COEL: A Cloud-Based Reaction Network Simulator
- 1Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Belvaux, Luxembourg
- 2Department of Electrical and Computer Engineering, Portland State University, Portland, OR, USA
Chemical Reaction Networks (CRNs) are a formalism to describe the macroscopic behavior of chemical systems. We introduce COEL, a web- and cloud-based CRN simulation framework, which does not require a local installation, runs simulations on a large computational grid, provides reliable database storage, and offers a visually pleasing and intuitive user interface. We present an overview of the underlying software, the technologies, and the main architectural approaches employed. Some of COEL’s key features include ODE-based simulations of CRNs and multicompartment reaction networks with rich interaction options, a built-in plotting engine, automatic DNA-strand displacement transformation and visualization, SBML/Octave/Matlab export, and a built-in genetic-algorithm-based optimization toolbox for rate constants. COEL is an open-source project hosted on GitHub (doi: 10.5281/zenodo.46544), which allows interested research groups to deploy it on their own sever. Regular users can simply use the web instance at no cost at coel-sim.org. The framework is ideally suited for a collaborative use in both research and education.
Code available at: 10.5281/zenodo.46544
1. Introduction and Related Work
In this paper, we introduce COEL, the first web- and cloud-based framework for modeling and simulating Chemical Reaction Networks (CRNs). COEL’s web client is accessible without any installation or download. Simulations are performed on a grid rather than the client’s machine, which allows to run significantly larger systems very quickly. Remote teams can share and manipulate chemical models in real time. Data are stored remotely and safely in COEL’s database, which is backed up daily. In developing COEL, we emphasized platform-wide visualization, allowing quick and easy data analysis for users. The platform also focuses on collaboration and transparency. Individuals can work on different facets of the same project and see each other’s modifications in real time. This has allowed us to study the same system and modify its simulation dynamics from separate campuses. We have successfully applied COEL (Banda et al., 2014a) as a sole tool to model and evaluate various chemical perceptrons (Banda et al., 2013, 2014b; Banda and Teuscher, 2014b), chemical delay lines, and time-series learners (Banda and Teuscher, 2014a; Moles et al., 2015).
A study by Vines et al. (2014) found that 80% of scientific data are lost within two decades, disappearing into old email addresses and obsolete storage devices. Alarmingly, the authors found that the average rate of data loss is 17% per year. Similarly, only 11% of the academic research was reproducible by the original research groups, as reported by Begley and Ellis (2012). To address the “data loss” and “reproducibility crisis,” the way we conduct research must dramatically change. We believe that COEL is a step in that direction: storing all experiments as well as the final and intermediate models in a centralized database that provides reliable and long-term storage will help to address the challenges. In addition, the data and models can easily be made accessible to the collaborators and to the public.
COEL is an open-source project hosted on GitHub: https://github.com/peterbanda/coel. That allows interested research groups to deploy it on their own sever. Regular users can simply use the web instance at no cost at http://coel-sim.org. The framework is ideally suited for a collaborative use in both research and education. Since COEL’s user interface is entirely web-based, it is platform-independent and supports any operating system with a web browser, including tablets and smartphones.
Out of all CRN simulators, COPASI (Hoops et al., 2006) is probably the most advanced and widely used tool. It simulates a variety of chemical objects and provides many options for the design of experiments and the statistical analysis. Other tools include the MATLAB Systems Biology Toolbox (Schmidt and Jirstrand, 2006) and CellDesigner (Funahashi et al., 2008). The majority of these tools, including COEL, share support for the SBML language (Hucka et al., 2003) for describing chemical systems, which enable cross-platform migration. Beyond deterministic ODE integration of CRNs and stochastic reactions, it is common to offer parameter optimization to help in the design of the networks themselves. COPASI and CellDesigner can simulate a number of other biochemical objects, such as cellular compartments, and support various chemical kinetic models. The primary reason for developing COEL was not to introduce new simulation algorithms or methods of analysis but to integrate the most common and practical features among other CRN simulators into an intuitive and modern web-based package.
