- 1Chest Research Foundation, Pune, India
- 2Vadu Rural Health Program, KEM Hospital Research Centre, Pune, India
- 3Microbial Culture Collection, National Centre for Cell Science, Pune, India
- 4INDEPTH Network, Accra, Ghana
Health and demographic surveillance system (HDSS) is a population-based health and vital event registration system that monitors demographic and health events in a geographically defined population at regular intervals. Human microbiome research in the past decade has been the field of increasingly intense research much due to its demonstrated impact upon various health conditions including human chronic airway diseases such as asthma and chronic obstructive pulmonary disease (COPD). Many confounding factors have been revealed to play a role in shaping the microbiome in chronic airway diseases. Asthma and COPD follows a typical pattern of disease progression, which includes stable and exacerbation state in which the microbiota is known to vary. However, many such studies lack extensive and longitudinal sampling with inadequate metadata, which has resulted in the inconsistencies in the observations. HDSS provides such a platform, which can offer a deeper understanding of the role of the microbiome in human health. In this review, we highlight opportunities and limitations in microbiome research with the help of studies conducted on chronic airway diseases like asthma and COPD. In addition, we also emphasize on the benefits of HDSS and future directions in lung microbiome research.
Introduction
Health and demographic surveillance system (HDSS) is a population-based health and vital event registration system that collects and monitors demographic and health events information in a geographically defined population at regular intervals (1). HDSS is usually comprised of a large team of qualified professionals and volunteers involved in large-scale data collection, monitoring, analysis, and report generation in a specific population for a long period of time. HDSS sites, because of their reliable information, can help policy makers to prioritize health issues and accordingly allocate resources more efficiently. It can act as a platform for intervention studies and provides feedback on programs’ effectiveness, which consequently helps in improving policies (2). INDEPTH is a global network of HDSS sites comprising of 42 member centers, which observe through 47 HDSS field sites (http://www.indepth-network.org/). These sites monitor life events of over three million individuals in 18 low and middle-income countries from Africa, Asia, and Oceania (3). Unlike cohort studies, HDSS follows the entire population in a defined geographic area for an ongoing period of time. During its initial census, HDSS registers all households and individuals who are then followed by frequent documentation of events such as births, deaths, marriages, and migrations in subsequent census rounds. Furthermore, information pertaining to health, social, and economic aspects of the population under surveillance is also collected, which eventually serves as a comprehensive metadata for any study on human populations. As a result, HDSS provides a platform for various research activities and offers a ready-made sampling frame for tracking the population. Apart from demographic information, any other information relevant to the particular study can be easily augmented and linked with other datasets in HDSS program (4).
We are increasingly able to understand that microbes contribute in many aspects of human health and physiology. Many intrinsic and extrinsic factors like mode of birth, infant feeding patterns, antibiotic usage, sanitary habits, age, genetics, diet, geography, socioeconomic status, and disease condition have been shown to alter the microbiome, which in turn affects human health (5–9). Extensive studies have been conducted across the globe focusing on the influence of microbiome in chronic diseases like asthma, chronic obstructive pulmonary disease (COPD), inflammatory bowel disease, obesity, diabetes, and cardiovascular diseases (10–15). Apart from genetics, many environmental factors are known to be associated with these diseases (16). Based on recent studies of the microbiome, it is clear that a robust research framework accompanied by a collection of comprehensive metadata is essential in building our knowledge of this tripartite relationship between humans, environment, and the microbes.
Asthma and COPD are common chronic airway diseases with high prevalence and mortality rates across the world (17, 18). Although several studies have linked various factors such as genetic predisposition, smoking, air pollution, and occupational exposures to these diseases; microbes are also known to play a vital role (19–22). In the past decade, the discovery of the human respiratory tract microbiome has steered growing number of studies to understand the role of the microbiome in asthma and COPD (23). Understanding why and how these diseases develop and progress over time requires long-term follow-up. Many cohorts have been established worldwide, such as ECLIPSE, CanCOLD, Birmingham, MicroCOPD (24–27). Although advancements have been made regarding descriptions of the main characteristics of chronic airway diseases, much is still to be explored about the factors associated with changes in the microbiome and the relevance of these changes in host-specific pathogenesis of the disease (23).
In this review, we discuss microbiome studies on chronic airway diseases like asthma and COPD to understand how HDSS can aid in human microbiome research. In addition to this, we also highlight opportunities and limitations in human microbiome research and also show how HDSS can benefit this field in future.
Microbiome in Asthma
Many factors, both environmental and genetic are responsible for the onset of asthma but no single cause has been identified so far. Although many studies including a study by Hilty et al. suggests a role for the microbiome in the etiology of asthma (28), the association is still not clearly understood (29). Evidence of colonization or infection with specific microbial species has been linked with the development or presence of asthma (30). The existence of abundant and diverse microbiota in airways suggests that they can modulate airway function and immunological responses (31, 32). In this context, members of phylum Proteobacteria like Haemophilus spp., Tropheryma whipplei, Moraxella catarrhalis, and Neisseria spp. have shown a strong correlation between the presence of asthma and its exacerbations (28, 33, 34). Genus Pseudomonas and Lactobacillus of phylum Proteobacteria and Firmicutes, respectively, dominate the oropharynx in asthmatics (35).
