- 1Beijing Key Laboratory of Ornamental Plants Germplasm Innovation & Molecular Breeding, National Engineering Research Center for Floriculture, Beijing Laboratory of Urban and Rural Ecological Environment, Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants of Ministry of Education, School of Landscape Architecture, Beijing Forestry University, Beijing, China
- 2Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, Beijing Forestry University, Beijing, China
Rosa chinensis, which is a famous traditional flower in China, is a major ornamental plant worldwide. Long-term cultivation and breeding have resulted in considerable changes in the number of rose petals, while most wild Rosaceae plants have only one whorl consisting of five petals. The petals of double flowers reportedly originate from stamens, but the underlying molecular mechanism has not been fully characterized. In this study, we observed that the number of petals of R. chinensis ‘Old Blush’ flowers increased and decreased in response to low- and high-temperature treatments, respectively, similar to previous reports. We characterized these variations in further detail and found that the number of stamens exhibited the opposite trend. We cloned an APETALA2 homolog, RcAP2. A detailed analysis of gene structure and promoter cis-acting elements as well as RcAP2 temporospatial expression patterns and responses to temperature changes suggested that RcAP2 expression may be related to the number of petals from stamen origin. The overexpression of RcAP2 in Arabidopsis thaliana transgenic plants may induce the transformation of stamens to petals, thereby increasing the number of petals. Moreover, silencing RcAP2 in ‘Old Blush’ plants decreased the number of petals. Our results may be useful for clarifying the temperature-responsive mechanism involved in petaloid stamen production, which may be relevant for the breeding of new rose varieties with enhanced flower traits.
Introduction
The China rose (Rosa chinensis), which is one of the most popular ornamental plants, produces flowers with diverse patterns. The considerable ornamental value of R. chinensis is at least partly due to the variable double-flower trait with different colors. Double-flower trait describes varieties of flowers with extra petals, and is the earliest documented form of floral abnormality, which was first recognized 2,000 years ago (Meyerowitz et al., 1989). Most wild Rosaceae species have only one whorl of five petals, while R. chinensis produces a double flower, which may originate from the stamen (Reynolds and Tampion, 1983). These petals evolved from the stamens, with the filaments widening to form narrow petals (Kramer and Irish, 1999; Cronk, 2001). A study of a rose hybrid population revealed that the double-flower phenotype is a dominant qualitative trait that is controlled by a single gene (Debener, 1999). However, the degree of double flowering, that is, the number of petals produced by secondary-effect genes, has not been fully characterized (Morey, 1959; Hibrand-Saint Oyant et al., 2008). In addition to genetic factors, environmental cues (e.g., temperature) also affect the number of petals (Garrod and Harris, 1974; Chmelnitsky et al., 2001). Low temperatures increase the number of petals and decrease the number of stamens, resulting in the bullhead rose phenotype (Zieslin, 1968, 1992). The development of petals from stamen origin represents the main factor involved in increasing the number of rose petals. The underlying mechanism is likely relatively complex.
Elucidating the mechanism regulating petal formation is one of the most important objectives of floral organ research. The floral organ model has been extensively studied, and the ABCE model has gradually been accepted (Soltis et al., 2006). This model adopts the basic theory of the classical ABC model, while simultaneously emphasizing the importance of the class E genes regarding floral organ development. Specifically, the class E genes are expressed in the four whorls of floral organs, and are indispensable in the development of sepals, petals, stamens, and carpels, although there is some functional redundancy. In the ABCE model, class A + E genes determine the formation of sepals, class A + B + E genes mediate the formation of petals, class B + C + E genes contribute to the formation of stamens, and class C + E genes are involved in the formation of carpels (Bowman et al., 1991; Coen and Meyerowitz, 1991; Causier et al., 2010). Several homeotic genes have been identified in each class, including APETALA1 (AP1) and APETALA2 (AP2) in class A, APETALA3 (AP3) and PISTILLATA (PI) in class B, AGAMOUS (AG) in class C, and SEPALLATA1 (SEP1), SEP2, SEP3, and SEP4 in class E (Pelaz et al., 2000). Based on an analysis of flower morphology in dicotyledonous plants, such as buttercup, and monocotyledons, including lily and tulip, the shifting border model has been proposed (Van Tunen et al., 1993; Bowman, 1997; Kramer et al., 1998). In this model, the tissues in which the class A and C genes are expressed are relatively fixed, but because of the expansion of the class B genes, the outer sepals can develop into petals (Kanno et al., 2007). The expansion of the class B genes has also been observed in other dicotyledonous plants, such as Sagittaria and Camellia species (Kramer et al., 1998). Moreover, recent studies regarding Macleaya and Rosaceae species have revealed that the tissues in which the class A and C genes are expressed may be variable (Ronse De Craene, 2003; Dubois et al., 2010). However, there are relatively few examples of a change in the tissues in which class A genes are expressed. Although the shifting border model can complement the ABCE model to some extent, it should be studied in greater detail. For example, additional floral organ development genes and plant species should be investigated to further characterize the role of the shifting border model during floral organ development.
Most of the studies on the double-flower phenotype have focused on the class C genes. In Arabidopsis thaliana, AG regulates stamen and carpel properties. The stamens are transformed into petals to form double flowers in the ag mutant. Earlier investigations confirmed that AG interacts with the transcription factor WUSCHEL (WUS), which controls the differentiation of the meristem, and then determines the floral organ properties (Lenhard et al., 2001; Lohmann et al., 2001). A previous study indicated that the Japanese gentian double-flower mutant is the result of the insertion of a 5,150-bp LTR retrotransposon in the sixth intron of GsAG1 (Nakatsuka et al., 2015). Meanwhile, the double-flower phenotype of Prunus lannesiana is due to the absence of a PrseAG exon (Liu et al., 2013). Additionally, the RhAG (Rosa hybrida cv. Vendela) expression level is low in the double flowers of cultivated rose species. The results of an in situ hybridization indicated that RhAG expression is concentrated in the central region of meristematic tissue, resulting in an increase in the number of petals and a decrease in the number of stamens (Dubois et al., 2010). Additional research confirmed that exposure to low temperatures alters the methylation level of a specific region of the RhAG promoter to regulate expression (Ma et al., 2015).
The mechanism that determines the number of petals has not been fully characterized, especially in rose species. First, RhAG is not located at the Blfo locus, the marker closest to the gene for double flowers (Debener and Mattiesch, 1999; Dugo et al., 2005), which may control whether single or double flowers are produced, or at the locus associated with the number of petals (Spiller et al., 2011). This suggests that RhAG may function as a downstream regulatory gene (Dubois et al., 2010). Second, the restricted spatial expression of RhAG may be insufficient for the transition of stamens to petals, necessitating the participation of class A genes (Spiller et al., 2011). The third whorl of the floral organ develops as stamens or petals depending on the balance between AP2 and AG (Wollmann et al., 2010). In A. thaliana, AP2 determines the sepal and petal characteristics, while limiting the tissues in which the class C genes are expressed, and regulating AG expression to upregulate the expression of class B genes (Liu and Meyerowitz, 1995; Jack, 2004). A lack of AP2 function will lead to stamens replacing petals, while the ectopic expression of AP2 will result in the production of double flowers (Theissen et al., 2000). Thus, the regulatory effect of AP2 on the number of rose petals cannot be ignored.
‘Old Blush’ is a diploid R. chinensis cultivar that originated in China over 1,000 years ago. This rose cultivar exhibits recurrent flowering and produces double flowers as well as other traits that are very important for modern rose breeding (Dubois et al., 2012). We observed that the number of petals in ‘Old Blush’ flowers differs considerably depending on whether plants are exposed to low or high temperatures. We identified an AP2 homolog (RcAP2) in ‘Old Blush’ based on previous studies. This homolog has an alternative splice variant (RcAP2v). We completed detailed analyses of genetic structures and subcellular localization. We also determined that the RcAP2 expression pattern is correlated with petal type and the degree of double flowering, especially under low or high temperatures. Additionally, RcAP2 was heterologously expressed and transiently silenced in A. thaliana and rose, respectively. Our results may help to clarify the regulatory mechanism associated with rose floral organ formation and contribute to the molecular breeding of rose species for the production of desirable flower types.