2. Features and Functionality
COEL’s main feature is the definition and simulation of CRNs. A CRN consists of a finite set of chemicals and reactions. The state of a CRN is represented by a vector of chemical species concentrations. Reactions can also involve catalysts or inhibitors, which accelerate or slow down the reaction without being consumed. Reaction rates define the strength or speed of reactions, as prescribed by kinetic laws: Michaelis–Menten kinetics for catalytic reactions, non-competitive inhibitory kinetics for inhibitory reactions, and mass-action kinetics otherwise. Based on the reaction type, COEL automatically computes appropriate rate functions with given numeric rate constants. But it also allows to define arbitrary rate functions using custom expressions over species labels giving users a full control over the system’s dynamics. Reactions can be uni- or bidirectional. Bidirectional reactions can have independent forward and backward rates.
Inspired by biochemical cells and membranes, COEL’s CRNs support hierarchical, tree-like compartmentalization. Each compartment hosts an independent reaction set and vector of chemical concentrations. Compartments communicate with each other through permeation, which are formalized and implemented in the form of “channels.”
A unique feature of COEL is the so-called interaction series. An interaction series allows the user to directly manipulate concentrations of species in the CRN, while the ODE solver carries out an execution. This feature is analogous to – though more capable than – the automatic chemical injections into a reaction chamber. For compartment-extended CRNs, interaction series can be identically hierarchical, thus allowing for specific interaction with each component in the network. Concentrations can be modified multiple times, as opposed to just being set initially. For example, it is useful to define a set of periodic injections for iterative processes. In specifying interactions, a user can define custom concentration-setting expressions based on the extended Java Math Expression Parser syntax with custom predefined variables. A basic expression validation is also implemented to prevent errors. The interaction series expressions are in fact a scripted language that can describe a variety of complicated experimental scenarios without modifying the underlying framework code (Figure 1). As a consequence, end users have the ability to manipulate the chemical system in an easy, flexible, and dynamic way. COEL’s basic tool for interpreting and observing CRN’s is the “translation series.” A single translation is a straightforward function of the current concentrations and of any predefined constants with a Boolean or numeric output.
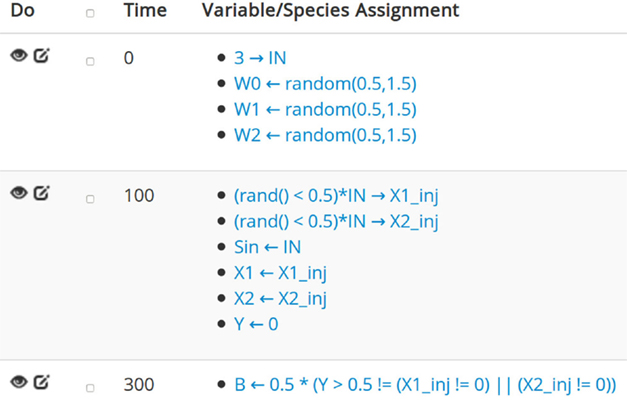
Figure 1. Details of a COEL interaction series showing its programing capabilities. Left arrows denote the setting of species concentrations and right arrows indicate assignments of user-defined variables. The interaction at time 100 does the following (note that at time 0, the variable IN is set to 3): first, the variables X1_inj and X2_inj are randomly set to 0 or 3 with equal probability. The concentration of Sin is set to 3. Then, both the concentrations of X1 and X2 are set to their respective injection variables. Finally, Y is flushed from the system. The bottommost interaction injects species B at concentration 0.5 if the output species Y does not match AND of the original input concentrations X1_inj and X2_inj.
COEL has a convenient web interface for visualizing DNA strands specified by the Microsoft Visual DSD syntax (Lakin et al., 2011). By implementing the methods of Soloveichik et al. (2010), COEL can transform any CRN based on mass-action kinetics into a DNA-strand displacement circuit. In strand displacement systems, the populations of single-stranded DNA molecules interact with double-stranded gate complexes, which mediate transformations between free signals. Once applied to a reaction set, the transformation produces a new CRN, describing displacements of single strands from partial or full double strands, along with the DNA structure of each species with numerically labeled domains. This allows for an automatic translation of abstract systems into wet chemistries.
Other features include performance evaluation, system dynamics analysis, evolutionary optimization of rate constants, random CRNs, Octave/Matlab export, and platform-wide features, such as CSV export and integrated web charting.
