- 1Institute of Plant Biotechnology, Development Center of Plant Germplasm Resources, College of Life and Environment Sciences, Shanghai Normal University, Shanghai, China
- 2College of Pharmacy, Zhejiang Chinese Medical University, Hangzhou, China
Tanshinones, one group of bioactive diterpenes, were widely used in the treatment of cardiovascular diseases. WRKYs play important roles in plant metabolism, but their regulation mechanism in Salvia miltiorrhiza remains elusive. In this study, one WRKY transcription factor SmWRKY1 was isolated and functionally characterized from S. miltiorrhiza. Multiple sequence alignment and phylogenetic tree analysis showed SmWRKY1 shared high homology with other plant WRKYs such as CrWRKY1. SmWRKY1 was found predominantly expressed in leaves and stems, and was responsive to salicylic acid (SA), methyl jasmonate (MeJA), and nitric oxide (NO) treatment. Subcellular localization analysis found that SmWRKY1 was localized in the nucleus. Over-expression of SmWRKY1 significantly elevated the transcripts of genes coding for enzymes in the MEP pathway especially 1-deoxy-D-xylulose-5-phosphate synthase (SmDXS) and 1-deoxy-D-xylulose-5-phosphate reductoisomerase (SmDXR), resulted in over fivefold increase in tanshinones production in transgenic lines (up to 13.7 mg/g DW) compared with the control lines. A dual-luciferase (Dual-LUC) assay showed that SmWRKY1 can positively regulate SmDXR expression by binding to its promoter. Our work revealed that SmWRKY1 participated in the regulation of tanshinones biosynthesis and acted as a positive regulator through activating SmDXR in the MEP pathway, thus provided a new insight to further explore the regulation mechanism of tanshinones biosynthesis.
Introduction
Salvia miltiorrhiza Bunge, belonging to the Lamiaceae family, is a famous and traditional Chinese herbal plant that has been widely used for the treatment of cardiovascular and cerebrovascular diseases (Zhang et al., 2010; Kai et al., 2011). The abietane-type diterpenes in S. miltiorrhiza are the liposoluble tanshinones including dihydrotanshinone, tanshinone I, tanshinone IIA and cryptotanshinone with the same mother ring structure (Supplementary Figure S1), which exert a variety of biological activities such as antioxidant, heart-protection, antibacteria, and antitumor (Xu et al., 2010, 2015; Zhang et al., 2011; Chen et al., 2012; Gong et al., 2012). However, serious quality decrease and the low content of tanshinones in cultivated S. miltiorrhiza greatly limited the increasing market need (Hao et al., 2015; Zhou Y. et al., 2016). Therefore, it is important to improve the content of tanshinones by genetic engineering, which relies on deep understanding of the tanshinone biosynthetic pathway to S. miltiorrhiza (Liao et al., 2009; Zhou et al., 2016a, 2017).
Tanshinones represent a class of diterpenes the synthesis of which might depend on the plastidic 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway, or on the cytosolic mevalonate (MVA) pathway that both built up the universal C5 precursors isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) (Ge and Wu, 2005; Kai et al., 2011) (Supplementary Figure S2). 1-Deoxy-D-xylulose 5-phosphate synthase (DXS) catalyzes the condensation of pyruvate and glyceraldehyde 3-phosphate to yield 1-deoxy-D-5-phosphate (DXP), which is then converted to MEP by DXP reductoisomerase (DXR). DXS and DXR are frequently considered as analyzing the rate-limiting steps in the MEP pathway, and corresponding genes from S. miltiorrhiza have been cloned by our group (Liao et al., 2009; Yan et al., 2009). Subsequently, the C20 precursor geranylgeranyl diphosphate is synthesized by geranylgeranyl diphosphate synthase (GGPPS) (Kai et al., 2010). Finally, diverse diterpenoids are synthesized through terpene synthases/cylases including copalyl diphosphate synthase (CPS), kaurene synthase-like (KSL), miltiradiene oxidase CYP76AH1 and other modifying enzymes (Gao et al., 2009; Guo et al., 2013; Hao et al., 2015). The isolation and characterization of the above-mentioned tanshinone biosynthetic genes made it possible to improve tanshinone production at higher concentrations in S. miltiorrhiza through genetic manipulation (Kai et al., 2011; Shi et al., 2014, 2016a). Currently, some downstream steps of tanshinone formation still remain unknown for the moment, which limits metabolic engineering (Zhou et al., 2017).
Transcription factors (TFs) that are often capable of coordinately regulating multiple biosynthetic pathway genes, showed great promise to improve the production of active compounds by metabolic engineering. Therefore, molecular dissection of the regulatory mechanism of tanshinones biosynthesis may provide clues for activating the pathway and for producing tanshinones of high yield. Recently, heterologous expression of maize TF C1 in S. miltiorrhiza hairy roots elevated the accumulation of tanshinones through activating mevalonate 5-diphosphate decarboxylase (SmMDC) gene as the target (Zhao et al., 2015). The two kinds of TFs SmMYC2 and SmMYB9b have been isolated and functionally identified involved in the regulation of tanshinone biosynthesis (Zhou Y. et al., 2016; Zhang et al., 2017). However, less is known about WRKY TFs and their regulation mechanism in S. miltiorrhiza (Li et al., 2015).
WRKY TFs are one of the largest gene families unique to plants, which are involved in responses to biotic and abiotic stress and plant secondary metabolism (Suttipanta et al., 2011; Phukan et al., 2016). The significant feature of the WRKY TF is their WRKY domain which is approximately 60-amino acids long with the highly conserved amino acid sequence WRKYGQK being located at the N-terminal, and a non-typical zinc-finger-like motif C2HC (C–X7–C–X23–H–X1–C) or C2H2(C–X4-5–C–X22-23–H–X1–H) at the C-terminus (Xu et al., 2004; Lu et al., 2015). WRKY proteins can bind to the W-box cis-elements (T)TGAC(C/T) in the promoter region of some defense-related genes (Xu et al., 2004; Rushton et al., 2010; Liu et al., 2016). WRKY TFs can be separated into three sub-groups in accordance with the number of specific WRKY domains and zinc-finger-like motifs: Group I contains two WRKY domains and one C2H2 motif, Groups II has one WRKY domain and a C2H2 motif and Group III possesses one WRKY domain and a C2HC motif (Eulgem et al., 2000; Rushton et al., 2010). WRKYs have been found as being implicated in multiple physiological activities, including stress resistance, trichome development and secondary metabolism (Ma et al., 2009; Jiang et al., 2016; Liu et al., 2017). For example, Gossypium arboreum WRKY1 (GaWRKY1) was found to participate in the regulation of sesquiterpene biosynthesis in cotton by regulating the target gene (+)Δ-cadinene synthase (CAD1) (Xu et al., 2004). WRKY27, being responsive to various abiotic stresses interacted with GmMYB174, and then cooperatively inhibited GmNAC29 expression, facilitating stress-tolerance of drought and cold in soybean (Wang et al., 2015). Recently, genes coding for WRKY have been isolated from some medicinal plants including Catharanthus roseus (Suttipanta et al., 2011), Artemisia annua (Chen et al., 2017), and Withania somnifera (Singh et al., 2017). Overexpression of C. roseus WRKY1 (CrWRKY1) led to an up to threefold increase in serpentine levels in engineered hairy roots through binding to the W-box elements of the promoter tryptophan decarboxylase (TDC) involved in the indole alkaloid (TIA) biosynthetic pathway (Suttipanta et al., 2011). The WRKY TF GLANDULAR TRICHOME-SPECIFIC WRKY 1 (AaGSW1) positively regulated the expression of AaCYP71AV1 and AaORA by conjunction to the W-box motifs of the promoters (Chen et al., 2017). A WRKY TF from W. somnifera bound to the W-box region in the promoters of squalene synthase and squalene epoxidase, regulating the accumulation of triterpenoids in W. somnifera including phytosterols and withanolides (Singh et al., 2017). However, functional WRKYs related to secondary metabolism of tanshinones or salvianolic acids in S. miltiorrhiza have not been reported.
