- 1Department of Agricultural and Food Sciences (DISTAL), University of Bologna, Bologna, Italy
- 2Ethiopian Institute of Agricultural Research (EIAR), Addis Ababa, Ethiopia
- 3Department of Agricultural, Food and Environmental Sciences, Polytechnic University of Marche, Ancona, Italy
- 4Department of Biotechnology, University of Verona, Verona, Italy
- 5Research Group on Food, Nutritional Biochemistry and Health, Universidad Europea del Atlántico, Santander, Spain
Downy mildew caused by Plasmopara viticola is one of the most devastating diseases of grapevine, attacking all green parts of the plant. The damage is severe when the infection at flowering stage is left uncontrolled. P. viticola management consumes a significant amount of classical pesticides applied in vineyards, requiring efficient and environmentally safe disease management options. Spray-induced gene silencing (SIGS), through the application of exogenous double-stranded RNA (dsRNA), has shown promising results for the management of diseases in crops. Here, we developed and tested the potential of dsRNA targeting P. viticola Dicer-like (DCL) genes for SIGS-based crop protection strategy. The exogenous application of PvDCL1/2 dsRNA, a chimera of PvDCL1 and PvDCL2, highly affected the virulence of P. viticola. The reduced expression level of PvDCL1 and PvDCL2 transcripts in infected leaves, treated with PvDCL1/2 dsRNA, was an indication of an active RNA interference mechanism inside the pathogen to compromise its virulence. Besides the protective property, the PvDCL1/2 dsRNA also exhibited a curative role by reducing the disease progress rate of already established infection. Our data provide a promising future for PvDCL1/2 dsRNA as a new generation of RNA-based resistant plants or RNA-based agrochemical for the management of downy mildew disease in grapevine.
Introduction
Grapevine (Vitis vinifera L.) is an important fruit crop cultivated worldwide for fresh and dry fruit consumption and for wine production. Wine production trend is increasing yearly with the world wine trade worth getting about US $40 billion in the year 2018; Italy, France, and Spain being the largest wine-producing countries, contributing half of the world production.1 Grapevine production is affected by several pre- and post-harvest pathogens that affect quality during production and processing. Some of the economically important diseases of the crop are gray mold, powdery mildew, and downy mildew caused by Botrytis cinerea, Erysiphe necator, and Plasmopara viticola, respectively. The obligate biotrophic oomycete P. viticola attacks all green parts of grapevine, and the damage is severe if the infection occurring during flowering is not managed. Surprisingly, all cultivated European V. vinifera cultivars are susceptible to the pathogen (Armijo et al., 2016), which makes the management of downy mildews in vineyard and other crops rely on synthetic fungicides. As a result, its management, together with powdery mildew, consumes about two-thirds of all synthetic fungicides sprayed for disease management of crops in the European Union (Eurostat., 2007). With such heavy reliance on agrochemicals to control P. viticola, not only pathogen strains have developed resistance to several fungicides (Gisi and Sierotzki, 2008), but there also exist social concerns about environment and human health, which makes it urgent to find alternative control strategies.
The findings that exogenous small RNAs (sRNA) and double-stranded RNA (dsRNA) trigger posttranscriptional gene silencing (Fire et al., 1998; Hamilton and Baulcombe, 1999) have opened new avenues to exploit the gene silencing mechanism as a new class of regulatory molecules during plant–pathogen interaction. The gene silencing occurs via RNA interference (RNAi) machinery, a natural biological process conserved in most eukaryotes where sRNA molecules regulate gene expression by targeting specific endogenous messenger RNA molecules in a sequence-specific manner (Vaucheret and Fagard, 2001; Castel and Martienssen, 2013). The silencing signals of sRNA are bidirectional cross-kingdom, moving from the host to its interacting organism, and vice versa (Tomilov et al., 2008; Weiberg et al., 2015; Wang et al., 2016; Cai et al., 2018).
The involvement of sRNAs in the crosstalk between plant hosts and their fungal and oomycete pathogens has also been suggested (Weiberg et al., 2013; Brilli et al., 2018), implying that exploiting the RNAi mechanisms of both the hosts and the pathogens can represent a new strategy in fungal and oomycete disease management. Transgene-derived artificial sRNAs inducing gene silencing, called host-induced gene silencing (HIGS), have been observed providing resistance to plants against fungi (Nowara et al., 2010; Koch et al., 2013; Zhu et al., 2017) and oomycetes (Vega-Arreguin et al., 2014; Jahan et al., 2015). Interestingly, recent findings revealed that the external application of dsRNA also conferred host plant resistance to fungal pathogens by silencing targeted genes (Koch et al., 2016; Wang et al., 2016; McLoughlin et al., 2018; Nerva et al., 2020), an approach referred to as spray-induced gene silencing (SIGS).
