- 1Laboratory of Cotton Disease, Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences, c/o Key Laboratory of Agro-products Quality and Safety Control in Storage and Transport Process, Ministry of Agriculture, Beijing, China
- 2Crop Improvement and Protection Research Unit, United States Department of Agriculture, Agricultural Research Service, Salinas, CA, United States
- 3Department of Plant Pathology, University of California, Davis, Davis, CA, United States
Verticillium wilt caused by Verticillium dahliae results in severe losses in cotton, and is economically the most destructive disease of this crop. Improving genetic resistance is the cleanest and least expensive option to manage Verticillium wilt. Previously, we identified the island cotton NBS-LRR-encoding gene GbaNA1 that confers resistance to the highly virulent V. dahliae isolate Vd991. In this study, we expressed cotton GbaNA1 in the heterologous system of Arabidopsis thaliana and investigated the defense response mediated by GbaNA1 following inoculations with V. dahliae. Heterologous expression of GbaNA1 conferred Verticillium wilt resistance in A. thaliana. Moreover, overexpression of GbaNA1 enabled recovery of the resistance phenotype of A. thaliana mutants that had lost the function of GbaNA1 ortholog gene. Investigations of the defense response in A. thaliana showed that the reactive oxygen species (ROS) production and the expression of genes associated with the ethylene signaling pathway were enhanced significantly following overexpression of GbaNA1. Intriguingly, overexpression of the GbaNA1 ortholog from Gossypium hirsutum (GhNA1) in A. thaliana did not induce the defense response of ROS production due to the premature termination of GhNA1, which lacks the encoded NB-ARC and LRR motifs. GbaNA1 therefore confers Verticillium wilt resistance in A. thaliana by the activation of ROS production and ethylene signaling. These results demonstrate the functional conservation of the NBS-LRR-encoding GbaNA1 in a heterologous system, and the mechanism of this resistance, both of which may prove valuable in incorporating GbaNA1-mediated resistance into other plant species.
Introduction
Plant resistance (R) genes encode products that play a central role in directly or indirectly recognizing effector proteins from pathogens, or in triggering downstream signaling in the innate immune systems in plants (Jones and Dangl, 2006; Zipfel, 2008). Superfamily of R proteins are primarily delineated by the presence, or lack thereof, of a few structural motifs or domains, such as a nucleotide-binding site (NBS), leucine-rich repeat (LRR), Toll/Interleukin-1 receptor (TIR), coiled-coil (CC), transmembrane (Martin et al., 2003; Joshi and Nayak, 2011). Over one hundred R genes have been cloned and characterized from a diversity of plant species, collectively conferring resistance to 122 pathogens (Whitham et al., 1994; Hinsch and Staskawicz, 1996; Anderson et al., 1997; Feuillet et al., 1999, 2003; Shen et al., 2007; Sanseverino et al., 2012; Periyannan et al., 2013; Wang et al., 2015; Zhu et al., 2017). Most of these encode nucleotide binding (NB) and C- terminal leucine-rich repeat (LRR) domains, and hence these types of proteins belong to the so-called NB-LRR protein family (Tameling and Takken, 2008; Collier and Moffett, 2009).
Two subclasses of plant NB-LRRs have been characterized and their names are derived from the domain structure at their N-termini. Those that possess a Toll and human interleukin-1 receptor (TIR) domain are referred to as TIR-NB-ARC-LRR or TNL proteins, while those carrying a predicted coiled-coil (CC) domain are classified as CC-NB-ARC-LRR, or CNL proteins (McHale et al., 2006). The two structural units of ARC1 and ARC2, constitute an ARC subdomain in plant NB-LRRs, and combine with the NB domain to form a NB pocket (Tameling et al., 2002; Albrecht and Takken, 2006; Rairdan and Moffett, 2006). The NB-LRR proteins exist in an auto-inhibited state unless the plant is challenged with a pathogen elicitor. NB-LRRs may recognize effector proteins from the pathogens through direct physical interaction (Deslandes et al., 2003; Catanzariti et al., 2010; Krasileva et al., 2010), or indirectly, by detecting modifications of host target proteins that are induced by the effector (Axtell and Staskawicz, 2003; Mackey et al., 2003; van der Hoorn and Kamoun, 2008). The NB-ARC domain of NB-LRRs functions as a molecular switch wherein the ADP-bound state represents the “off” and the ATP-bound state as the “on” state (Moffett et al., 2002; Takken et al., 2006; Collier and Moffett, 2009; Lukasik and Takken, 2009; Slootweg et al., 2013). The conformational change in the NB-ARC domain coincides with the exchange of bound ADP for ATP leading to a stabilization of the active conformation, and subsequent activation of immune signaling pathways (Collier and Moffett, 2009; Lukasik and Takken, 2009; Eitas and Dangl, 2010).
Cultivated cotton is susceptible to Verticillium wilt, a vascular disease that can result in devastating losses of yield and quality. The leaves on infected plants turn yellow or defoliate, and eventually die following infection by Verticillium dahliae. In some years, more than 50% of the cotton acreage is affected by Verticillium wilt, significantly reducing the fiber quality and yield (National Cotton Council of America-Disease Database). Efforts to understand the molecular mechanisms of Verticillium wilt caused by V. dahliae have been made, including characterization of several genes that contribute to defense responses such as GbTLP1 (Munis et al., 2010), GbCAD1 and GbSSI2 (Gao et al., 2013), GbRLK (Zhao et al., 2013), GbSTK (Zhang et al., 2013a), GhPAO (Mo et al., 2015), GbSBT1 (Duan et al., 2016), GbNRX1 (Li et al., 2016), GbRVd (Yang et al., 2016), GaRPL18 (Gong et al., 2017), and GhPGIP1 (Liu N. et al., 2017).
The receptor-like protein encoded by Ve1 (Verticillium resistance gene 1)-like genes, including GbVe, GbVe1, Gbvdr5, GbaVd1, and GbaVd2, are homologous to the well-characterized major resistance genes first described in tomato (Zhang et al., 2011, 2012; Yang et al., 2015; Chen et al., 2017). By definition, Ve homologs confer resistance to race 1 isolates of V. dahliae, which encode the secreted effector Ave1 (de Jonge et al., 2012). The resistance genes activate diverse defense responses following infection by V. dahliae, including the regulation of defense hormone (salicylic acid, ethylene, etc.) levels that are involved in spermine and camalexin signaling, enhancing reactive oxygen species scavenging capacity and oxidative stress tolerance, activating the expression of the pathogenesis-related genes, and accelerating phytoalexin (gossypol) synthesis (Gao et al., 2013; Zhang et al., 2013a; Mo et al., 2015; Duan et al., 2016; Yang et al., 2016; Gong et al., 2017; Liu H. et al., 2017). For instance, silencing of GbNRX1, a thioredoxin, resulted in defective dissipation of apoplastic ROS, which led to higher ROS accumulation within protoplasts and hence critical for the apoplastic immune response (Li et al., 2016).
Similar to other plant species, the NBS-LRRs comprise a protein superfamily in cotton which encodes at least 300 nucleotide-binding site (NBS) domains and many of these are found encoded in gene clusters in genome (Paterson et al., 2012; Li et al., 2014; Chen et al., 2015). Most (about 76.7%) NBS-encoding genes have undergone striking mutations that reflect an ongoing plant–pathogen “arms race” (Paterson et al., 2012). Comparative genomic analyses showed that tandem duplications may have played a significant role in the expansion of the NBS-encoding gene family in G. raimondii (nearly immune to the pathogen) following its divergence from G. arboretum (highly susceptible to the pathogen). Correlation analysis revealed that the resistance genes cluster around known Verticillium wilt resistance QTLs, and several of these contain NBS-LRR domains (Chen et al., 2015). However, few NBS-LRR proteins have been reported to function as Verticillium wilt resistance in cotton, except for GbRVd (Yang et al., 2016).
The NBS-LRR class gene GbaNA1 is in the Verticillium wilt resistance locus VdRL08, and confers resistance to the non-race 1 Verticillium dahliae isolate Vd991 (Li N.Y. et al., 2017). The GbaNA1 homolog in Gossypium hirsutum prematurely terminates and is non-functional, and is the underlying reason for the susceptibility of G. hirsutum (Chen et al., 2015; Li N.Y. et al., 2017). In this study, we further investigated the Verticillium wilt resistance function of GbaNA1 by heterologous expression in Arabidopsis thaliana. The main objectives of the current study were to: (1) study the role of the functional GbaNA1 in Verticillium wilt resistance in A. thaliana; (2) detect whether GbaNA1 has the ability to recover the function of GbaNA1 ortholog mutant in A. thaliana; (3) explore the defense responses mediated by GbaNA1 in A. thaliana; and (4) use the transgenic Arabidopsis to confirm the loss of resistance gene function owing to the truncation of GbaNA1 homolog in G. hirsutum.
Materials and Methods
Culture Condition and Inoculation Method
Arabidopsis thaliana seedlings were grown in pots with potting soil (PINDSTRUP, Denmark) including 20% vermiculite in a greenhouse at temperatures of 24°C during the day and 20°C at night, 60–70% relative humidity, and under a 16/8 light-dark photoperiod. The highly virulent V. dahliae strain Vd991 (used in all experiments) was cultured in potato dextrose broth (PDB) medium at 25°C for 7 days on a shaker. Conidia were harvested by centrifugation and washed with sterile water; the final concentration was adjusted to 5 × 106 conidia/mL using a hemocytometer. For inoculations with V. dahliae, A. thaliana seedlings were uprooted, and the roots were dipped in V. dahliae conidial suspension for 2 min followed by replanting into vermiculite soil. Verticillium wilt symptoms were recorded 3 weeks after inoculation.
For fungal biomass quantification, stems of three inoculated plants per gene target (one per replicate) were harvested at 21 days post-inoculation. qPCR was performed using a SYBR premix Ex Taq II kit (TaKaRa, Japan) with primers specific to the A. thaliana ubiquitin extension protein 1 (UBQ1, NM_115119.4) and V. dahliae elongation factor 1-α (EF-1α) (Supplementary Table S1).