In this study, the gene SmWRKY1 coding for a WRKY TF (named as SmWRKY1) has been isolated and functionally characterized. Phylogenetic analysis showed that SmWRKY1 shared high homology with AtWRKY70, CrWRKY1, and GaWRKY1. Multiple sequence alignment revealed that the nucleus-localized SmWRKY1 contained one WRKY domain and a C2HC motif and could be classified into group III WRKY TFs. Introduction of SmWRKY1 into S. miltiorrhiza hairy roots increased the transcripts of SmDXS and SmDXR involved in the MEP pathway, resulting in higher level of tanshinones in transgenic lines compared with the control lines (2.175 mg/g DW). The highest content of tanshinones was produced in SmWRKY1-3 line at 13.731 mg/g DW, which was 5.3-fold higher than the control. A Dual-LUC assay revealed that SmWRKY1 activated the expression of SmDXR by directly binding to its promoter. Taken together, our work revealed that SmWRKY1 positively elevated the accumulation of tanshinones, which provides a new insight to further excavate the regulation mechanism of tanshinones biosynthesis.
Materials and Methods
Plant Materials and Elicitor Treatments
Salvia miltiorrhiza seedlings were cultivated as we reported before (Kai et al., 2011; Shi et al., 2014). N. benthamiana were prepared according to previous reports (Shi et al., 2016b; Zhou Y. et al., 2016).
For elicitor induction treatments, methyl jasmonate (MeJA) and salicylic acid (SA) were prepared as before (Kai et al., 2012; Hao et al., 2015). For NO elicitation, firstly 100 mM [Na2]Fe(CN)5NO]x2 H2O (SNP) solution was obtained, and then applied to cultures to 100 μM. All the above-mentioned solutions were sterilized through 0.22 μm filters (Pall Corporation, United States). And solvent of the equivalent volume was added into the control group.
Isolation and Bioinformatics Analysis of SmWRKY1
A local transcription database of S. miltiorrhiza was built up as reported previously (Shi et al., 2016b). One partial WRKY sequence with high homology to other plants WRKY genes, albeit lacking a part of 3′-terminal was chosen for further study. Gene-specific forward primer SmWRKY1-F605 was designed to amplify the 3′ end of SmWRKY1 by combination with the reverse primer AUAP using rapid amplification of cDNA ends (RACE) as reported before (Liao et al., 2009; Kai et al., 2010; Zhang et al., 2011). 5′-sequence and 3′-terminal products was aligned and assembled to obtain the full-length cDNA sequence of the putative SmWRKY1 gene. Primer pairs SmWRKY1-KF and SmWRKY1-KR were synthesized for amplification of the full ORF of SmWRKY1 according to the procedure as described below: initial denaturation at 94°C for 10 min, 35 cycles of 94°C for 45 s, 55°C for 45 s and 72°C for 90 s, followed by a final extension at 72°C for 10 min. All primers used for identification of SmWRKY1 were listed in Supplementary Table S1. The characteristics of SmWRKY1 were further analyzed by a series of bioinformatics tools. Nucleotide blast, protein blast and ORF Finder were used to analyze nucleotide sequence and the complete open reading frame1. MEGA 6 was applied to construct a phylogenetic tree by the neighbor-joining (NJ) method and 1000 replications were performed for bootstrap values. Multiple sequences alignment between SmWRKY1 and other plant WRKYs were performed on amino acids level by using Clustal X with default parameters (Shi et al., 2016b; Zhou Y. et al., 2016).
Expression Pattern of SmWRKY1 in Different Tissues and Under Various Elicitors Treatments
Different tissues including taproot, stem, leaf, flower, and seed were gathered from 2-year-old S. miltiorrhiza plants in maturation. Elicitor treatments were conducted on S. miltiorrhiza hairy roots sub-cultured for 60 days infected with Agrobacterium C58C1. Hairy roots were harvested at selected time points (0, 0.5, 1, 2, and 4 h) after MJ treatment. And for SA and NO induction, hairy roots were collected at 0, 3, 4, 6, 9, and 12h after treatment. All the treated samples were immediately frozen in liquid nitrogen and stored for analyzing the expression profiles of SmWRKY1.
Subcellular Localization of SmWRKY1
To analyze the subcellular localization of SmWRKY1, PCR products of SmWRKY1 ORF with BglII and KpnI restriction sites were digested with BglII and KpnI and cloned into the vector pMON530 to generate the vector pMON530-SmWRKY1-GFP. The constructed expression vector was transferred into Agrobacterium strain ASE and injected into 40-day-old tobacco leaves. GFP fluorescence was observed after 48 h cultivation using the confocal microscope (Carl Zeiss) (Shi et al., 2016b; Zhou et al., 2016a).
Generation of Transgenic SmWRKY1 Hairy Roots
The full-length coding sequence of SmWRKY1 with restriction sites SpeI and BstEII was cloned and inserted into modified pCAMBIA2300sm vector (having replaced the small fragment digested by EcoRI and HindIII with the corresponding GFP-GUSA gene expression cassette from pCAMBIA1304) under the control of the CaMV 35S promoter to generate pCAMBIA2300sm-SmWRKY1. Agrobacterium rhizogenes strain C58C1 containing pCAMBIA2300sm-SmWRKY1 was used to infect the aseptic explants and the empty pCAMBIA2300sm was regarded as the control. The transformation procedure was the same as in our previous study (Kai et al., 2011; Shi et al., 2014, 2016a,b; Zhou Y. et al., 2016). Hairy roots in good state were sub-cultured and primer pairs CaMV35S-F23 and SmWRKY1-QR were used to identify positive colonies by polymerase chain reaction (PCR) analysis. Genomic DNA was isolated from individual hairy root sample by the cetyltrimethyl ammonium bromide method as previously reported (Zhou et al., 2016a,b). Identified positive-colonies were cut into segments of 4 cm for further shake-flask culture in 100 ml 1/2 MS medium and cultured at 25°C on an orbital shaker shaking at the speed of 100 rpm in darkness (Shi et al., 2016a,b). Primer sequences were listed in Supplementary Table S1.
Total RNA Isolation and Relative Expression Analysis via qRT-PCR
Expression profiles of SmWRKY1 and several key enzyme genes involved in tanshinones biosynthetic pathway were investigated by real-time quantitative PCR analysis (qRT-PCR). Total RNA for reverse transcription (RT) reaction was extracted as described before (Shi et al., 2016b). qRT-PCR was carried out by gene-specific primers (listed in Supplementary Table S1) using previously reported procedure (Shi et al., 2016b).
Dual-Luciferase (Dual-LUC) Assay
For the dual-luciferase (Dual-LUC) assay, the promoters of SmDXR and SmDXS2 with KpnI and XhoI restriction sites were cloned into pGREEN 0800 to drive the luciferase reporters, respectively. The complete ORF of SmWRKY1 was inserted into the pCAMBIA2300sm vector as effector. The pCAMBIA2300sm-SmWRKY1 and pCAMBIA2300sm empty plasmid were transferred into A. tumefaciens strain GV3101 individually. The pGREEN-pSmDXR, pGREEN-pSmDXS2 was each co-transformed with the helper plasmid pSoup19 into GV3101, and the assay was conducted as described before (Zhang et al., 2015). The pCAMBIA2300sm empty plasmid was used as a negative control. The 35S promoter-driven gene for Renilla (reniformis) luciferase was taken as an internal control. Each measurement was repeated for three biological times. The reporter and the effector strains were mixed in a ratio of 1:1, followed by injection of tobacco leaves. Two days after injection the leaf samples were collected for the dual-LUC assay following the manufacturer’s (Promega) protocol.
Tanshinones Analysis
The 60-day-old hairy roots were dried at 50°C to constant weight in an oven. Approximate 200 mg dried hairy roots were ground into powder and immersed in 16 mL methanol/dichloromethane (3:1, v/v) for extraction of tanshinones as described earlier (Hao et al., 2015). HPLC analysis was performed by using an Agilent 1260 apparatus equipped with a Waters reversed-phase C18 symmetry column, and the detection conditions were as described previously (Shi et al., 2016b).
Statistical Analysis
All experiments including identification of positive hairy roots lines, qRT-PCR, HPLC analysis of tanshinones, Dual-LUC assay were performed three times and the results are shown as means values of three independent repeats ± standard deviation. Statistical significance was determined by the one sample t-test and one-way analysis of variance using SPSS 11.5 software (SPSS).
