The Identification of Three Cancer Stem Cell Subpopulations within Moderately Differentiated Lip Squamous Cell Carcinoma
- 1Gillies McIndoe Research Institute, Wellington, New Zealand
- 2Wellington Regional Plastic, Maxillofacial and Burns Unit, Hutt Hospital, Wellington, New Zealand
Aim: To identify and characterize cancer stem cells (CSCs) in moderately differentiated lip squamous cell carcinoma (MDLSCC).
Method: MDLSCC samples underwent 3,3-diaminobenzidine (DAB) immunohistochemical (IHC) staining for squamous cell carcinoma marker EMA, CSC marker CD44 and embryonic stem cell markers NANOG, octamer-binding transcription factor 4 (OCT4), spalt-like transcription factor 4 (SALL4), sex-determining region Y-box 2 (SOX2), and phosphorylated signal transducer and activator of transcription 3 (pSTAT3). Immunofluorescent IHC staining was performed on two MDLSCC samples. Western blotting (WB) was used to confirm the expression of the aforementioned proteins and their transcription activation was investigated using NanoString and RT-qPCR.
Results: IHC staining demonstrated the presence of (1) an EMA+/CD44+/SALL4+/NANOG+/pSTAT3+/SOX2+/OCT4− CSC subpopulation within the tumor nests (TNs); (2) a CD44+/SALL4+/NANOG+/pSTAT3+/SOX2+/OCT4− CSC subpopulation; and (3) a CD44+/SALL4+/NANOG+/pSTAT3+/SOX2+/OCT4+ CSC subpopulation within the stroma, between the TNs. NanoString and RT-qPCR confirmed the presence of mRNA for CD44, SALL4, STAT3, SOX2, and OCT4, and WB confirmed the presence of NANOG, pSTAT3, SOX2, and OCT4.
Conclusion: This study demonstrates three putative CSC subpopulations within MDLSCC.
Introduction
Lip cancer constitutes a subsite of oral cavity cancer with more than 80% being squamous cell carcinoma (SCC) (1). There is a higher incidence of lip SCC in males in North America (12.7/100,000 per annum), Europe (12.0/100,000 per annum), and Oceania (13.5/100,000 per annum) (2) and the incidence is rising among females (2).
The etiological factors for oral cavity SCC (OCSCC) including lip SCC include allelic imbalance involving tumor suppressor genes (3), oncogenes (3), and carcinogen metabolizing enzymes (4), and the risk factors include tobacco use (5), immune deficiency (6), UV exposure (6), and human papilloma virus infection (7, 8). An overall 5-year survival rate of 88–97% has been reported (1, 9), and this is reduced to 78% in the presence of nodal metastasis and local recurrence (9–11). While surgery and radiotherapy are equally effective for the treatment of early lip SCC, combined treatment is required for advanced lesions (12).
There is growing evidence supporting the hierarchical model of carcinogenesis, which proposes that the development, growth, and spread of cancer are driven by a small population of cancer stem cells (CSCs) (13, 14). Various methods used for identification of CSCs in OCSCC include Hoechst dye exclusion, sphere forming assays, aldehyde dehydrogenase activity, and CSC marker identification (such as CD44, CD133, E-cadherin, keratins, and integrins) (14–16). More recent studies have utilized the embryonic stem cell (ESC) markers, sex-determining region Y (SRY)-box 2 (SOX2) (17), octamer-binding transcription factor 4 (OCT4) (17, 18), phosphorylated signal transducer and activator of transcription 3 (pSTAT3) (19), spalt-like transcription factor 4 (SALL4) (20), and Homeobox protein NANOG (18) to identify CSCs within OCSCC (21, 22).
CD44 is a transmembrane glycoprotein, a receptor for the glycosaminoglycan hyaluronan (23), and a CSC marker associated with cell proliferation, migration, and differentiation (24). SOX2 is a member of the SOX (SRY-related HMG Box) gene (25), encoding transcription factors with a single HMG DNA-binding domain and its suppression is considered vital to maintain stem cell pluripotency (17, 26). OCT4 is a POU domain transcription factor that works synergistically with SOX2 to regulate ESC pluripotency (27). SALL4 is an ESC marker that plays a role in multiple cancer types by regulating proliferation, apoptosis, chemoresistance, and maintenance of CSCs (23, 28), as well as modulating expression of pSTAT3, which is required for tumor formation, growth, and suppression of apoptosis (29). NANOG is a transcription factor that plays an essential role in maintaining stemness of ESC and controls cell proliferation, migration, and invasion (27, 30).
We have recently identified and characterized CSC subpopulations within oral tongue (21) and buccal mucosal (22) SCC. Although there are a number of reports on the presence and role of CSCs in OCSCC (21), there is paucity of data on the presence of CSC in lip SCC. This study aimed to identify and characterize the CSC subpopulations within moderately differentiated lip SCC (MDLSCC) using the SCC marker EMA, the CSC marker CD44, and the ESC markers SOX2, OCT4, pSTAT3, SALL4, and NANOG, at both the transcriptional and translational levels.
Materials and Methods
Tissue Samples
Previously untreated primary MDLSCC samples from one female and nine male patients, aged 46–94 years (mean, 64.4 years), were sourced from the Gillies McIndoe Research Institute and used for this study, which was approved by the Central Health and Disabilities Ethics Committee (ref. no. 12/CEN/74).