Gene Cloning
To clone GbaNA1 (MF078620), 3-week-old seedlings of Gossypium barbadense cv. Hai7124 were inoculated with 5 mL of 5 × 106 conidia/mL conidial suspension, and root samples were collected 72 h after inoculation. Total RNA was extracted using a Plant RNA Purification Kit (Tiangen, Beijing, China), and cDNA was synthesized by using a RevertAidTM First Strand cDNA Synthesis Kit from MBI (Fermentas, Glen Burnie, Maryland, MA, United States). Primers were designed according to the full open reading frame (ORF) of the gene Gorai.007323100.1 in the G. raimondii reference genome (Paterson et al., 2012; Supplementary Table S1). Primers were used to amplify the target fragment from genomic DNA and cDNA. The PCR conditions consisted of an initial 94°C denaturation step for 10 min, followed by 36 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min. PCR products were cloned into the pGEM-T-Easy vector (Promega, Madison, WI, United States) and confirmed by sequencing. GhNA1 (MF078621) from G. hirsutum was sequenced using the same method.
Sequence Analysis
The ORFs of GbaNA1 were determined using ORF Finder1, and the protein sequences were deduced on the basis of codon sequences. The conserved domains of GbaNA1 were predicted using the InterProScan database (Version 5.21) as described (Li N.Y. et al., 2017). The protein coding region of the NBS-LRR gene (AT4G27220.1), orthologous to GbaNA1 in G. barbadense, was acquired by BLASTp analysis using GbaNA1 as a query against A. thaliana proteins. The typical motifs of known NB-ARC and LRR domains in AT4G27220.1, including P-loop, RNBS-A, Kinase 2, RNBS-B, RNBS-C, GLPL, RNBS-D and MHD, and that the LRR domain contained 12 imperfect LRRs, were determined by protein sequence alignment with the NB-LRR protein GbaNA1. ClustalX 1.83 software was used for the multiple sequence alignment (Thompson et al., 1997).
Generation and Analysis of Transgenic A. thaliana
The ORF fragments from GbaNA1 and GhNA1 (GbaNA1 allelic gene in G. hirsutum) were amplified with primers containing Sac I and BstB I enzyme sites and were integrated into the binary vector pFAST-G02 under the control of the CaMV35S promoter. The recombinant plasmid (pFAST-G02::GbaNA1) was transformed into Agrobacterium tumefaciens (strain LBA4404) and introduced into 4-week-old A. thaliana (ecotype Col-0) plants using an agrobacterium-mediated floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS medium containing 50 mg/L Basta, and the T3 homozygous transgenic plants were identified with PCR and RT-PCR using the genomic DNA and cDNA samples, respectively; wild-type gDNA and cDNA were used as controls. The amplification conditions consisted of an initial 94°C denaturation step for 10 min, which was followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and UBQ1 was used as a control. GbaNA1 was also introduced into the A. thaliana mutant At4g27270 (na1, SALK_ 127692, the GbaNA1 ortholog in A. thaliana) as described above. The phenotypes of transgenic plants resistant to V. dahliae Vd991 were assayed using a root-dip method as described above with 5 mL of V. dahliae Vd991 conidial suspension (2 × 106 conidia/mL). The development of fungal biomass in plant tissue was determined by absolute quantification using a method similar to that previously described (Santhanam et al., 2013). In this study, qPCR was performed using SYBR premix Ex Taq II kit (TaKaRa, Japan) with primers specific to the A. thaliana UBQ1 gene and V. dahliae elongation factor 1-α (EF-1α).
ROS Accumulation Detection with DAB Staining
ROS accumulation was detected in transgenic A. thaliana and wide-type (Col-0) leaves from 3-week-old plants 12 h after infiltration with 50 μL V. dahliae (strain Vd991) conidia suspension (2 × 106 conidia/mL) using 3′3-diaminobenzidine (DAB) solution as previously described (Bindschedler et al., 2006). A sterile water treatment was used as the control. Briefly, the leaves were treated with 1 mg/ml DAB containing 0.05% v/v Tween 20 and 10 mM sodium phosphate buffer (pH 7.0). Then the leaves were incubated at 25°C in the dark, and infiltrated under gentle vacuum. The reaction was terminated at 10–12 h post-inoculation and the DAB solution was removed with a distilled water rinse. Ethanol (75%) was then added to the leaves to remove the chlorophyll and placed in 30% glycerol after the decolorization. Six leaves per treatment were included in each of the three replicates. Samples were observed using a SMZ18 stereo microscope (Nikon, Japan) and the percent of brown pixels of every image from the six leaves examined for each treatment and replication of the same size and resolution was included in obtaining the statistics using the Matlab software.
Relative Gene Expression Analysis
For the expression analysis of GhNA1 in cotton, 3-week-old seedlings of G. hirsutum Junmian No.1 were inoculated with 5 mL of conidial suspension (5 × 106 conidia/mL) of V. dahliae Vd991 using a root-dip method. The inoculated root samples were collected at six time points (2, 6, 12, 24, 48, and 72 h) after inoculation, with three seedlings for each sample. For the expression analysis of ethylene signaling-associated genes and defense response genes in transgenic A. thaliana, wild-type (Col-0), GbaNA1-overexpression transgenic line (OE1), GbaNA1 ortholog gene mutant (na1), and the transgenic line of na1 mutant with complemented GbaNA1 were inoculated with 5 × 106 conidia/mL of V. dahliae (strain Vd991) conidia suspension using a root-dip method. Three root samples from each treatment were collected at 24 h after inoculation. RT-qPCR analyses were performed using the SYBR Premix Ex Taq kit (Takara) and a QuantStudio 6 Flex Real Time PCR System (Applied Biosystems, Foster City, CA, United States). PCR conditions consisted of an initial denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. The A. thaliana UBQ1 was used as endogenous control. All detections were carried out with three independent biological replicates. The relative expression levels of genes were evaluated using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Results
Heterologous Overexpression of GbaNA1 Enhanced Verticillium Wilt Resistance in A. thaliana
In our previous study, silencing of island cotton NBS-LRR gene GbaNA1 impaired resistance to the non-race 1 V. dahliae isolate Vd991 in cotton (Li N.Y. et al., 2017). To investigate the role of GbaNA1 in the defense against V. dahliae, the gene was heterologously transferred into the A. thaliana genome using an Agrobacterium tumefaciens-mediated transformation method (Clough and Bent, 1998). The overexpression transformation construct GbaNA1 driven by the CaMV35S (35S) promoter (P35S:GbaNA1) was transferred into A. thaliana (ecotype Col-0) via Agrobacterium tumefaciens-mediated transformation (Figure 1A). Of the independent T3 transgenic lines obtained, the introduced gene could be detected in all eight GbaNA1-overexpression lines (OE1 – OE8) using PCR primers specific to GbaNA1 (Figure 1B). Reverse transcription-PCR (RT-PCR) analysis further confirmed that the integrated genes were successfully expressed, as the GbaNA1 transcript could be detected in the transgenic lines but not in the wild type Col-0 (Figure 1C). For Verticillium wilt resistance tests, 4-week-old seedlings of three transgenic lines were arbitrarily selected for inoculation with a highly virulent V. dahliae strain Vd991. Relative to the wild-type Col-0, GbaNA1-overexpressing lines exhibited significantly enhanced resistance to V. dahliae Vd991, as indicated by a reduction of leaf chlorosis and withering (Figure 1D). Furthermore, quantitative PCR (qPCR) analysis of fungal biomass demonstrated significantly less V. dahliae biomass (<20%) in transgenic plants than in the wild-type plants (Figure 1E). These results suggested that the island cotton NBS-LRR gene GbaNA1 conferred resistance to strain Vd991 after interfamily transfer into A. thaliana ecotype Col-0.
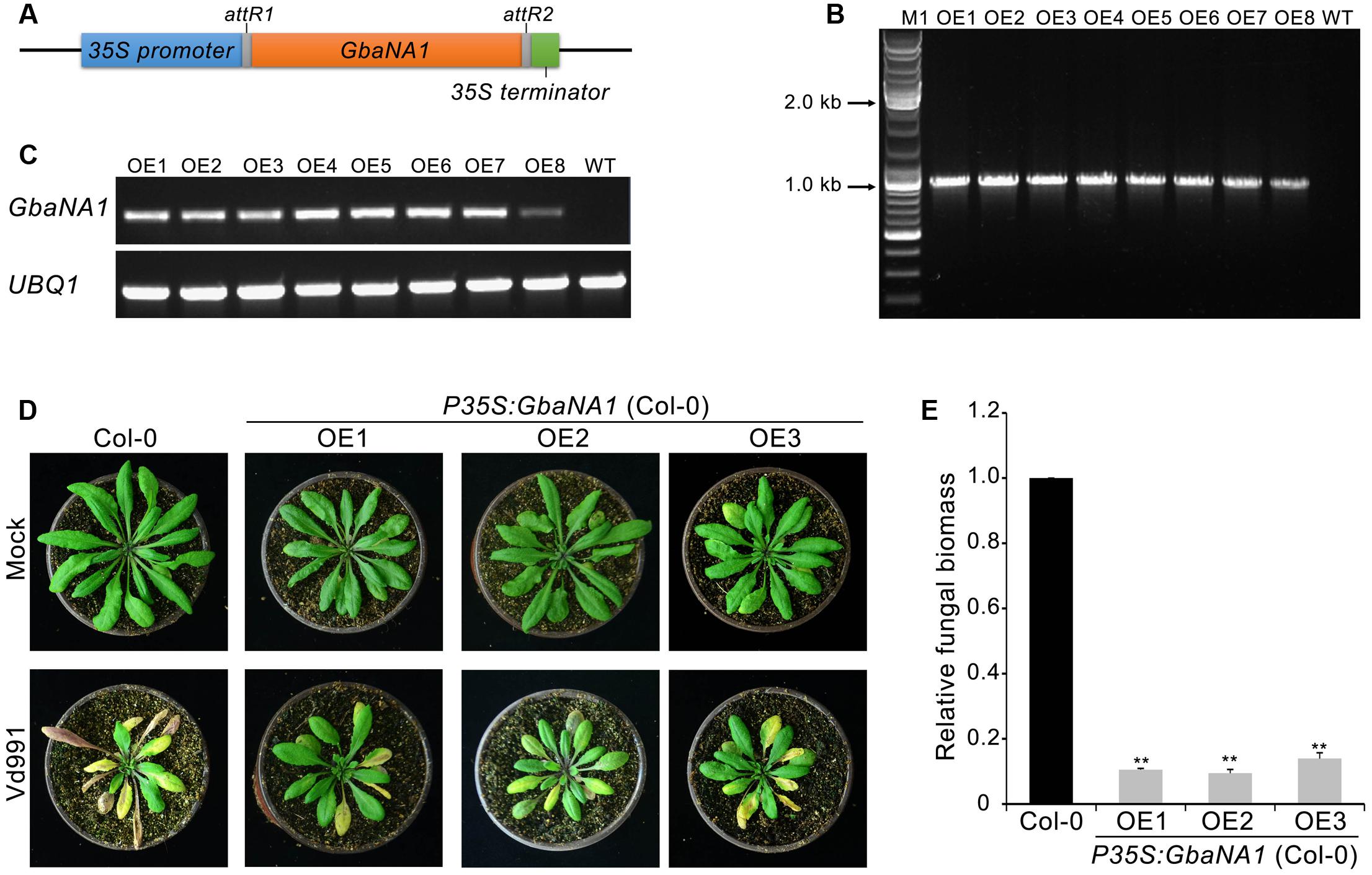
FIGURE 1. Transgenic expression of GbaNA1 enhances Verticillium wilt resistance in Arabidopsis thaliana. (A) Diagram of the GbaNA1-overexpressing transformation vector pFAST:GbaNA1. (B) PCR products targeting a fragment of GbaNA1 amplified from DNA extracted from GbaNA1-overexpressing transgenic lines of A. thaliana ecotype Col-0. (C) RT-PCR amplification of GbaNA1 cDNA in the same transgenic A. thaliana. UBQ1 is shown as a control. (D) Phenotype assay of GbaNA1 transgenic A. thaliana inoculated with V. dahliae, strain Vd991. A. thaliana plants were engineered to express cotton CaMV 35S-driven GbaNA1 (P35S:GbaNA1). Three-week-old seedling of wild-type (Col-0) and transgenic lines (OE1, OE2, and OE3) inoculated with V. dahliae strain Vd991 via root-dipping in a suspension of 5 × 106 conidia/mL or sterile water (Mock). Disease symptoms were observed 21 days after inoculation. (E) Quantification of V. dahliae biomass in GbaNA1 transgenic A. thaliana plants (OE1, OE2, and OE3) compared to the wild-type (Col-0). Relative fungal biomass was determined using quantitative real-time PCR and genomic DNA extracted from total plants of three plants 21 days after inoculation. V. dahliae elongation factor 1-α (EF-1α) was used to quantify fungal colonization, and A. thaliana UBQ1 was used as endogenous plant control. Error bars represent standard errors of three biological replicates, ∗∗indicates statistical significance (P < 0.01), according to unpaired Student’s t-tests.