Materials and Methods
Plant Materials and Growth Conditions
Rosa chinensis ‘Old Blush’ and A. thaliana ‘Columbia’ plants were grown in a greenhouse under a 12-h light [photosynthetic photon flux density (PPFD) of 800 μmol m-2 s-1]: 12-h dark photoperiod. The plants were incubated at 24°C (light): 14°C (dark) with 60% relative humidity. Rose flower development was divided into 11 stages based on the phenotype as well as the results of an earlier analysis of paraffin sections (Jackson, 1991). The major developments at each stage were as follows: S1: vegetation cone and leaf primordia formed; S2: floral meristem formed; S3: sepal primordia appeared; S4: petal primordia appeared and sepal primordia continued to develop; S5: stamen primordia appeared and developed; S6: pistil primordia appeared and other types of primordia continued to develop; S7: small buds appeared (approximately 2 mm diameter); S8: small buds increased in size (approximately 4 mm diameter); S9: buds differentiated, but sepals remained intact; S10: flowers started to open; and S11: flowers were fully open, with carpels and stamens visible. All materials were collected at 12:00 am (25°C). Ten buds were collected at S1–S6, while six buds or flowers were harvested at S7–S11. The collected samples were pooled and immediately frozen in liquid nitrogen. Sample collections were completed with three biological replicates.
RNA and DNA Extraction
Total RNA was extracted using the SV Total RNA Isolation System (Promega, Madison, WI, United States), and used as the template for the first-strand cDNA synthesis using the PrimeScriptTM RT Reagent Kit with gDNA Eraser (Takara Bio Inc., Shiga, Japan). Additionally, DNA was extracted from R. chinensis ‘Old Blush’ leaves using the Plant Genomic DNA kit (Omega Bio-tek Inc., Norcross, GA, United States).
Gene and Promoter Isolation
Primers RcAP2-F1 and RcAP2-R1 were designed based on the conserved sequences of AP2 homologs to PCR amplify the RcAP2 gene. The cDNA samples appropriate for rapid amplification of cDNA ends (RACE) were synthesized using the SMARTerTM RACE cDNA Amplification Kit (Clontech, United States). The 5′-GSP primers for the 5′-RACE were designed according to the sequences obtained from the 3′-RACE experiment. The Universal Primer Mix from the 5′- and 3′-RACE PCR kit was used for a touchdown PCR, which was completed with the following program: five cycles of 94°C for 30 s and 72°C for 3 min; five cycles of 94°C for 30 s, 70°C for 30 s, and 72°C for 3 min; 27 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min. Primers RcAP2-F2 and RcAP2-R2 were designed based on the 5′- and 3′-ends of the candidate sequence, and used to amplify the full-length gene with the following program: 94°C for 5 min; 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 3 min; 72°C for 10 min. The RhAG sequence (RHU43372) was used to design primers RcAG-F and RcAG-R for the amplification of the RcAG DNA sequence. The RcAP2 promoter was obtained using the Genome Walking Kit (Takara). Details regarding primers proRcAP2-SP1, proRcAP2-SP2, and proRcAP2-SP3 are provided in Supplementary Table S1. Amplified cDNA or promoter fragments were purified with the MiniBEST Agarose Gel DNA Extraction Kit (Takara). The purified products were ligated into the pEASY-blunt cloning vector (TransGen Biotech, China) and subsequently transformed into Escherichia coli DH5α cells. Positive colonies were sequenced by SinoGenoMax (China). Details regarding the RcAP2 CDS (GenBank ID KX371592), DNA sequence (GenBank ID MF773427), splice variants (GenBank ID MF773425), and promoter sequence as well as the RcAG sequence (GenBank ID MF773426) have been deposited in the NCBI database1. Details regarding all primers are provided in Supplementary Table S4.
Bioinformatics Analysis
Sequence analysis and contig assembly were completed using DNAMAN software, while the RcAP2 open reading frame (ORF) was predicted using ORF Finder2. Multiple sequences were aligned using ClustalX with default parameters. The alignments were edited and marked using GeneDoc software. Phylogenetic analyses were conducted with MEGA7 using the Neighbor-Joining method and the bootstrap consensus tree was inferred from 1000 replicates. The function prediction of transcription factor using PlantTFDB online tool3, the RegRNA 2.0 online tool4 (Chang et al., 2013) was used to identifying the functional motifs and sites of RcAP2. The cis-acting element analyses were completed using online software PlantCARE5 (Lescot et al., 2002) and SoftBerry6 (Solovyev et al., 2010).
The GenBank ID of AP2 protein sequences showed in alignment analyses were as follows: AAL57045.2 Malus domestica; AKN10311.1 Eriobotrya japonica; AJT39804.1 Prunus mume; AEB92231.1 Prunus persica; ACO52508.1 Vitis vinifera; EOX91494.1 Theobroma cacao; AEL29576.1 Betula platyphylla; EXC24730.1 Morus notabilis; AGM20693.1 Populus tomentosa; EEE91165.1 Populus trichocarpa; KYP44918.1 Cajanus cajan; KHN36247.1 Glycine soja; AIS71911.1 Nicotiana tabacum; and AAC13770.1 Arabidopsis thaliana.
The GenBank ID of AP2 sequences showed in phylogenetic analyses were as follows: HQ637468.1 Brassica napus; JN247401.1 Actinidia deliciosa; JN247408.1 Betula platyphylla; JF683605.2 Prunus persica; U12546.1 Arabidopsis thaliana; DQ988341.1 Larix marschlinsii; AB495344.1 Nymphaea hybrid; JQ398741.1 Camellia sinensis; HQ113365.1 Actinidia deliciosa; EU883665.1 Poncirus trifoliata; FJ809943.1 Vitis vinifera; DQ988340.1 Larix marschlinsii; KP405836.1 Prunus mume; AB495343.1 Nymphaea hybrid; AF134116.3 Hyacinthus orientalis; AY128328.1 Arabidopsis thaliana; AY069953.1 Hordeum vulgare; AF253971.1 Picea abies; AF253970.1 Picea abies; EU677382.1 Solanum lycopersicum; KT163121.1 Thermopsis textitturcica; KJ482645.1 Brassica juncea; EF216863.2 Ipomoea nil; DQ119837.1 Dendrobium crumenatum; AB195246.1 Gnetum parvifolium; AB195244.1 Ginkgo biloba; AB195242.1 Cycas revoluta; AB101587.1 Pinus thunbergii; AB101586.1 Pinus thunbergii; KM386630.1 Fagopyrum esculentum; KM386629.1 Fagopyrum esculentum; KM386628.1 Fagopyrum esculentum; and GU983666.1 Malus domestica.
qPCR Validation
The qPCR assay was completed with the following program: 95°C for 30 s; 40 cycles of 95°C for 5 s and 60°C for 30 s; 95°C for 15 s, 60°C for 1 min and 95°C for 15 s for the melting-curve analysis. The qPCR solution consisted of 2 μL first-strand cDNA, 10 μL SYBR Premix Ex Taq (Takara Bio Inc., Shiga, Japan), 0.4 μL 10 μM forward and reverse primers, and 7.2 μL sterile distilled water. Each sample was assessed with three technical replicates. The expression data were normalized using RcActin or AtActin as the internal controls according to the 2-ΔΔCt method (Young et al., 2010). The qPCR primers are listed in Supplementary Table S4. The qPCR assay was conducted with three biological replicates. Gene expression diagrams were prepared with the Origin 8 program (OriginLab, Northampton, MA, United States).
