- 1National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Jiamusi Branch of Heilongjiang Academy of Agricultural Sciences, Jiamusi, China
Phytophthora root rot (PRR) caused by Phytophthora sojae is a major soybean disease that causes severe economic losses worldwide. Using soybean cultivars carrying a Rps resistance gene is the most effective strategy for controlling this disease. We previously detected a novel Phytophthora resistance gene, RpsZS18, on chromosome 2 of the soybean cultivar Zaoshu18. The aim of the present study was to identify and finely map RpsZS18. We used 232 F2:3 families generated from a cross between Zaoshu18 (resistant) and Williams (susceptible) as the mapping population. Simple sequence repeat (SSR) markers distributed on chromosome 2 were used to map RpsZS18. First, 12 SSR markers linked with RpsZS18 were identified by linkage analyses, including two newly developed SSR markers, ZCSSR33 and ZCSSR46, that flanked the gene at distances of 0.9 and 0.5 cM, respectively. Second, PCR-based InDel markers were developed based on sequence differences between the two parents and used to further narrow down the mapping region of RpsZS18 to 71.3 kb. Third, haplotype analyses were carried out in the RpsZS18 region using 14 soybean genotypes with whole-genome resequencing. We detected six genes with unique haplotype sequences in Zaoshu18. Finally, quantitative real-time PCR assays of the six genes revealed an EF-hand calcium-binding domain containing protein encoding gene (Glyma.02g245700), a pfkB carbohydrate kinase encoding gene (Glyma.02g245800), and a gene with no functional annotation (Glyma.02g246300), are putative candidate PRR resistance genes. This study provides useful information for breeding P. sojae-resistant soybean cultivars.
Introduction
Phytophthora root rot (PRR), caused by the soil-borne oomycete Phytophthora sojae (Kanfman and Gerdemann), is a devastating disease in most soybean-growing regions. P. sojae can infect soybean plants at all growth stages under saturated soil conditions (Schmitthenner, 1985). PRR usually causes yield losses of 30–50% under favorable conditions, but during severe epiphytotic outbreaks infections can result in 100% yield loss (Li and Ma, 1999; Zhang et al., 2010). In China, PRR was first detected in Heilongjiang Province in 1989, and now has spread to most soybean-producing areas (Zhu et al., 2003; Chen and Wang, 2017). The use of resistant cultivars is the most economical and environmentally safe method to control this disease.
Two types of resistance to P. sojae have been identified in soybean, namely complete and partial resistance (Sugimoto et al., 2012). Complete resistance is race-specific, and is the result of a single dominant resistance gene (Rps) that confers immunity or near immunity. Partial resistance is controlled by major and minor genes and limits pathogen colonization and spread (Tooley and Grau, 1984; Dorrance et al., 2003b, 2004; Sugimoto et al., 2012). Currently, the best method to control PRR involves growing soybean cultivars with complete resistance because partial resistance to P. sojae is ineffective under conditions of high disease pressure (Schmitthenner, 1999; Dorrance et al., 2003b). The first Rps gene (Rps1a) was identified in soybean in the 1950s (Bernard et al., 1957), and since then, more than 20 Rps genes have been identified and mapped to nine chromosomes, namely chromosomes 2, 3, 7, 10, 13, 14, and 16–19 (Sugimoto et al., 2012; Lin et al., 2013; Zhang et al., 2013a,b; Sun et al., 2014; Ping et al., 2015; Li et al., 2016; Cheng et al., 2017; Li Y. et al., 2017; Niu et al., 2017; Qin et al., 2017; Sahoo et al., 2017; Zhong et al., 2017). Some soybean cultivars and lines carrying Rps genes, including Rps1a, Rps1c, and Rps1k, have been used widely in commercial breeding programs to reduce yield losses caused by PRR (Dorrance and Schmitthenner, 2000; Sugimoto et al., 2012). Among the identified Rps genes, only Rps1k has been cloned and characterized. Rps1k was mapped in a 125-kb region through the construction of high resolution genetic and physical maps (Kasuga et al., 1997). A gene family encoding a nucleotide binding site-leucine-rich repeat (NBS-LRR) structure has been isolated from the region of Rps1k by screening bacterial artificial chromosome (BAC) libraries, and two NBS-LRR-encoding genes (Rps1k-1 and Rps1k-2) were cloned (Gao et al., 2005; Gao and Bhattacharyya, 2008). However, the exact physical location of Rps1k remains unknown in the soybean Williams 82 (carries Rps1k) reference genome (Lin et al., 2013; Sun et al., 2014; Li et al., 2016), and this has also hindered studies of the mechanism of soybean resistance to P. sojae.
Previous studies have described a typical gene-for-gene interaction between soybean and P. sojae. More than 200 P. sojae physiological races or pathotypes have been identified since 1955 (Schmitthenner et al., 1994; Ryley et al., 1998; Dorrance et al., 2003a; Sugimoto et al., 2012; Stewart et al., 2014, 2015; Xue et al., 2015). In China, there is considerable diversity in the virulence of isolates from different regions, especially those from the Huanghe-Huaihe and Yangtze basins (Zhu et al., 2003). Among the known Rps genes, only Rps1c, Rps1k, and RpsYD25 can control PRR in the major soybean-producing regions of China (Zhu et al., 2003; Fan et al., 2009; Zhang et al., 2010; Li Y. et al., 2017). However, no Rps gene that confers resistance to all P. sojae races or pathotypes has been found in China to date (Zhu et al., 2003; Cui et al., 2010; Zhang et al., 2010, 2014). Therefore, it is imperative that novel Rps genes effective against new P. sojae races or pathotypes are identified and incorporated into commercial cultivars. Developing markers closely linked to Rps genes is an efficient way to use marker-assisted selection to breed cultivars with stable and durable resistance to P. sojae.
The sequencing and assembly of the soybean Williams 82 reference genome and the refinement of gene annotations have facilitated the development of more molecular markers and the discovery of candidate Rps genes (Grant et al., 2009; Schmutz et al., 2010; Zhang et al., 2013a,b; Li et al., 2016; Song et al., 2016; Cheng et al., 2017; Li Y. et al., 2017). High-density simple sequence repeat (SSR) markers were developed based on the Williams 82 reference genome by Song et al. (2010), and since then, many Rps genes has been identified and finely mapped including RpsJS, Rps11, and RpsHN (Lin et al., 2013; Sun et al., 2014; Ping et al., 2015; Niu et al., 2017). Based on the published Williams 82 genomic sequence information, new SSR markers have been developed to build higher-resolution linkage maps, and novel Rps genes like RpsYD29 and Rps10 have been identified (Zhang et al., 2013a,b). In recent years, the reduced costs of sequencing and application of bioinformatic analysis based on next-generation sequencing, have allowed single nucleotide polymorphism (SNP) and insertion/deletion (InDel) markers to be identified and applied to the mapping population (Li et al., 2016; Li Y. et al., 2017).
Zaoshu18 is an elite soybean cultivar, which was grown in the Huanghuaihai region of China during the 1980s. In previous studies, we found that Zaoshu18 exhibited excellent resistance to PRR (Zhu et al., 2006; Chen et al., 2008; Zhang et al., 2014). Our initial study revealed that Zaoshu18 resistance to P. sojae was controlled by a single dominant gene, RpsZS18, which preliminarily mapped on chromosome 2 (Yao et al., 2010). However, because of the lack of polymorphic markers and genomic sequence information, only a few SSR markers with distant genetic distances were found, so an accurate genetic linkage map could not be constructed. Further, in the wide mapping interval of the previous map (Yao et al., 2010), none of the typical NBS-LRR-encoding disease resistance genes were found. Therefore, a novel Rps gene structure with a different P. sojae resistance mechanism may exist on soybean chromosome 2. The objectives of this study were (1) to develop markers closely linked to RpsZS18 conferring PRR resistance, (2) to finely map RpsZS18 on chromosome 2, and (3) to analyze RpsZS18 candidate gene(s) based on whole-genome resequencing of 10 soybean genotypes, including two contrasting parents.
Materials and Methods
Plant Materials and P. sojae Isolates
Phenotyping was performed for Zaoshu18 and 20 differential cultivars containing a single Rps gene. Four cultivars, Williams, Zhonghuang13, Zhonghuang47, and Jikedou2, were used as susceptible controls. The F1 soybean seeds used in this study were produced from a cross between the susceptible cultivar Williams and the resistant cultivar Zaoshu18, which carries the PRR resistance gene RpsZS18. The F1 plants were self-pollinated to produce an F2 population consisting of 232 individuals. Each F2 plant was self-crossed and threshed individually to generate seeds for 232 families, which were used for phenotypic and genotypic evaluations.
The phenotype test was conducted using a total of 14 P. sojae isolates with different pathotypes. All the different isolates were cultured on V8 juice agar medium. The P. sojae isolates PsFJ2, PsFJ3, Ps41-1, and PsUSAR2 were used for phenotypic evaluations of the parents and the F2:3 population.
Evaluation of PRR in the F2:3 Population
We planted 15–20 seeds of each cultivar in 10-cm diameter paper cups, with 14 pots for each cultivar. After 12 days, the seedlings were inoculated with 14 P. sojae isolates, one for each pot, using a modified hypocotyl-inoculation technique described by Zhang et al. (2013a). Briefly, a 5-mm incision was made in the hypocotyls, into which the slurry of P. sojae inoculum, made from a 7-day-old V8 juice agar culture, was inserted. Inoculated seedlings were placed in a mist chamber at 24°C with 100% relative humidity for 2 days. The plants were then moved to a greenhouse and grown at 23–26°C. After a 4-day incubation, the numbers of dead seedlings were recorded. Cultivars were considered resistant if all the seedlings were alive with no expanded lesions. Cultivars were considered susceptible if all the seedlings were dead.