The Orthologous GbaNA1 Mutant in A. thaliana Is Susceptible to V. dahliae
The A. thaliana NBS-LRR gene (AT4G27220.1) is the ortholog of GraNA1 (Gorai.007G323100.1) in G. raimondii (Paterson et al., 2012), and GraNA1 and GbaNA1 are allelic between G. raimondii and G. barbadense (Li N.Y. et al., 2017). BLASTp analysis using GbaNA1 as a query against A. thaliana proteins returned AT4G27220.1 as the best hit (Identities = 238/785, 30%; Positives = 389/785, 49%), suggesting that GbaNA1, G. raimondii Gorai.007323100.1 and AT4G27220.1 are orthologous genes. AT4G27220.1 also belongs to the NB-ARC domain-containing disease resistance protein family. Protein sequence alignment showed that although the sequences display significant differences because cotton and A. thaliana are phylogenetically divergent, many residues (especially the residues associated with the NB-ARC and LRR domains) were conserved between GbaNA1 and AT4G27220.1 (Figure 2A), and contained the typical motifs of known NB-ARC and LRR domain-containing proteins, including P-loop, RNBS-A, Kinase 2, RNBS-B, RNBS-C, GLPL, RNBS-D and MHD, and the LRR domain containing 12 imperfect LRRs (Figure 2A).
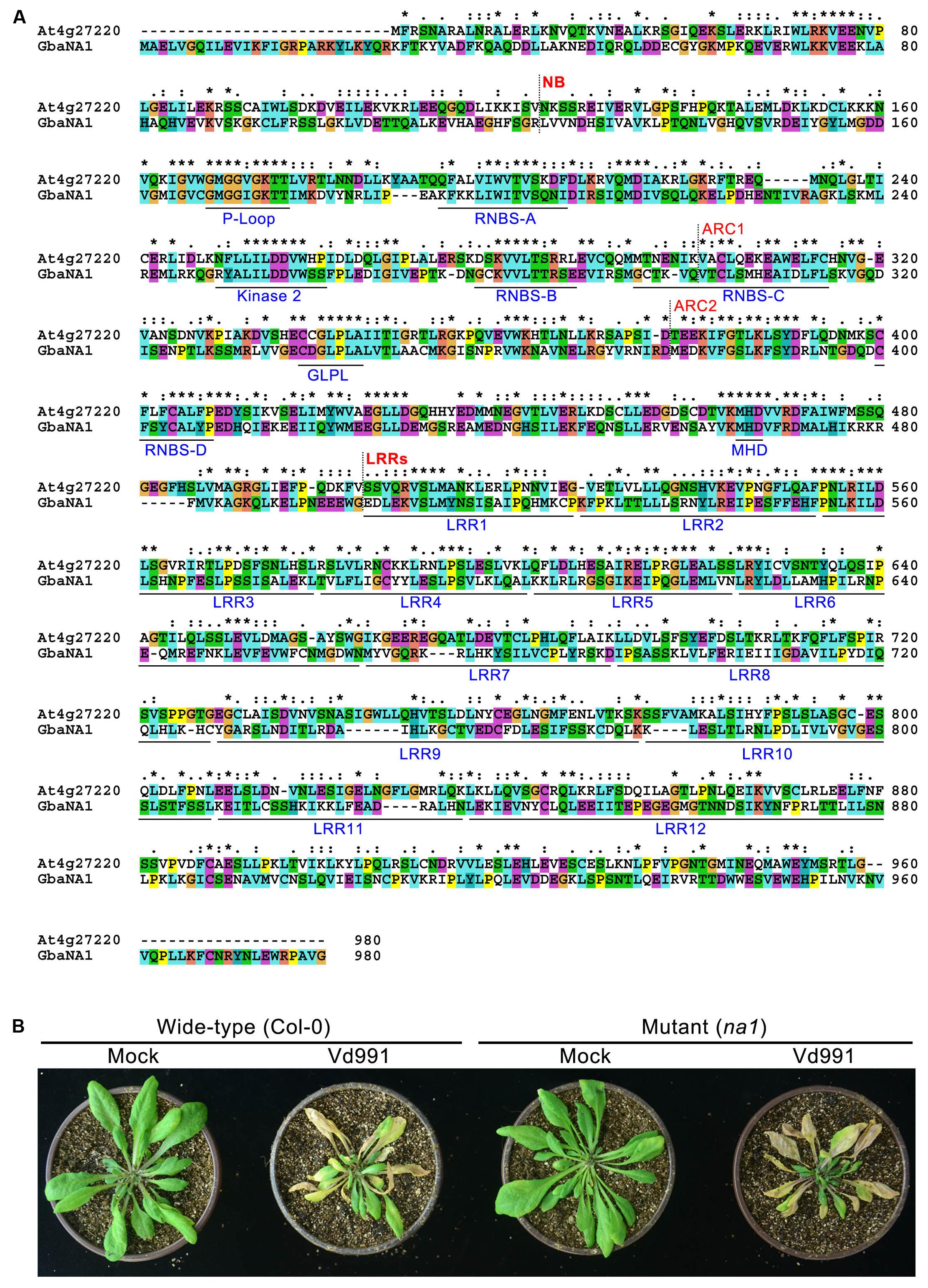
FIGURE 2. Characteristics of GbaNA1 orthologous proteins in Arabidopsis thaliana. (A) Protein sequence alignment of GbaNA1 and its orthologous sequence in A. thaliana Col-0. The alignment was performed by Clustal X2 with a GONNET 80 protein weight matrix. Asterisks represent conserved resides. (B) Verticillium wilt phenotype A. thaliana line At4g27220, a T-DNA mutant of the gene orthologous to GbaNA1 (NA1). Two-week-old seedlings of At4g27220 (genotype na1) and the wild-type (Col-0) were inoculated with V. dahliae strain Vd991 by root-dipping in a suspension of 5 × 106 conidia/mL. Roots were dipped in sterile water as controls (Mock). Phenotypes were investigated 14 days after inoculation.
To further confirm whether the GbaNA1 ortholog gene, AT4G27220.1 is involved in Verticillium wilt resistance, the response of mutant AT4G27220.1 (Germplasm/Stock in TAIR: SALK_127692, hereinafter named na1) to V. dahliae isolate Vd991 was tested using the root dip inoculation method. The pathogenicity assay showed that the mutant line na1 displayed greater sensitivity to V. dahliae compared with the wild-type Col-0 ecotype, indicated by a significant increase in leaf chlorosis and wilting 2 weeks after inoculation (Figure 2B). Investigation of the fungal biomass by qPCR analysis suggested rapid multiplication in the na1 lines compared to wild-type Col-0 ecotype (Figure 3B). Together, these results showed that the GbaNA1 ortholog gene AT4G27220.1 conferred Verticillium wilt resistance in A. thaliana.