The exogenous application of dsRNAs targeting Dicer-like (DCL), lanosterol 14α-demethylase, chitin synthase, and elongation factor genes of B. cinerea negatively affected its pathogenicity in multiple hosts (Wang et al., 2016; Nerva et al., 2020). Similarly, spraying of dsRNA targeting three cytochrome P450 genes of Fusarium graminearum inhibited fungal growth at sprayed and distal parts of detached barley leaves (Koch et al., 2016). While these research findings provided proof that SIGS-based plant protection is effective against targeted pathogens, there is also indication that the effects of dsRNA can be reproduced on closely related pathogens based on sequence homology (McLoughlin et al., 2018). According to McLoughlin et al. (2018), dsRNA targeting SS1G_05899 and SS1G_02495 genes of Scelerotinia sclerotiorum, both involved in redox reaction, restricted the progress of the pathogen on a susceptible Brassica napus cultivar. Remarkably, the cultivar was also resistant to B. cinerea when treated with dsRNA targeting BC1G_01592 and BC1G_04955, the B. cinerea homologs to SS1G_05899 and SS1G_02495, respectively. Such results provide compelling evidence about the adaptability and flexibility of SIGS technology in crop disease management. In this study, we investigate the potential of dsRNA targeting P. viticola DCL genes for SIGS-based crop protection strategy. We show that the application of dsRNA targeting PvDCL1/2 extremely reduces the pathogenicity of P. viticola and the expression level of the targeted genes, indicating that RNAi-based control strategy can indeed represent a promising alternative to hazardous agrochemical application to manage downy mildew disease of grapevine.
Materials and Methods
Design and Production of dsRNA and Rate of Application
Plasmopara viticola genes encoding two Dicer-like proteins, as defined by the presence of a Dicer dimerization domain, corresponding to PVITv1_T038441 and PVITv1_T003331, hereafter referred to as PvDCL1 and PvDCL2, respectively (Brilli et al., 2018), were selected. For RNAi, 258- and 257-bp fragments of PvDCL1 and PvDCL2 sequences, respectively (Supplementary Table 1), were chosen as target, and the corresponding chimeric dsRNA molecule (PvDCL1/2, 515 bp) was chemically synthesized by AgroRNA (Genolution Inc., Seoul, Republic of Korea; Supplementary Figure 1). DsRNA targeting B. cinerea DCL 1 and 2 genes, BcDCL1/2 (490 bp; Wang et al., 2016), produced in the same way, was used as the negative control. After assaying different dsRNA concentrations, 75, 100, or 125 ng μl–1 concentrations of dsRNA were used for spot inoculation in a total volume of 50 μl.
Plant Material and Plasmopara viticola Inoculation
Seedlings of V. vinifera cv. Trebbiano were raised in growth chamber at 22°C ± 1°C and 12/12 h light cycle. P. viticola (strain 465, belonging to University of Bologna collection) was maintained on grapevine leaves at 22°C ± 1°C and 12/12 h of photoperiod. Sporangia were harvested in distilled water and filtered through cheesecloth. Sporangia concentration was determined using hemocytometer.
Fully expanded third and fourth leaves from 6–8-week-old grapevine seedlings were detached and immediately placed on wet absorbing paper in a plastic box. Detached leaves were surface sterilized for 1 min with 70% ethanol and then rinsed three times with sterile water. For assaying dsRNA as preventive treatment, the abaxial side of each leaf was treated with three droplets of 50 μl of either dsRNA or water. After 2 h, 7.5 μl of a 1 × 105 ml–1 sporangia solution was placed on top of the droplets. Disease progress was evaluated until 14 days post inoculation (dpi) in five biological replicates. A single leaf was considered a biological replicate.
For assaying dsRNA as curative treatment, each leaf was first challenged by the pathogen by applying four droplets of 7.5 μl of a 1 × 105 ml–1 sporangia solution, and after 7 dpi, when a visible sign of P. viticola was observed, 50 μl of either dsRNA or water was placed on top of each spot of the progressing pathogen. Disease progress was evaluated until 14 dpi, i.e., 7 days post treatment (dpt) of either dsRNA or water, in three biological replicates.
To assess the progress of the pathogen, leaf area covered by P. viticola (in square millimeters) was measured from the digital images using the free software ImageJ program.2 Leaf area covered by the pathogen, area under disease progress curve (AUDPC), and disease progress rate data were analyzed using analysis of variance. Means were separated by Tukey’s honestly significant difference test.
RNA Extraction and Quantitative PCR Analysis
Leaves that were treated in the preventive assay were collected at 7 dpi, in three replicates, immediately frozen in liquid nitrogen, and kept at −80°C until use. RNA was extracted using a rapid cetyltrimethylammonium bromide (CTAB) method (Gambino et al., 2008). First-strand cDNA was synthesized from 1 μg of total RNA, pretreated with TURBO DNA-free KitTM (Invitrogen, CA, United States), using ImProm-II Reverse Transcriptase (Promega), following the manufacturer’s guide. Quantitative PCR (qPCR) was performed in an MX3000 thermocycler (Stratagene, CA, United States) using 0.25 μl of cDNA and 200 nM of specific forward and reverse primers (Supplementary Table 2) in a total volume of 12.5 μl using Maxima® SYBR Green/ROX qPCR Master Mix (Fermentas). Each amplification reaction was run in duplicate. The cycling parameters were as follows: 5 min at 95°C, 40 cycles of 15 s at 95°C, 25 s at 61°C, and 30 s at 72°C. A melting curve was established from 55°C to 90°C by changing 0.5°C every 10 s. For normalization, P. viticola elongation factor eIF1b was used. Each primer pair’s amplification efficiency was calculated using LinReg (Ruijter et al., 2009). The amplification efficiency value obtained was used to calculate the relative quantity (RQ) and normalized RQ (NRQ) according to Hellemans et al. (2007). Statistical analyses of the qPCR results were made after log2(NRQ) transformation (Rieu and Powers, 2009). Statistical significance was calculated by Tukey’s honestly significant difference test.