3. Architecture and Technology
COEL was implemented as a Java Enterprise application with a modular architecture that uses a strict separation of the business logic and technological application aspects. The absence of strict inter-modular/inter-layer dependencies enables quick and easy customization of components and providers.
With our decomposed application, only the domain objects, the data holders of business data, implemented as POJOs (Plain Java Objects), are shared among all application parts and layers. Figure 2 presents a high-level overview of COEL’s architecture with call request and pathways. On the very top, we have two clients representing the only entry points to the application: the web client backed by Grails (Official Grails website, 2015) framework (discussed in Section 3.3) and the plain console client implemented in standard Java for “headless” scripting.
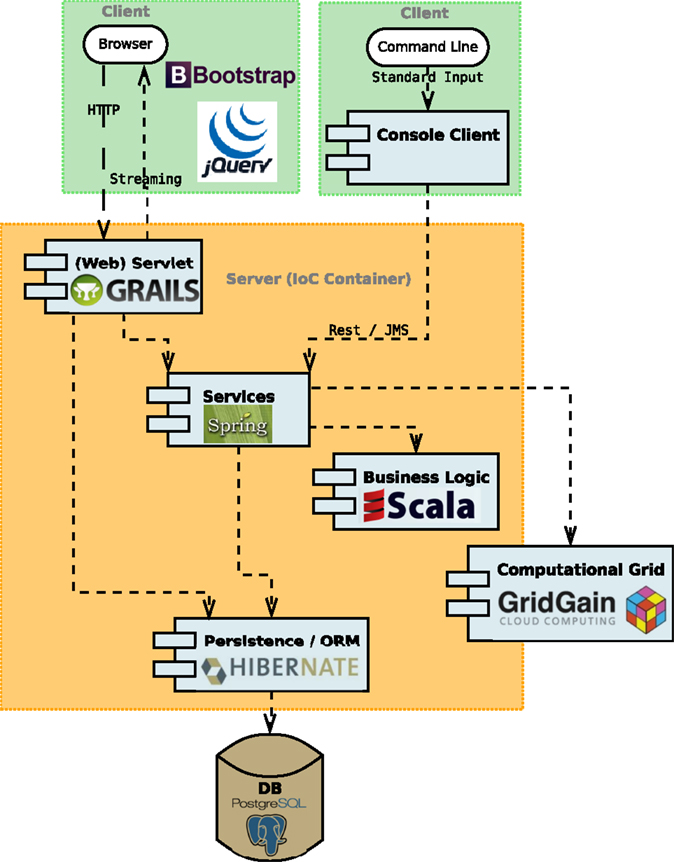
Figure 2. A high-level overview of COEL’s architecture. It consists of web and console clients, web servlet, services, business logic, persistence layer, and computational grid. The application (IoC) container holding the server side of the application is implemented in the Spring framework.
Based on user’s requests, the clients call the services, such as ArtificialChemistryService and EvolutionService, living in the application container (Section 3.1), which then redirects either to a computational grid implemented on the top of the GridGain In-Memory Data Fabric (Grid Gain website, 2015) (Section 3.2) or to the persistence layer with Data-Access Objects (DAOs) and Object-Relation Mapping (ORM) provided by Hibernate (Official Hibernate website, 2015) (Section 3.4). In addition, the web client controllers have a direct link to the persistence layer, which is beneficial especially for basic CRUD operations. At the bottom, a PostgreSQL (Official PostgreSQL website, 2015) database stores and provides data on demand of the persistence layer. The business logic, such as the chemistry simulation and the GA optimization, is implemented mainly in the Scala language, leveraging both object-oriented and functional programing approaches. All technologies and libraries integrated into COEL are either open source or free to use.
COEL’s components, such as services, views, database tables, and computational tasks, are easily extensible. For instance, to add a new function called flattenCompartment, we need to first add a function signature to the service interface ArtificialChemistryService and then add an implementation to ArtificialChemistryServiceImpl. To access the compartments stored in the COEL database, we can use the get method of acCompartmentDAO that is already provided in the service class through dependency injection (Section 3.1). The service function ArtificialChemistryService.flattenCompartment can then be called in a client’s controller with appropriate views, such as AcCompartmentController.