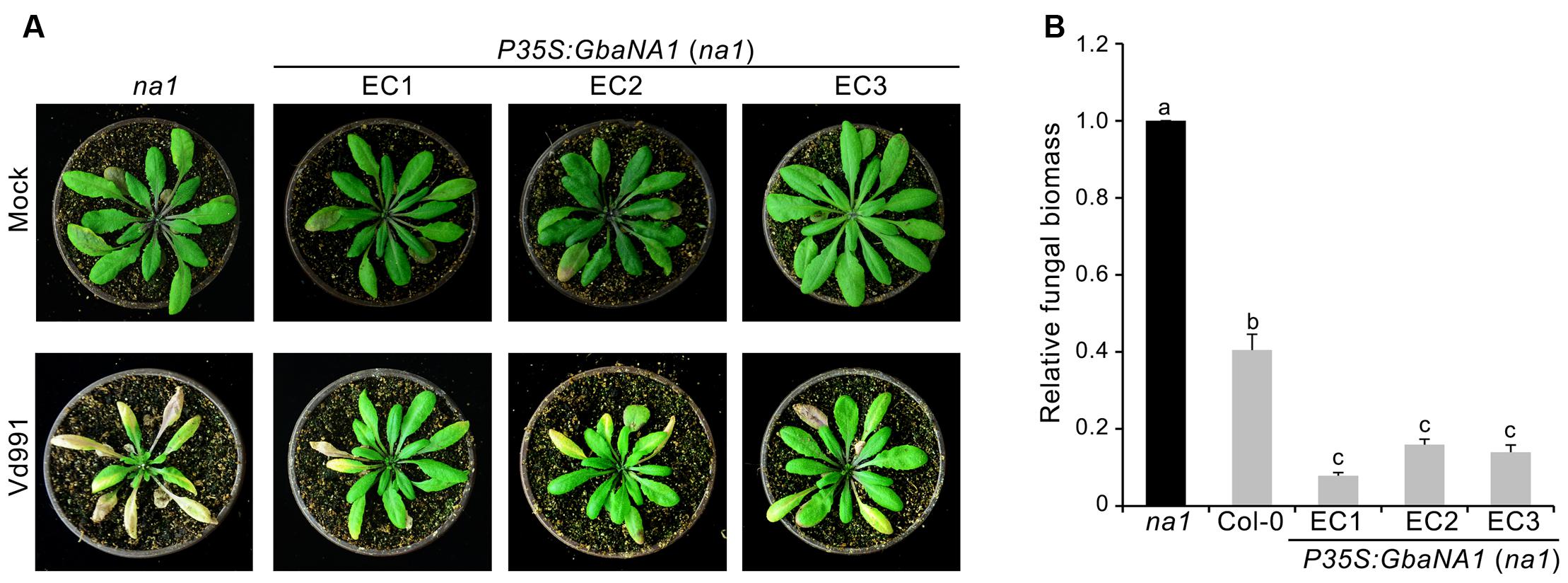
FIGURE 3. Overexpression of cotton GbaNA1 in Arabidopsis thaliana mutant At4g27220 (na1) enhances the Verticillium wilt resistance of A. thaliana. (A) Enhancement of the Verticillium wilt resistance of na1 A. thaliana mutants by GbaNA1 overexpression. Overexpression of GbaNA1 was driven by the CaMV 35S promoter. Three-week-old transgenic lines (EC1, EC2 and EC3) were subjected to a root-dip inoculation in a suspension of 5 × 106 conidia/mL of V. dahliae, strain Vd991. Verticillium wilt symptoms were assessed 21 d after inoculation. Sterile water root dips were used as controls (Mock). (B) Quantification of V. dahliae biomass in transgenic GbaNA1 overexpression lines of na1 mutants, na1 mutant and the wild-type (Col-0). Relative fungal biomass was determined using quantitative real-time PCR and genomic DNA extracted from total plants 21 days after inoculation. V. dahliae elongation factor 1-α (EF-1α) was used to quantify fungal colonization, and A. thaliana UBQ1 was used as an endogenous plant control. Error bars represent standard errors of three biological replicates. Different letters above the bars indicate significant differences (P < 0.01).
Overexpression of GbaNA1 Reduced Symptom Severity in the AT4G27220 Mutant
To further investigate the involvement of GbaNA1 in Verticillium wilt defense responses, the sensitivity to V. dahliae were assessed in the mutant na1 of A. thaliana after receiving the gene GbaNA1 driven by the 35S promoter. The ectopic transformants were verified by PCR and the expression of GbaNA1 was detected by RT-PCR, and several independent ectopic transformants were obtained, of which three were used for further analysis in this study (Supplementary Figures S1A,B). Inoculation of three separate transgenic na1 lines complemented with genes GbaNA1 displayed significantly less chlorosis and wilting compared with the na1 mutant (Figure 3A). Quantification of fungal biomass by qPCR demonstrated that na1 complemented with GbaNA1 developed significantly less fungal biomass than the non-complemented na1 mutants (Figure 3B), consistent with constitutive over-expression of GbaNA1 driven by the 35S promoter in the wild-type Col-0 ecotype (Figure 1D). These results indicated that GbaNA1 can restore the Verticillium wilt resistance mediated by the ortholog AT4G27220.1 in A. thaliana, and further confirms the significant role of GbaNA1 in reducing V. dahliae colonization and symptom severity.
Overexpression of GbaNA1 Enhanced the Defense Response of ROS Activation
To explore the V. dahliae defense responses mediated by GbaNA1, ROS accumulation was assessed in leaves of A. thaliana ecotype Col-0, GbaNA1 transgenic Col-0 mutants, na1 mutants, and na1 mutants complemented with GbaNA1 after leaf infiltration with a conidia suspension of V. dahliae strain Vd991. Leaves of the wild-type Col-0 displayed more ROS accumulation around infiltration sites (indicated by dark brown deposits visible in leaves) 12 h post-infiltration with conidial suspension, compared to infiltration with sterile water (Figures 4A,C). Col-0 GbaNA1 transgenic plants displayed significantly more ROS accumulation relative to the wild-type Col-0 plants (Figures 4A,C). Similarly, overexpression of GbaNA1 in na1 mutant plants resulted in higher ROS accumulation than the levels observed in the na1 mutant plants following inoculation (Figures 4B,D). These results demonstrated that GbaNA1 has the ability to enhance the defense response to V. dahliae through GbaNA1-mediated ROS activation.
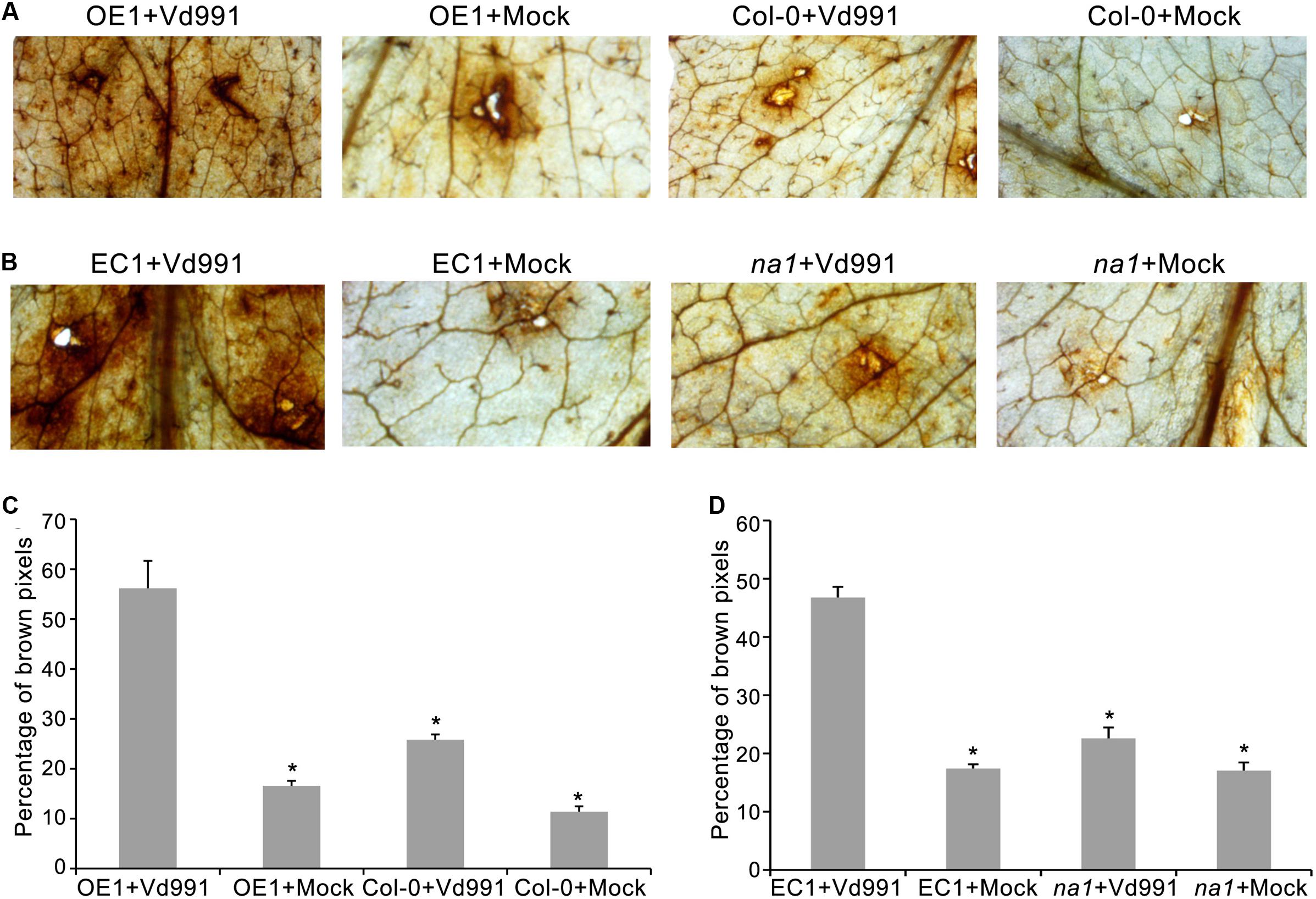
FIGURE 4. Cotton GbaNA1 modulates ROS accumulation in Arabidopsis thaliana. (A) The ROS-inducing activities of GbaNA1 transgenic A. thaliana and wide type (Col-0) plants inoculated with V. dahliae, strain Vd991. ROS accumulation was assessed in GbaNA1 transgenic A. thaliana and wide type (Col-0) leaves from 3-week-old plants 12 h after infiltration with a 50 μL suspension (5 × 106 conidia/mL) of V. dahliae, strain Vd991. Sterile water treatments were used as controls (Mock). (B) Detection of ROS-inducing activities of A. thaliana na1 mutants and na1 mutants after introduction of GbaNA1. (C) The percent of brown pixels of GbaNA1 transgenic A. thaliana (OE1) and wild-type (Col-0) plants inoculated with V. dahliae strain Vd991. ROS accumulation level was assessed in GbaNA1 transgenic A. thaliana and wide type (Col-0) leaves from 3-week-old plants 12 h after infiltration with a 50 μL conidial suspension (5 × 106 conidia/mL) of V. dahliae, strain Vd991 followed by staining with DAB. Sterile water treatments were used as controls (Mock). (D) Detection of ROS-inducing level of A. thaliana na1 mutants and na1 mutants after the introduction of GbaNA1. EC1, the transgenic lines of overexpression GbaNA1 in na1 mutant.
