The Potential Link between Thermal Resistance and Virulence in Salmonella: A Review
- 1Cell and Molecular Biology Program, University of Arkansas, Fayetteville, AR, United States
- 2Center for Food Safety, University of Arkansas, Fayetteville, AR, United States
- 3Department of Food Science, University of Arkansas, Fayetteville, AR, United States
- 4Department of Poultry Science, University of Arkansas, Fayetteville, AR, United States
In some animals, the typical body temperature can be higher than humans, for example, 42°C in poultry and 40°C in rabbits which can be a potential thermal stress challenge for pathogens. Even in animals with lower body temperatures, when infection occurs, the immune system may increase body temperature to reduce the chance of survival for pathogens. However, some pathogens can still easily overcome higher body temperatures and/or rise in body temperatures through expression of stress response mechanisms. Salmonella is the causative agent of one of the most prevalent foodborne illnesses, salmonellosis, and can readily survive over a wide range of temperatures due to the efficient expression of the heat (thermal) stress response. Therefore, thermal resistance mechanisms can provide cross protection against other stresses including the non-specific host defenses found within the human body thus increasing pathogenic potential. Understanding the molecular mechanisms associated with thermal responses in Salmonella is crucial in designing and developing more effective or new treatments for reducing and eliminating infection caused by Salmonella that have survived heat stress. In this review, Salmonella thermal resistance is assessed followed by an overview of the thermal stress responses with a focus on gene regulation by sigma factors, heat shock proteins, along with the corresponding thermosensors and their association with virulence expression including a focus on a potential link between heat resistance and potential for infection.
Introduction
Salmonella is a Gram-negative foodborne pathogen that is a major concern for the food industry and public health authorities because of its capability to cause both widespread contamination and infection within the United States (US) and worldwide (1–5). An estimated one million cases of Salmonella-related illnesses occur annually within the US. For example, in 2014, Salmonella was responsible for 10 multistate outbreaks with approximately 1,000 reported outbreak cases (3, 6). Numerous strategies have been implemented to reduce Salmonella transmission, contamination, and infection. Salmonella infections are most commonly acquired through ingestion of contaminated foods such as eggs and poultry meat (7). Salmonella can colonize the small intestines of poultry birds, along with the cecum, without demonstrating any symptoms related to Salmonella infections. Therefore, poultry serves as an efficient vector of transmission for multiple serovars of Salmonella to humans through consumption of contaminated food products.
In order for Salmonella to survive and colonize the human body, it must overcome multiple non-specific host defenses encountered within the host such as low pH, limited nutrient availability and in poultry birds, a high body temperature (42°C). Due to the wide temperature range that Salmonella may grow in, it must possess specific mechanisms that can overcome thermal stress to proliferate and survive. However, prior to ingestion, Salmonella is already preexposed to a higher core body temperature in poultry compared to humans (37°C). During infection, one of the primary defenses of the innate immunity is an increase in body temperature through pyrogens (antigens that stimulate fever) such as lipopolysaccharide found in the cell wall of Gram-negative bacteria (8). This preexposure could increase the potential of Salmonella to establish infection of the host due to adaption to higher temperatures. Therefore, the aim of this review is to provide an overview of phenotypic and molecular responses to temperature changes as it relates to poultry, thermal stress regulation, and how this increases pathogenic potential of Salmonella.
Thermal and Non-Thermal Stresses
With over 2,500 serovars of Salmonella, several have developed the ability to overcome high temperatures allowing for survival through thermal processing; however, this is strain specific (9–11). O’Bryan et al. (12) reviewed the thermal resistance of Salmonella species and other foodborne pathogens associated with meat and poultry. They concluded that a variety of factors and parameters are involved in the thermal resistance and inactivation of those pathogens and spoilage microorganisms such as various temperature exposures, growth phase, and the intrinsic conditions of the food product. Strains of the same microbial species were found to be capable of responding differently to the same treatments possibly due to specific variations in gene composition for each respective strain. Likewise, the stages of growth, the age of the culture, and the conditions of bacterial growth have yielded various outcomes regarding heat inactivation or destruction of microorganisms, which could contribute to determining the best methods to reduce microbial growth and contamination within these products (12).
There are several factors that allow Salmonella strains to survive the food processing environment and overcome thermal treatment. For example, preexposure to stress and growth conditions prior to thermal treatment could increase survival capability during processing. Specifically, S. Senftenberg was found to survive in broiler litter for up to 24 h at 80°C (13). Microorganisms tested against heat are known to elicit different responses in regard to prior growth conditions with stationary phase cells being more resistant to heat than log phase cells (14–16). In addition, stressed cells such as those exposed to temperatures slightly above an organism’s optimal growth range (heat shocked cells), those grown on limited carbon sources, those experiencing desiccation, and those undergoing starvation stress prior to heat treatments have been shown to exhibit more thermal tolerance (17–21).
