- 1Department of Biotechnology, National Agri-Food Biotechnology Institute, Mohali, India
- 2Department of Biotechnology, Panjab University, Chandigarh, India
- 3Copenhagen Plant Science Centre, PLEN, University of Copenhagen, Copenhagen, Denmark
Enhancement of micronutrient bioavailability is crucial to address the malnutrition in the developing countries. Various approaches employed to address the micronutrient bioavailability are showing promising signs, especially in cereal crops. Phytic acid (PA) is considered as a major antinutrient due to its ability to chelate important micronutrients and thereby restricting their bioavailability. Therefore, manipulating PA biosynthesis pathway has largely been explored to overcome the pleiotropic effect in different crop species. Recently, we reported that functional wheat inositol pentakisphosphate kinase (TaIPK1) is involved in PA biosynthesis, however, the functional roles of the IPK1 gene in wheat remains elusive. In this study, RNAi-mediated gene silencing was performed for IPK1 transcripts in hexaploid wheat. Four non-segregating RNAi lines of wheat were selected for detailed study (S3-D-6-1; S6-K-3-3; S6-K-6-10 and S16-D-9-5). Homozygous transgenic RNAi lines at T4 seeds with a decreased transcript of TaIPK1 showed 28–56% reduction of the PA. Silencing of IPK1 also resulted in increased free phosphate in mature grains. Although, no phenotypic changes in the spike was observed but, lowering of grain PA resulted in the reduced number of seeds per spikelet. The lowering of grain PA was also accompanied by a significant increase in iron (Fe) and zinc (Zn) content, thereby enhancing their molar ratios (Zn:PA and Fe:PA). Overall, this work suggests that IPK1 is a promising candidate for employing genome editing tools to address the mineral accumulation in wheat grains.
Introduction
Mineral malnutrition is among the most critical worldwide challenges to humankind. In developing countries ∼50% of the children are estimated to be micronutrient deficient (WHO, 2007). This condition might be a result of crop production in areas with low mineral phytoavailability and/or consumption of staple crops with inherently low tissue mineral concentrations (Welch and Graham, 2005). The mineral malnutrition can be addressed through dietary enhancement, mineral supplementation, food fortification, or increasing the concentrations and/or bioavailability of mineral elements. However, methodologies to expand dietary enhancement, mineral supplementation, and food fortification have not generally been very effective (White and Broadley, 2005). Thus, biofortification of crops by decreasing the concentration of antinutrients and other secondary metabolites can be considered as an important means to increase the bioavailability of mineral elements.
Cereal crops are rich sources of mineral ions, but the presence of certain anti-nutritional components limits their bioavailability (Raboy, 2009). Phytic acid (myo-inositol hexakisphosphate, PA, IP6) is one of the major anti-nutritional component in grains of wheat, rice, soybean, and other crops, that limit the bioavailability of micronutrients. In grains PA is major storage form of phosphorus (P). The developing grains accumulate PA in vacuoles as phytate, that chelates important micronutrients and thereby causing drastic reduction in the bioavailability of mineral ions (Garcìa-Estepa et al., 1999). Additionally, PA cannot be utilized efficiently by monogastric animals due to lack of phytase enzyme. Furthermore, excretion of undigested PA in animal waste contributes to environmental phosphate pollution, by accelerating soil eutrophication (Stevenson-Paulik et al., 2005). Therefore, reducing the PA content in cereal grains is a desired goal for the genetic improvement of several crops to address the dual issues of micronutrient bioavailability and eutrophication (Raboy, 2009; Secco et al., 2017).
Phytic acid biosynthesis pathway genes have been reported from multiple crop species (Stevenson-Paulik et al., 2002; Suzuki et al., 2007; Sweetman et al., 2007; Stiles et al., 2008). PA in plants can be synthesized by a lipid dependent or independent pathway depending on the primary inositol source (Stevenson-Paulik et al., 2005; Raboy, 2009). Lipid-independent pathway is predominant in seeds of cereals and legumes. The first committed step performed by myo-inositol-3-phosphate synthase (MIPS) involves the formation of inositol-3-phosphate (Ins3P) from glucose-6-phosphate. Subsequent steps involves sequential and ordered phosphorylation at the remaining five positions of the inositol ring through various enzymes (Stiles et al., 2008; Rasmussen et al., 2010). The enzymes catalyzing these phosphorylation reactions include inositol monophosphatase (IMP), inositol tris/tetraphosphate kinase (ITPK), inositol polyphosphate kinase (IPK2) and inositol-pentakisphosphate 2-kinase (IPK1) (Raboy, 2009; Rasmussen et al., 2010).
Based on the physiological properties, low phytic acid (lpa) mutants in maize have been characterized as lpa1, lpa2 or lpa3. Maize lpa1 mutant was shown to be defective in a multi-drug resistance-associated protein (MRP) ABC transporter with unknown function (Shi et al., 2007). Maize lpa2 mutant is caused by mutation in an inositol phosphate kinase gene (Shi et al., 2003). The maize lpa3 mutant had mutation in a gene that encodes myo-inositol kinase enzyme (Shi et al., 2005). Barley lpa2 mutant lines (M635 and M955) had enhanced free phosphate (Pi) and decreased PA levels due to loss of inositol phosphate kinase activity (Dorsch et al., 2003). Utilizing reverse genetics in Arabidopsis led to the identification of genes that are necessary for PA accumulation in seed (Kim and Tai, 2011). These observations indicated that most of the PA pathway genes could be functionally conserved among multiple crops.
The hexaploid wheat (Triticum aestivum L.), is one of the most important staple food crop providing nearly 55% of the carbohydrates and 20% of the food calories consumed globally (Breiman and Graur, 1995). The initial lack of information about PA biosynthesis pathway genes from wheat and inefficient genetic transformation were limiting factors hindering the PA associated research. To address this issue, we reported PA biosynthesis pathway genes and putative PA transporter from hexaploid wheat that suggested to be potential candidates for gene suppression (Bhati et al., 2014; Aggarwal et al., 2015). Further silencing of wheat ABCC13, a putative transporter of PA resulted in reduced PA without significantly affecting the levels of zinc (Zn) and iron (Fe) in the grains. Moreover, other morphological changes suggest a multifunctional aspect of this transporter (Bhati et al., 2016). The pleiotropic effects and compromised spike development in TaABCC13 silenced plants encouraged to explore alternative approaches for achieving low PA. Studies have indicated that genes involved during the early and late biosynthesis pathway could be the potential targets for achieving low PA (Raboy, 2009; Rasmussen et al., 2010). This has been successfully demonstrated by targeting genes related to PA biosynthesis in barley (Ye et al., 2011), maize (Shi et al., 2007), Arabidopsis (Stevenson-Paulik et al., 2005) and rice (Ali et al., 2013a,b). Although enhanced micronutrient content was reported in these low PA crops, no molecular basis was accounted for such trait.
Previously, TaIPK1 was shown to be a functionally active that is differentially expressed at the transcript level and putatively catalyzes the last step of PA biosynthesis (Bhati et al., 2014). Transcript abundance of TaIPK1 derived from either B or D genomes were highly expressed when compared to the A genome. Subsequently, IPK1 was targeted for RNAi-mediated gene silencing and its effects were studied in hexaploid wheat (C306). Multiple non-segregating RNAi lines (S3-D-6-1; S6-K-3-3; S6-K-6-10 and S16-D-9-5) were selected for detailed characterization of the IPK1 transcript reduction effect. Silencing of IPK1 resulted in significant lowering of grain PA along with increase in levels of free Pi. The decrease of phytate in wheat grains was accompanied by the increase in the content of micronutrients like Fe and Zn. In addition, lowering of the wheat IPK1 transcripts also showed pleiotropic effects that includes decreased grain yield.
Materials and Methods
Plant Material and Growth Conditions
Hexaploid wheat (Triticum aestivum) variety, C306, was used for this study. For genetic transformation, surface sterilized seeds of C306 were sown in pots with sterile vermiculite and kept in a plant growth chamber (Conviron, Canada) under a 16 h photoperiod at 400 μmol⋅m-2⋅s-1, 70% relative humidity and 25°C/18°C (day/night). For wheat transformation tagged spikes were harvested at 12–16 DAA. To study gene expression, the main individual spikes of the transgenic wheat and respective controls were tagged at the first DAA. Subsequently seeds from the tagged spikes were harvested at 14 or 21 DAA in liquid nitrogen for RNA extraction.
