- Department of Plant Science, McGill University, Sainte-Anne-de-Bellevue, QC, Canada
The epigenetic modulatory SAGA complex is involved in various developmental and stress responsive pathways in plants. Alternative transcripts of the SAGA complex's enzymatic subunit GCN5 have been identified in Brachypodium distachyon. These splice variants differ based on the presence and integrity of their conserved domain sequences: the histone acetyltransferase domain, responsible for catalytic activity, and the bromodomain, involved in acetyl-lysine binding and genomic loci targeting. GCN5 is the wild-type transcript, while alternative splice sites result in the following transcriptional variants: L-GCN5, which is missing the bromodomain and S-GCN5, which lacks the bromodomain as well as certain motifs of the histone acetyltransferase domain. Absolute mRNA quantification revealed that, across eight B. distachyon accessions, GCN5 was the dominant transcript isoform, accounting for up to 90% of the entire transcript pool, followed by L-GCN5 and S-GCN5. A cycloheximide treatment further revealed that the S-GCN5 splice variant was degraded through the nonsense-mediated decay pathway. All alternative BdGCN5 transcripts displayed similar transcript profiles, being induced during early exposure to heat and displaying higher levels of accumulation in the crown, compared to aerial tissues. All predicted protein isoforms localize to the nucleus, which lends weight to their purported epigenetic functions. S-GCN5 was incapable of forming an in vivo protein interaction with ADA2, the transcriptional adaptor that links the histone acetyltransferase subunit to the SAGA complex, while both GCN5 and L-GCN5 interacted with ADA2, which suggests that a complete histone acetyltransferase domain is required for BdGCN5-BdADA2 interaction in vivo. Thus, there has been a diversification in BdGCN5 through alternative splicing that has resulted in differences in conserved domain composition, transcript fate and in vivo protein interaction partners. Furthermore, our results suggest that B. distachyon may harbor compositionally distinct SAGA-like complexes that differ based on their histone acetyltransferase subunit.
Introduction
The acetylation of lysine residues on histone tails is an epigenetic mark that is involved in regulating important plant processes such as flowering time, flower development and abiotic stress responses (Bertrand et al., 2003; He et al., 2003; Cohen et al., 2009; Moraga and Aquea, 2015). GCN5 is a conserved histone acetyltransferase that acts as one of the enzymatic subunits of the multi-protein SAGA complex, and has been shown to associate with up to 40% of Arabidopsis thaliana gene promoters (Benhamed et al., 2008; Koutelou et al., 2010). It is known to acetylate histones present at the promoter of genes, an epigenetic mark that is correlated with increased transcription rates (Sterner and Berger, 2000; Robert et al., 2004; Rosaleny et al., 2007). GCN5's enzymatic capability is conferred by a HAT domain, which may be subdivided into four motifs, arranged as C, D, A, and B (Dyda et al., 2000; Sterner and Berger, 2000). Acetyl-CoA provides the acetyl group required for GCN5's enzymatic function, and the binding of this coenzyme occurs in the cleft delineated by motifs A and B (Dyda et al., 2000; Sterner and Berger, 2000). Thus, the motifs that makeup the GCN5 HAT domain are responsible for its ability to influence gene expression.
The acetylation of histone H3 and H4 tails by GCN5 occurs on multiple lysine residues in a stepwise fashion, H3K14 being the primary site of acetylation (Kuo et al., 1996; Grant et al., 1997; Benhamed et al., 2006; Earley et al., 2007; Kuo and Andrews, 2013; Cieniewicz et al., 2014; Mahrez et al., 2016). The bromodomain is GCN5's second conserved protein domain, responsible for recognizing and binding acetylated lysine residues, and is required for proper GCN5-dependent deposition order (Hassan et al., 2007; Zeng et al., 2008; Cieniewicz et al., 2014). The bromodomain is also involved in promoter binding and targeting by GCN5, and increases the acetylation of H3 tails when the other H3 in the same nucleosome is acetylated (Hassan et al., 2002; Benhamed et al., 2008). Therefore, the bromodomain may influence the targeting and extent of GCN5's enzymatic activity.
Across all the organisms in which GCN5 has been characterized, it is physically linked to the remainder of the SAGA complex through an interaction with the transcriptional adaptor ADA2 (Candau and Berger, 1996; Candau et al., 1996, 1997; Stockinger et al., 2001; Bhat et al., 2003; Fan et al., 2004; Mao et al., 2006; Gamper et al., 2009). The region required for the GCN5-ADA2 interaction differs among organisms. In yeast, this interaction requires the region present between the HAT and bromodomain, while in A. thaliana, which contains two copies of ADA2, the required region of interaction is exclusively the HAT domain (Candau and Berger, 1996; Candau et al., 1997; Mao et al., 2006). Such an example displays the differences in the composition of the SAGA complex across different organisms. However, in all known instances, GCN5 interacts with one or more ADA2 protein(s) in order to link it to the remainder of the SAGA complex.
In addition to its role as the physical link between GCN5 and the SAGA complex, ADA2 increases GCN5's substrate specificity to include histones in a nucleosomal context, compared to free histones only, when GCN5 is alone (Grant et al., 1997, 1999; Sterner and Berger, 2000; Gamper et al., 2009). Furthermore, ADA2 allows for GCN5 to be recruited to genomic loci indirectly through interactions that occur between other SAGA complex members and transcription factors, which complement the direct transcription factor interactions GCN5 exhibits (Belotserkovskaya et al., 2000; Kuo et al., 2000; Brown et al., 2001; Lang et al., 2001; Stockinger et al., 2001; Barbaric et al., 2003; Bhat et al., 2004; Gao et al., 2007; Nagy and Tora, 2007; Hirsch et al., 2015; Setiaputra et al., 2015). SGF29, another SAGA complex member, recognizes H3K4me2/3 and increases SAGA recruitment and GCN5-dependent histone acetylation at sites bearing such epigenetic marks (Bian et al., 2011; Schram et al., 2013; Ringel et al., 2015). Thus, association with the SAGA complex expands GCN5's function.
Alternative splicing is a widespread process that may increase the functional diversity of genes; in A. thaliana an estimated 61% of multi-exon containing genes may undergo alternative splicing (Marquez et al., 2012). This cellular process is known to functionally regulate certain epigenetic modulators in plants (Shen et al., 2016). Of further interest, a GCN5 splice variant has been identified in Humans, which includes an N-terminal extension to its sequence that represents a PCAF domain (Smith et al., 1998). Interestingly, the only known GCN5 transcript present in Drosophila melanogaster is homologous to the entire region of the longer human GCN5 splice variant, containing the PCAF homology domain in addition to the HAT and bromodomain sequences (Smith et al., 1998). This additional PCAF domain is believed to be responsible for the capability to interact with the CBP and p300 transcriptional coactivators (Yang et al., 1996; Smith et al., 1998). Such examples of alternative splicing allows for a single gene to generate functionally distinct protein products. However, this is not always the case, as certain splice variants are actively degraded through a ribosomal-dependent cellular process termed nonsense-mediated decay (NMD) (Lareau et al., 2007; Saltzman et al., 2008; Kwon et al., 2014). Therefore, alternative splicing is known to influence the functional diversity of epigenetic modulators.
This study describes the functional characterization of B. distachyon GCN5, as well as of two novel GCN5 splice variants, which differ from the former based on the presence and integrity of conserved protein domains.
Materials and Methods
Plant Material
Unless otherwise noted, B. distachyon accession Bd21 was used for all experiments. Seeds were imbibed for 4 h and subsequently surface sterilized by first removing the lemma, treating for 30 s with 70% ethanol, washing twice in sterile water, treating for 3 min with 1.3% bleach and washing three times in sterile water prior to stratification in the dark at 4°C for 5–7 days. Plants were grown at 22°C in an E15 Conviron growth cabinet with a 16/8 h light/dark photoperiod and 130 μmol/ms of light.
For A. thaliana, ecotype Col-0 was used for all experiments. Seeds were sterilized by inverting in 70% ethanol for 30 s, 1.3% bleach for 5 min and rinsed four times with sterile water. Sterile seeds were stratified at 4°C for 2–4 days in the dark prior to planting.
Sequence Analysis of BdGCN5 Alternative Transcripts
Nucleic acid sequences of GCN5 (XM_003573876.3), S-GCN5 (MG552853), and L-GCN5 (MG552854) were aligned using the Clustal Omega multiple sequence alignment tool (Sievers et al., 2011).