To build COEL and to maintain its library dependencies, we use Apache Maven (Official Apache Maven website, 2015). For a new application version, we run a set of JUnit tests, which guarantee that the core functionality works as expected. If users want to report a production issue or recommend a new feature, they can submit a report through a Jira issue tracking system. COEL is deployed to the Tomcat application server.
3.1. Application Container
The Spring Framework (Walls, 2014; Official Spring Project website, 2015) provides COEL’s core application infrastructure. Spring is a leading enterprise solution for Java consisting of several sub-projects, which can be used separately or together. Spring does not enforce strict dependencies, and it detaches technical and business concerns.
The Inversion of Control (IoC) container is a central part of the Spring Framework. It controls the creation, the number of instances (singleton and prototype scopes), the lifecycle, interdependencies (loose-coupling or wiring), and the general configuration of application components, modules, adapters, specific utility classes, or in general any POJO, the creation and use of which should be maintained in the application context. Spring IoC is a simple and transparent integrator of various components and frameworks.
IoC encourages the best practices of programing with interfaces, i.e., a bean (object in the IoC container) should consist of an interface and an implementation class. Therefore, each bean knows that it can talk to a different bean that does something specific, but not which type of object, how its functionality is implemented, or how the call is carried out. The IoC container injects the dependencies at runtime, and so beans take care only about their business purpose, not the creation and maintenance of their relationships. This approach is superior to the factory design pattern because all dependencies get injected through the application container and configured by annotations and/or XML. Beans are not aware of the container’s existence, i.e., unlike the factory pattern they do not need to call the application container in order to get their dependencies. IoC is often described with the Hollywood principle: “Don’t call us, we call you.”
Other popular containers include GUICE and Pico, which are not as feature-rich and instead strictly focus on IoC and dependency injection. We opted for Spring because it offers other functionality, e.g., for persistence and services, which we could easily integrate out of the box.
The IoC abstraction results in modular, lightweight, and layered architecture with loose coupled and pluggable components. We also aimed to implement beans as thread-safe and stateless if possible, so several callers could safely query the same component without worrying about timing and/or call history. Spring IoC enables COEL to become a truly test-driven project. Because of dependency injections, our JUnit tests could switch to test (rather than production) application context and substitute for, e.g., classes that require remote access to production systems with mock objects.
3.2. Cloud Computing
COEL’s computational grid has been built on top of the GridGain In-Memory Computing Platform (Grid Gain website, 2015), which implements a scalable, low-latency, and zero task-deployment computational grid fitting seamlessly into our Spring-backed IoC container (Section 3.1).
Complex tasks, such as evolutionary optimizations of rate constants, performance evaluations, and dynamics analyses, might run for months on a single core. To address that challenge, COEL uses a scalable computational grid that currently consists of 30 nodes with about 800 cores. The grid, which is partially shared with researchers and students in Teuscher’s group, is maintained and hosted by the Computer Action Team (CAT) at Portland State University (PSU). The grid technology also allows us to add any geographically remote resource because the communication is carried out by TCP/IP protocol with optimized serialization of exchanged data. We plan to utilize existing grid technology to pool the resources with other remote teams.
COEL’s grid acts transparently, as a single computing resource. GridGain enables COEL’s users to be more productive by eliminating the complexity of distributed computing. Regardless of a geographic location, users can add tasks to the grid from COEL’s web client. When a user submits a task, the request is received by the grid master and the task splits into many partial jobs, which are distributed over the grid.
GridGain provides zero task-deployment technology that can stream a new task’s byte code, i.e., a compiled class, to the nodes without restart if it is not provided in the user libraries. Further, the grid’s topology might change freely during its lifetime. A new (slave) node can be added to the grid on-the-fly by registering the IP address with the master node. COEL’s grid supports several enterprise features, contributing to the effective and robust execution of jobs. The grid keeps track of various node statistics, such as CPU performance, execution time, and availability, which are constantly updated and utilized for adaptive job distribution, thus leading to an increased overall job throughput. Also, if a node disconnects from the grid, the exception is noted by a periodic heartbeat. The node’s jobs are then redistributed across the grid. If a node finishes its execution sooner than expected, it attempts to steal jobs from other nodes.