Designing of RNAi Constructs
Previously reported RNAi vector pMCG161 (TAIR stock-CD3-459) was used for the genetic transformation of wheat as mentioned earlier (Zalewski et al., 2010; Gasparis et al., 2011; Bhati et al., 2016). The T-DNA of the vector contained bar gene (also called pat, phosphinothricin acetyl transferase) as a selectable marker that confers resistance to herbicide BASTA and allows the selection of transgenic plants over the optimized concentration. RNAi cassette is expressed under the control of CaMV 35S promoter, while monocot specific UBI intron promoter controls expression of bar gene. Cloning of 320 bp fragment of IPK1 in sense and anti-sense orientation was done in pMCG161 vector using two restriction sites separated by a rice waxy intron. Primers were designed from the conserved region of all the homoeologs, i.e., TaIPK1:2AL-TRIAE_CS42_ 2AL_TGACv1_095050_AA0306410, TaIPK1:2BL-TRIAE_CS42_ 2BL_TGACv1_129504_AA0386330, TaIPK1:2DL-TRIAE_ CS42_2DL_TGACv1_159606_AA0540500 (Supplementary Figure S1) with AscI/AvrII overhangs in forward primer and SpeI/SgfI overhangs in reverse primer. The sequences of primers were given in Supplementary Table S1. Restriction analysis using multiple enzymes and sequencing confirmed the cloning of sense and anti-sense sequences in silencing cassette. The confirmed RNAi construct was transformed into the AGL1 strain of Agrobacterium tumefaciens and subsequently used for wheat transformation.
Agrobacterium-Mediated Wheat Transformation
A single clone of Agrobacterium confirmed for the presence of TaIPK1:pMCG161 RNAi construct was used for wheat transformation. Wheat transformation was performed as described earlier with minor modifications (Przetakiewicz et al., 2004; Gasparis et al., 2011; Bhati et al., 2016). In brief, seeds harvested from the 14–16 DAA tagged spikes were surface sterilized with NaClO (1.2% v/v in 10% ethyl alcohol). Approximately 700 immature (1 to 1.2 mm) embryos were isolated by dissecting the seeds aseptically and then cultured on callus induction media consisting of MS medium with 2 mg L-1 of 2,4-dichlorophenoxyacetic acid (2,4-D). The callus inducing plates carrying 25–30 immature embryos were incubated for 72 h at 22°C/dark. After incubation, embryos were infected with co-cultivation suspension of AGL1 strain (OD600 0.3–0.5) harboring RNAi construct and the suspension containing Silwet (0.001% v/v) was spotted drop by drop on each growing calli (Gasparis et al., 2011). Co-cultivation step was followed by washing with autoclaved MilliQ water containing Cefotaxime (350 mg L-1). Washed calli were transferred onto the callus induction media having Cefotaxime (250 mg L-1). The plates were then incubated at 22°C/dark for three cycles of 12 days with sub culturing to fresh media after each cycle. Healthy calli developed on Post-infection callus induction media were then subjected to herbicide selection by subculturing it on 2 mg L-1 BASTA along with growth supplement zeatin, 1 mg L-1. These plates were then incubated at 22°C/light and sub-culturing was performed for three cycles of 12 days each. The developing plantlets were then further cultured on half- strength MS medium containing 2.5mg L-1 BASTA for two cycles. Later, plantlets with healthy rooting were transferred to vermiculite pots for hardening. The shoots regenerated from independent calli were considered as independent T-DNA integration events and assigned as T0 putative plants.
Transgene Integration and Segregation Analysis
The plants those survived the hardening were checked for gene integration through PCR. Total genomic DNA was isolated from young leaves of putative transgenic and non-transformed (control) plants using the DNeasy Plant Mini Kit–QIAGEN following manufacturer protocol. Primers were designed for amplification of the bar gene and OCS region of RNAi cassette (Supplementary Table S1). Leaves from the individual tiller of each T0 plant were also screened by PCR to eliminate the possibility of chimeric plants. Amplicons generated from PCR-positive T0 plants were purified and sequenced for confirmation. Subsequently, seeds collected from the PCR-positive tillers of T0 plants were subjected to segregation analysis on 2.5 mg L-1 BASTA that was optimized for cultivar C306 (control wheat) in hydroponic media. These plantlets were transferred to soil pots, after collecting data for survival ratio. Further, the selection fidelity was confirmed with PCR by amplifying and sequencing of a bar fragment gene. Plants that are positive in PCR amplification and follows Mendelian segregation ratio were cultivated up to the T2 and T3 generations. For T4 generation, ten plants from three non-segregating independent transgenic lines (S3, S6, and S16) were propagated and used for further analysis.
Gene Expression Analysis by Using Quantitative RT-PCR
For the confirmation of gene silencing, SYBR Green (Quanti-Tect SYBR Green RT-PCR Master mix, QIAGEN) based reactions were performed on ABI PRISM 7500 Fast Real-Time Platform (Applied Biosystems). The silencing of the gene was confirmed in the immature seeds of the developing grains at T4. During the wheat transformation wheat lines derived from callus showing no integration of the vector were subsequently propagated along with the transgenic wheat as a control plants for comparative analysis. The main individual spikes of selected T3 plants were tagged at the first DAA and tissue samples were harvested at 14 DAA in liquid nitrogen. RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, United States), following manufacturer’s instructions. gDNA free cDNA was prepared using 2 μg of RNA. Transcriptor First Strand cDNA Synthesis Kit RT-PCR (Roche, Indianapolis, IN, United States) was used for cDNA preparation following the manufacturer’s guidelines. Further, level of silencing was checked using real time PCR primers targeting TaIPK1 CDS region other than RNAi fragments. The sequences of primers are listed in Supplementary Table S1. Relative expression level was quantified using 2-ΔΔCt method after normalizing Ct values against wheat ADP-ribosylation factor 1 (ARF1) and 18S rRNA genes as an internal control (Livak and Schmittgen, 2001).
Morphological Analyses of Transgenic Plants
TaIPK1:RNAi lines cultivated in plant growth chamber were observed for spike characteristics that include spikelet count, spike length, and awn length. Spikes from each T3 line (only from primary tiller) that showed reduction in TaIPK1 transcript level were considered for above observations along with their respective controls. The spikes of the transgenic wheat were observed for the head sterility under the light microscope (Leica 2500LCD). Seed count was done from the primary spikes of the C306 and RNAi lines. Average seed weight was measured by taking 50 random seeds from C306 and RNAi lines. Further, grain morphology was also observed for C306 and transgenic RNAi lines using light microscope.
Germination Assays
The germination pattern of the TaIPK1:RNAi lines and non-transformed plants were observed to check any effect of the integration of the plasmid or transgenic events. Seeds were surface sterilized with NaClO (1.2% v/v in 10% ethyl alcohol). Seeds were then transferred on distilled water-soaked filter paper, kept at the base in Petri dishes. The plates were kept in a plant growth chamber maintained at 25°C/18°C (day/night) and 70% relative humidity. Pictures of seeds were taken after 48 and 72 h post-germination.
PA and Inorganic Phosphate (Pi) Estimation
Total PA was estimated in mature seeds of T3 and T4 lines along with controls using K-PHYT kit (Megazyme Inc, Bray, Ireland) as per the manufacturer’s protocol. Briefly, the seeds were grounded to fine powder and extracted using 0.66 N HCl with continuous stirring overnight. The supernatant was then used for colorimetric development as mentioned in the manufacturer instruction booklet. For free phosphate estimation, mature seeds from selected TaIPK1:RNAi lines were used along with control plants. Free phosphate was measured using ascorbate and ammonium molybdate method (Ames, 1966).
Micronutrient and Total Phosphorus (P) Analyses Using ICP-MS
Total seed phosphorus and metal analysis was performed using Inductive Coupled Plasma-MS (ICP-MS). Mature seeds of T3 and T4 lines along with control plants were grounded to fine powder and subjected to the microwave-digested with HNO3 (SuraPureTM, Merck). Subsequently, it was analyzed as described previously (Bhati et al., 2014). For analysis in T4 seeds, three to four plants of the respective transgenic lines were chosen for calculating an average accumulation of micronutrients.
In Silico Analysis
The expression pattern for the TaIPK1 homoeologs were extracted as transcripts per million (TPMs) from Wheat Expression Browser expVIP: http://www.wheat-expression.com/ (Borrill et al., 2015). Expression was performed during different developmental stages of grain, leaf, root, spike, and stem at the mentioned Zadoks scale (Zadoks et al., 1974). Five different grain tissues (whole endosperm, starchy endosperm, transfer cells, aleurone, and aleurone+endosperm) and three developmental stages consisting of 10, 20, and 30 DPA (days post-anthesis) for wheat was utilized to check the expression of TaIPK1 homoeologs.