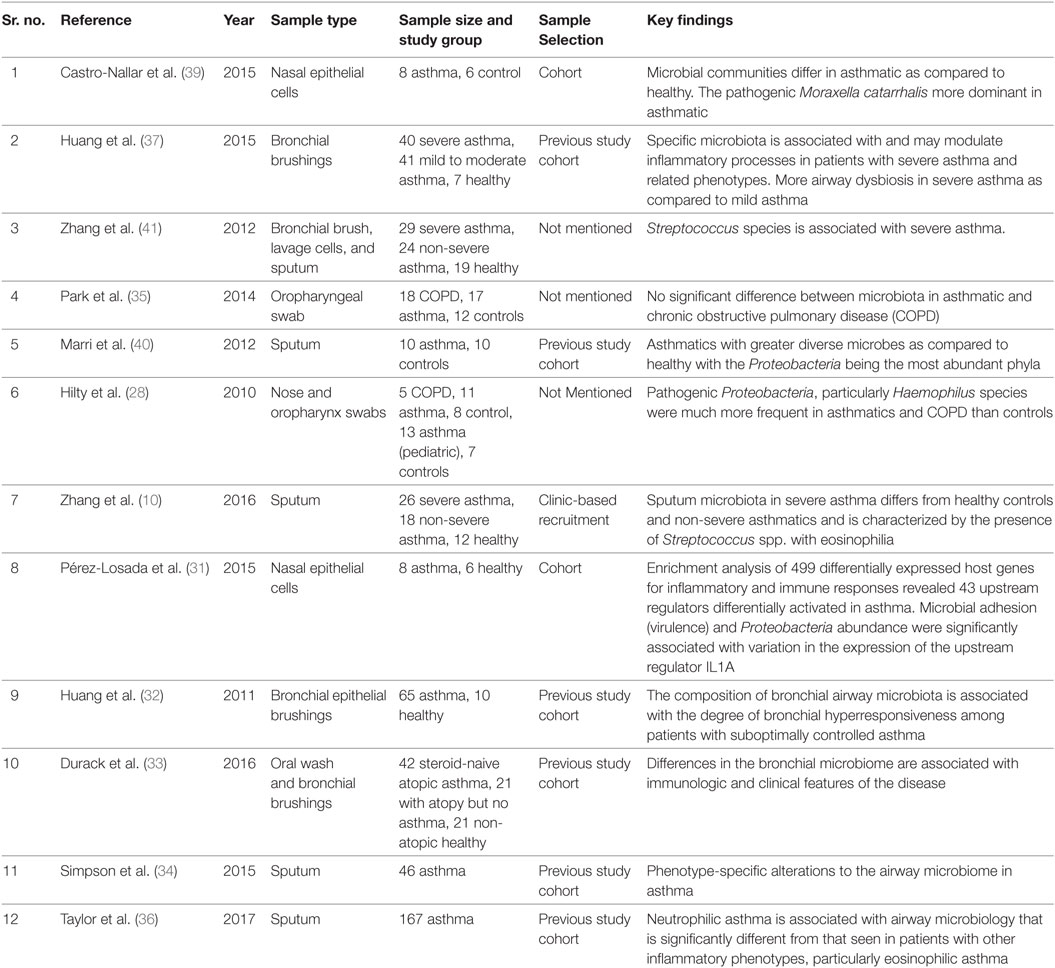
Recently, it has been reported that specific bacteria are associated with different stages of asthma severity and phenotypes (33, 36, 37). Castro-Nallar et al. and Depner et al. have shown that nasal microbial communities are different in asthmatic children compared to healthy children and pathogenic M. catarrhalis are present in abundance in asthmatic children (38, 39). Similarly, Marri et al. found higher diversity among microbial communities in asthma compared to healthy with Proteobacteria being the most abundant phyla but showed similar microbial composition in mild and severe asthma (40). Another study in asthma and COPD have shown the statistically non-significant difference in overall airway microbiota between two groups. But they found that Pseudomonas spp. and Lactobacillus spp. were highly abundant in both groups (35). Zhang et al. have shown that the presence of Streptococcus spp. in the airway is associated with severe asthma (41). Huang et al. revealed that specific bacteria are associated with asthma severity and observed airway microbial dysbiosis in subjects with severe asthma as compared to mild asthma (37). Summary of bacterial groups found to be altered and factors known to be associated with microbial dysbiosis in asthma subjects is given in Figure 1.
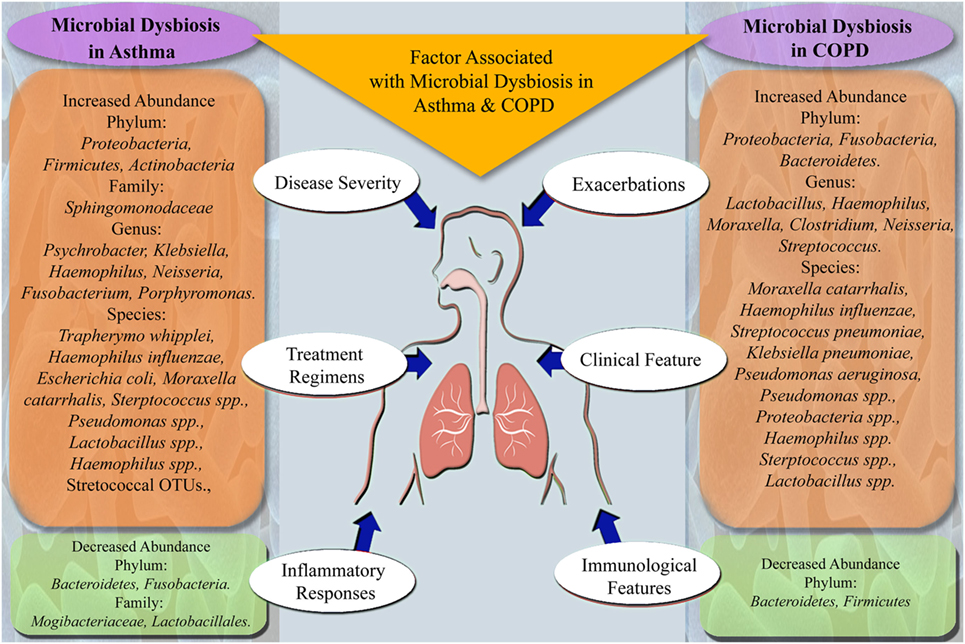
Figure 1. Summary of microbes altered in microbial dysbiosis and factors associated with microbial dysbiosis in asthma and chronic obstructive pulmonary disease (COPD) subjects.