3.3. Web Client
COEL’s web client is implemented in Grails (Official Grails website, 2015), which is a powerful web 2.0 framework based on the Groovy dynamic language for the JVM. JVM compatibility means that Java, Groovy, and Scala source code compiles into Java byte code and are therefore inter-callable. Grails follows the “convention over configuration” approach, which emphasizes standard naming, binding, and data flow. The structure of the application is simply implied if it is not explicitly configured. This approach is heavily utilized in a function called scaffolding. Based on a domain object structure, Grails generates the controller with basic CRUD operations and associated web pages dynamically at runtime.
The web front-end relies on Javascript provided by the jQuery library (jQuery website, 2015), which makes the UI interactive and intuitive. It also moves a part of the data processing and visualization directly to the web browser. For instance, although COEL runs all simulations server side, if a user wishes to see a chart, e.g., of concentration traces, COEL sends raw data to the client, which is then transformed into a chart by using the Google Charts API. For styling and for some widgets, we used the Twitter Bootstrap library (Bootstrap website, 2015).
3.4. Persistence
The persistence layer consists of Data-Access Objects (DAOs), which wrap storing, retrieving, deleting, and filtering functionality for domain objects. To map an object-oriented domain model to a traditional, relational database we use Hibernate (Official Hibernate website, 2015), an Object-Relational Mapper (ORM) for the Java language. DAOs and Hibernate are widely supported by Spring, which offers hooks for a fast integration.
Hibernate solves object-relational impedance mismatch by replacing direct persistence-related database accesses with high-level object handling functions. Hibernate provides a declarative strategy for data persistence. We defined a mapping of columns, reference metadata, and inheritance strategy mapping. Based on that information, Hibernate handles details about persistence implementation, such as the generation of SQL statements, JDBC connection, and the translation from the POJO to the JDBC result set, automatically. Hibernate also supports various advanced features, such as cache and DB access optimization.
COEL’s approach is to store data in a structured database, thereby enabling prompt retrieval, searching, and post-processing. We have chosen PostgreSQL (Official PostgreSQL website, 2015) because it is a mature open-source database providing standard SQL/PLSQL language support, a comprehensive console as well as a graphical-based administration UI (PgAdmin), and support for array data types, which is handy for storing scientific vector data. The database model currently contains 83 tables. To assure compatibility for each version of COEL, we migrate data by a set of SQL scripts. Also, every day, the entire database is backed up.
4. Conclusion
COEL is a web-based chemistry simulation framework with a modern layered architecture. It includes a scalable computational grid, a stylish and interactive web UI, and the safe and transactive persistence of chemistry models and results. We paid particular attention to the overall usability, the fluid layout, and to the embedded data visualization. COEL can be accessed without installation from any web browser. As such, it is easier to start using and has a larger potential target audience than existing CRN desktop applications. Also, keeping COEL in the cloud allows for easy collaboration and sharing of models and results.
In addition to chemical simulations, COEL also offers limited capabilities to simulate more generic unconventional compute models that are based on complex, spatial, random, and layered networks with configurable node functions and interaction series.
Future work will include the addition of priorities and user privileges for running tasks and for sharing models and results. We are exploring the possibility to expose certain services through a RESTful API that 3rd party applications could call and tailor COEL’s functionality as needed.
Author Contributions
PB and CT wrote the paper. PB developed the presented framework (COEL). CT supervised the project.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This material is based upon work supported by the National Science Foundation under grant no. 1028120.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/article/10.3389/frobt.2016.00013
References
Banda, P., Blount, D., and Teuscher, C. (2014a). “COEL: a web-based chemistry simulation framework,” in CoSMoS 2014: Proceedings of the 7th Workshop on Complex Systems Modelling and Simulation, eds Stepney S. and Andrews P. (Frome: Luniver Press), 35–60.