Histochemical and Immunohistochemical (IHC) Staining
Hematoxylin and eosin (H&E) staining was performed on 4-μm thick formalin-fixed paraffin-embedded blocks of 10 MDLSCC samples that were subsequently analyzed by an anatomical pathologist (Helen D. Brasch) to confirm the presence of SCC and the histological grading. 3,3-Diaminobenzidine (DAB) IHC staining for NANOG (1:100; cat# ab80892, Abcam, Cambridge, MA, USA), SOX2 (1:200; cat# PA1-094, Thermo Fisher Scientific, Waltham, MA, USA), SALL4 (1:30; cat# CM385M-16, Cell Marque, Rocklin, CA, USA), pSTAT3 (1:100; cat# 9145, Cell Signaling Technology, Danvers, MA, USA), OCT4 (1:1,000; cat# ab109183, Abcam), CD44 (1:1,500; cat# MRQ-13, Cell Marque), and epithelial membrane antigen (EMA, ready-to-use; cat# PA0035, Leica) diluted with Bond™ primary antibody diluent (Leica AR9352) was performed on the tissue sections using the Leica Bond Rx auto-stainer (Leica) as previously described (21, 31).
Immunofluorescent (IF) IHC staining was performed on two representative MDLSCC samples from the original cohort used for DAB IHC staining to investigate protein co-expression. Vectafluor Excel anti-rabbit 594 (ready-to-use; cat# VEDK-1594, Vector Laboratories, Burlingame, CA, USA) and Alexa Fluor anti-mouse 488 (1:500; cat# A21202, Thermo Fisher Scientific) were used to detect combinations that included NANOG, SOX2, or pSTAT3 and VectaFluor Excel anti-mouse (ready-to-use; cat# VEDK2488, Vector Laboratories) and Alexa Fluor anti-rabbit 594 (1:500; cat# A21207, Thermo Fisher Scientific) were used to detect combinations that included OCT4 or SALL4. All IF IHC-stained slides were mounted in Vectashield HardSet Antifade mounting medium with 4′,6-diamidino-2-phenylindole (cat#H-1500, Vector Laboratories).
Positive human control tissues used for the primary antibodies were skin for EMA, tonsil for CD44, pSTAT3 and SOX2, and seminoma for NANOG, SALL4, and OCT4. Negative antibody control was performed on one MDLSCC sample per antibody staining run.
Image Analysis
DAB IHC-stained slides were viewed using an Olympus BX53 light microscope (Tokyo, Japan) and the images were captured with the CellSens 2.0 software (Olympus). IF IHC-stained slides were viewed and the images were captured using an Olympus FV1200 biological confocal laser-scanning microscope and processed with the CellSens Dimension 1.11 software using 2D deconvolution algorithm (Olympus).
NanoString Gene Expression Analysis
Total RNA was isolated and quantified as previously described (31) from six snap-frozen MDLSCC samples from the original cohort of 10 patients used for DAB IHC staining. They were subjected to the NanoString nCounter gene expression assay (NanoString Technologies, Seattle, WA, USA) by New Zealand Genomics (Dunedin, New Zealand). Briefly, total RNA was extracted using the MagJET RNA kit (Thermo Fisher Scientific) with the protocol adapted for tissue and run on a KingFisher Duo machine (Thermo Fisher Scientific). RNA samples were then quantitated on a Qubit® 2.0 fluorometer (Thermo Fisher Scientific) and were subject to RNA integrity analysis via the 2100 Bioanalyzer Instrument (Agilent Technologies). Probes for the genes encoding CD44 (NM_001001392.1), NANOG (NM_024865.2), OCT4 (NM_002701.4), STAT3 (NM_139276.2), and the housekeeping genes glucuronidase beta (GUSB) (NM_000181.1), clathrin heavy chain (CLTC) (NM_4859.2), and hypoxanthine phosphoribosyltransferase 1 (NM_000194.1) were designed and manufactured by NanoString Technologies. Raw data were analyzed using nSolver™ software (NanoString Technologies) using standard settings and normalized against the housekeeping gene.
RT-qPCR
Total RNA was isolated from six snap-frozen MDLSCC samples from the original cohort of 10 patients used for DAB IHC staining using the RNeasy mini Kit (Qiagen) with a DNase digest and the QIAcube system (Qiagen). Total RNA quantity and quality were assessed using NanoDrop 2000 (Thermo Fisher Scientific). Reverse transcription reactions were performed using the iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad, Hercules, CA, USA). The expression of stem cell markers was detected using gene-specific TaqMan primers-probe sets (SOX2: Hs01053049_s1; SALL4: Hs00360675_m1) with the Rotor-Gene Multiplex RT-PCR Kit (Qiagen). All measurements were performed in triplicate and relative mRNA expression was determined by the ΔΔCt method. GAPDH was used as the endogenous control and probe-specific gBlocks gene sequences (IDT technologies) were used as calibrators. All samples with a threshold cycle ≥35.0 were considered negative. Graphs were generated with Microsoft Excel and results are shown as relative expression.