Results
Isolation and Bioinformatics Analysis of SmWRKY1
By searching our local transcriptome database, a WKRY fragment with 5′ untranslated region (UTR) but lacking a part of 3′ terminal sequence was chosen for further research because it showed high homology with GaWRKY1 and CrWRKY1 as well as Arabidopsis thaliana WKRY70. By 3′-RACE, an about 432 bp PCR products was obtained in which a 3′-UTR of 238 bp was found downstream from the stop codon. After sequence assembly, the full-length gene was cloned and designated as SmWRKY1. Its sequence consists of 17 bp 5′UTR, a complete 789 bp ORF, encoding a 262 amino acid protein, along with 238 bp 3′ UTR.
To further figure out the biological characteristics and phylogenetic relationship of SmWRKY1, a series of bioinformatic analyses were performed. Multiple alignment of SmWRKY1 with related WRKY proteins from other plant species revealed that SmWRKY1, AaWRKY1, and CrWRKY1 all contained a conserved WRKY domain (WRKYGQK) and a special zinc-finger like motif in its C-terminal, which is typical of the group III of WRKY TFs (Figure 1A), indicative of their similar function. The amino acid sequence alignment of SmWRKY1 with (other) plant WRKYs allowed for constructing a neighbor-joining tree (Figure 1B). The results revealed that SmWRKY1 shared 62, 49, 37, and 29% identities with EgWRKY70, NtWRKY70, CrWRKY1, and AaWRKY1, respectively.
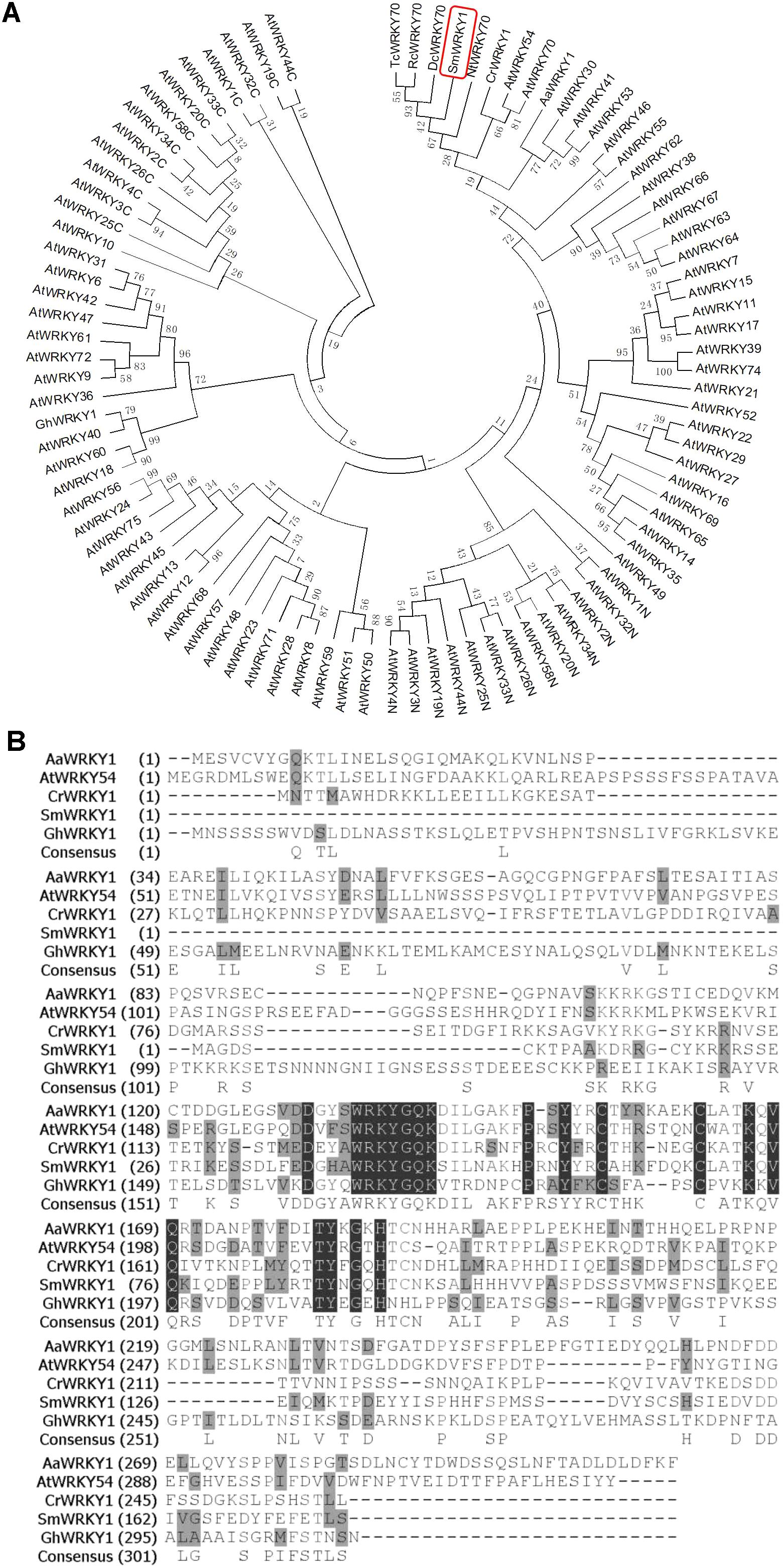
FIGURE 1. (A) Multiple alignment of SmWRKY1 with related WRKY proteins from other plant species. Black boxes indicate identical residues; gray boxes indicate identical residues for at least three of the sequences. (B) Phylogenic tree analysis of SmWRKY1 and WRKY TFs from Arabidopsis thaliana, Artemisia annua, Catharanthus roseus, Nicotiana tabacum, etc. Phylogenic tree was constructed on MEGA6.0 by using neighbor-joining method and the bootstrap values were obtained for 1000 replications.
Tissue and Induction Expression Profiles of SmWRKY1
To investigate the tissue expression pattern of SmWRKY1, roots, stems, leaves, flowers, and seeds from 2-year-old S. miltiorrhiza plants were analyzed. SmWRKY1 showed high expression in leaves and stems, low expression in flowers and roots, and its transcript was barely detected in seeds (Figure 2A). This result indicated that SmWRKY1 is not constitutively expressed in tissue.
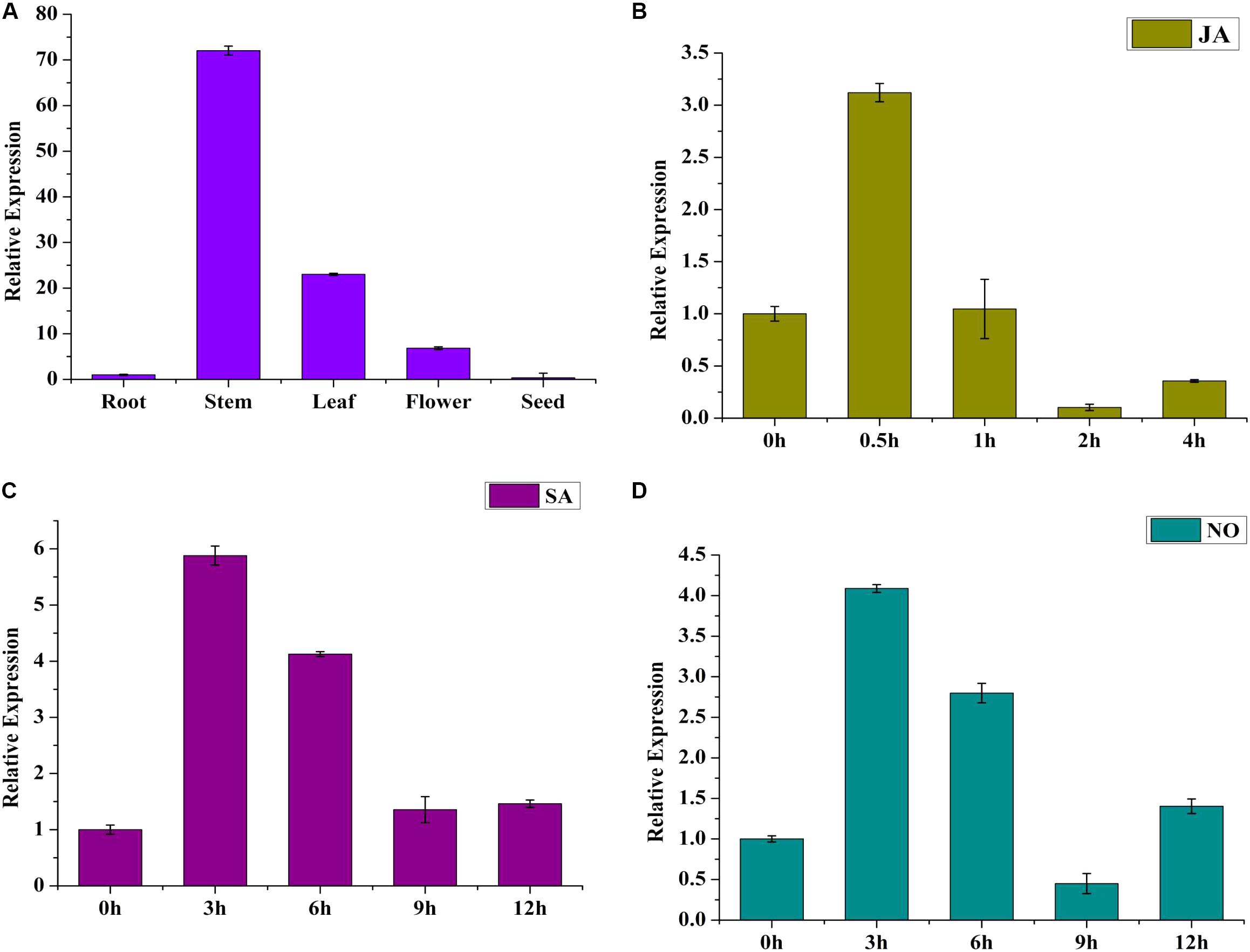
FIGURE 2. (A) Expression pattern of SmWRKY1 in different tissues. Each tissue was obtained from several individual 2-year-old S. miltiorrhiza plants in nature. Transcript abundance of SmWRKY1 is normalized to actin by the method of 2-ΔΔCt. (B) Time course of the expression level of SmWRKY1 after SA treatment as determined by qRT-PCR. (C) The expression level of SmWRKY1 after SA treatment for selected time points measured by qRT-PCR. (D) The expression level of SmWRKY1 after NO treatment for selected points by qRT-PCR analysis, respectively. Values are means ± standard deviation of triplicate analyses.