For phenotypic evaluation of the F2:3 population, we planted 25–30 seeds from each of the 232 families in 10-cm diameter paper pots, with two pots per family. The parents were planted separately as controls. After a 4-day incubation, the numbers of dead seedlings were recorded. Families with 0–20, 80–100, and 21–79% dead seedlings were considered homozygous resistant, homozygous susceptible, and segregating, respectively (Gordon et al., 2006; Lin et al., 2013; Zhang et al., 2013a,b; Ping et al., 2015). All cultivars, parents, and families were tested twice. If the results were inconsistent in the two tests, a third test was conducted to confirm the accuracy of the phenotype.
SSR Marker Development and Analyses
Equal amounts of leaf tissue from 25 to 30 seedlings of each family and parental cultivars were collected and pooled together before inoculation. Each of 25–30 individual plants from one family was ensured to be harvested leaf tissue. Genomic DNA of each family was extracted using a Plant Genomic DNA Kit (Tiangen, Beijing, China).
To map the RpsZS18 gene, 28 published SSR markers from SoyBase (https://www.soybase.org/) and 154 new SSR markers were used to screen for polymorphisms between parents according to the rough mapping interval of Yao et al. (2010). The 154 new SSR markers were designed based on the chromosome 2 sequence of Williams82 (Glycine max V2.0) in SoyBase (https://www.soybase.org). Novel SSR motifs were screened using the SSR Hunter 1.3 program, and SSR primers were designed with Primer Premier 5.0. All SSR primer pairs were synthesized by Sangon Biotech (Beijing, China). Subsequently, polymorphic SSR markers were used to analyze the genotypes of the 232 F2:3 families.
Each polymerase chain reaction (PCR) was completed in a 10-μl sample consisting of 30 ng genomic DNA, 5 μl 2 × Taq PCR MasterMix (Tiangen), and each primer (0.2 μM). The PCRs were conducted in a thermal cycler (Biometra, Gottingen, Germany) using the following program: 95°C for 3 min; 35 cycles of 95°C for 45 s, 52–58°C for 45 s, and 72°C for 45 s; 72°C for 7 min. Samples were cooled to 10°C. The PCR products were mixed with 2 μl 6 × loading buffer (0.25% bromophenol blue, 25% xylene cyanol FF, and 40% sugar) and separated in 8% non-denaturing polyacrylamide gels.
Data Analyses and Linkage Map Construction
The segregation of phenotypes and molecular markers in the F2:3 population was evaluated for goodness-of-fit to Mendelian segregation ratios using the Chi-squared test. Linkage analyses were completed using MAPMAKER/EXP version 3.0 (Lincoln et al., 1993). Genetic distances were calculated according to the Kosambi mapping function (Kosambi, 1943). Linkage groups were determined using a log-likelihood threshold of 3.0. The genetic linkage map of molecular markers linked to RpsZS18 was prepared using MapDraw (Liu and Meng, 2003).
Whole-Genome Resequencing of Soybean Cultivars
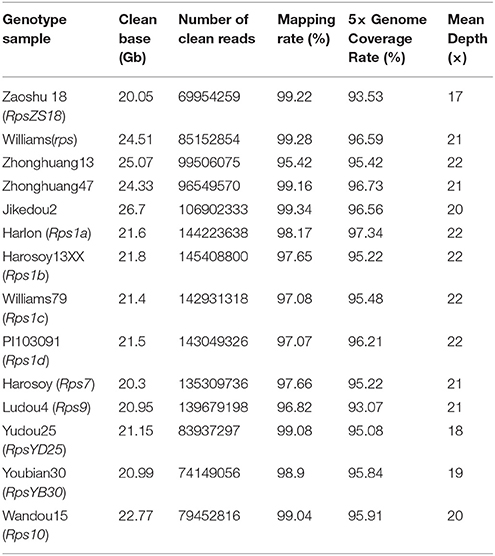
We performed whole-genome resequencing (WGRS) of the two parent cultivars, Zaoshu18 and Williams, and 12 different soybean cultivars with or without a known Rps gene. Genomic DNA was extracted from the 14 cultivars to construct Illumina libraries, which were sequenced by Biomarker Technologies and Annoroad Gene Technology (Beijing, China). The raw 150 paired-end reads data were filtered to produce clean reads as follows: (1) removal of reads with adapters, reads containing more than 15% bases with a Phred quality score <19, and reads with more than 5% undetermined bases; (2) alignment of filtered clean reads to the Williams82 reference genome (Glycine max V2.0) using the genome-alignment-software BWA; and (3) calculation of mapping rate, 5× genome coverage, and mean depth (Li and Durbin, 2009).
SNPs and InDels were detected using the SNP analyses software GATK (McKenna et al., 2010). ANNOVAR software and the existing genomic annotation file (gff/gtf) were used to annotate the SNPs (Wang et al., 2010). DELLY (Rausch et al., 2012) was used for the analysis of chromosome structure variation (SV). All potential SV sites were detected and filtered for genomewide testing. By comparing the reads with the reference genome, we identified all variants, including SNPs, InDels, and SVs, among the samples.
Fine Mapping with InDels
Based on the genetic linkage map constructed with SSR markers, the physical region of RpsZS18 was determined according to the reference genome sequence of Williams 82 (Glycine max V2.0). InDels in the RpsZS18 mapping region were identified between the parental cultivars Williams and Zaoshu18. PCR-based InDel markers were developed in the mapping region of RpsZS18 using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). F2:3 families and parental cultivars were genotyped based on the InDel markers. PCR reactions were conducted as described above for the SSR markers. New recombination sites in the F2:3 population were discovered using the InDel markers, and the candidate region for RpsZS18 was shortened.
Resistance-Related Variation Identification and Candidate Gene Prediction
To identify the RpsZS18 disease resistance candidate gene, the Homozygous SNPs, InDels and high confident SVs in the physical mapping interval of RpsZS18 corresponding to the 14 different soybean cultivars with whole-genome sequences were selected for haplotype analyses. The identified variants were used to develop reference-based assembly of corresponding RpsZS18 allelic regions among the 14 sequenced soybean cultivars by substituting the bases with confident variant calls in the reference genome (Singh et al., 2016a,b). Haplotype sequences of the 15 soybean cultivars were compared to each other to identify specific variations of Zaoshu18. Website GeneScan (http://genes.mit.edu/GENSCAN.html) was used to predict whether these variations constitute additional gene models.
Expression Analyses of Candidate Genes
Zaoshu18 and Williams seedlings were inoculated with the isolate PsUSAR2 when the first pair of true leaves had expanded completely. We collected 1-cm samples from above and below the treated hypocotyl tissues at 0, 6, 12, 24, 48, and 72 h post inoculation, and stored them at −80°C. For the analysis of tissue-specific transcript abundance, the roots, stems and leaves were also collected at 0 h post inoculation. Allele expression patterns were tested for the 14 sequenced soybean cultivars and Williams at 24 h post inoculation. Three individuals were mixed at each time point for total RNA extraction. Total RNA was isolated using an RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China), and cDNA was synthesized using a PrimeScript™RT Reagent Kit (TaKaRa, Japan). Specific primers for the quantitative real-time PCR (qRT-PCR) were designed according to the conserved coding region between the haplotype sequences using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The housekeeping gene Actin11 was used as an internal control. Gene expression was determined by qRT-PCR using SYBR® Premix Ex TaqII (TliRNaseH Plus) (TaKaRa, Japan) on an ABI 7500 platform (Applied Biosystems, USA). The 2−ΔΔCT method was used to calculate the relative expression levels of candidate genes (Livak and Schmittgen, 2001). Three biological replicates with their respective three technical replicates were conducted for each sample.