Results
Homeologous Dependent Expression Pattern of TaIPK1
The expression patterns for IPK1 homoeolog genes was performed in wheat tissues, by using the Wheatexp; Wheat Expression Browser (Choulet et al., 2014; Pfeifer et al., 2014; Borrill et al., 2015). The expression for homoeolog-specific transcript of TaIPK1 gene was determined in five different tissues, i.e., grain, leaf, root, spike, stem and at three developmental stages (Z71, Z75, and Z85). Our analysis suggested expression from all the three homoeologous alleles. A higher transcript level for TaIPK1-2B homoeolog was observed in most of the examined tissues (Supplementary Figure S2A). Furthermore, identical expression of TaIPK1 homoeologs was observed in different grain tissues suggesting the importance of all the gene transcripts during grain development (Supplementary Figure S2B). The expression validation for TaIPK1 homoeologs was performed in C306, an Indian hexaploid wheat variety. qRT-PCR showed high transcript level of TaIPK1-2B at different developmental stages (14 and 21 DAA) (Figure 1).
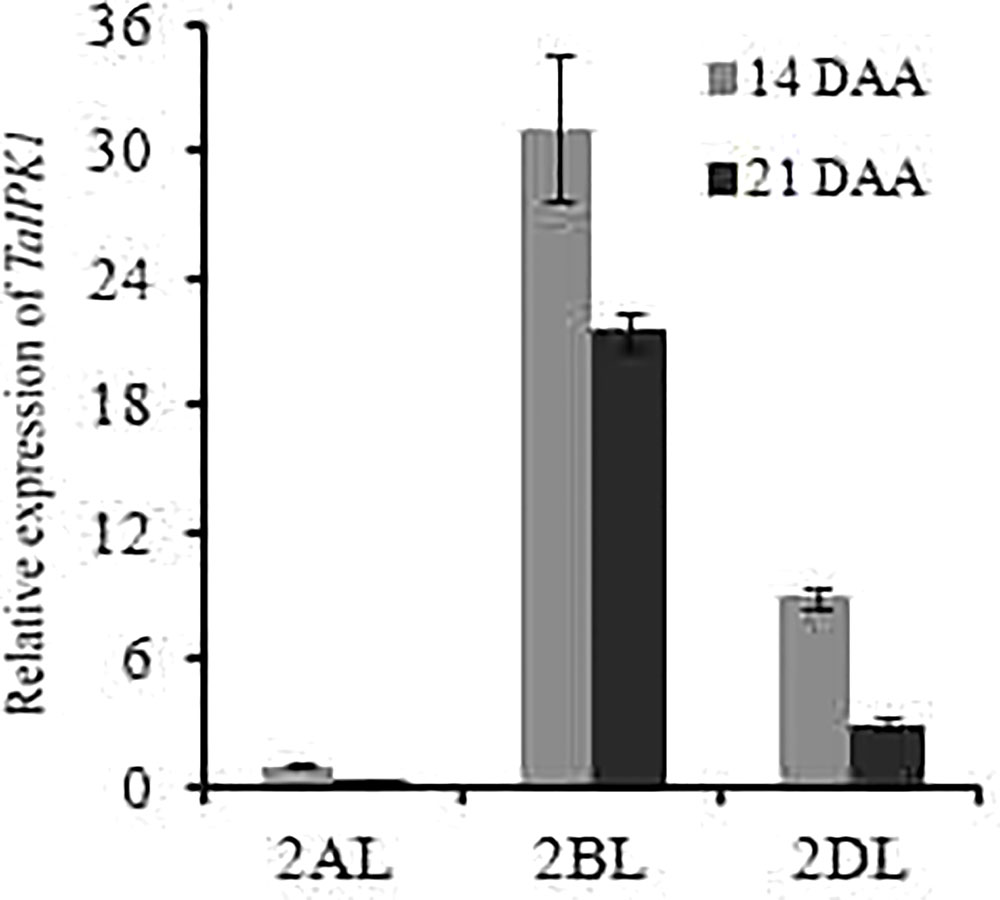
FIGURE 1. Differential expression analysis of three homoeologs of TaIPK1 at two seed developmental stages (14 and 21 DAA). Transcript specific primers were designed for 2AL, 2BL, 2DL of TaIPK1 homoeologs based on genomic information available at IWGSC. gDNA free cDNA was prepared using 2 μg of RNA. qRT-PCR assays were performed using SYBR green and Ct values were normalized against wheat ADP-ribosylation factor 1 (ARF1) as an internal control. The indicated error bars represents the standard deviation from three independent replicates.
Preparation of RNAi Construct, Wheat Transformation and Screening of T0 Transgenic Plants
RNAi construct was designed to achieve reduced endogenous TaIPK1 mRNA. A schematic representation of the RNAi construct is presented (Figure 2A). The Agrobacterium strain AGL1 expressing TaIPK1:pMCG161 RNAi construct was used for wheat transformation. A total of 16 plantlets originating from independent calli were obtained after herbicide (BASTA) selection (Supplementary Figures S3A,B). Only eleven plantlets survived during the hardening procedure in the soilrite containing pots. Since, these plantlets were derived from distinct callus, they were considered as independent transgenic events for pMCG161:TaIPK1 RNAi T-DNA insertion. T0 plants surviving the hardening procedure in soil were named as S1–S6, S8–S11 and S16. Further, screening of these plants was performed by PCR amplification of bar and OCS1 terminator sequences that resulted in the confirmation of T-DNA insertion in nine plantlets (Figure 2B). During various stages of screening, the PCR fragments were sequenced to confirm the presence of bar or OCS1 sequences (Supplementary Figure S4). Three independent events (S3, S6, and S16) showed healthy growth under selection and screening procedures were chosen for further analysis. Progenies of these three events were propagated till the T3/T4 generation.
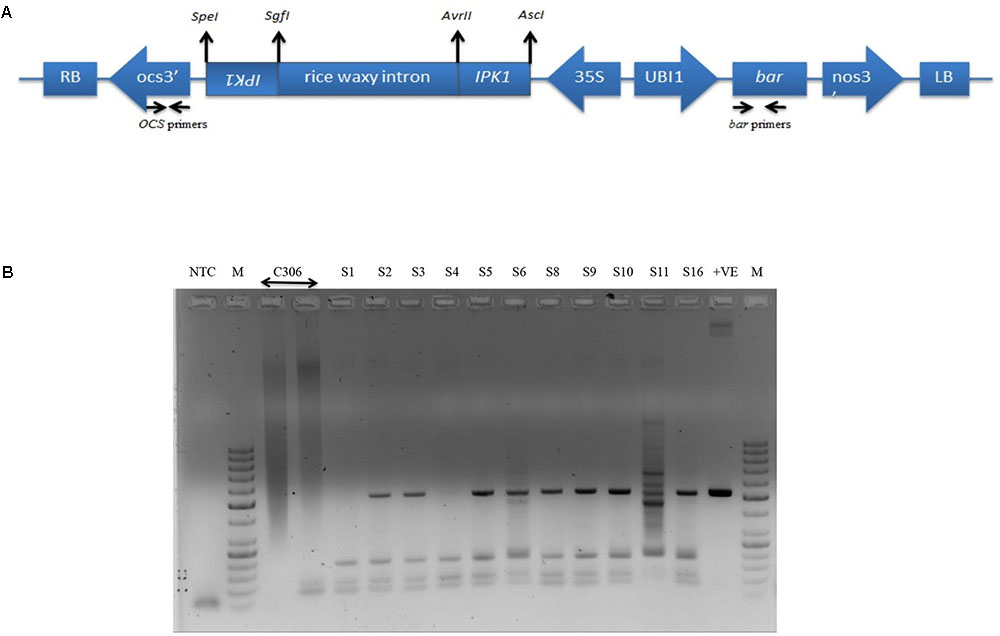
FIGURE 2. Schematic representation and confirmation of the RNAi construct in wheat for targeted gene silencing of TaIPK1. (A) Vector backbone of pMCG161 was utilized to clone fragments of TaIPK1 gene in the sense and antisense orientations. bar gene was used as a plant selection marker (B) Representative picture for the screening of the putative transgenics to confirm the genomic integration of the RNAi constructs in wheat. PCR amplification of bar gene from the genomic DNA of 11 independent integration events (T0 stage) recovered after hardening procedure (S1–S6, S8–S11, S16 are putative transgenic plants, C306, control plant, +ve, plasmid pMCG161, NTC, no template control).
Silencing of TaIPK1 Enhances Free Pi and Decreases Total PA
The lpa2 mutant phenotype is generally accompanied by enhanced accumulation of free Pi (Raboy, 2009; Yuan et al., 2012). Therefore, estimation for Pi was performed in the multiple lines of T3 seeds. Our analysis, showed a significant increase in the Pi level in most of the transgenic lines (S3-D-6, S6-K-3, and S6-K-6) as compared to control. In general, an increase in the transgenic grain Pi ranged from ∼1.2- to 1.7-fold, as compared to the control (Figure 3A). These lines were selected for further study.