Western Blotting (WB)
Six snap-frozen MDLSCC samples from the original cohort of 10 patients included in DAB IHC staining underwent WB as previously described (32), using primary antibodies for NANOG (1:1,000; cat# ab47102, Abcam), OCT4 (1:2,000; cat# ab109183, Abcam), pSTAT3 (1:1,000; cat# 9145, Cell Signaling Technology), SOX2 (1:1,000; cat# PA1-094, Thermo Fisher Scientific), and β-actin (1:1,000; ab8226, Abcam). Two primary antibodies were used for SALL4 (1:1,000, cat# ab57577 and 1:1,000, cat# ab157172, both from Abcam). Secondary antibodies used were goat anti-rabbit horseradish peroxidase (HRP) conjugate (1:10,000; cat# A16110, Thermo Fisher Scientific) or Alexa Fluor® 647 rabbit anti-mouse (1:2,000; cat# A21239, Thermo Fisher Scientific) as appropriate. HRP conjugated secondary antibody detection was achieved using Clarity™ Western ECL substrate (Bio-Rad). Membranes were imaged using a ChemiDoc MP imaging system (Bio-Rad).
Results
Histochemical and IHC Staining
Hematoxylin and eosin staining confirmed the diagnosis and histological grading of MDLSCC in all 10 samples. Staining patterns for the CSC markers in all 10 samples are shown in Table S1 in Supplementary Material.
DAB IHC staining demonstrated that some cells within the tumor nests (TNs) stained positively with the SCC marker EMA (data not shown), as expected. There was also membranous staining of the CSC marker CD44 (Figure 1A, brown) that was localized to cells within the TNs and the stroma. Staining of NANOG (Figure 1B, red), pSTAT3 (Figure 1C, brown), SALL4 (Figure 1D, brown), and SOX2 (Figure 1E, brown) was localized to cells within the TNs and the stroma. Immunoreactivity for OCT4 (Figure 1F, red) was primarily cytoplasmic and focal in cells within the stroma.
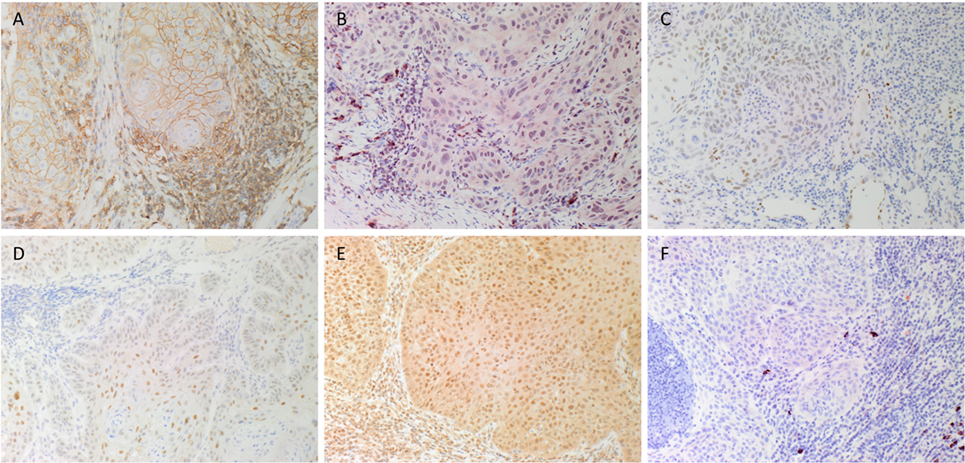
Figure 1. Representative DAB immunohistochemical-stained sections of moderately differentiated lip squamous cell carcinoma demonstrating nuclear membrane staining of CD44 [(A), brown] on cells within the stroma and cell membrane staining on cells within the tumor nests (TNs). Nuclear staining of NANOG [(B), red] was seen in cells within the TNs and the stroma. Patchy areas of weak staining for phosphorylated signal transducer and activator of transcription 3 [(C), brown] was detected on cells within the TNs. Focal moderate expression of spalt-like transcription factor 4 [(D), brown] on cells within the TNs and weak staining on cells within the stroma. Widespread and strong staining of sex-determining region Y-box 2 [(E), brown] was seen on cells within the TNs and the stroma. Staining for octamer-binding transcription factor 4 [(F), red] was limited to cells within the stroma. Original magnification: 200×.
Expected staining patterns for CD44, NANOG, pSTAT3, SALL4, SOX2, and OCT4 (Figure S1 in Supplementary Material) were demonstrated in the respective positive controls. An appropriate negative control by the omission of the primary antibody in MDLSCC samples provided a control for the secondary antibody (Figure S2 in Supplementary Material).
Immunofluorescent IHC staining performed on two representative MDLSCC samples from the original cohort of 10 patients used for DAB IHC staining demonstrated expression of the SCC marker EMA (Figure 2A, green) by cells within the TNs, while SOX2 (Figure 2A, red) was localized to cells within both the TNs and the stroma. Dual staining of SOX2 (Figures 2B,C, red) with CD44 (Figure 2B, green) and SALL4 (Figure 2C, green) showed that SOX2 was localized to cells within the TNs and the stroma that also expressed CD44 and SALL4. Co-staining of NANOG (Figure 2D, red) and pSTAT3 (Figure 2E, red) with CD44 (Figures 2D,E, green) demonstrated that the CD44+ cells within the TNs and the stroma also expressed NANOG and pSTAT3. Interestingly, the expression of OCT4 (Figure 2F, green) was confined only to a proportion of SOX2+ (Figure 2F, red) cells within the stroma. Images of the individual stains are presented in Figure S3 in Supplementary Material.
