- 1National Key Laboratory of Crop Genetic Improvement and National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan, China
- 2Section of Cell and Developmental Biology, University of California, San Diego, San Diego, CA, United States
A well-developed root system in rice and other crops can ensure plants to efficiently absorb nutrients and water. Auxin is a key regulator for various aspect of root development, but the detailed molecular mechanisms by which auxin controls crown root development in rice are not understood. We show that overexpression of a YUC gene, which encodes the rate-limiting enzyme in auxin biosynthesis, causes massive proliferation of crown roots. On the other hand, we find that disruption of TAA1, which functions upstream of YUC genes, greatly reduces crown root development. We find that YUC overexpression-induced crown root proliferation requires the presence of the transcription factor WOX11. Moreover, the crown rootless phenotype of taa1 mutants was partially rescued by overexpression of WOX11. Furthermore, we show that WOX11 expression is induced in OsYUC1 overexpression lines, but is repressed in the taa1 mutants. Our results indicate that auxin synthesized by the TAA/YUC pathway is necessary and sufficient for crown root development in rice. Auxin activates WOX11 transcription, which subsequently drives crown root initiation and development, establishing the YUC-Auxin-WOX11 module for crown root development in rice.
Introduction
Roots determine the amount of nutrients and water available for plant growth and development, direct impacting yield and other agriculturally important traits. Rice root system consists of seminal roots and postembryonic shoot-borne crown roots with lateral roots branching off from both (Mai et al., 2014). Because of its agronomic importance, rice root system has been studied extensively using both genetic and genomic approaches. The emerging picture is that auxin plays an essential role in almost every aspect of rice root growth and development. Disruption of auxin biosynthesis, metabolism, transport, or signaling has a profound impact on rice root development.
Genetic screens for mutants that display altered patterns and/or morphology of root systems identified multiple loci (Mai et al., 2014). Molecular cloning and characterization of the rice root mutants clearly demonstrated the essential roles of auxin in root development. For example, gain-of-function mutations in the domain II of OsIAA11 and OsIAA13, which encodes negative regulators of auxin signaling, abolish lateral development (Kitomi et al., 2012; Zhu et al., 2012). Gain-of-function mutations in OsIAA23 lead to a dramatic reduction of crown roots and lateral roots (Jun et al., 2011). Moreover, OsIAA23 is required for QC maintenance (Jun et al., 2011). Other auxin signaling components such as OsTIR1/AFB2 (Xia et al., 2012), OsCAND1 (Wang et al., 2011), OsCYP2 (Kang et al., 2013), LATERAL ROOTLESS2 (LRT2) (Jing et al., 2015) are also required for crown root and lateral development. Forward genetic screens have isolated six crown rootless (crl) mutants, which either did not develop any crown roots or had dramatically reduced number of crown roots (Inukai et al., 2001). CRL1 encodes the OsLBD3-2, which is transcriptionally regulated by the Auxin Response Factor 16, suggesting that CRL1 is also part of the auxin regulated network required for crown root development (Inukai et al., 2001; Coudert et al., 2015). The crl2 and crl3 mutants are defective in crown root primordia development and cell elongation (Inukai et al., 2001; Kitomi et al., 2008), but the molecular identities of CRL2 and CRL3 have not been determined. The crl4 phenotypes were caused by a mutation in the GNOM1 gene, which encodes a ADP-ribosylation factor, and which is implicated in trafficking of the auxin efflux carrier PIN-FORMED (PIN) proteins, suggesting that polar auxin transport is also required for rice root development (Liu et al., 2009). Overexpression of OsPID, which was proposed as a regulator of PIN polarity, also affects root development (Morita and Kyozuka, 2007). Other auxin transport-related genes including OsPIN1 (Xu et al., 2005), OsPIN2 (Chen et al., 2012), and OsAUX1 (Zhao H. et al., 2015) have been implicated in crown root development as well. CRL5 encodes a member of the large AP2/ERF transcription factor family (Kitomi et al., 2011). The crl5 mutant produced fewer crown roots and displayed impaired initiation of crown root primordia (Kitomi et al., 2011). CRL5 is also part of the auxin regulated network because exogenous auxin treatment induced CRL5 expression without de novo protein biosynthesis. Auxin-induced CRL5 expression requires the degradation of AUX/IAA proteins. OsARF1 binds to the CRL5 promoter, and CRL5 controls the cytokinin signaling pathway via type-A response regulators (ARRs) (Kitomi et al., 2011). CRL6 encodes a member of the large chromodomain, helicase/ATPase, and DNA-binding domain (CHD) family protein (Wang et al., 2016). CRL6 influences crown root formation by regulating primordial initiation and development. It was shown that the expressions of OsIAA genes were down-regulated in crl6, linking CRL6 to auxin regulatory network (Wang et al., 2016).
Auxin is mainly synthesized by the TAA/YUC pathway, which is highly conserved throughout the plant kingdom (Zhao, 2012). TAA aminotransferases convert Trp to Indole-3-pyruvate (IPA), which subsequently undergoes oxidative decarboxylation catalyzed by the YUC flavin monooxygenases to produce IAA (Mashiguchi et al., 2011; Won et al., 2011). It was shown that auxin synthesized by the TAA/YUC pathway plays critical roles in root development in Arabidopsis (Stepanova et al., 2008; Chen et al., 2014). Disruption of various combinations of YUC genes and/or TAA genes can cause moderate to very severe root defects in Arabidopsis. For example, the taa1 tar1 tar2 triple mutants in Arabidopsis fail to make root meristem during embryogenesis (Stepanova et al., 2008). Similar phenotypes were observed in yuc1 yuc4 yuc10 yuc11 quadruple mutants (Cheng et al., 2007). Some other yuc combinations in Arabidopsis such as yuc3 yuc5 yuc7 yuc8 yuc9 quintuple mutants (yucQ) have very short and agravitropic roots (Chen et al., 2014). In rice, the taa1 mutants, also known as fib1(fish bone 1) display pleotropic phenotypes including agravitropic roots, long seminal roots, few crown roots, and a lack of lateral roots (Yoshikawa et al., 2014). Other TAA homologs in rice such as OsTAR1 is likely also involved in auxin biosynthesis (Kakei et al., 2017). Overexpression of OsYUC1 increased IAA levels and led to characteristic auxin overproduction phenotypes including thick hairy roots, ectopic crown roots developed from elongated node, and defective leaf growth (Yamamoto et al., 2007). Inhibition of OsYUC1 expression by antisense construct leads to severe shoot dwarfism and defective root formation (Yamamoto et al., 2007). However OsYUC4 RNAi lines did not displayed abnormal phenotype (Yamamoto et al., 2007). Both the OsCOW1 (CONSTITUTIVELY WILTED 1) and OsNAL7 (NARROW LEAF 7) are the OsYUC8 gene (Woo et al., 2007; Fujino et al., 2008). The Osyuc8 mutants greatly reduced the amount of roots (Woo et al., 2007; Fujino et al., 2008). It has been evident that auxin synthesized by the TAA/YUC pathway plays important roles in rice root development.
Besides auxin, the transcription factor WOX11 appears to play a paramountly important role in root development (Zhao et al., 2009). Crown root development is inhibited in the loss-of-function wox11 mutants, whereas overexpression of WOX11 stimulates crown root growth and the development of crown roots from the upper stem nodes (Zhao et al., 2009). It was shown that WOX11 interacts with ERF3 and binds to the RR2 promoter to directly regulate the crown root development (Zhao Y. et al., 2015). WOX11 is also involved in lateral root initiation, root hair formation, and abiotic stress-responsive development (Zhao Y. et al., 2015; Cheng et al., 2016). Furthermore, it has been suggested that WOX11 can recruit ADA2-GCN5 histone acetyltransferase module to activate downstream target genes in crown root development (Zhou et al., 2017). In Arabidopsis, WOX11 was induced by exogenous IAA application and it controls the first-step of cell fate transition during callus initiation (Liu et al., 2014; Hu and Xu, 2016). WOX11 also mediated the primary root development by regulating the WOX5/7 expression with or without auxin induction (Sheng et al., 2017).