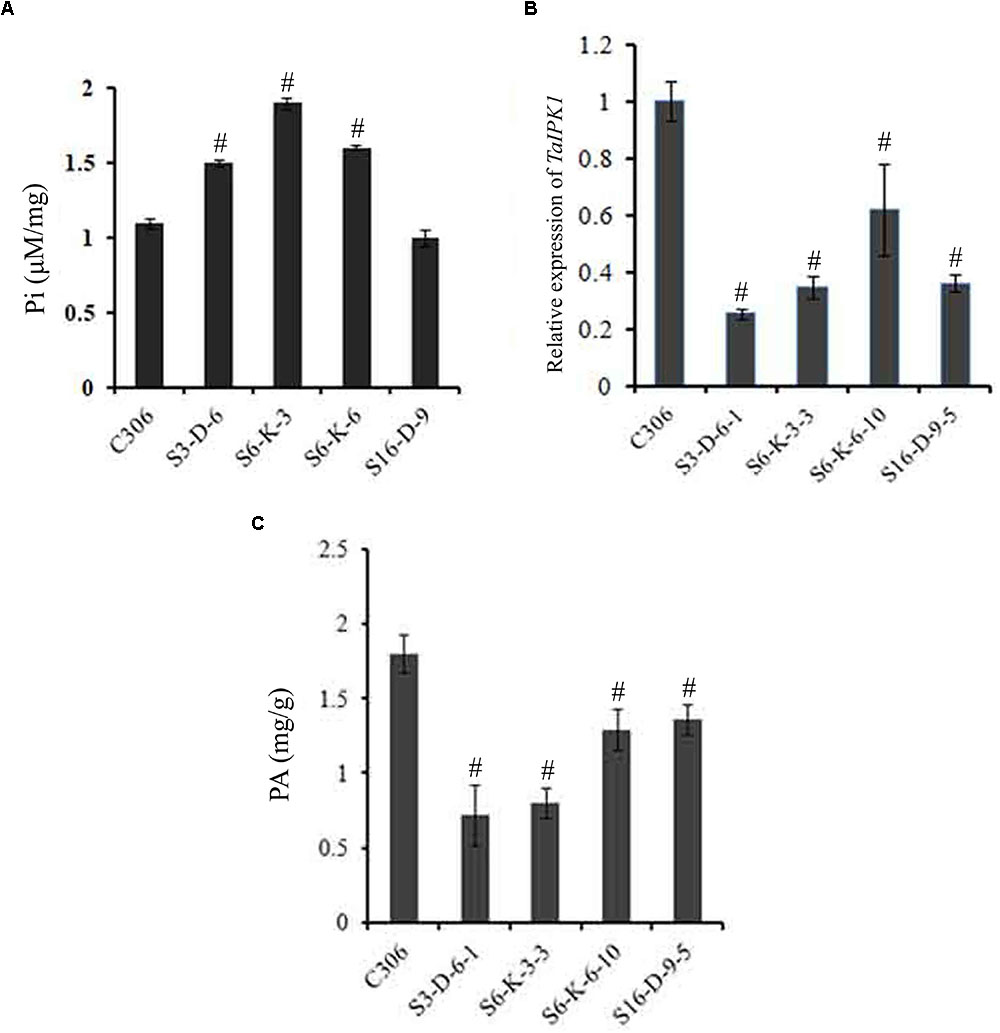
FIGURE 3. Confirmation of silencing in TaIPK1: RNAi lines in T4 seeds. (A) Free Pi content of control C306 and TaIPK1:RNAi lines were estimated using colorimetric based assays. (B) Relative fold change of TaIPK1 expression in wheat transgenic lines. RNAi lines from three independent events were subjected to expression analysis at 14 DAA stage. The cDNA templates were prepared from 2 μg of DNase free RNA. qRT-PCR assays were performed using SYBR green and Ct values were normalized against wheat ADP-ribosylation factor 1 (ARF1) as an internal control. (C) Total phytic acid in mature wheat grains of transgenic lines (T4). PA was measured in the mature seeds collected from the primary tiller of each line. #Indicates significant differences at p < 0.05.
The relative quantification of gene silencing was performed using qRT-PCR in the multiple lines of the T4 seeds (14 DAA). Expression data indicated consistent and significant decrease in the transcript levels of the TaIPK1 in the T4 seeds as represented in Supplementary Figure S5. This suggests an effective silencing of wheat IPK1 in the selected lines. Overall, silencing resulted in ∼40 to 65% reduction of TaIPK1 transcript in the seed tissue (Figure 3B and Supplementary Figure S5). The transgenic lines, S3-D-6-1; S6-K-3-3; S6-K-6-10 and S16-D-9-5, representing distinct events showing the significant level of silencing were therefore selected for analysis. Significant reduction of PA ∼28–56% in the mature T4 grains was observed when compared to the control seeds (Figure 3C). The maximal PA reduction was observed was for S3-D6-1 and S6-K-3-3 lines, when compared to control seeds. Overall, our data suggested that silencing of IPK1 in wheat causes significant lowering of PA and thereby, enhancing free Pi.
Phenotypic Changes in the Spikes of Transgenic Wheat
The germination pattern of the T2 seeds was studied to check any effect of lowering of PA or the integration of the plasmid. In general, no significant difference was observed for the percentage germination of seeds for the transgenic and non-transgenic control. Additionally, no delay in the germination for the transgenic seeds were observed (Supplementary Figure S6). This might suggest that the T-DNA integration does not hamper the viability of transgenic wheat seeds.
The morphological traits of the selected events in T3/4 generation were compared with the non-transgenic plants. Multiple physiological and phenotypic parameters contributing to the grain yield (grains count per spike, spikelet count, seed weight, spike length, and awn length) were determined. Slight head sterility was observed in a few of the transgenic lines compared to non-transgenic control plants (Figure 4A). Transgenic wheat grains showed insignificant alteration in the grain morphology (Figure 4B). None of the changes in the glume arrangement or spike length were observed, suggesting the typical spikelet arrangement (Figures 4C,D). The awn length was found to be almost similar except for the line S3-D-6-1 as compared to non-transgenic control plants (Figure 5A). Developing spikes of transgenic lines did not show any significant changes in the spikelet count, except for the line S6-K-3-3, wherein a reduction of 12–14% was observed (Figure 5B). Seed count and seed weight were measured in the mature grains of non-transgenic wheat and TaIPK1:RNAi lines and observations were noted. Although, no significant differences were observed between the average seed weight of TaIPK1:RNAi lines and control, but had a substantial effect on total seed count (Figures 5C,D).
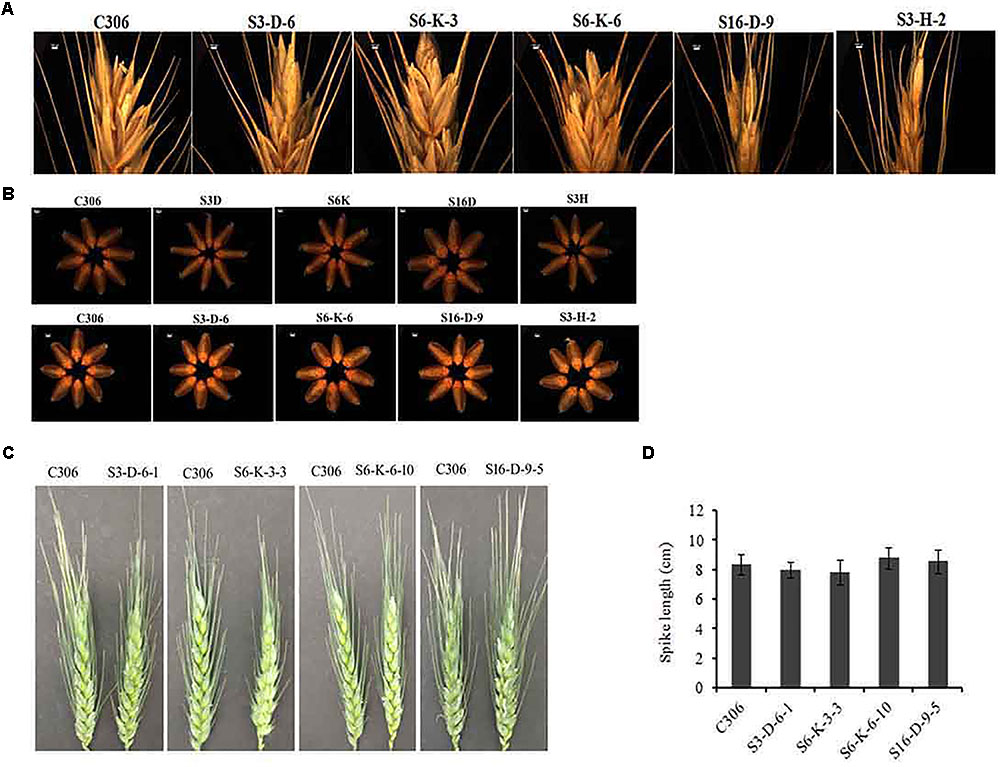
FIGURE 4. Phenotypic characteristics of TaIPK1:RNAi lines. (A) Representative pictures for grain filling at the spike head on wheat RNAi lines and control (C306) and (B) Representative pictures of seeds collected from wheat RNAi lines and control (C306) at T2 and T3 stages. Eight seeds were selected randomly from C306 and TaIPK1:RNAi lines and images were captured using a light microscope (Leica Microscope). (C) Representative pictures of wheat caryopsis on the onset of flowering of control C306 and TaIPK1:RNAi lines at T4. (D) Spike length of the primary tiller of transgenic and control plants representing T4 stage. Each bar indicates the mean of eight biological replicates.