Temperature Treatments of Rosa chinensis ‘Old Blush’
Two-year-old ‘Old Blush’ plants grown in pots under the conditions described in the Section “Plant materials and growth conditions” were used to determine the effects of temperature on the number of rose petals. Two artificial climate chambers were set as follows: 800 μmol m-2 s-1 PPFD; 12-h light: 12-h dark cycle; 60% relative humidity; and one of two temperature regimens [i.e., 16°C (light): 6°C (dark); 24°C: 14°C; or 32°C: 22°C]. Each temperature treatment involved 30 rose plants. The samples which collected at 24°C: 14°C for RcAP2 expression analysis were as control, and using the same data for indicating RcAP2 expression levels at S1–S11 and comparing with high-or low-temperature treatments. All buds at the beginning of S1 were observed. The buds and flowers collected at all floral development stages were frozen in liquid nitrogen until analyzed. We counted the number of floral organs (i.e., sepals, petals, stamens, and carpels) at S11. Twenty randomly selected flowers were examined for each treatment. The petals derived from stamens were considered as petals. Significant differences were determined according to Fisher’s LSD (P < 0.05) and Student’s t-test.
Heterologous Expression in Arabidopsis thaliana
The RcAP2 cDNA sequence was amplified using primers RcAP2-F3 and RcAP2-R3 (Supplementary Table S4), and the PCR product was cloned into the SalI/SpeI-cleaved pSuper1300 vector using the In-Fusion® HD Cloning Kit (Clontech, United States). The resulting pSuper1300-RcAP2 vector, in which RcAP2 was under the control of the cauliflower mosaic virus 35S promoter, was introduced into A. thaliana (Columbia) cells according to the floral-dip method involving Agrobacterium tumefaciens EHA105 (Clough and Bent, 1998). The T0 seeds were selected on MS medium containing 50 mg/L hygromycin B (Roche, Germany). The positive lines were confirmed by RT-PCR using primers RT-RcAP2-F/R and RT-AtActin-F/R. Choose three lines with highest expression of RcAP2 and identified their T3 pure lines with RcAP2 high expression levels using RT-PCR and qPCR methods. The floral phenotypes of T3 transgenic plants were observed and the flowers were collected. Ten flowers were analyzed, with three technical replicates. Significant differences were determined according to Fisher’s LSD (P < 0.05) and Student’s t-test.
RcAP2 Silencing in ‘Old Blush’ Plants
To silence RcAP2 in ‘Old Blush’ plants, a 405-bp RcAP2 CDS fragment was amplified using primers pTRV2-RcAP2-F/R (Supplementary Table S4). The PCR product was cloned into the BamHI/EcoRI-cleaved TRV2 vector (Liu et al., 2002) using the In-Fusion® HD Cloning Kit (Clontech, United States). The pTRV1, pTRV2, and pTRV2::RcAP2 plasmids were inserted into A. tumefaciens AGL0 cells. The silencing of RcAP2 was achieved using a graft-accelerated VIGS method (Yan et al., 2017). Thirty flowers bloomed after the A. tumefaciens-mediated insertion of pTRV1 + pTRV2 or pTRV1 + pTRV2::RcAP2. The floral phenotypes of the transgenic plants were observed and the flowers were collected. In pTRV1 + pTRV2::RcAP2 treatments, twelve sprouts appeared petal reduction phenotype after flowering. Each four flowers mixed-samples as a line for phenotypes observed and expression analysis. Samples were frozen in liquid nitrogen for a subsequent qPCR analysis of RcAP2 expression.
Subcellular Localization of RcAP2
The RcAP2 cDNA sequence was amplified using primers RcAP2-F4 and RcAP2-R4. The PCR product was cloned into the EcoRI/HindIII-cleaved pEZS-NL transient expression vector (provided by D. Ehrhardt, Carnegie Institution, Stanford, CA, United States). To examine the subcellular localization of RcAP2, isolated maize protoplasts were transformed with the prepared vector using an established protocol (Sheen et al., 1995). The transformed protoplasts were incubated for 16 h and then stained with 0.1 mg/mL 4′,6-diamino-2-phenylindole (DAPI; Sigma-Aldrich) in phosphate-buffered saline for 10 min. The RcAP2 protein was localized based on GFP fluorescence, which was detected with a laser confocal microscope (Leica TCS SP8, Germany). The GFP and DAPI were excited at 488 and 408 nm, respectively.
Results
RcAP2 Is an Ortholog of APELATA2 and Has an Alternative Splice Variant (RcAP2v)
To identify the R. chinensis AP2 ortholog, we obtained the RcAP2 DNA and mRNA sequences as well as the sequences of the promoter and a splice variant, RcAP2v. The encoded RcAP2 protein, which consists of 535 amino acids, was aligned with the AP2 sequences from 13 different species, all of which contained two conserved AP2 domains (Figure 1A). We constructed a phylogenetic tree based on the full-length cDNA sequences to assess the relationships among RcAP2 and the other AP2 genes available in the NCBI database (Figure 1B). The R. chinensis AP2 was most closely related to the AP2 genes from Prunus persica, Prunus mume, and Malus domestica (i.e., Rosaceae species), followed by the Betula platyphylla AP2 gene.
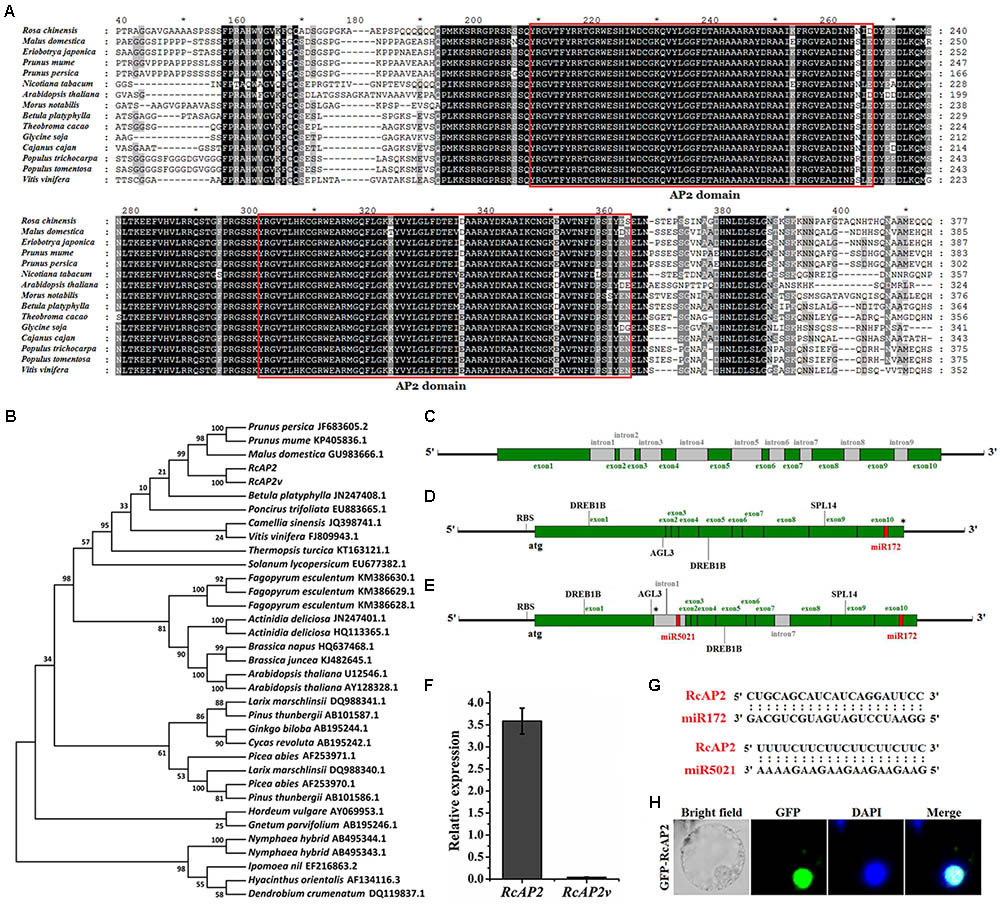
FIGURE 1. Bioinformatic analysis of RcAP2. (A) Sequence alignments of AP2 amino acid sequences among 15 different species. Deduced amino acid sequences of RcAP2 and another 14 AP2s were aligned using ClustalX with default settings and edited using GeneDoc. Boxes show two predicted AP2 domains. Each asterisk in (A) indicated the middle position of its left and right numbers. (B) Phylogenetic analysis of AP2 homologs from various species. The phylogenetic trees were constructed using MEGA7 software. The GenBank ID of each AP2 has been labeled. (C) Gene structure of RcAP2 DNA sequence. Green boxes, exons; gray boxes, introns. (D) Gene structure diagram of RcAP2 cDNA sequence. Four functional motifs (two DREB1B, one AGL3, and one SPL14) and one miR172 binding site were identified using RegRNA 2.0 online tool. RBS, ribosomal binding site. Asterisk marks termination codon. (E) Gene structure of RcAP2v cDNA sequence. Gray boxes, introns. One miR5021 binding site is shown in red. RBS, ribosomal binding site. Asterisk marks termination codon. (F) qPCR analysis of RcAP2 and RcAP2v transcript levels in whole flowers at 24°C (light): 14°C (dark). (G) Sequence alignments of two predicted miRNA-binding sequences and RcAP2. (H) Subcellular localization of RcAP2. RcAP2 and GFP fusion protein localized to nucleus of maize protoplasts. Bars = 10 μm.