In spite of the relatively large body of epidemiologic literature on the association of microbiome and asthma, the evidence overall remains inconclusive (19). It was learned from above studies that it is difficult to discriminate between resident and transient microbes, hence studies discussed here do not give the real picture of the microbes associated with a disease condition. This could be because many intrinsic and extrinsic factors are known to play a key role in shaping the microbiome, which necessitates collection and analysis of relevant metadata to understand their effect on the microbiome.
Microbiome in COPD
Various risk factors are associated with COPD, including tobacco smoke, household air pollution, occupational exposure, etc. However, the role of microbial infection in COPD exacerbations is debated (29). Like asthma, the role of the microbiome in COPD was first shown by Hilty et al. (28). Among the most studied phylum, Proteobacteria, Haemophilus spp. are frequently found in bronchi of COPD subjects and members of phylum Bacteroidetes are more commonly found in healthy airways (42, 43). Previous studies do not suggest any correlation of conventionally known bacterial pathogens with COPD exacerbations. However, a study by Aguirre et al. identified pathogenic genus Haemophilus and Moraxella in COPD, which could not be detected previously by culture-dependent approach (44).
Erb-downward et al. suggested lower microbial diversity in patients with decreased lung function (45). Similarly, Garcia-Nuñez et al. showed that decrease in microbial diversity is associated with disease severity in COPD (46). On the contrary, Pragman et al. indicated that there is a significant increase in microbial diversity with the development of COPD (47). The latest study also mentioned that age is associated with increased microbial diversity in COPD, unlike the previous studies where age factor was not considered. Erb-downward et al. showed that in never-smokers, smokers, and COPD, microbiota is similar; however, it differs at microanatomic sites in lungs (45). Similarly, Cabrera-Rubio et al. showed the existence of different microbiota in the upper and lower compartments of the respiratory tract in COPD subjects (48). Pragman et al. revealed that anaerobic bacteria from the phylum Fusobacteria and Bacteroidetes and genus Clostridium are present in the air-rich environment of lungs of COPD subjects (47). Zakharkina et al. showed the varying prevalence of various genera such as Moraxella, Neisseria, etc., in COPD and healthy individuals (49). Sze et al. mentioned that there is a detectable bacterial community within lung tissue, which is different in subjects with very severe COPD as compared to healthy (50). Wu et al. showed a significant increase in abundance of Streptococcus pneumoniae, Klebsiella pneumoniae, and Pseudomonas aeruginosa in COPD as compared to healthy (51). Recently, this was further confirmed by Garcia-Nuñez et al. that potentially pathogenic microorganisms other than P. aeruginosa can be the cause of exacerbations in COPD, and hence antibiotic treatments against the activity of other pathogens need to be considered while treating COPD (52).
In addition to microbial profiling, a study by Lee et al. on metatranscriptomic profiles in COPD patients showed inconsistencies with the results obtained by microbial profiling (53). Furthermore, functional metagenomics of lower and upper respiratory tract microbiome in COPD patients showed the role of the microbiome in disease pathogenesis (54, 55). Studies have shown that exacerbations of COPD are associated with changes in the microbiome with an increase in specific taxa, which are related to typical COPD pathogens and decrease in microbes that contribute to compositional and functional homeostasis (56–58). Figure 1 summarizes the microbial taxa and factors found to be associated with microbial dysbiosis in COPD subjects.
Although adequate research has been carried out in COPD as compared to asthma, these studies also lack consistency in the results. Along with longitudinal studies and inclusion of comprehensive metadata, studies on genetically related and unrelated populations are also essential in these diseases as genetic factors have also shown to play a role in the development of COPD (22). Although disease-specific cohorts are available in COPD, no comprehensive longitudinal study considering genetic factors and other related metadata is available.
Current Methodology for Lung Microbiome Research
Along with the culture-based techniques, many other approaches such as cloning, DGGE, PhyloChip microarrays, and T-RFLP were used to study the microbial communities in the human body (32, 37, 38, 42, 49–51, 57, 59). However, in the past decade, the field of microbial identification has changed enormously due to advancement in metagenomics research with the substantial support of the sequencing technologies. The majority of the studies conducted in asthma and COPD population explored microbiota by amplifying 16S rRNA gene by PCR followed by high throughput sequencing (10, 11, 33–36, 40, 43–48, 52, 55, 56, 60–62). These studies provided profiling of microbiota in the diseased condition. However, a study also performed predictive functional analysis based on the 16S rRNA gene sequences (63). In some studies, the bacterial burden was quantified with quantitative PCR based on the 16S rRNA gene (34, 36, 50, 51, 60, 64). Metagenomics studies involving transcriptomic and metatranscriptomic of both host and microbiota were also performed to understand the functional changes associated with asthma and COPD development (31, 39, 53–55, 58). These modern techniques have advanced the human microbiome research substantially.
Opportunities and Limitations in Microbiome Studies for Asthma and COPD
The human microbiome research has expounded the important role of microbes in chronic diseases, including infectious diseases by exploring the functional capacities of microbes. It is clear from the previous studies that many factors are associated with changes in the microbiome like the severity of the disease, age, sampling site and size, treatment type, and antibiotic usage. Together with a disease condition, temporal studies considering these factors will help in better understanding of changes in microbial communities in asthma and COPD. Along with this, many other unexplored factors like occupational hazards, geographical location, hygiene and sanitation, social habits, and dietary habits also have a role to play in shaping the microbiome. Proper gathering of information of all the confounders associated with these chronic diseases is necessary to get deeper insights in human microbiome, but this is often a challenge because of lack of robust metadata and set up for longitudinal follow-up of the population.