Exposure to non-thermal stress may also have an impact on the capability of Salmonella to respond to thermal threats. For example, Milillo and colleagues concluded that combining organic acids with heat can effectively reduce Salmonella over a short period of time (22, 23). They applied mild thermal treatments and organic acids with a 1-min exposure time. Sodium propionate in combination with heating was demonstrated to be the most significantly effective for reducing viable Salmonella (22). In a follow-up study, Milillo et al. (23) conducted microarray experiments to explore the specific response of S. Typhimurium to organic acids in combination with heat. Exposure to sodium acetate with heat (55°C) and sodium propionate with heat (55°C) led to differentially 288 (124 upregulated and 168 downregulated genes) and 319 (131 upregulated and 181 downregulated genes) gene expression level changes, respectively. Numerous heat shock genes including dnaK, hptJ, dnaJ, grpE, clpP, and hscAB were repressed by both treatments. They concluded that this synergism may be attributed to damage in the synthesis of heat shock genes of S. Typhimurium due to membrane damage. Given the potential for such synergism among otherwise unrelated interventions, there may be opportunities for optimizing hurdle technologies in the food industry and demonstrating the utility of using genomic screening to develop application approaches for these technologies.
Thermosensors
In order for Salmonella to overcome and adapt to an ever-changing environment it must overcome stressors encountered during its travel through the host; therefore, adaptation through sensory mechanisms is imperative. Thermosensors are considered the cell’s “thermometer” by utilizing various types of biological systems to detect temperature fluctuations within the cell. There are four different groups of thermosensors including proteins, lipids and membrane fluidity, RNA’s that are temperature responsive, and DNA structure and topology. Thermosensors play a major role in temperature detection and are found within the 5′ UTR region, which can regulate gene expression to produce adaptive heat stress responses. When temperature decreases or increases to harmful levels, stress responses (cold and heat shock) are needed to protect the bacterial cell and are thoroughly dependent on bacterial signal transduction mechanisms (24). Genes involved in these mechanisms are regulated at different genetic stages beginning from transcription through translation and into posttranslational levels (25, 26).
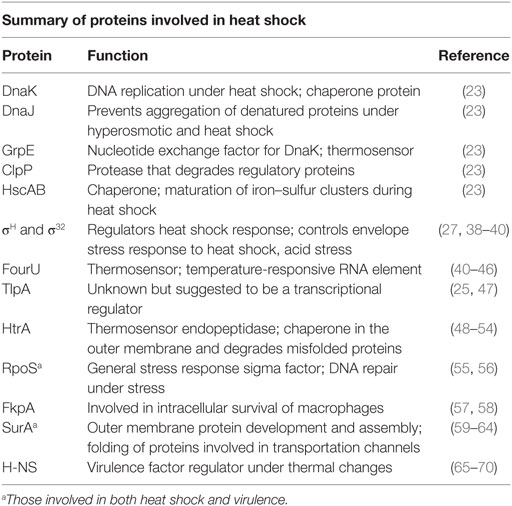
As a protective reaction, misfolded and unfolded proteins are present in considerable numbers in the cytoplasmic membrane and the outer membrane during exposure to higher than optimal temperatures which, in turn, initiates the expression of heat shock proteins (HSPs) through the regulation of the heat shock factor σH (27–31). Proteins involved in heat shock are summarized in Table 1. Induction of HSP formation is accomplished through the production of chaperones, proteases, and small heat shock proteins (s-HSPs). These function in protection, refolding salvaged proteins, removing damaged proteins, and repairing degrading protein aggregation (32–37).
FourU is a family of thermosensors located at the untranslated region (5′-UTR). This temperature-responsive RNA element contains a stretch of four uridine nucleotides within the ribosomal binding site. It pairs with a sequence of AGGA and was initially discovered in S. Typhimurium as the small heat shock gene agsA, aggregation suppression A (40–46). Afterward, a similar RNA thermometer was also confirmed to be associated with Yersinia virulence through the induction of the transcriptional activator lcrF (44, 71).
TlpA, TIR-like protein A, is considered one of the first reported proteins with thermosensory gene regulation activity to the high temperature response (HTR) encoded by Salmonella enteric virulence plasmid, pSLT (25, 47). It is a robust homolog to a eukaryotic protein family known as tropomyosin, and the structure of TlpA is in a dimer form with an unusually long alpha-helical coiled coil structure (72). It consists of an N-terminal DNA binding domain and exhibits transcriptional autoregulatory repression activity. At temperatures below 30°C, the transcription of tlpA is low and the TlpA repression activity is high. The TlpA exists in two forms, as a dimeric α-helical (folded) coiled coil oligomer at low temperature (28°C) and an unfolded (non-functional) monomer at high temperature (37°C) that leads to increased transcription (25, 73, 74). Although the function of this protein is still unidentified, it was demonstrated that this transcriptional regulator was not essential for virulence of Salmonella using a mouse infection model (75).