In this paper, we investigate the relationship between auxin and WOX11, the two important regulators of crown root development in rice. We show that overexpression of the YUC genes leads to massive over proliferation of crown roots. However, in the absence of WOX11, overexpression of YUC genes did not stimulate crown root development. On the other hand, overexpression of WOX11 in the taa1/fib1 mutant background, which fails to produce crown roots, restored the crown root development. Our results demonstrate that auxin synthesized by the TAA/YUC pathway is necessary and sufficient for crown root initiation and development. Moreover, we show that WOX11 functions downstream TAA/YUC pathway and that auxin-induced crown root development is largely controlled by WOX11.
Materials and Methods
Phylogenetic Analysis of OsYUCs and AtYUCs and Structural Analysis of OsYUC Genes
Multiple-alignment was performed using Clustal Omega (McWilliam et al., 2013) and the resulting sequence alignments were then used to construct the unrooted phylogenetic tree by the neighbor joining method with a bootstrap analysis of 1000 replicates using MEGA 7.0 (Kumar et al., 2016). The exon-intron structure of each OsYUC gene was identified by using the Gene Structure Display Server (Hu et al., 2015).
Genotyping of oswox11-1 and ostaa1/fib1
The mutant oswox11-1 was obtained from Zhao et al. (2009). The insertions were confirmed by PCR using WOX11-specific primers WOX11-F2 and WOX11-R2 and the T-DNA left side primer L2. The primer sequences were shown in Supplementary Table S2.
To genotype the taa1/fib1 mutants generated by CRISPR, we amplified about 500 bp DNA-fragment that covers the CRISPR target sequence using the primers Taa1-seqF/Taa1-seqR (Supplementary Table S2). The PCR products were sequenced directly using the Taa1-F primer.
Cloning of the DNA Constructs for OsYUCs and OsWOX11 Overexpression
The cDNAs of OsYUC1, OsYUC5, OsYUC6, OsYUC7, and OsWOX11 were amplified from the rice cultivar Zhonghua 11 (ZH11) and then cloned into the pCAMBIA1301U-HPT. The cDNAs were placed under the control of the maize Ubiquitin promoter. We also cloned the genomic fragments of OsYUC3, OsYUC4, OsYUC8, OsYUC10, OsYUC11, and OsYUC14 from the rice cultivar Zhonghua 11 (ZH11) into the pCAMBIA1301U-HPT to overexpress them.
Construct CRISPR Mutants of Rice TAA1
The binary vector pCXUN (Chen et al., 2009) was used for making the CRISPR/Cas9 backbone vector pCXUN-Cas9 (He et al., 2017). Specifically, it was constructed by inserting the rice codon-optimized Cas9 between the two XcmI sites under control of the maize UBIQUITIN promoter. The TAA1-specific guide RNA was produced by the rice U3 promoter.
Rice Transformation
Rice cultivar ZH11 (Oryza sativa L. ssp. japonica) was obtained from the rice collection of the National Key Laboratory of Crop Genetic Improvement, Wuhan, China. ZH11 was transformed via Agrobacterium tumefaciens (EHA105)-mediated callus transformation as previously described (Hiei et al., 1994). Overexpressing constructs of OsYUC1, OsYUC3, OsYUC4, OsYUC5, OsYUC6, OsYUC7, OsYUC8, OsYUC10, OsYUC11, and OsYUC14 were transformed into ZH11. The OsYUC1 overexpression construct was also transformed into Hwa (used as wild type variety) and oswox11-1 mutant. The OsWOX11 overexpression construct was transformed into ostaa1fib1 mutant calli.
RNA Isolation and RT-PCR Analysis
Total RNAs were isolated using TRIzol reagent (Invitrogen). Complementary DNAs were made by reverse-transcription according to the manufacturer’s instructions (Invitrogen). RT-qPCRs were performed using gene-specific primers (Supplementary Table S2) and SYBR Premix Ex-Taq on a real-time PCR 7500 system (Applied Biosystems). Data were collected using the ABI PRISM 7500 sequence detection system following the manufacturer’s instruction. The rice ACTIN1 gene was used as the internal control. At least three biological replicates and three technical repeats were conducted.
Auxin Measurements
The extraction and measurement of auxin in rice were conducted using a previously reported method (Liu et al., 2012).
Results
Induction of Ectopic Crown Root Development by Overexpressing of OsYUC1
OsYUC1 was previously implicated in auxin biosynthesis. It was reported that abnormal roots were produced at the regeneration stage and rooting stage when OsYUC1 overexpression construct was transformed into rice calli through Agrobacteria-mediated transformation (Yamamoto et al., 2007). We overexpressed OsYUC1 in rice as part of our effort to understand auxin biosynthesis in rice. Although it is known that overexpression of OsYUC1 stimulates root development, we were still surprised by the massive proliferation of the root system induced by elevated OsYUC1 expression (Figure 1A). The OsYUC1 overexpression lines initiated massive number of roots from mesocotyls, leaf sheath, and other shoot parts. The lush root hairs and adventitious roots/crown roots cover almost the entire shoots (Figure 1A). The expression levels of OsYUC1 correlate well with the severity of the phenotypes observed in the OsYUC1 overexpression lines (Figure 1B). We further analyzed the IAA concentrations in some of the OsYUC1 overexpression lines. As expected, overexpression of OsYUC1 led to increased auxin concentrations (Figure 1C).
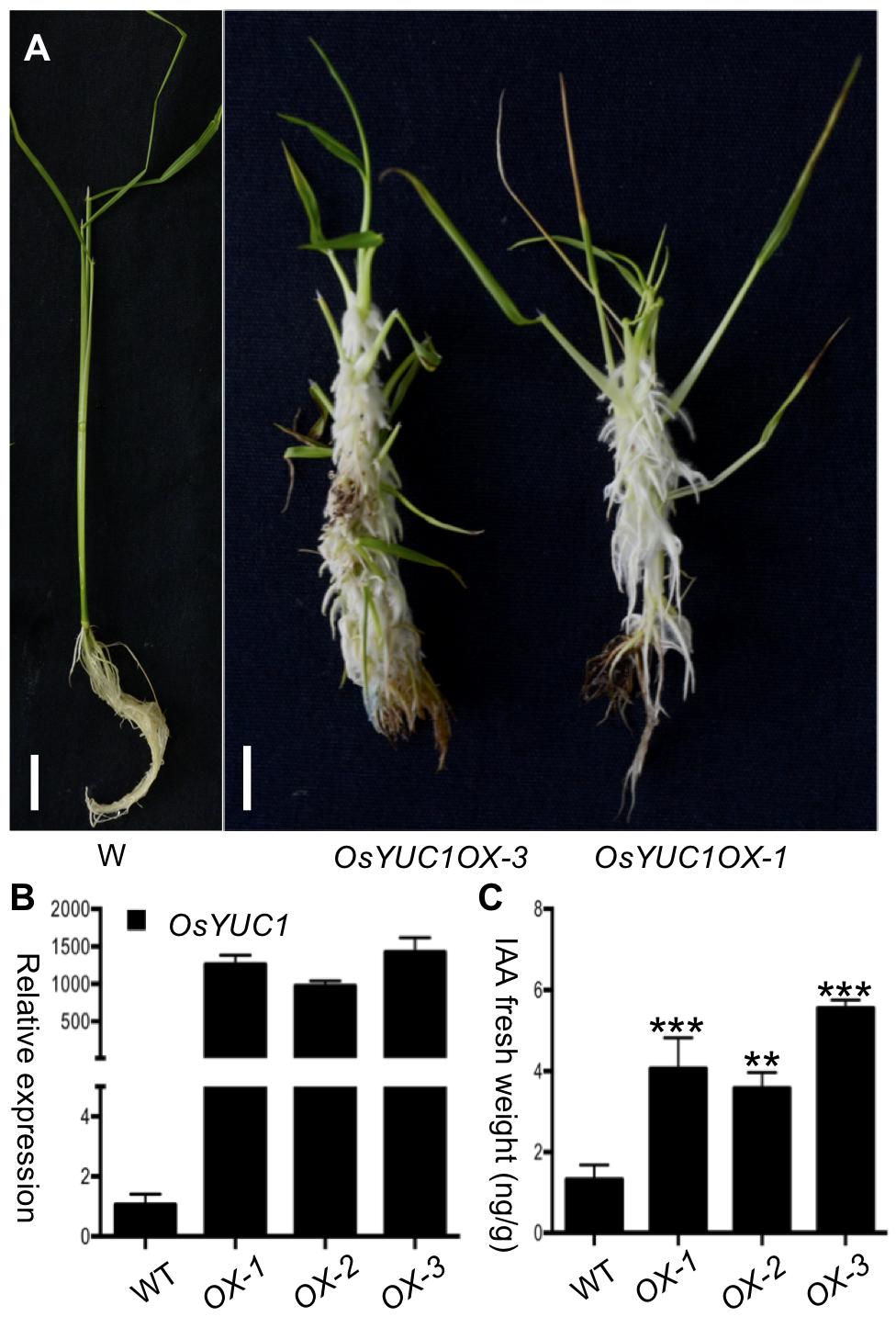
FIGURE 1. Overexpression of OsYUC1 promotes crown root proliferation and auxin overproduction. (A) Massive crown root proliferation was observed when OsYUC1 was overexpressed (the right two plants). The overexpression lines initiated many crown roots from the shoots. The crown roots also had more and longer root hairs compared to the wild type plant (left). OsYUC1OX refers to OsYUC1 overexpression under the control of the strong maize UBIQUITIN promoter. OsYUC1OX-1 and OsYUC1OX-3 were two independent TO plants. Bar = 1 cm. (B) Relative transcript levels (folds) of OsYUC1 in the independent overexpression lines. OX refers to OsYUC1 overexpression. WT refers to WT. Note that expression levels in the OX lines were increased hundreds fold. (C) Comparison of IAA contents in whole seedlings of WT and OsYUC1 overexpression tines. The data are presented as mean ±SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (Student’s t-test).