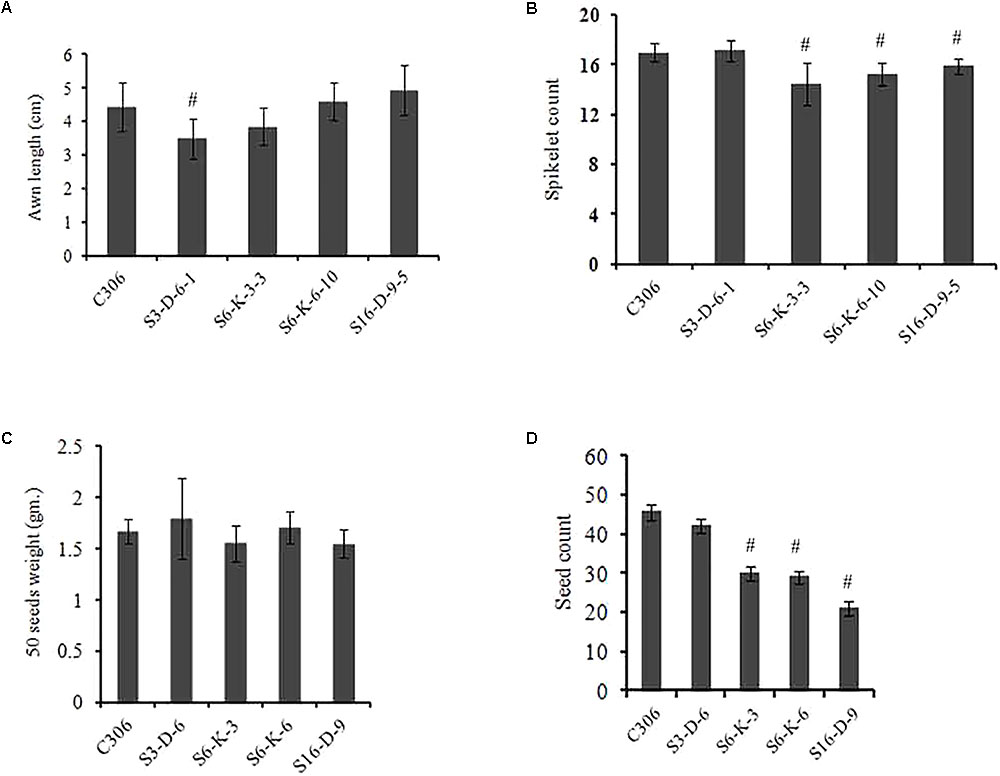
FIGURE 5. Spike characteristics of wheat transgenic plants. (A) Awn length from primary tiller of control C306 and TaIPK1:RNAi lines. (B) Spikelet count from the primary tiller of TaIPK1:RNAi lines and control C306 and at T4 stage. (C) Seed weight of control C306 and TaIPK1:RNAi lines. Average seed weight was measured by weighing 50 random seeds from each line. The data shown here were collected from T3 progenies. Each bar indicates the mean of three biological replicates (three technical replicates). (D) Seed count from the primary tiller of control C306 and TaIPK1:RNAi lines for T3 progenies. Each bar indicates the mean of three biological replicates. The data in (A,B) indicate the means of eight biological replicates. #Indicates significant differences at p < 0.05.
Reduction of PA in Grains Enhances Micronutrient Content and Perturb Expression of Phosphate Transporters
In order to check if wheat IPK1 transgenic RNAi lines showed any altered accumulation of micronutrients, transgenic seeds were analyzed for metal content. Analysis was performed using ICP-MS that suggested a significant increase in the accumulation of Zn and Fe in grains of multiple transgenic lines of T3 as compared to non-transgenic seeds (Supplementary Figure S7). Although, the Fe content was variable among the transgenic events, the accumulation of Zn was consistently high. The results showed ∼1.2- to 1.7-fold increase in the levels of Fe and ∼1.3- to 2.2-fold increase in Zn. Maximum accumulation of Fe and Zn was observed in S6-K-3, S6-K-6, and S16-D-9 lines. Similar observation was recorded for the T4 grains, where increased accumulation of Fe and Zn was observed (Table 1). Overall, this data suggested that lowering of PA in wheat, mediated by silencing of IPK1 resulted in enhanced accumulation of Zn and Fe. Increased molar ratio of Zn/PA and Fe/PA was observed in multiple lines that negatively correlated with PA levels in wheat grain (Figures 6A,B). The improved metal to PA molar ratio was consistent in both T3 and T4 generation seeds.
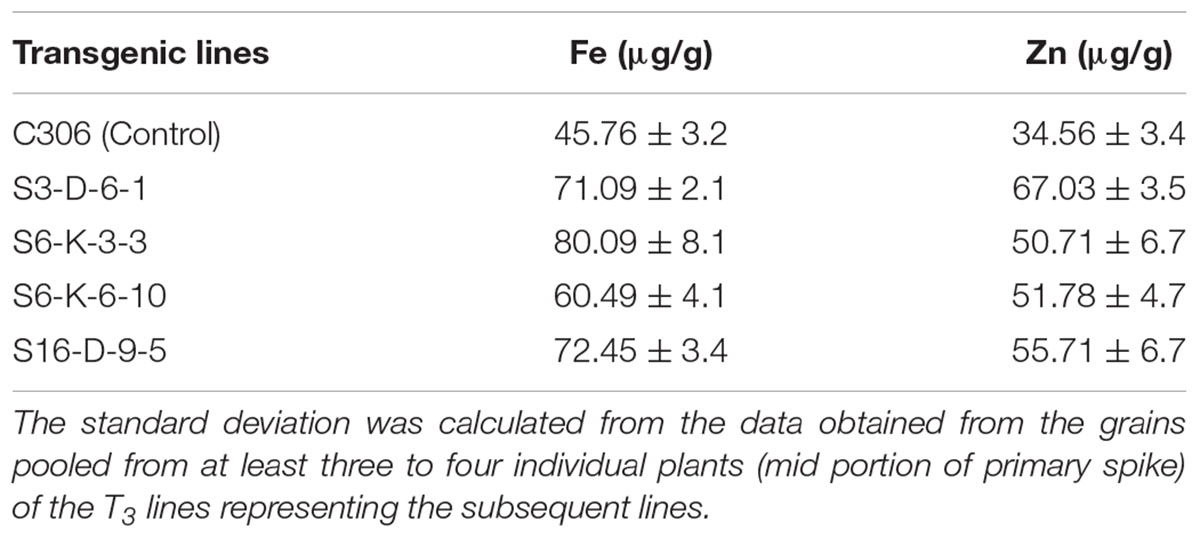
TABLE 1. Metal concentration of Fe and Zn in the T4 mature grains of wheat transgenic and non-transgenic control (C306).