Results
Phenotypic Evaluation
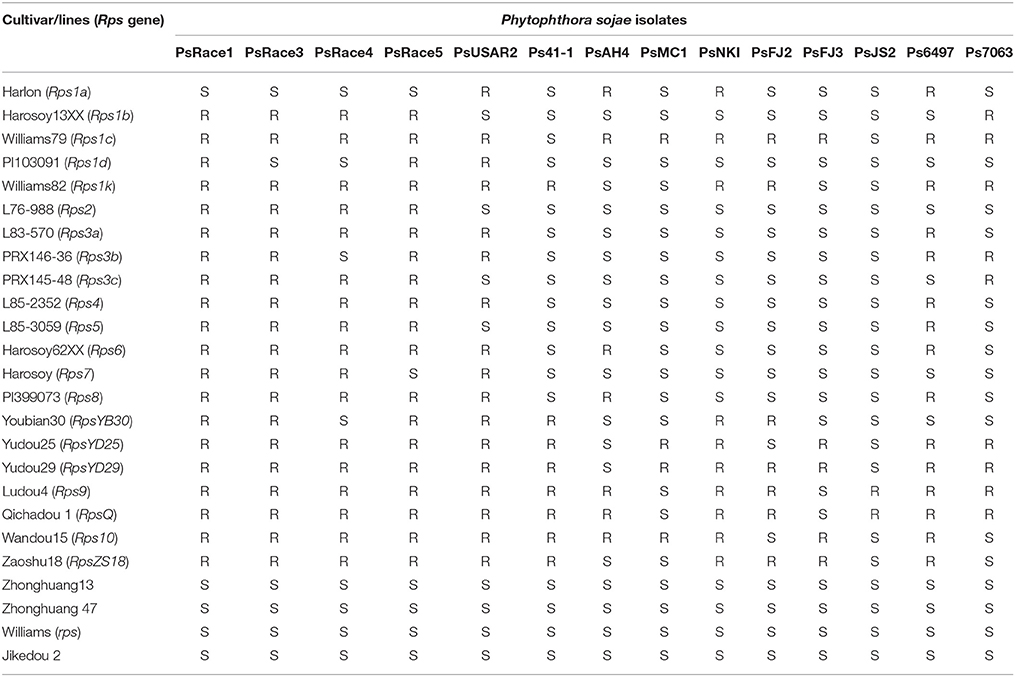
All the plants of the four susceptible controls, Williams, Zhonghuang13, Zhonghuang47, and Jikedou2, were dead after being inoculated individually with each of the 14 P. sojae isolates. The other 20 resistant cultivars carrying a single known Rps gene were resistant to 3–12 isolates. Zaoshu18 was resistant to 10 of the 14 P. sojae isolates (Table 1). A total of 17 reaction types among the 25 soybean cultivars were formed in response to the 14 P. sojae isolates, and Zaoshu18 showed a distinct reaction type. This suggested that Zaoshu18 contained a novel Rps gene or genes combination.To test the phenotypes of the F2:3 population, Four P. sojae isolates, PsFJ2, PsFJ3, PsUSAR2, and Ps41-1, were selected for inoculation. Six days after being inoculated with the four isolates, all the Williams soybean plants (susceptible parent) were dead, whereas all the Zaoshu18 plants (resistant parent) were alive and exhibited no disease symptoms (Figure 1). Among the 232 F2:3 families of the mapping population, the ratio of homozygous resistant to segregating to homozygous susceptible for four isolates PsFJ2, PsFJ3, PsUSAR2, and Ps41-1 was 54:112:66, which fitted the expected 1:2:1 ratio (Table 2). Thus, resistance to the four isolates PsFJ2, PsFJ3, PsUSAR2, and Ps41-1 in Zaoshu18 was confirmed to be controlled by a single dominant gene.

Figure 1. Phytophthora resistance phenotype of the soybean cultivars Zaoshu18 and Williams were tested with isolate PsUSAR2 using the hypocotyl-inoculation technique for 6 days. All the Williams soybean plants (susceptible parent) were fallen and dead, whereas all the Zaoshu18 plants (resistant parent) were alive and exhibited no disease symptoms. The red arrow represents the inoculation site, in which the lesion invades and spreads along the hypocotyl in the susceptible cultivar Williams, whereas the resistant cultivar Zaoshu18 shows an immune response with no spread of the lesion.
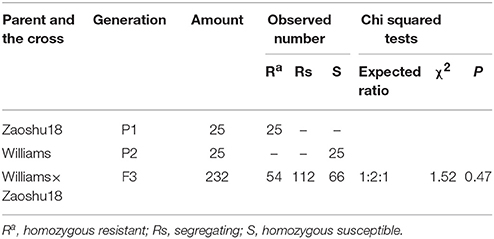
Table 2. Genetic segregation in response to Phytophthora sojae isolates in 232 F2:3 families derived from a cross between soybean cultivars Zaoshu18 and Williams.
Linkage Analyses and Genetic Mapping
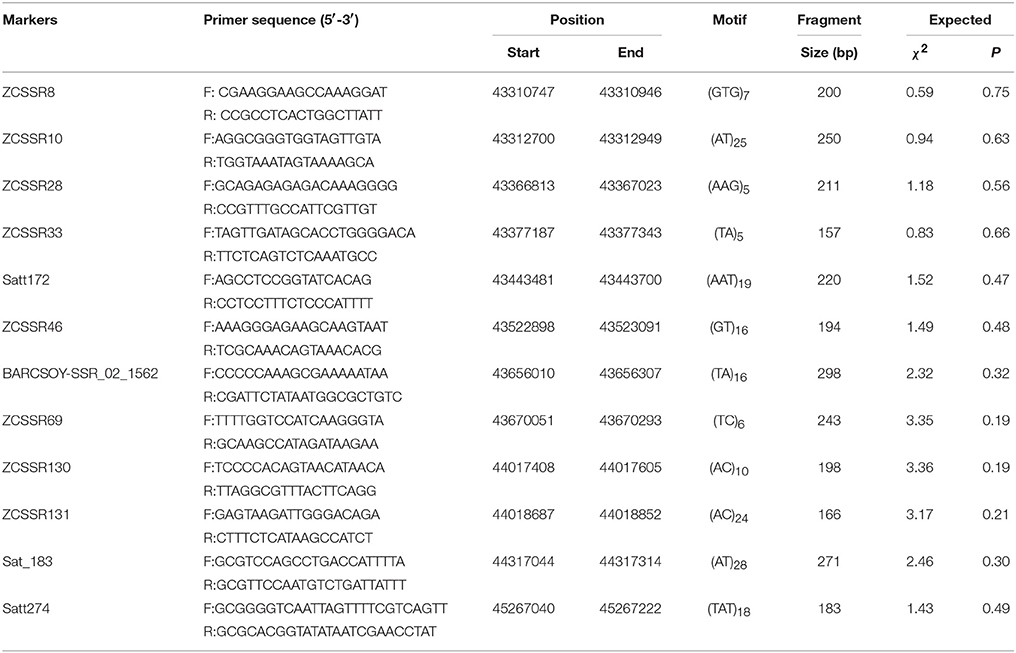
We selected 28 published SSR markers distributed on chromosome 2 to genotype the Zaoshu18 and Williams. Four of the 28 SSR markers, namely Satt172, Sat_183, Satt274, and BARCSOYS_02_1562, were polymorphic (Table 3). Subsequently, 154 SSR markers were developed in the target region, and eight of them (ZCSSR8, ZCSSR10, ZCSSR28, ZCSSR33, ZCSSR46, ZCSSR69, ZCSSR130, and ZCSSR131) were polymorphic between the parents, Zaoshu18 and Williams. A total of 12 polymorphic SSR markers were used to genotype the F2:3 population for linkage analyses (Figure 2A). All 12 markers were linked to RpsZS18; ZCSSR33, and ZCSSR46 flanked RpsZS18 at genetic distances of 0.9 and 0.5 cM, and Satt172 co-segregated with RpsZS18 (Figure 2A).
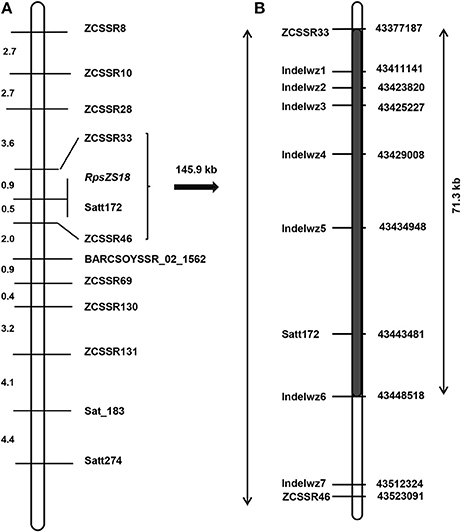
Figure 2. Genetic and physical map of the RpsZS18 region. (A) Linkage map of 12 polymorphic SSR markers linked to RpsZS18 on soybean chromosome 2. Genetic distances, in cM, are shown on the left, and the locations of the markers and RpsZS18 are indicated on the right. (B) The physical map of the RpsZS18 which was constructed by genotyping of InDel markers between ZCSSR33 and ZCSSR46. The black bar represents the 71.3-kb region in which no recombination events happened in the F2:3 population. The locations of InDel markers are on the left, and the physical position of the chromosome 2 are provided on the right.
InDel Marker Analyses in the RpsZS18 Mapping Region
RpsZS18 was mapped between SSR markers ZCSSR33 and ZCSSR46, which covered ~145.9 kb in the Williams82 reference genome sequence (Glycine max V2.0) (Table 3). According to the Glyma 2.0 annotations, there are 19 genes in the target region (Table S1). To narrow the mapped region of RpsZS18 and identify the candidate genes controlling PRR resistance, PCR-based InDel makers were developed in the RpsZS18 mapping region based on the InDels identified by WGRS between the two parents, Williams and Zaoshu18. Seven InDel markers could clearly distinguish polymorphisms among the 232 F2:3 families using polyacrylamide gels (Table S2). Among the seven InDel markers, five (Indelwz1, Indelwz2, Indelwz3, Indelwz4, and Indelwz5) co-segregated with RpsZS18 and no recombination events were found in the 232 F2:3 families (Figure 2B). SSR marker ZCSSR33 and InDel maker Indelwz6 identified two and one recombinants in the mapping population, respectively, and identifed a 71.3-kb physical interval of RpsZS18.