Rice Has 14 YUC Genes for Auxin Biosynthesis
In order to further study the roles of YUC-mediated auxin biosynthesis in root development, we conducted in silico analyses of YUC genes in rice and compared them with the Arabidopsis YUC genes. Previous bioinformatics analyses identified 7 YUC genes in rice (Yamamoto et al., 2007), which can be divided into four sub-groups (Figure 2A). We used the Arabidopsis YUC1 protein sequence (AtYUC1) as the query for phylogenetic analysis of YUCs in rice and identified 14 OsYUC genes that share significant homology with the AtYUC1 (Figure 2A). We can divide the OsYUCs into four groups as well (Figure 2A). All of the putative OsYUC enzymes contain the conserved motifs for binding FAD and NADPH cofactors (Supplementary Figure S1) (Hou et al., 2011), suggesting that they are probably functional flavin monooxygenases. All of the OsYUC genes have several introns with variable lengths (Figure 2B). In contrast, some of the Arabidopsis YUC genes such as YUC8 and YUC5 do not have any introns (Chen et al., 2014).
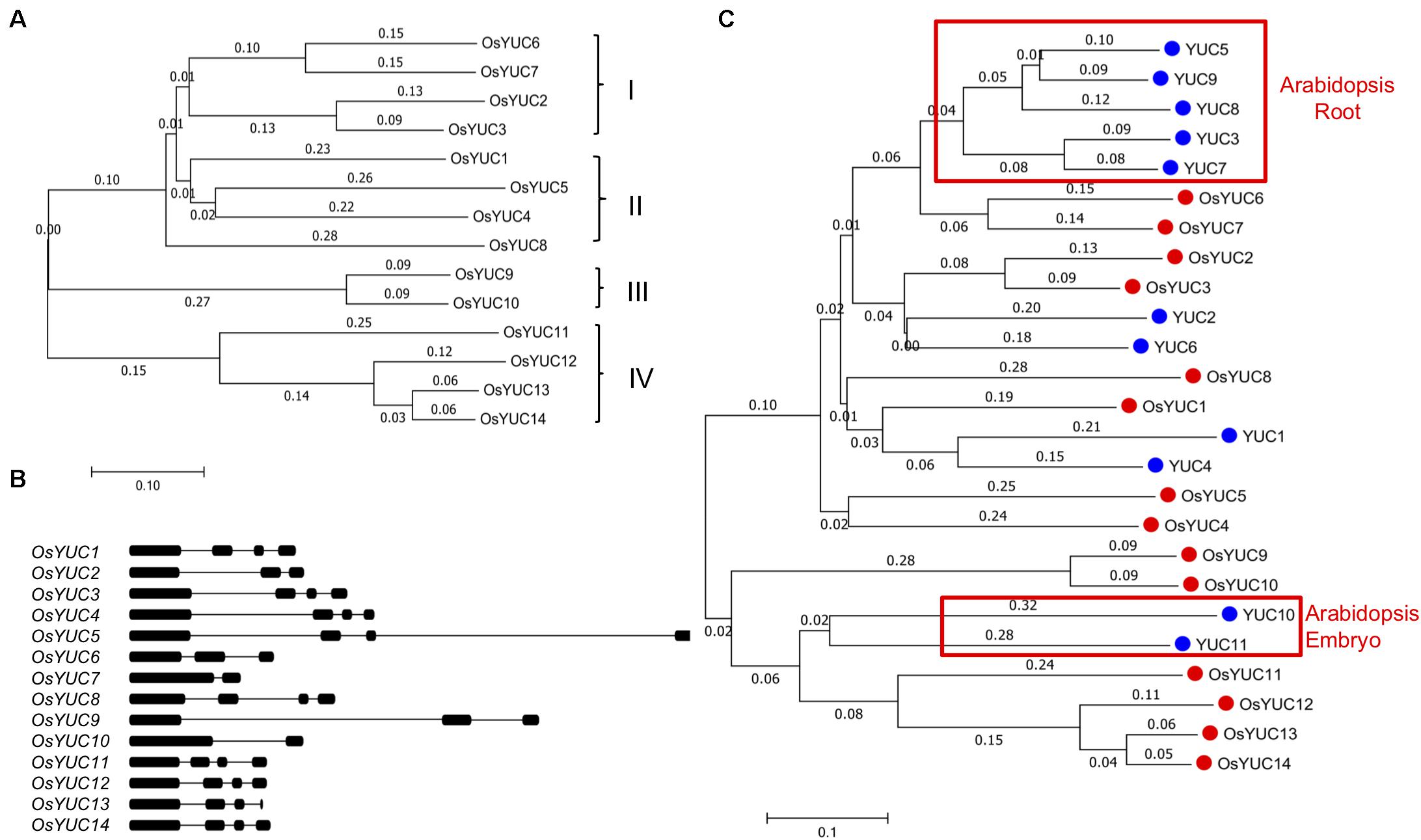
FIGURE 2. Identification and analysis of putative rice YUC genes. (A) Phylogenic analysis of OsYUCs. The phylogenetic trees were constructed using Mega7.0 program. Based on the analysis, OsYUCs were divided into four sub-groups. (B) Gene structures of the OsYUCs. Thin lines represent introns, dark bars refer to exons. (C) Phylogenic comparison of rice YUCs and Arabidopsis YUCs. Note that Arabidopsis appears to have more root YUCs whereas rice seems to have expanded embryo YUCs.
We investigated the phylogenetic relationship between rice and Arabidopsis YUC genes by comparing the full-length protein sequences (Figure 2C). We had two interesting observations: (1) Rice appeared to have dramatically reduced the YUC genes grouped to the Arabidopsis root YUC genes. In Arabidopsis, five YUC genes (YUC3, YUC5, YUC7, YUC8, and YUC9) were shown to play important roles in Arabidopsis root development (Chen et al., 2014). Rice only has the OsYUC6 and OsYUC7 that belong to this group. (2) Rice has expanded the YUC genes related to embryogenesis and endosperm development. Arabidopsis YUC10 and YUC11 along with YUC1 and YUC4 are required for embryogenesis (Cheng et al., 2006). Rice has six YUC genes that are closely related to AtYUC10 and AtYUC11 (Figure 2C).
Overexpression of Most OsYUC Genes Leads to Auxin Overproduction
To functionally characterize the OsYUC genes in rice, we tried to overexpress all of the OsYUC genes (either cDNA or genomic sequences) using the strong maize UBIQUITIN promoter. Overexpression of any OsYUC genes except four caused the obvious auxin overproduction phenotypes. The roles of OsYUC2, OsYUC9, OsYUC12, and OsYUC13 in auxin biosynthesis have not been experimentally determined yet because we ran into some difficulties in generating their overexpression constructs. Nevertheless, we showed that at least 10 YUC genes in rice had the capacity to synthesize auxin. Moreover, overexpression of the YUC genes resulted in very similar phenotypes. Representative phenotypes of overexpression of OsYUC1, OsYUC3, OsYUC5, OsYUC6, OsYUC7, OsYUC8, and OsYUC11 were shown (Figures 1, 3). Overall, overexpression of OsYUC genes caused the development of ectopic adventitious root, shortened seminal roots, and over proliferation of root hairs (Figures 1, 3). For example, overexpression of OsYUC8 led to phenotypes very similar to those observed in OsYUC1 overexpression.