Results
Spray-Induced Gene Silencing of Plasmopara viticola DCL Genes Hampers Disease Development
Preliminary inoculation assay was conducted to determine a baseline concentration of PvDCL1/2 dsRNA that could affect P. viticola DCL1 and DCL2 genes and consequently inhibit its germination and/or colonization of grapevine leaves. After the treatment with 10 and 50 ng μl–1 PvDCL1/2 dsRNA and water, as control, detached grapevine leaves were challenged with P. viticola sporangia. Inoculated leaves were monitored for 2 weeks. Sign of P. viticola infection was conspicuous around the inoculation spot starting from the fifth dpi, mostly on control and on leaves treated with 10 ng μl–1 PvDCL1/2 dsRNA, where white fluffy growth of sporangiophores and sporangia appeared. At 14 dpi, the rate of disease progress was relatively slower in leaves that received 50 ng μl–1 of PvDCL1/2 dsRNA (Supplementary Figure 2), indicating that pathogen control efficiency can increase with higher concentrations.
Therefore, the ability of PvDCL1/2 dsRNA to control P. viticola growth in preventive treatment was assessed using higher concentrations (i.e., 75, 100, and 125 ng μl–1). Treatments with BcDCL1/2 targeting B. cinerea DCL1 and DCL2 genes and water were used as controls. As shown in Figure 1A, the fluffy growth of sporangiophores was quite visible on control leaves treated with either water or BcDCL1/2 dsRNA at the three different concentrations. On the contrary, the pathogen’s progress was substantially low or null on leaves that received PvDCL1/2 dsRNA. As a consequence, the area covered by P. viticola and the AUDPC values at 7, 10, and 14 dpi were significantly and consistently lower in leaves treated with PvDCL1/2 dsRNA than in those treated with BcDCL1/2 dsRNA or water (Figure 1B), confirming that PvDCL1/2 dsRNA hampered P. viticola growth. To confirm that the inhibition of P. viticola growth by PvDCL1/2 dsRNA was due to the downregulation of PvDCL1 and PvDCL2 genes, their expression, normalized to P. viticola elongation factor eIF1b, was quantified at 7 dpi using qPCR. We found that the relative