Haplotype Analyses of RpsZS18 Candidate Genes
To identify the candidate genes conferring the resistance to PRR in Zaoshu18, based on homozygous SNPs, InDels, and high confident SVs identified by the WGRS, the 145.9-kb RpsZS18 allelic regions of the 14 sequenced cultivars were constructed by reference-based assembly (Table S3, Supplementary Materials S1). Each soybean cultivar generated 20–25 Gb of clean base data with 70–145 million paired-end 150-bp reads, which resulted in an average depth of 17–22 × genome coverage. Each of these samples had a 5× genome coverage rate higher than 95% (Table 4). In this interval, no SV was found among the 14 sequenced cultivars. Totally, 447 SNPs and 141 InDels were identified in the candidate region of RpsZS18 among the 15 soybean genotypes. Haplotype sequences of the corresponding RpsZS18 region were constructed for the 14 sequenced soybean cultivars. As analyzed the haplotype sequences among the 15 soybean genotypes, no additional, extended or truncated genes were identified. Haplotype sequences of the 14 soybean genotypes and Williams82 were aligned to identify the haplotype specific candidate genes for RpsZS18. The haplotype sequence of Zaoshu18 was compared with the other 13 soybean genotypes and the reference genotypes Williams82, among the 19 annotated gene models, 13 gene sequences with flanking 1-kb region of Zaoshu18 were consistent with the at least one haplotype sequences of the genotypes, while the other six genes are unique haplotype sequences of Zaoshu18, and all of six genes were in the 71.3-kb interval of RpsZS18 flanked by ZCSSR33 and Indelwz6 (Figure 3). The six gene models containing Zaoshu18-specific sequences were predicted as candidate genes of RpsZS18. Among them, Glyma.02g245700 and Glyma.02g245900 encodes elongation factor (EF)-hand calcium-binding domain containing protein; Glyma.02g245800 encodes a sugar kinase; Glyma.02g246000 encodes a serine/threonine protein kinase with a leucine-rich repeat (STK-LRR) structure; Glyma.02g246200 and Glyma.02g246300 have no annotations so far.
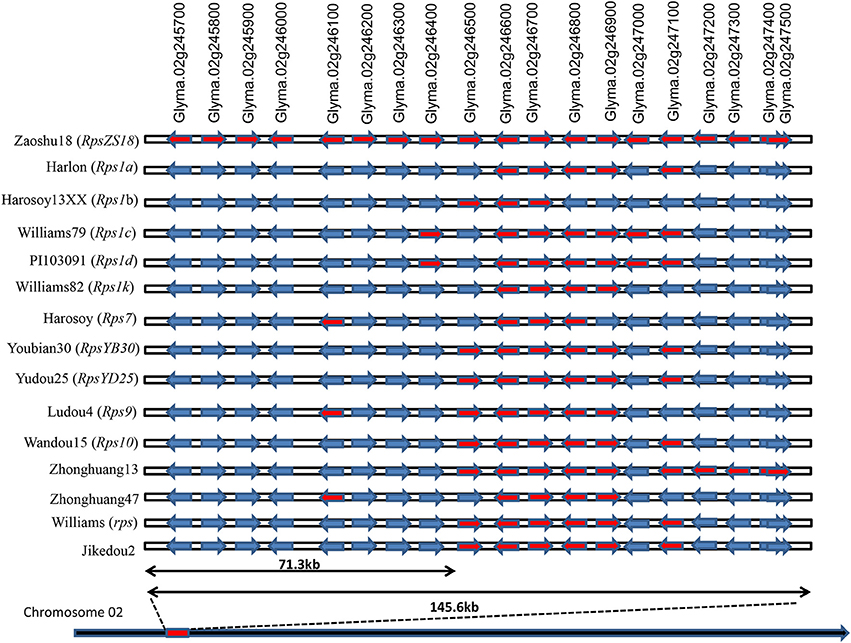
Figure 3. Haplotype sequences comparing between Zaoshu18 and other soybean genotypes in the corresponding RpsZS18 mapping interval. Gene models marked red were the genes which shared the same haplotype sequences with Zaoshu18, while ones marked blue were the genes with the different haplotype sequences compared to Zaoshu18.
Expression Analyses of Candidate Genes
To confirm which of these genes are related to resistance to P. sojae, qRT-PCRs were performed to analyze the expression pattern of the six genes in Zaoshu18 and Williams. In order to detect the expression pattern of genes more effectively, we selected two pairs of primers for each gene to confirm the gene expression pattern (Table S4). However, only three of the six genes were detected; the other three genes were not detected due to low or no expression, no matter in root, stem, or leaf tissues using the two pairs of primers or redesigned primers. The three genes (Glyma.02g245700, Glyma.02g245800, and Glyma.02g246300) had different expression levels in different tissues and at all the time points between Zaoshu18 and Williams (Figure 4, Figure S1). The expression levels in root, stem and leaf of the three genes in Zasoshu18 were higher than that in Williams at 0 h post inoculation (Figures 4A–C). The relative expression level of Glyma.02g245700 and Glyma.02g245800 in stem were higher than that in root and leaf. The expression level of Glyma.02g246300 in leaf is higher in root and stem. At all six time points, the expression levels of all three genes were higher in Zaoshu18 compared with in Williams (Figures 4D–F). In Zaoshu18, the highest expression level of Glyma.02g245700 was at 6 h after inoculation, whereas in Williams, Glyma.02g245700 expression was highest at 6 and 12 h and then reduced at 24, 48, and 72 h after treatment. The expression level of Glyma.02g245800 in Zaoshu18 was up-regulated at 6, 12, 24, and 48 h after treatment. Overall, the expression trend of Glyma.02g246300 in Zaoshu18 was down-regulated compared with at 0 h, but the expression level in Williams was so low that it was almost undetectable. These results show that these three genes had different expression levels in Zaoshu18 and Williams and were up- or down-regulated after P. sojae infection. Therefore, the three genes that have different expression levels between resistant and susceptible parents were preferred as resistance candidate gene. Allele expression patterns were tested for the 14 sequenced soybean cultivars and the reference genotype Williams82 at 24 h post inoculation. The expression levels of these three candidate genes in Zaoshu18 were significantly higher than those in the other 14 soybean cultivars (Figure 5).
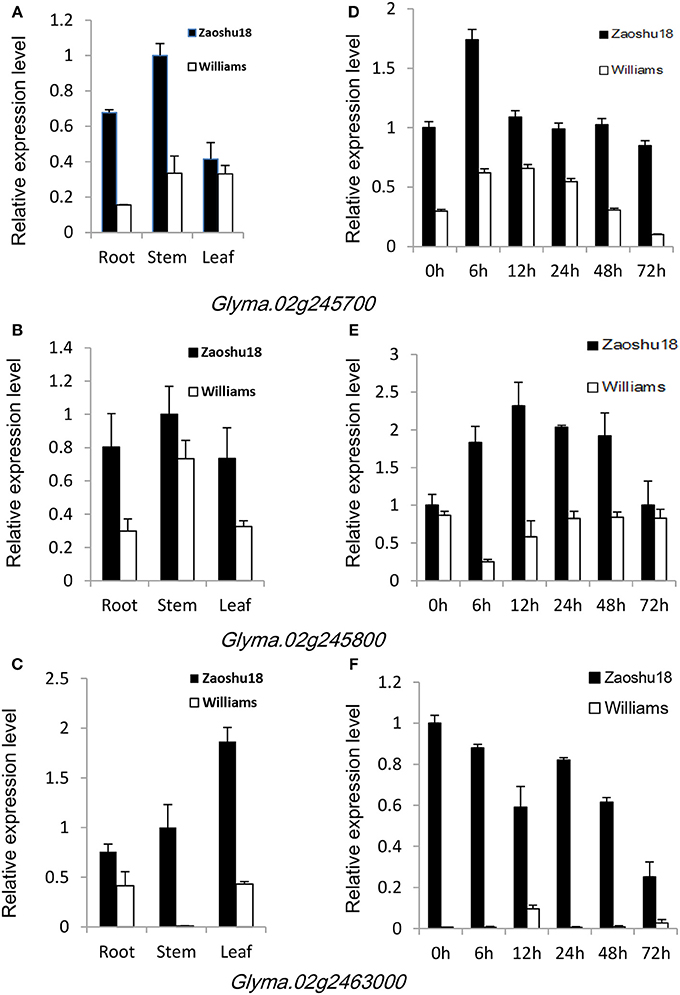
Figure 4. Expression patterns of Glyma.02g245700, Glyma.02g245800, and Glyma.02g246300 in Zaoshu18 and Williams. Each sample was mixed with three individuals at each time point. Three biological replicates with their respective three technical replicates were performed. Bars indicated standard error of the mean. (A) Expression patterns of Glyma.02g245700 in various tissues of Zaoshu18 and Williams. Roots, stems, and leaves were harvested from 12-day-old seedlings without inoculation. (B) Relative expression levels of Glyma.02g245700 in Zaoshu18 and Williams. Infected samples were collected at 0, 6, 12, 24, 48, and 72 h post-inoculation with P. sojae isolate PsUSAR2. (C) Expression patterns of Glyma.02g245800 in various tissues of Zaoshu18 and Williams. (D) Relative expression levels of Glyma.02g245800 in Zaoshu18 and Williams. (E) Expression patterns of Glyma.02g246300 in various tissues of Zaoshu18 and Williams. (F) Relative expression levels of Glyma.02g246300 in Zaoshu18 and Williams.