Asthma and COPD follows a typical pattern of disease progression, which includes stable and exacerbation state in which the microbiota is also shown to vary. Hence, proper documentation and sample collection of the same patients at different stages of progression of the disease is necessary to understand the role of the microbiome in the disease condition. There is a need to re-examine the differences in the pattern of microbiota; before, during, and after the onset of exacerbations with detailed documentation of other important factors. This necessitates long-term follow-up of the population under study, which is a challenging task in the field of human microbiome research due to the paucity of comprehensive metadata and system for longitudinal follow-up of the population. The treatment regime of patients also alters the microbiota and many studies conducted so far lack the information on the use of antibiotics and medication. A study with COPD diagnosed patients, with and without any treatment may help in better understanding of the disease progression and role of microbes.
Although alterations in microbial communities are associated with severity of the disease, it is not clear whether they have a causative or exacerbating effect. This gives a unique opportunity to explore these associations with studies focusing on the establishment and development of microbiome and their role in normal functioning of the airway. This would eventually lead us to understand the succession and inoculation of microbiota in normal to the diseased condition. The understanding of the transient or permanent existence of microbiota in specific locations within airway would also lead to more information on the chronic nature of airway diseases. However, studies of short- and long-term antibiotic usage and its effect on the composition of microbiota in the context of airway microbiota need more attention.
Even though it is known that host’s genetic predisposition plays a role, many microbiome studies have not considered this factor. Proper control samples with comprehensive metadata from first-degree relatives of patients can help in exploring the effect of genetics in these chronic diseases. Moreover, studies on airway microbiome conducted so far include samples from genetically homogenous populations originating from the same geographic zone. As genetics is known a key factor in these diseases, similar studies on different populations are required to give a holistic picture. It is worth conducting such studies on a heterogeneous population with different ethnicity, heterogeneous diet, lifestyle, and climatic/geographic variation.
Most of the studies on asthma and COPD in this review lack extensive and longitudinal sampling with inadequate metadata collection. One of the reasons for this could be that these studies were either hospital-based or a part of related observational studies (Tables 1 and 2). There is a need to design broader microbiome research in asthma and COPD population with long-term follow-up, comprehensive metadata, and statistically significant sampling size. This will help in improving our knowledge of pathophysiology of airway diseases. This will also open the door for prebiotics, probiotics, or new antibiotics treatment in chronic airway disease management. This profiling of microbes in subjects with chronic airway diseases would also help physicians for diagnostic purpose with the help of advanced molecular tools of microbiome research. Some other reasons for inconsistencies in the results might be because of technical pitfalls (65). These factors are not discussed here as they are out of the scope of this review.
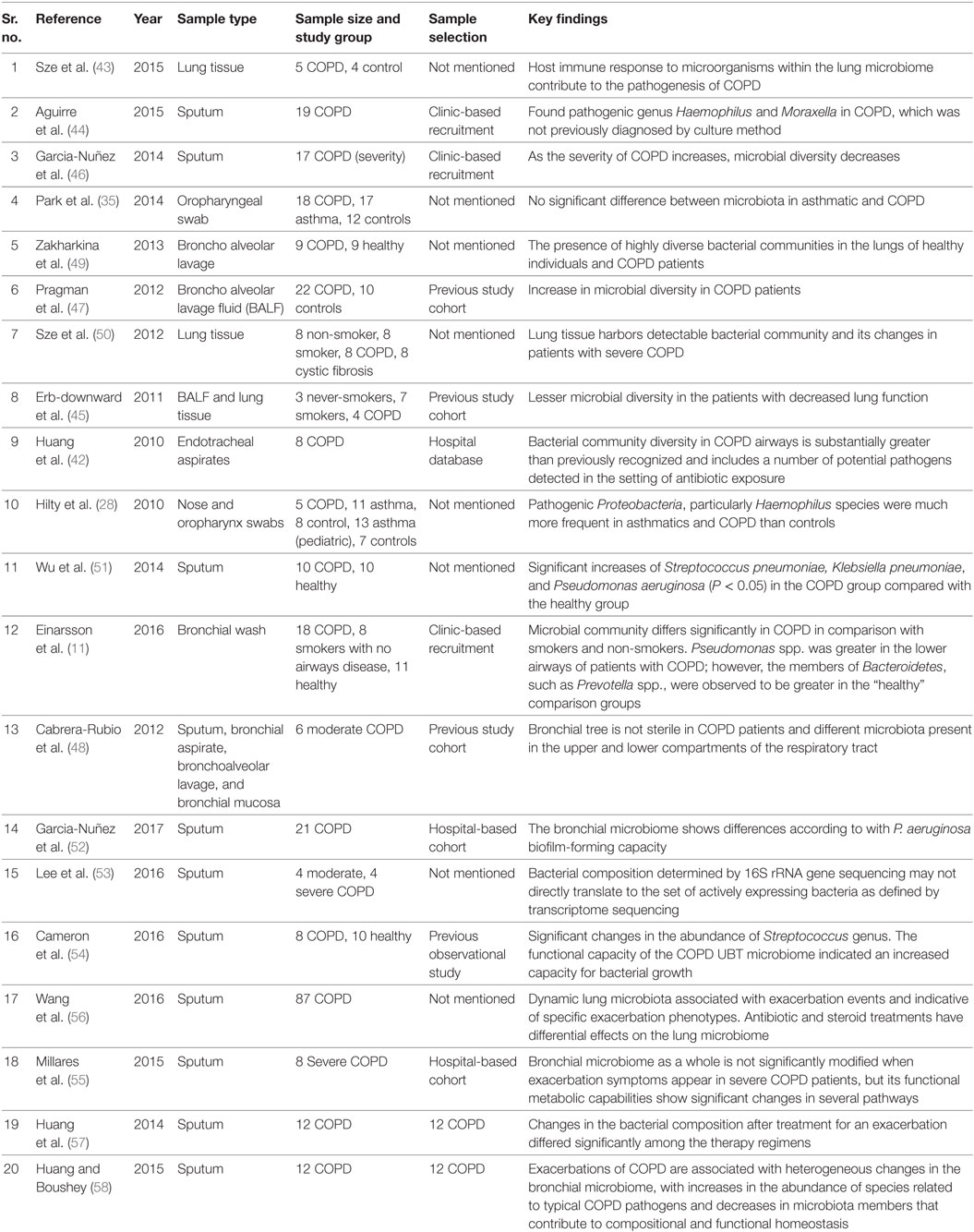
Table 2. List of microbiome studies conducted in chronic obstructive pulmonary disease (COPD) population.
