- 1Department of Epidemiology and Environmental Health, School of Public Health and Health Professions, State University of New York (SUNY) at Buffalo, Buffalo, NY, United States
- 2Department of Epidemiology, Fielding School of Public Health, University of California at Los Angeles (UCLA), Los Angeles, CA, United States
- 3Shanxi Tumor Hospital, Taiyuan, Shanxi, China
- 4Taiyuan City Center for Disease Control and Prevention (CDC), Taiyuan, Shanxi, China
- 5School of Public Health, Fudan University, Shanghai, China
Background: The ataxia telangiectasia-mutated (ATM) gene has a key role in DNA repair including activation and stabilization of p53, which implicates the importance of ATM polymorphisms in the development of cancer. This study aims to investigate the association of two ATM single-nucleotide polymorphisms (SNPs) with lung cancer, as well as their potential interaction with p53 gene and other known risk factors of lung cancer.
Methods: A population-based case–control study was conducted in Taiyuan city, China with 399 cases and 466 controls matched on the distribution of age and sex of cases. The two ATM gene SNPs, ATMrs227060 and ATMrs228589 as well as p53 gene SNP, p53rs1042522 were genotyped using Sequenom platform. Unconditional logistic regression models were used to estimate crude and adjusted odds ratios (aOR) and 95% confidence intervals (CIs). Adjusted models controlled for age, sex, and smoking status.
Results: The study showed that TT genotype of ATMrs227060 (aOR = 1.58, 95% CI: 1.06–2.35) and AA genotype of ATMrs228589 were significantly associated with lung cancer (aOR = 1.50, 95% CI: 1.08–2.08) in a recessive model. Additionally, carrying variant genotypes of ATMrs227060 (TT), ATMrs228589 (AA), and p53rs1042522 (CC) concomitantly was associated with much higher risk (aOR = 3.68, 95% CI: 1.43–9.45) of lung cancer than carrying variant genotypes of any one of the above three SNPs. We also found multiplicative and additive interaction between tea drinking and ATMrs227060 in association with lung cancer.
Conclusion: This study indicates that ATM gene variants might be associated with development of lung cancer in Chinese population. These results need to be validated in larger and different population samples.
Introduction
Lung cancer has been increasing both in incidence and mortality in China (1, 2). Smoking and air pollution are major risk factors of lung cancer in China. However, the disproportion between incidence of lung cancer and prevalence of smoking indicates that genetic factors may play an important role in the development of lung cancer in Chinese population (3).
The ataxia telangiectasia-mutated (ATM) gene is a Ser/Thr protein kinase of the phosphoinositidyl 3-kinase family located on long arm of chromosome 11q22–q23 (4). It is a key gene involved in DNA repair, mainly in double-strand breaks (5). It is rapidly recruited to the sites of DNA damage where it modulates downstream effectors [e.g., p53, BRCA-1, checkpoint kinase 2 (CHK2), RAD50, and NBS1] through phosphorylation of protein kinases and substrates. Cells with mutated ATM gene lose the ability to cope with genotoxic stresses due to lack of a coordinated DNA damage response (DDR) and loss of regulation of cell-cycle damage checkpoints (6). The principal component of ATM-mediated DDR involves the tumor-suppressor protein p53. p53 gene regulates the transcription of other genes responsible for cellular antitumor responses such as cell-cycle arrest, apoptosis, senescence, genetic stability, and suppression of angiogenesis (7). Functional polymorphisms of p53 gene alter protein activity and have been shown to be associated with risk of development of several human cancers including lung cancer (8–11). ATM mediates post-translational modifications of p53 that lead to its activation and transcription of genes involved in cell-cycle arrest and apoptosis. In addition, modulation of proteins CHK2 and MDM2 (which inhibits p53 activation) results in rapid stabilization and accumulation of p53 at the DDR sites (12). More recent studies suggest a wider role for ATM apart from involvement in DDR functions such as response to other forms of genotoxic stress like oxidative stress and maintaining cellular homeostasis including various cellular signaling pathways, insulin signaling, and mitochondrial homeostasis (6, 12, 13). This further indicates that polymorphisms of the ATM gene may be involved in cancer initiation and progression through multiple pathways.
Ataxia telangiectasia-mutated gene polymorphisms have been associated with several cancers including breast, oral, cervical, and lung (14–22). Among the ATM single-nucleotide polymorphisms (SNPs), we identified two intron variant tagging SNPs with r2 > 0.6 and minor allele frequency >5% (MAF), ATMrs227060 (MAF: 32.3%) and ATMrs228589 (MAF: 46.1%) (23–25). While these SNPs showed positive associations with other cancers, limited studies examined their association with lung cancer with inconsistent results. In previous studies, ATMrs227060 was positively associated with non-Hodgkin’s lymphoma while ATMrs228589 showed positive association with breast cancer in Ashkenazi Jewish women as well as chronic myeloid leukemia (26–28). In relation to lung cancer, a Caucasian study showed positive association of ATMrs227060 with non-small cell lung cancer (NSCLC) recent studies in Taiwanese population did not find an association of either SNP with lung cancer (15, 22). In the current study, we investigated the association of these two ATM SNPs with lung cancer in a Chinese population. In addition, we explored potential interaction between the ATM gene SNPs and the most studied p53 gene SNP, p53rs1042522 in which, Arginine (Arg) is substituted by Proline (Pro) at codon 72 (Arg72Pro) (29). We also explored potential interaction between the ATM SNPs and smoking, alcohol intake, and tea drinking behaviors in association with lung cancer.