FIGURE 3. Effects of overexpression of OsYUC genes on rice development. (A) Overexpression of the OsYUC3 leads to short seminal root and more crown roots. The shoots of the overexpression lines (left three plants) appear shorter as well. Similar phenotypes were also observed when OsYUC5 (B), OsYUC6 (C), OsYUC7 (D), OsYUC8 (E), and OsYUC11 (F) were overexpressed. All of the OsYUC overexpression lines showed ectopic crown roots. OsYUCsOX refers to OsYUCs overexpression under the control of the strong maize UBIQUITIN promoter. Bar = 3 cm. (G) The mature stage plants of WT and OsYUC3 overexpression lines. Very few tillers were developed in the overexpression lines (the right two plants). OsYUC3OX-8 and OsYUC3OX-18 were two independent T0 plants. Bar = 10 cm.
The strong OsYUC overexpression lines died and never produced any seeds. Some moderately overexpression lines were able to reach to adult stage and produce some seeds. The relatively weaker OsYUC overexpression lines developed just slightly increased number of crown roots, but the lines also had dramatically reduced tiller numbers. Sometimes, only one single tiller was produced (Figure 3G).
Auxin Synthesized by the TAA/YUC Pathway Is Required for Crown Root Development
Because of the existence of 14 YUC genes in rice and some of which are likely have redundant functions, it is difficult to study loss-of-function yuc mutants in rice. To generate partial auxin deficient mutants in rice, we turned to the TAA1/FIB genes in rice, which have less redundancy and which function upstream of YUC (Yoshikawa et al., 2014). Disruption of the OsTAA1 gene caused pleiotropic phenotypes and decreased IAA content by half compared to wild type (Yoshikawa et al., 2014). We generated taa1/fib1 mutants using the CRISPR/Cas9 gene editing technology in order to study the roles of auxin in root development (Figure 4A). The taa1/fib1 homozygous mutants showed phenotypes similar to those reported previously (Yoshikawa et al., 2014). The taa1/fib1 showed characteristic auxin deficient phenotypes including severe dwarf, fewer and smoother crown roots with few lateral roots and longer seminal roots compared to wild type (Figures 4B–F). Our results demonstrate that auxin synthesized by the TAA/YUC pathway is required for normal root development in rice.
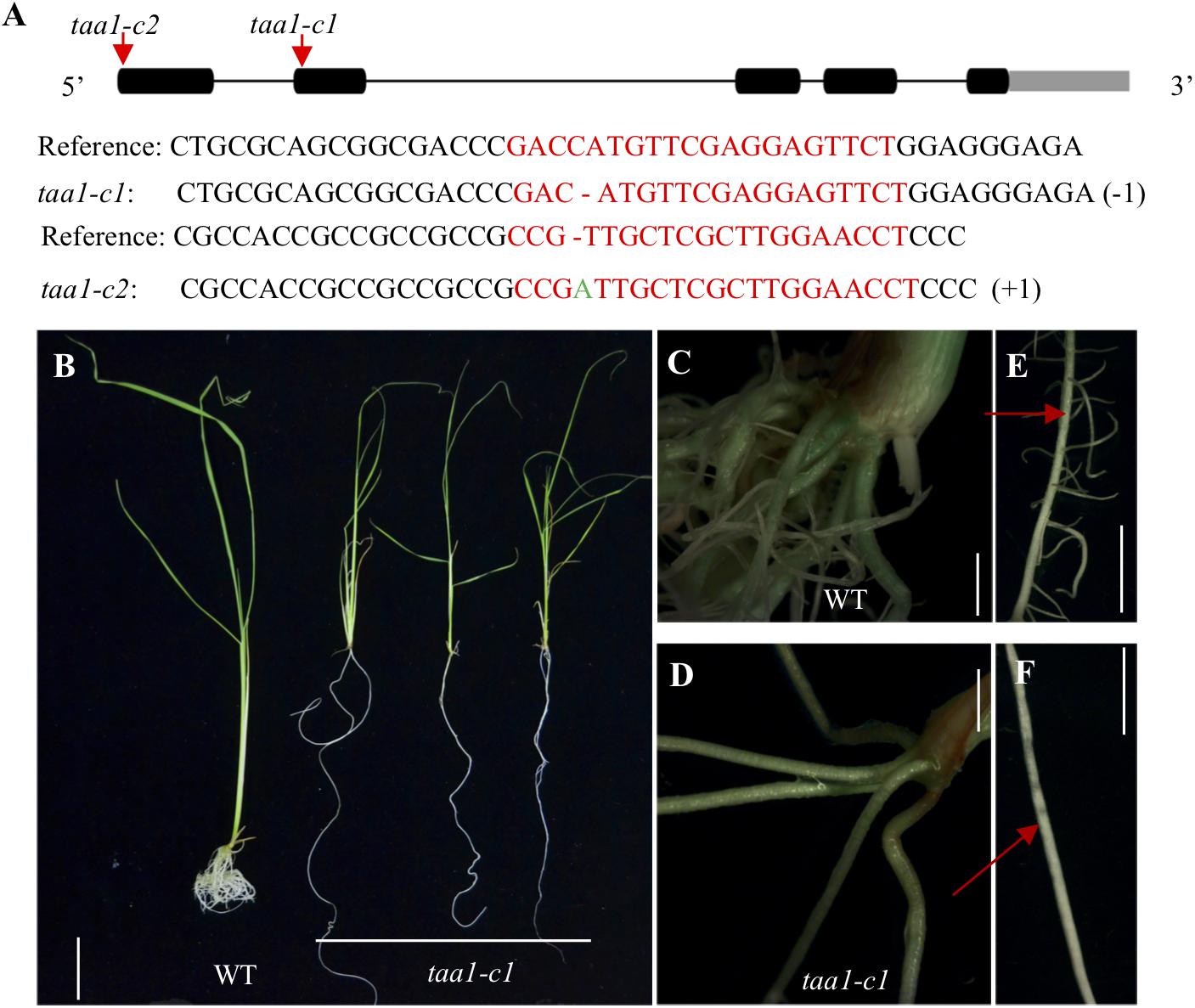
FIGURE 4. Generation of CRISPR mutations in the TAA1 gene in rice and characterization of the root defects of the taa1 mutants. (A) Two independent alleles of taa mutants (taa1-c1 and taa1-c2, c stands for CRISPR) were generated using CRISPR/Cas9 gene-editing technology as described in the Materials and Methods. The mutations were located inside the TAA1/FIB1 target sequence. The arrows indicate CRISPR/Cas9 target sites, which were located inside the first and the second exon, respectively. The taa1-c1 had one 1 bp deletion and the taa1-c2 contained 1 bp insertion. (B) Comparison of the seedlings of taa1-c1 homozygous mutants with WT. The taa1-c1 mutant had longer seminal root and fewer crown roots compared to WT. Bar = 3 cm. (C–D) The detailed root images of WT and taa1-c1 homozygous mutants, respectively. The taa1-c1 mutant failed to develop crown roots. Bar = 0.2 cm. WT plants developed lateral roots (E) whereas taa1-c1 did not have lateral roots (F). Bar = 0.5 cm.
Overexpression of OsYUC Genes Leads to Transcriptional Activation of WOX11
Development of ectopic crown roots in the OsYUC overexpression lines (Figures 1, 3) resembled the phenotypes observed in the OsWOX11 overexpression lines (Zhao et al., 2009), though the OsWOX11 overexpression phenotypes were much weaker. We hypothesized that auxin produced by YUCs might up-regulate WOX11 expression, consequently stimulating root development. It was previously reported that auxin treatments induced WOX11 expression in Arabidopsis (Liu et al., 2014). We determined the transcript levels of rice WOX11 in the OsYUC1 overexpression lines (Figure 5A). Overexpression of OsYUC1 led to significant increases of WOX11 transcripts, suggesting a regulatory module of YUC-Auxin-WOX11 in rice root development.