Ethylene Signaling Is Critical for GbaNA1-Mediated Resistance against V. dahliae
Previous results of the expression pattern of GbaNA1 after treatment with V. dahliae strain Vd991 and ET were similar in cotton (Li N.Y. et al., 2017), suggesting that defense responses mediated by GbaNA1 were also associated with ethylene signaling in A. thaliana. To test this association, the relative expression of seven genes (ERF3, ERF4, ERF13, ERF104, EIN2, ETR2) in the ethylene signaling pathway was measured using reverse transcription-quantitative PCR (RT-qPCR) in Col-0, GbaNA1 transgenic Col-0 mutants, na1 mutants, and na1 mutants complemented with GbaNA1 24 h post-inoculation with V. dahliae strain Vd991. Expression levels of six genes (except for ERF13) in GbaNA1 transgenic Col-0 mutants were significantly up-regulated relative to the wild-type plants after inoculation with V. dahliae strain Vd991 (Figure 5). As expected, the expression of six genes suppressed in na1 mutant plants (lacking of AT4G27220.1) were restored to the relatively high expression level after complementation of the ortholog of GbaNA1 (Figure 5). The relative expression ERF13 in the ethylene signaling pathway could still be significantly activated by GbaNA1 after inoculation with V. dahliae strain Vd991, but was not affected by the ortholog AT4G27220.1 in A. thaliana (Supplementary Figure S2). Furthermore, the defense response genes SCL14 and PR5 were significantly up-regulated after overexpression of GbaNA1 in the wild-type Col-0 and na1 mutant plants, but were suppressed in the na1 mutant plants due to the lack of GbaNA1 ortholog in A. thaliana. These results suggested that ethylene signaling is crucial for GbaNA1-mediated defense responses against V. dahliae strain Vd991.
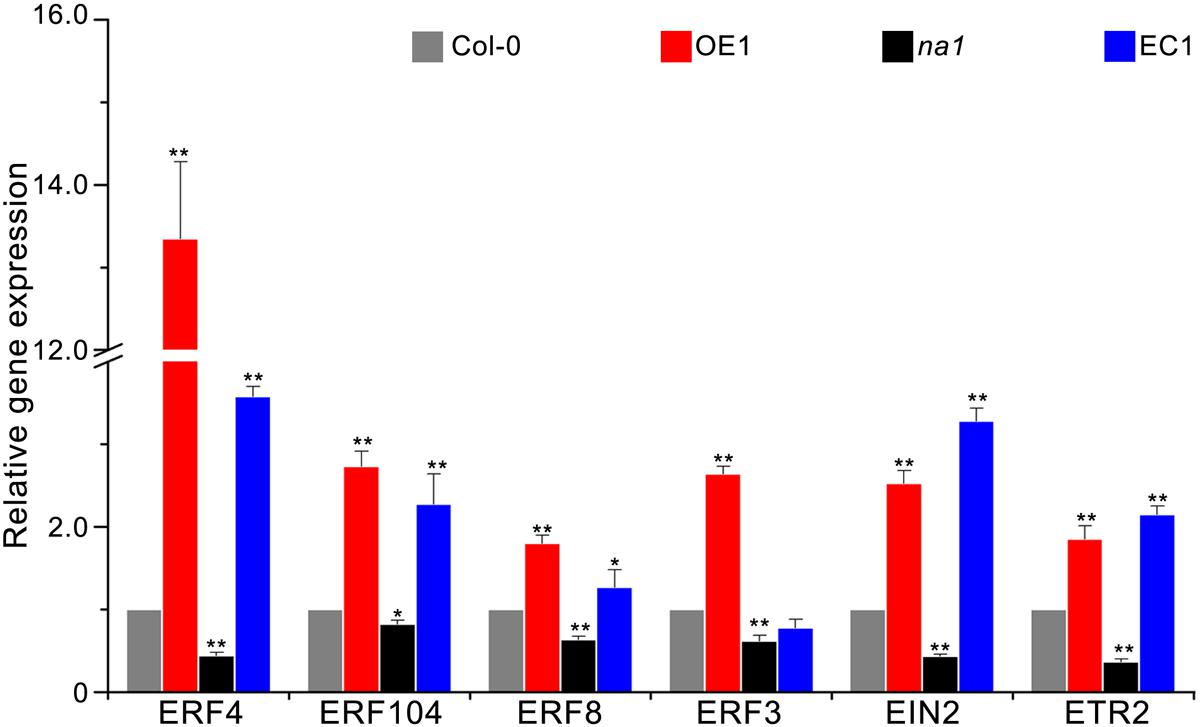
FIGURE 5. GbaNA1 regulates the expression levels of ethylene signaling-associated genes. Relative expression of six ethylenesignaling-associated genes in A. thaliana lines. Wild type Col-0, GbaNA1-overexpression transgenic line OE1, GbaNA1 ortholog mutant na1, and a GbaNA1 overexpression transgenic line of na1 were inoculated with a suspension of 5 × 106 conidia/mL of V. dahliae (strain Vd991) using a root-dip method. Root samples were collected for RNA isolation and cDNA synthesis 24 h after inoculation. Relative expression of ethylene signaling-associated genes was assessed by reverse transcription-quantitative PCR using the comparative threshold 2-ΔΔCT method and A. thaliana UBQ1 as a reference. Values represent averages of three independent biological replicates. Error bars represent standard errors. Asterisks (∗) and double asterisks (∗∗) represents statistical significance of P < 0.05 and P < 0.01, respectively, according to an unpaired Student’s t-tests.
Overexpression of GbaNA1 Homolog in G. hirsutum Results in Loss of Resistance Gene Function
A previous study revealed high allelic divergence in GbaNA1, between the homologs of the Verticillium wilt susceptible G. hirsutum and the resistant G. barbadense (Li N.Y. et al., 2017). The premature termination of the protein encoded by GbaNA1 homologs in the G. hirsutum accessions results in a lack of the most conserved motifs in the NB-ARC domain (Li N.Y. et al., 2017). Cloning the GbaNA1 homolog (GhNA1) from G. hirsutum accessions following RT-PCR confirmed that the coding sequence consisted of 756 bp (Figure 6A) which would encode a product of 251 aa residues in length. RT-qPCR analysis showed that GhNA1 was not responsive to infection with V. dahliae strain Vd991 (Figure 6B). To assess the relationship between transgenic GhNA1 plants and Verticillium wilt resistance, GhNA1-overexpressing A. thaliana transgenic lines were generated, and the positive ectopic transformants, and those expressing the gene, were determined by PCR and RT-PCR (Supplementary Figures S1C,D), respectively. Unlike the GbaNA1-overexpressing lines, which displayed enhanced the resistance to V. dahliae, the GhNA1-overexpressing lines displayed no enhanced resistance against V. dahliae strain Vd991, and the disease symptom severity and in planta fungal biomass were not significantly different from the wild-type Col-0 inoculated with V. dahliae (Figures 6C,D). In addition, the accumulation of ROS in the GhNA1 transgenic lines was also similar to the wild-type Col-0 12 h after infiltration with a suspension of V. dahliae conidia (Figure 6E and Supplementary Figure S3). These results suggested that GhNA1 of G. hirsutum no longer confers resistance to Verticillium wilt due to its premature termination, unlike the allele from G. barbadense cultivars, which would yield the full-length protein product.