expression of both PvDCL1 and PvDCL2 was reduced as compared to the controls (Figure 2). Compared with water and BcDCL1/2-treated leaves, the NRQs of PvDCL1 and PvDCL2 transcripts in leaves treated with 100 ng μl–1 concentration of PvDCL1/2 dsRNA were reduced on average by 48 and 44%, respectively, which is in line with the concept of RNAi-based sequence-specific silencing via SIGS.
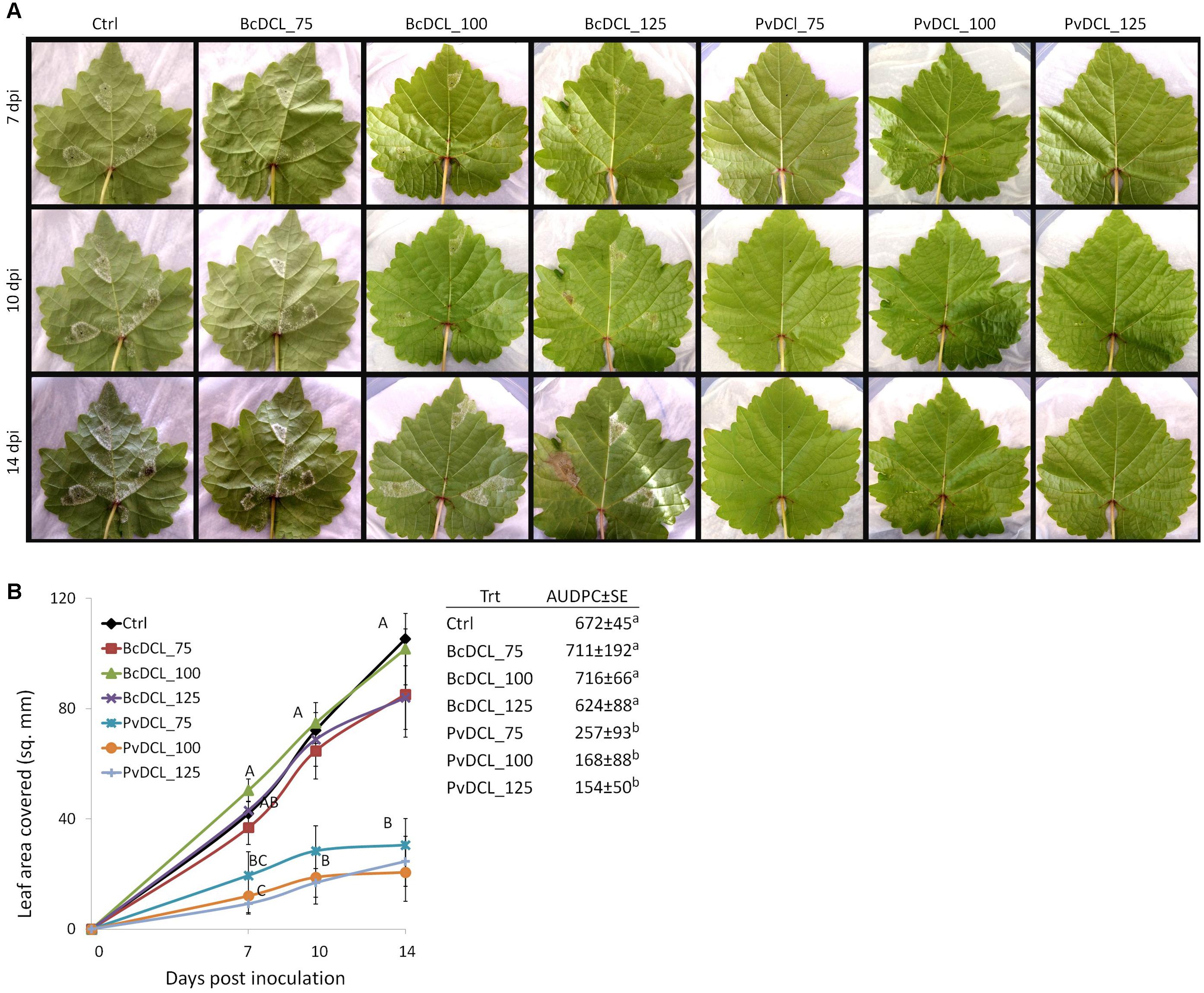
Figure 1. Externally applied PvDCL1/2 double-stranded RNA (dsRNA) on detached grapevine leaves and Plasmopara viticola infection. (A) Progress of P. viticola on grapevine leaves at 7, 10, and 14 days post inoculation (dpi). Leaves were treated with 50 μl of water (ctrl) or dsRNA [75, 100, or 125 ng μl– 1 of dsRNA of BcDCL1/2 (BcDCL_75/100/125) and PvDCL1/2 (PvDCL_75/100/125)] before being inoculated with 7.5 μl of a 1 × 105 ml– 1 sporangia. (B) Disease progression of P. viticola expressed as leaf area covered and as area under the disease progress curve (AUDPC ± SE, mm2 × day) through 14 dpi. Error bars indicate standard error. Means at each dpi and AUDPC followed by a common letter are significantly not different according to Tukey’s honestly significant difference test (P ≤ 0.05).
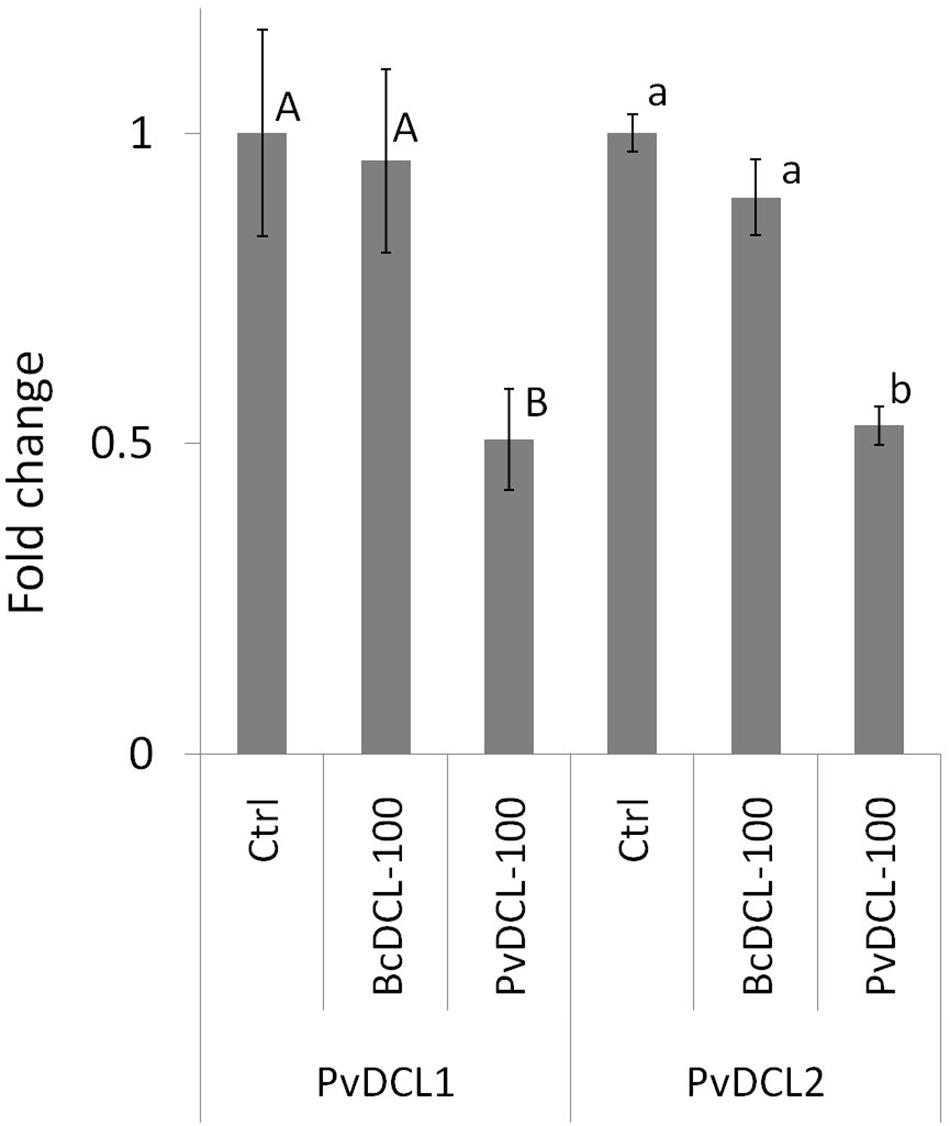
Figure 2. Expression profiles of PvDCL1 and PvDCL2 following Plasmopara viticola inoculation on leaf samples treated with 50 μl of water (ctrl) or 100 ng μl– 1 of double-stranded RNA (dsRNA) of either BcDCL1/2 (BcDCL-100) or PvDCL1/2 (PvDCL-100). Gene expression level was determined by quantitative PCR (qPCR). Bars represent fold change of dsRNA-treated sample relative to ctrl sample at 7 days post inoculation. Normalization based on the expression levels of elongation factor, PveIF1b, was carried out before calculating fold changes. Error bar represents standard error of the mean of three biological replicates. Expression values followed by a common letter are significantly not different among samples, according to Tukey’s honestly significant difference test (P ≤ 0.05), using one-way ANOVA of log2 [normalized relative quantity (NRQ)].