Gene Expression Analysis
Absolute RNA Quantification
Plasmid DNA was extracted from Escherichia coli strain DH5-α carrying pGEM®-T Easy (Promega) constructs containing GCN5, S-GCN5, or L-GCN5 using the EZ-10 Spin Column Plasmid DNA Minipreps Kit (Bio Basic Inc.). DNA concentration was quantified using a ND-1000 spectrophotometer (NanoDrop). A dilution series was prepared in order to generate a standard curve ranging in concentration from 10−13 to 10−20 mol/μl.
Aerial tissues from 2-week-old Bd21, Bd3-1, Bd18-1, Bd2-3, Bd21-3, Bd1-1, Bd30-1, and Bd29-1 B. distachyon individuals were collected and frozen in liquid nitrogen. RNA extractions were performed using EZ-10 Spin Column Plant RNA Mini-Preps Kit (Bio Basic Inc.). cDNA synthesis was performed using iScript Advanced cDNA Synthesis Kit (BioRad Laboratories Inc.). qPCR was performed using Green-2-Go qPCR Mastermix (Bio Basic Inc.) and gene-specific primers in a CFX Connect™ (BioRad Laboratories Inc.). All genes were normalized to both SamDC and Ef1-α (Hong et al., 2008). Primers used in this study are listed in Supplementary Table 1. Statistical analyses were performed using the JMP software. Three biological replicates were used for the absolute quantification as well as for all other gene expression analyses.
Nonsense-Mediated Decay Assays
Aerial tissues of 2 week-old B. distachyon accession Bd21 or 2 week-old A. thaliana ecotype Col-0 were treated by adding them to 1X MS in 15 ml polypropylene tubes (Sarstedt Inc.) supplemented with 0.005% Silwet-77, and either cycloheximide (Sigma-Aldrich) to a final concentration of 20 μM dissolved in DMSO, or DMSO only (mock). Polypropylene tubes were continually inverted for 4 h prior to sampling and freezing in liquid nitrogen. RNA extractions and qPCR were performed as described above.
Collection of B. distachyon tissues for BdGCN5 spatial expression analysis: Bd21 plants were grown in soil and aerial or crown tissues were collected at 3 weeks of age and immediately frozen in liquid nitrogen. RNA extractions and qPCR were performed as described above.
Abiotic Stress Treatments
All treatments were performed in sterile media (1X MS, 0.9% agar), with 2-week-old plants grown in culture tubes. Heat exposure: plants were transferred to 42°C and samples were collected at 30 min, 1 and 4 h post-treatment. Cold exposure: plants were transferred to 4°C and samples were collected at 1, 2, 4, 6, 8, 10, 12, 24 h post-treatment. Untreated plants grown at 22°C were concurrently collected as controls. All replicates of abiotic stress treatments were applied at the same time of day.
Primer Design
Gene-specific primers were designed using NCBI's Primer-Blast software and ordered from Integrated DNA Technologies Inc.
Subcellular Localization
GCN5, S-GCN5, and L-GCN5 coding sequences were cloned into the BglII sites of the pAVA321 localization vector in E. coli DH5-α, resulting in an N-terminal GFP tag (von Arnim et al., 1998). All plasmid constructs were validated by Sanger sequencing. Plasmid DNA was isolated for each of the following pAVA321 constructs GCN5, S-GCN5, L-GCN5, and empty vector using the EZ-500 Spin Column Plasmid DNA Maxipreps Kit (Bio Basic Inc.). Isolated DNA was diluted to a concentration of 3 ng/μl and used for biolistic particle bombardment as described below.
M-17 tungsten microcarriers (BioRad Laboratories Inc.) were prepared by suspending 100 mg in 1 ml of 100% ethanol. Microcarriers were centrifuged at 14,000 rpm for 5 min and re-suspended in 1 ml of 100% ethanol. This washing procedure was repeated 5 times. Following the final wash step, the microcarriers were re-suspended in 1 ml of 100% ethanol and stored at −20°C. Prior to use, prepared microcarriers were sonicated at 4°C for 1 min at the maximum setting using a Bioruptor UCD-200 (Diagenode). To adsorb DNA, 2 μl of plasmid DNA was mixed with 2 μl of 100% ethanol and vortexed for 10 s. Eight microliters of 100% ethanol was added to the above mixture and vortexed for 10 s. Twenty microliters of prepared tungsten microcarriers were added to the above mixture and vortexed for 15 s. Eight microliters of the DNA-adsorbed microcarriers were loaded onto Swinnex filter holders (Merck Millipore) and allowed to dry for 2 min.
Onion epidermal peels were placed on 1X MS, 1.5% agar plates and bombarded using a particle inflow gun (Vain et al., 1993). The solenoid timer was set to 50 ms and the helium pressure to 85 psi. Following bombardment, onion samples were placed in the dark at 28°C for 16–24 h then visualized using a V20 Discovery Stereomicroscope (Zeiss) equipped with an X-Cite Series 120Q UV lamp (Lumen Dynamics Group Inc.).
Bimolecular Fluorescence Complementation
The pDOE set of vectors was used for this BiFC assay (Gookin and Assmann, 2014). The coding sequence of B. distachyon ADA2 was cloned into the BamHI-SpeI sites of MCS1, while the coding sequences of GCN5, S-GCN5, and L-GCN5 were cloned into the AatII-RsrII sites of MCS3 and maintained in E. coli DH5-α. All plasmid constructs were validated by Sanger sequencing.
Plasmid DNA from positive clones were extracted using the EZ-10 Spin Column Plasmid DNA Minipreps Kit (Bio Basic Inc.) and electroporated into competent Agrobacterium tumefaciens strain GV3101::pMP90 using an Eppedorf Multiporator® and the suggested parameters for A. tumefaciens (Eppendorf Multiporator® Protocol No. 4308 915.502−12/2001). Positive A. tumefaciens were confirmed by PCR at both the MCS1 and MCS3 sites.
To perform the infiltration, confirmed A. tumefaciens clones were grown overnight at 28°C in LB media, supplemented with 30 μg/ml gentamycin and 50 μg/ml kanamycin. The following day, cultures were supplemented with 150 μM of acetosyringone and incubated for 2 h. Subsequently, cells were pelleted and resuspended in infiltration solution (10 mM MES, 10 mM MgCl2, and 150 μM acetosyringone) to a final OD600 of 0.7. This solution was infiltrated into the abaxial surface of Nicotiana benthaminana leaves and the plants were grown in the dark for 48 h. Two hours prior to visualization, N. benthamiana leaves were infiltrated with a solution of 1 mg/ml DAPI, 0.5% Triton X-100, 10 mM MES, and 10 mM MgCl2. Leaf epidermal cells were visualized using a AxioImager.Z1 (Zeiss).
Results
B. distachyon Has at Least Three Distinct GCN5 Alternative Transcripts
Efforts to clone the GCN5 transcript from B. distachyon accession Bd21 led to the identification of two previously unknown splice variants. The first alternative transcript, termed S-GCN5 due to its coding region being the shorter of the two splice variants, results from an alternative splice site that shortens exon 5 (Figure 1A). The second, termed L-GCN5 due to its coding region being the longer of the two splice variants, results from two alternative splice sites that include a supplementary exon in the transcript, between the locations of exons 7 and 8 in the GCN5 transcript (Figure 1A). Both S-GCN5 and L-GCN5 result in earlier stop codons when compared to GCN5, reducing the length of the coding region and expected protein products (Figure 1A).