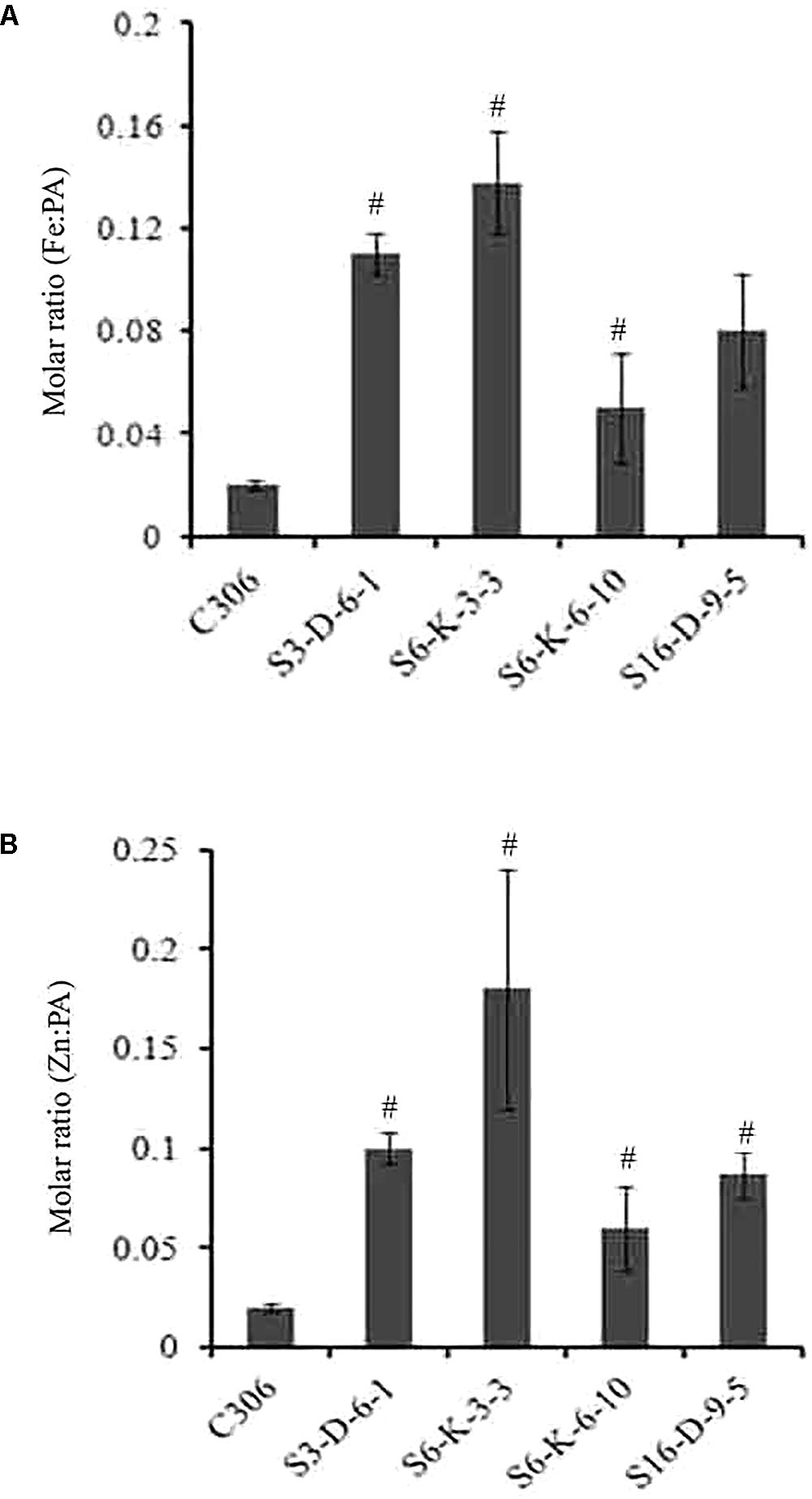
FIGURE 6. Molar ratio of Fe:PA and Zn:PA in different transgenic lines of wheat. (A,B) Molar ratios of Fe:PA and Zn:PA was calculated of four different transgenic wheat lines and were compared to the control (C306) seeds. The standard bar indicates average of four technical plants for each of the respective lines. Mean values showed a significant difference at p < 0.05 (#) with respect to their control.
Since, an increase in the free Pi was observed, we also studied the expression of previously identified phosphate transporter (PHT1 sub-family) genes in the transgenic lines (Shukla et al., 2016). Many of the wheat PHTs were differentially expressed in these transgenic lines suggesting that lowering of PA impacts their transcript levels (Supplementary Figure S8). Taken together, our data conclude that changes in the content of Pi in the grain tissue impacts expression pattern of PHTs.
Discussion
The present investigation deciphers functional importance of IPK1 for its role in the accumulation of PA in hexaploid wheat. IPK1 had been a target of choice to achieve low phytate crops (Stevenson-Paulik et al., 2005; Shukla et al., 2009; Ali et al., 2013a). Our study also demonstrated that lowering of PA in hexaploid wheat could result in enhanced micronutrient accumulation of Zn and Fe. Therefore, this study indicates that IPK1 is a good candidate of wheat to alter PA content and thereby enhancing micronutrient bioavailability.
IPK1 as a Candidate Gene for Developing Low Phytate Crops
Earlier, a strong correlation was observed between expression of IPK1 and PA accumulation during the early phase of wheat grain development (7–28 DAA) (Bhati et al., 2014). This reinforces that, the IPK1 enzyme plays important role in the PA biosynthesis during grain maturation. Subsequently, current study observed that all the three homoeologs of IPK1 were expressed in all seed tissues including vegetative parts (Figure 1 and Supplementary Figure S2). The functionality of IPK1 gene was demonstrated in multiple crops by enzymatic characterization or by utilizing multiple yeast mutants to rescue their defective growth phenotypes (Sweetman et al., 2006; Sun et al., 2007; Suzuki et al., 2007). Wheat IPK1 could rescue ScΔipk1 mutant thermo-sensitivity and therefore confirms its active function in the heterologous system (Bhati et al., 2014). Moreover, TaIPK1 also had 83.5 and 53.2% similarity to OsIPK1 and AtIPK1 reinforcing the suitability of this candidate gene for developing lpa wheat. Therefore, it could be speculated that like rice, wheat IPK1 is a suitable candidate for utilizing recent biotechnological tool like genome editing. Our further investigation led to the identification of another IPK1-like gene (TRIAE_CS42_4BL_TGACv1_320862_AA1050490.1) located on chromosome 4. The 270 aa sequence lacks the prerequisite domains required for a functional kinase activity (González et al., 2010). Therefore, it is possible that the identified new wheat IPK1 lacks kinase activity. In this study, to overcome the limitation of functional redundancy in hexaploid wheat, conserved region from three homoeologs of TaIPK1 was targeted. Subsequently, a significant decrease in the transcript of IPK1 (40–65%) was achieved. In addition to that, reduction in mRNA of TaIPK1 homoeologous was also observed in these transgenics (Supplementary Figure S5B). This emplies that targeting the conserved region of the sequences in wheat is an excellent strategy for functional validation studies (Gasparis et al., 2011; Bhati et al., 2016).
During this study, transgenic wheat lines with low PA show increased Pi content that is typical for lpa2 phenotype. Previously, lpa2 phenotype was observed in rice, maize, Arabidopsis, and soybeans (Raboy et al., 2000; Shi et al., 2003; Stevenson-Paulik et al., 2005; Yuan et al., 2012; Ali et al., 2013a). A summary of such approaches to reduce PA has been reflected in Supplementary Table S2. In our case, transgenic lines showed increased Pi (upto 1.6-fold) as compared to control plants. Likewise, Arabidopsis and soybean ipk1 mutant showed increase of free Pi (Stevenson-Paulik et al., 2005; Yuan et al., 2012). In wheat, 56% reduction of PA was observed, as compared to 32% decrease during the earlier study (Bhati et al., 2016). This suggests that IPK1 is a valuable candidate to significantly reduce PA in wheat grains. Although, candidate genes like MIPS, ITPKs are yet to be tested in wheat, but IPK1 is certainly one of the promising candidate for developing lpa trait. In another study mutagenized M2 wheat lines caused 37% reduction in PA content in seeds (Guttieri et al., 2004). However, lpa mutants of barley and maize showed approximately 50–95% reduction in PA (Raboy et al., 2000; Dorsch et al., 2003). Thus, one could speculate that complex hexaploid wheat genome confers a greater buffering capacity as compared to diploid genomes of barley and maize. Additionally, one could measure the enzymatic activity of wheat IPK1 arising from multiple genomes, i.e., A, B, and D.
IPK1 Function Is Conserved across Species
Earlier studies showed that lowering of PA is often associated with some pleiotropic effects (Guttieri et al., 2004; Stevenson-Paulik et al., 2005; Bhati et al., 2016). Therefore, comprehensive analysis was performed for various morphological traits such as seed weight, seed count, and germination ability. Silencing of IPK1 in rice did not show any negative effects during seed germination or other agronomic traits (Ali et al., 2013a). In contrast, our data indicated that lowering of PA by targeting TaIPK1 impacts seed count (Figure 5D). The transgenic wheat in our study did not show any pleiotropic changes for seed weight and germination (Figure 5 and Supplementary Figure S6). Taken together, this study and previous support validate the possibility that plant IPK1 as an important candidate gene for developing reduced-phytate trait in multiple crop species (Shi et al., 2007; Shukla et al., 2009; Ali et al., 2013a).
Very limited studies have been performed to study the changes in global gene expression for lpa mutants during seed development. Earlier, it was shown that lpa mutations effects global gene expression pattern for those involved in apoptosis, inositol phosphate synthesis, oligopeptide, and transmembrane transporters (Redekar et al., 2015; Zhang et al., 2016). Our study suggested that over-accumulation of Pi in seeds could cause differential changes in the wheat PHTs. In Arabidopsis, IPK1 were linked to a phosphate sensing and phytate production in seeds (Stevenson-Paulik et al., 2005). Most of the wheat transgenic lines obtained in this study showed high total phosphorus (Supplementary Figure S7). Herein, our data confirmed that silencing of IPK1 not only resulted in increased free Pi but also impact the expression of wheat PHTs and thereby impacting the dynamics of Pi transporter in lpa wheat grains.