Spray-Induced Gene Silencing of PvDCLs Shows a Curative Effect Against Plasmopara viticola
The observed protective effect of PvDCL1/2 dsRNA prompted us to check whether the dsRNA also has a curative effect against P. viticola. Detached leaves were initially inoculated with P. viticola sporangia, and then, once the infection has been established (i.e., 7 dpi), dsRNA was applied [i.e., the time of either dsRNA or water application is marked as 0 day post treatment (dpt)]. At each inoculation spot, 50 μl of dsRNA or water was added on top of the growing mycelia. At 4 dpt, the progress of the pathogen stagnated in most of the treatments, with more pronounced effect on leaves that received 100 and 125 ng μl–1 of PvDCL1/2 dsRNA (Figure 3A). After 4 dpt, recovering of pathogen growth was more apparent on all leaves. At 7 dpt, the disease advanced more on leaves treated with BcDCL1/2 and water than on those treated with PvDCL1/2, especially at the highest concentration (Figure 3A). Computing the rate of disease progress, taking diseased area at 7 dpi (0 dpt) as a reference, the disease progress rate was relatively slower on leaves treated with PvDCL1/2, with more pronounced effect at 7 dpt (Figure 3B). The result shows that the PvDCL1/2 dsRNA can also hamper the expansion of already established downy mildew disease.
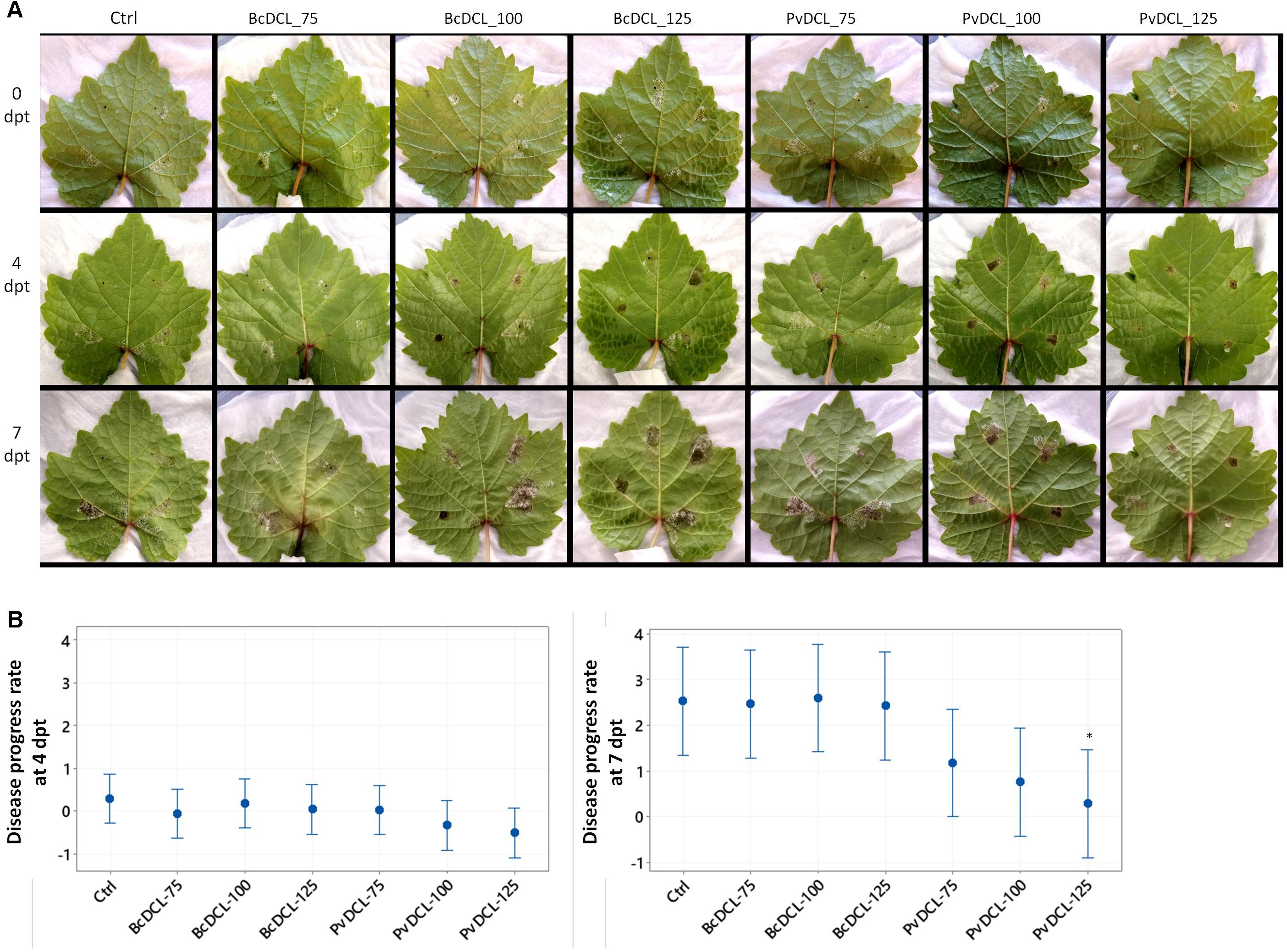
Figure 3. Progress of Plasmopara viticola on grapevine leaves after being treated with PvDCL1/2 double-stranded RNA (dsRNA). (A) Progress of already established P. viticola infection after receiving dsRNA treatments. Leaves were treated with 50 μl of water (ctrl) or dsRNA [75, 100, or 125 ng μl– 1 of dsRNA of BcDCL1/2 (BcDCL_75/100/125) or PvDCL1/2 (PvDCL_75/100/125)] 7 days after being inoculated with 7.5 μl of a 1 × 105 ml– 1 sporangia [i.e., 0 days post treatment (dpt) of dsRNA]. (B) Disease progress rate at 4 and 7 dpt, computed by taking leaf area covered by P. viticola at 7 dpi (0 dpt) as a reference. Bars are 95% confidence interval, and asterisks (∗) indicate statistically significant differences according to Tukey’s honestly significant difference test (P ≤ 0.05).
Compared to the preventive application, where all the three concentrations of PvDCL1/2 inhibited the growth of the pathogen significantly, when used as curative treatment, the rate of the pathogen growth was reduced significantly only at the highest concentration of PvDCL1/2 dsRNA. These data show that the exogenously applied dsRNA targeting PvDCL1/2 has both promising protective and curative effects.