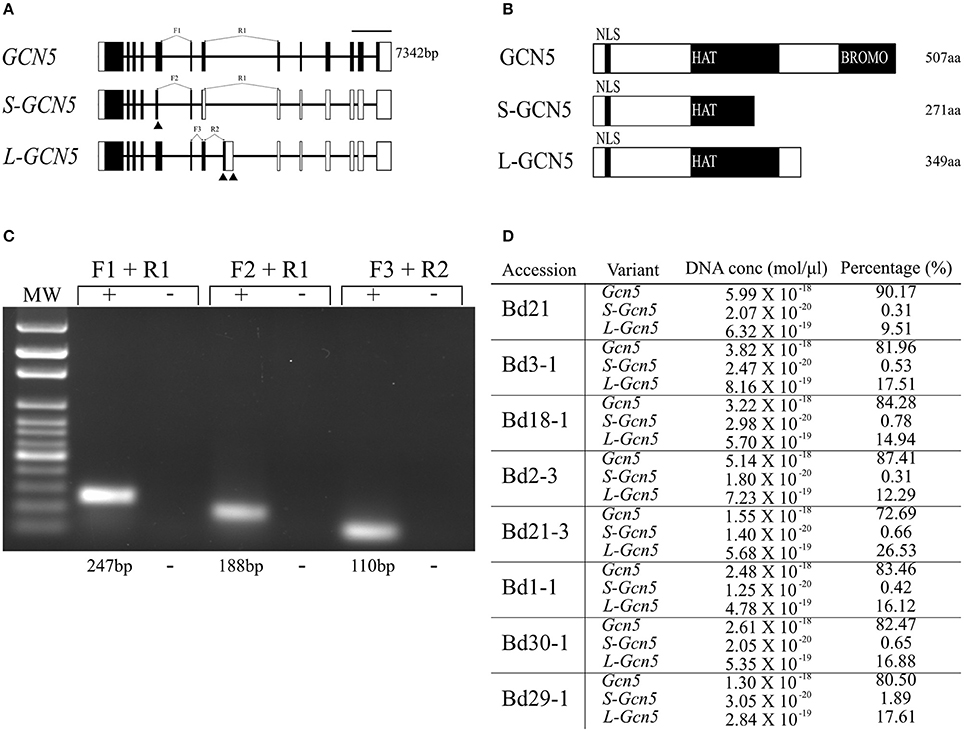
Figure 1. BdGCN5 splice variants differ in their conserved domain composition and absolute transcript abundance. (A) Genomic representation of GCN5 alternative transcripts. Exons are depicted as black boxes, UTRs are depicted as white boxes. Forward (F) and reverse (R) primers used for qPCR are presented at their appropriate locations. Black arrows represent alternative splice sites. Scale bar = 1,000 bps. (B) Predicted protein product representation of BdGCN5 variants. Black boxes represent conserved protein domains. NLS, nuclear localization signal; HAT, histone acetyltransferase domain; and BROMO, Bromodomain. (C) Confirmation of specific detection of BdGCN5 splice variants through qPCR, using the primer pairs depicted in (A), MW, molecular weight marker. (D) Absolute RNA quantification of each BdGCN5 variant is presented as an absolute value in mol/μl of input cDNA (the cDNA reaction contained 75 ng/μl of input RNA from aerial tissue extractions), as well as the percentage of the total BdGCN5 transcript pool in each tested accession.
Multiple sequence alignments and analyses of the predicted domains of the protein isoforms demonstrated that the alternative splicing events altered the conserved domain composition of the variants, when compared to GCN5 (Figure 1B, Supplementary Figure 1). The protein sequence of both splice variants was missing the bromodomain, however the L-GCN5 protein retained an integral HAT domain, while the S-GCN5 protein only retained a part of the HAT domain (Figure 1B). More specifically, of the four motifs that make up the HAT domain, the S-GCN5 protein sequence contained the complete C and D motifs, and part of the A motif, while the B motif was missing (Supplementary Figure 1). Therefore, at least three distinct BdGCN5 alternative transcripts are present in B. distachyon, which differ based on the composition and integrity of their conserved protein domains.
GCN5 Is the Dominant Transcript Isoform
Absolute mRNA quantification was performed to gain insight regarding the abundance of each BdGCN5 alternative transcript in aerial tissues of eight B. distachyon accessions (Bd21, Bd3-1, Bd18-1, Bd2-3, Bd21-3, Bd1-1, Bd30-1, and Bd29-1) known to display natural diversity in many phenotypic characteristics as well as photoperiod and vernalization requirements (Colton-Gagnon et al., 2014; Ream et al., 2014; Tyler et al., 2014). Variant-specific primers were validated and standard curves were generated in order to quantify absolute RNA levels (Figure 1C, Supplementary Figure 2). GCN5 was the dominant transcript, accounting for 73–90% of the entire BdGCN5 transcript pool, followed by L-GCN5 at 9–26% and S-GCN5 at <2% (Figure 1D). Across all accessions, Bd21 had the highest proportion of GCN5 RNA as well as the lowest of both S-GCN5 and L-GCN5, while Bd21-3 had the highest proportion of L-GCN5 and Bd29-1 of S-GCN5. Thus, the quantity that each alternative BdGCN5 transcript accounts for within aerial tissues was variable among accessions of B. distachyon grown under control conditions. However, GCN5 was always dominant, followed by L-GCN5 then S-GCN5.
S-GCN5 Is Degraded through the Nonsense-Mediated Decay Pathway
Alternative splicing that results in transcript variants with early stop codons and long 3′ UTRs may be targets of nonsense-mediated decay (NMD), a ribosome-dependent, splice variant degradation pathway that is involved in actively suppressing transcript abundance levels. Both S-GCN5 and L-GCN5 have such sequence features and were therefore selected for NMD analysis. Since the NMD pathway has yet to be investigated in B. distachyon, sequence homology analysis of known NMD-requiring components from A. thaliana was performed in B. distachyon sequence databases, in order to confirm that this organism contains the appropriate machinery. B. distachyon encodes homologs of all currently known NMD-requiring proteins characterized in A. thaliana, with protein sequence similarities ranging from 56 to 82% (Supplementary Figure 3).
A cycloheximide treatment inhibits the ribosome, and thus also inhibits the NMD pathway. Analysis of cycloheximide-treated B. distachyon aerial tissues indicated that the S-GCN5 transcript is actively degraded by NMD under normal conditions, while the L-GCN5 transcript is not thusly affected (Figures 2B,C). As there are no currently known NMD targets in B. distachyon, a positive control for the effectiveness of the treatment was performed on a previously identified NMD target in A. thaliana, which confirmed the effectiveness of the assay (Figure 2A).
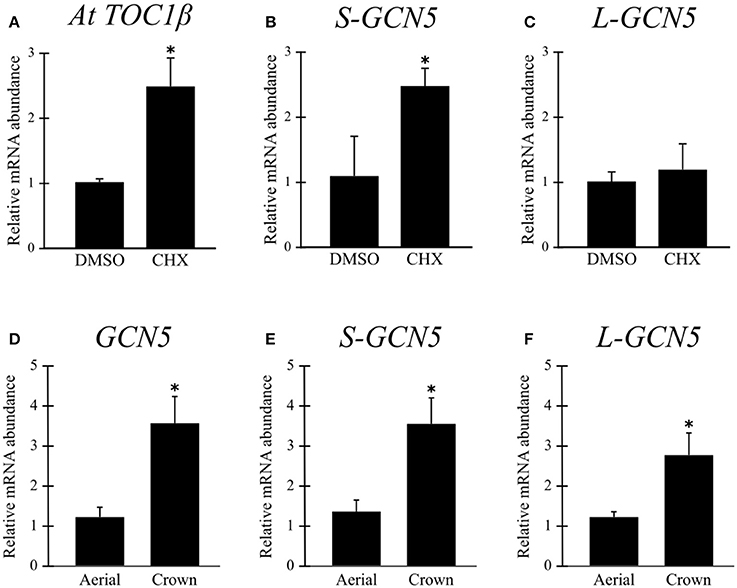
Figure 2. The S-GCN5 transcript is degraded by the nonsense-mediated decay pathway. (A–C) Relative transcript abundance of AtTOC1β (positive control), S-GCN5, and L-GCN5 following cycloheximide (CHX) or control (DMSO) treatments. (D–F) Relative transcript abundance of GCN5, S-GCN5, and L-GCN5 in aerial and crown tissues. Error bars represent the standard deviation of three biological replicates. An asterisk indicates a statistically significant difference (P < 0.05).
BdGCN5 Variants Are Differentially Expressed in Different B. distachyon Tissues and Following Exposure to Various Abiotic Stresses
Relative expression data indicated that mRNA abundance levels of all the alternative BdGCN5 transcripts are significantly higher in crown tissue, when compared to aerial tissue (Figures 2D–F). Exposure of B. distachyon individuals to low (4°C) temperatures demonstrated that the apparent circadian variation in relative transcript abundance of all BdGCN5 variants is repressed, mainly between 6 and 12 h following cold treatment (Figure 3A). In contrast, exposure to high (42°C) temperatures indicated that the abundance of all alternative transcripts was significantly increased following 30 min and 1 h of heat exposure and returns to control levels after 4 h of the applied heat treatment (Figure 3B). All BdGCN5 transcripts followed similar trends across tested treatments and tissues.