RcAP2 contains 10 exons and 9 introns (Figure 1C and Supplementary Table S1). When we cloned the RcAP2 full-length coding sequence (CDS) from rose cDNA, we also obtained a CDS that comprised the first and seventh intron. This additional CDS may represent an intron retention splice variant of RcAP2 (Figure 1E). We then predicted the transcriptional regulatory motifs and miRNA-binding sites using the RegRNA 2.0 online tool. Four functional motifs were detected in RcAP2 and RcAP2v (i.e., two DREB1B, one AGL3, and one SPL14) (Supplementary Table S2). Additionally, we identified one miR172-binding site in the 10th exon of RcAP2, and one miR5021-binding site in the retained first intron of RcAP2v (Figures 1D,E). We designed the primers in the first intron-retaining region to detect the abundance of RcAP2v. However, the expression of RcAP2v was very low compared with RcAP2 (Figure 1F). The sequences of miR172-binding site and miR5021-binding site were complementary to RcAP2 sequences (Figure 1G). An analysis of the RcAP2-green fluorescent protein (GFP) fluorescence indicated that RcAP2 was localized to the nucleus of maize protoplasts (Figure 1H).
Temporospatial Expression of RcAP2 in Rosa chinensis
We divided the rose flower development process into 11 stages (S1–S11). Stages S1 to S6 comprise the main period of floral organ differentiation, during which elongating shoots begin to appear (Figure 2A). The initiation of floral organs and the formation of various primordia were observed at each stage using paraffin sections (Figure 2B). The primordia developed as follows. The vegetation cone and leaf primordia formed (S1). The vegetation cone was converted to the floral meristem (S2) and the outermost sepal primordia formed (S3). This was followed by the formation of the inner petal primordia (S4), stamen primordia (S5), and pistil primordia (S6). During S7 and S8, small buds became visible, while the internal floral organs continued to differentiate and develop. The flower buds were fully developed at S9, and started to bloom (Figure 2C).
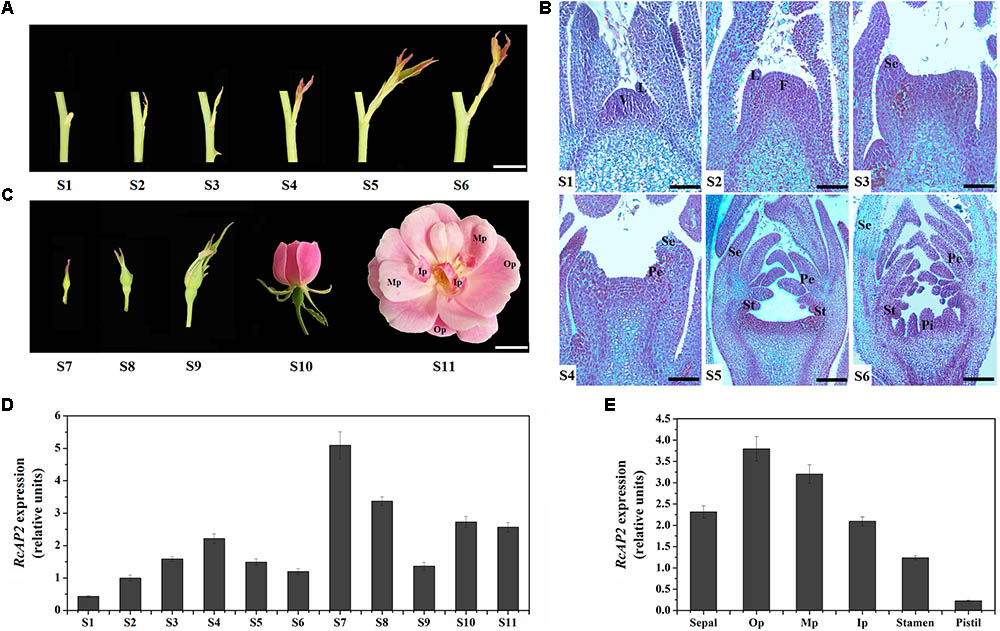
FIGURE 2. Spatiotemporal pattern of RcAP2 expression in ‘Old Blush’ flowers. (A) Diagram of S1–S6 developmental stages of ‘Old Blush’ shoot. S1: stage 1, vegetation cone and leaf primordia formation; S2: stage 2, floral meristem formation; S3: stage 3, sepal primordia appear; S4: stage 4, petal primordia appear and sepal primordia continue to develop; S5: stage 5, stamen primordia appear and develop; S6: stage 6, pistil primordia appear and other types of primordia continue to develop. Bars = 10 mm. (B) Observation of S1–S6 paraffin sections. V, vegetation cone; L, leaf primordia; F, floral meristem; Se, sepal primordia; Pe, petal primordia; St, stamen primordia; Pi, pistil primordia. Bars = 80 μm (S1, S2), 120 μm (S3, S4), or 200 μm (S5, S6). (C) Phenotypes of S7–S11 developmental stages of ‘Old Blush’ flowers. S7: stage 7, small buds appear with diameter about 2 mm; S8: stage 8, small buds with diameter about 4 mm; S9: stage 9, bud differentiation is complete and sepals have not cracked; S10: stage 10, flower is partially open; S11: stage 11, flower is fully open, carpels and stamens appear. Op, outer whorl petals; Mp, middle whorl petals; Ip, inner whorl petals. Bars = 10 mm. (D) qPCR analysis of RcAP2 transcript levels in 11 developmental stages of rose flower. S1–S11 corresponds to stages in A–C. (E) qPCR analysis of RcAP2 transcript levels different floral organs and Op, Mp, and Ip as defined in C.
RcAP2 was expressed during all R. chinensis floral development stages. However, the RcAP2 expression level was relatively low from S1 to S6 before rapidly increasing and peaking at S7. The RcAP2 expression level gradually decreased as buds expanded. The opening of flowers was accompanied by an increase in RcAP2 expression (Figure 2D). The distribution of RcAP2 expression in different floral organs exhibited some regularity. We defined the first whorl with five petals as Op (i.e., outer whorl petals), the narrow petals closest to the stamens as Ip (i.e., inner whorl petals), and the remaining petals as Mp (i.e., middle whorl petals) (Figure 2C). RcAP2 was most highly expressed in the Op. The RcAP2 expression level gradually decreased from the Op to the stamen, with the lowest level detected in the pistil (Figure 2E).