How HDSS Can Benefit Microbiome Studies
Inclusion and documentation of all the mentioned perplexing factors, which are known to alter the microbiome in chronic diseases can be achieved in the HDSS environment. HDSS can play an important role in risk factor surveillance and provide health information, which reflects the prevailing disease burden on populations more accurately. This would enable us to identify specific causes and their impacts on microbial composition during different phases of the disease. Many hurdles so far experienced in microbiome research, such as statistically significant sample size, long-term follow-up, and collection of comprehensive metadata can be overcome with HDSS setup. In microbiome research, it would be advantageous to collect maximum metadata on various confounding factors to interpret the data more accurately (9). For example, data on exposure to biomass and tobacco smoke, dust, pesticides, particulate matter, and other occupational or geographic factors may help in more accurate data interpretation in asthma and COPD rather than focus only on directly related factors such as disease condition and its severity. Such factors are shown to be a potential confounder in microbiome studies and controlling these factors in study design will help to increase reproducibility of results in future microbiome studies (9). HDSS provides a framework, which may consist of such information for various time points, thus, strengthening the metadata required for conducting human microbiome studies. HDSS provides an appropriate sample frame as the population covered under HDSS is longitudinally followed up and information on aspects like age, gender, and other sociodemographic characters is readily available (Figure 2).
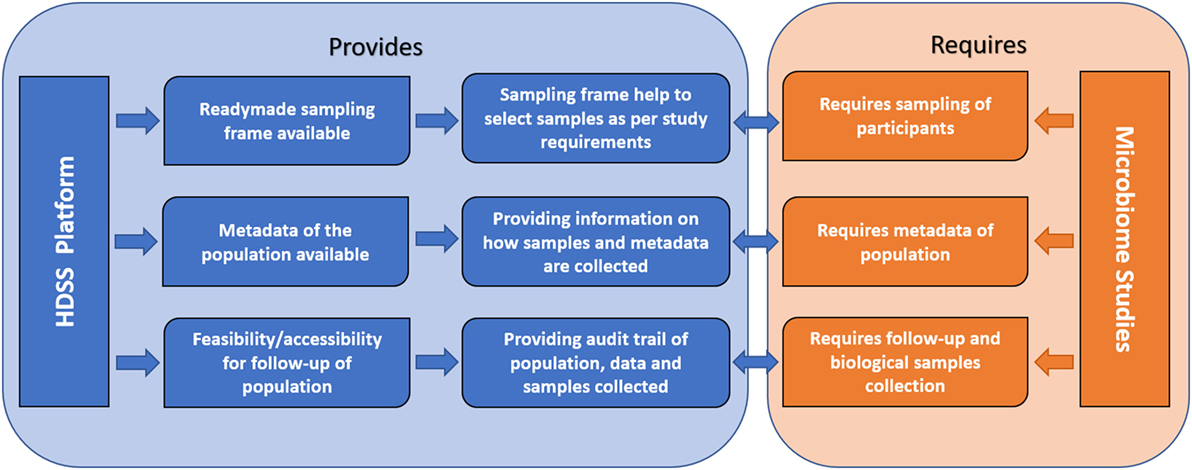
Figure 2. Conceptual framework showing how health and demographic surveillance system (HDSS) benefit human microbiome studies.
Microbiome research on chronic diseases with known genetic basis requires comparison within and between distinct homogenous populations. The homogenous population, followed by HDSS in a defined geographic area would be consistent with environmental conditions and may also have a similar diet. As HDSS helps to generate a baseline data for comparison with the case of future studies, it is relatively easy to identify such prospective cohorts within HDSS population. This would help in addressing the issues of adjusting confounding factors for a particular microbiome study. Along with this appropriate control samples with comparable metadata for genetically related individuals of the patients can help in exploring the effect of genetics on microbiome in these chronic diseases.
In HDSS setup, after the provision for initial sampling, additional metadata and assessment of intervention can be added at any stage of the study. Additionally, compliance for retention of subjects in longitudinal studies in HDSS area can be significantly higher. Genetic history and pedigree can be of much importance in microbiome studies, which can be gathered easily in HDSS. Studies on first- and second-degree relatives, homogeneity and heterogeneity, endogamous (inbreeding) population, identical twins, and multi-generational studies are possible in HDSS.
Besides the points mentioned in this review, HDSS can assist in monitoring and tracking of new health threats, such as emerging and re-emerging infectious disease and drug resistance and alert the health community to prepare a response. A key advantage of HDSS over traditional cross-sectional studies is their ability to measure trends over time and, therefore, to provide a clearer picture of demographic and health developments. This approach is useful in characterizing populations at the community level and provides important information for health planning and intervention.
Health and demographic surveillance system sites have already shown its potential to conduct many epidemiological studies in chronic diseases such as hypertension, diabetes, and cancer (3, 4). Many HDSS sites are now also involved in genomic research and developing their health-related genomic research capacity through the Human Heredity and Health in Africa (H3Africa) consortium primarily, in Africa. In this way, this consortium brings genomic dimension to longitudinal population-based studies (66, 67). Developing such consortium for microbiome studies may add value for epidemiological and microbiome studies at HDSS sites.
Human microbiome research is relatively new and promising field, which requires an expensive infrastructure and expertise. HDSS sites can evolve and build capacity through a partnership with local, national, and international advanced institutions to enhance knowledge generation apart from the traditional observational, epidemiological, and interventional studies to systematic studies aimed at understanding the molecular mechanisms of the disease. Collaboration at national and international level can benefit both HDSS and human microbiome research.