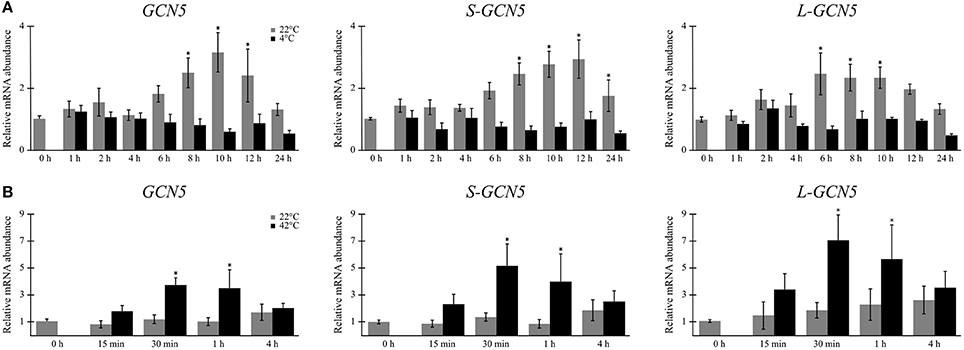
Figure 3. BdGCN5 alternative transcripts are differentially expressed following exposure to temperature extremes. Relative expression profiles of each BdGCN5 alternative transcript in aerial tissues following exposure to: (A) cold (4°C) or (B) heat (42°C) for the indicated amounts of time. Error bars represent the standard deviation of at least three biological replicates. An asterisk indicates a statistically significant difference (P < 0.05).
All BdGCN5 Isoforms Localize to the Nucleus
To perform their function, histone acetyltransferase enzymes and other epigenetic modulators must localize to the nucleus. All BdGCN5 transcript variants retain a single nuclear localization signal (Figure 1B, Supplementary Figure 1B). A localization assay using a N-terminal GFP tag in onion cells was performed to determine the in vivo localization of the predicted protein products. Note that NMD did not affect recombinant protein accumulation as exclusively the coding region was used to generate the constructs. All BdGCN5 variants' predicted protein products localized to the nucleus (Figure 4).
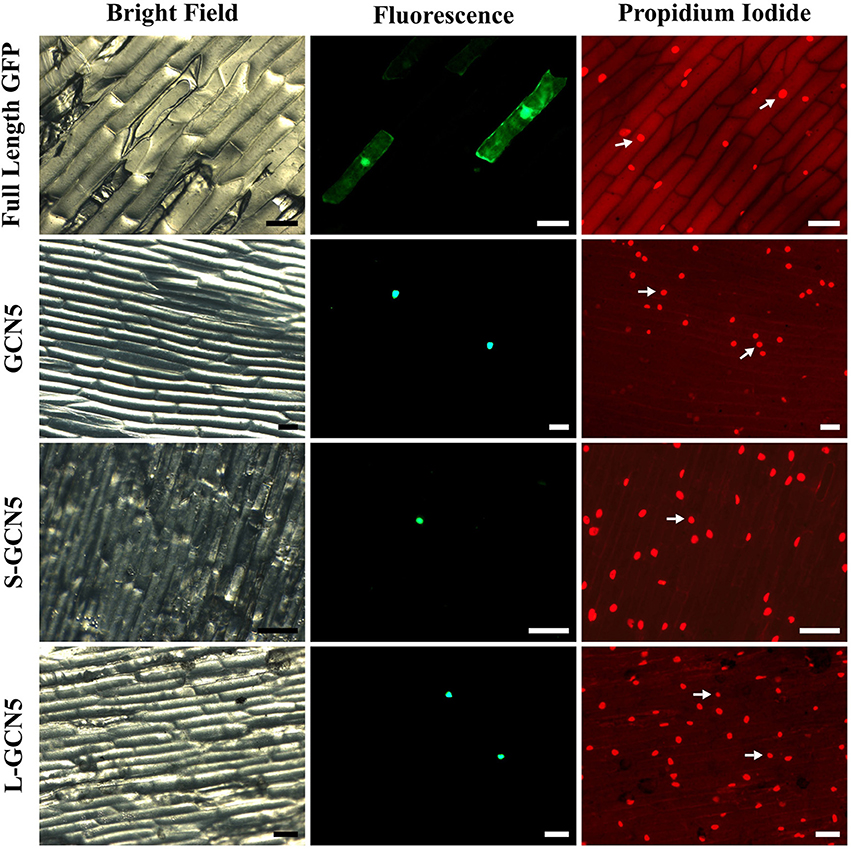
Figure 4. All BdGCN5 isoforms localize to the nucleus. GFP was N-terminally tagged to GCN5, S-GCN5, and L-GCN5 using the pAVA321 localization vector. Untagged GFP, as well as the N-terminally GFP-tagged constructs, were used for particle bombardment of onion scale cells. Images of bright field, fluorescence (GFP filter) and following propidium iodide staining are presented. Arrows indicate nuclei detected under both propidium iodide and GFP filters. Scale bar = 100 μm.
GCN5 Isoforms Differ in Their Ability to Interact with the SAGA Complex Member ADA2
In all organisms in which GCN5 has been characterized, it has always been in the context of multi-protein complexes (Candau and Berger, 1996; Candau et al., 1996, 1997; Stockinger et al., 2001; Bhat et al., 2003; Fan et al., 2004; Mao et al., 2006; Gamper et al., 2009). Therefore, the ability for BdGCN5 variants to interact with ADA2, a transcriptional adaptor that physically links GCN5 to the remainder of the SAGA complex in other organisms, was assessed through a bimolecular fluorescence complementation experiment using the pDOE set of vectors (Gookin and Assmann, 2014). Infiltration into N. benthamiana demonstrated that exclusively GCN5 and L-GCN5 interacted with ADA2 in vivo, while S-GCN5 was incapable of such an interaction (Figure 5). Of note, the main difference between the L-GCN5 and S-GCN5 amino acid sequences is the presence of a complete HAT domain in L-GCN5, whereas S-GCN5's HAT domain is truncated (Figure 1, Supplementary Figure 1B). This information, coupled to the protein interaction results, indicated that a complete HAT domain was required for BdGCN5-ADA2 interaction in B. distachyon. This experiment was repeated with similar results with other pDOE vector constructs that tag the split-YFP sections in differing orientations. No fluorescence was observed when the appropriate controls were tested. The observed GCN5/L-GCN5—ADA2 interaction occurred in the nucleus, which further confirms the subcellular localization of the BdGCN5 protein variants determined in Figure 4. Therefore, GCN5 and L-GCN5 were capable of multi-protein complex formation through direct protein interaction with ADA2, and this interaction required a complete HAT domain.
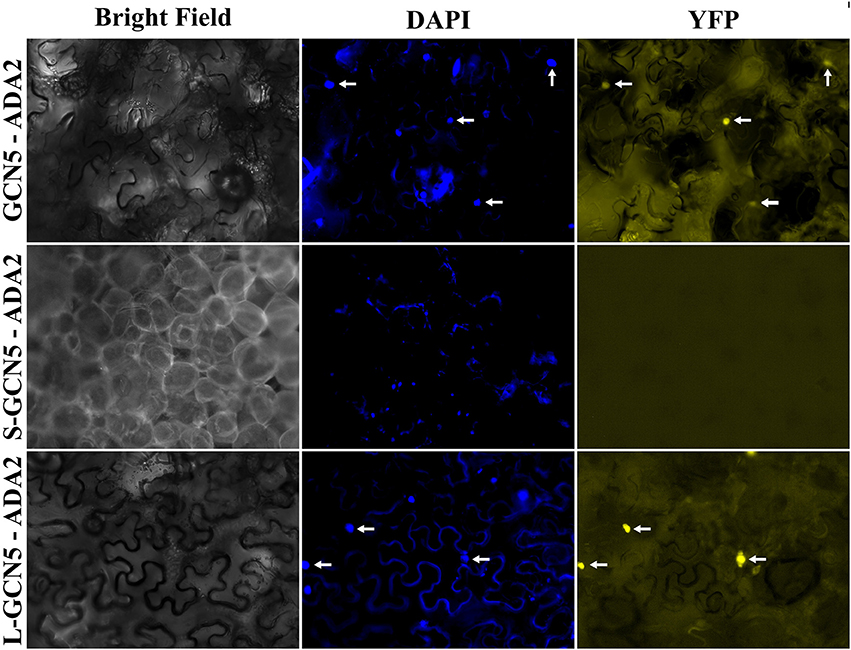
Figure 5. GCN5 and L-GCN5 interact with ADA2 in vivo. Representative images of N. benthamiana epidermal cells infiltrated with A. tumefaciens harboring the pDOE-8 bimolecular fluorescence complementation vector that contained either GCN5, S-GCN5 or L-GCN5, and ADA2. Images of bright field and fluorescence (YFP filter) and following DAPI staining are presented. Arrows indicate nuclei detected under both DAPI and YFP filters.