Discussion
In grapevine cultivation, downy mildew, caused by P. viticola, is among the major diseases requiring repeated applications of pesticides within a growing season. In this study, we show that external application of long non-coding dsRNA, 515 bp long, targeting the two DCL genes of P. viticola reduced the progress of the pathogen on grapevine leaves. Transcript level reduction of the target genes, PvDCL1 and PvDCL2, suggests specific RNA silencing effect triggered by PvDCL1/2 dsRNA. To our knowledge, this is the first report showing the potential of exogenously applied RNAi molecules as an effective strategy for oomycete management in crops. The results presented further support the use of SIGS-based strategy for fungal pathogen management (Koch et al., 2016; Wang et al., 2016; McLoughlin et al., 2018; Nerva et al., 2020).
Non-coding sRNA molecules derived from plant pathogens could play a role in suppressing host immunity (Weiberg et al., 2013; Brilli et al., 2018) and hence could be regarded as additional classes of effectors, besides protein coding effector genes studied so far. It has been demonstrated that B. cinerea sRNAs (Bc-sRNAs) triggered the silencing of Arabidopsis and tomato targets involved in host immunity, such as mitogen-activated protein kinase 1 (MPK1), MPK2, peroxiredoxin, and cell wall-associated kinase genes. Once they have entered the plant cell, Bc-sRNAs hijack the host’s RNAi machinery, binding to Argonaute 1 (AGO1) protein and directing the silencing of host immunity genes (Weiberg et al., 2013). Accordingly, the ago1 mutant Arabidopsis exhibited reduced susceptibility to B. cinerea, and the expression of sRNAs that target B. cinerea DCL1 and DCL2 in Arabidopsis and tomato led to the silencing of the BcDCL genes and affected the fungal pathogenicity and growth, also when exogenously applied on different organs and tissues (Weiberg et al., 2013; Wang et al., 2016). In addition, dcl1 dcl2 B. cinerea double mutant, which is unable to produce sRNAs, displayed a stunted pathogenicity on several hosts (Weiberg et al., 2013; Wang et al., 2016). In a recent study, it was observed that during V. vinifera–P. viticola interaction, the sRNA profile of P. viticola showed enrichment in 21- and 25-nt sRNAs, which were also abundantly expressed in sporangia (Brilli et al., 2018). According to the study, the presence of DCLs, AGOs, and RNA-dependent RNA polymerase confirms the existence of RNA silencing machinery in P. viticola, which is active during its interaction with grapevine (Brilli et al., 2018).