Materials and Methods
Study Population
A population-based case–control study was conducted in Taiyuan city, the capital of Shanxi Province, China. Incident cases were recruited between 2005 and 2007 from Shanxi Tumor Hospital, which catered to 70% of all cancer patients in Taiyuan city, Shanxi Province, China. Eligibility criteria for cases included age 20 years or older, residence in Taiyuan city for 10 years or more, medically stable condition and willingness to participate in the study. Controls were recruited from 13 randomly selected communities in Taiyuan to match cases according to distribution of age and gender. Eligible controls were aged 20 years or older, lived in Taiyuan city for 10 years or more and had no history of cancer or any other serious chronic disease. A total of 399 cases (89% response rate) and 466 controls (85% response rate) participated in the study. Detailed methodology for this population-based case–control study was previously published (30). Half of the participants were males and 56% of participants were aged 55 years or older. Around 47% of the participants were smokers, 25% of them reported alcohol intake, and 58% of them were current tea drinkers.
Data Collection
Participants were interviewed face to face by study personnel using a structured questionnaire regarding their demographic characteristics, smoking, alcohol drinking and tea drinking habits, diet during the past 1 year, cooking and indoor heating methods, and individual and family medical history. Ever smokers were defined as those with a lifetime exposure of at least 100 cigarettes.
Biospecimen Collection and Processing
Approximately 98% of cases and 99% of controls provided peripheral blood samples. Serum and blood clot were separated immediately and stored in a −80°C freezer. Genomic DNA was isolated for analysis using a modified phenol–chloroform method (31).
Primer Design and Genotyping
Genotyping for ATM (rs227060 and rs228589) and p53 (rs1042522) SNPs was performed using Sequenom platform (Sequenom, Inc., San Diego, CA, USA) in the Center for Genomics and Personalized Medicine Research at Wake Forest School of Medicine (Winston-Salem, NC, USA). Polymerase chain reaction (PCR) and extension primers were designed using MassARRAY Assay Design 3.1 software (Sequenom, Inc., San Diego, CA, USA). We followed the manufacturer’s iPLEX Application Guide (Sequenom, Inc., San Diego, CA, USA) in performing genotyping procedures. PCR reactions were carried out in 96-well plates. Two negative control (water) samples and two randomly selected replicated samples were randomly plated in each plate. The concordance rate (percentages of agreement) in quality control pairs was over 99%. Technicians were blinded to sample case/control status. The three SNPs conformed to Hardy–Weinberg equilibrium.
Statistical Analysis
We used unconditional logistic regression models to analyze allele and genotype associations (in codominant, dominant, and recessive models) of the ATM SNPs with lung cancer by estimating crude and adjusted odds ratios (aOR) and 95% confidence intervals (CIs). In addition, we estimated crude and adjusted associations of risk genotype combinations of ATMrs227060, ATMrs228589, and p53rs1042522 with lung cancer. Adjusted models were controlled for age, sex, and smoking status (never smokers/ever smokers) of the participants. We performed internal validation for the genetic effect using a bootstrap method to obtain bias corrected estimates based on 1,000 replication data sets created by random sampling with replacement. Stratified analyses were used to assess the association of the SNPs with lung cancer among subgroups of age, sex, smoking, alcohol intake, tea drinking, and histological subtypes of lung cancer. For histological subtypes, the cases in each subgroup were compared to overall controls. In the current study, we assessed additive and multiplicative interaction of the each ATM SNP with p53 SNP (gene–gene) as well as with other risk factors (gene–environment) like smoking, alcohol drinking, and tea drinking exposures. Multiplicative interaction was assessed by introducing a product term in logistic regression models. Additive interaction was assessed by estimating relative excess risk due to interaction (RERI). All analyses were performed using SAS 9.3 software. Microsoft Excel was used additionally for estimating RERI.
