, 239–244. doi: 10.1016/j.tplants.2007.04.004
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wasternack, C., and Song, S. (2017). Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321. doi: 10.1093/jxb/erw443
Withers, J., Yao, J., Mecey, C., Howe, G. A., Melotto, M., and He, S. Y. (2012). Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 20148–20153. doi: 10.1073/pnas.1210054109
Wu, H., Ye, H., Yao, R., Zhang, T., and Xiong, L. (2015). OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 232, 1–12. doi: 10.1016/j.plantsci.2014.12.010
Wu, K., Zhang, L., Zhou, C., Yu, C. W., and Chaikam, V. (2008). HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 59, 225–234. doi: 10.1093/jxb/erm300
Xiao, B., Huang, Y., Tang, N., and Xiong, L. (2007). Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115, 35–46. doi: 10.1007/s00122-007-0538-9
Xie, K., and Yang, Y. (2013). RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant 6, 1975–1983. doi: 10.1093/mp/sst119
Xu, L. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. doi: 10.1105/tpc.003368
Xu, L., Liu, F., Wang, Z., Peng, W., Huang, R., Huang, D., et al. (2001). An Arabidopsis mutant cex1 exhibits constant accumulation of jasmonate-regulated AtVSP, Thi2.1 and PDF1.2. FEBS Lett. 494, 161–164. doi: 10.1016/S0014-5793(01)02331-6
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yang, D. L., Yao, J., Mei, C. S., Tong, X. H., Zeng, L. J., Li, Q., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109, E1192–E1200. doi: 10.1073/pnas.1201616109
Yang, W., Guo, Z., Huang, C., Duan, L., Chen, G., Jiang, N., et al. (2014). Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 5:5087. doi: 10.1038/ncomms6087
Ye, H., Du, H., Tang, N., Li, X., and Xiong, L. (2009). Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 71, 291–305. doi: 10.1007/s11103-009-9524-8
Zhu, Z., An, F., Feng, Y., Li, P., Xue, L. A. M., Jiang, Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 12539–12544. doi: 10.1073/pnas.1103959108
Keywords: jasmonates, abscisic acid, JAZ protein, crosstalk, drought resistance
Citation: Fu J, Wu H, Ma S, Xiang D, Liu R and Xiong L (2017) OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 8:2108. doi: 10.3389/fpls.2017.02108
Received: 17 October 2017; Accepted: 27 November 2017;
Published: 11 December 2017.
Edited by:
Hanwei Mei, Shanghai Agrobiological Gene Center, ChinaReviewed by:
Tibor Janda, Centre for Agricultural Research (MTA), HungaryGuangxiao Yang, Huazhong University of Science and Technology, China
Copyright © 2017 Fu, Wu, Ma, Xiang, Liu and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lizhong Xiong, lizhongx@mail.hzau.edu.cn
 Jie Fu
Jie Fu Hua Wu
Hua Wu Siqi Ma
Siqi Ma Denghao Xiang
Denghao Xiang Ruyi Liu
Ruyi Liu Lizhong Xiong
Lizhong Xiong