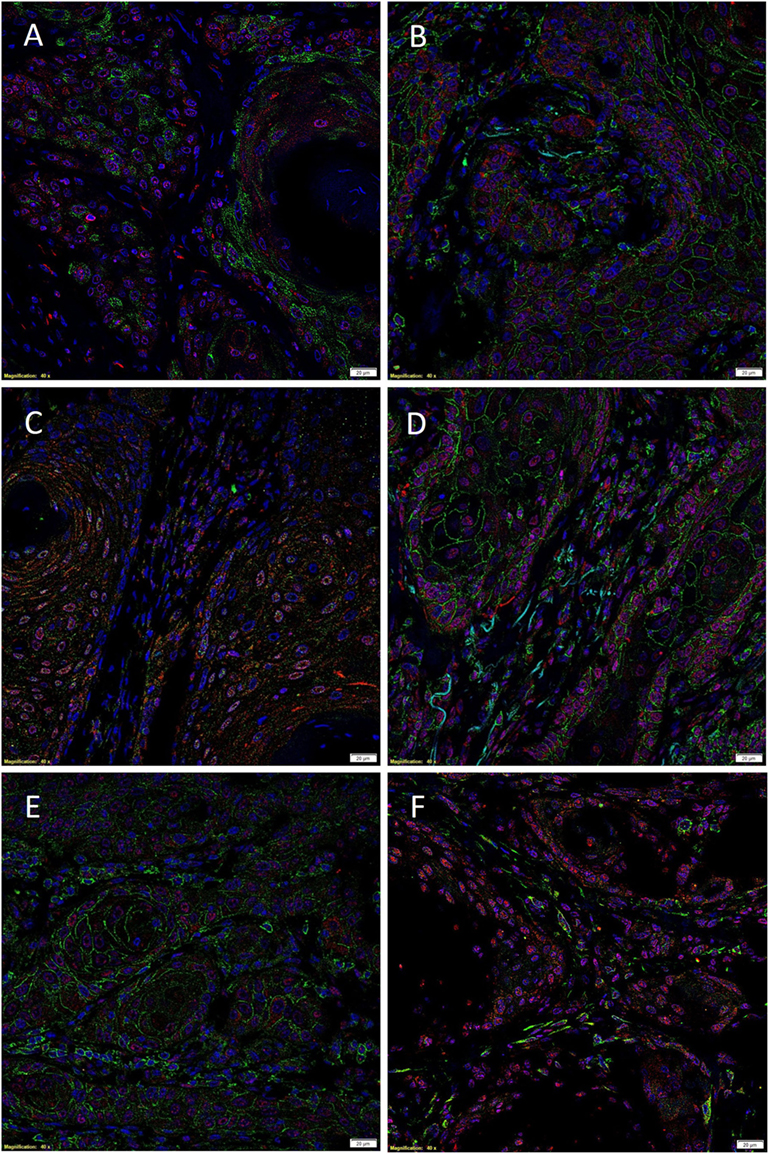
Figure 2. Representative immunofluorescent immunohistochemical-stained sections of moderately differentiated lip squamous cell carcinoma demonstrating the expression of sex-determining region Y-box 2 (SOX2) [(A), red] by the EMA+ [(A), green] cells within the TNs and the stroma. SOX2 [(B,C), red] was expressed by cells within both the TNs and the stroma that expressed CD44 [(B), green] and spalt-like transcription factor 4 [(C), green]. Expression of NANOG [(D), red] and phosphorylated signal transducer and activator of transcription 3 [(E), red] was seen in cells within the TNs and the stroma that expressed CD44 [(D,E), green]. Octamer-binding transcription factor 4 [(F), green] was expressed in a proportion of cells within the stroma that expressed SOX2 [(F), red]. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole [(A–F), blue]. Scale bars: 20 μm.
NanoString Gene Expression Analysis
NanoString transcriptional profiling of the six MDLSCC samples from the original cohort of 10 patients used for DAB IHC staining normalized against the housekeeping genes GUSB, CLTC, and HPRT1 confirmed the relative abundance of mRNA for CD44 and STAT3 in all the six samples. OCT4 was detected in four out of the six samples, while NANOG was undetectable in all six samples (Figure 3A).
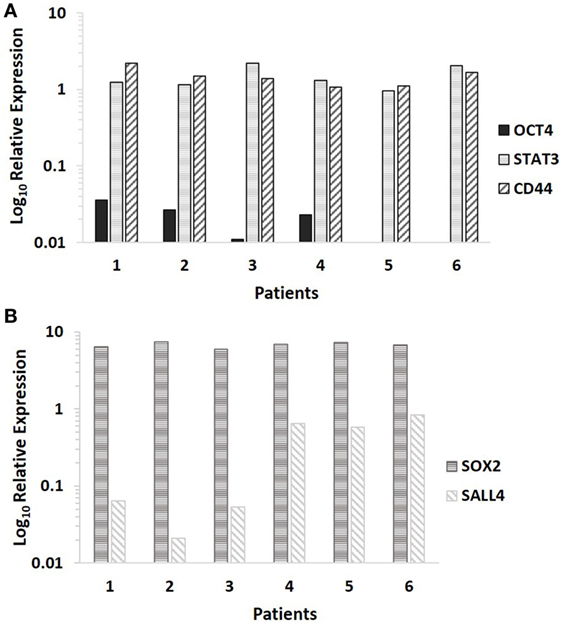
Figure 3. Log10 relative expression of cancer stem cell-related mRNA transcripts in six moderately differentiated lip squamous cell carcinoma samples analyzed by NanoString (A) and RT-qPCR (B). Expression is depicted relative to the clathrin heavy chain housekeeping gene (A) and the GAPDH housekeeping gene (B). CD44 and STAT3 were detected in all six samples, NANOG was undetectable in all six samples, while octamer-binding transcription factor 4 was detected in four out of six samples (A). Sex-determining region Y-box 2 and spalt-like transcription factor 4 were detected in all six samples (B).