Results
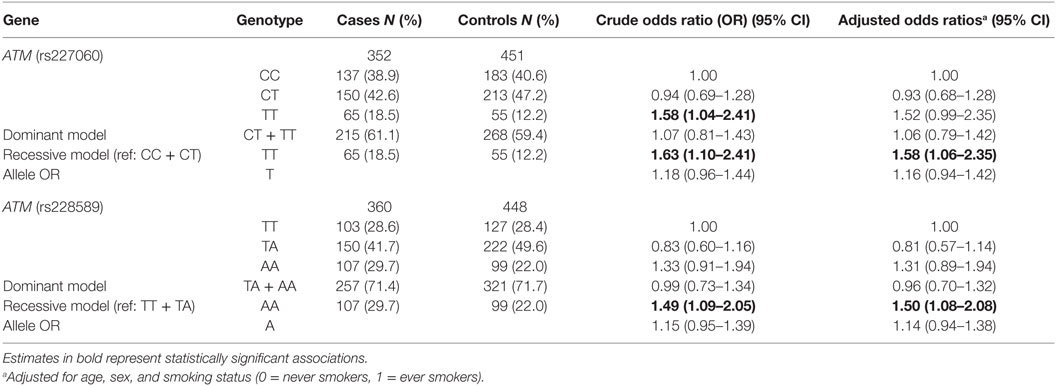
The description of sociodemographic and behavioral characteristics of cases and controls has been presented in detail in a previous publication (30). Briefly, a higher proportion of cases were heavy smokers and tea non-drinkers. Education level, BMI, and average household annual income 10 years prior were all higher among controls. Analysis of association of the ATM SNPs with lung cancer has been shown in Table 1. The frequency of C and T alleles of ATMrs227060 was 64.2 and 35.8% in controls and the frequency of T and A alleles ATMrs228589 was 53.1 and 46.9% in controls, respectively. Lung cancer was significantly associated with TT genotype of ATMrs227060 in a recessive model (aOR = 1.58, 95% CI: 1.06–2.35). AA genotype of ATMrs228589 was also significantly associated with lung cancer in a recessive model (aOR = 1.50, 95% CI: 1.08–2.08). Among the haplotypes identified from ATMrs227060 and ATMrs228589 (data not shown), TTAA haplotype was more frequent among cases compared to controls but failed to show an association with lung cancer (aOR = 1.53, 95% CI: 0.95–1.47). Internal validation using a bootstrap procedure with 1,000 replicated data sets confirmed the statistically significant association estimates between the ATM SNPs and lung cancer (results not shown).
Figure 1 shows the association of ATMrs227060 and ATMrs228589 with lung cancer among risk factor subgroups. ATMrs227060 (aOR = 2.23, 95% CI: 1.30–3.80) and ATMrs228589 (aOR = 1.65, 95% CI: 1.07–2.55) showed a significant association with lung cancer among tea non-drinkers. Among histological subtypes of lung cancer, ATMrs227060 was significantly associated with small cell carcinoma (SmCC) while ATMrs228589 was significantly associated with squamous cell carcinoma (SCC) and SmCC.
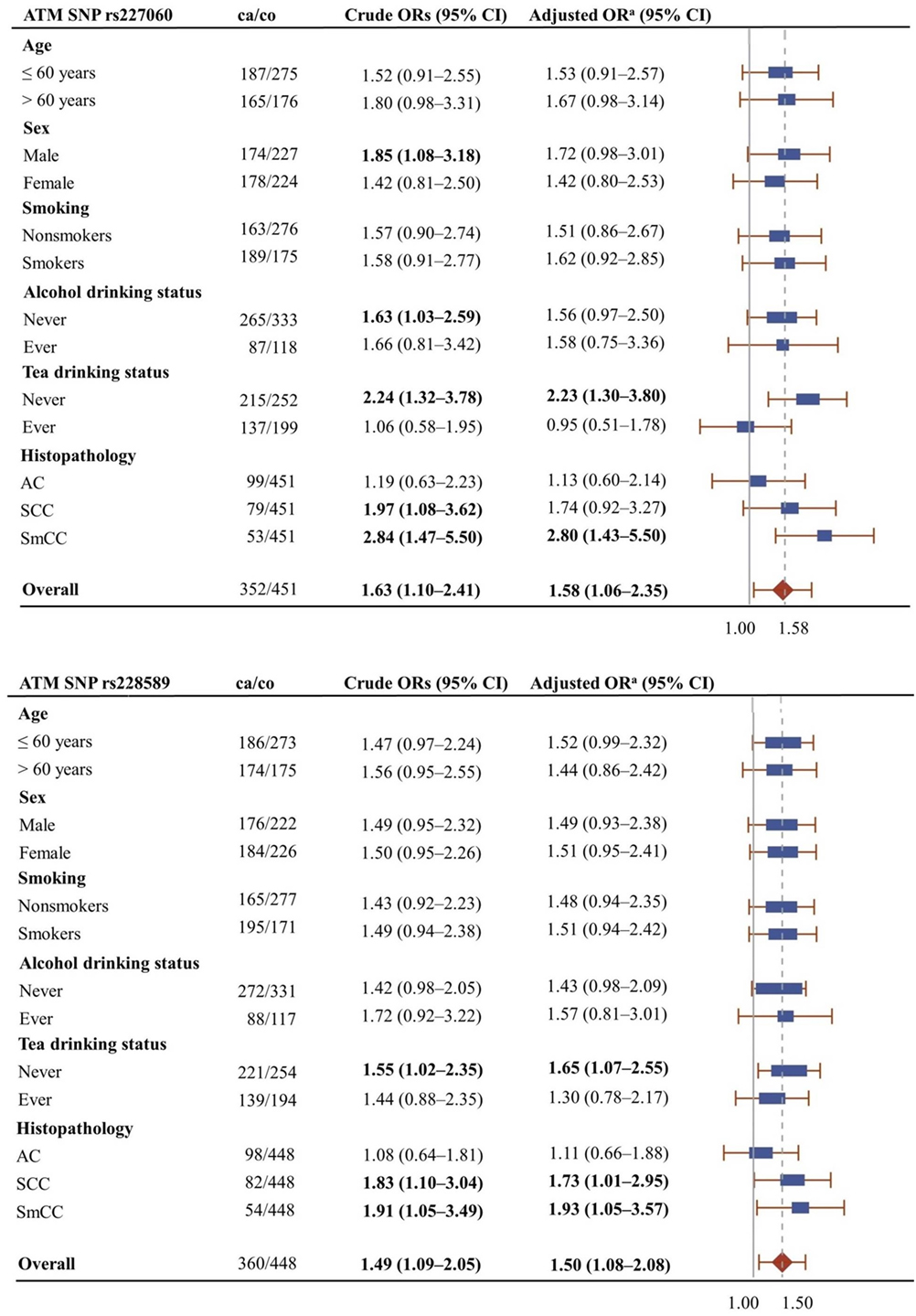
Figure 1. Forest plots showing the association of ATMrs227060 and ATMrs228589 with lung cancer among different subgroups. Plot shows adjusted odds ratios. aAdjusted for age, sex, and smoking status (0 = never smokers, 1 = ever smokers). ca/co, number of cases/controls; AC, adenocarcinoma; SCC, squamous cell carcinoma; SmCC, small cell carcinoma. The solid line in the plot represents null value and the broken line represents overall odds ratio (OR). Size of the square represents the inverse of the variance of the log OR. ORs in bold represent statistically significant associations.