Discussion
Alterative splicing of a histone demethylase was recently demonstrated to occur in Medicago truncatula, where alternative splice sites result in early stop codons and loss of predicted amino acid sequence length (Shen et al., 2016). While functional differences were not investigated in this study, a subset of the alternative isoforms are favored following cold exposure, suggestive of a role in low temperature responses (Shen et al., 2016). Additionally, alternative splicing in GCN5 has previously been observed in humans, where an isoform representative of the B. distchyon GCN5 sequence exists, as well as an isoform that contains an additional N-terminal extension that is homologous to a region of the human PCAF protein, another histone acetyltransferase (Smith et al., 1998). This additional domain allows for interaction with CBP or p300, two transcriptional co-activators (Yang et al., 1996). The only known GCN5 in D. melanogaster is homologous to this longer human GCN5 sequence, as it contains the N-terminal region of PCAF homology (Smith et al., 1998). Previous cases of alternative splicing in epigenetic modifiers have thus already been demonstrated. Here, alternative GCN5 transcripts identified in B. distachyon are presented. The splicing in these variants show similarities with the histone demethylase identified in M. truncatula, as they result in early stop codons and the loss of conserved protein domains (Figure 1A,B, Supplementary Figure 1B), as opposed to the previously identified GCN5 variants in human that result in the gain of an additional domain.
The three transcriptional BdGCN5 variants all retain an identical N-terminal region (amino acids 1–247; Figure 1B, Supplementary Figure 1). This region contains the only NLS present in the sequences and the nuclear localization of all BdGCN5 isoforms was determined using a GFP-tagging assay in onion cells (Figure 4). This suggests that, regardless of the functional differences among BdGCN5 variants, all isoforms that accumulate to significant levels in B. distachyon perform a cellular function in the context of a nuclear environment, characteristic of epigenetic modifiers.
The region responsible for the enzymatic capability of the BdGCN5 proteins is the HAT domain and is present in its entirety in both the GCN5 and L-GCN5 sequences, but is truncated in the S-GCN5 variant. HAT domains may be further subdivided into four distinct motifs, organized in the following order: C, D, A, and B (Supplementary Figure 1B; Tercero et al., 1992; Neuwald and Landsman, 1997; Dutnall et al., 1998; Wolf et al., 1998; Dyda et al., 2000). The A and B motifs display the highest level of sequence conservation throughout acetyltransferase enzymes, while the C motif displays the lowest, and is dispensable for activity in certain histone acetyltransferases (Dyda et al., 2000; Tyler et al., 2006). This high level of conservation in motifs A and B is most likely due to their essential function in forming an acetyl-CoA binding site and the presence of known catalytic residues (Dyda et al., 2000; Toleman et al., 2004). The motifs missing in the S-GCN5 sequence encompass the entirety of the B motif and a significant section of the A motif. The S-GCN5 protein product is therefore missing regions involved in the binding of an essential catalytic substrate and containing key catalytic residues. This result is not unprecedented, as many other enzymes have alternatively spliced isoforms that lose catalytic function (Kelemen et al., 2013).
The final domain in the BdGCN5 sequences is the bromodomain, which is only present in the GCN5 sequence (Figure 1B, Supplementary Figure 1B). While the bromodomain is not enzymatically active, it may influence the protein's enzymatic capability as its function is to recognize and bind to acetylated lysine residues (Ornaghi et al., 1999; Owen et al., 2000; Sterner and Berger, 2000; Yang, 2004; Hassan et al., 2007; Cieniewicz et al., 2014). Furthermore, promoter binding of the GCN5 protein characterized in other organisms is partially dependent on the presence of an intact bromodomain (Hassan et al., 2002; Benhamed et al., 2008). In A. thaliana, truncation of GCN5's bromodomain resulted in loss of association with 11% of the promoters tested (Benhamed et al., 2008). Moreover, yeast GCN5 proteins lacking bromodomain function displayed increased initial acetylation rates and an altered order of lysine acetylation on histone tails (Cieniewicz et al., 2014). Therefore, the lack of a bromodomain in both of the BdGCN5 splice variants may indicate a diversification of this histone acetyltransferase's function in terms of catalytic activity as well as histone lysine substrate preference.
Absolute RNA quantification of the BdGCN5 alternative transcripts indicate that, across all accessions analyzed, GCN5 is the dominant isoform, followed by L-GCN5, while S-GCN5 contributes little to the total BdGCN5 transcript pool. Additionally, the B. distachyon accessions selected for absolute transcript analysis are known to display phenotypic and stress responsive variability, processes that involve epigenetic mechanisms (He, 2009; Colton-Gagnon et al., 2014; Ream et al., 2014; Tyler et al., 2014; Mayer et al., 2015). While no direct link is hereby suggested, it is interesting to note that there is observable variation in the absolute accumulation of BdGCN5 transcriptional variants across B. distachyon accessions.
The low absolute transcript values of the L-GCN5 and S-GCN5 splice variants are likely due to favoring of the splicing machinery for the GCN5 isoform. Another contributing factor may be the nonsense-mediated decay pathway, which is known to degrade a subset of alternatively spliced transcripts (Lareau et al., 2007; Saltzman et al., 2008; Kwon et al., 2014). The NMD mechanism is conserved across many eukaryotic organisms and targets certain splice variants with early stop codons and long 3′ UTRs, characteristics that both of the BdGCN5 splice variants share (Figure 1A; Conti and Izaurralde, 2005; Drechsel et al., 2013). Cycloheximide assays, which inhibit the ribosome's function and thus NMD, determined that S-GCN5 transcript levels are indeed under the repression of the NMD pathway, while the L-GCN5 transcript is not (Figures 2B,C).
In terms of relative transcript abundance, all BdGCN5 variants follow similar trends, be it across tissues or following exposure to temperature extremes (Figures 2D–F, 3). Therefore, for the tested tissues and conditions, there is no favoring of the splicing machinery for one variant, as has been observed with other alternatively spliced genes in plants (Filichkin et al., 2010; Shen et al., 2016). The higher observed relative transcript abundance of all BdGCN5 variants in crown tissue, when compared to aerial tissues, suggests a putative role in meristem-dependent processes (Figures 2D–F). This observation is not surprising as GCN5 is known to be involved in developmental mechanisms in other plant species (Bertrand et al., 2003; Vlachonasios et al., 2003; Cohen et al., 2009; Kornet and Scheres, 2009; Poulios and Vlachonasios, 2016; Chen et al., 2017). Following exposure to low temperatures, the apparent circadian increase in gene expression is repressed in all BdGCN5 alternative transcripts (Figure 3A). Exposure of plants to cold is known to result in such a profile for certain transcripts that are not cold-inducible (Bieniawska et al., 2008). While relative transcript accumulation levels of the BdGCN5 isoforms is not suggestive of roles in cold stress responses, the recruitment to, and subsequent activity at gene promoters by BdGCN5 protein products may not be ruled out. Notably, A. thaliana GCN5 is known to interact with CBF1, a key transcription factor involved in the cold acclimation process (Stockinger et al., 2001). Furthermore, in both A. thaliana and B. distachyon, the SAGA complex member ADA2 has been shown to interact with CBF1 as well (Stockinger et al., 2001; Demone, 2012). As for high temperature exposure, the observed up-regulation of BdGCN5 transcripts within the first hour of heat stress (Figure 3B) is consistent with observations made of A. thaliana's GCN5, as well as of the role of GCN5 in heat responses in A. thaliana, and more recently in B. distachyon (Hu et al., 2015; Martel, 2017).