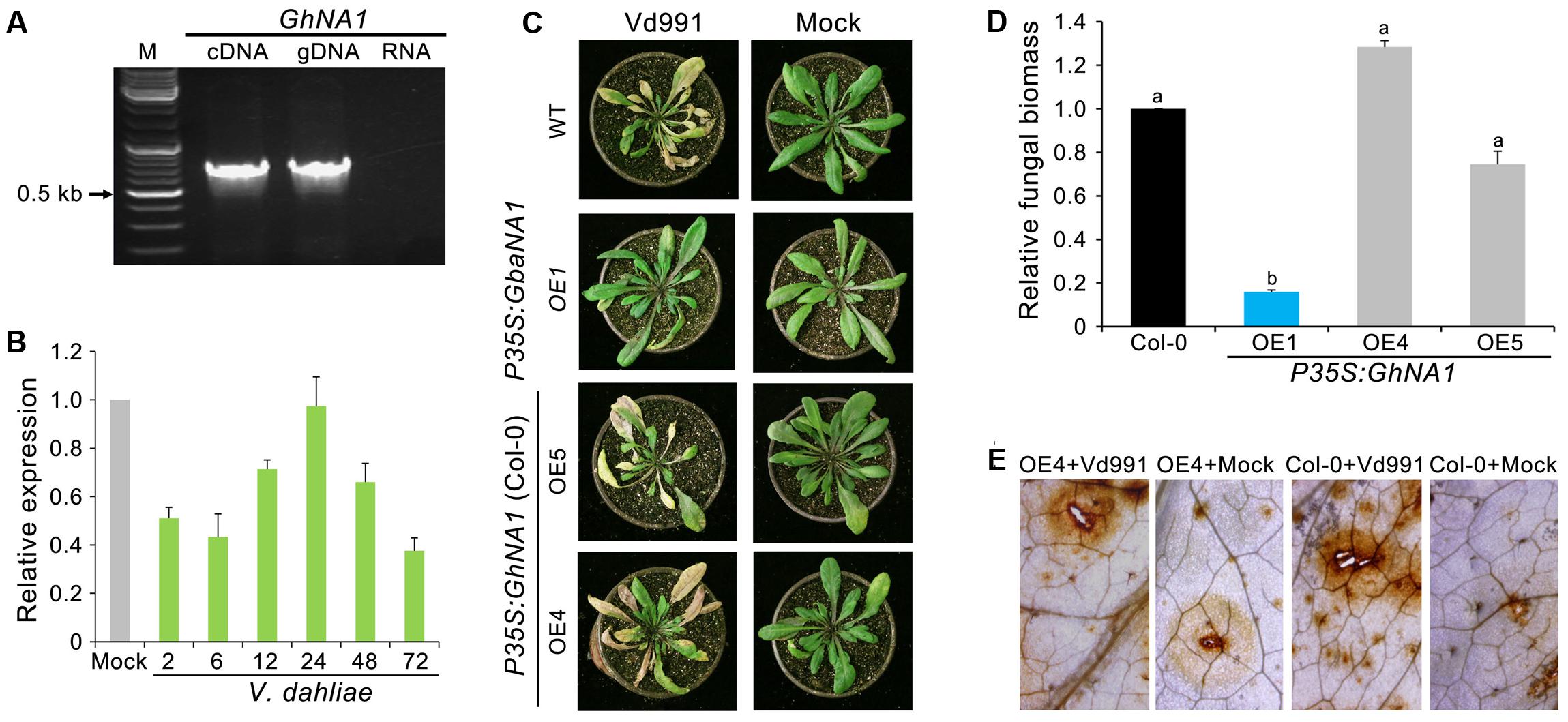
FIGURE 6. The GbaNA1 homolog from G. hirsutum does not enhance Verticillium resistance in Arabidopsis thaliana. (A) Cloning GhNA1 from the Verticillium wilt susceptible G. hirsutum cv. Junmian No. 1. GhNA1 was cloned by reverse transcription-PCR using the cDNA template and genomic DNA, respectively. DNA contamination in the RNA sample was assayed by PCR (RNA lane). (B) Expression analysis of GhaNA1 in G. hirsutum cv. Junmian No. 1 after inoculation with V. dahliae Vd991. Three-week-old cotton seedlings were root-dipped (2 × 106 conidia/mL) collected over a time-course and RNA was extracted from roots. The relative expression levels of GhNA1 were assessed by quantitative reverse transcriptase PCR, using the A. thaliana UBQ1 gene as a reference. Plants treated with sterile water were used as controls (Mock). Error bars represent standard errors of three biological replicates. (C) Verticillium wilt symptoms of GhNA1 transgenic A. thaliana inoculated with V. dahliae. Two independent transgenic lines of 3-week-old Col-0 seedlings were inoculated with V. dahliae (strain Vd991) via root-dipping in a suspension of 5 × 106 conidia/mL. The GbaNA1 transgenic line (OE1) and wide type (Col-0) served as positive and negative controls, respectively. Mock inoculations were performed with sterile water. (D) Quantification of V. dahliae biomass in GhNA1 transgenic A. thaliana plants (OE4, OE5, and OE6) compared to the wild type (Col 0). Fungal biomass was determined by quantitative real-time PCR using genomic DNA extracted from whole plants 21 days after inoculation. Error bars represent standard errors of three biological replicates. Columns with different letters indicate statistical significance (P < 0.01), according to unpaired Student’s t-tests. (E) Detecting the ROS-inducing activities of GhNA1 transgenic A. thaliana. Leaves from 3-week-old plants were visualized 12 h after infiltration with 50 μL V. dahliae, strain Vd991 (5 × 106 conidia/mL). Sterile water treatment served as a control (Mock). Leaves were stained with DAB.
Discussion
Improving genetic resistance is the preferred method to manage Verticillium wilt in most crops (Schaible et al., 1951; Putt, 1964; Simko et al., 2004; Bolek et al., 2005; Zebrowska et al., 2006), but is also the most difficult to implement because of the general lack of effective resistance genes against this disease. In our previous study, we identified an island cotton NBS-LRR gene GbaNA1, which conferred resistance to the non-race 1 V. dahliae strain Vd991, and found that the GbaNA1 homolog in susceptible G. hirsutum displayed premature termination and was therefore non-functional (Li N.Y. et al., 2017). In this study, we investigated the Verticillium wilt resistance function of GbaNA1 by ectopic expression in A. thaliana, and found that ROS activation and ethylene signaling were critical for GbaNA1-mediated resistance against V. dahliae.
Several NB-LRR genes have been identified to function as R genes in A. thaliana, including RPM1, RPS1, RPS2, RPS4, and RPS5 (Lee and Yeom, 2015). In cotton, the genome has an expanded repertoire of NBS-encoding genes (Xiang et al., 2017), even notable in the diploid genome of G. raimondii that encodes more than 300 of these types of genes (Paterson et al., 2012). Comparative genomic analysis showed that the NBS-encoding genes are significantly expanded in G. raimondii, which is nearly immune to Verticillium wilt, as compared to G. arboretum, which is susceptible to Verticillium wilt (Li et al., 2014). Large scale transcriptome analysis of a cotton response to V. dahliae revealed that the NBS-encoding genes may be involved in Verticillium wilt resistance (Xu et al., 2011, 2014; Sun et al., 2013; Zhang et al., 2013b, 2017; Shao et al., 2015). However, few of the candidate NBS-encoding genes involved in Verticillium wilt resistance have been cloned or studied in cotton, except for the NBS-LRR-encoding GbRVd (Yang et al., 2016).
Identification of effective Verticillium wilt resistance genes is difficult because of the complexity of the cotton genome and aggressive pathogenicity of V. dahliae on most cultivars, although at least 80 different Verticillium wilt resistance quantitative trait loci (QTLs) have been reported on cotton (Wang et al., 2007, 2008, 2012; Yang et al., 2008; Jiang et al., 2009; Zhao et al., 2014). At present, the homology-based cloning or differential expression screening are generally employed to clone Verticillium wilt resistance genes, and several genes have been identified and proven to play important roles during V. dahliae infection on cotton (Munis et al., 2010; Gao et al., 2013; Zhang et al., 2013b; Zhao et al., 2013; Mo et al., 2015; Duan et al., 2016; Li et al., 2016; Yang et al., 2016; Gong et al., 2017; Liu H. et al., 2017). However, only a few of these have the R gene characteristics.
Recently, several genes possessing the R gene characteristics of the receptor-like proteins were cloned from G. barbadense using homology-based cloning methods (Zhang et al., 2011, 2012; Yang et al., 2015; Chen et al., 2017), following the discovery of the tomato receptor like protein Ve1 that specifically mediates the resistance to the V. dahliae race 1 strain (Kawchuk et al., 2001; Fradin et al., 2009). In our previous study, 26 Verticillium dahliae resistance loci (VdRLs) were identified in G. barbadense by the bioinformatics-driven method based on the resistance gene analogue (RGAs) clusters and their transcriptome (Chen et al., 2015). Finally, we obtained the NBS-LRR gene (GbaNA1) from the VdRL08 locus that is involved in Verticillium wilt resistance (Li N.Y. et al., 2017). Compared with the resistant G. barbadense, the premature termination of the protein encoded by GbaNA1 homologs in the Verticillium wilt susceptible G. hirsutum accessions resulted in a truncated protein that lacks the most conserved motifs in the NB-ARC domain (Li N.Y. et al., 2017). These conserved motifs are important for NB-LRR function in disease resistance (Tameling et al., 2002; Tornero et al., 2002; Williams et al., 2011; Wang et al., 2015). To our knowledge, GbaNA1 is the first typical NBS-LRR protein to be involved in Verticillium wilt resistance in cotton, and an ortholog can be functional in another plant family, as confirmed in this study using A. thaliana transgenic lines.
The recognition of a specific pathogen effector (elicitor) by a corresponding R protein, including the NBS-LRR protein encoded by R genes, can initiate a cascade of defense responses, including a hypersensitive response, ROS production, hormone synthesis and signaling transport, and activation of defense-related genes (De Young and Innes, 2006; Caplan et al., 2008; Elmore et al., 2011). In the case of NBS-LRR proteins, tomato Mi-1.1 and 1.2 have been shown to play dual regulatory roles in regulating host cell death (Lukasik-Shreepaathy et al., 2012). The overexpression of VaRGA1 in Nicotiana benthamiana conferred enhanced resistance to Phytophthora parasitica through the activation of salicylic acid (SA) signaling and phenylpropanoid pathways (Li X. et al., 2017). In cotton, defense responses including hormone signaling, ROS scavenging and activation of the pathogenesis-related gene expression, were all proven to play roles in Verticillium wilt resistance (Gao et al., 2013; Zhang et al., 2013a; Mo et al., 2015; Duan et al., 2016; Yang et al., 2016; Gong et al., 2017; Liu H. et al., 2017). The NBS-LRR protein GbaNA1 can be significantly induced following treatment with the ethylene (Li N.Y. et al., 2017), and several genes encoding ethylene-responsive element-binding factor were significantly up-regulated after GbaNA1 overexpression in A. thaliana. Furthermore, ROS production was also significantly increased in GbaNA1-overexpressing A. thaliana lines compared with the wild-type plants. Ethylene and ROS are important signaling molecules mediating numerous important biological processes (Zhang et al., 2016). In A. thaliana, the crosstalk between the ethylene and ROS accumulation leads to stomatal closure and associated immunity after infection with Pseudomonas syringae (Mersmann et al., 2010). Activation of ethylene signaling pathways also enhances disease resistance by regulating ROS and phytoalexin production in rice during infection by Magnaporthe oryzae (Yang et al., 2017). Defense responses of ethylene signaling activation and increased ROS production can be mediated by the NBS-LRR proteins. For instance, overexpression of rice NBS-LRR resistance gene, OsBIHD1, resulted in enhanced expression of the ethylene synthesis genes involved in ethylene-mediated immunity (Liu H. et al., 2017); and the wheat NBS-LRR protein TaRCR1 regulating certain reactive oxygen species (ROS)-scavenging and production, play important roles in plant defense responses to the necrotrophic fungal pathogen, Rhizoctonia cerealis (Zhu et al., 2017). The activation of ethylene signaling is also evidenced by the negligible growth of transgenic lines relative to the wild-type plants (Figure 1D). Ethylene is a developmental regulator that is involved in manifold physiological processes throughout the plant life cycle (Iqbal et al., 2017). Together, this study of GbaNA1 in transgenic A. thaliana supports the hypothesis that ethylene signaling and increased ROS production are important for GbaNA1-mediated resistance against V. dahliae.