A previous publication reported the association of p53rs1042522 SNP with lung cancer (32). Since the role of ATM gene in DNA repair closely relates to the function of p53 gene, in the current study, we analyzed the association of lung cancer with different genotype combinations of ATMrs227060, ATMrs228589, and p53rs1042522. The results are presented in Table 2, which shows a statistically significant association with lung cancer in presence of risk genotypes of all three SNPs (aOR = 3.68, 95% CI = 1.43–9.45). Test for linear trend showed statistical significance (ptrend = 0.002). Similar to the results in Table 1, internal validation using the bootstrap procedure described before confirmed the statistically significant associations in Table 2 (results not shown).
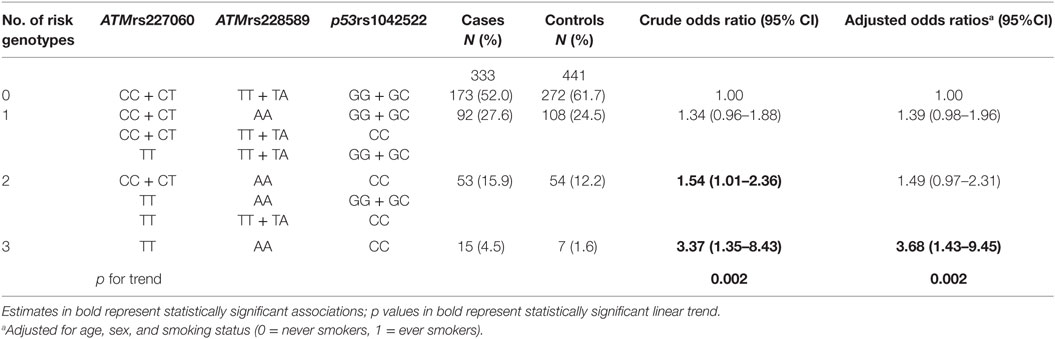
Table 2. Association of combined genotypes of ATMrs227060, ATMrs228589, and p53rs1042522 with lung cancer.
Based on the results from Table 2, we tested for gene–gene interaction between each of the ATM SNPs and the p53 SNP. We did not find statistically significant multiplicative or additive interaction between ATMrs227060–p531042522 and ATMrs228589–p531042522 (Table 3). We observed statistically significant mul-tiplicative and additive interaction between ATMrs227060 and tea drinking but not smoking and alcohol drinking in association with lung cancer. Smoking, alcohol drinking, or tea drinking did not modify the association between ATMrs228589 and lung cancer (Table 3).