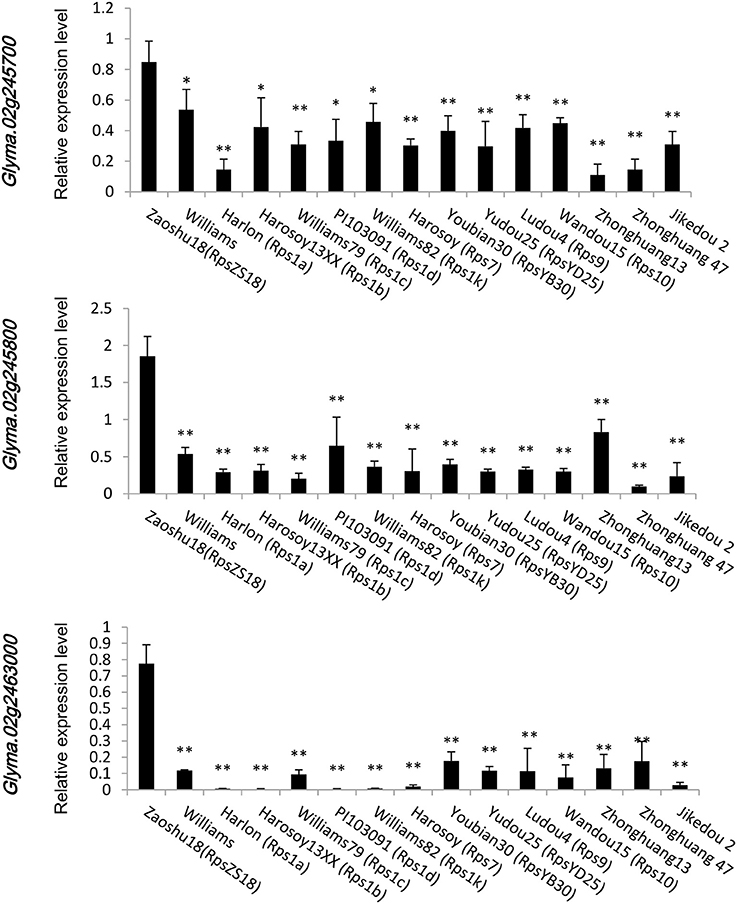
Figure 5. Allele expression patterns of Glyma.02g245700, Glyma.02g245800, and Glyma.02g246300 in the 14 sequenced soybean cultivars and the reference genotype Williams82 at 24 h post inoculation. Three biological replicates with their respective three technical replicates were performed and statistically analyzed using Student's t-test (*P < 0.05, **P < 0.01). Bars indicated standard error of the mean.
Discussion
Using soybean cultivars that express resistance genes is the safest and most economical way to reduce yield losses caused by PRR (Sugimoto et al., 2012). However, disease resistance provided by the expression of an Rps gene generally lasts for 8–15 years because of the emergence of new P. sojae pathotypes (Schmitthenner, 1985). Therefore, researchers continue to search for new Rps genes. Furthermore, mapping genes and developing closely linked markers can greatly facilitate marker-assisted pyramiding of Rps genes. Previous studies have demonstrated that the soybean cultivar Zaoshu18 exhibited elite resistance to PRR (Zhu et al., 2006; Yao et al., 2010), and that RpsZS18 provided resistance against more P. sojae pathotypes in China than Rps1c and Rps1k (Zhang et al., 2014).
In this work, we found that the reaction of Zaoshu18 to the 14 isolates was distinct from that of the other soybean cultivars tested (Table 1). Genetic analyses of resistance against four P. sojae isolates, PsFJ2, PsFJ3, PsUSAR2, and Ps41-1, revealed resistance to PRR in Zaoshu18 was controlled by a single dominant gene, which validated our previous result (Yao et al., 2010). The combination of the four isolates can defeat all the identified Rps genes except RpsYD29, however, RpsYD29 was identified on chromosome 3 (Zhang et al., 2013a) (Table 1). From the phenotypic evaluation, we concluded that Zaoshu18 contained a novel Rps gene. Four P. sojae isolates differing in virulence were used to assess a mapping population to obtain accurate PRR phenotypes. We mapped the likely location of the RpsZS18 gene in the soybean genome to a 145.9-kb fragment between SSR markers ZCSSR33 (0.9 cM) and ZCSSR46 (0.5 cM) on chromosome 2, and created a genetic linkage map with higher resolution than the previous map of Yao et al. (2010). The order of all of the SSR markers distributed in our linkage map was consistent with the order in the Williams 82 physical map (Schmutz et al., 2010; Song et al., 2010). A SSR marker, Satt172, was found to co-segregate with RpsZS18.
More than 20 Rps genes conferring resistance to PRR have been identified on nine soybean chromosomes, but RpsZS18 is the only Rps gene that has been identified on chromosome 2. On chromosome 2, three quantitative trait loci (QTL) for PRR resistance have been identified, but these QTLs are not close to the region containing RpsZS18, as determined in this study (Burnham et al., 2003; Han et al., 2008; Li et al., 2010). All the three QTLs are closely linked to the marker Sattt579 on chromosome 2, but the location of the marker on the reference genome Williams82 is at the position 19688108-19688299 which is far away the genomic location of RpsZS18. Some studies have reported several disease resistance genes to soybean mosaic virus (SMV) and soybean cyst nematode on soybean chromosome 2 (Hayes et al., 2000; Wang et al., 2004; Zhan et al., 2006; Yuan et al., 2008; Ma et al., 2017). QTLs for insect-resistance have also been detected on chromosome 2, including four QTLs effective against budworms, one QTL conferring resistance against Megacopta cribraria, and one QTL that offers protection from Prodenia litura (Rector et al., 1998; Terry et al., 2000; Xing et al., 2008). But the locations of these resistance genes or QTLs were far away from RpsZS18. These results indicated that RpsZS18 is a novel disease-resistance locus.
Identification of candidate genes for disease resistance is imperative for analyzing the mechanism of resistance to pathogens in plants and will be useful in future cloning of resistance genes. However, mapped target regions generally contain many genes with different functional annotations, and a lack of SNP markers which can be used to detect gene recombination in mapping population makes it difficult to confirm Rps candidate genes. Generally, only genes that encode proteins with predicted disease resistance-related structures are selected as resistance genes, and there is no way to exclude other types of genes in a target region (Zhang et al., 2013b; Li Y. et al., 2017; Niu et al., 2017). Among the identified Rps genes, only Rps1k has been cloned and characterized. A NBS-LRR-encoding gene cluster was identified in the region of Rps1k (Gao et al., 2005; Gao and Bhattacharyya, 2008). Among all known types of R genes, more than 80% of characterized R genes are located close to NBS-LRR-encoding genes, including Rps1k (Shao et al., 2016). However, in this study, no typical NBS-LRR resistance gene was found in the mapped region of RpsZS18, which may indicate that RpsZS18 is an Rps gene with a novel Phytophthora resistance mechanism. To further investigate this idea, we applied WGRS analyses. Because the four P. sojae isolates used to evaluated the phenotypes of the mapping population were able to defeat all the sequenced soybean cultivars containing known Rps genes except RpsZS18 in this study, it suggested that the other sequenced Rps-containing cultivars do not contain RpsZS18. Hence, RpsZS18 haplotype is absent from the other resistant varieties. Fourteen RpsZS18 allelic haplotypes sequence of soybean genotypes were reconstructed by substituting the bases with confident variant calls in the reference genome. Identification of varients in gene region including promoter, UTR, extron or intron region is an effective way of discovering candidate genes, and this approach has been used to identify candidate genes for multiple plant diseases in recent years (Silva et al., 2012; Sudheesh et al., 2015; Singh et al., 2016a,b; Li B. et al., 2017; Li W. et al., 2017; Pandey et al., 2017). Allele haplotype sequences containing the respective SNPs and InDels are aligned with each other. The soybean cultivars that were re-sequenced did not contain Rps genes on chromosome 2, so, when the corresponding RpsZS18 alleles were compared among the other sequenced soybean genotypes, genes that contained Zaoshu18-specific haplotype sequences were likely to be Rps candidate genes. By haplotype analyses, we identified six gene models that contained Zaoshu18-specific haplotype sequences, and all of them were in the 71.3-kb candidate genomic region of RpsZS18. These two results revealed that these six genes co-segregated with RpsZS18 in the F2:3 population and formed a specific RpsZS18 haplotype.
To further confirm which of these genes were expressed differently between the parental cultivars after treatment with P. sojae, qRT-PCR analysis was applied, and differential expression patterns of three genes were detected at different time points between Zaoshu18 and Williams. Differentially expressed genes in soybean that are responsive to P. sojae were found to peak at 24 h after inoculation or infection (Moy et al., 2004; Zhao et al., 2015). Further analysis of allelic genes showed that the expression levels of the three candidate genes were significantly higher than those of other sequenced soybean genotypes, indicating that these three specific haplotype genes have particular expression patterns in Zaoshu18. Glyma.02g245700 encodes a protein with an EF-hand calcium-binding domain, which is important in the regulation of calcium signaling. Calcium is the most widely accepted messenger and plays an important role in diverse physiological stimuli, stresses, and pathogen attack, including Phytophthora (Yoshioka et al., 2003; Gao et al., 2014; Chen et al., 2015). Plants contain two major immune responses to defend against pathogen infections: pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl, 2006; Boller and He, 2009). The EF-hand domain can bind calcium directly and trigger PAMPs or effectors, which activate complex downstream responses (Lecourieux et al., 2006; Gao et al., 2014). The expression of the gene models in Zaoshu18 was significantly higher than their expression in the susceptible parent Williams, suggesting that Zaoshu18 could react more effectively through the calcium regulatory network.