The fact that the external application of PvDCL1/2 dsRNA extremely reduced the pathogenicity of P. viticola, coupled with the observed reduction in PvDCL1 and PvDCL2 transcript levels, might suggest that the pathogen can take up external dsRNA and that the RNAi machinery is active during the infection process. Similarly, reduced disease symptoms and sequence-specific silencing of target genes were also observed in B. cinerea, F. graminearum, and S. sclerotiorum (Koch et al., 2016; Wang et al., 2016; McLoughlin et al., 2018), following the external application of dsRNA.
Reduced pathogenicity of plant pathogens due to sRNA and dsRNA has put forward the considerations of RNAi-based technology as a new plant protection method, at least for those pathogens having bona fide RNA silencing machinery. In planta gene silencing of pathogen target genes, a mechanism known as HIGS, has also been reported (Nowara et al., 2010; Koch et al., 2013; Vega-Arreguin et al., 2014; Jahan et al., 2015; Zhu et al., 2017). Furthermore, for vegetatively propagated crops like grapevine, HIGS can be exploited to obtain RNAi-based rootstocks, which can produce sRNA able to move to a grafted untransformed scion and protect it from pathogen infection, as sRNAs have high mobility between shoot and root (Gouil and Lewsey, 2021; Li et al., 2021). In addition, in planta expressed RNAi sequences do not encode for protein products and are designed against specific genes of target pathogens or susceptibility factor without affecting other non-target organisms. All these features together could reduce data requirements for risk assessment of such RNAi-based plants (Limera et al., 2017; Arpaia et al., 2020).
In addition to the HIGS potential application, the results of this research confirm the potential of the gene silencing technology also to develop new RNAi-based fungicides, known as SIGS. To ensure sustainable food production, European Union and global sustainability policies emphasize the need to replace contentious pesticides with safe, efficient, and cost-effective alternatives (Taning et al., 2020). The high selectivity of RNAi-based products, due to sequence-specific modes of action, compared with other conventional pesticides, makes them a promising solution to substitute or reduce reliance on contentious pesticides. Yet there are still relevant aspects to be clarified, such as local and remote translocation and environmental stability of applied sRNAs, before pushing forward SIGS as an alternative solution to toxic pesticides. Despite many solutions reported to stabilize the RNA molecules and make their administration in the field easy and effective, more effort should be taken on the risk assessment studies in order to clarify the risks associated with the use of these molecule for the farmers, consumers, and environment and proceed with the necessary regulatory protocols in order for them to reach the market.
In this study, we demonstrated that dsRNA specifically designed to silence PvDCL1 and PvDCL2 genes efficiently controls downy mildew disease caused by P. viticola on grapevine, a disease that forces to consume significant amounts of pesticides that are applied every year on vineyards. Although the mechanism behind the uptake and transport of the externally applied dsRNA needs further studies, the presented data give important scientific information on such new-generation RNA-based fungicides, which are environmentally safe and sustainable. So far, externally applied RNAi-based disease suppression data are limited on plant pathogens from Ascomycetes, but with our findings, we extended the possibility of using externally applied dsRNA for managing devastating plant pathogen oomycetes like Phytophthora and Pythium species.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
ZH made the experiments and wrote the manuscript. DG made the cloning and plasmid constructs together with LC. BMo and FN contributed to the experiment to confirm gene silencing. MC helped with infections. SS and BMe provided support with the RNAi experiment and participated actively in manuscript writing. EB provided funding and general supervision to the experiments and writing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This publication is based upon work from COST Action iPlanta, supported by COST (European Cooperation in Science and Technology), www.cost.eu.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.667539/full#supplementary-material
Footnotes
References
Armijo, G., Schlechter, R., Agurto, M., Muñoz, D., Nuñez, C., and Arce-Johnson, P. (2016). Grapevine pathogenic microorganisms: understanding infection strategies and host response scenarios. Front. Plant Sci. 7:382. doi: 10.3389/fpls.2016.00382
Arpaia, S., Christiaens, O., Giddings, K., Jones, H., Mezzetti, B., Moronta-Barrios, F., et al. (2020). Biosafety of GM crop plants expressing dsRNA: data requirements and EU regulatory considerations. Front. Plant Sci. 11:940. doi: 10.3389/fpls.2020.00940
Brilli, M., Asquini, E., Moser, M., Bianchedi, P. L., Perazzolli, M., and Si-Ammour, A. (2018). A multi-omics study of the grapevine-downy mildew (Plasmoparaviticola) pathosystem unveils a complex protein coding- and noncoding-based arms race during infection. Sci. Rep. 8:757. doi: 10.1038/s41598-018-19158-8
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F. M., Palmquist, J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. doi: 10.1126/science.aar4142
Castel, S. E., and Martienssen, R. A. (2013). RNA interference in the nucleus: roles for small RNAs in transcription, epi- genetics and beyond. Nat. Rev. Genet. 14, 100–112. doi: 10.1038/nrg3355
Eurostat. (2007). The Use of Plant Protection Products in the European Union, Data 1992–2003. Luxembourg: Office for Official Publications of the European Communities.
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811.