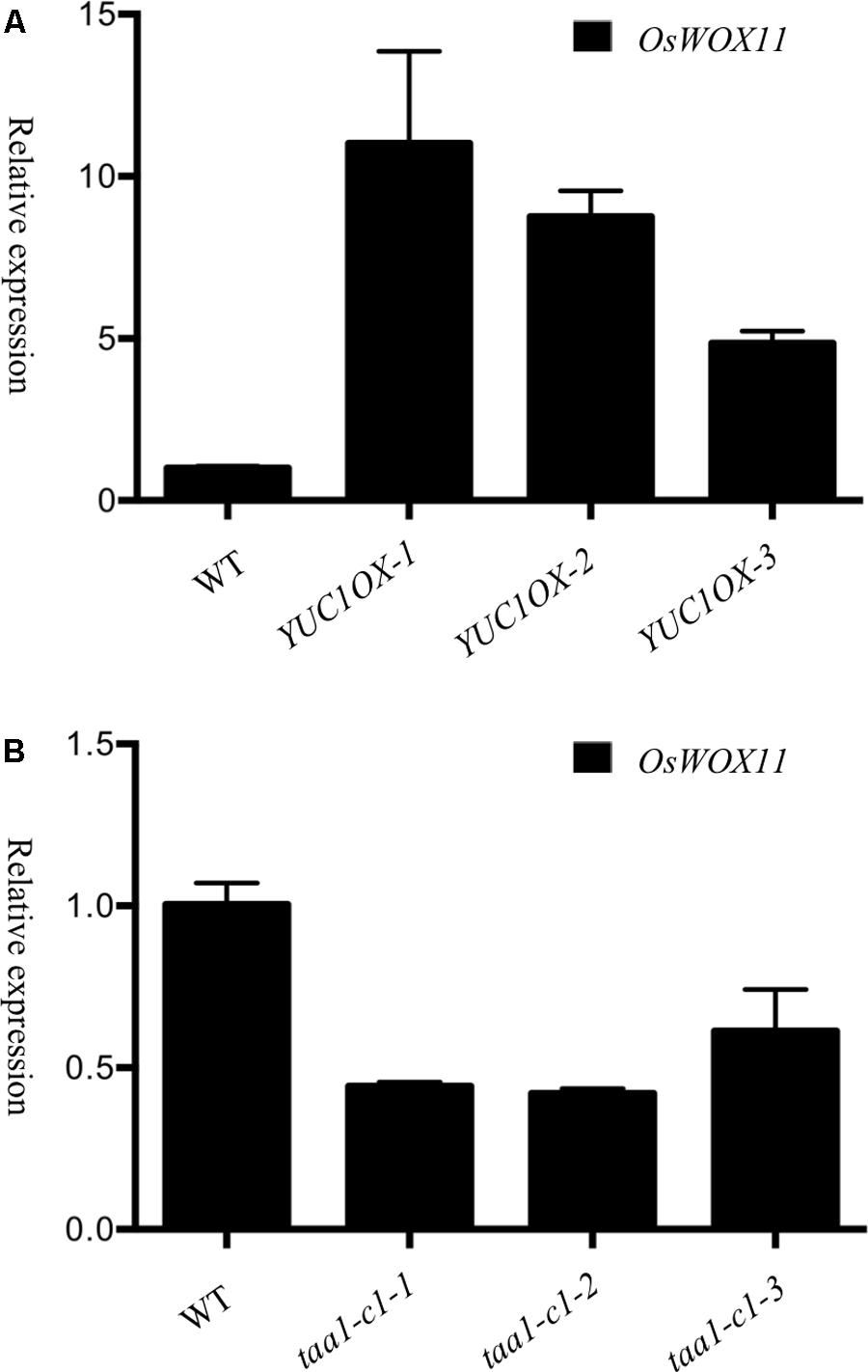
FIGURE 5. WOX11 expression is regulated by the TAA/YUC auxin biosynthesis pathway. (A) Overexpression of OsYUC1 induced WOX11 expression. The relative expression levels of WOX11 from three independent lines were shown. The OsYUC1 expression levels and IAA concentrations of’ the three lines were shown in Figure 1B. (B) OsWOX11 expression was repressed in taa mutants. Relative transcript levels (fold) of OsWOX11 were decreased in the taa1-cl homozygous mutants, taa1-cl-1, taa1-cl-2 and taal-cI-3 were individual plants.
We also analyzed the transcript levels of OsWOX11 in taa1/fib1 mutants, which contain much less auxin compared to wild type (Yoshikawa et al., 2014). If WOX11 is indeed regulated by auxin at transcription level, we would expect that the expression levels of WOX11 would be much reduced. Indeed, OsWOX11 expression were significantly reduced in our taa1/fib1 mutants (Figure 5B).
Over-Proliferation of Ectopic Roots in OsYUC1 Overexpression Lines Is Abolished in the wox11-1 Mutant
If the root phenotypes of the OsYUC overexpression lines are mediated by WOX11, disruption of WOX11 in the OsYUC overexpression lines would suppress the root proliferation phenotypes. We introduced our OsYUC1 overexpression construct (UBIQUITIN:OsYUC1), which was able to induce massive root development in wild type Zhonghua 11 (Figure 1), into the homozygous wox11-1 mutant that is in the background Hwa. We found that UBIQUITIN:OsYUC1 was able to cause the auxin overproduction phenotypes in Hwa as well (Figure 6A and Supplementary Figure S2). However, in the wox11-1 mutants, the same construct did not cause the auxin overproduction phenotypes (Figures 6B,C and Supplementary Figure S2). We analyzed more than 120 independent UBIQUITIN:OsYUC1 wox11-1 T0 plants and did not observe any obvious auxin overproduction phenotypes. We analyzed the expression levels of OsYUC1 in the UBIQUITIN:OsYUC1 wox11-1 and indeed the OsYUC1 expression levels were increased. Our results clearly demonstrated that the root development phenotypes of OsYUC overexpression lines are dependent on the presence of WOX11 and that WOX11 is likely downstream of OsYUC genes.