Banda, P., Teuscher, C., and Stefanovic, D. (2014b). Training an asymmetric signal perceptron through reinforcement in an artificial chemistry. J. R. Soc. Interface 11. doi:10.1098/rsif.2013.1100
Banda, P., and Teuscher, C. (2014a). “An analog chemical circuit with parallel-accessible delay line for learning temporal tasks,” in ALIFE 14: The Fourteenth Conference on the Synthesis and Simulation of Living Systems, Vol. 14, eds Sayama H., Rieffel J., Risi S., Doursat R., and Lipson H. (Cambridge, MA: MIT Press), 482–489.
Banda, P., and Teuscher, C. (2014b). “Learning two-input linear and nonlinear analog functions with a simple chemical system,” in Unconventional Computing and Natural Computing Conference, Vol. 8553, eds Ibarra O. H., Kari L., and Kopecki S. (Switzerland: Springer International Publishing), 14–26. Lecture Notes in Computer Science.
Banda, P., Teuscher, C., and Lakin, M. R. (2013). Online learning in a chemical perceptron. Artif. Life 19, 195–219. doi:10.1162/ARTL_a_00105
Begley, C. G., and Ellis, L. M. (2012). Drug development: raise standards for preclinical cancer research. Nature 483, 531–533. doi:10.1038/483531a
Bootstrap website. (2015). Available at: http://getbootstrap.com
Funahashi, A., Matsuoka, Y., Jouraku, A., Morohashi, M., Kikuchi, N., and Kitano, H. (2008). CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE 96, 1254–1265. doi:10.1109/JPROC.2008.925458
Grid Gain website. (2015). Available at: http://www.gridgain.com
Hoops, S., Sahle, S., Gauges, R., Lee, C., Pahle, J., Simus, N., et al. (2006). COPASI – a complex pathway simulator. Bioinformatics 22, 3067–3074. doi:10.1093/bioinformatics/btl485
Hucka, M., Finney, A., Sauro, H. M., Bolouri, H., Doyle, J. C., Kitano, H., et al. (2003). The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19, 524–531. doi:10.1093/bioinformatics/btg015
jQuery website. (2015). Available at: http://jquery.com
Lakin, M. R., Youssef, S., Polo, F., Emmott, S., and Phillips, A. (2011). Visual DSD: a design and analysis tool for DNA strand displacement systems. Bioinformatics 27, 3211–3213. doi:10.1093/bioinformatics/btr543
Moles, J., Banda, P., and Teuscher, C. (2015). Delay line as a chemical reaction network. Parallel Process. Lett. 25, 1540002. doi:10.1142/S0129626415400022
Official Apache Maven website. (2015). Available at: http://maven.apache.org
Official Grails website. (2015). Available at: http://www.grails.org
Official Hibernate website. (2015). Available at: http://hibernate.org
Official PostgreSQL website. (2015). Available at: http://www.postgresql.org
Official Spring Project website. (2015). Available at: http://spring.io
Schmidt, H., and Jirstrand, M. (2006). Systems biology toolbox for MATLAB: a computational platform for research in systems biology. Bioinformatics 22, 514–515. doi:10.1093/bioinformatics/bti799
Soloveichik, D., Seelig, G., and Winfree, E. (2010). DNA as a universal substrate for chemical kinetics. Proc. Natl. Acad. Sci. U.S.A. 107, 5393–5398. doi:10.1073/pnas.0909380107
Vines, T. H., Albert, A. Y. K., Andrew, R. L., Débarre, F., Bock, D. G., Franklin, M. T., et al. (2014). The availability of research data declines rapidly with article age. Curr. Biol. 24, 94–97. doi:10.1016/j.cub.2013.11.014
Keywords: COEL, chemical reaction network, chemical modeling tool, web tool, computational grid, DNA-strand displacement, code: java,scala,groovy
Citation: Banda P and Teuscher C (2016) COEL: A Cloud-Based Reaction Network Simulator. Front. Robot. AI 3:13. doi: 10.3389/frobt.2016.00013
Received: 07 December 2015; Accepted: 14 March 2016;
Published: 04 April 2016
Edited by:
Fabrizio Riguzzi, Università di Ferrara, ItalyReviewed by:
Nicos Angelopoulos, Imperial College London, UKAmanda Clare, Aberystwyth University, UK
Copyright: © 2016 Banda and Teuscher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Banda, peter.banda@uni.lu
 Peter Banda
Peter Banda Christof Teuscher
Christof Teuscher