From previous studies, it is evident that the PA had a high affinity for micronutrients such as Zn, Fe, Ca, etc. (Vohra et al., 1965; Persson et al., 1998; Iwai et al., 2012) and renders these minerals unavailable for gut absorption. It seems that unless significant lowering of PA is achieved, no effective changes in the seed micronutrient content will be observed (Bhati et al., 2016). In the current study, reduced wheat PA content (upto 56%) in grains exhibited a concomitant increase of Fe and Zn levels (Figures 3C, 6). It signifies that it is necessary to target genes that could potentially suppress PA biosynthesis efficiently. In another study wheat lines generated achieved Fe and Zn as high as 2.1- and 3.7-fold by overexpressing OsNAS2 (Singh et al., 2017). Enhanced micronutrient levels do not correspond to metal bioavailability. However, the metal to PA molar ratios could be an indirect evidence for enhanced bioavailability (Morris and Ellis, 1989; Ma et al., 2005). Our data showed an increase in the Fe:PA and Zn:PA molar ratios. This suggests that lowering of PA enhances micronutrient molar ratio, and thus, possibly increasing the mineral bioavailability. Similarly, lowering of PA in wheat-Aegilops derivatives and Triticum monococcum enhances the bioavailability of zinc (Salunke et al., 2012). The strong validity of the observed bioavailability of micronutrients with the phytate:mineral molar ratios in the lpa mutants reinforce the significance of utilizing these parameters for the micronutrient biofortification programs.
This study is an important step to achieve high micronutrients in grains of the hexaploid wheat. Recently researchers have also focused on understanding Zn and Fe transport as a novel route to improved loading in developing grains (Rouached, 2013; Grillet et al., 2014). Such parallel approaches for enhancing micronutrients have been recently evaluated by overexpressing certain metal transporters, phytases, etc (Abid et al., 2017; Connorton et al., 2017; Singh et al., 2017). Altogether, such strategies reinforce the importance of achieving enhanced bioavailable Fe and Zn in cereal crops to address malnutrition.
Author Contributions
AP conceived the idea and designed the experiments. SA performed most of the experiment. KB, AK, and ST helped in the experiments dealing with wheat transformation. VS and GK performed the experiments related to transporters. AP, SA, KB, and ST wrote the article. All the authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Executive Director, NABI for facilities and support. Part of this research was funded by the Department of Biotechnology (DBT), Government of India (BT/PR5989/AGII/106/867/2012) to AP and ST. SA acknowledges ICMR for Senior Research Fellowships. Fellowship for AK was supported by NABI-CORE grant. VS fellowship was supported by CSIR. Technical assistance from Dr. Pankaj Pandey is highly appreciated. Authors thank Pratima Pandey, IISER-Mohali for critical reading of the manuscript and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00259/full#supplementary-material
Abbreviations
ABC-MRP, ATP binding cassette multi drug resistance protein; DAA, day after anthesis; lpa, low phytic acid; PA, phytic acid.
References
Abid, N., Khatoon, A., Maqbool, A., Irfan, M., Bashir, A., Asif, I., et al. (2017). Transgenic expression of phytase in wheat endosperm increases bioavailability of iron and zinc in grains. Transgenic Res. 26, 109–122. doi: 10.1007/s11248-016-9983-z
Aggarwal, S., Shukla, V., Bhati, K., Kaur, M., Sharma, S., Singh, A., et al. (2015). Hormonal regulation and expression profiles of wheat genes involved during phytic acid biosynthesis pathway. Plants 4, 298–319. doi: 10.3390/plants4020298
Ali, N., Paul, S., Gayen, D., Sarkar, S. N., Datta, K., and Datta, S. K. (2013a). Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene (IPK1). PLoS One 8:e68161. doi: 10.1371/journal.pone.0068161
Ali, N., Paul, S., Gayen, D., Sarkar, S. N., Datta, S. K., and Datta, K. (2013b). RNAi mediated down regulation of myo-inositol-3-phosphate synthase to generate low phytate rice. Rice 6:12. doi: 10.1186/1939-8433-6-12
Ames, B. N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8, 115–118. doi: 10.1016/0076-6879(66)08014-5
Bhati, K. K., Aggarwal, S., Sharma, S., Mantri, S., Singh, S. P., Bhalla, S., et al. (2014). Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Sci. 224, 74–85. doi: 10.1016/j.plantsci.2014.04.009
Bhati, K. K., Alok, A., Kumar, A., Kaur, J., Tiwari, S., and Pandey, A. K. (2016). Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 67, 4379–4389. doi: 10.1093/jxb/erw224
Borrill, P., Ramirez-Gonzalez, R., and Uauy, C. (2015). expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol. 15, 2172–2186. doi: 10.1104/pp.15.01667
Breiman, A., and Graur, B. (1995). Wheat evolution. Isr. J. Plant Sci. 43, 85–98. doi: 10.1080/07929978.1995.10676595
Choulet, F., Alberti, A., Theil, S., Glover, N., Barbe, V., Daron, J., et al. (2014). Structural and functional partitioning of bread wheat chromosome 3B. Science 345:1249721. doi: 10.1126/science.1249721
Connorton, J. M., Jones, E. R., Rodriguez-Ramiro, I., Fairweather-Tait, S., Uauy, C., and Balk, J. (2017). Vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. 174, 2434–2444. doi: 10.1104/pp.17.00672
Dorsch, J. A., Cook, A., Young, K. A., Anderson, J. M., Bauman, A. T., Volkmann, C. J., et al. (2003). Seed phosphorus and inositol phosphate phenotype of barley low phytic acid genotypes. Phytochemistry 62, 691–706. doi: 10.1016/S0031-9422(02)00610-6
Garcìa-Estepa, R. M., Guerra-Hernández, E., and Garcìa-Villanova, B. (1999). Phytic acid content in milled cereal products and breads. Food Res. Int. 32, 217–221. doi: 10.1016/S0963-9969(99)00092-7
Gasparis, S., Orczyk, W., Zalewski, W., and Nadolska-Orczyk, A. (2011). The RNA-mediated silencing of one of the Pin genes in allohexaploid wheat simultaneously decreases the expression of the other, and increases grain hardness. J. Exp. Bot. 62, 4025–4036. doi: 10.1093/jxb/err103
González, B., Baños-Sanz, J. I., Villate, M., Brearley, C. A., and Sanz-Aparicio, J. (2010). Inositol 1,3,4,5,6-pentakisphosphate 2-kinase is a distant IPK member with a singular inositide binding site for axial 2-OH recognition. Proc. Natl. Acad. Sci. U.S.A. 107, 9608–9613. doi: 10.1073/pnas.0912979107
Grillet, L., Mari, S., and Schmidt, W. (2014). Iron in seeds-loading pathways and subcellular localization. Front. Plant Sci. 4:535. doi: 10.3389/fpls.2013.00535
Guttieri, M., Bowen, D., Dorsch, J. A., Raboy, V., and Souza, E. (2004). Identification and characterization of a low phytic acid wheat. Crop Sci. 44, 418–424. doi: 10.2135/cropsci2004.4180
Iwai, T., Takahashi, M., Oda, K., Terada, Y., and Yoshida, K. T. (2012). Dynamic changes in the distribution of minerals in relation to phytic acid accumulation during rice seed development. Plant Physiol. 160, 2007–2014. doi: 10.1104/pp.112.206573
Kim, S.-I., and Tai, T. H. (2011). Identification of genes necessary for wild-type levels of seed phytic acid in Arabidopsis thaliana using a reverse genetics approach. Mol. Genet. Genomics 286, 119–133. doi: 10.1007/s00438-011-0631-2
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, G., Jin, Y., Piao, J., Kok, F., Guusje, B., and Jacobsen, E. (2005). Phytate, calcium, iron, and zinc contents and their molar ratios in foods commonly consumed in China. J. Agric. Food Chem. 53, 10285–10290. doi: 10.1021/jf052051r
Morris, E. R., and Ellis, R. (1989). Usefulness of the dietary phytic acid/zinc molar ratio as an index of zinc bioavailability to rats and humans. Biol. Trace Elem. Res. 19, 107–117. doi: 10.1007/BF02925452
Persson, H., Türk, M., Nyman, M., and Sandberg, A.-S. (1998). Binding of Cu2+, Zn2+, and Cd2+ to Inositol Tri-, Tetra-, Penta-, and Hexaphosphates. J. Agric. Food Chem. 46, 3194–3200. doi: 10.1021/jf971055w
Pfeifer, M., Kugler, K. G., Sandve, S. R., Zhan, B., Rudi, H., Hvidsten, T. R., et al. (2014). Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345:1250091. doi: 10.1126/science.1250091
Przetakiewicz, A., Karas, A., Orczyk, W., and Nadolska-Orczyk, A. (2004). Agrobacterium-mediated transformation of polyploid cereals. The efficiency of selection and transgene expression in wheat. Cell Mol. Biol. Lett. 9, 903–917.