To study whether SmWRKY1 could respond to exogenous hormone treatment, 60-day-old S. miltiorrhiza hairy roots were treated with MeJA for different time points, where that at 0 h was used as control and expression was detected by qRT-PCR. The result indicated that exogenous MeJA induced SmWRKY1 expression (Figure 2B), reaching a peak at 0.5 h, arising to about threefold. Thereafter, the transcript level declined rapidly within 2 h. Treatment with SA and NO did also induce SmWRKY1 expression, with a maximum peak being reached at 3 h, followed by a gradual decrease over 12 h (Figures 2C,D).
Subcellular Localization of SmWRKY1
To experimentally confirm the subcellular localization of SmWRKY1, SmWRKY1 was cloned into the pMON530 vector to fuse with green fluorescent protein (GFP) reporter gene to generate vector pMON530-SmWRKY1-GFP. Then, the constructed vector and the pMON530 (used as the control) was transformed into the ASE strain and expressed in tobacco leaves, respectively. In the leaves of control vector transformed plant, the fluorescence of GFP was detected in the cytoplasm and nucleus (Figure 3). In contrast (to this), the fluorescence of SmWRKY1/GFP fusion protein was only found in nucleus. The expression pattern was consistent with the character of SmWRKY1 as a TF.
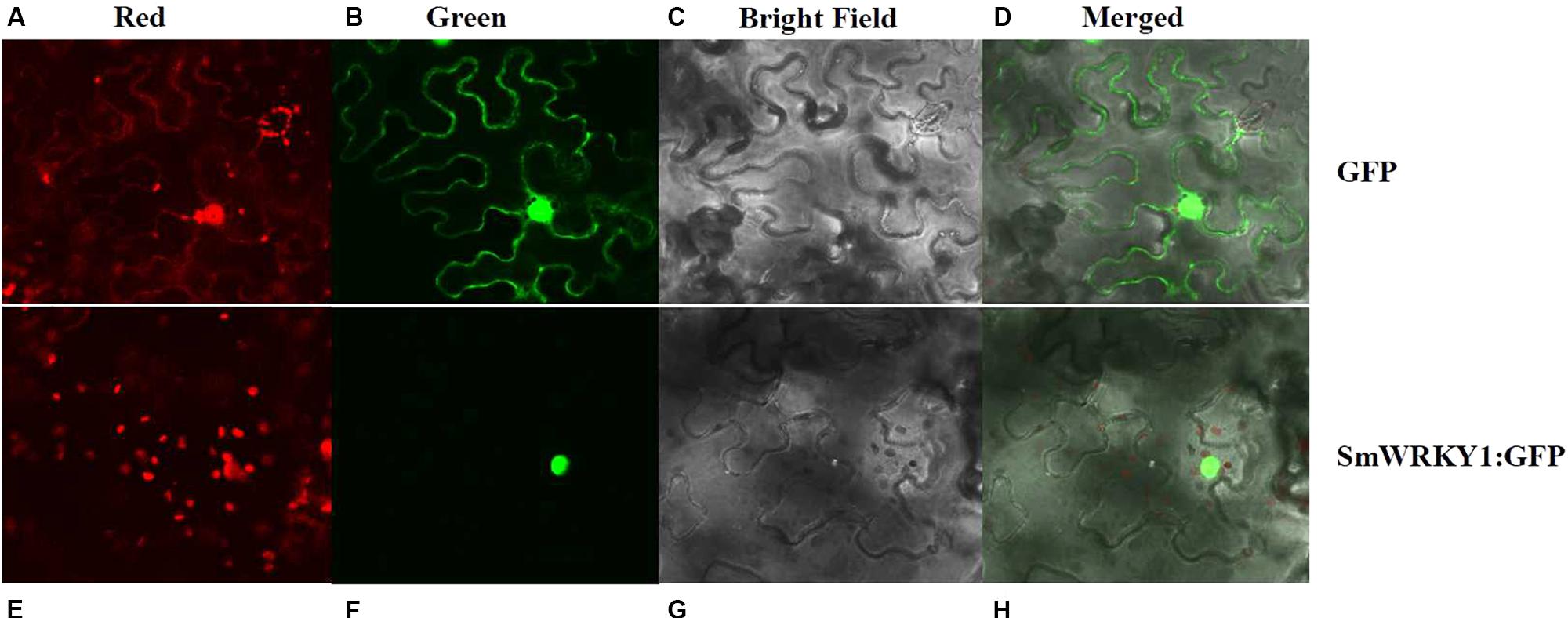
FIGURE 3. Subcellular localization of SmWRKY1. (A–D) The free GFP expressed in N. benthamiana leaves. (E–H) SmWRKY1:: GFP expressed in N. benthamiana leaves.
Acquisition of SmWRKY1 Transgenic Hairy Roots
To further investigate the function of SmWRKY1 in S. miltiorrhiza, we inserted SmWRKY1 into a modified pCAMBIA2300sm vector (Supplementary Figure S4). Then the recombinant overexpression vector pCAMBIA2300sm-SmWRKY1 was introduced into A. rhizogenes strain C58C1 and used to infect S. miltiorrhiza explants, and the empty vector pCAMBIA2300sm was used as control. After 2–3 weeks the fresh hairy roots differentiated from the stem and leaf explant as shown in Figure 4. The positive lines carrying the SmWRKY1 gene were verified by PCR. The positive rate was 20.5% among the 39 samples (Figure 5). qRT-PCR analysis of the expression of SmWRKY1 in over-expression lines found that SmWRKY1 expressed 20–48 times higher than in the empty vector control transformed lines (Figure 6A). The three high expression lines including 1, 2, and 32 (designated as 3) were chosen for further analysis.

FIGURE 4. Generation of transgenic hairy root of S. miltiorrhiza. (A) S. miltiorrhiza explants on ½ MS medium. (B) The growing hairy root on the infected S. miltiorrhiza explants. (C) Monoclone of hairy root. (D) Hairy roots culture in ½ MS medium.