RcAP2 Expression Was Associated With Changes in the Number of Rose Petals at Low and High Temperatures
The temperatures ‘Old Blush’ plants are exposed to during cultivation influence the number of petals that are produced, with more petals at low temperatures. To characterize this phenomenon, we analyzed plants exposed to different temperatures. Similarly growing 2-year-old rose plants were incubated under one of the following conditions: 16°C (light): 6°C (dark); 24°C (light): 14°C (dark); or 32°C (light): 22°C (dark). The resulting phenotype and number of floral organs were analyzed (Figures 3A–D). There were no significant differences in the total number of floral organs among the three treatments. However, the flowers that underwent the low-temperature treatment (16°C: 6°C) produced more petals than the flowers exposed to normal temperatures (24°C: 14°C), while the flowers that underwent the high-temperature treatment (32°C: 22°C) produced fewer petals. The number of stamens exhibited the opposite pattern (Figure 3D). The excess petals were slender, and differed from the Op petals (Figure 3A). We also observed variability in the number of petals derived from stamens at different temperatures.
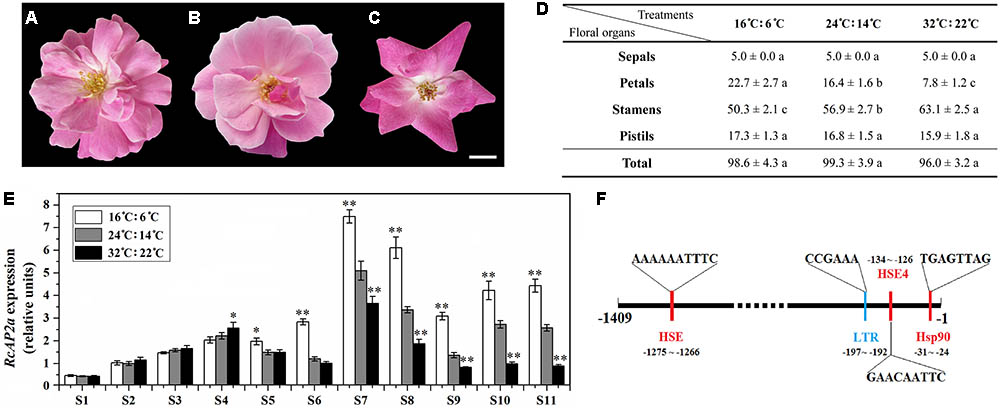
FIGURE 3. Effect of temperature on petal number of ‘Old Blush’ flowers. (A–C) Phenotypes of rose flowers at S11 under different temperatures. All buds were observed from beginning of S1. (A) 16°C: 6°C (light: dark). (B) 24°C: 14°C (light: dark). (C) 32°C: 22°C (light: dark). Bar = 1 cm. (D) Statistics of number of S11 floral organs under different temperatures. Values are means + SD of three biological replicates. Different lowercase letters indicate significant differences (Fisher’s LSD, P < 0.05). (E) RcAP2 transcript levels in rose flowers in response to different temperature treatments. RcAP2 transcript levels under 16°C: 6°C (light: dark) (white columns), under 24°C: 14°C (light: dark) (gray columns), and under 32°C: 22°C (light: dark) (black columns). Flowers from S1 to S11 were collected and analyzed. Values are means + SD of three biological replicates. ∗ and ∗∗ represent significant differences at P < 0.05 and P < 0.01, respectively (Student’s t-test). Flowers grown under 24°C: 14°C (light: dark) served as control. (F) Analysis of cis-acting elements in RcAP2 promoter using PlantCARE (http:// bioinformatics.psb.ugent.be/webtools/plantcare/html/) and SoftBerry (http://linux1.softberry.com/berry.phtml?topic=nsitep&group=programs&subgroup=promoter). Locations and sequences of four cis-acting elements that respond to temperature are shown: red, responsive to high temperature; blue, responsive to low temperature.
We analyzed the RcAP2 expression level at all floral development stages under three temperature conditions (Figure 3E). The expression level under the normal temperature treatment (24°C: 14°C) was used as the control. The RcAP2 mRNA level under the control conditions increased starting at S6, peaking at S7. Additionally, the low-temperature treatment was associated with the highest RcAP2 expression levels from S5 onwards. The high-temperature treatment inhibited the increase in RcAP2 expression from S7 until the flower blooming stage. We analyzed the RcAP2 promoter sequence using the PlantCARE database and SoftBerry program. The promoter contained four cis-acting elements related to temperature responses, three heat shock elements, and one low-temperature response element (Figure 3F and Supplementary Table S3).
Manipulation of RcAP2 Ectopic Expression Modulated the Ratio of Petals to Stamens in Arabidopsis thaliana
We transformed A. thaliana (Columbia) plants with the 35S::RcAP2 plasmid to generate transgenic plants over expressing RcAP2. Transgenic lines in which RcAP2 was highly expressed were identified by a reverse transcription polymerase chain reaction (RT-PCR) assay. We selected lines 2, 5, and 9 for subsequent analyses (Figure 4). The second whorl of line 5 flowers was obviously different from that of the wild-type flowers (Figure 4B). The number of petals increased from four (i.e., normal) to about seven, while the number of stamens decreased. Some stamens developed abnormally. Some formed as slender petals, while some formed as shortened filaments and anthers with no pollen attached, similar to transitional-state anthers. The slender petals were counted as petals. Abnormal stamens were also detected in line 2 (Figure 4C). There were almost no differences between the wild-type and transgenic plants regarding the total number of floral organs, including the number of sepals and pistils. The increase in the number of petals was offset by the decrease in the number of stamens (Figure 4F). The ectopic expression of RcAP2 in A. thaliana may induce the conversion of stamens to petals, with no changes to the total number of floral organs.
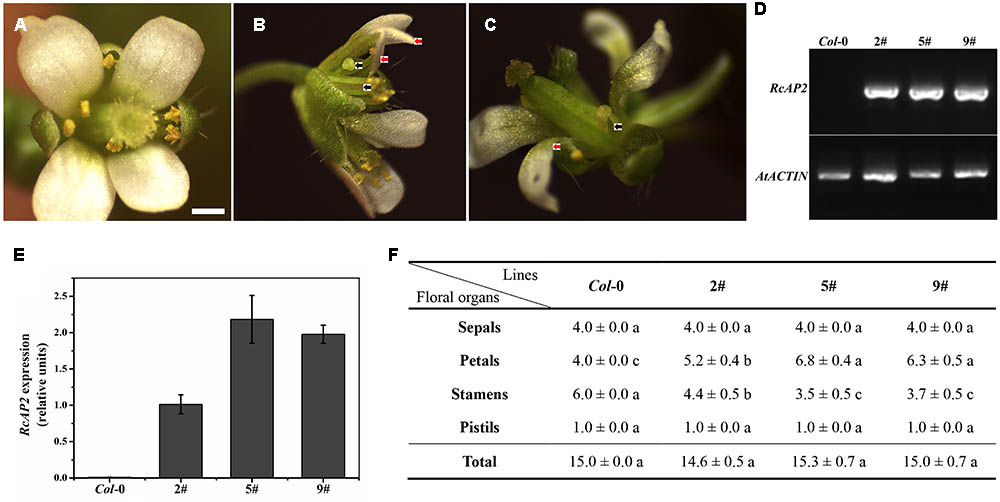
FIGURE 4. RcAP2 ectopic expression in Arabidopsis thaliana. (A) Flower of wild-type Arabidopsis thaliana (Columbia). Bar = 0.5 mm. (B) Flower phenotype of 5# and 9# transgenic lines of RcAP2. (C) Flower phenotype of 2# transgenic line of RcAP2. (B,C) Red arrows indicate abnormal stamens resembling slender petals; black arrows indicate abnormal stamens with shortened filaments, flat anthers, and no pollen attached, similar to transitional-state stamens. (D) RT-PCR detection of RcAP2 transcript levels in Arabidopsis transgenic lines. Wild-type Col-0 served as control. PCR-amplified RcAP2 fragment was 418 bp (base pair) and AtACTIN fragment was 373 bp. The original gel image of (D) was provided in Supplementary Figure S2. (E) qPCR detection of RcAP2 transcripts in Arabidopsis transgenic lines. Wild-type Col-0 served as control. (F) Statistics of floral organ numbers in wild-type and Arabidopsis transgenic lines. Values are means + SD of three biological replicates. Different lowercase letters indicate significant differences (Fisher’s LSD, P < 0.05).