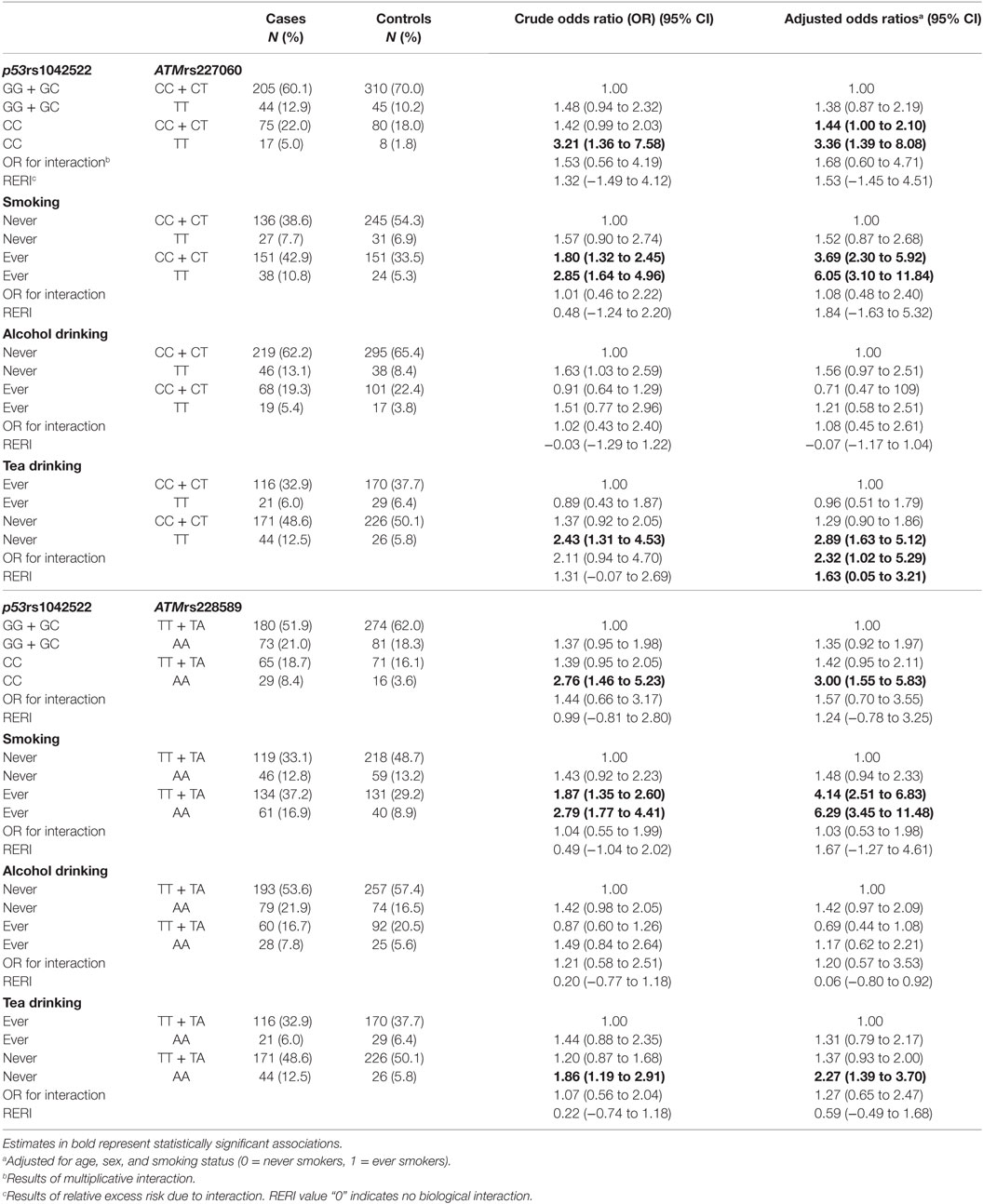
Table 3. Potential gene–gene and gene–environment interactions of ATM gene polymorphisms in association with lung cancer.
Discussion
In the current study, ATMrs227060 (TT) and ATMrs228589 (AA) were positively associated with lung cancer in recessive models. In addition, both showed positive association with lung cancer in tea non-drinkers and with SmCC subtype. Bonnen et al. earlier reported high linkage disequilibrium (LD) in the ATM gene locus (33). We verified this information from publicly available databases (HapMap and 1000 Genomes Project) for ATMrs227060 and ATMrs228589 (r2 range: 61.9–75.2) (23, 24) in Chinese population samples. This may explain the similar associations of the two ATM SNPs with lung cancer in our study. Both these SNPs may have a role in modifying the ATM gene function and consequently altering DNA repair mechanisms. A previous study suggested that ATMrs227060 may potentially regulate PI3 kinase activity because of its proximity to the ATM PI3 kinase domain (22). Another study showed that ATMrs228589 modulates DNA repair capacity of UV damaged DNA, which may be due to defects in ATM signaling (34). However, it is also possible that these tagging SNPs may not be the causal polymorphisms but merely represent other functional polymorphisms that are in high LD with the current SNPs [e.g., rs189037 (35, 36), rs652311 (15), rs170548 (22), rs664143 (16)].
In the presence of p53rs1042522 variant genotype (CC) in addition to variant genotypes of ATMrs227060 (TT) and ATMrs228589 (AA), the aOR for developing lung cancer was 3.68 (95% CI: 1.43–9.45), which was 2.65 times higher than with the presence of variant genotype of any one of the three SNPs (aOR = 1.39, 95% CI: 0.98–1.96). Analysis for linear trend indicated significant gene-dosage effects of the risk genotypes of the three SNPs in association with lung cancer. This suggests that the risk of development of lung cancer associated with polymorphisms in ATM gene variants is much higher in a presence of coexisting mutations in the p53 gene.