Another thermosensing gene known as htrA, high temperature requirement A, is a member of the serine proteases group within the endoproteases family and is regulated by sigma factor E (48–52). It is a highly conserved gene in numerous microorganisms and was first discovered in Escherichia coli as degP. At low temperature, the protein HtrA (DegP) functions as a chaperone in the outer membrane; however, at high temperatures, it acts as a protease to degrade misfolded proteins with ATP-independent activity and other cofactors (53, 54). An earlier study also linked the activity of this gene to its sensitivity to thermal stress (76). A strain with a mutation in this gene exhibited an inability to grow at high temperature characterized by the inability to degrade unfolded proteins in the periplasmic space. S. Typhimurium was less affected by the sigma factor E mutation than E. coli (77–79).
Cellular Responses and Regulation to Heat Stress
Salmonella can proliferate either in a planktonic form, floating freely within a liquid medium, or attach and grow while immobilized to a solid medium. A large number of proteins form the family of s-HSP that consists of proteins with up to 50 amino acids, which are considered energy free and universally found in numerous microorganisms with diverse group and variable molecular weights. These proteins possess chaperone-like functions and commonly maintain protein homeostasis. The s-HSPs are active primarily during stress to stabilize cell proteins at diverse cellular activities (metabolism, translation, transcription, and others), binding unfolded proteins and forming a complex that blocks non-specific irreversible aggregation (80–83). With the detection of heat stress, the adaptive regulation of genes is initiated with the expression of sigma factors. Two sigma factors are generally expressed: a cytoplasmic thermal stress response regulated by heat shock sigma factor, σH or σ32, and an extracytoplasmic thermal stress response regulated by the extracytoplasmic function sigma factor, σE or σ24, also known as extreme heat stress sigma factor (84–88).
Sigma factors comprise a large group of genes expressing proteins with critical mechanisms associated with the RNA polymerase holoenzyme complex that function as guidance for core RNA polymerases to recognize their promoters and initiate transcription. The sigma factors are primarily divided into two categories, sigma factor 70 family (σ70) that coordinates the transcriptional activities in various stress responses, also known as σA in Bacillus subtilis and other bacterial species (51, 89–91), and a second identified family of sigma factors encoded by rpoN, known as sigma factor 54 (σ54/N) (92–94), identified in Campylobacter jejuni, Enterococcus faecalis, Listeria monocytogenes, and Pseudomonas spp. (29, 95–97).
Heat shock responses are regulated by the alternative sigma factors σ32/H and σ24/E. These two factors make up the third and fourth subgroups of sigma factors encoded by rpoH and rpoE genes, respectively (98, 99). rpoH regulates the transcription of heat shock genes and is itself regulated during translation. When the temperature is at an optimal microbial growth range, the translation of the rpoH gene is blocked. The stem III and I of the rpoH mRNA secondary structure is liberated with increasing temperatures (42°C), facilitating the ribosomal binding and enhancing the efficiency of translation (27, 38–40). Sigma factors associated with heat stress response have been demonstrated to regulate over a 100 genes. Of those, sigma factor σ32/H controls more than 30 proteins, most of which are associated chaperones and proteases (30, 31, 100–103). A more recent study by Lim et al. (104) made it clear that σ32/H is not just localized at the bacterial cytoplasm but is also found in the inner membrane through a direct interaction with the signal recognition particle and its signal receptor.
Proteases expressed by sigma factor σ32/H can control and decrease the expression of the membrane HSPs to a level as needed by the cell to withstand environmental stresses. For instance, FtsH is one of the ATP-dependent proteases, which possesses numerous cellular functions and has been demonstrated to be very critical to E. coli viability (105–108). In addition, FtsH functions as a protein qualifying protease and has a role in membrane protein degradation activities primarily those with SsrA-tagged cytoplasmic proteins at their carboxy terminal (109, 110). FtsH degrades MgtC, a membranous protein with five transmembrane domains. MgtC, a virulence factor, has been identified as being required for survival inside macrophages (111). Katz and Ron (108) demonstrated a maintenance role of FtsH for lipopolysaccharide biosynthesis with a shielding permeability function (108, 112–114). Although σ32/H and σ24/E are alternative sigma factors, σ32/H regulates HSPs for the cytoplasmic components and σ24/E regulates the extracytoplasmic (cell envelope) proteins in response to high temperatures and other envelope stress factors (50, 52, 88, 115–118). An interesting finding is that one of the four promoters of rpoH gene expression is regulated by σ24/E for additional coordination of thermal responses requiring both cytoplasmic and extracytoplasmic components (119–124).