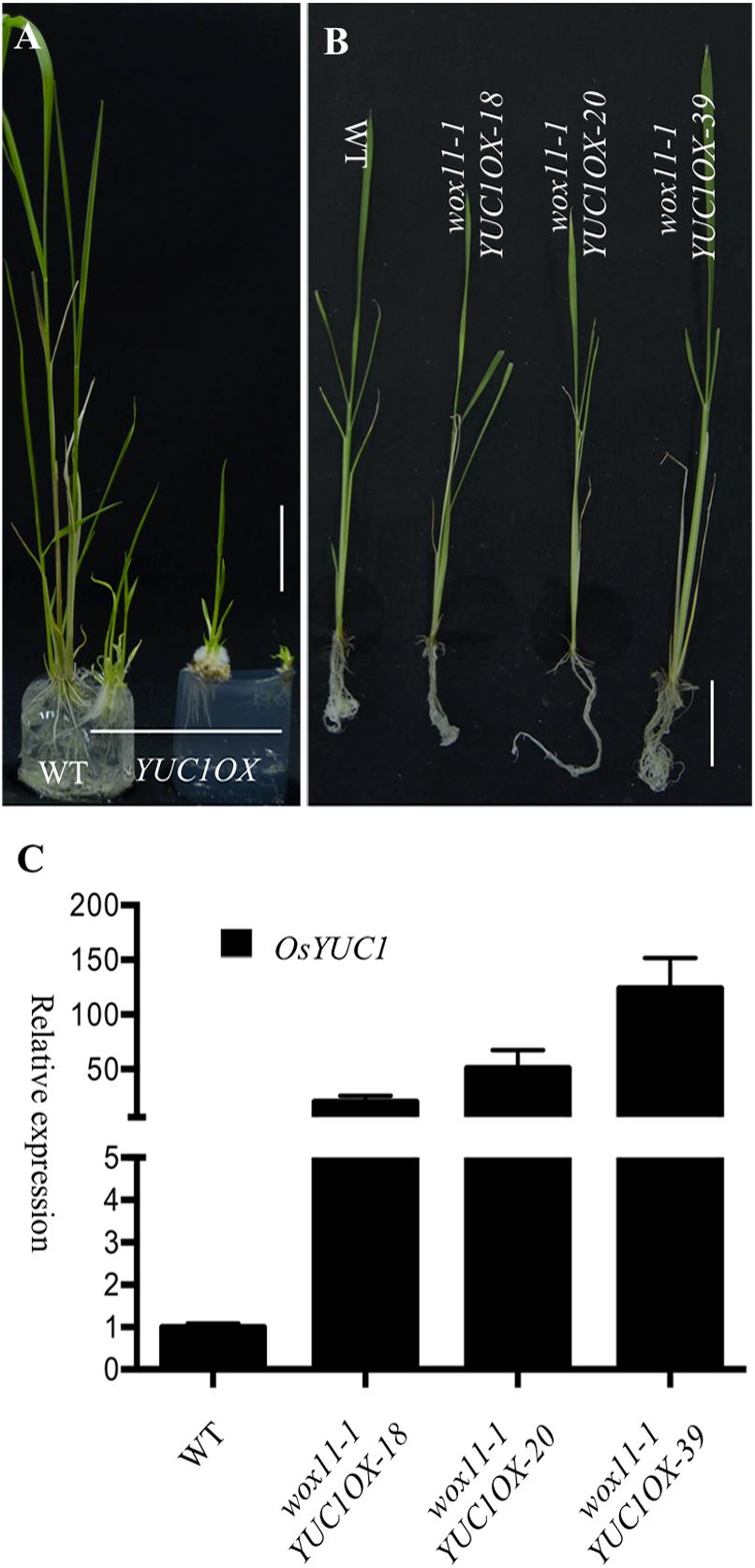
FIGURE 6. The crown root proliferation in OsYUC1 overexpression lines is dependent on the presence of WOX11. (A) Overexpression of OsYUC1 in Hwa cultivar, which was the background of wox11-1, promoted root development. YUC1OX refers to OsYUC1 overexpression. Bar = 3 cm. (B) Overexpression of OsYUC1 in wox11-1 mutant background was not able to induce crown root development. No ectopic crown roots were observed in OsYUC1 overexpression lines. wox11-1OsYUC1OX-18, wox11-1OsYUC1OX-20, and wox11-1OsYUC1OX-39 were three independent T0 plants. Bar = 4 cm. (C) Relative transcript levels of OsYUC1 in wox11-1 mutant lines. wox11-1OsYUC1OX-18, wox11-1OsYUC1OX-20, and wox11-1OsYUC1OX-39 had much elevated OsYUC1 transcripts levels.
Overexpression OsWOX11 Suppresses Crown Root Defects in taa1/fib1 Mutant
To further investigate whether OsWOX11 functions downstream of YUC/TAA pathway, we overexpressed OsWOX11 in taa1/fib1, which lacked the capacity to develop crown roots. Because homozygous taa1/fib1 mutants are sterile, we used the seeds from heterozygous taa1/fib1 plants to produce calli for transformation. T0 plants were genotyped to determine the zygosity of taa1/fib1 mutation. Overexpression of OsWOX11 in WT or heterozygous taa1/fib1 produced ectopic crown roots as previously reported (Figure 7 and Supplementary Figure S3). Interestingly, overexpression of OsWOX11 in the taa1/fib1 background also showed phenotypes similar to those observed in the OsYUC overexpression lines (Figures 7A left, 7C and Supplementary Figure S3). Our observation that overexpression of OsWOX11 stimulated crown root development in taa1/fib1 mutant strongly indicates that WOX11 functions downstream of TAA/YUC in crown root development.
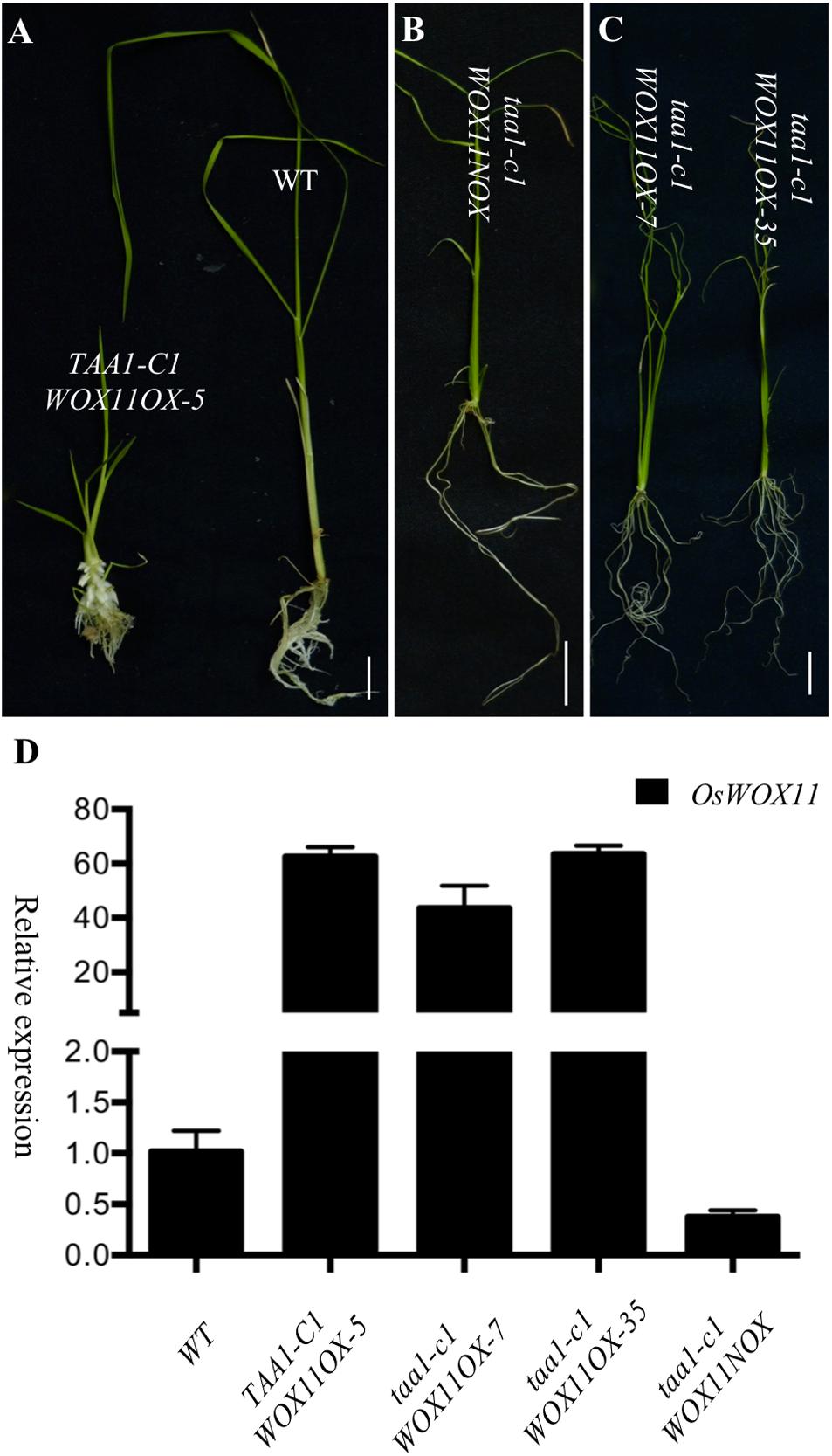
FIGURE 7. Rescue crown root defects in taa1-c1 by overexpressing OsWOX11. (A) Overexpression of WOX11 stimulates ectopic crown root development (left plant) compared to WT (right). OsWOX11 refers to OsWOX11 overexpression under the control of the strong maize UBIQVITIN promoter. Very few crown roots were developed in taa1-c1 mutants (B). But overexpression of WOX11 stimulates crown root development in taal mutant background (C). (D) The transcript levels (fold) of OsWOX11 in the transgenic plants.
Discussion
Our genetic analysis of OsYUC overexpression lines and the taa1/fib1 mutants demonstrated that auxin synthesized by the TAA/YUC pathway is necessary and sufficient for crown root development in rice. We further showed that auxin-induced crown root initiation and elongation are mediated by the transcription factor WOX11, establishing a YUC-Auxin-WOX11 module for crown root development in rice.