RT-qPCR
RT-qPCR analysis of six snap-frozen MDLSCC samples from the original cohort of 10 patients used for DAB IHC staining demonstrated abundance of mRNA transcripts for SOX2 and SALL4 in all six samples (Figure 3B).
Western Blotting
Western blot analysis of six snap-frozen MDLSCC samples from the original cohort of 10 patients included in DAB IHC staining showed that NANOG was detected in all six samples (Figure 4A). OCT4 was detected in two of the six samples (Figure 4B), pSTAT3 was detected in five of the six samples (Figure 4C), and SOX2 was detected in all six samples (Figure 4D). A band corresponding to the expected size of SALL4 was undetectable by WB in any of the six samples (data not shown).
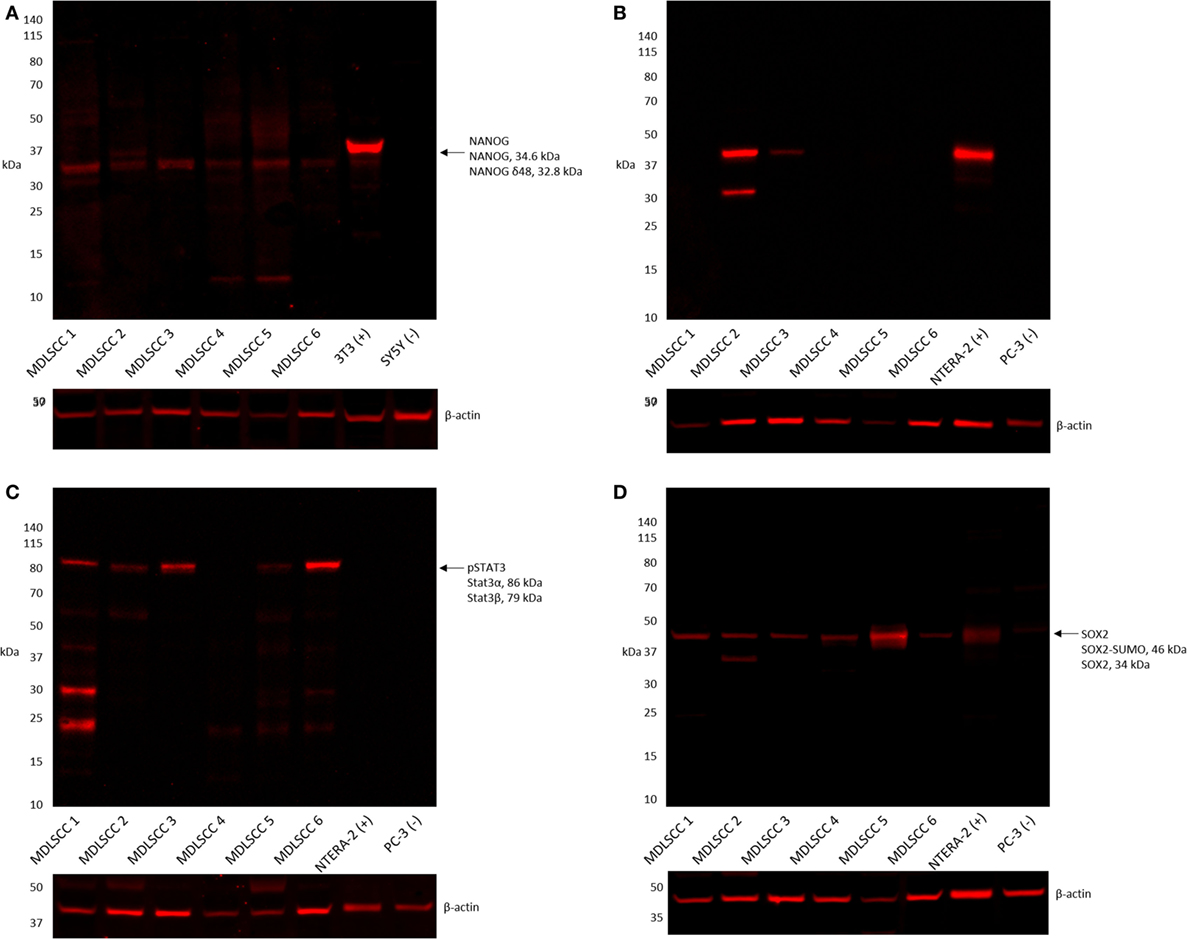
Figure 4. Representative western blot analysis performed on six moderately differentiated lip squamous cell carcinoma samples demonstrating detection of NANOG across all six samples (A), octamer-binding transcription factor 4 in two samples (B), phosphorylated signal transducer and activator of transcription 3 in five samples (C), and sex-determining region Y-box 2 in all six samples (D).