ADA2 is a transcriptional co-activator that interacts with GCN5 in all organisms in which this interaction has been assessed, and is also a member of the SAGA complex (Candau and Berger, 1996; Candau et al., 1996, 1997; Stockinger et al., 2001; Bhat et al., 2003; Fan et al., 2004; Mao et al., 2006; Gamper et al., 2009). A. thaliana contains two copies of ADA2 (ADA2a and ADA2b), however B. distachyon retains only one ADA2 sequence in its genome, homologous to A. thaliana's ADA2a (Stockinger et al., 2001; Demone, 2012). Therefore, a protein-protein interaction study was performed in N. benthamiana between B. distachyon ADA2, and BdGCN5 variants in order to determine whether each isoform is capable of multi-protein complex formation. Understanding this interaction is important as incorporation into such complexes would allow for the indirect recruitment of the histone acetyltransferase to additional genomic loci and the expansion of its catalytic capabilities by other SAGA-complex subunits (Belotserkovskaya et al., 2000; Kuo et al., 2000; Brown et al., 2001; Lang et al., 2001; Stockinger et al., 2001; Barbaric et al., 2003; Bhat et al., 2004; Gao et al., 2007; Nagy and Tora, 2007; Bian et al., 2011; Schram et al., 2013; Hirsch et al., 2015; Setiaputra et al., 2015). Exclusively GCN5 and L-GCN5 were capable of interacting with ADA2, indicating that these two isoforms would gain the increased function resulting from the SAGA complex association, whereas S-GCN5 would not (Figure 5). However, the low accumulation levels of the S-GCN5 transcript (Figure 1D), in conjunction with the observation that it is degraded by NMD (Figures 2B,C), puts into question whether any S-GCN5 protein product accumulates in B. distachyon tissues. Previous examples are known to this effect, where splice variants that lose important catalytic domains do not display detectable protein accumulation levels (Schneider et al., 2005). Conversely, there are known examples of enzymes losing catalytic activity, including an acetyltransferase, through alternative splicing patterns, many of which are believed to modulate the level of active protein present in the cell (Kim et al., 2005; Schneider et al., 2005; Kelemen et al., 2013). However, while there is a single example that demonstrates that an NMD targeted transcript may be stabilized in order to be translated into protein, the predominant consensus is that transcripts targeted by NMD may not produce significant levels of protein product (Lareau et al., 2007; Saltzman et al., 2008; Colak et al., 2013; Kwon et al., 2014). Therefore, there are three possible fates for the S-GCN5 transcript: protein product may accumulate and function in the plant, this splice variant's role may be limited to transcriptional regulation, or it may simply be an aberrant transcript for which there is no significant function.
The confirmed interaction between GCN5/L-GCN5 and ADA2, but not between S-GCN5 and ADA2 may shed light upon the differences in higher-level protein organization of plant GCN5-containing complexes, when compared to those of other eukaryotic organisms. In yeast, the region required for the GCN5—ADA2 interaction is that present between the HAT and bromodomain (Candau and Berger, 1996; Candau et al., 1997). This region is missing in L-GCN5, which indicates that the interaction requirement is different in B. distachyon than in yeast. Furthermore, GCN5—ADA2 interaction studies performed with A. thaliana homologs (both ADA2a and ADA2b were assessed) indicated that this region was not required for protein-protein interaction (Mao et al., 2006). This A. thaliana study also demonstrated that the HAT domain was sufficient for GCN5—ADA2(a/b) interaction, which corroborates the B. distachyon interaction results where L-GCN5 associates with ADA2, while S-GCN5 does not (Figure 5; Mao et al., 2006). Taken together, these results suggest that the region necessary for the GCN5-ADA2 interaction in plants, and therefore its conformation, is different from that of yeast.
The above interaction study suggests that the B. distachyon SAGA complex could contain GCN5, L-GCN5, or a combination thereof in its HAT module. Furthermore, the proportion of GCN5-containing complexes would outnumber the L-GCN5 containing complexes if the absolute mRNA quantification data accurately correlates to protein accumulation levels. Therefore, B. distachyon most likely has alternative compositions of the SAGA complex that differ based on, at least, the BdGCN5 variant it harbors in its HAT module. These related complexes may differ significantly in their enzymatic capability and their preferred order of lysine acetylation, as was observed when a non-functional bromodomain was generated in yeast GCN5 (Cieniewicz et al., 2014). In addition, the loss of the bromodomain may significantly alter the sites at which L-GCN5-containing complexes may be recruited to, as was observed when the bromodomain was removed from GCN5 of yeast and A. thaliana (Hassan et al., 2002; Benhamed et al., 2008). If any S-GCN5 protein accumulates to significant levels in B. distachyon, its role would not gain functionality through interaction with the SAGA complex, which would limit its enzymatic capability and substrate specificity. In addition, as the HAT motifs S-GCN5 is missing are those that are most highly conserved and contain catalytic residues, an enzymatic function would be unlikely. However, NMD results indicate that S-GCN5 protein accumulation is unlikely, suggestive of a transcriptional role or S-GCN5 being an aberrant splicing product. Therefore, these findings provide insight regarding the diversification of different BdGCN5 isoforms, a subset of which may act as a part of SAGA-like complexes. Further investigation is required to understand the functional differences that may be displayed among SAGA complexes containing different BdGCN5 isoforms.
Author Contributions
AM and J-BC: designed all experiments. HB: identified the alternative transcripts. AM: performed all experiments and analyses. BM: contributed to the BiFC experiment. AM, BM, and J-BC: prepared the manuscript.
Funding
This work was supported by the Natural Sciences and Engineering Council of Canada (NSERC) Discovery grant 06679 to J-BC. AM was supported by an Alexander-Graham-Bell scholarship from NSERC and a Master's Research Scholarship from the Fonds de Recherche du Québec—Nature et Technologies (FRQNT). BM was supported by a Vanier Canada Graduate Scholarship from NSERC. The authors acknowledge the support of Centre SEVE—FRQNT.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Dr. Youssef Chebli from the Geitmann laboratory at McGill University for help with a subset of the presented microscope images. We also thank Dr. Olivia Wilkins for her thoughtful discussions and contributions regarding this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02176/full#supplementary-material
Supplementary Table 1. Primers used in this study.
Supplementary Figure 1. BdGcn5 nucleotide and amino acid sequence alignment.
Supplementary Figure 2. Standard curves for absolute RNA quantification.
Supplementary Figure 3. B. distachyon nonsense-mediated decay gene homologs.