Silencing RcAP2 by RNA Interference Decreased the Number of Petals in ‘Old Blush’ Flowers
The R. chinensis RcAP2 gene was functionally characterized by virus-induced gene silencing (VIGS) using a graft-accelerated method (Figure 5A). The pTRV1 + pTRV2::RcAP2 treatment resulted in an obvious decrease in the expression of RcAP2. Moreover, the number of petals decreased in the resulting flowers (Figures 5B,C). The outer whorl petals were preserved, the missing petals were petals from stamen origin. Compared with the effects of the pTRV1 + pTRV2 treatment control, the changes in RcAP2 expression and the phenotype induced by the pTRV1 + pTRV2::RcAP2 treatment confirmed that RcAP2 had been partially silenced (Figure 5D). Additionally, the RcAP2 transcript level was directly related to the number of petals in ‘Old Blush’ flowers.
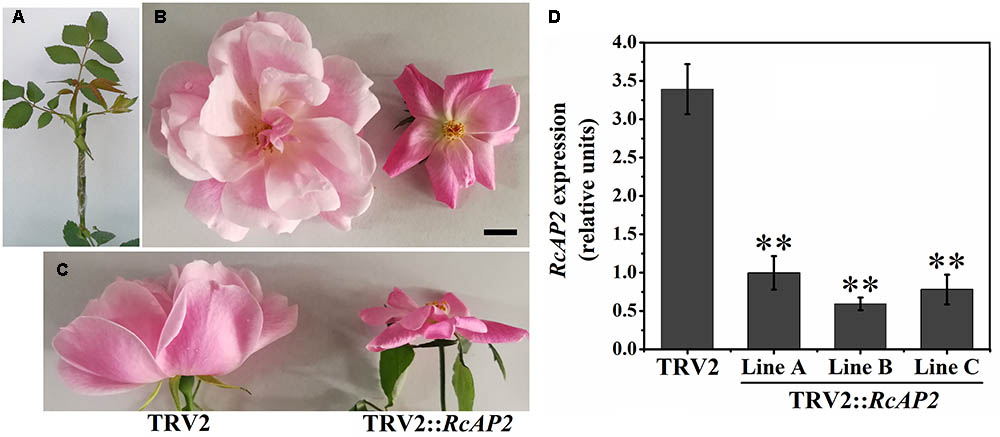
FIGURE 5. RcAP2 gene silencing in ‘Old Blush.’ (A) Shoots surviving after infection with A. tumefaciens and grafting. (B,C) Flower phenotype of control (left) and RcAP2-silenced (right) flowers. (C) Side view of flowers. TRV2 represents A. tumefaciens-mediated insertion of pTRV1 + pTRV2 (control); TRV2::RcAP2 represents pTRV1 + pTRV2::RcAP2. Bar = 1 cm. (D) qPCR detection of RcAP2 transcript levels in VIGS-treated flowers. Gray bars represent RcAP2 transcript levels relative to RcActin transcript levels. Lines A, B, and C represent three mixed samples, each consisting of four phenotypically obvious flowers. Values are means + SD of three technical replicates. ∗∗P < 0.01, (Student’s t-test).
Discussion
To elucidate the functions of R. chinensis AP2, we analyzed its sequence in detail. We observed that RcAP2 contains two AP2 domains, and exhibits characteristics typical of AP2. However, our phylogenetic analysis revealed the considerable genetic distance between the AP2 of Rosaceae species and that of other plant species. Thus, AP2 may exhibit different functions in different species. The subcellular localization of RcAP2 indicated this protein is present in the nucleus, which is consistent with the localization of transcription factors. In addition to the 3′ region, there are many miRNA-binding sites located in the 5′ and CDS regions for inhibiting translation (Hausser et al., 2013). We predicted the presence of a miR172-binding site in the CDS region of RcAP2. Recent studies concluded that miRNAs are important for the development of plant floral organs. The binding of miRNAs to specific sites in the target mRNA sequences affect floral development and regulate floral organ traits (Jones-Rhoades et al., 2006). Additionally, miR172 not only controls AG expression, it also restricts AP2 expression in the third and fourth whorls of floral organs during the early flowering stages, degrades AP2 mRNA, and inhibits AP2 translation. A previous study revealed that miR172 and its AP2 target are highly conserved components related to the regulation of plant development (Zhu and Helliwell, 2010). However, another study concluded that the regulatory activities of miR172 are insufficient for explaining the contradictory relationship between AP2 expression and its genetic activity (Wollmann et al., 2010). We also detected a miR5021-binding site in the retained intron of RcAP2v, but the function of miR5021 in plants is unknown. The translation of RcAP2v ends in the first intron, and the generated protein does not contain two AP2 domains. The variable splicing of different exons sometimes results in the early termination of a protein, and may function as a molecular switch (Kelemen et al., 2013). Thus, RcAP2v may contribute to the regulation of RcAP2 activity, although, it has a very low expression at the normal temperature. Temperature-dependent alternative splicing has been first described for the MADS-box gene FLM involved in the control of flowering time (Posé et al., 2013), and further identified for a large number of genes (Verhage et al., 2017). RcAP2 appears as a likely candidate to have an alternative splice variant whose levels depend on temperature which needs further study. Our sequence analyses suggest RcAP2 may be functionally complex.
In A. thaliana, AP2 mRNA is distributed throughout various types of floral primordia and vegetative tissues (e.g., stems and leaves). Additionally, the AP2 promoter is active in all floral whorls (Jofuku et al., 1994; Würschum et al., 2006). The AP2 orthologs in petunia, apple, and strawberry are also ubiquitously expressed (Maes et al., 2001; Su et al., 2004; Shengmei et al., 2006). Although AP2 is widely expressed, there are obvious differences in mRNA accumulation among floral parts. The AP2 orthologs exhibit very specific expression patterns in the floral primordia and inflorescences of maize and snapdragon (Keck et al., 2003; Chuck et al., 2008). In A. thaliana, AP2 mRNA accumulates predominantly in the outer floral whorls (Wollmann et al., 2010). We observed that RcAP2 expression in each floral organ whorl underwent changes as flowers developed. It is noteworthy that RcAP2 expression increased significantly from S6 to S7. This is the period in which the primordia of all floral organs are formed, but they have not developed into their corresponding floral organs. We speculate that floral organ development is affected by many factors, including intrinsic gene regulatory activities as well as environmental stimuli. Petals from stamen origin are the main source of rose petals. An increase in the petaloid nature of stamens was associated with a gradual transition of the stamens to petals, along with upregulated RcAP2 expression. These results imply that RcAP2 may be involved in the development of petals from stamen origin in R. chinensis.