Discussion
This study adds to the growing body of evidence for the presence of CSCs in OCSCC. Our study demonstrates the presence of three distinct CSC subpopulations within MDLSCC (1): a CD44+/SALL4+/NANOG+/pSTAT3+/SOX2+/OCT4− subpopulation within the TNs (2); a CD44+/SALL4+/NANOG+/pSTAT3+/SOX2+/OCT4−, and (3) a CD44+/SALL4+/NANOG+/pSTAT3+/SOX2+/OCT4+ CSC subpopulation within the stroma of MDLSCC. It is exciting to speculate that the interactions between the CSCs within the TNs and the adjacent stromal environment may promote migration of CSCs away from the TNs, into the stroma, giving rise to the CSC subpopulations within the stroma, potentially via an epithelial–mesenchymal transition process (33–35). Alternatively, the primitive subpopulations within the stroma may represent normal “resident” stem cells, although this is a topic of further investigation.
The novel identification of more than one subpopulation of CSCs within MDLSCC parallels our recent findings in oral tongue (21) and buccal mucosa (22) SCC. Determination of which CSC subpopulation within the stroma possess the capacity for epithelial–mesenchymal transition (36) is a subject of further work, and we believe that a new paradigm opens up in the investigation of the biology of this tumor. The putative interplay between the stromal and TN CSC subpopulations raises the possibility of the CSC subpopulation within the TN giving rise to the CSC subpopulations within the stroma, which possess the ability to migrate away to establish local and distant TNs, although this is the subject of further investigation.
NANOG and SOX2 were expressed on cells within the TNs and the stroma. Overexpression of SOX2 in tumors has been correlated with increased tumor thickness and invasion, metastasis in esophageal cancer, drug resistance, and decreased survival in tumors such as breast cancer and lung adenocarcinoma (26, 37). Similarly, overexpression of NANOG is associated with unfavorable tumor features and poor survival of OCSCC patients (38).
Interestingly, OCT4 is expressed exclusively by the CSC subpopulation within the stroma. It is intriguing that the expression of OCT4 demonstrated by IHC staining in the stroma of all 10 MDLSCC samples was supported by NanoString and WB analyses in only two out of six samples. This may be due to sampling bias and/or antibody specificity. Investigating the expression of OCT4 in head and neck SCC samples of 119 patients, Koo et al. (39) demonstrate no staining and weak staining of OCT4 in 5% and 34% of the samples, respectively. Higher expression of OCT4 correlates significantly with poor histologic grade and worse overall and disease-specific survival in head and neck SCC (39).
Wu et al. (40) examined 156 pancreatic cancer tissue samples and found that high nuclear expression of pSTAT3 was associated with higher tumor grade and shorter median survival, compared to those with low expression of pSTAT3. Our IHC staining results showing weak expression of pSTAT3 by cells in both the TNs and the stroma is interesting and may reflect a tumor with relatively better prognosis. The relatively moderate expression of SALL4 as detected by IHC staining may be explained by the fact that only MDLSCC was analyzed in this study. High SALL4 expression levels have been correlated with poor overall survival of patients with aggressive tumors such as hepatocellular, endometrial cancer, gastric, and esophageal cancer (28).
This report demonstrates three putative subpopulations of CSCs within MDLSCC. It adds to the increasing support of the hierarchical concept of cancer. Further study may provide insights into the role CSCs may play in the biology of MDLSCC. It is exciting to speculate that CSCs may be a potential novel therapeutic target.
Limitations
1. This study included a relatively small sample size. A larger study is needed to confirm the observed expression pattern.
2. Further work is needed on well and poorly differentiated lip SCC lesions to compare the expression patterns observed in this study.
3. Functional cell culture work is needed to demonstrate the ability for the three putative CSC subpopulations to form an orthoptic model for this cancer.
Author Contributions
TI and ST formulated the study hypothesis and designed the study. RR, HB, TI, and ST interpreted the DAB and IF IHC data. JD performed WB analysis. JD, TI, PD, and ST interpreted the WB data. TI and ST interpreted the NanoString data. RR, TI, and ST drafted the manuscript. All authors commented on and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. TI, PD, and ST are inventors of the PCT patent application (No. PCT/NZ2015/050108) cancer diagnosis and therapy.
Acknowledgments
We thank Ms. Liz Jones and Ms. Alice M. Chibnall of the Gillies McIndoe Research Institute for their assistance in IHC staining and performing tissue processing for NanoString and analysis of the data, respectively. TI was supported, in part, by the Pacific Emerging Researcher First Grant, Health Research Council of New Zealand (grant no. HRC 16/434). Aspects of this work were presented at the Australia and New Zealand Head & Neck Cancer Society Annual Scientific Meeting and the International Federation of Head and Neck Oncologic Societies 2016 World Tour, 25-27 October 2016, Auckland, New Zealand.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/article/10.3389/fsurg.2017.00012/full#supplementary-material.
Abbreviations
DAB, 3,3-diaminobenzidine; CSC, cancer stem cell; ESC, embryonic stem cell; H&E, hematoxylin and eosin; IF, immunofluorescent; IHC, immunohistochemical; MDLSCC, moderately differentiated lip squamous cell carcinoma; OCSCC, oral cavity squamous cell carcinoma; SCC, squamous cell carcinoma; TN, tumor nest; WB, Western blotting.
References
1. Maruccia M, Onesti MG, Parisi P, Cigna E, Troccola A, Scuderi N. Lip cancer: a 10-year retrospective epidemiological study. Anticancer Res (2012) 32(4):1543–6.