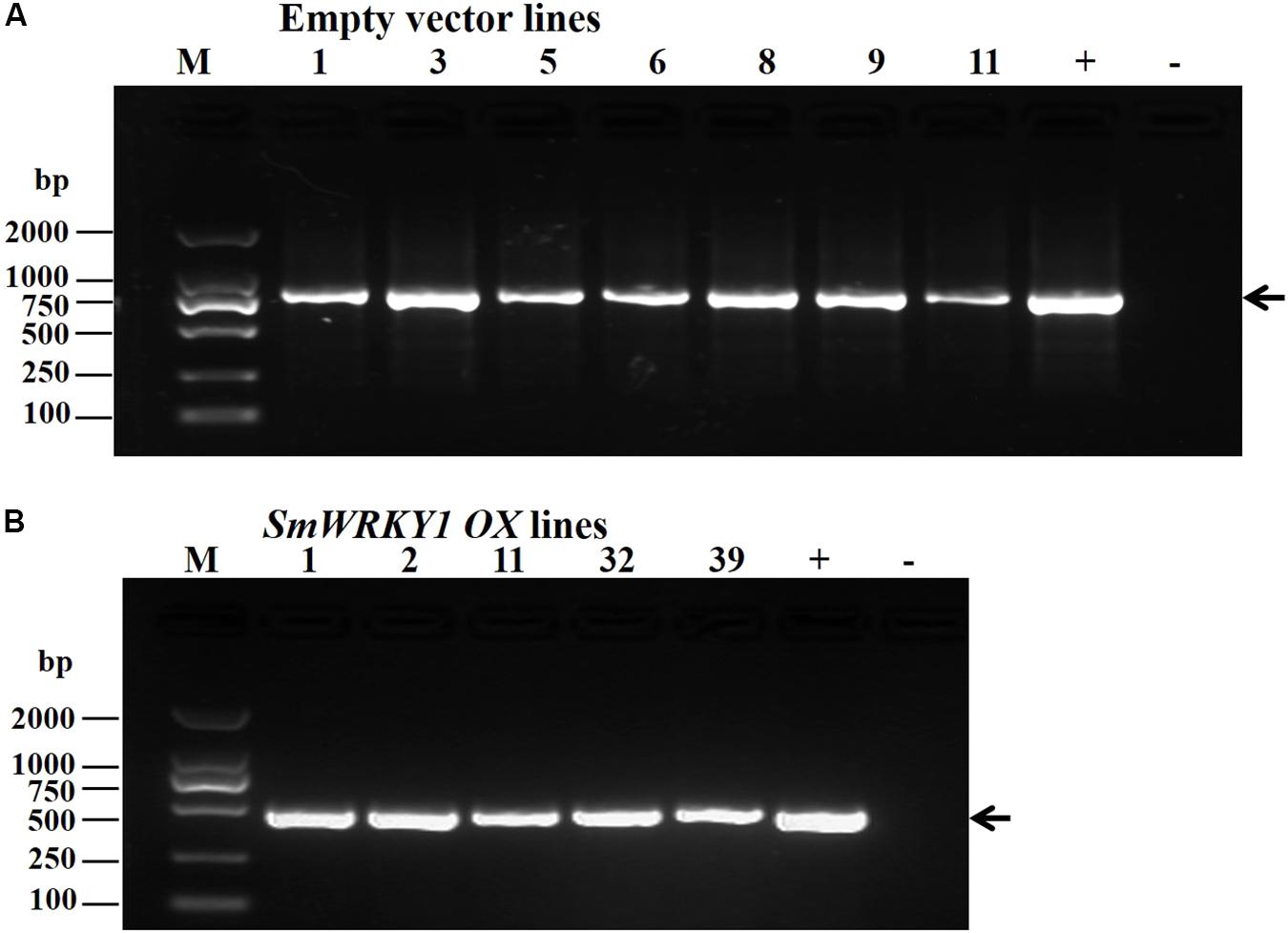
FIGURE 5. (A) Identification of positive transgenic hairy root lines by PCR. (GusA-F and GusA-R were used to identify empty vector pCAMBIA2300sm transformed lines (B) Primers CaMV35S-F23 and SmWRKY1-QR were used to identify the positive colony of SmWRKY1 overexpression transgenic lines).
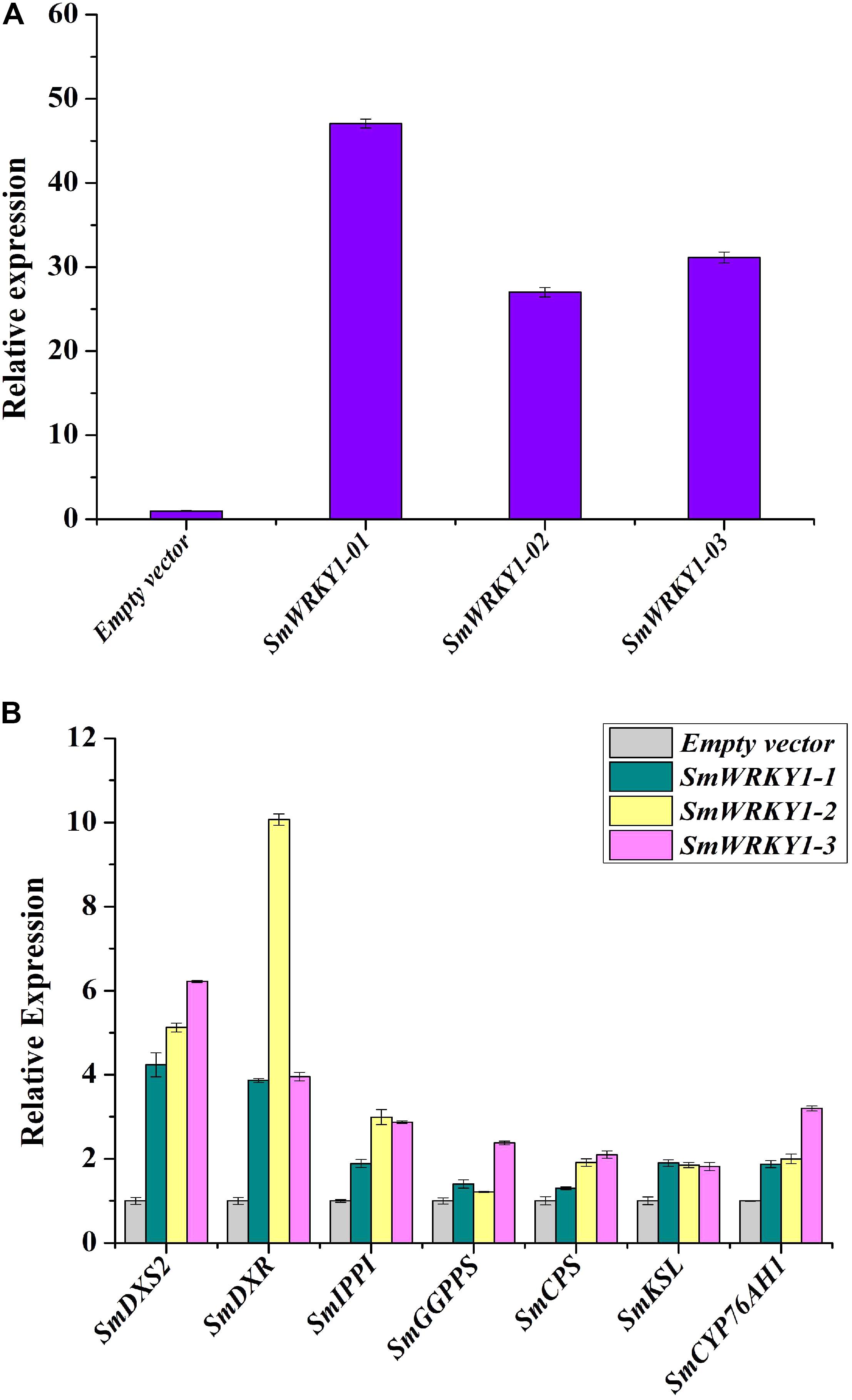
FIGURE 6. Transcript levels of SmWRKY1 and genes related to tanshinones biosynthesis in SmWRKY1 transgenic hairy roots. Expression of SmWRKY1 was analyzed by qRT-PCR. All data are means of three replicates, with error bars indicating Standard Deviation. (A) The transcript level of SmWRKY1 in SmWRKY1 transgenic hairy roots. (B) The transcript level of the key tanshinones biosynthesis genes in SmWRKY1 transgenic hairy roots.