Glyma.02g245800 encodes a sugar kinase that belongs to the pfkB-type carbohydrate kinase family. pfkB-type carbohydrate kinases are involved in energy and carbohydrate metabolism (Gilkerson et al., 2012). But in a study of plant and pathogen interactions, a pfkB-type carbohydrate kinase was up-regulated in the non-host resistance reaction of Oryza sativa to wheat leaf rust fungus (Li et al., 2012). pfkB-type carbohydrate kinase was also up-regulated in the susceptible host–pathogen interaction between wheat and leaf rust fungus (Rampitsch et al., 2006). A proteomics study in soybean lines resistant and sensitive to P. sojae indicated that the percentage of differentially abundant protein involved in energy was significantly higher in resistant lines than in susceptible lines (Zhang et al., 2011). These results suggested that pfkB-type carbohydrate kinase may play an important role in the energy regulation in soybean during pathogenic infection. We found that the expression level of Glyma.02g245800 was significantly higher in Zaoshu18 than in the susceptible cultivar Williams at all six time points, probably because expression of this gene effectively improved energy production to help the plant resist the invasion of P. sojae.
Glyma.02g246300 had no annotation. BLAST searches against NCBI databases did not identify any annotated genes with high similarity to Glyma.02g246300. The expression level of Glyma.02g246300 was reduced after P. sojae inoculation in Zaoshu18, but was almost undetectable in Williams. It may be that Glyma.02g246300 expression was inhibited by the pathogen P. sojae, but lost its function in the susceptible cultivar Williams. Glyma.02g246300 may be a novel gene, and further cloning experiment are needed to clarify its structure and function.
The other three genes were not detected by the qRT-PCRs because of their low expression levels. However, two of the three genes were annotated as related to resistance plant pathogen. Glyma.02g2459000 is calcium-binding EF-hand protein like Glyma.02g245700, but the gene model only contains one 255-bp extron. Glyma.02g246000 encodes a STK-LRR, and STK-LRRs are involved in transmembrane signal transductions and are important for plant disease resistance (Hardie, 1999; Fluhr and Kaplan-Levy, 2002). Many resistance genes in plants encode enzymes with structures similar to those of STKs, including Xa21, Xa21D, Xa26, and Lr10 (Song et al., 1995; Feuillet et al., 1997; Wang et al., 1998; Sun et al., 2004). Recently, two genes encoding STK-LRRs were predicted as the Rps candidate genes, RpsQ and RpsHN (Li Y. et al., 2017; Niu et al., 2017). Based on these results, we consider the two genes may also contribute to the resistance of RpsZS18.
Although we identified three genes that were differentially expressed in resistant and susceptible species, and confirmed these candidate genes were likely to be involved in the resistance of Zaoshu18, the WGRS did not account for all the genomic information, and the annotation of the Williams82 reference genome information did not fully represent the annotation of the Zaoshu18 genome. WGRS can detect only SNPs and InDels, and cannot detect the insertion and deletion of large fragments; Different coverage of SNPs may indicate that some of the duplications have not been identified in this region (Figures S2, S3). Therefore, there may be other resistance genes in the locus that were not detected by re-sequencing. Only the screening and sequencing of BAC clones will be able to clear the genomic information in the mapping interval of RpsZS18.
RpsZS18 is the only Rps gene on chromosome 2, so it should be relatively easy to combine it with other Rps genes in soybean breeding programs. Doing so will produce cultivars with more durable resistance to PRR, and perhaps produce cultivars with resistance to broad-spectrum pathogens. The Zaoshu18 soybean cultivar is an elite parent, and has been used to generate several soybean varieties (Liu et al., 2013). Related varieties may contain the same Rps genes as Zaoshu18 (Chen et al., 2008; Xia et al., 2011). The SSR and InDel markers that co-segregated with RpsZS18 can be applied in molecular marker-assisted selection to breed more excellent soybean cultivars. These markers for Zaoshu18 will provide an effective tool as a better application for breeding materials.
In conclusion, we finely mapped the RpsZS18 gene and identified six candidate genes, three of which were shown to be differently expressed between resistant and susceptible parental cultivars. The co-segregated markers may facilitate the tracking of RpsZS18 in progenies for marker-assisted selection in soybean breeding programs. The candidate genes also provide novel fundamental details regarding the PRR resistance mechanism, and may be useful in future cloning experiments for the functional characterization of the RpsZS18 gene.
Author Contributions
ZZ: conceived and designed the experiments; CZ, SS, LY, JD, and CD: performed the experiments; CZ: analyzed data and wrote the manuscript; ZZ: revised the paper; All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31771822), the Special Fund for Agroscientific Research in the Public Interest (201303018), The National Key Research and Development Program of China (2016YFD0100201), the National Infrastructure for Crop Germplasm Resources (NICGR2017-008), and the Scientific Innovation Program of Chinese Academy of Agricultural Sciences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professors Lijuan Qiu and Tianfu Han in the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences for supplying the soybean cultivars tested, and Prof. Yuanchao Wang in Nanjing Agricultural University and Shuzhen Zhang in Northeast Agricultural University for providing the Phytophthora sojae isolates used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00044/full#supplementary-material
References
Bernard, R. L., Smith, P. E., Kaufmann, M. J., and Schmitthenner, A. F. (1957). Inheritance of resistance to Phytophthora root and stem rot in the soybean. Agron. J. 49, 391.
Boller, T., and He, S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. doi: 10.1126/science.1171647
Chen, C., Sun, X., Duanmu, H., Zhu, D., Yu, Y., Cao, L., et al. (2015). GsCML27, a gene encoding a calcium-binding EF-hand protein from Glycine soja, plays differential roles in plant responses to bicarbonate, salt and osmotic stresses. PLoS ONE 10:e0141888. doi: 10.1371/journal.pone.0141888
Chen, X. L., Zhu, Z. D., Wang, X. M., Xiao, Y. N., and Wu, X. F. (2008). Postulation of Phytophthora resistance genes in soybean cultivars or lines. Sci. Agric. Sin. 41, 1227–1234.
Chen, X., and Wang, Y. (2017). “Phytophthora sojae,” in Biological Invasions and Its Management in China, eds F. Wan, M. Jiang, and A. Zhan (Singapore: Springer), 199–223.
Cheng, Y., Ma, Q., Ren, H., Xia, Q., Song, E., Tan, Z., et al. (2017). Fine mapping of a Phytophthora-resistance gene RpsWY in soybean (Glycine max L.) by high-throughput genome-wide sequencing. Theor. Appl. Genet. 130, 1041–1051. doi: 10.1007/s00122-017-2869-5
Cui, L., Yin, W., Tang, Q., Dong, S., Zheng, X., Zhang, Z., et al. (2010). Distribution, pathotypes, and metalaxyl sensitivity of Phytophthorasojae from Heilongjiang and Fujian provinces in China. Plant Dis. 94, 881–884. doi: 10.1094/PDIS-94-7-0881
Dorrance, A. E., Jia, H., and Abney, T. S. (2004). Evaluation of soybean differentials for their interaction with Phytophthora sojae. Plant Health Prog. 207–209. doi: 10.1094/PHP-2004-0309-01-RS
Dorrance, A. E., McClure, S. A., and DeSilva, A. (2003a). Pathogenic diversity of Phytophthora sojae in Ohio soybean fields. Plant Dis. 87, 139–146. doi: 10.1094/PDIS.2003.87.2.139
Dorrance, A. E., McClure, S. A., and St. Martin, S. K. (2003b). Effect of partial resistance on Phytophthora stem rot incidence and yield of soybean in Ohio. Plant Dis. 87, 308–312. doi: 10.1094/PDIS.2003.87.3.308
Dorrance, A. E., and Schmitthenner, A. F. (2000). New sources of resistance to Phytophthora sojae in the soybean plant introductions. Plant Dis. 84, 1303–1308. doi: 10.1094/PDIS.2000.84.12.1303
Lecourieux, D., Ranjeva, R., and Pugin, A. (2006). Calcium in plant defence-signalling pathways. New Phytol. 171, 249–269. doi: 10.1111/j.1469-8137.2006.01777.x
Fan, A. Y., Wang, X. M., Fang, X. P., Wu, X. F., and Zhu, Z. D. (2009). Molecular identification of Phytophthora resistance gene in soybean cultivar Yudou 25. Acta Agron. Sin. 35, 1844–1850. doi: 10.3724/SP.J.1006.2009.01844
Feuillet, C., Schachermayr, G., and Keller, B. (1997). Molecular cloning of a new receptor-like kinase gene encoded at the Lrl0 disease resistance locus of wheat. Plant J. 11, 45–52. doi: 10.1046/j.1365-313X.1997.11010045.x
Fluhr, R., and Kaplan-Levy, R. N. (2002). “Plant disease resistance: commonality and novelty in multicellular innate immunity,” in Toll-Like Receptor Family Members and Their Ligands, eds B. Beutler and H. Wagner (Berlin; Heidelberg: Springer), 23–46.