2. Moore S, Johnson N, Pierce A, Wilson D. The epidemiology of lip cancer: a review of global incidence and aetiology. Oral Dis (1999) 5(3):185–95. doi:10.1111/j.1601-0825.1999.tb00300.x
3. Murugan AK, Munirajan AK, Tsuchida N. Ras oncogenes in oral cancer: the past 20 years. Oral Oncol (2012) 48(5):383–92. doi:10.1016/j.oraloncology.2011.12.006
4. Grimm M, Cetindis M, Lehmann M, Biegner T, Munz A, Teriete P, et al. Association of cancer metabolism-related proteins with oral carcinogenesis – indications for chemoprevention and metabolic sensitizing of oral squamous cell carcinoma? J Transl Med (2014) 12:208. doi:10.1186/1479-5876-12-208
7. Wei W, Zhang BY, Song YT, Sun JY, Chen C, Yu WB, et al. The analysis of human papillomavirus infection in lip squamous cell carcinoma patients. Chin J Exp Clin Virol (2012) 26(5):356–8.
8. Scully C, Kirby J. Statement on mouth cancer diagnosis and prevention. Br Dent J (2014) 216(1):37–8. doi:10.1038/sj.bdj.2013.1235
9. Zitsch RP III, Park CW, Renner GJ, Rea JL. Outcome analysis for lip carcinoma. Otolaryngol Head Neck Surg (1995) 113(5):589–96. doi:10.1177/019459989511300510
10. Califano L, Zupi A, Massari PS, Giardino C. Lymph-node metastasis in squamous cell carcinoma of the lip. A retrospective analysis of 105 cases. Int J Oral Maxillofac Surg (1994) 23(6 Pt 1):351–5. doi:10.1016/S0901-5027(05)80053-0
11. Australian Institute of Health and Welfare. Cancer survival and prevalence in Australia: period estimates from 1982 to 2010. Asia Pac J Clin Oncol (2013) 9(1):29–39. doi:10.1111/ajco.12062
12. Kerawala C, Roques T, Jeannon J, Bisase B. Oral cavity and lip cancer: United Kingdom national multidisciplinary guidelines. J Laryngol Otol (2016) 130(Suppl 2):S83–9. doi:10.1017/s0022215116000499
13. González-Moles MA, Scully C, Ruiz-Ávila I, Plaza-Campillo JJ. The cancer stem cell hypothesis applied to oral carcinoma. Oral Oncol (2013) 49(8):738–46. doi:10.1016/j.oraloncology.2013.04.002
14. Kaveh K, Nathan L, Reigh-Yi L. The role of cancer stem cells in head and neck squamous cell carcinoma and its clinical implications. In: Dmitry Bulgin E, editor. New Aspects in Molecular and Cellular Mechanisms of Human Carcinogenesis. Saint Louis, MO: InTech (2016). p. 97–113.
15. Costea DE, Tsinkalovsky O, Vintermyr OK, Johannessen AC, Mackenzie IC. Cancer stem cells – new and potentially important targets for the therapy of oral squamous cell carcinoma. Oral Dis (2006) 12(5):443–54. doi:10.1111/j.1601-0825.2006.01264.x
16. Krishnamurthy S, Nör JE. Head and neck cancer stem cells. J Dent Res (2012) 91(4):330–40. doi:10.1177/0022034511423393
17. Huang C-F, Xu X-R, Wu T-F, Sun Z-J, Zhang W-F. Correlation of ALDH1, CD44, OCT4 and SOX2 in tongue squamous cell carcinoma and their association with disease progression and prognosis. J Oral Pathol Med (2014) 43(7):492–8. doi:10.1111/jop.12159
18. Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res (2008) 14(13):4085–95. doi:10.1158/1078-0432.ccr-07-4404
19. Fasanaro E, Staffieri C, Cappellesso R, Marino F, Ottaviano G, Val M, et al. Prognostic significance of serine-phosphorylated STAT3 expression in pT1-T2 oral tongue carcinoma. Clin Exp Otorhinolaryngol (2015) 8(3):275–80. doi:10.3342/ceo.2015.8.3.275
20. Miettinen M, Wang Z, McCue PA, Sarlomo-Rikala M, Rys J, Biernat W, et al. SALL4 expression in germ cell and non-germ cell tumors: a systematic immunohistochemical study of 3215 cases. Am J Surg Pathol (2014) 38(3):410–20. doi:10.1097/pas.0000000000000116
21. Baillie R, Itinteang T, Yu HH, Brasch HD, Davis PF, Tan ST. Cancer stem cells in moderately differentiated oral tongue squamous cell carcinoma. J Clin Pathol (2016) 69(8):742–4. doi:10.1136/jclinpath-2015-203599
22. Yu HH, Featherston T, Tan ST, Chibnall AM, Brasch HD, Davis PF, et al. Characterization of cancer stem cells in moderately differentiated buccal mucosal squamous cell carcinoma. Front Surg (2016) 3(46):1–9. doi:10.3389/fsurg.2016.00046
23. Bradshaw A, Wickremsekera A, Tan ST, Peng L, Davis PF, Itinteang T. Cancer stem cell hierarchy in glioblastoma multiforme. Front Surg (2016) 3(21):1–15. doi:10.3389/fsurg.2016.00021