Future Directions for Lung Microbiome Research
In the past decade, sequencing-based techniques have generated abundant data, which has associated microbes with various respiratory health conditions including asthma and COPD. However, there is a paucity of direct functional and mechanistic data, which hinders clinical applications. Additionally, there are inconsistencies in the results, which can be correlated with various perplexing factors and technical challenges. These can be overcome through longitudinal, clinical, and in vitro studies in a defined population with similar environmental conditions. Cross-sectional studies conducted till date were hampered by their inability to explain the large variation in bacterial communities between individual samples and across subjects. Microbiome alterations/manipulation has a potential in disease control, which can be achieved through microbiota transplantation, dietary and lifestyle modifications, targeted antibiotics, interventions through probiotics and prebiotics. As this science progresses, it can open new therapeutic options for respiratory diseases. As there are complex association/interactions between microbes and humans, it is essential to study organisms and their actual roles in disease progression. Studies on larger cohorts with comprehensive metadata will help in deciding the directions, especially for personalized medicine.
Literature Survey
We have reviewed the work done until June 2017 by searching PubMed, Science Direct, HighwirePress, and Google Scholar. The keywords for the search were airway microbiome, airway microbiota, asthma, COPD, metagenomics, microbiome, microbiota, obstructive airway diseases, epidemiology, HDSS, and combinations thereof.
Ethics Statement
This paper is part of literature search done for a larger study titled “Exploring the Human Oral and Nasal Microbiota in Healthy and Diseased Population—A Study of Rural Population in Pune, India,” which is approved by KEM Hospital Research Centre Ethics Committee. A study was conducted following good clinical practices of research in the human population.
Author Contributions
DA: initial draft and rewriting of all drafts to finalization. RP: initial draft and review of drafts. DD: review of drafts and rewriting of all drafts to finalization. YS and SS: review of drafts. SJ: conceptualization and review of drafts.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the authors of previous studies for providing valuable information and data without which this review was not possible. We would also like to thanks, Dr. Monica Jain (Ph.D. English), Assistant Professor, BJS College, Pune, India for language editing.
Funding
This review is supported by the Vadu Rural Health Program, KEM Hospital Research Centre, Pune.
References
1. International Development Research Centre. Population and Health in Developing Countries. Vol. 1 Population, Health, and Survival at INDEPTH Sites, 2002. Ottawa, ON: International Development Research Centre (2009). p. 1–292.
2. Habte D. Population and Health in Developing Countries Volume 1: Population, Health and Survival at INDEPTH Sites. Ottawa: IDRC (2002). 1 p.
3. Sankoh O, Byass P. The INDEPTH Network: filling vital gaps in global epidemiology. Int J Epidemiol (2012) 41:579–88. doi:10.1093/ije/dys081
4. Ng N, Van Minh H, Juvekar S, Razzaque A, Huu Bich T, Kanungsukkasem U, et al. Using the INDEPTH HDSS to build capacity for chronic non-communicable disease risk factor surveillance in low and middle-income countries. Glob Health Action (2009) 2:7–18. doi:10.3402/gha.v2i0.1984
5. Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature (2012) 488:178–84. doi:10.1038/nature11319
6. Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One (2008) 3:e3064. doi:10.1371/journal.pone.0003064
7. Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, et al. The firmicutes/bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol (2009) 9:123. doi:10.1186/1471-2180-9-123
8. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A (2010) 107:14691–6. doi:10.1073/pnas.1005963107
9. Chen J, Ryu E, Hathcock M, Ballman K, Chia N, Olson JE, et al. Impact of demographics on human gut microbial diversity in a US Midwest population. PeerJ (2016) 4:e1514. doi:10.7717/peerj.1514
10. Zhang Q, Cox M, Liang Z, Brinkmann F, Cardenas PA, Duff R, et al. Airway microbiota in severe asthma and relationship to asthma severity and phenotypes. PLoS One (2016) 11(4):e0152724. doi:10.1371/journal.pone.0152724
11. Einarsson GG, Comer DM, Mcilreavey L, Parkhill J, Ennis M, Tunney MM, et al. Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax (2016) 71(9):795–803. doi:10.1136/thoraxjnl-2015-207235
12. Wilson Tang WH, Hazen SL. The gut microbiome and its role in cardiovascular diseases. Circulation (2017) 135:1008–10. doi:10.1161/CIRCULATIONAHA.116.024251
13. Amar J, Serino M, Lange C, Chabo C, Iacovoni J, Mondot S, et al. Involvement of tissue bacteria in the onset of diabetes in humans: evidence for a concept. Diabetologia (2011) 54(12):3055–61. doi:10.1007/s00125-011-2329-8
14. Karlsson F, Tremaroli V, Nielsen J. Assessing the human gut microbiota in metabolic diseases. Diabetes (2013) 62:3341–9. doi:10.2337/db13-0844
15. Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A (2011) 109(2):594–9. doi:10.1073/pnas.1116053109
16. Benziger CP, Roth GA, Moran AE. The Global Burden of Disease Study and the preventable burden of NCD. Glob Heart (2016) 11:393–7. doi:10.1016/j.gheart.2016.10.024
17. Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med (2015) 3:2011–30. doi:10.1371/journal.pmed.0030442
18. Mosenifar Z. Differentiating COPD from asthma in clinical practice. Postgrad Med (2009) 121:105–12. doi:10.3810/pgm.2009.05.2009
19. Armann J. Do bacteria have a role in asthma development? Eur Respir J (2010) 36(3):469–71. doi:10.1183/09031936.00041410
21. Sze MA, Hogg JC, Sin DD. Bacterial microbiome of lungs in COPD. Int J Chron Obstruct Pulmon Dis (2014) 9:229–38. doi:10.2147/COPD.S38932
22. Brashier BB, Kodgule R. Risk factors and pathophysiology of chronic obstructive pulmonary disease (COPD). J Assoc Physicians India (2012) 60:17–21.