Gao, H., and Bhattacharyya, M. K. (2008). The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol. 8:29 doi: 10.1186/1471-2229-8-29
Gao, H., Narayanan, N. N., Ellison, L., and Bhattacharyya, M. K. (2005). Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol. Plant Microbe Interact. 18, 1035–1045 doi: 10.1094/MPMI-18-1035
Gao, X., Cox, K. L., and He, P. (2014). Functions of calcium-dependent protein kinases in plant innate immunity. Plants 3, 160–176. doi: 10.3390/plants3010160
Gilkerson, J., Perez-Ruiz, J. M., Chory, J., and Callis, J. (2012). The plastid-localized pfkB-type carbohydrate kinases FRUCTOKINASE-LIKE 1 and 2 are essential for growth and development of Arabidopsis thaliana. BMC Plant Biol. 12:102. doi: 10.1186/1471-2229-12-102
Gordon, S. G., St Martin, S. K., and Dorrance, A. E. (2006). Rps8 maps to a resistance gene rich region on soybean molecular linkage group F. Crop Sci. 46, 168–173. doi: 10.2135/cropsci2004.04-0024
Grant, D., Nelson, R. T., Cannon, S. B., and Shoemaker, R. C. (2009). SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 38, 843–846. doi: 10.1093/nar/gkp798
Han, Y., Teng, W., Yu, K., Poysa, V., Anderson, T., Qiu, L., et al. (2008). Mapping QTL tolerance to Phytophthora root rot in soybean using microsatellite and RAPD/SCAR derived markers. Euphytica 162, 231–239. doi: 10.1007/s10681-007-9558-4
Hardie, D. G. (1999). Plant protein serine/threonine kinases: classification and functions. Annu. Rev. Plant. Biol 50, 97–131. doi: 10.1146/annurev.arplant.50.1.97
Hayes, A. J., Ma, G., Buss, G. R., and Maroof, M. A. (2000). Molecular marker mapping of RSV4, a gene conferring resistance to all known strains of soybean mosaic virus. Crop Sci. 40, 1434–1437. doi: 10.2135/cropsci2000.4051434x
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kasuga, T., Salimath, S. S., Shi, J., Gijzen, M., Buzzell, R. I., and Bhattacharyya, M. K. (1997). High resolution genetic and physical mapping of molecular markers linked to the Phytophthora resistance gene Rps1-k in soybean. Mol. Plant Microbe Interact. 10, 1035–1044. doi: 10.1094/MPMI.1997.10.9.1035
Burnham, K. D., Dorrance, A. E., VanToai, T. T., and St Martin, S. K. (2003). Quantitative trait loci for partial resistance to in soybean. Crop Sci. 43, 1610–1617. doi: 10.2135/cropsci2003.1610
Kosambi, D. D. (1943). The estimation of map distances from recombination values. Ann. Eugenics 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Li, B. Y., and Ma, S. M. (1999). Studies of phytophthora root rot of soybean on incidence and control in Heilongjiang province. Chin. J. Oil Crop Sci. 21, 47–50.
Li, B., Zhao, Y., Zhu, Q., Zhang, Z., Fan, C., Amanullah, S., et al. (2017). Mapping of powdery mildew resistance genes in melon (Cucumismelo L.) by bulked segregant analysis. Sci. Hortic. e220, 160–167. doi: 10.1016/j.scienta.2017.04.001
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Goodwin, P. H., Han, Q., Huang, L., and Kang, Z. (2012). Microscopy and proteomic analysis of the non-host resistance of Oryza sativa to the wheat leaf rust fungus, Pucciniatriticina f. sp. tritici. Plant Cell Rep. 31, 637–650. doi: 10.1007/s00299-011-1181-0
Li, L., Lin, F., Wang, W., Ping, J., Fitzgerald, J., Zhao, M., et al. (2016). Fine mapping and candidate gene analysis of two loci conferring resistance to Phytophthorasojae in soybean. Theor. Appl. Genet. 129, 2379–2386 doi: 10.1007/s00122-016-2777-0
Li, W., Zhu, Z., Chern, M., Yin, J., Yang, C., Ran, L., et al. (2017). A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170, 114–126. doi: 10.1016/j.cell.2017.06.008
Li, X., Han, Y., Teng, W., Zhang, S., Yu, K., Poysa, B., et al. (2010). Pyramided QTL underlying tolerance to Phytophthora root rot in mega-environments from soybean cultivars ‘Conrad’ and ‘Hefeng 25’. Theor. Appl. Genet. 121, 651–658. doi: 10.1007/s00122-010-1337-2
Li, Y., Sun, S., Zhong, C., Wang, X., Wu, X., and Zhu, Z. (2017). Genetic mapping and development of co-segregating markers of RpsQ, which provides resistance to Phytophthorasojae in soybean. Theor. Appl. Genet. 130, 1223–1233. doi: 10.1007/s00122-017-2883-7
Lin, F., Zhao, M., Ping, J., Johnson, A., Zhang, B., Abney, T. S., et al. (2013). Molecular mapping of two genes conferring resistance to Phytophthorasojae in a soybean landrace PI 567139B. Theor. Appl. Genet. 126, 2177–2185. doi: 10.1007/s00122-013-2127-4
Lincoln, S. E., Daly, M. J., and Lander, E. S. (1993). Constructing Genetic Maps with MAPMAKER/EXP Version 3.0: a Tutorial and Reference Manual. Whitehead Inst Biomed Res Tech Rpt, 3rd edn. Whitehead Institute for Biomedical Research, Cambridge, 97.
Liu, R. H., and Meng, J. L. (2003). MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25, 317–321.
Liu, Z., Sun, S., Li, W., Chen, L., Chang, R., and Qiu, L. (2013). Analysis of parental relationship for soybean cultivars released from 1983 to 2010 in Beijing. Acta. Agron. Sin. 39, 1693–1700. doi: 10.3724/SP.J.1006.2013.01693
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, F. F., Wu, M., Liu, Y. N., Feng, X. Y., Wu, X. Z., Chen, J. Q., et al. (2017). Molecular characterization of NBS–LRR genes in the soybean Rsv3 locus reveals several divergent alleles that likely confer resistance to the soybean mosaic virus. Theor. Appl. Genet. doi: 10.1007/s00122-017-2999-9. [Epub ahead of print].
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Moy, P. D., Qutob, B. P., Chapman, I., Atkinson, M., and Gijzen (2004). Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol. Plant Microbe Interact. 17, 1051–1062. doi: 10.1094/MPMI.2004.17.10.1051
Niu, J., Guo, N., Sun, J., Li, L., Cao, Y., Li, S., et al. (2017). Fine mapping of a resistance gene RpsHN that controls Phytophthora sojae using recombinant inbred lines and secondary populations. Front. Plant Sci. 8:538. doi: 10.3389/fpls.2017.00538
Pandey, M. K., Khan, A. W., Singh, V. K., Vishwakarma, M. K., Shasidhar, Y., Kumar, V., et al. (2017). QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.). Plant Biotechnol. J. 15, 927–941. doi: 10.1111/pbi.12686
Ping, J., Fitzgerald, J. C., Zhang, C., Lin, F., Bai, Y., Wang, D., et al. (2015). Identification and molecular mapping of Rps11, a novel gene conferring resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 129, 445–451. doi: 10.1007/s00122-015-2638-2
Qin, J., Song, Q., Shi, A., Li, S., Zhang, M., and Zhang, B. (2017). Genome-wide association mapping of resistance to Phytophthora sojae in a soybean [Glycine max (L.) Merr.] germplasm panel from maturity groups IV and V. PLoS ONE 12:e0184613. doi: 10.1371/journal.pone.0184613
Rampitsch, C., Bykova, N. V., McCallum, B., Beimcik, A., and Ens, W. (2006). Analysis of the wheat and Pucciniatriticina (leaf rust) proteomes during a susceptible host-pathogen interaction. Proteomics 6, 1897–1907. doi: 10.1002/pmic.200500351
Rausch, T., Zichner, T., Schlattl, A., Stütz, A. M., Benes, V., and Korbel, J. O. (2012). DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339. doi: 10.1093/bioinformatics/bts378
Rector, B. G., All, J. N., Parrott, W. A., and Boerma, H. R. (1998). Identification of molecular markers linked to quantitative trait loci for soybean resistance to corn earworm. Theor. Appl. Genet. 96, 786–790. doi: 10.1007/s001220050803
Ryley, M. J., Obst, N. R., Irwin, J. A. G., and Drenth, A. (1998). Changes in the racial composition of Phytophthora sojae in Australia between 1979 and 1996. Plant Dis. 82, 1048–1054. doi: 10.1094/PDIS.1998.82.9.1048
Sahoo, D. K., Abeysekara, N. S., Cianzio, S. R., Robertson, A. E., and Bhattacharyya, M. K. (2017). A novel Phytophthorasojae resistance Rps12 gene mapped to a genomic region that contains several Rps genes. PLoS ONE 12:e0169950. doi: 10.1371/journal.pone.0169950
Schmitthenner, A. F. (1985). Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 69, 362–368. doi: 10.1094/PD-69-362
Schmitthenner, A. F. (1999). “Phytophthora rot of soybean,” in Compendium of Soybean Diseases, 4th Edn., eds G. L. Hartman, J. B. Sinclair, and J. C. Rupe (St. Paul, MN: The American Phytopathological Society Press), 39–42.