A recent case–control study in Taiwanese population (358 cases and 716 controls) examined the association of ATM polymorphisms, including ATMrs227060 and ATMrs228589 with lung cancer. The distribution of variant genotypes among controls for both the ATM SNPs [ATMrs227060 (TT: 14.3%) and ATMrs228589 (AA: 21.0%)] was similar to the controls in the current study [ATMrs227060 (TT: 12.2%) and ATMrs228589 (AA: 22.0%)]. However, the Taiwanese study did not find a difference in proportion of the variant genotypes of either ATMrs227060 or ATMrs228589 between lung cancer cases and healthy controls (15). We are uncertain whether differences in participant selection or other additional factors contributed to the variation in findings between the two studies. Another study in a Caucasian population investigated the association of ATM SNPs with NSCLC (556 cases and 556 controls). The allele frequencies of the variant genotypes of the two ATM SNPs in the Caucasian study were different from the current study [ATMrs227060 (TT: 7.6%) and ATMrs228589 (AA: 31.5%)]. The Caucasian study reported a significant association between ATMrs227060 (C>T) and lung cancer (OR = 1.55, 95% CI: 1.02–2.35) after controlling for age, gender, and smoking status, which was similar to findings in the current study for ATMrs227060 (C>T) (OR = 1.58, 95% CI: 1.06–2.35). No significant association was reported for ATMrs228589 in association with NSCLC in the Caucasian study (22). All three existing studies including the current study that investigated the association of the ATM SNPs (rs227060 and rs228589) and lung cancer had relatively small sample size. This may have contributed to the inconsistency in findings between the studies, requiring further evaluation of these associations in larger and different populations.
In the stratified analysis, the association between ATMrs227060 or ATMrs228589 and lung cancer in our study population was similar among smokers and non-smokers as evidenced by the slightly elevated ORs of similar magnitude in these subgroups. These results were different from the Caucasian study, which reported a significant association of ATMrs227060 only among former smokers (22). In the current study, ATMrs227060 was significantly associated with lung cancer among tea non-drinkers (aOR = 2.23, 95% CI: 1.30–3.80) but not among tea ever drinkers (aOR = 0.95, 95% CI: 0.51–1.78). Tea is a popular beverage in China. In our study population, 42% of participants currently/formerly consumed tea. Tea drinking was found to be significantly protective against lung cancer in current drinkers (aOR = 0.54, 95% CI: 0.37–0.80; results not shown) in our study population. The flavins and catechins—mainly epigallocatechin gallate—in tea, especially green tea, contribute to its antitumor properties such as inhibiting oxidative damage, modulating cell signaling pathways to prevent tumorigenesis, and promoting apoptosis and cell-cycle arrest in cancer cells (37). In our exploratory interaction analysis, we observed statistically significant multiplicative and additive interaction between ATMrs227060 and tea drinking in association with lung cancer where carrying the variant genotype ATMrs227060TT and not consuming tea was associated with a higher risk of lung cancer compared to carrying ATMrs227060TT or not drinking tea alone. These results suggest that the association between ATM SNPs and lung cancer may be modified by tea drinking. However, given the small sample size among the tea drinking subgroups, the results we observed might be due to chance. When stratified by histological type of lung cancer, we observed that both ATMrs227060 and ATMrs228589 were significantly associated with SCC (a subtype of NSCLC) and SmCC. We did not find statistically significant additive or multiplicative interaction between the either of the ATM SNPs and smoking or alcohol drinking factors.
Ataxia telangiectasia-mutated gene serves as a crucial player in DDR mainly by regulating the stabilization and activation of p53 (12, 38). Activated p53 modulates the expression of genes involved in either activation of cell-cycle checkpoints or apoptosis (39). Cell-cycle arrest provides time to repair damaged DNA and if the damage is irreparable, p53 promotes apoptosis (12). Mutations in the ATM gene could disrupt this finely regulated process and cause persistent and unrepaired DNA damage. This damage leads to chromosomal aberrations, which increase the risk of cancer development. In vitro studies have reported the importance of ATM-mediated phosphorylation in IR-induced checkpoint activation (40). Previous studies reported higher genetic instability and lower ATM protein expression in lung cancer patients compared to controls (41). Our results support the experimental study findings on the role of ATM SNPs in development of lung cancer especially in individuals with homozygous variant alleles.
A limitation of our study is the relatively small sample size. In particular, the association of ATM haplotypes and ATM SNPs with lung cancer among risk factor strata may have been affected by the smaller subgroup samples and these results should be interpreted cautiously. However, the associations that we found in our study contribute to the understanding of the potential role of ATM gene polymorphisms in the development of lung cancer and justify the need to pursue validation in larger studies.
In conclusion, our results indicate that having variant genotypes of ATMrs227060 (C>T) and ATMrs228589 (T>A) are independently associated with increased risk of developing lung cancer in a Chinese population. Further, carriers of variant genotypes of ATMrs227060, ATMrs228589, and p53rs1042522 (G>C) have a significantly increased risk for lung cancer compared to carriers of variant genotypes of the SNPs individually.
Ethics Statement
Before initiation of participant recruitment, IRB approvals were obtained from Fudan University (IRB#04-10-0022) and UCLA (IRB#11-003153), respectively. A written informed consent was obtained from each participant.
Author Contributions
LM: design and implementation of the study as well as supervision of analysis and manuscript preparation. AM: data analysis and manuscript preparation. S-CC: sample processing, genotyping, and editing the manuscript. RN, LL, BZ, JS, XH, LM: participant recruitment and data collection. JL, JS, and LL: monitoring data collection and study implementation. SY: study implementation. Z-FZ: study design and implementation.
Conflict of Interest Statement
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported in part by the National Nature Science Foundation of China grant award to LM (NSFC-30500417). The work is also partially supported by NIH grants (ES06718, CA09142, DA11386) and the Alper Research Center for Environmental Genomics of the UCLA Jonsson Comprehensive Cancer Center.