23. Martin C, Burgel P, Lepage P, Andréjak C, de Blic J, Bourdin A, et al. Host–microbe interactions in distal airways: relevance to chronic airway diseases. Eur Respir Rev (2014) 24:78–91. doi:10.1183/09059180.00011614
24. Agusti A, Calverley PM, Celli B, Coxson HO, Edwards LD, Lomas D, et al. Characterisation of COPD heterogeneity in the ECLIPSE cohort. Respir Res (2010) 11:122. doi:10.1186/1465-9921-11-122
25. Bourbeau J, Tan WC, Benedetti A, Aaron SD, Chapman KR, Coxson HO, et al. Canadian Cohort Obstructive Lung Disease (CanCOLD): fulfilling the need for longitudinal observational studies in COPD. COPD (2012) 11(2):125–32. doi:10.3109/15412555.2012.665520
26. Enocson A, Dickens AP, Jordan R, Fitzmaurice D, Adab P. The birmingham chronic obstructive pulmonary disease (copd) cohort study: preliminary baseline characteristics of ever-smokers and never-smokers. Am J Respir Crit Care Med (2014) 189:A2947.
27. Grønseth R, Haaland I, Wiker HG, Martinsen EMH, Leiten EO, Husebø G, et al. The Bergen COPD microbiome study (MicroCOPD): rationale, design, and initial experiences. Eur Clin Respir J (2014) 1:1–8. doi:10.3402/ecrj.v%vi%i.26196
28. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One (2010) 5(1):e8578. doi:10.1371/journal.pone.0008578
29. Yayan J. No direct association between asthma and the microbiome based on currently available techniques. Medicine (Baltimore) (2014) 93:e199. doi:10.1097/MD.0000000000000199
30. Huang YJ, Lynch SV. The emerging relationship between the airway microbiota and chronic respiratory disease: clinical implications. Expert Rev Respir Med (2011) 5(6):809–22. doi:10.1586/ers.11.76
31. Pérez-losada M, Castro-Nallar E, Bendall ML, Freishtat RJ, Crandall KA. Dual transcriptomic profiling of host and microbiota during health and disease in pediatric asthma. PLoS One (2015) 10(6):e0131819. doi:10.1371/journal.pone.0131819
32. Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol (2010) 127:372–81.e1–3. doi:10.1016/j.jaci.2010.10.048
33. Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol (2016) 140(1):63–75. doi:10.1016/j.jaci.2016.08.055
34. Simpson JL, Daly J, Baines KJ, Yang IA, Upham JW, Reynolds PN, et al. Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur Respir J (2016) 47:792–800. doi:10.1183/13993003.00405-2015
35. Park H, Shin JW, Park S, Kim W. Microbial communities in the upper respiratory tract of patients with asthma and chronic obstructive pulmonary disease. PLoS One (2014) 9:e109710. doi:10.1371/journal.pone.0109710
36. Taylor SL, Leong LEX, Choo JM, Wesselingh S, Yang IA, Upham JW, et al. Inflammatory phenotypes in severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol (2017). doi:10.1016/j.jaci.2017.03.044
37. Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol (2015) 136(4):874–84. doi:10.1016/j.jaci.2017.03.044
38. Depner M, Ege MJ, Cox MJ, Dwyer S, Walker AW, Birzele LT, et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J Allergy Clin Immunol (2015) 139(3):826–34.e13. doi:10.1016/j.jaci.2016.05.050
39. Castro-Nallar E, Shen Y, Freishtat RJ, Pérez-losada M, Manimaran S, Liu G, et al. Integrating microbial and host transcriptomics to characterize asthma-associated microbial communities. BMC Med Genomics (2015) 8:50. doi:10.1186/s12920-015-0121-1
40. Marri PR, Stern DA, Wright AL, Billheimer D. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol (2012) 131:346–52.e1–3. doi:10.1016/j.jaci.2012.11.013
41. Zhang Q, Cox MJ, Liang Z, Brinkmann F, Duff R, Gibeon D, et al. Altered airway microbiome in severe asthma. Am J Respir Crit Care Med (2012) 185:A6717. doi:10.1164/ajrccm-conference.2012.185.1_MeetingAbstracts.A6717
42. Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, et al. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS (2010) 14:9–59. doi:10.1089/omi.2009.0100
43. Sze MA, Dimitriu PA, Suzuki M, McDonough JE, Campbell JD, Brothers JF, et al. The host response to the lung microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med (2015) 192:438–45. doi:10.1164/rccm.201502-0223OC
44. Aguirre E, Galiana A, Mira A, Guardiola R, Sánchez-Guillén L, Garcia-Pachon E, et al. Analysis of microbiota in stable patients with chronic obstructive pulmonary disease. APMIS (2015) 123(5):427–32. doi:10.1111/apm.12363
45. Erb-downward JR, Thompson DL, Han MK, Freeman CM, Schmidt LA, Young VB, et al. Analysis of the lung microbiome in the “Healthy” smoker and in COPD. PLoS One (2011) 6: e16384. doi:10.1371/journal.pone.0016384
46. Garcia-Nuñez M, Millares L, Pomares X, Ferrari R, Pérez-Brocal V, Gallego M, et al. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J Clin Microbiol (2014) 52:4217–23. doi:10.1128/JCM.01967-14
47. Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One (2012) 7:e47305. doi:10.1371/journal.pone.0047305
48. Cabrera-Rubio R, Garcia-Núñez M, Setó L, Antó JM, Moya A, Monsó E, et al. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol (2012) 50:3562–8. doi:10.1128/JCM.00767-12