References
Barbaric, S., Reinke, H., and Hörz, W. (2003). Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23, 3468–3476. doi: 10.1128/MCB.23.10.3468-3476.2003
Belotserkovskaya, R., Sterner, D. E., Deng, M., Sayre, M. H., Lieberman, P. M., and Berger, S. L. (2000). Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20, 634–647. doi: 10.1128/MCB.20.2.634-647.2000
Benhamed, M., Bertrand, C., Servet, C., and Zhou, D. X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for liqht-responsive gene expression. Plant Cell 18, 2893–2903. doi: 10.1105/tpc.106.043489
Benhamed, M., Martin-Magniette, M. L., Taconnat, L., Bitton, F., Servet, C., De Clercq, R., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56, 493–504. doi: 10.1111/j.1365-313X.2008.03606.x
Bertrand, C., Bergounioux, C., Domenichini, S., Delarue, M., and Zhou, D. X. (2003). Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278, 28246–28251. doi: 10.1074/jbc.M302787200
Bhat, R. A., Borst, J. W., Riehl, M., and Thompson, R. D. (2004). Interaction of maize Opaque-2 and the transcriptional co-activators GCN5 and ADA2, in the modulation of transcriptional activity. Plant Mol. Biol. 55, 239–252. doi: 10.1007/s11103-004-0553-z
Bhat, R. A., Riehl, M., Santandrea, G., Velasco, R., Slocombe, S., Donn, G., et al. (2003). Alteration of GCN5 levels in maize reveals dynamic responses to manipulating histone acetylation. Plant J. 33, 455–469. doi: 10.1046/j.1365-313X.2003.01642.x
Bian, C., Xu, C., Ruan, J., Lee, K. K., Burke, T. L., Tempel, W., et al. (2011). Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30, 2829–2842. doi: 10.1038/emboj.2011.193
Bieniawska, Z., Espinoza, C., Schlereth, A., Sulpice, R., Hincha, D. K., and Hannah, M. A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147, 263–279. doi: 10.1104/pp.108.118059
Brown, C. E., Howe, L., Sousa, K., Alley, S. C., Carrozza, M. J., Tan, S., et al. (2001). Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292, 2333–2337. doi: 10.1126/science.1060214
Candau, R., and Berger, S. L. (1996). Structural and functional analysis of yeast putative adaptors: evidence for an adaptor complex in vivo. J. Biol. Chem. 271, 5237–5245. doi: 10.1074/jbc.271.9.5237
Candau, R., Moore, P. A., Wang, L., Barlev, N., Ying, C. Y., Rosen, C. A., et al. (1996). Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16, 593–602. doi: 10.1128/MCB.16.2.593
Candau, R., Zhou, J., Allis, C. D., and Berger, S. L. (1997). Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16, 555–565. doi: 10.1093/emboj/16.3.555
Chen, F., Fu, B., Pan, Y., Zhang, C., Wen, H., Weng, Y., et al. (2017). Fine mapping identifies CsGCN5 encoding a histone acetyltransferase as putative candidate gene for tendril-less1 mutation (td-1) in cucumber. Theor. Appl. Genet. 130, 1549–1558. doi: 10.1007/s00122-017-2909-1
Cieniewicz, A. M., Moreland, L., Ringel, A. E., MacKintosh, S. G., Raman, A., Gilbert, T. M., et al. (2014). The bromodomain of gcn5 regulates site specificity of lysine acetylation on histone H3. Mol. Cell. Proteomics 13, 2896–2910. doi: 10.1074/mcp.M114.038174
Cohen, R., Schocken, J., Kaldis, A., Vlachonasios, K. E., Hark, A. T., and McCain, E. R. (2009). The histone acetyltransferase GCN5 affects the inflorescence meristem and stamen development in Arabidopsis. Planta 230, 1207–1221. doi: 10.1007/s00425-009-1012-5
Colak, D., Ji, S. J., Porse, B. T., and Jaffrey, S. R. (2013). Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell 153, X1252–X1265. doi: 10.1016/j.cell.2013.04.056
Colton-Gagnon, K., Ali-Benali, M. A., Mayer, B. F., Dionne, R., Bertrand, A., Do Carmo, S., et al. (2014). Comparative analysis of the cold acclimation and freezing tolerance capacities of seven diploid Brachypodium distachyon accessions. Ann. Bot. 113, 681–693. doi: 10.1093/aob/mct283
Conti, E., and Izaurralde, E. (2005). Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17, 316–325. doi: 10.1016/j.ceb.2005.04.005
Demone, J. (2012). Characterizing the Role of the Transcriptional Adaptor ADA2: An Integrating Node in the Cold Response Mechanism of Brachypodium distachyon. MSc, McGill University.
Drechsel, G., Kahles, A., Kesarwani, A. K., Stauffer, E., Behr, J., Drewe, P., et al. (2013). Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25, 3726–3742. doi: 10.1105/tpc.113.115485
Dutnall, R. N., Tafrov, S. T., Sternglanz, R., and Ramakrishnan, V. (1998). Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell 94, 427–438. doi: 10.1016/S0092-8674(00)81584-6
Dyda, F., Klein, D. C., and Hickman, A. B. (2000). GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29, 81–103. doi: 10.1146/annurev.biophys.29.1.81
Earley, K. W., Shook, M. S., Brower-Toland, B., Hicks, L., and Pikaard, C. S. (2007). In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J. 52, 615–626. doi: 10.1111/j.1365-313X.2007.03264.x
Fan, Q., An, L., and Cui, L. (2004). PfADA2, a Plasmodium falciparum homologue of the transcriptional coactivator ADA2 and its in vivo association with the histone acetyltransferase PfGCN5. Gene 336, 251–261. doi: 10.1016/j.gene.2004.04.005
Filichkin, S. A., Priest, H. D., Givan, S. A., Shen, R., Bryant, D. W., Fox, S. E., et al. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58. doi: 10.1101/gr.093302.109
Gamper, A. M., Kim, J., and Roeder, R. G. (2009). The STAGA subunit ADA2b Is an important regulator of human GCN5 catalysis. Mol. Cell. Biol. 29, 266–280. doi: 10.1128/MCB.00315-08
Gao, M. J., Hegedus, D. D., Sharpe, A. G., Robinson, S. J., Lydiate, D. J., and Hannoufa, A. (2007). Isolation and characterization of a GCN5-interacting protein from Arabidopsis thaliana. Planta 225, 1367–1379. doi: 10.1007/s00425-006-0446-2
Gookin, T. E., and Assmann, S. M. (2014). Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J. 80, 553–567. doi: 10.1111/tpj.12639
Grant, P. A., Duggan, L., Côté, J., Roberts, S. M., Brownell, J. E., Candau, R., et al. (1997). Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an ada complex and the saga (spt/ada) complex. Gene Dev. 11, 1640–1650. doi: 10.1101/gad.11.13.1640
Grant, P. A., Eberharter, A., John, S., Cook, R. G., Turner, B. M., and Workman, J. L. (1999). Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895–5900. doi: 10.1074/jbc.274.9.5895
Hassan, A. H., Awad, S., Al-Natour, Z., Othman, S., Mustafa, F., and Rizvi, T. A. (2007). Selective recognition of acetylated histones by bromodomains in transcriptional co-activators. Biochem. J. 402, 125–133. doi: 10.1042/BJ20060907
Hassan, A. H., Prochasson, P., Neely, K. E., Galasinski, S. C., Chandy, M., Carrozza, M. J., et al. (2002). Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111, 369–379. doi: 10.1016/S0092-8674(02)01005-X
He, Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant 2, 554–564. doi: 10.1093/mp/ssp005
He, Y., Michaels, S. D., and Amasino, R. M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302, 1751–1754. doi: 10.1126/science.1091109
Hirsch, C. L., Akdemir, Z. C., Wang, L., Jayakumaran, G., Trcka, D., Weiss, A., et al. (2015). Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Gene Dev. 29, 803–816. doi: 10.1101/gad.255109.114
Hong, S. Y., Seo, P. J., Yang, M. S., Xiang, F., and Park, C. M. (2008). Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8:112. doi: 10.1186/1471-2229-8-112
Hu, Z., Song, N., Zheng, M., Liu, X., Liu, Z., Xing, J., et al. (2015). Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J. 84, 1178–1191. doi: 10.1111/tpj.13076
Kelemen, O., Convertini, P., Zhang, Z., Wen, Y., Shen, M., Falaleeva, M., et al. (2013). Function of alternative splicing. Gene 514, 1–30. doi: 10.1016/j.gene.2012.07.083
Kim, K., Ryu, J. H., Park, J. W., Kim, M. S., and Chun, Y. S. (2005). Induction of a SSAT isoform in response to hypoxia or iron deficieny and its protective effects on cell death. Biochem. Biophys. Res. Commun. 331, 78–85. doi: 10.1016/j.bbrc.2005.03.121
Kornet, N., and Scheres, B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21, 1070–1079. doi: 10.1105/tpc.108.065300
Koutelou, E., Hirsch, C. L., and Dent, S. Y. R. (2010). Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 22, 374–382. doi: 10.1016/j.ceb.2010.03.005
Kuo, M. H., Brownell, J. E., Sobel, R. E., Ranalli, T. A., Cook, R. G., Edmondson, D. G., et al. (1996). Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272. doi: 10.1038/383269a0
Kuo, M. H., vom Baur, E., Struhl, K., and Allis, C. D. (2000). Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6, 1309–1320. doi: 10.1016/S1097-2765(00)00129-5
Kuo, Y. M., and Andrews, A. J. (2013). Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PLoS ONE 8:e54896. doi: 10.1371/annotation/b2bf9c2e-90a9-4228-9b38-2f1bc977a437
Kwon, Y. J., Park, M. J., Kim, S. G., Baldwin, I. T., and Park, C. M. (2014). Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 14:136. doi: 10.1186/1471-2229-14-136
Lang, S. E., McMahon, S. B., Cole, M. D., and Hearing, P. (2001). E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 276, 32627–32634. doi: 10.1074/jbc.M102067200
Lareau, L. F., Brooks, A. N., Soergel, D. A., Meng, Q., and Brenner, S. E. (2007). The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv. Exp. Med. Biol. 623, 190–211. doi: 10.1007/978-0-387-77374-2_12
Mahrez, W., Arellano, M. S. T., Moreno-Romero, J., Nakamura, M., Shu, H., Nanni, P., et al. (2016). H3K36ac is an evolutionary conserved plant histone modification that marks active genes. Plant Physiol. 170, 1566–1577. doi: 10.1104/pp.15.01744
Mao, Y., Pavangadkar, K. A., Thomashow, M. F., and Triezenberg, S. J. (2006). Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim. Biophys. Acta Gene Struct. Exp. 1759, 69–79. doi: 10.1016/j.bbaexp.2006.02.006
Marquez, Y., Brown, J. W. S., Simpson, C., Barta, A., and Kalyna, M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22, 1184–1195. doi: 10.1101/gr.134106.111
Martel, A. (2017). Functional Characterization of Gcn5 Splice Variants and Elucidation of the Role of Histone Acetylation in Brachypodium distachyon Temperature Stress Responses. MSc, McGill University.