Auxin is the primary hormone that controls root development. In tissue culture, it is well known that high auxin/cytokinin ratio is necessary to stimulate root growth. Recently, it was shown that the cell fate transition during de novo root organogenesis in Arabidopsis requires YUC-mediated auxin biosynthesis (Chen et al., 2016). On the other hand, Arabidopsis auxin overproduction mutants superroot1 (Mikkelsen et al., 2004), superroot2 (Pacurar et al., 2014) and auxin overproduction transgenic lines including iaaM overexpression lines (Cheng et al., 2006), CYP79B2 overexpression lines (Zhao et al., 2002) and YUC overexpression lines (Zhao et al., 2001) all have root phenotypes. SUR1 and SUR2 produce adventitious roots from hypocotyls whereas iaaM, CYP79B2, and YUC overexpression lines have shorter primary roots and more root hairs (Zhao et al., 2001; Zhao et al., 2002). Auxin overproduction in rice by overexpressing OsYUC genes also promotes adventitious/crown root development and produces more root hairs, indicating that auxin is sufficient for the development of adventitious roots in both rice and Arabidopsis (Yamamoto et al., 2007). In Arabidopsis, mutations in TAAs or YUCs can completely eliminate root development or greatly reduce root elongation and lateral root development (Cheng et al., 2007; Stepanova et al., 2008). In rice, the taa1/fib1 fails to make crown roots and produced fewer lateral roots (Yoshikawa et al., 2014). It is clear that the TAA/YUC biosynthesis pathway is conserved between Arabidopsis and rice. Moreover, the roles of auxin produced by TAA/YUC in root development are also very similar in Arabidopsis and rice.
WOX11, a WUSCHEL (WUS)-related Homeobox (WOX) gene, is sufficient to stimulate crown root development in rice (Zhao et al., 2009). Interestingly, the functions of WOX11 in root development appeared to be conserved between Arabidopsis and rice as well. WOX11 along with its close homolog WOX12 controls the first-step cell fate transition during de novo root organogenesis in Arabidopsis (Liu et al., 2014). Much is known about how WOX11 regulates root development. OsWOX11 physically interacts with the ERF3 and regulates the cytokinin-responsive gene RR2, which plays a role in crown root development (Zhao Y. et al., 2015). Interestingly, CRL5 is also an ERF protein and also regulates cytokinin signaling (Kitomi et al., 2011). It will be interesting to test whether CRL5 and WOX11 physically interact with each other and whether ERF3 and CRL5 have overlapping functions. AtWOX11 was shown to physically interact with LBD16 and the WOX11-LBD16 was shown to promote the root primordium-like identity during Arabidopsis tissue culture. Shoot regeneration needs suppression of LBD16 expression in Arabidopsis (Liu et al., 2018). Interestingly, the crl1, which encodes the OsLBD3-2, fails to produce any crown roots (Inukai et al., 2001; Coudert et al., 2015), suggesting that CRL1 may also interact with WOX11 and the OsLBD3-2-WOX11 may have functions in rice similar to those of WOX11-LBD16 in Arabidopsis. Comparison of transcription of wox11 and WT in conjugation of WOX11 binding sites have revealed that WOX11 target genes, which are mainly involved in cytokinin homeostasis/signaling, stress response, and redox metabolic processes (Jiang et al., 2017).
Several previous observations led us to connect WOX11 to auxin synthesized by the TAA/YUC pathway. First, both YUC-mediated auxin biosynthesis and WOX11 are required for cell fate transition occurring during de novo root organogenesis in Arabidopsis (Chen et al., 2016). Second, WOX11 is induced by auxin treatment in Arabidopsis (Liu et al., 2014). More importantly, overexpression of OsYUC1 produced phenotypes similar to those of WOX11 overexpression lines. In this work, we established that WOX11 is required for auxin-mediated crown root development. Moreover, we demonstrated that WOX11 functions downstream of auxin produced by TAA/YUC.
There is still a missing link between auxin produced by YUC/TAA and WOX11. We hypothesized that auxin triggers the degradation of AUX/IAA repressors, subsequently ARFs can activate downstream signaling components including WOX11. This hypothesis is consistent with previous findings that dominant IAA mutants severely affected crown root development (Jun et al., 2011; Kitomi et al., 2012; Zhu et al., 2012). We identified four putative ARF binding sites (TGTCTC/ACAGAG) in the OsWOX11 promoter region (Supplementary Table S1), providing a potential mechanism for auxin to regulate WOX11 expression through ARFs. It will be interesting to find which ARF may bind to the auxin response elements in WOX11 promoter region. Based on our genetic analysis, we propose that developmental and environmental signals activate the TAA/YUC auxin biosynthesis pathway to produce auxin, which subsequently triggers a signal transduction pathway to activate WOX11 expression and root development.
Accession Numbers
Sequence data of rice genes in this article are accessible in the GenBank/EMBL data libraries with the following accession numbers: OsYUC1, LOC_Os01g45760; OsYUC2, LOC_Os05g45240; OsYUC3, LOC_Os01g53200; OsYUC4, LOC_Os01g12490; OsYUC5, LOC_Os12g32750; OsYUC6, LOC_Os07g25540; OsYUC7, LOC_Os04g03980; OsYUC8, LOC_Os03g06654; OsYUC9, LOC_Os01g16714; OsYUC10, LOC_Os01g16750; OsYUC11, LOC_Os12g08780; OsYUC12, LOC_Os02g17230; OsYUC13, LOC_Os11g10140; OsYUC14, LOC_Os11g10170; OsTAA1, LOC_Os01g07500; OsWOX11, LOC_Os07g48560.
Sequence data of Arabidopsis genes can be found in the GenBank/EMBL data libraries using the following accession numbers: YUCCA1, At4g32540; YUCCA2, At4g13260; YUCCA3, At1g04610; YUCCA4, At5g11320; YUCCA5, At5g43890; YUCCA6, At5g25620; YUCCA7, At2g33230; YUCCA8, At4g04610; YUCCA9, At1g04180; YUCCA10, At1g48910; YUCCA11, At1g21430.
Author Contributions
TZ and YZ conceived the study and designed the experiments. TZ, RL, JX, LY, and RW performed the experiments. TZ and YZ wrote the manuscript.