Schmitthenner, A. F., Hobe, M., and Bhat, R. G. (1994). Phytophthorasojae races in Ohio over a 10-year interval. Plant Dis. 78, 269–276. doi: 10.1094/PD-78-0269
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Shao, Z.-Q., Xue, J.-Y., Wu, P., Zhang, Y.-M., Wu, Y., Hang, Y.-Y., et al. (2016). Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 170, 2095–2109. doi: 10.1104/pp.15.01487
Silva, J., Scheffler, B., Sanabria, Y., De Guzman, C., Galam, D., Farmer, A., et al. (2012). Identification of candidate genes in rice for resistance to sheath blight disease by whole genome sequencing. Theor. Appl. Genet. 124, 63–74 doi: 10.1007/s00122-011-1687-4
Singh, V. K., Khan, A. W., Jaganathan, D., Thudi, M., Roorkiwal, M., Takagi, H., Garg, V., et al. (2016a). QTL-seq for rapid identification of candidate genesfor 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol. J. 14, 2110–2119. doi: 10.1111/pbi.12567
Singh, V. K., Khan, A. W., Saxena, R. K., Kumar, V., Kale, S. M., Sinha, P., et al. (2016b). Next-generation sequencing for identification of candidate genes for Fusarium wilt and sterility mosaic disease in pigeonpea (Cajanuscajan). Plant Biotechnol. J. 14, 1183–1194. doi: 10.1111/pbi.12470
Song, Q., Jenkins, J., Jia, G., Hyten, D. L., Pantalone, V., Jackson, S. A., et al. (2016). Construction of high resolution genetic linkage maps to improve the soybean genome sequence assembly Glyma1.01. BMC Genomics 17:33. doi: 10.1186/s12864-015-2344-0
Song, Q., Jia, G., Zhu, Y., Grant, D., Nelson, R. T., Hwang, E. Y., et al. (2010). Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci. 50, 1950–1960. doi: 10.2135/cropsci2009.10.0607
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Stewart, S., Abeysekara, N., and Robertson, A. E. (2014). Pathotype and genetic shifts in a population of Phytophthora sojae under soybean cultivar rotation. Plant Dis. 98, 614–624. doi: 10.1094/PDIS-05-13-0575-RE
Stewart, S., Robertson, A., Wickramasinghe, D., Draper, M., Michel, A., and Dorrance, A. E. (2015). Population structure among and within Iowa, Missouri, Ohio, and South Dakota populations of Phytophthorasojae. Plant Dis. 100, 367–379. doi: 10.1094/PDIS-04-15-0437-RE
Sudheesh, S., Lombardi, M., Leonforte, A., Cogan, N. O., Materne, M., Forster, J. W., et al. (2015). Consensus genetic map construction for field pea (Pisum sativum L.), trait dissection of biotic and abiotic stress tolerance and development of a diagnostic marker for the er1 powdery mildew resistance gene. Plant Mol. Boil. Rep. 33, 1391–1403. doi: 10.1007/s11105-014-0837-7
Sugimoto, T., Kato, M., Yoshida, S., Matsumoto, I., Kobayashi, T., Kaga, A., et al. (2012). Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breed. Sci. 61, 511–522. doi: 10.1270/jsbbs.61.511
Sun, J., Li, L., Zhao, J., Huang, J., Yan, Q., Xing, H., et al. (2014). Genetic analysis and fine mapping of RpsJS, a novel resistance gene to Phytophthorasojae in soybean [Glycine max (L.) Merr.]. Theor. Appl. Genet. 127, 913–919. doi: 10.1007/s00122-014-2266-2
Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–527. doi: 10.1046/j.1365-313X.2003.01976.x
Terry, L. I., Chase, K., Jarvik, T., Orf, J., Mansur, L., and Lark, K. G. (2000). Soybean quantitative trait loci for resistance to insects. Crop Sci. 40, 375–382. doi: 10.2135/cropsci2000.402375x
Tooley, P. W., and Grau, C. R. (1984). The relationship between rate-reducing resistance to Phytophthora megasperma f. sp. glycinea and yield of soybean. Phytopathology 74, 1209–1216. doi: 10.1094/Phyto-74-1209
Wang, G.-L., Ruan, D.-L., Song, W.-Y., Sideris, S., Chen, L., Pi, L.-Y., et al. (1998). Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10, 765–779. doi: 10.1105/tpc.10.5.765
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, 164–164. doi: 10.1093/nar/gkq603
Wang, Y. J., Dong, Y., Wang, X. Q., Yang, Y. L., Yu, D. Y., Gai, J. Y., et al. (2004). Mapping of five genes resistant to SMV strains in soybean. Acta. Genet. Sin. 31, 87–90.
Xia, C. J., Zhang, J. Q., Wang, X. M., Wu, X. F., Liu, Z. X., and Zhu, Z. D. (2011). Analyses of resistance genes to Phytophthora root rot in soybean germplasm. Chin. J. Oil Crop Sci. 33. 396–401.
Xing, G. N., Zhou, B., and Zhao, T. J. (2008). Mapping QTLs of resistance to Megacotacri braria (Fabricius) in soybean. Acta. Agron. Sin. 34, 361–368.
Xue, A. G., Marchand, G., Chen, Y., Zhang, S., Cober, E. R., and Tenuta, A. (2015). Races of Phytophthorasojae in Ontario, Canada, 2010–2012. Can. J. Plant Pathol. 37, 376–383. doi: 10.1080/07060661.2015.1052562
Yao, H. Y., Wang, X. M., Wu, X. F., Xiao, Y. N., and Zhu, Z. D. (2010). Molecular mapping of Phytophthora resistance gene in soybean cultivar zaoshu18. J. Plant Genet. Resour. 11, 213–217.
Yoshioka, H., Numata, N., Nakajima, K., Katou, S., Kawakita, K., Rowland, O., et al. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15, 706–718. doi: 10.1105/tpc.008680
Yuan, C. P., Lu, W. G., Liu, Z. X., Li, Y. H., Li, W. D., Guan, R. X., et al. (2008). SSR analysis of new developed soybean lines resistant to soybean cyst nematode (Heterodera Glycines Ichinohe) Race4. Acta. Agron. Sin. 34, 1858–1864. doi: 10.3724/SP.J.1006.2008.01858
Zhan, Y., Yu, D. Y., Chen, S. Y., and Gai, J. Y. (2006). Inheritance and gene mapping of resistance to SMV strain SC-7 in soybean. Acta. Agron. Sin. 32, 936–938.
Zhang, J., Xia, C., Wang, X., Duan, C., Sun, S., Wu, X., et al. (2013a). Genetic characterization and fine mapping of the novel Phytophthora resistance gene in a Chinese soybean cultivar. Theor. Appl. Genet. 126, 1555–1561. doi: 10.1007/s00122-013-2073-1
Zhang, J., Sun, S., Wang, G., Duan, C., Wang, X., Wu, X., et al. (2014). Characterization of Phytophthora resistance in soybean cultivars/lines bred in Henan province. Euphytica 196, 375–384. doi: 10.1007/s10681-013-1040-x
Zhang, J., Xia, C., Duan, C., Sun, S., Wang, X., Wu, X., et al. (2013b). Identification and candidate gene analysis of a novel Phytophthora resistance gene Rps10 in a Chinese soybean cultivar. PLoS ONE 8:e69799. doi: 10.1371/journal.pone.0069799
Zhang, S. Z., Xu, P. F., Wu, J. J., Zhang, J. X., Li, W. B., Chen, C., et al. (2010). Races of Phytophthorasojae and their virulences on soybean cultivars in Heilongjiang, China. Plant Dis. 94, 87–91. doi: 10.1094/PDIS-94-1-0087
Zhang, Y., Zhao, J., Xiang, Y., Bian, X., Zuo, Q., Shen, Q., et al. (2011). Proteomics study of changes in soybean lines resistant and sensitive to Phytophthora sojae. Proteome Sci. 9:52. doi: 10.1186/1477-5956-9-52
Zhao, M., Cai, C., Zhai, J., Lin, F., Li, L., Shreve, J., et al. (2015). Coordination of microRNAs, phasiRNAs, and NB-LRR Genes in response to a plant pathogen: insights from analyses of a set of soybean Rps gene near-isogenic lines. Plant Genome 8, 1–13. doi: 10.3835/plantgenome2014.09.0044
Zhong, C., Sun, S., Li, Y., Duan, C., and Zhu, Z. (2017). Next-generation sequencing to identify candidate genes and develop diagnostic markers for a novel Phytophthora resistance gene, RpsHC18, in soybean. Theor. Appl. Genet. doi: 10.1007/s00122-017-3016-z. [Epub ahead of print].
Keywords: Phytophthora root rot, Phytophthora sojae, resistance gene, RpsZS18, soybean
Citation: Zhong C, Sun S, Yao L, Ding J, Duan C and Zhu Z (2018) Fine Mapping and Identification of a Novel Phytophthora Root Rot Resistance Locus RpsZS18 on Chromosome 2 in Soybean. Front. Plant Sci. 9:44. doi: 10.3389/fpls.2018.00044
Received: 19 September 2017; Accepted: 09 January 2018;
Published: 30 January 2018.
Edited by:
Nicolas Rispail, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Hai Nian, South China Agricultural University, ChinaChang-Jie Jiang, National Institute of Agrobiological Sciences, Japan
Jack Vossen, Wageningen University & Research, Netherlands
Copyright © 2018 Zhong, Sun, Yao, Ding, Duan and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhendong Zhu, zhuzhendong@caas.cn
 Chao Zhong
Chao Zhong Suli Sun1
Suli Sun1 Liangliang Yao
Liangliang Yao Zhendong Zhu
Zhendong Zhu