Expression of Tanshinone Biosynthetic Genes in SmWRKY1 Transgenic Hairy Roots
To study whether SmWRKY1 participated in the regulation of tanshinone biosynthesis, transcript levels of several genes related to tanshinones biosynthesis in SmWRKY1 transgenic hairy root were analyzed by qRT-PCR. Several tanshinone biosynthesis pathway genes were up-regulated in the SmWRKY1-overexpressing hairy roots (Figure 6B), the most striking ones were SmDXS2 and SmDXR genes, which increased 4–6-fold and 4–10-fold as compared with the control, respectively. Though the expression of SmIPPI, SmGGPPS, SmCPS, SmKSL, and SmCYP76AH1 was a little lower than that of SmDXS and SmDXR, their expression in overexpression lines was two–fourfold higher than in the control lines. In contrast, the expression of all these seven tanshinones biosynthesis pathway genes was significantly decreased in the knock-down lines. All these results suggested that SmWRKY1 may be a positive regulator in tanshinones biosynthesis.
SmWRKY1 Activates the Transcription of SmDXR in Vivo
Expression profiles showed that SmWRKY1 significantly promotes the expression of SmDXR and SmDXS2, coding for enzymes being in charge of pivotal catalytic steps of tanshinone accumulation. By analyzing the sequence of SmDXR and SmDXS2 promoters, we found a W-box in the promoter of SmDXR (Figure 7A). Then the dual luciferase (Dual-LUC) method was employed to verify whether SmWRKY1 protein activates the transcription of SmDXR and SmDXS2 or not. The results showed that SmWRKY1 over-expression led to an increase by 6 fold of SmDXR expression (Figure 7B).
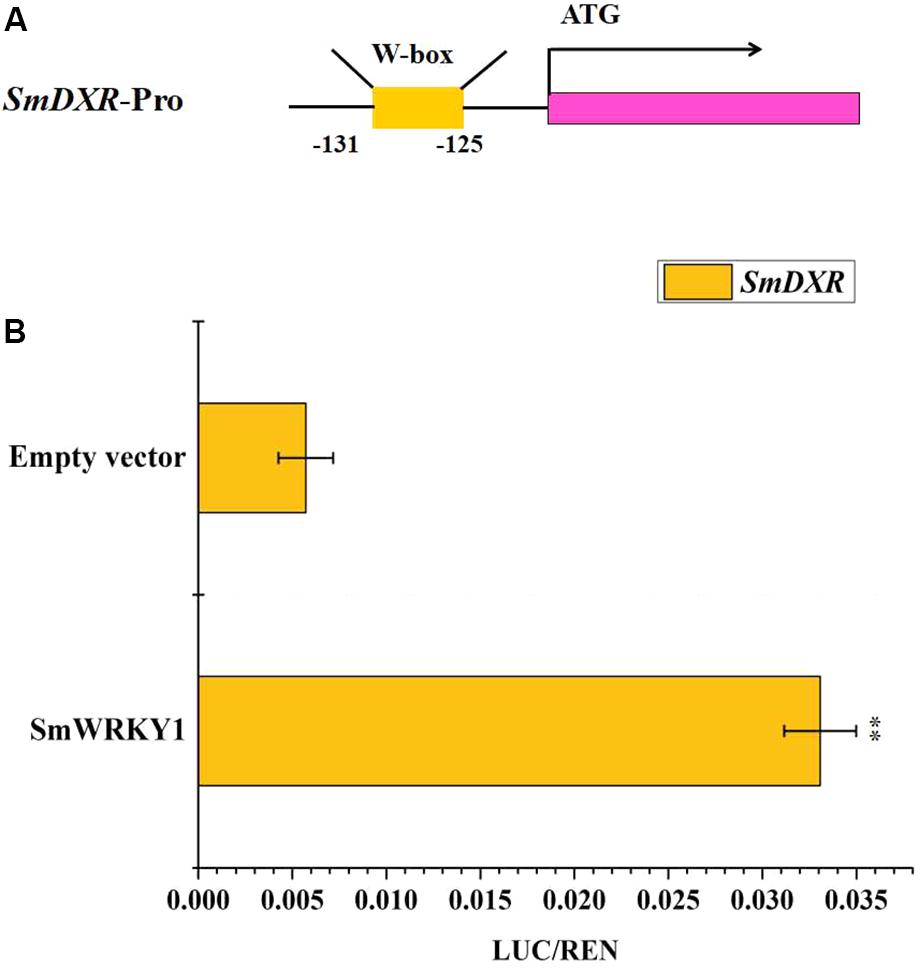
FIGURE 7. The SmDXR promoter was fused to the luciferase (LUC) reporter and the promoter activity was determined by a transient dual-LUC assay in tobacco. The value of LUC activity/Renilla (REN) luciferase was regarded as the activating activity. Error bars indicate SD (n = 3). Student’s t-test: ∗P < 0.05, ∗∗P < 0.01. (A) The analysis of motifs in SmDXR promoter. (B) The Dual-luciferase assay that SmWRKY1 activates the promoter of SmDXR.
Accumulation of Tanshinone Was Obviously Affected by SmWRKY1
Based on the quantitative data, we wanted to further evaluate whether the expression of SmWRKY1 in transgenic hairy roots affects the content of tanshinone. Three overexpression lines and two knock-down lines were used to examine four monomers of tanshinones, including cryptotanshinone, dihydrotanshinone I, tanshinone I, tanshinone IIA in hairy roots by HPLC (Supplementary Figure S3). The results showed that the content of cryptotanshinone (2.4–3.8 mg/g DW), dihydrotanshinone I (2.0–3.0 mg/g DW), tanshinone I (4.5–6.4 mg/g DW), and tanshinone IIA (0.4–0.6 mg/g DW) were significantly up-regulated and the total tanshinone had risen to 9.4–13.7 mg/g DW in overexpression lines. Among them pCAMBIA2300sm-SmWRKY1-3 lines accumulated the highest content of total tanshinone, which was 6.3 times of the control (Figure 8). These results further confirmed the positive role of SmWRKY1 in the regulation of tanshinone biosynthesis.
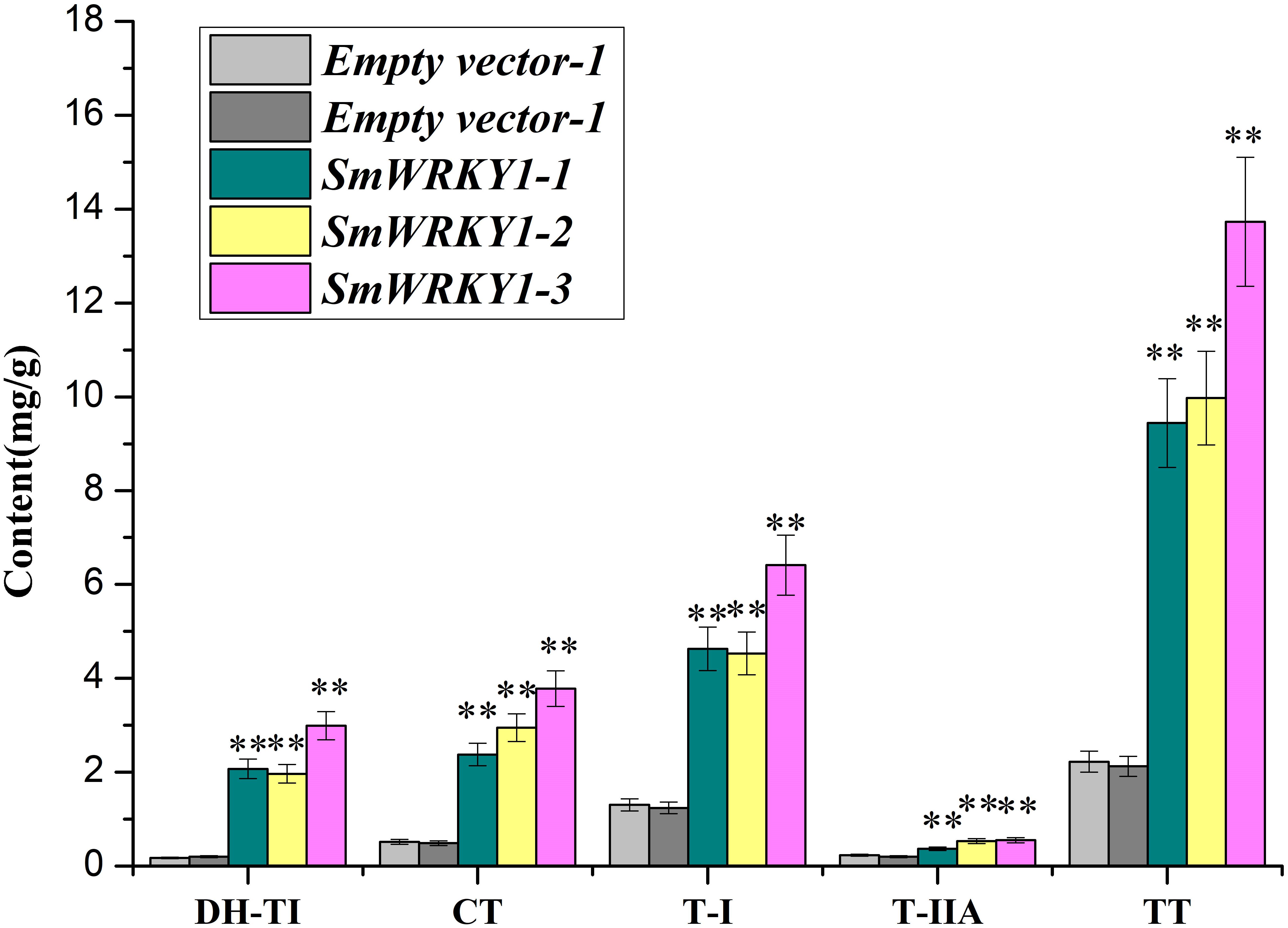
FIGURE 8. The production of tanshinone in SmWRKY1 transgenic hairy roots compared with control tissue, detected by HPLC. All data are means of three replicates, with error bars indicating Standard Deviation. ∗, significant at P < 0.05, ∗∗, highly significant at P < 0.01.