Conclusion
Our study found that heterologous overexpression of GbaNA1 enhanced Verticillium wilt resistance in A. thaliana, resulting in the activation of defense responses of ROS accumulation and genes associated with the ethylene signaling pathway. In A. thaliana, the NBS-LRR gene AT4G27220.1 is orthologous to GbaNA1, and the AT4G27220.1 mutant (na1) was susceptible to V. dahliae. Overexpression of GbaNA1 in na1 mutant plants restored activation of the expression of ethylene signaling pathway-related genes and increased ROS production, resulting in the restoration of A. thaliana resistance to V. dahliae. Moreover, heterologous expression of the GbaNA1 homolog from G. hirsutum (GhNA1) in A. thaliana could not activate the defense responses and enhance Verticillium wilt resistance, due to the truncation of GhNA1 and its product that lacks the conserved NB-ARC domain and LRR domain motifs. These results indicate that GbaNA1 encodes a structural R protein that confers Verticillium wilt resistance by mechanisms that are conserved across some plant families.
Author Contributions
X-FD, J-YC, and KS conceived and designed the experiments. N-YL, LZ, D-DZ, T-GL, and Y-JG performed the experiments. Z-QK, W-QZ, J-JL, and X-FM prepared biological material. X-FD and J-YC wrote the original draft. SK, DS, and KS edited and re-wrote parts of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2017YFD0200601 and 2017YFD0201900), the National Natural Science Foundation of China (31671986, 31471759, 31772245, and 31501600), the Special Public Welfare Industry Research on Agriculture (201503109), an Agricultural Science and Technology Innovation Program grant to X-FD, the Fundamental Research Funds for Central Non-profit Scientific Institution (Y2016CG11, S2016JC05, and S2016CG01).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00119/full#supplementary-material
Footnotes
References
Albrecht, M., and Takken, F. L. (2006). Update on the domain architectures of NLRs and R proteins. Biochem. Biophys. Res. Commun. 339, 459–462. doi: 10.1016/j.bbrc.2005.10.074
Anderson, P. A., Lawrence, G. J., Morrish, B. C., Ayliffe, M. A., Finnegan, E. J., and Ellis, J. G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9, 641–651. doi: 10.1105/tpc.9.4.641
Axtell, M. J., and Staskawicz, B. J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. doi: 10.1016/S0092-8674(03)00036-9
Bindschedler, L. V., Dewdney, J., Blee, K. A., Stone, J. M., Asai, T., Plotnikov, J., et al. (2006). Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 47, 851–863. doi: 10.1111/j.1365-313X.2006.02837.x
Bolek, Y., El-Zik, K. M., Pepper, A. E., Bell, A. A., Magill, C. W., Thaxton, P. M., et al. (2005). Mapping of Verticillium wilt resistance genes in cotton. Plant Sci. 168, 1581–1590. doi: 10.1016/j.plantsci.2005.02.008
Caplan, J., Padmanabhan, M., and Dinesh-Kumar, S. P. (2008). Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3, 126–135. doi: 10.1016/j.chom.2008.02.010
Catanzariti, A. M., Dodds, P. N., Ve, T., Kobe, B., Ellis, J. G., and Staskawicz, B. J. (2010). The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol. Plant Microbe Interact. 23, 49–57. doi: 10.1094/MPMI-23-1-0049
Chen, J., Li, N., Ma, X., Gupta, V. K., Zhang, D., Li, T., et al. (2017). The ectopic overexpression of the cotton Ve1 and Ve2-Homolog sequences leads to resistance response to Verticillium wilt in Arabidopsis. Front. Plant Sci. 8:844. doi: 10.3389/fpls.2017.00844
Chen, J. Y., Huang, J. Q., Li, N. Y., Ma, X. F., Wang, J. L., Liu, C., et al. (2015). Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biol. 15:148. doi: 10.1186/s12870-015-0508-3
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Collier, S. M., and Moffett, P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14, 521–529. doi: 10.1016/j.tplants.2009.08.001
de Jonge, R., van Esse, H. P., Maruthachalam, K., Bolton, M. D., Santhanam, P., Saber, M. K., et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 5110–5115. doi: 10.1073/pnas.1119623109
De Young, B. J., and Innes, R. W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249. doi: 10.1038/ni1410
Deslandes, L., Olivier, J., Peeters, N., Feng, D. X., Khounlotham, M., Boucher, C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100, 8024–8029. doi: 10.1073/pnas.1230660100
Duan, X., Zhang, Z., Jin, W., and Zuo, K. (2016). Characterization of a novel cotton subtilase gene GbSBT1 in response to extracellular stimulations and its role in Verticillium resistance. PLOS ONE 11:e153988. doi: 10.1371/journal.pone.0153988
Eitas, T. K., and Dangl, J. L. (2010). NB-LRR proteins: Pairs, pieces, perception, partners and pathways. Curr. Opin. Plant Biol. 13, 472–477. doi: 10.1016/j.pbi.2010.04.007
Elmore, J. M., Lin, Z. J., and Coaker, G. (2011). Plant NB-LRR signaling: upstreams and downstreams. Curr. Opin. Plant Biol. 14, 365–371. doi: 10.1016/j.pbi.2011.03.011
Feuillet, C., Travella, S., Stein, N., Albar, L., Nublat, A., and Keller, B. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. U.S.A. 100, 15253–15258. doi: 10.1073/pnas.2435133100
Feuillet, J. G., Lawrence, G. J., Luck, J. E., and Dodds, P. N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506. doi: 10.1105/tpc.11.3.495
Fradin, E. F., Zhao, Z., Ayala, J. C. J., Castroverde, C. D. M., Nazar, R. N., Robb, J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. doi: 10.1104/pp.109.136762
Gao, W., Long, L., Zhu, L. F., Xu, L., Gao, W. H., Sun, L. Q., et al. (2013). Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell Proteomics 12, 3690–3703. doi: 10.1074/mcp.M113.031013
Gong, Q., Yang, Z., Wang, X., Butt, H. I., Chen, E., He, S., et al. (2017). Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 2017:59. doi: 10.1186/s12870-017-1007-5
Hinsch, M., and Staskawicz, B. (1996). Identification of a new Arabidopsis disease resistance locus, RPs4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant Microbe Interact. 9, 55–61. doi: 10.1094/MPMI-9-0055
Iqbal, N., Khan, N. A., Ferrante, A., Trivellini, A., Francini, A., and Khan, M. I. R. (2017). Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant Sci. 8:475. doi: 10.3389/fpls.2017.00475
Jiang, F., Zhao, J., Zhou, L., Guo, W. Z., and Zhang, T. Z. (2009). Molecular mapping of Verticillium wilt resistance QTL clustered on chromosomes D7 and D9 in upland cotton. Sci. China C Life Sci. 52, 872–884. doi: 10.1007/s11427-009-0110-8
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Joshi, R. K., and Nayak, S. (2011). Functional characterization and signal transduction ability of nucleotide-binding site-leucine-rich repeat resistance genes in plants. Genet. Mol. Res. 10, 2637–2652. doi: 10.4238/2011.October.25.10
Kawchuk, L. M., Hachey, J., Lynch, D. R., Kulcsar, F., van Rooijen, G., Waterer, D. R., et al. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 6511–6515. doi: 10.1073/pnas.091114198
Krasileva, K. V., Dahlbeck, D., and Staskawicz, B. J. (2010). Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22, 2444–2458. doi: 10.1105/tpc.110.075358
Lee, H. A., and Yeom, S. I. (2015). Plant NB-LRR proteins: tightly regulated sensors in a complex manner. Brief. Funct. Genomics 14, 233–242. doi: 10.1093/bfgp/elv012
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, N. Y., Ma, X. F., Short, D. P. G., Li, T. G., Zhou, L., Gui, Y. J., et al. (2017). The island cotton NBS-LRR gene GbaNA1 confers resistance to the non-race 1 Verticillium dahliae isolate Vd991. Mol. Plant Pathol. doi: 10.1111/mpp.12630 [Epub ahead of print].
Li, X., Zhang, Y., Yin, L., and Lu, J. (2017). Overexpression of pathogen-induced grapevine TIR-NB-LRR gene VaRGA1 enhances disease resistance and drought and salt tolerance in Nicotiana benthamiana. Protoplasma 254, 957–969. doi: 10.1007/s00709-016-1005-8
Li, Y. B., Han, L. B., Wang, H. Y., Zhang, J., Sun, S. T., Feng, D. Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Liu, H., Dong, S., Gu, F., Liu, W., Yang, G., Huang, M., et al. (2017). NBS-LRR protein Pik-H4 interacts with OsBIHD1 to balance rice blast resistance and growth by coordinating ethylene-brassinosteroid pathway. Front. Plant Sci. 8:127. doi: 10.3389/fpls.2017.00127
Liu, N., Zhang, X., Sun, Y., Wang, P., Li, X., Pei, Y., et al. (2017). Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 7:39840. doi: 10.1093/bfgp/elv012
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lukasik, E., and Takken, F. L. (2009). STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 12, 427–436. doi: 10.1016/j.pbi.2009.03.001
Lukasik-Shreepaathy, E., Slootweg, E., Richter, H., Goverse, A., Cornelissen, B. J., and Takken, F. L. (2012). Dual regulatory roles of the extended N terminus for activation of the tomato MI-1.2 resistance protein. Mol. Plant Microbe Interact. 25, 1045–1057. doi: 10.1094/MPMI-11-11-0302
Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R., and Dangl, J. L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. doi: 10.1016/S0092-8674(03)00040-0
Martin, G. B., Bogdanove, A. J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. doi: 10.1146/annurev.arplant.54.031902.135035
McHale, L., Tan, X., Koehl, P., and Michelmore, R. W. (2006). Plant NBS-LRR proteins: adaptable guards. Genome Biol. 7:212. doi: 10.1186/gb-2006-7-4-212
Mersmann, S., Bourdais, G., Rietz, S., and Robatzek, S. (2010). Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154, 391–400. doi: 10.1104/pp.110.154567
Mo, H., Wang, X., Zhang, Y., Zhang, G., Zhang, J., and Ma, Z. (2015). Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 83, 962–975. doi: 10.1111/tpj.12941
Moffett, P., Farnham, G., Peart, J., and Baulcombe, D. C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519. doi: 10.1093/emboj/cdf453
Munis, M. F., Tu, L., Deng, F., Tan, J., Xu, L., Xu, S., et al. (2010). A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem. Biophys. Res. Commun. 393, 38–44. doi: 10.1016/j.bbrc.2010.01.069
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1111/j.1462-5822.2008.01260.x
Periyannan, S., Moore, J., Ayliffe, M., Bansal, U., Wang, X., Huang, L., et al. (2013). The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–788. doi: 10.1126/science.1239028
Putt, E. D. (1964). Breeding behavior of resistance to leaf mottle or Verticillium in sunflower. Crop Sci. 4, 177–179. doi: 10.2135/cropsci1964.0011183X000400020016x
Rairdan, G. J., and Moffett, P. (2006). Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18, 2082–2093. doi: 10.1105/tpc.106.0427
Sanseverino, W., Hermoso, A., D’Alessandro, R., Vlasova, A., Andolfo, G., Frusciante, L., et al. (2012). PRGdb 2.0: towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res. 41, 1167–1171. doi: 10.1093/nar/gks1183
Santhanam, P., van Esse, H. P., Albert, I., Faino, L., Nürnberger, T., and Thomma, B. P. (2013). Evidence for functional diversification within a fungal NEP1-like protein family. Mol. Plant Microbe Interact. 26, 278–286. doi: 10.1094/MPMI-09-12-0222-R
Schaible, L., Cannon, O. S., and Waddoups, V. (1951). Inheritance of resistance to Verticillium wilt in a tomato cross. Phytopathology 41, 986–990.