Previous studies uncovered the changes in the number of rose petals induced by low temperatures (Zieslin, 1968, 1992). Additionally, low temperatures also modify the methylation status of a specific region in the RhAG promoter (Ma et al., 2015). In our study, the number of petals from stamen origin increased and decreased in response to low- and high-temperature treatments, respectively. The RcAP2 expression level was also affected by temperature changes. In A. thaliana, one model for the antagonistic relationship between class A and C genes involves AP2 and AG during the regulation of perianth formation. Additionally, ap2 mutants reportedly exhibit enhanced AG activity, and the outer floral organs develop into reproductive organs (Bowman et al., 1991). However, in stamen primordia, AP2 and AG expression overlap. Additionally, AP2 mRNA can be detected in all four whorls of the ag mutant, implying AG does not regulate AP2 at the transcript level (Bomblies et al., 1999; Wollmann et al., 2010). Some 35S::AP2 Arabidopsis transgenic lines with more stamens than the wild type, and in 35S::AP2m1 lines, a mutant with six mismatches to miRNA172, presented the transformation of stamens to petals, which was consistent with the ability of AP2 to negatively regulate AG (Chen, 2004). In A. thaliana, AP2 can bind the non-canonical AT-rich target sequence, TTTGTT, in the second intron of AG to directly regulate AG expression (Dinh et al., 2012). We analyzed the full-length RcAG sequence and detected three TTTGTT sequences in the first intron (Supplementary Figure S1). These conserved target sequences may represent a link between RcAP2 and RcAG.
In A. thaliana, the ap2 mutants exhibit structural deformities in the four whorls of floral organs. Specifically, whorl 1 is transformed into pistils, there is a decrease in the number of whorls 2 and 3, and the carpels do not fuse (Jofuku et al., 1994). Furthermore, transgenic Nicotiana benthamiana plants expressing the A. thaliana miR172-resistant AP2 produce flowers with an abnormal number of petals and stamens (Mlotshwa et al., 2006). Meanwhile, the expression of the water lily AP2 homolog in A. thaliana plants leads to a significant increase in the number of petals (Luo et al., 2012). A previous study confirmed that AP2 can promote ectopic floral organ formation in a process that may depend at least partially on a stem cell transcription factor, WUS (Zhao et al., 2007). In the current study, we observed a decrease in the number of stamens as well as changes in stamen morphology in RcAP2-expressing transgenic A. thaliana plants. This suggests the R. chinensis AP2 homolog inhibits the expression of class C genes and limits the tissues in which these genes are expressed, ultimately disrupting the third whorl of floral organs. The ectopic expression of RcAP2 is accompanied by the production of abnormal stamens, leading to an increase in the number of petals, which is similar to the effect of low temperatures on R. chinensis flowers. Silencing RcAP2 in ‘Old Blush’ plants via VIGS decreased the number of petals derived from stamens, but did not affect the five petals of the outermost whorl. Therefore, the RcAP2 transcript level is directly related to the number of petals (i.e., the number of petals derived from stamens). Temperature-induced changes to RcAP2 expression and the ectopic expression of RcAP2 in A. thaliana or RcAP2 silencing in rose (i.e., human manipulation) alter the number of petals in flowers. The presence of temperature-related cis-acting elements in the RcAP2 promoter also serves as evidence that RcAP2 expression is responsive to temperature changes. The temperature-induced phenotypic changes to rose flowers are very important for the ornamental flower industry, and are associated with the differentiation and development of rose floral organs. Our results suggest that RcAP2 may be an important regulator of the number of petals, and may also mediate the temperature-related formation of petals derived from stamens.
Author Contributions
QZ and YH conceived and designed the study. YH and AT performed most of the experiments. YH analyzed the data and wrote the manuscript. HW, TZ, TC, JW, WY, and HP provided help with the experiments and data analysis.
Funding
This work was financially supported by the Fundamental Research Funds for the Central Universities (No. 2016ZCQ02), the Beijing Natural Science Foundation (No. 6174045), the China Postdoctoral Science Foundation (No. 2017M620650), and the Special Fund for Beijing Common Construction Project (No. 2016GJ-03).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00481/full#supplementary-material
Footnotes
- ^http://www.ncbi.nlm.nih.gov/genbank/
- ^http://www.ncbi.nlm.nih.gov/projects/gorf/
- ^http://planttfdb.cbi.pku.edu.cn/prediction.php
- ^http://regrna2.mbc.nctu.edu.tw/detection.html
- ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^http://linux1.softberry.com/berry.phtml?topic=nsitep&group=programs&subgroup=promoter
References
Bomblies, K., Dagenais, N., and Weigel, D. (1999). Redundant enhancers mediate transcriptional repression of AGAMOUS by APETALA2. Dev. Biol. 216, 260–264. doi: 10.1006/dbio.1999.9504
Bowman, J. L. (1997). Evolutionary conservation of angiosperm flower development at the molecular and genetic levels. J. Biosci. 22, 515–527. doi: 10.1007/BF02703197
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20.
Causier, B., Schwarz-Sommer, Z., and Davies, B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21, 73–79. doi: 10.1016/j.semcdb.2009.10.005
Chang, T. H., Huang, H. Y., Hsu, J. B., Weng, S. L., Horng, J. T., and Huang, H. D. (2013). An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformat. 14(Suppl. 2):S4. doi: 10.1186/1471-2105-14-S2-S4
Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
Chmelnitsky, I., Colauzzi, M., Algom, R., and Zieslin, N. (2001). Effects of temperature on phyllody expression and cytokinin content in floral organs of rose flowers. Plant Growth Regulat. 35, 207–214. doi: 10.1023/A:1014465128264
Chuck, G., Meeley, R., and Hake, S. (2008). Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135, 3013–3019. doi: 10.1242/dev.024273
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Cronk, Q. C. B. (2001). Plant evolution and development in a post-enomic context. Nat. Rev. Genet. 2, 607–619. doi: 10.1038/35084556
Debener, T. (1999). Genetic analysis of horticulturally important morphological and physiological characters in diploid roses. Gartenbauwissenschaft 64, 14–20.
Debener, T., and Mattiesch, L. (1999). Construction of a genetic linkage map for roses using RAPD and AFLP markers. Theor. Appl. Genet. 99, 891–899. doi: 10.1007/s001220051310
Dinh, T. T., Girke, T., Liu, X., Yant, L., Schmid, M., and Chen, X. (2012). The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139, 1978–1986. doi: 10.1242/dev.077073
Dubois, A., Carrere, S., Raymond, O., Pouvreau, B., Cottret, L., Roccia, A., et al. (2012). Transcriptome database resource and gene expression atlas for the rose. BMC Genomics 13:638. doi: 10.1186/1471-2164-13-638
Dubois, A., Raymond, O., Maene, M., Baudino, S., Langlade, N. B., Boltz, V., et al. (2010). Tinkering with the C-function: a molecular frame for the selection of double flowers in cultivated roses. PLoS One 5:e9288. doi: 10.1371/journal.pone.0009288
Dugo, M. L., Satovic, Z., Millan, T., Cubero, J. I., Rubiales, D., Cabrera, A., et al. (2005). Genetic mapping of QTLs controlling horticultural traits in diploid roses. Theor. Appl. Genet. 111, 511–520. doi: 10.1007/s00122-005-2042-4
Garrod, J. F., and Harris, G. P. (1974). Studies on the glasshouse carnation: effects of temperature and growth substances on petal number. Ann. Bot. 38, 1025–1031. doi: 10.1093/oxfordjournals.aob.a084892
Hausser, J., Syed, A. P., Bilen, B., and Zavolan, M. (2013). Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 23, 604–615. doi: 10.1101/gr.139758.112
Hibrand-Saint Oyant, L., Crespel, L., Rajapakse, S., Zhang, L., and Foucher, F. (2008). Genetic linkage maps of rose constructed with new microsatellite markers and locating QTL controlling flowering traits. Tree Genet. Genomes 4, 11–23. doi: 10.1007/s11295-007-0084-2
Jack, T. (2004). Molecular and genetic mechanisms of floral control. Plant Cell 16, 16–17. doi: 10.1105/tpc.017038
Jackson, D. (1991). Molecular Plant Pathology: A Practical Approach. Oxford: Oxford University Press.