Discussion
The regulation of isoprenoid biosynthesis in plants has been in the focus of many laboratories around the world starting latest in about 1950. The plant S. miltiorrhiza is of some medical interest due to the presence of various diterpenoids of the tanshinone type. Thus, genetic engineering has become an effective and important way to increase the accumulation of active ingredients in S. miltiorrhiza. In the past decade, much progress has been made in isolation of tanshinone biosynthetic genes and metabolic regulation of tanshinone biosynthesis by expression of single or multiple target genes with various combination (Kai et al., 2011; Shi et al., 2014, 2016a). Overexpression of SmDXS in transgenic hairy root lines can significantly enhance the production of tanshinones (Zhou Y. et al., 2016). In the meantime SmDXR also been identified as an important enzyme gene in tanshinone biosynthesis as its over-expression led to significantly improve the production of tanshinones in hairy root lines (Shi et al., 2014). Co-expression of SmHMGR and SmGGPPS increased tanshinone production significantly in a transgenic S. miltiorrhiza hairy root line HG9 (Kai et al., 2011). In addition, introduction of SmHMGR and SmDXR into S. miltiorrhiza hairy roots enhanced the content of tanshinones to 3.25 mg/g DW (Shi et al., 2014). Simultaneous introduction of SmGGPPS and SmDXSII into S. miltiorrhiza hairy root significantly improved the production of tanshinones and increased production of carotenoids, gibberellins and chlorophyll in A. thaliana plants (Shi et al., 2016a). However, tanshinones engineering may be impeded by the fact that the multiple steps in the downstream part of tanshinone biosynthesis are only partially characterized, although the identification of those enzymes is ongoing. Therefore, the attempts undertaken were not always crowned by a big success due to the huge and complex regulatory network.
Currently, people began to realize that it would be more effective to use TFs that might activate whole pathways, instead of individually over-expressing genes that encode putatively rate-limiting enzymes. TFs are divided into different families in plants, such as bHLH, MYB, ERF, WRKY, and so on. Each of them has their specific functions in the regulation of secondary metabolism of plants. Recently, Zhang et al. (2017) reported that overexpression of SmMYB9b enhances tanshinone concentration in S. miltiorrhiza hairy roots (2.2-fold improvement over the control), but the direct target gene in tanshinones biosynthetic pathway was not identified. In addition, Zhou Y. et al. (2016) reported on ectopic RNA interference (RNAi)-mediated knockdown experiments, which suggested that the accumulation was impaired by the loss of function in SmMYC2a/b. However, the opposite effect by OE of SmMYC2a/b had not been studied yet, and the identification of direct target genes in tanshinone biosynthesis is also awaiting. In addition, to our knowledge, no WRKY TF from S. miltiorrhiza has been functionally characterized yet to date. In present study, we functionally identified a new TF SmWRKY1 from S. miltiorrhiza and found that OE of SmWRKY1 can obviously promote the accumulation of tanshinones in transgenic lines. The highest tanshinone production was observed in transgenic SmWRKY1-3 with the concentration of 13.7 mg/g DW (5.3-fold improvement than the control), to our knowledge, this is the highest tanshinone content achieved through genetic engineering.
Meanwhile, expression profile analysis showed that overexpression of SmWRKY1 can promote the transcripts level of several biosynthetic genes especially SmDXR, and previous studies have proved that WRKY TFs could directly bind to the W-box of related genes from different signal pathways (Eulgem et al., 2000). Thus we scanned the promoter of SmDXR, and a W-box motif was found, which implied that it can be bound and regulated by SmWRKY1. Thereafter, the Dual-LUC assay proved that SmWRKY1 can transcriptionally activate SmDXR expression by directly binding to its promoter region, in good agreement with that SmDXR has been identified an important target gene in tanshinone biosynthesis (Shi et al., 2014). In some other medicinal plants including C. roseus (Suttipanta et al., 2011), A. annua (Chen et al., 2017), and W. somnifera (Singh et al., 2017), WRKYs have also been found to activate the transcripts of different target genes by binding to the W-box in their promoters, suggesting that the W-box was their recognition cis-element.
Conclusion
Our work functionally identified a new TF SmWRKY1 gene from S. miltiorrhiza and found that overexpression of SmWRKY1 can obviously promote the accumulation of tanshinone in transgenic lines. The Dual-LUC assay indicated that SmWRKY1 positively regulated tanshinone biosynthesis through a direct target gene SmDXR involved in the MEP pathway. Our study may provide a new insight by genetic engineering strategy with functional TFs to improve the yield of target compounds in S. miltiorrhiza and also a certain basis to analyze the entire regulation network of tanshinone biosynthesis.
Author Contributions
GK conceived and designed the study. WC, YaW, MS, XH, and WZ performed the experiments and analyzed the data. WC, MS, JR, and YuW drafted the manuscript, and GK revised the manuscript.
Funding
This work was supported by National Natural Science Fund (31270007, 81522049, and 31571735), Zhejiang Provincial Key University Project on the Construction of First-Class Subjects, New Century Talent Project (NECT-13-0902), Shanghai Science and Technology Committee Project (17JC1404300 and 15430502700), the “Dawn” Program of Shanghai Education Commission (16SG38), and Shanghai Engineering Research Center of Plant Germplasm Resources (17DZ2252700).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very grateful for the two reviewers’ useful comments and careful corrections.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00554/full#supplementary-material
Footnotes
References
Chen, J., Shi, D. Y., Liu, S. L., and Zhong, L. (2012). Tanshinone IIA induces growth inhibition and apoptosis in gastric cancer in vitro and in vivo. Oncol. Rep. 27, 523–528. doi: 10.3892/or.2011.1524
Chen, M., Yan, T., Shen, Q., Lu, X., Pan, Q., Huang, Y., et al. (2017). GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 214, 304–316. doi: 10.1111/nph.14373
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206.
Gao, W., Hillwig, M. L., Huang, L., Cui, G., Wang, X., Kong, J., et al. (2009). A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org. Lett. 11, 5170–5173. doi: 10.1021/ol902051v
Ge, X., and Wu, J. (2005). Induction and potentiation of diterpenoid tanshinone accumulation in Salvia miltiorrhiza hairy roots by β-aminobutyric acid. Appl. Microbiol. Biotechnol. 68, 183–188. doi: 10.1007/s00253-004-1873-2
Gong, Y., Li, Y., Abdolmaleky, H. M., Li, L., and Zhou, J. R. (2012). Tanshinones inhibit the growth of breast cancer cells through epigenetic modification of Aurora A expression and function. PLoS One 7:e33656. doi: 10.1371/journal.pone.0033656
Guo, J., Zhou, Y. J., Hillwig, M. L., Shen, Y., Yang, L., Wang, Y., et al. (2013). CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. U.S.A. 110, 12108–12113. doi: 10.1073/pnas.1218061110
Hao, X., Shi, M., Cui, L., Xu, C., Zhang, Y., and Kai, G. (2015). Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol. Appl. Biochem. 62, 24–31. doi: 10.1002/bab.1236
Jiang, W., Fu, X., Pan, Q., Tang, Y., Shen, Q., Lv, Z., et al. (2016). Overexpression of AaWRKY1 leads to an enhanced content of artemisinin in Artemisia annua. Biomed Res. Int. 2016:7314971. doi: 10.1155/2016/7314971
Kai, G. Y., Liao, P., Xu, H., Wang, J., Zhou, C. C., Zhou, W., et al. (2012). Molecular mechanism of elicitor-induced tanshinone accumulation in Salvia miltiorrhiza hairy root cultures. Acta Physiol. Plant. 34, 1421–1433.