References
1. Chen W, Zhang S, Zou X. Evaluation on the incidence, mortality and tendency of lung cancer in China. Thorac Cancer (2010) 1(1):35–40. doi: 10.1111/j.1759-7714.2010.00011.x
2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer (2010) 127(12):2893–917. doi:10.1002/ijc.25516
3. Lam WK, White NW, Chan-Yeung MM. Lung cancer epidemiology and risk factors in Asia and Africa. Int J Tuberc Lung Dis (2004) 8(9):1045–57.
4. Abraham RT. PI 3-kinase related kinases: ’big’ players in stress-induced signaling pathways. DNA Repair (2004) 3(8–9):883–7. doi:10.1016/j.dnarep.2004.04.002
5. Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer (2003) 3(3):155–68. doi:10.1038/nrc1011
6. Ambrose M, Gatti RA. Pathogenesis of ataxia-telangiectasia: the next generation of ATM functions. Blood (2013) 121(20):4036–45. doi:10.1182/blood-2012-09-456897
7. Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer (2009) 9(2):95–107. doi:10.1038/nrc2584
8. Bellini MF, Cadamuro AC, Succi M, Proenca MA, Silva AE. Alterations of the TP53 gene in gastric and esophageal carcinogenesis. J Biomed Biotechnol (2012) 2012:891961. doi:10.1155/2012/891961
9. Francisco G, Menezes PR, Eluf-Neto J, Chammas R. Arg72Pro TP53 polymorphism and cancer susceptibility: a comprehensive meta-analysis of 302 case-control studies. Int J Cancer (2011) 129(4):920–30. doi:10.1002/ijc.25710
10. Grochola LF, Zeron-Medina J, Meriaux S, Bond GL. Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol (2010) 2(5):a001032. doi:10.1101/cshperspect.a001032
11. Yan L, Zhang D, Chen C, Mao Y, Xie Y, Li Y, et al. TP53 Arg72Pro polymorphism and lung cancer risk: a meta-analysis. Int J Cancer (2009) 125(12):2903–11. doi:10.1002/ijc.24603
12. Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol (2013) 14(4):197–210. doi:10.1038/nrm3546
13. Canman CE, Lim DS. The role of ATM in DNA damage responses and cancer. Oncogene (1998) 17(25):3301–8. doi:10.1038/sj.onc.1202577
14. Bau DT, Chang CH, Tsai MH, Chiu CF, Tsou YA, Wang RF, et al. Association between DNA repair gene ATM polymorphisms and oral cancer susceptibility. Laryngoscope (2010) 120(12):2417–22. doi:10.1002/lary.21009
15. Hsia TC, Tsai CW, Liang SJ, Chang WS, Lin LY, Chen WC, et al. Effects of ataxia telangiectasia mutated (ATM) genotypes and smoking habits on lung cancer risk in Taiwan. Anticancer Res (2013) 33(9):4067–71.
16. Kim JH, Kim H, Lee KY, Choe KH, Ryu JS, Yoon HI, et al. Genetic polymorphisms of ataxia telangiectasia mutated affect lung cancer risk. Hum Mol Genet (2006) 15(7):1181–6. doi:10.1093/hmg/ddl033
17. Lee KM, Choi JY, Park SK, Chung HW, Ahn B, Yoo KY, et al. Genetic polymorphisms of ataxia telangiectasia mutated and breast cancer risk. Cancer Epidemiol Biomarkers Prev (2005) 14(4):821–5. doi:10.1158/1055-9965.EPI-04-0330
18. Lo YL, Hsiao CF, Jou YS, Chang GC, Tsai YH, Su WC, et al. ATM polymorphisms and risk of lung cancer among never smokers. Lung Cancer (2010) 69(2):148–54. doi:10.1016/j.lungcan.2009.11.007
19. Oliveira S, Ribeiro J, Sousa H, Pinto D, Baldaque I, Medeiros R. Genetic polymorphisms and cervical cancer development: ATM G5557A and p53bp1 C1236G. Oncol Rep (2012) 27(4):1188–92. doi:10.3892/or.2011.1609
20. Wang CH, Wu KH, Yang YL, Peng CT, Tsai FJ, Lin DT, et al. Association between ataxia telangiectasia mutated gene polymorphisms and childhood leukemia in Taiwan. Chin J Physiol (2011) 54(6):413–8. doi:10.4077/CJP.2011.AMM106
21. Wang HC, Chang WS, Tsai RY, Tsai CW, Liu LC, Su CH, et al. Association between ataxia telangiectasia mutated gene polymorphisms and breast cancer in Taiwanese females. Anticancer Res (2010) 30(12):5217–21.