Gambino, G., Perrone, I., and Gribaudo, I. (2008). A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 525, 520–525. doi: 10.1002/pca.1078
Gisi, U., and Sierotzki, H. (2008). Fungicide modes of action and resistance in downy mildews. Eur. J. Plant Pathol. 122, 157–167. doi: 10.1007/s10658-008-9290-5
Gouil, Q., and Lewsey, M. G. (2021). Small RNAs shoot for the root. Nat. Plants 7, 2–3. doi: 10.1038/s41477-020-00836-3
Hamilton, A. J., and Baulcombe, D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. doi: 10.1126/science.286.5441.950
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., and Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. doi: 10.1186/gb-2007-8-2-r19
Jahan, S. N., Åsman, A. K. M., Corcoran, P., Fogelqvist, J., Vetukuri, R. R., and Dixelius, C. (2015). Plant-mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans. J. Exp. Bot. 66, 2785–2794. doi: 10.1093/jxb/erv094
Koch, A., Biedenkopf, D., Furch, A., Weber, L., Rossbach, O., Abdellatef, E., et al. (2016). An RNAi-Based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 12:e1005901. doi: 10.1371/journal.ppat.1005901
Koch, A., Kumar, N., Weber, L., Keller, H., Imani, J., and Kogel, K. H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. U. S. A. 110, 19324–19329. doi: 10.1073/pnas.1306373110
Li, S., Wang, X., Xu, W., Liu, T., Cai, Ch, Chen, L., et al. (2021). Unidirectional movement of small RNAs from shoots to roots in interspecific heterografts. Nat. Plants 7, 50–59. doi: 10.1038/s41477-020-00829-2
Limera, C., Sabbadini, S., Sweet, J. B., and Mezzetti, B. (2017). New biotechnological tools for the genetic improvement of major moody fruit species. Front. Plant Sci. 8:1418. doi: 10.3389/fpls.2017.01418
McLoughlin, A. G., Wytinck, N., Walker, P. L., Girard, I. J., Rashid, K. Y., de Kievit, T., et al. (2018). Identification and ap- plication of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci. Rep. 8:7320. doi: 10.1038/s41598-018-25434-4
Nerva, L., Sandrini, M., Gambino, G., and Chittara, W. (2020). Double Stranded RNAs (dsRNAs) as a Sustainable Tool against Gray Mold (Botrytis cinerea) in Grapevine: effectiveness of different application methods in an open-air environment. Biomolecules 10:200. doi: 10.3390/biom10020200
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-Induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Rieu, I., and Powers, S. J. (2009). Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 2, 1031–1033. doi: 10.1105/tpc.109.066001
Ruijter, J. M., Ramakers, C., Hoogaars, W. M., Karlen, Y., Bakker, O., van den Hoff, M. J., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. doi: 10.1093/nar/gkp045
Taning, C. N. T., Mezzetti, B., Kleter, G., Smagghe, G., and Baraldi, E. (2020). Does RNAi-Based Technology Fit within EU Sustainability Goals? Trends Biotechnol. doi: 10.1016/j.tibtech.2020.11.008 [Epub Online ahead of print].
Tomilov, A. A., Tomilova, N. B., Wroblewski, T., Michelmore, R., and Yoder, J. I. (2008). Trans-specific gene silencing between host and parasitic plants. Plant J. 56, 389–397. doi: 10.1111/j.1365-313X.2008.03613.x
Vaucheret, H., and Fagard, M. (2001). Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17, 29–35. doi: 10.1016/S0168-9525(00)02166-1
Vega-Arreguin, J. C., Jalloh, A., Bos, J. I., and Moffett, P. (2014). Recognition of an Avr3a homologue plays a major role in mediating nonhost resistance to Phytophthora capsici in Nicotiana species. Mol. Plant Microbe Interact. 27, 770–780. doi: 10.1094/MPMI-01-14-0014-R
Wang, M., Weiberg, A., Lin, F. M., Thomma, B. P., Huang, H. D., and Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2:16151. doi: 10.1038/NPLANTS.2016.151
Weiberg, A., Bellinger, M., and Jin, H. (2015). Conversations between kingdoms: small RNAs. Curr.Opin. Biotechnol. 32, 207–215. doi: 10.1016/j.copbio.2014.12.025
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Keywords: Dicer-like genes, double-stranded RNA (dsRNA), Plasmopara viticola, spray-induced gene silencing, Vitis vinifera
Citation: Haile ZM, Gebremichael DE, Capriotti L, Molesini B, Negrini F, Collina M, Sabbadini S, Mezzetti B and Baraldi E (2021) Double-Stranded RNA Targeting Dicer-Like Genes Compromises the Pathogenicity of Plasmopara viticola on Grapevine. Front. Plant Sci. 12:667539. doi: 10.3389/fpls.2021.667539
Received: 13 February 2021; Accepted: 06 April 2021;
Published: 18 May 2021.
Edited by:
Andreia Figueiredo, University of Lisbon, PortugalReviewed by:
Atanas Ivanov Atanassov, Joint Genomic Center, BulgariaBasavaprabhu L. Patil, Indian Institute of Horticultural Research (ICAR), India
Copyright © 2021 Haile, Gebremichael, Capriotti, Molesini, Negrini, Collina, Sabbadini, Mezzetti and Baraldi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elena Baraldi, elena.baraldi@unibo.it
 Zeraye Mehari Haile
Zeraye Mehari Haile Daniel Endale Gebremichael
Daniel Endale Gebremichael Luca Capriotti
Luca Capriotti Barbara Molesini
Barbara Molesini Francesca Negrini
Francesca Negrini Marina Collina1
Marina Collina1 Silvia Sabbadini
Silvia Sabbadini Elena Baraldi
Elena Baraldi