Kai, G. Y., Liao, P., Zhang, T., Zhou, W., Wang, J., Xu, H., et al. (2010). Characterization, expression profiling, and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotechnol. Bioprocess Eng. 15, 236–245.
Kai, G. Y., Xu, H., Zhou, C., Liao, P., Xiao, J., Luo, X., et al. (2011). Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 13, 319–327. doi: 10.1016/j.ymben.2011.02.003
Li, J. B., Luan, Y. S., and Liu, Z. (2015). Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol. Plant. 155, 248–266. doi: 10.1111/ppl.12315
Liao, P., Zhou, W., Zhang, L., Wang, J., Yan, X. M., Zhang, Y., et al. (2009). Molecular cloning, characterization and expression analysis of a new gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from Salvia miltiorrhiza. Acta Physiol. Plant. 31, 565–572. doi: 10.1016/j.jplph.2010.06.008
Liu, Q., Liu, Y., Tang, Y., Chen, J., and Ding, W. (2017). Overexpression of NtWRKY50 increases resistance to Ralstonia solanacearum and alters salicylic acid and jasmonic acid production in tobacco. Front. Plant Sci. 8:1710. doi: 10.3389/fpls.2017.01710
Liu, X., Song, Y., Xing, F., Wang, N., Wen, F., and Zhu, C. (2016). GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 253, 1265–1281. doi: 10.1007/s00709-015-0885-3
Lu, M., Wang, L. F., Du, X. H., Yu, Y. K., Pan, J. B., Nan, Z. J., et al. (2015). Molecular cloning and expression analysis of jasmonic acid dependent but salicylic acid independent LeWRKY1. Genet. Mol. Res. 14, 15390–15398. doi: 10.4238/2015.November.30.16
Ma, D., Pu, G., Lei, C., Ma, L., Wang, H., Guo, Y., et al. (2009). Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50, 2146–2161. doi: 10.1093/pcp/pcp149
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Shi, M., Luo, X., Ju, G., Li, L., Huang, S., Zhang, T., et al. (2016a). Enhanced diterpene tanshinone Accumulation and bioactivity of transgenic Salvia miltiorrhiza hairy roots by pathway engineering. J. Agric. Food Chem. 64, 2523–2530. doi: 10.1021/acs.jafc.5b04697
Shi, M., Luo, X., Ju, G., Yu, X., Hao, X., Huang, Q., et al. (2014). Increased accumulation of the cardio-cerebrovascular disease treatment drug tanshinone in Salvia miltiorrhiza hairy roots by the enzymes 3-hydroxy-3-methylglutaryl CoA reductase and 1-deoxy-D-xylulose 5-phosphate reductoisomerase. Funct. Integr. Genomics 14, 603–615. doi: 10.1007/s10142-014-0385-0
Shi, M., Zhou, W., Zhang, J., Huang, S., Wang, H., and Kai, G. (2016b). Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins. Sci. Rep. 6:20919. doi: 10.1038/srep20919
Singh, A. K., Kumar, S. R., Dwivedi, V., Rai, A., Pal, S., Shasany, A. K., et al. (2017). A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 215, 1115–1131. doi: 10.1111/nph.14663
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., and Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Wang, F., Chen, H. W., Li, Q. T., Wei, W., Li, W., Zhang, W. K., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Xu, H., Zhang, L., Zhou, C. C., Xiao, J. B., Liao, P., and Kai, G. Y. (2010). Metabolic regulation and genetic engineering of pharmaceutical component tanshinone biosynthesis in Salvia miltiorrhiza. J. Med. Plants Res. 4, 2591–2597.
Xu, M., Hao, H., Jiang, L., Long, F., Wei, Y., Ji, H., et al. (2015). In vitro inhibitory effects of ethanol extract of Danshen (Salvia miltiorrhiza) and its components on the catalytic activity of soluble epoxide hydrolase. Phytomedicine 22, 444–451. doi: 10.1016/j.phymed.2015.02.001
Xu, Y. H., Wang, J. W., Wang, S., Wang, J. Y., and Chen, X. Y. (2004). Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol. 135, 507–515. doi: 10.1104/pp.104.038612
Yan, X. M., Zhang, L., Wang, J., Liao, P., Zhang, Y., Zhang, R., et al. (2009). Molecular characterization and expression of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) gene from Salvia miltiorrhiza. Acta Physiol. Plant. 31, 1015–1022.
Zhang, G., Tian, Y., Zhang, J., Shu, L., Yang, S., Wang, W., et al. (2015). Hybrid de novo genome assembly of the Chinese herbal plant danshen (Salvia miltiorrhiza Bunge). Gigascience 4:62. doi: 10.1186/s13742-015-01043
Zhang, J., Zhou, L., Zheng, X., Zhang, J., Yang, L., Tan, R., et al. (2017). Overexpression of SmMYB9b enhances tanshinone concentration in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 36, 1297–1309. doi: 10.1007/s00299-017-2154-8
Zhang, L., Yan, X. M., Wang, J., Li, S. S., Liao, P., and Kai, G. Y. (2011). Molecular cloning and expression analysis of a new putative gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Salvia miltiorrhiza. Acta Physiol. Plant. 33, 953–961.
Zhang, Y., Li, X., and Wang, Z. (2010). Antioxidant activities of leaf extract of Salvia miltiorrhiza Bunge and related phenolic constituents. Food Chem. Toxicol. 48, 2656–2662. doi: 10.1016/j.fct.2010.06.036
Zhao, S., Zhang, J., Tan, R., Yang, L., and Zheng, X. (2015). Enhancing diterpenoid concentration in Salvia miltiorrhiza hairy roots through pathway engineering with maize C1 transcription factor. J. Exp. Bot. 66, 7211–7226. doi: 10.1093/jxb/erv418
Zhou, W., Huang, F., Li, S., Wang, Y., Zhou, C., Shi, M., et al. (2016a). Molecular cloning and characterization of two 1-deoxy-D-xylulose-5-phosphate synthase genes involved in tanshinone biosynthesis in Salvia miltiorrhiza. Mol. Breed. 36:124. doi: 10.1007/s11032-016-0550-3
Zhou, W., Huang, Q., Wu, X., Zhou, Z., Ding, M., Shi, M., et al. (2017). Comprehensive transcriptome profiling of Salvia miltiorrhiza for discovery of genes associated with the biosynthesis of tanshinones and phenolic acids. Sci. Rep. 7:10554. doi: 10.1038/s41598-017-10215-2
Zhou, W., Wu, S., Ding, M., Li, J., Shi, Z., Wei, W., et al. (2016b). Mapping of Ppd-B1, a major candidate gene for late heading on wild emmer chromosome arm 2bs and assessment of its interactions with early heading QTLs on 3AL. PLoS One 11:e0147377. doi: 10.1371/journal.pone.0147377
Keywords: Salvia miltiorrhiza, hairy roots, SmWRKY1, MEP pathway, tanshinones, metabolic engineering
Citation: Cao W, Wang Y, Shi M, Hao X, Zhao W, Wang Y, Ren J and Kai G (2018) Transcription Factor SmWRKY1 Positively Promotes the Biosynthesis of Tanshinones in Salvia miltiorrhiza. Front. Plant Sci. 9:554. doi: 10.3389/fpls.2018.00554
Received: 15 December 2017; Accepted: 09 April 2018;
Published: 27 April 2018.
Edited by:
Laigeng Li, Shanghai Institutes for Biological Sciences (CAS), ChinaCopyright © 2018 Cao, Wang, Shi, Hao, Zhao, Wang, Ren and Kai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoyin Kai, kaiguoyin@163.com; guoyinkai@yahoo.com
†Co-first authors
 Wenzhi Cao1†
Wenzhi Cao1† Guoyin Kai
Guoyin Kai