Funding
This work was supported by a National Transgenic Science and Technology Program (2016ZX08010002) to RW and a startup fund from the Huazhong Agricultural University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Profs. Li-Jia Qu, Lizhong Xiong, and Guoliang Wang for providing plasmids. We thank Dr. Hongbo Liu for IAA measurement. We also thank Yu Zhao for providing wox11-1 mutants.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00523/full#supplementary-material
References
Chen, L., Tong, J., Xiao, L., Ruan, Y., Liu, J., Zeng, M., et al. (2016). YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 67, 4273–4284. doi: 10.1093/jxb/erw213
Chen, Q., Dai, X., De-Paoli, H., Cheng, Y., Takebayashi, Y., Kasahara, H., et al. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55, 1072–1079. doi: 10.1093/pcp/pcu039
Chen, S., Songkumarn, P., Liu, J., and Wang, G. L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121. doi: 10.1104/pp.109.137125
Chen, Y., Fan, X., Song, W., Zhang, Y., and Xu, G. (2012). Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 10, 139–149. doi: 10.1111/j.1467-7652.2011.00637.x
Cheng, S., Zhou, D. X., and Zhao, Y. (2016). WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 11:e1130198. doi: 10.1080/15592324.2015.1130198
Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Cheng, Y., Dai, X., and Zhao, Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439. doi: 10.1105/tpc.107.053009
Coudert, Y., Le, V. A., Adam, H., Bes, M., Vignols, F., Jouannic, S., et al. (2015). Identification of CROWN ROOTLESS1-regulated genes in rice reveals specific and conserved elements of postembryonic root formation. New Phytol. 206, 243–254. doi: 10.1111/nph.13196
Fujino, K., Matsuda, Y., Ozawa, K., Nishimura, T., Koshiba, T., Fraaije, M. W., et al. (2008). NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genomics 279, 499–507. doi: 10.1007/s00438-008-0328-3
He, Y., Zhang, T., Yang, N., Xu, M., Yan, L., Wang, L., et al. (2017). Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. J. Genet. Genomics 44, 469–472. doi: 10.1016/j.jgg.2017.08.003
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Hou, X., Liu, S., Pierri, F., Dai, X., Qu, L. J., and Zhao, Y. (2011). Allelic analyses of the Arabidopsis YUC1 locus reveal residues and domains essential for the functions of YUC family of flavin monooxygenases. J. Integr. Plant Biol. 53, 54–62. doi: 10.1111/j.1744-7909.2010.01007.x
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, X., and Xu, L. (2016). Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 172, 2363–2373. doi: 10.1104/pp.16.01067
Inukai, Y., Miwa, M., Nagato, Y., Kitano, H., and Yamauchi, A. (2001). Characterization of rice mutants deficient in the formation of crown roots. Breed. Sci. 51, 123–129. doi: 10.1270/jsbbs.51.123
Jiang, W., Zhou, S., Zhang, Q., Song, H., Zhou, D. X., and Zhao, Y. (2017). Transcriptional regulatory network of WOX11 is involved in the control of crown root development, cytokinin signals, and redox in rice. J. Exp. Bot. 68, 2787–2798. doi: 10.1093/jxb/erx153
Jing, H., Yang, X., Zhang, J., Liu, X., Zheng, H., Dong, G., et al. (2015). Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat. Commun. 6:7395. doi: 10.1038/ncomms8395
Jun, N., Gaohang, W., Zhenxing, Z., Huanhuan, Z., Yunrong, W., and Ping, W. (2011). OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 68, 433–442. doi: 10.1111/j.1365-313X.2011.04698.x
Kakei, Y., Nakamura, A., Yamamoto, M., Ishida, Y., Yamazaki, C., Sato, A., et al. (2017). Biochemical and chemical biology study of rice OsTAR1 revealed that tryptophan aminotransferase is involved in auxin biosynthesis: identification of a potent OsTAR1 inhibitor, Pyruvamine2031. Plant Cell Physiol. 58, 598–606. doi: 10.1093/pcp/pcx007
Kang, B., Zhang, Z., Wang, L., Zheng, L., Mao, W., Li, M., et al. (2013). OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J. 74, 86–97. doi: 10.1111/tpj.12106
Kitomi, Y., Inahashi, H., Takehisa, H., Sato, Y., and Inukai, Y. (2012). OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci. 190, 116–122. doi: 10.1016/j.plantsci.2012.04.005
Kitomi, Y., Ito, H., Hobo, T., Aya, K., Kitano, H., and Inukai, Y. (2011). The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 67, 472–484. doi: 10.1111/j.1365-313X.2011.04610.x
Kitomi, Y., Tano, H. K., and Inukai, Y. (2008). Map ping of the CRO WN ROO TLESS3 gene, CRL3, in rice. Rice Genet. Newsl. 24, 31–33.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liu, H., Li, X., Xiao, J., and Wang, S. (2012). A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8:2. doi: 10.1186/1746-4811-8-2
Liu, J., Hu, X., Qin, P., Prasad, K., Hu, Y., and Xu, L. (2018). The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in Arabidopsis tissue culture. Plant Cell Physiol. doi: 10.1093/pcp/pcy010 [Epub ahead of print].
Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z., Huang, H., et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26, 1081–1093. doi: 10.1105/tpc.114.122887
Liu, S., Wang, J., Wang, L., Wang, X., Xue, Y., Wu, P., et al. (2009). Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 19, 1110–1119. doi: 10.1038/cr.2009.70
Mai, C. D., Phung, N. T., To, H. T., Gonin, M., Hoang, G. T., Nguyen, K. L., et al. (2014). Genes controlling root development in rice. Rice 7:30. doi: 10.1186/s12284-014-0030-5
Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H., Natsume, M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517. doi: 10.1073/pnas.1108434108
McWilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y. M., Buso, N., et al. (2013). Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600.
Mikkelsen, M. D., Naur, P., and Halkier, B. A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37, 770–777. doi: 10.1111/j.1365-313X.2004.02002.x
Morita, Y., and Kyozuka, J. (2007). Characterization of OsPID, the rice ortholog of PINOID, and its possible involvement in the control of polar auxin transport. Plant Cell Physiol. 48, 540–549. doi: 10.1093/pcp/pcm024
Pacurar, D. I., Pacurar, M. L., Bussell, J. D., Schwambach, J., Pop, T. I., Kowalczyk, M., et al. (2014). Identification of new adventitious rooting mutants amongst suppressors of the Arabidopsis thaliana superroot2 mutation. J. Exp. Bot. 65, 1605–1618. doi: 10.1093/jxb/eru026
Sheng, L., Hu, X., Du, Y., Zhang, G., Huang, H., Scheres, B., et al. (2017). Non-canonical WOX11-mediated root branching contributes to plasticity in Arabidopsis root system architecture. Development 144, 3126–3133. doi: 10.1242/dev.152132
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L. M., Xie, D. Y., Dolezal, K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. doi: 10.1016/j.cell.2008.01.047
Wang, X. F., He, F. F., Ma, X. X., Mao, C. Z., Hodgman, C., Lu, C. G., et al. (2011). OsCAND1 is required for crown root emergence in rice. Mol. Plant 4, 289–299. doi: 10.1093/mp/ssq068
Wang, Y., Wang, D., Gan, T., Liu, L., Long, W., Wang, Y., et al. (2016). CRL6, a member of the CHD protein family, is required for crown root development in rice. Plant Physiol. Biochem. 105, 185–194. doi: 10.1016/j.plaphy.2016.04.022
Won, C., Shen, X., Mashiguchi, K., Zheng, Z., Dai, X., Cheng, Y., et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523. doi: 10.1073/pnas.1108436108
Woo, Y.-M., Park, H.-J., Su’udi, M., Yang, J.-I., Park, J.-J., Back, K., et al. (2007). Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 65, 125–136. doi: 10.1007/s11103-007-9203-6
Xia, K., Wang, R., Ou, X., Fang, Z., Tian, C., Duan, J., et al. (2012). OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One 7:e30039. doi: 10.1371/journal.pone.0030039
Xu, M., Zhu, L., Shou, H., and Wu, P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. doi: 10.1093/pcp/pci183
Yamamoto, Y., Kamiya, N., Morinaka, Y., Matsuoka, M., and Sazuka, T. (2007). Auxin biosynthesis by the YUCCA Genes in Rice. Plant Physiol. 143, 1362–1371. doi: 10.1104/pp.106.091561
Yoshikawa, T., Ito, M., Sumikura, T., Nakayama, A., Nishimura, T., Kitano, H., et al. (2014). The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. 78, 927–936. doi: 10.1111/tpj.12517
Zhao, H., Ma, T., Wang, X., Deng, Y., Ma, H., Zhang, R., et al. (2015). OsAUX1 controls lateral root initiation in rice (Oryza sativa L.). Plant Cell Environ. 38, 2208–2222. doi: 10.1111/pce.12467
Zhao, Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338. doi: 10.1093/mp/ssr104
Zhao, Y., Cheng, S., Song, Y., Huang, Y., Zhou, S., Liu, X., et al. (2015). The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 27, 2469–2483. doi: 10.1105/tpc.15.00227
Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D., et al. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. doi: 10.1126/science.291.5502.306
Zhao, Y., Hu, Y., Dai, M., Huang, L., and Zhou, D. X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21, 736–748. doi: 10.1105/tpc.108.061655
Zhao, Y., Hull, A. K., Gupta, N. R., Goss, K. A., Alonso, J., Ecker, J. R., et al. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16, 3100–3112. doi: 10.1101/gad.1035402
Zhou, S., Jiang, W., Long, F., Cheng, S., Yang, W., Zhao, Y., et al. (2017). Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell 29, 1088–1104. doi: 10.1105/tpc.16.00908
Keywords: auxin, WOX11, crown root, YUCCA, TAA
Citation: Zhang T, Li R, Xing J, Yan L, Wang R and Zhao Y (2018) The YUCCA-Auxin-WOX11 Module Controls Crown Root Development in Rice. Front. Plant Sci. 9:523. doi: 10.3389/fpls.2018.00523
Received: 02 March 2018; Accepted: 04 April 2018;
Published: 23 April 2018.
Edited by:
Hong Qiao, The University of Texas at Austin, United StatesReviewed by:
Yanhai Yin, Iowa State University, United StatesYuling Jiao, Institute of Genetics and Developmental Biology (CAS), China
Copyright © 2018 Zhang, Li, Xing, Yan, Wang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunde Zhao, yundezhao@ucsd.edu; yzhao@biomail.ucsd.edu
 Tao Zhang
Tao Zhang Ruonan Li1
Ruonan Li1 Lang Yan
Lang Yan Yunde Zhao
Yunde Zhao