Raboy, V. (2009). Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 177, 281–296. doi: 10.1016/j.plantsci.2009.06.012
Raboy, V., Gerbasi, P. F., Young, K. A., Stoneberg, S. D., Pickett, S. G., Bauman, A. T., et al. (2000). Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 124, 355–368. doi: 10.1104/pp.124.1.355
Rasmussen, S. K., Ingvardsen, C. R., and Torp, A. M. (2010). Mutations in genes controlling the biosynthesis and accumulation of inositol phosphates in seeds. Biochem. Soc. Trans. 38, 689–694. doi: 10.1042/BST0380689
Redekar, N. R., Biyashev, R. M., Jensen, R. V., Helm, R. F., Grabau, E. A., and Maroof, M. A. S. (2015). Genome-wide transcriptome analyses of developing seeds from low and normal phytic acid soybean lines. BMC Genomics 16:1074. doi: 10.1186/s12864-015-2283-9
Rouached, H. (2013). Recent developments in plant zinc homeostasis and the path toward improved biofortification and phytoremediation programs. Plant Signal. Behav. 8:e22681. doi: 10.4161/psb.22681
Salunke, R., Rawat, N., Tiwari, V. K., Kumari, N., Randhawa, G. S., Dhaliwal, H. S., and Roy, P. (2012). Determination of bioavailable-zinc from biofortified wheat using a coupled in vitro digestion/Caco-2 reporter-gene based assay. J. Food Compos. Anal. 25, 149–159. doi: 10.1016/j.jfca.2011.09.006
Secco, D., Bouain, N., Rouached, A., Prom-U-Thai, C., Hanin, M., Pandey, A. K., et al. (2017). Phosphate, phytate and phytases in plants: from fundamental knowledge gained in Arabidopsis to potential biotechnological applications in wheat. Crit. Rev. Biotechnol. 37, 898–910. doi: 10.1080/07388551.2016.1268089
Shi, J., Wang, H., Hazebroek, J., Ertl, D. S., and Harp, T. (2005). The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 42, 708–719. doi: 10.1111/j.1365-313X.2005.02412.x
Shi, J., Wang, H., Schellin, K., Li, B., Faller, M., Stoop, J. M., et al. (2007). Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 25, 930–937. doi: 10.1038/nbt1322
Shi, J., Wang, H., Wu, Y., Hazebroek, J., Meeley, R. B., and Ertl, D. S. (2003). The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 131, 507–515. doi: 10.1104/pp.014258
Shukla, V., Kaur, M., Aggarwal, S., Bhati, K. K., Kaur, J., Mantri, S., et al. (2016). Tissue specific transcript profiling of wheat phosphate transporter genes and its association with phosphate allocation in grains. Sci. Rep. 6:39293. doi: 10.1038/srep39293
Shukla, V. K., Doyon, Y., Miller, J. C., DeKelver, R. C., Moehle, E. A., Worden, S. E., et al. (2009). Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459, 437–441. doi: 10.1038/nature07992
Singh, S. P., Keller, B., Gruissem, W., and Bhullar, N. K. (2017). Rice NICOTIANAMINE SYNTHASE 2 expression improves dietary iron and zinc levels in wheat. Theor. Appl. Genet. 130, 283–292. doi: 10.1007/s00122-016-2808-x
Stevenson-Paulik, J., Bastidas, R. J., Chiou, S.-T., Frye, R. A., and York, J. D. (2005). Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. U.S.A. 102, 12612–12617. doi: 10.1073/pnas.0504172102
Stevenson-Paulik, J., Odom, A. R., and York, J. D. (2002). Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J. Biol. Chem. 277, 42711–42718. doi: 10.1074/jbc.M209112200
Stiles, A. R., Qian, X., Shears, S. B., and Grabau, E. A. (2008). Metabolic and signaling properties of an itpk gene family in Glycine max. FEBS Lett. 582, 1853–1858. doi: 10.1016/j.febslet.2008.04.054
Sun, Y., Thompson, M., Lin, G., Butler, H., Gao, Z., Thornburgh, S., et al. (2007). Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from maize: molecular and biochemical characterization. Plant Physiol. 144, 1278–1291. doi: 10.1104/pp.107.095455
Suzuki, M., Tanaka, K., Kuwano, M., and Yoshida, K. T. (2007). Expression pattern of inositol phosphate-related enzymes in rice (Oryza sativa L.): implications for the phytic acid biosynthetic pathway. Gene 405, 55–64. doi: 10.1016/j.gene.2007.09.006
Sweetman, D., Johnson, S., Caddick, S. E. K., Hanke, D. E., and Brearley, C. A. (2006). Characterization of an Arabidopsis inositol 1,3,4,5,6-pentakisphosphate 2-kinase (AtIPK1). Biochem. J. 394, 95–103. doi: 10.1042/BJ20051331
Sweetman, D., Stavridou, I., Johnson, S., Green, P., Caddick, S. E. K., and Brearley, C. A. (2007). Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase 4 (AtITPK4) is an outlier to a family of ATP-grasp fold proteins from Arabidopsis. FEBS Lett. 581, 4165–4171. doi: 10.1016/j.febslet.2007.07.046
Vohra, P., Gray, G. A., and Kratzer, F. H. (1965). Phytic acid-metal complexes. Proc. Soc. Exp. Biol. Med. 120, 447–449. doi: 10.3181/00379727-120-30559
Welch, R. M., and Graham, R. D. (2005). Agriculture: the real nexus for enhancing bioavailable micronutrients in food crops. J. Trace Elem. Med. Biol. 18, 299–307. doi: 10.1016/j.jtemb.2005.03.001
White, P. J., and Broadley, M. R. (2005). Biofortifying crops with essential mineral elements. Trends Plant Sci. 10, 586–593. doi: 10.1016/j.tplants.2005.10.001
WHO (2007). Conclusions and recommendations of the WHO Consultation on prevention and control of iron deficiency in infants and young children in malaria-endemic areas. Food Nutr. Bull. 28, S621–S627. doi: 10.1177/15648265070284S414
Ye, H., Zhang, X.-Q., Broughton, S., Westcott, S., Wu, D., Lance, R., et al. (2011). A nonsense mutation in a putative sulphate transporter gene results in low phytic acid in barley. Funct. Integr. Genomics 11, 103–110. doi: 10.1007/s10142-011-0209-4
Yuan, F.-J., Zhu, D.-H., Tan, Y.-Y., Dong, D.-K., Fu, X.-J., Zhu, S.-L., et al. (2012). Identification and characterization of the soybean IPK1 ortholog of a low phytic acid mutant reveals an exon-excluding splice-site mutation. Theor. Appl. Genet. 125, 1413–1423. doi: 10.1007/s00122-012-1922-7
Zadoks, J. C., Chang, T. T., and Konza, C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x
Zalewski, W., Galuszka, P., Gasparis, S., Orczyk, W., and Nadolska-Orczyk, A. (2010). Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J. Exp. Bot. 61, 1839–1851. doi: 10.1093/jxb/erq052
Keywords: phytic acid, Triticum aestivum, inositol pentakisphosphate kinase, gene silencing, wheat transformation
Citation: Aggarwal S, Kumar A, Bhati KK, Kaur G, Shukla V, Tiwari S and Pandey AK (2018) RNAi-Mediated Downregulation of Inositol Pentakisphosphate Kinase (IPK1) in Wheat Grains Decreases Phytic Acid Levels and Increases Fe and Zn Accumulation. Front. Plant Sci. 9:259. doi: 10.3389/fpls.2018.00259
Received: 12 December 2017; Accepted: 14 February 2018;
Published: 06 March 2018.
Edited by:
Hatem Rouached, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Ping Lan, Institute of Soil Science (CAS), ChinaSebastian Gasparis, Plant Breeding and Acclimatization Institute (IHAR), Poland
Kulvinder S. Gill, Washington State University, United States
Copyright © 2018 Aggarwal, Kumar, Bhati, Kaur, Shukla, Tiwari and Pandey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ajay K. Pandey, pandeyak@nabi.res.in; pandeyak1974@gmail.com
 Sipla Aggarwal
Sipla Aggarwal Anil Kumar
Anil Kumar Kaushal K. Bhati
Kaushal K. Bhati Gazaldeep Kaur
Gazaldeep Kaur Vishnu Shukla
Vishnu Shukla Siddharth Tiwari
Siddharth Tiwari Ajay K. Pandey
Ajay K. Pandey