Shao, B. X., Zhao, Y. L., Chen, W., Wang, H. M., Guo, Z. J., Gong, H. Y., et al. (2015). Analysis of upland cotton (Gossypium hirsutum) response to Verticillium dahliae inoculation by transcriptome sequencing. Genet. Mol. Res. 14, 13120–13130. doi: 10.4238/2015.October.26.8
Shen, Q. H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1130. doi: 10.1126/science.1136372
Simko, I., Costanzo, S., Haynes, K. G., Christ, B. J., and Jones, R. W. (2004). Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor. Appl. Genet. 108, 217–224. doi: 10.1007/s00122-003-1431-9
Slootweg, E. J., Spiridon, L. N., Roosien, J., Butterbach, P., Pomp, R., Westerhof, L. A., et al. (2013). Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol. 162, 1510–1528. doi: 10.1104/pp.113.218842
Sun, Q., Jiang, H., Zhu, X., Wang, W., He, X., Shi, Y., et al. (2013). Analysis of sea-island cotton and upland cotton in response to Verticillium dahliae infection by RNA sequencing. BMC Genomics 14:852. doi: 10.1186/1471-2164-14-852
Takken, F. L., Albrecht, M., and Tameling, W. I. (2006). Resistance proteins: molecular switches of plant defence. Curr. Opin. Plant Biol. 9, 383–390. doi: 10.1016/j.pbi.2006.05.009
Tameling, W. I., Elzinga, S. D., Darmin, P. S., Vossen, J. H., Takken, F. L., Haring, M. A., et al. (2002). The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14, 2929–2939. doi: 10.1105/tpc.005793
Tameling, W. I., and Takken, F. L. (2008). Resistance proteins: scouts of the plant innate immune system. Eur. J. Plant Pathol. 121, 243–255. doi: 10.1007/s10658-007-9187-8
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tornero, P., Chao, R. A., Luthin, W. N., Goff, S. A., and Dangl, J. L. (2002). Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell 14, 435–450. doi: 10.1105/tpc.010393
van der Hoorn, R. A. L., and Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
Wang, F. R., Liu, R. Z., Wang, L. M., Zhang, C. Y., Liu, G. D., Liu, Q. H., et al. (2007). Molecular markers of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.) cultivar and their effects on assisted phenotypic selection. Cotton Sci. 19, 424–430.
Wang, G. F., Ji, J., El-Kasmi, F., Dangl, J. L., Johal, G., and Balint-Kurti, P. J. (2015). Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLOS Pathog. 11:e1004674. doi: 10.1371/journal.ppat.1004674
Wang, H. M., Lin, Z. X., Zhang, X. L., Chen, W., Guo, X. P., Nie, Y. C., et al. (2008). Mapping and QTL analysis of Verticillium wilt resistance genes in cotton. J. Integr. Plant Biol. 50, 174–182. doi: 10.1111/j.1744-7909.2007.00612.x
Wang, K. B., Wang, Z. W., Li, F. G., Ye, W. W., Wang, J. Y., Song, G. L., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Whitham, S., Dineshkumar, S. P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115. doi: 10.1016/0092-8674(94)90283-6
Williams, S. J., Sornaraj, P., DeCourcy-Ireland, E., Menz, R. I., Kobe, B., Ellis, J. G., et al. (2011). An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol. Plant Microbe Interact. 24, 897–906. doi: 10.1094/MPMI-03-11-0052
Xiang, L., Liu, J., Wu, C., Deng, Y., Cai, C., Zhang, X., et al. (2017). Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species. BMC Genomics 18:292. doi: 10.1186/s12864-017-3682-x
Xu, L., Zhang, W., He, X., Liu, M., Zhang, K., Shaban, M., et al. (2014). Functional characterization of cotton genes responsive to Verticillium dahliae through bioinformatics and reverse genetics strategies. J. Exp. Bot. 65, 6679–6692. doi: 10.1093/jxb/eru393
Xu, L., Zhu, L., Tu, L., Liu, L., Yuan, D., Jin, L., et al. (2011). Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. doi: 10.1093/jxb/err245
Yang, C., Guo, W. Z., Li, G. Y., Feng, G., Lin, S. S., and Zhang, T. Z. (2008). QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci. 174, 290–298. doi: 10.1016/j.plantsci.2007.11.016
Yang, C., Li, W., Cao, J., Meng, F., Yu, Y., Huang, J., et al. (2017). Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 89, 338–353. doi: 10.1111/tpj.13388
Yang, J., Ma, Q., Zhang, Y., Wang, X., Zhang, G., and Ma, Z. (2016). Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene 575, 687–694. doi: 10.1016/j.gene.2015.09.046
Yang, Y., Ling, X., Chen, T., Cai, L., Liu, T., Wang, J., et al. (2015). A cotton Gbvdr5 gene encoding a leucine-rich-repeat receptor-like protein confers resistance to Verticillium dahliae in transgenic Arabidopsis and upland cotton. Plant Mol. Biol. Rep. 33, 987–1001. doi: 10.1007/s11105-014-0810-5
Zebrowska, J., Hortyäski, J., Cholewa, T., and Honcz, K. (2006). Resistance to Verticillium dahliae (Kleb.) in the strawberry breeding lines. Commun. Agric. Appl. Biol. Sci. 71, 1031–1036.
Zhang, B., Yang, Y., Chen, T., Yu, W., Liu, T., Li, H., et al. (2012). Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLOS ONE 7:e51091. doi: 10.1371/journal.pone.0051091
Zhang, M., Smith, J. A., Harberd, N. P., and Jiang, C. (2016). The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 91, 651–659. doi: 10.1007/s11103-016-0488-1
Zhang, W., Zhang, H., Liu, K., Jian, G., Qi, F., and Si, N. (2017). Large-scale identification of Gossypium hirsutum genes associated with Verticillium dahliae by comparative transcriptomic and reverse genetics analysis. PLOS ONE 12:e0181609. doi: 10.1371/journal.pone.0181609
Zhang, Y., Wang, X., Li, Y., Wu, L., Zhou, H., Zhang, G., et al. (2013a). Ectopic expression of a novel Ser/Thr protein kinase from cotton (Gossypium barbadense), enhances resistance to Verticillium dahliae infection and oxidative stress in Arabidopsis. Plant Cell Rep. 32, 1703–1713. doi: 10.1007/s00299-013-1481-7
Zhang, Y., Wang, X., Yang, S., Chi, J., Zhang, G., and Ma, Z. (2011). Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep. 30, 2085–2096. doi: 10.1007/s00299-011-1115-x
Zhang, Y., Wang, X. F., Ding, Z. G., Ma, Q., Zhang, G. R., Zhang, S. L., et al. (2013b). Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genomics 14:637. doi: 10.1186/1471-2164-14-637
Zhao, J., Gao, Y., Zhang, Z., Chen, T., Guo, W., and Zhang, T. (2013). A receptor-like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought-stress tolerance in Arabidopsis. BMC Plant Biol. 13:110. doi: 10.1186/1471-2229-13-110
Zhao, Y., Wang, H., Chen, W., and Li, Y. (2014). Genetic structure, linkage disequilibrium and association mapping of Verticillium wilt resistance in elite cotton (Gossypium hirsutum L.) germplasm population. PLOS ONE 9:e86308. doi: 10.1371/journal.pone.0086308
Zhu, X., Lu, C., Du, L., Ye, X., Liu, X., Coules, A., et al. (2017). The wheat NB-LRR gene TaRCR1 is required for host defence response to the necrotrophic fungal pathogen Rhizoctonia cerealis. Plant Biotechnol. J. 15, 674–687. doi: 10.1111/pbi.12665
Keywords: Verticillium wilt resistance, NBS-LRR, Arabidopsis thaliana, R gene, transgenic, ethylene signaling, ROS production
Citation: Li N-Y, Zhou L, Zhang D-D, Klosterman SJ, Li T-G, Gui Y-J, Kong Z-Q, Ma X-F, Short DPG, Zhang W-Q, Li J-J, Subbarao KV, Chen J-Y and Dai X-F (2018) Heterologous Expression of the Cotton NBS-LRR Gene GbaNA1 Enhances Verticillium Wilt Resistance in Arabidopsis. Front. Plant Sci. 9:119. doi: 10.3389/fpls.2018.00119
Received: 08 December 2017; Accepted: 22 January 2018;
Published: 06 February 2018.
Edited by:
Jesús Mercado-Blanco, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Sabina Berne, University of Ljubljana, SloveniaMaria De La O. Leyva Perez, Aarhus University, Denmark
Copyright © 2018 Li, Zhou, Zhang, Klosterman, Li, Gui, Kong, Ma, Short, Zhang, Li, Subbarao, Chen and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Feng Dai, daixiaofeng_caas@126.com Jie-Yin Chen, chenjieyin@caas.cn Krishna V. Subbarao, kvsubbarao@ucdavis.edu
†These authors have contributed equally to this work.
 Nan-Yang Li
Nan-Yang Li Lei Zhou
Lei Zhou Dan-Dan Zhang1†
Dan-Dan Zhang1† Zhi-Qiang Kong
Zhi-Qiang Kong Krishna V. Subbarao
Krishna V. Subbarao Jie-Yin Chen
Jie-Yin Chen Xiao-Feng Dai
Xiao-Feng Dai