Jofuku, K. D., den Boer, B. G., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Jones-Rhoades, M. W., Bartel, D. P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
Kanno, A., Nakada, M., Akita, Y., and Hirai, M. (2007). Class B gene expression and the modified ABC model in nongrass monocots. Sci. World J. 7, 268–279. doi: 10.1100/tsw.2007.86
Keck, E., McSteen, P., Carpenter, R., and Coen, E. (2003). Separation of genetic functions controlling organ identity in flowers. EMBO J. 22, 1058–1066. doi: 10.1093/emboj/cdg097
Kelemen, O., Convertini, P., Zhang, Z., Wen, Y., Shen, M., Falaleeva, M., et al. (2013). Function of alternative splicing. Gene 514, 1–30. doi: 10.1016/j.gene.2012.07.083
Kramer, E. M., Dorit, R. L., and Irish, V. F. (1998). Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149, 765–783.
Kramer, E. M., and Irish, V. F. (1999). Evolution of genetic mechanisms controlling petal development. Nature 399, 144–148. doi: 10.1038/20172
Lenhard, M., Bohner, A., Jurgens, G., and Laux, T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by inerations between WUSHEL and AGAMOUS. Cell 05, 805–814. doi: 10.1016/S0092-8674(01)00390-7
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Liu, Z., and Meyerowitz, E. M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121, 975–991.
Liu, Z., Zhang, D., Liu, D., Li, F., and Lu, H. (2013). Exon skipping of AGAMOUS homolog PrseAG in developing double flowers of Prunus lannesiam (Rosaceae). Plant Cell Rep. 32, 227–237. doi: 10.1007/s00299-012-1357-2
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., et al. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. doi: 10.1016/S0092-8674(01)00384-1
Luo, H., Chen, S., Jiang, J., Teng, N., Chen, Y., and Chen, F. (2012). The AP2-like gene NsAP2 from water lily is involved in floral organogenesis and plant height. J. Plant Physiol. 169, 992–998. doi: 10.1016/j.jplph.2012.02.018
Ma, N., Chen, W., Fan, T., Tian, Y., Zhang, S., Zeng, D., et al. (2015). Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida). BMC Plant Biol. 15:237. doi: 10.1186/s12870-015-0623-1
Maes, T., Van de Steene, N., Zethof, J., Karimi, M., D’Hauw, M., Mares, G., et al. (2001). Petunia Ap2-like genes and their role in flower and seed development. Plant Cell Online 13, 229–244. doi: 10.1105/tpc.13.2.229
Meyerowitz, E. M., Smyth, D. R., and Bowman, J. L. (1989). Abnormal flowers and pattern formation in floral. Development 106, 209–217.
Mlotshwa, S., Yang, Z., Kim, Y., and Chen, X. (2006). Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol. Biol. 61, 781–793. doi: 10.1007/s11103-006-0049-0
Morey, D. (1959). Observations on the genetics of doubleness in roses. American Rose Annu. 44, 113–116.
Nakatsuka, T., Saito, M., Yamada, E., Fujita, K., Yamagishi, N., Yoshikawa, N., et al. (2015). Isolation and characterization of C-class MADS-box gene involved in the formation of double flowers in Japanese gentian. BMC Plant Biol. 15:182. doi: 10.1186/s12870-015-0569-3
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Posé, D., Verhage, L., Ott, F., Yant, L., Mathieu, J., Angenent, G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. doi: 10.1038/nature12633
Reynolds, J., and Tampion, J. (1983). Double Flowers: A Scientific Study. London: Polytechnic of Central London Press London.
Ronse De Craene, L. P. (2003). The evolutionary significance of homeosis in flowers: a morphological perspective. Int. J. Plant Sci. 164, 225–235. doi: 10.1086/376878
Sheen, J., Hwang, S., Niwa, Y., Kobayashi, H., and Galbraith, D. W. (1995). Green-fluorescent protein as a new vital marker in plant cells. Plant J. 8, 777–784. doi: 10.1046/j.1365-313X.1995.08050777.x
Shengmei, Z., Hongyan, S., Lei, W., Xiansheng, Z., and Huairui, S. (2006). Cloning and transformation of apetala2 homologues genes from apple. Acta Horticult. Sin. 33, 239–243.
Solovyev, V. V., Shahmuradov, I. A., and Salamov, A. A. (2010). Identification of promoter regions and regulatory sites. Methods Mol. Biol. 674, 57–83. doi: 10.1007/978-1-60761-854-6_5
Soltis, P. S., Soltis, D. E., Kim, S., Chanderbali, A., and Buzgo, M. (2006). Expression of floral regulators in basal angiosperms and the origin and evolution of ABC-function. Adv. Bot. Res. 44, 323–347. doi: 10.1016/S0065-2296(06)44012-X
Spiller, M., Linde, M., Hibrand-Saint Oyant, L., Tsai, C. J., Byrne, D. H., Smulders, M. J., et al. (2011). Towards a unified genetic map for diploid roses. Theor. Appl. Genet. 122, 489–500. doi: 10.1007/s00122-010-1463-x
Su, H., Zhou, S., Wang, L., Zhang, X. S., and Shu, H. R. (2004). Cloning and expression analysis of APETALA2 homologous genes in strawberry. Acta Bot. Boreali Occident. Sin. 25, 1937–1942.
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Münster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. doi: 10.1023/A:1006332105728
Van Tunen, A. J., Eikelboom, W., and Angenent, G. C. (1993). Floral organogenesis in Tulipa. Flower Newsl. 16, 33–37.
Verhage, L., Severing, E. I., Bucher, J., Lammers, M., Busscher-Lange, J., Bonnema, G., et al. (2017). Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS One 12:e0172950. doi: 10.1371/journal.pone.0172950
Wollmann, H., Mica, E., Todesco, M., Long, J. A., and Weigel, D. (2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633–3642. doi: 10.1242/dev.036673
Würschum, T., Groß-Hardt, R., and Laux, T. (2006). APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18, 295–307. doi: 10.1105/tpc.105.038398
Yan, H., Shi, S., Ma, N., Cao, X., Zhang, H., Qiu, X., et al. (2017). Graft-accelerated virus-induced gene silencing facilitates functional genomics in rose flowers. J. Integr. Plant Biol. 60, 34–44. doi: 10.1111/jipb.12599
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Zhao, L., Kim, Y., Dinh, T. T., and Chen, X. (2007). miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 51, 840–849. doi: 10.1111/j.1365-313X.2007.03181.x
Zhu, Q. H., and Helliwell, C. A. (2010). Regulation of flowering time and floral patterning by miR172. J. Exp. Bot. 2, 487–495. doi: 10.1093/jxb/erq295
Zieslin, N. (1968). The development and causes of petal blackening and malformation of Baccara rose flowers. Act Hortic 14, 149–156.
Keywords: APETALA2, Rosa chinensis, petals derived from stamens, petal number, temperature fluctuations
Citation: Han Y, Tang A, Wan H, Zhang T, Cheng T, Wang J, Yang W, Pan H and Zhang Q (2018) An APETALA2 Homolog, RcAP2, Regulates the Number of Rose Petals Derived From Stamens and Response to Temperature Fluctuations. Front. Plant Sci. 9:481. doi: 10.3389/fpls.2018.00481
Received: 22 January 2018; Accepted: 29 March 2018;
Published: 12 April 2018.
Edited by:
Kimberley Cathryn Snowden, The New Zealand Institute for Plant & Food Research Ltd., New ZealandReviewed by:
Marie Monniaux, UMR5667 Reproduction et Developpement des Plantes (RDP), FranceMichael Lenhard, University of Potsdam, Germany
Copyright © 2018 Han, Tang, Wan, Zhang, Cheng, Wang, Yang, Pan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qixiang Zhang, zqxbjfu@126.com
 Yu Han
Yu Han Aoying Tang
Aoying Tang Huihua Wan1
Huihua Wan1 Tengxun Zhang
Tengxun Zhang Tangren Cheng
Tangren Cheng