49. Zakharkina T, Heinzel E, Koczulla RA, Greulich T, Rentz K, Pauling JK, et al. Analysis of the airway microbiota of healthy individuals and patients with chronic obstructive pulmonary disease by T-RFLP and clone sequencing. PLoS One (2013) 8:e68302. doi:10.1371/journal.pone.0068302
50. Sze MA, Dimitriu PA, Hayashi S, Elliott WM, Mcdonough JE, Gosselink JV, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med (2012) 185:1073–80. doi:10.1164/rccm.201111-2075OC
51. Wu D, Hou C, Li Y, Zhao Z, Liu J, Lu X, et al. Analysis of the bacterial community in chronic obstructive pulmonary disease sputum samples by denaturing gradient gel electrophoresis and real-time PCR. BMC Pulm Med (2014) 14:179. doi:10.1186/1471-2466-14-179
52. Garcia-Nuñez M, Marti S, Puig C, Perez-Brocal V, Millares L, Santos S, et al. Bronchial microbiome, PA biofilm-forming capacity and exacerbation in severe COPD patients colonized by P. aeruginosa. Future Microbiol (2017) 12:379–92. doi:10.2217/fmb-2016-0127
53. Lee S, Kuan C, Wu LS, Weng JT. Metagenome and metatranscriptome profiling of moderate and severe COPD sputum in Taiwanese Han males. PLoS One (2016) 11(7):e0159066. doi:10.1371/journal.pone.0159066
54. Cameron SJS, Lewis KE, Huws SA, Lin W, Hegarty J, Lewis PD, et al. Metagenomic sequencing of the chronic obstructive pulmonary disease upper bronchial tract microbiome reveals functional changes associated with disease severity. PLoS One (2016) 11(2):e0149095. doi:10.1371/journal.pone.0149095
55. Millares L, Pérez-Brocal V, Ferrari R, Gallego M, Pomares X, García-Núñez M, et al. Functional metagenomics of the bronchial microbiome in COPD. PLoS One (2015) 10:e0144448. doi:10.1371/journal.pone.0144448
56. Wang Z, Bafadhel M, Haldar K, Spivak A, Mayhew D, Miller BE, et al. Lung microbiome dynamics in COPD exacerbations. Eur Respir J (2016) 47:1082–92. doi:10.1183/13993003.01406-2015
57. Huang YJ, Sethi S, Murphy T, Nariya S, Boushey HA, Lynch SV. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J Clin Microbiol (2014) 52:2813–23. doi:10.1128/JCM.00035-14
58. Huang YJ, Boushey HA. The sputum microbiome in chronic obstructive pulmonary disease exacerbations. Ann Am Thorac Soc (2015) 12(Suppl 2):S176–80. doi:10.1513/AnnalsATS.201506-319AW
59. Markus Hilty CB, Hilty M, Burke C, Pedro H, Cardenas P, Bush A, et al. Disordered microbial communities in asthmatic airways. PLoS One (2010) 5(1):e8578. doi:10.1371/journal.pone.0008578
60. Molyneaux PL, Mallia P, Cox MJ, Footitt J, Willis-owen SAG, Homola D, et al. Outgrowth of the bacterial airway microbiome after rhinovirus exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med (2013) 188(10):1224–31. doi:10.1164/rccm.201302-0341OC
61. Pragman AA, Baller JA, Reilly C, Isaacson RE, Wendt CH. The lung tissue microbiota in mild or moderate chronic obstructive pulmonary disease. Am J Respir Crit Care Med (2015) 191:6337.
62. Millares L, Ferrari R, Gallego M, Garcia-Nuñez M, Pérez-Brocal V, Espasa M, et al. Bronchial microbiome of severe COPD patients colonised by Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis (2014) 33:1101–11. doi:10.1007/s10096-013-2044-0
63. Millares L, Bermudo G, Perez-Brocal V, Domingo C, Garcia-Nunez M, Pomares X, et al. The respiratory microbiome in bronchial mucosa and secretions from severe IgE-mediated asthma patients. BMC Microbiol (2017) 17:20. doi:10.1186/s12866-017-0933-6
64. Simpson JL, Baines KJ, Horvat JC, Essilfie AT, Brown AC, Tooze M, et al. COPD is characterized by increased detection of Haemophilus influenzae, Streptococcus pneumoniae and a deficiency of Bacillus species. Respirology (2016) 21:697–704. doi:10.1111/resp.12734
65. Teeling H, Glöckner FO. Current opportunities and challenges in microbial metagenome analysis – a bioinformatic perspective. Brief Bioinform (2012) 13:728–42. doi:10.1093/bib/bbs039
66. Ramsay M, Sankoh O. African partnerships through the H3Africa Consortium bring a genomic dimension to longitudinal population studies on the continent. Int J Epidemiol (2015) 45(2):305–8. doi:10.1093/ije/dyv187
Keywords: asthma, chronic obstructive pulmonary disease, health and demographic surveillance system, microbiome, microbiota
Citation: Agarwal D, Dhotre D, Patil R, Shouche Y, Juvekar S and Salvi S (2017) Potential of Health and Demographic Surveillance System in Asthma and Chronic Obstructive Pulmonary Disease Microbiome Research. Front. Public Health 5:196. doi: 10.3389/fpubh.2017.00196
Received: 23 February 2017; Accepted: 20 July 2017;
Published: 04 August 2017
Edited by:
Margaret Ip, The Chinese University of Hong Kong, Hong KongReviewed by:
Isaac Kirubakaran Sundar, University of Rochester, United StatesThiago Soares De Souza Vieira, University of Massachusetts Medical School, United States
Copyright: © 2017 Agarwal, Dhotre, Patil, Shouche, Juvekar and Salvi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dhiraj Agarwal, dhiraj.agarwal99@gmail.com;
Dhiraj Dhotre, dhiraj@nccs.res.in;
Rutuja Patil, ru2.patil@gmail.com
 Dhiraj Agarwal
Dhiraj Agarwal Dhiraj Dhotre
Dhiraj Dhotre Rutuja Patil
Rutuja Patil Yogesh Shouche
Yogesh Shouche Sanjay Juvekar
Sanjay Juvekar Sundeep Salvi
Sundeep Salvi