22. Yang H, Spitz MR, Stewart DJ, Lu C, Gorlov IP, Wu X. ATM sequence variants associate with susceptibility to non-small cell lung cancer. Int J Cancer (2007) 121(10):2254–9. doi:10.1002/ijc.22918
23. Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics (2008) 24(24):2938–9. doi:10.1093/bioinformatics/btn564
24. Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics (2015) 31(21):3555–7. doi:10.1093/bioinformatics/btv402
25. Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res (2001) 29(1):308–11. doi:10.1093/nar/29.1.308
26. Gorre M, Mohandas PE, Kagita S, Cingeetham A, Vuree S, Jarjapu S, et al. Significance of ATM gene polymorphisms in chronic myeloid leukemia – a case control study from India. Asian Pac J Cancer Prev (2016) 17(2):815–21. doi:10.7314/APJCP.2016.17.2.815
27. Koren M, Kimmel G, Ben-Asher E, Gal I, Papa MZ, Beckmann JS, et al. ATM haplotypes and breast cancer risk in Jewish high-risk women. Br J Cancer (2006) 94(10):1537–43. doi:10.1038/sj.bjc.6603062
28. Rendleman J, Antipin Y, Reva B, Adaniel C, Przybylo JA, Dutra-Clarke A, et al. Genetic variation in DNA repair pathways and risk of non-Hodgkin’s lymphoma. PLoS One (2014) 9(7):e101685. doi:10.1371/journal.pone.0101685
29. Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol (1987) 7(2):961–3. doi:10.1128/MCB.7.2.961
30. Myneni AA, Chang SC, Niu R, Liu L, Ochs-Balcom HM, Li Y, et al. Genetic polymorphisms of TERT and CLPTM1L and risk of lung cancer – a case-control study in a Chinese population. Lung Cancer (2013) 80(2):131–7. doi:10.1016/j.lungcan.2013.01.021
31. Cao W, Hashibe M, Rao JY, Morgenstern H, Zhang ZF. Comparison of methods for DNA extraction from paraffin-embedded tissues and buccal cells. Cancer Detect Prev (2003) 27(5):397–404. doi:10.1016/S0361-090X(03)00103-X
32. Li Y, Chang SC, Niu R, Liu L, Crabtree-Ide CR, Zhao B, et al. TP53 genetic polymorphisms, interactions with lifestyle factors and lung cancer risk: a case control study in a Chinese population. BMC Cancer (2013) 13:607. doi:10.1186/1471-2407-13-607
33. Bonnen PE, Story MD, Ashorn CL, Buchholz TA, Weil MM, Nelson DL. Haplotypes at ATM identify coding-sequence variation and indicate a region of extensive linkage disequilibrium. Am J Hum Genet (2000) 67(6):1437–51. doi:10.1086/316908
34. Shin A, Lee KM, Ahn B, Park CG, Noh SK, Park DY, et al. Genotype-phenotype relationship between DNA repair gene genetic polymorphisms and DNA repair capacity. Asian Pac J Cancer Prev (2008) 9(3):501–5.
35. Liu J, Wang X, Ren Y, Li X, Zhang X, Zhou B. Effect of single nucleotide polymorphism Rs189037 in ATM gene on risk of lung cancer in Chinese: a case-control study. PLoS One (2014) 9(12):e115845. doi:10.1371/journal.pone.0115845
36. Shen L, Yin ZH, Wan Y, Zhang Y, Li K, Zhou BS. Association between ATM polymorphisms and cancer risk: a meta-analysis. Mol Biol Rep (2012) 39(5):5719–25. doi:10.1007/s11033-011-1381-2
37. Singh BN, Shankar S, Srivastava RK. Green tea catechin, epigallocatechin-3-gallate (EGCG): mechanisms, perspectives and clinical applications. Biochem Pharmacol (2011) 82(12):1807–21. doi:10.1016/j.bcp.2011.07.093
38. Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science (1998) 281(5383):1674–7. doi:10.1126/science.281.5383.1674
39. Sullivan KD, Gallant-Behm CL, Henry RE, Fraikin JL, Espinosa JM. The p53 circuit board. Biochim Biophys Acta (2012) 1825(2):229–44. doi:10.1016/j.bbcan.2012.01.004
40. Chen MJ, Lin YT, Lieberman HB, Chen G, Lee EY. ATM-dependent phosphorylation of human Rad9 is required for ionizing radiation-induced checkpoint activation. J Biol Chem (2001) 276(19):16580–6. doi:10.1074/jbc.M008871200
Keywords: lung cancer, ataxia telangiectasia-mutated gene, DNA repair, single-nucleotide polymorphisms, Chinese population
Citation: Myneni AA, Chang S-C, Niu R, Liu L, Zhao B, Shi J, Han X, Li J, Su J, Yu S, Zhang Z-F and Mu L (2017) Ataxia Telangiectasia-Mutated (ATM) Polymorphisms and Risk of Lung Cancer in a Chinese Population. Front. Public Health 5:102. doi: 10.3389/fpubh.2017.00102
Received: 31 December 2016; Accepted: 18 April 2017;
Published: 08 June 2017
Edited by:
Jimmy Thomas Efird, East Carolina University, United StatesReviewed by:
Yesilda Balavarca, Nationales Centrum für Tumorerkrankungen, GermanyMin Shi, National Institute of Environmental Health Sciences (NIH), United States
Copyright: © 2017 Myneni, Chang, Niu, Liu, Zhao, Shi, Han, Li, Su, Yu, Zhang and Mu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lina Mu, linamu@buffalo.edu
 Ajay A. Myneni
Ajay A. Myneni Shen-Chih Chang
Shen-Chih Chang Rungui Niu3
Rungui Niu3 Zuo-Feng Zhang
Zuo-Feng Zhang Lina Mu
Lina Mu