Mayer, B. F., Ali-Benali, M. A., Demone, J., Bertrand, A., and Charron, J. B. (2015). Cold acclimation induces distinctive changes in the chromatin state and transcript levels of COR genes in Cannabis sativa varieties with contrasting cold acclimation capacities. Physiol. Plant. 155, 281–295. doi: 10.1111/ppl.12318
Moraga, F., and Aquea, F. (2015). Composition of the SAGA complex in plants and its role in controlling gene expression in response to abiotic stresses. Front. Plant Sci. 6:865. doi: 10.3389/fpls.2015.00865
Nagy, Z., and Tora, L. (2007). Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26, 5341–5357. doi: 10.1038/sj.onc.1210604
Neuwald, A. F., and Landsman, D. (1997). GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22, 154–155. doi: 10.1016/S0968-0004(97)01034-7
Ornaghi, P., Ballario, P., Lena, A. M., Gonzàlez, A., and Filetici, P. (1999). The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J. Mol. Biol. 287, 1–7. doi: 10.1006/jmbi.1999.2577
Owen, D. J., Ornaghi, P., Yang, J. C., Lowe, N., Evans, P. R., Ballario, P., et al. (2000). The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 19, 6141–6149. doi: 10.1093/emboj/19.22.6141
Poulios, S., and Vlachonasios, K. E. (2016). Synergistic action of histone acetyltransferase GCN5 and receptor CLAVATA1 negatively affects ethylene responses in Arabidopsis thaliana. J. Exp. Bot. 67, 905–918. doi: 10.1093/jxb/erv503
Ream, R. S., Woods, D. P., Schwartz, C. J., Sanabria, C. P., Mahoy, J. A., Walters, E. M., et al. (2014). Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol. 164, 694–709. doi: 10.1104/pp.113.232678
Ringel, A. E., Cieniewicz, A. M., Taverna, S. D., and Wolberger, C. (2015). Nucleosome competition reveals processive acetylation by the SAGA HAT module. Proc. Natl. Acad. Sci. U.S.A. 112, E5461–E5470. doi: 10.1073/pnas.1508449112
Robert, F., Pokholok, D. K., Hannett, N. M., Rinaldi, N. J., Chandy, M., Rolfe, A., et al. (2004). Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16, 199–209. doi: 10.1016/j.molcel.2004.09.021
Rosaleny, L. E., Ruiz-García, A. B., García-Martínez, J., Pérez-Ortín, J. E., and Tordera, V. (2007). The Sas3p and Gcn5p histone acetyltransferases are recruited to similar genes. Genome Biol. 8:R119. doi: 10.1186/gb-2007-8-6-r119
Saltzman, A. L., Yoon, K. K., Pan, Q., Fagnani, M. M., Maquat, L. E., and Blencowe, B. J. (2008). Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 28, 4320–4330. doi: 10.1128/MCB.00361-08
Schneider, C., Boeglin, W. E., and Brash, A. R. (2005). Human cyclo-oxygenase-1 and an alternative splice variant: contrasts in expression of mRNA, protein and catalytic activities. Biochem. J. 385, 57–64. doi: 10.1042/BJ20041115
Schram, A. W., Baas, R., Jansen, P. W., Riss, A., Tora, L., Vermeulen, M., et al. (2013). A dual role for SAGA-associated factor 29 (SGF29) in ER stress survival by coordination of both histone H3 acetylation and histone H3 Lysine-4 trimethylation. PLoS ONE 8:e70035. doi: 10.1371/journal.pone.0070035
Setiaputra, D., Ross, J. D., Lu, S., Cheng, D. T., Dong, M. Q., and Yip, C. K. (2015). Conformational flexibility and subunit arrangement of the modular yeast Spt-Ada-Gcn5 acetyltransferase complex. J. Biol. Chem. 290, 10057–10070. doi: 10.1074/jbc.M114.624684
Shen, Y., Wu, X., Liu, D., Song, S., Liu, D., and Wang, H. (2016). Cold-dependent alternative splicing of a Jumonji C domain-containing gene MtJMJC5 in Medicago truncatula. Biochem. Biophys. Res. Commun. 474, 271–276. doi: 10.1016/j.bbrc.2016.04.062
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Smith, E. R., Belote, J. M., Schiltz, R. L., Yang, X. J., Moore, P. A., Berger, S. L., et al. (1998). Cloning of drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 26, 2948–2954. doi: 10.1093/nar/26.12.2948
Sterner, D. E., and Berger, S. L. (2000). Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459. doi: 10.1128/MMBR.64.2.435-459.2000
Stockinger, E. J., Mao, Y., Regier, M. K., Triezenberg, S. J., and Thomashow, M. F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29, 1524–1533. doi: 10.1093/nar/29.7.1524
Tercero, J. C., Riles, L. E., and Wickner, R. B. (1992). Localized mutagenesis and evidence for post-transcriptional regulation of MAK3: a putative N-acetyltransferase required for double-stranded RNA virus propagation in Saccharomyces cerevisiae. J. Biol. Chem. 267, 20270–20276.
Toleman, C., Paterson, A. J., Whisenhunt, T. R., and Kudlow, J. E. (2004). Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J. Biol. Chem. 279, 53665–53673. doi: 10.1074/jbc.M410406200
Tyler, L., Fangel, J. U., Fagerström, A. D., Steinwand, M. A., Raab, T. K., Willats, W. G. T., et al. (2014). Selection and phenotypic characterization of a core collection of Brachypodium distachyon inbred lines. BMC Plant Biol. 14:25. doi: 10.1186/1471-2229-14-25
Tyler, R. C., Bitto, E., Berndsen, C. E., Bingman, C. A., Singh, S., Lee, M. S., et al. (2006). Structure of Arabidopsis thaliana At1g77540 protein, a minimal acetyltransferase from the COG2388 family. Biochemistry 45, 14325–14336. doi: 10.1021/bi0612059
Vain, P., Keen, N., Murillo, J., Rathus, C., Nemes, C., and Finer, J. J. (1993). Development of the particle inflow gun. Plant Cell Tiss. Org. Cult. 33, 237–246. doi: 10.1007/BF02319007
Vlachonasios, K. E., Thomashow, M. F., and Triezenberg, S. J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15, 626–638. doi: 10.1105/tpc.007922
von Arnim, A. G., Deng, X. W., and Stacey, M. G. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. doi: 10.1016/S0378-1119(98)00433-8
Wolf, E., Vassilev, A., Makino, Y., Sali, A., Nakatani, Y., and Burley, S. K. (1998). Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell 94, 439–449. doi: 10.1016/S0092-8674(00)81585-8
Yang, X. J. (2004). Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26, 1076–1087. doi: 10.1002/bies.20104
Yang, X.-J., Ogryzko, V. V., Nishikawa, J.-I., Howard, B. H., and Nakatani, Y. (1996). A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. 382, 319–324. doi: 10.1038/382319a0
Keywords: alternative splicing, histone acetylation, GCN5, SAGA, Brachypodium distachyon, functional diversification
Citation: Martel A, Brar H, Mayer BF and Charron J-B (2017) Diversification of the Histone Acetyltransferase GCN5 through Alternative Splicing in Brachypodium distachyon. Front. Plant Sci. 8:2176. doi: 10.3389/fpls.2017.02176
Received: 28 September 2017; Accepted: 12 December 2017;
Published: 21 December 2017.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Konstantinos Vlachonasios, Aristotle University of Thessaloniki, GreeceShih-Long Tu, Institute of Plant and Microbial Biology, Academia Sinica, Taiwan
Copyright © 2017 Martel, Brar, Mayer and Charron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Benoit Charron, jean-benoit.charron@mcgill.ca
 Alexandre Martel
Alexandre Martel Jean-Benoit Charron
Jean-Benoit Charron