24. Chinn SB, Darr OA, Peters RD, Prince ME. The role of head and neck squamous cell carcinoma cancer stem cells in tumorigenesis, metastasis, and treatment failure. Front Endocrinol (2012) 3(90):1–6. doi:10.3389/fendo.2012.00090
25. Amini S, Fathi F, Mobalegi J, Sofimajidpour H, Ghadimi T. The expressions of stem cell markers: Oct4, Nanog, Sox2, nucleostemin, Bmi, Zfx, Tcl1, Tbx3, Dppa4, and Esrrb in bladder, colon, and prostate cancer, and certain cancer cell lines. Anat Cell Biol (2014) 47(1):1–11. doi:10.5115/acb.2014.47.1.1
26. Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med (2014) 3(1):19. doi:10.1186/2001-1326-3-19
27. Hadjimichael C, Chanoumidou K, Papadopoulou N, Arampatzi P, Papamatheakis J, Kretsovali A. Common stemness regulators of embryonic and cancer stem cells. World J Stem Cells (2015) 7(9):1150–84. doi:10.4252/wjsc.v7.i9.1150
28. Zhang X, Yuan X, Zhu W, Qian H, Xu W. SALL4: an emerging cancer biomarker and target. Cancer Lett (2015) 357(1):55–62. doi:10.1016/j.canlet.2014.11.037
29. Suiqing C, Min Z, Lirong C. Overexpression of phosphorylated-STAT3 correlated with the invasion and metastasis of cutaneous squamous cell carcinoma. J Dermatol (2005) 32(5):354–60. doi:10.1111/j.1346-8138.2005.tb00906.x
30. Zhang Z, Filho MS, Nor JE. The biology of head and neck cancer stem cells. Oral Oncol (2012) 48(1):1–9. doi:10.1016/j.oraloncology.2011.10.004
31. Tan EM, Chudakova DA, Davis PF, Brasch HD, Itinteang T, Tan ST. Characterisation of subpopulations of myeloid cells in infantile haemangioma. J Clin Pathol (2015) 68(7):812–8. doi:10.1136/jclinpath-2014-202846
32. Itinteang T, Dunne JC, Chibnall AM, Brasch HD, Davis PF, Tan ST. Cancer stem cells in moderately differentiated oral tongue squamous cell carcinoma express components of the renin-angiotensin system. J Clin Pathol (2016) 69(10):942–5. doi:10.1136/jclinpath-2016-203736
33. Peek EM, Li DR, Zhang H, Kim HP, Zhang B, Garraway IP, et al. Stromal modulation of bladder cancer-initiating cells in a subcutaneous tumor model. Am J Cancer Res (2012) 2(6):745–51.
34. Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res (2008) 68(3):918–26. doi:10.1158/0008-5472.can-07-5714
35. De Wever O, Mareel M. Role of tissue stroma in cancer cell invasion. J Pathol (2003) 200(4):429–47. doi:10.1002/path.1398
36. Xu H, Tian Y, Yuan X, Wu H, Liu Q, Pestell RG, et al. The role of CD44 in epithelial-mesenchymal transition and cancer development. Onco Targets Ther (2015) 8:3783–92. doi:10.2147/ott.s95470
37. Forghanifard MM, Ardalan Khales S, Javdani-Mallak A, Rad A, Farshchian M, Abbaszadegan MR. Stemness state regulators SALL4 and SOX2 are involved in progression and invasiveness of esophageal squamous cell carcinoma. Med Oncol (2014) 31(4):922. doi:10.1007/s12032-014-0922-7
38. Lee HJ, Kang YH, Lee JS, Byun JH, Kim UK, Jang SJ, et al. Positive expression of NANOG, mutant p53, and CD44 is directly associated with clinicopathological features and poor prognosis of oral squamous cell carcinoma. BMC Oral Health (2015) 15(1):153. doi:10.1186/s12903-015-0120-9
39. Koo BS, Lee SH, Kim JM, Huang S, Kim SH, Rho YS, et al. Oct4 is a critical regulator of stemness in head and neck squamous carcinoma cells. Oncogene (2015) 34(18):2317–24. doi:10.1038/onc.2014.174
Keywords: lip, oral cavity, squamous cell, carcinoma, cancer stem cell, cancer
Citation: Ram R, Brasch HD, Dunne JC, Davis PF, Tan ST and Itinteang T (2017) The Identification of Three Cancer Stem Cell Subpopulations within Moderately Differentiated Lip Squamous Cell Carcinoma. Front. Surg. 4:12. doi: 10.3389/fsurg.2017.00012
Received: 03 December 2016; Accepted: 13 February 2017;
Published: 06 March 2017
Edited by:
Vincent Vander Poorten, KU Leuven, BelgiumReviewed by:
A. B. Zulkiflee, University Malaya Medical Centre, MalaysiaTommaso Lombardi, Hôpitaux Universitaires de Genève, Switzerland
Copyright: © 2017 Ram, Brasch, Dunne, Davis, Tan and Itinteang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Swee T. Tan, swee.tan@gmri.org.nz
†Equal senior authors.
 Rachna Ram
Rachna Ram Helen D. Brasch
Helen D. Brasch Jonathan C. Dunne
Jonathan C. Dunne Paul F. Davis
Paul F. Davis Swee T. Tan
Swee T. Tan Tinte Itinteang
Tinte Itinteang