Heat Shock and Virulence
The adaptation of Salmonella to heat shock can also lead to a range of other effects, including an increase in virulence potential through gene regulatory mechanisms. Exposing Salmonella to thermal stress results in protective responses and can induce changes in gene expression levels of virulence genes. Numerous chaperones and proteases regulated by the alternative heat shock factors, σH/32, σE/24, and others such as σS (RpoS) are notably involved in bacterial virulence with several studies linking these proteins to Salmonella and E. coli virulence factors (51, 125–129). Although both σH/32 and σE/24 are regulators for heat shock stress, their molecular mechanisms for initiation responses are not similar.
Sirsat et al. (130) examined the effect of heat stress on S. Typhimurium gene expression using transcriptional profiling. Microarray analysis was applied to identify the thermal stress response of S. Typhimurium at a sublethal temperature of 42°C with 144 upregulated and 167 downregulated genes detected. These genes belonged to various functional categories, but primarily to the general stress response sigma factor S (RpoS) and HSPs, and to sigma factors H and E (RpoH and RpoE). The latter protein has been shown to be critical in the virulence of numerous pathogens (131–133). However, RpoS regulates genes responsible for lethality in mice where preadaptation through RpoS by increasing virulence potential of Salmonella cells that survive processing as suggested by Dodd and Aldsworth [(55); Ibanez-Ruiz et al. (56)]. Therefore, sigma factors and HSPs may increase pathogenic potential by overcoming various stressors and increasing pathogenic and colonization potential. Interestingly, research has indicated that RpoS can function as a DNA repair protein that is active under stressful conditions. Thermal stress can induce DNA damage suggesting that there is correlation between thermal stress, the general stress response, and virulence of Salmonella (56). However, more research is needed to confirm this. Generally, genes associated with stress and energy metabolism represent the first responses of the cells to tolerate heat stress. These genes may possibly give the pathogen cross-resistance to other stresses and result in more virulent cells. The study conducted by Sirsat et al. (130) was considered the first to report that sublethal heat stress-influenced Salmonella interaction with Caco-2 cells through the expression of fimbriae-associated genes. Genes of two Salmonella pathogenicity islands (SPI-2 and SPI-5) were upregulated, resulting in improved adhesion (SPI-5 only) and survival in the host while genes of SPI-1 were downregulated.
A loss of rpoE gene activity has also been shown to cause a defect in cell viability of E. coli and increase cell envelope stress (50, 118, 122, 134). In Salmonella, rpoE mutants were found to be less responsive to heat shock temperatures, exhibiting an intracellular defect in the survivability within a macrophage and becoming avirulent in a mouse infection model (126, 135–137). In addition, the rpoE gene has been shown to be essential in response to starvation stress (138), oxidative stress (92), antimicrobial peptide resistance (139), and osmotic and cold stresses (127). Lewis et al. (128) discovered that both functions of htrA, are important with the function of the proteases being most critical inside the host. A more recent study verified that HtrA protein activity is critical for S. Enteritidis persistence in egg whites at 42°C (129).
FkpA, an FKBP-type periplasmic peptidyl-prolyl cis/trans isomerase (PPIase), is involved in heat tolerance (116). This protein is comparable to proteins known as macrophage infectivity potentiators found in other pathogenic bacteria and improves the survivability and proliferation inside the macrophages and epithelial cells (140). Horne et al. (141) demonstrated that a mutation in fkpA causes the corresponding Salmonella strain to become avirulent; however, Humphreys et al. (57) argued that a single mutant deletion of fkpA was not enough to reach that conclusion. They observed that only when combining that mutation with one of the other σE regulated genes, surA or htrA, would the virulence of S. Typhimurium be disrupted (57, 58). In a more recent study reported by Weski and Ehrmann (142), they conducted a genetic analysis of chaperones and proteases of E. coli associated with the cell envelope, evaluating single and double mutant deletions under different growth conditions. A fkpA mutation was examined at 37 and 42°C using rich medium agar plates with and without 0.5 M NaCl with the corresponding mutants found to not exhibit any detectable defects under any of the conditions. However, when combining this strain with another mutation in dsbA, disulfide bond formation A, the strain displayed weak growth on the hyperosmolar media when incubated at 37°C, while no sign of growth was observed on the hyperosmolar media when incubated at 42°C with a minimal growth of dsbA single mutants at the latter condition (142, 143).
SurA, survival protein A, is also a PPIase. It is regulated by σE and contributes to thermotolerance fitness. This protein participates in outer membrane protein (OMPs) development and assembly and plays a role in the folding of transportation channels, known as porins (59–64). Sklar et al. (62) observed that the surA role is associated with the initial phases of OMP biosynthesis. Previously, Tormo et al. (144) had demonstrated that surA was critical to E. coli for survival during stationary phase. Tamae et al. (145) screened approximately 4,000 single mutant deletions, among them a ΔsurA that exhibited chemical sensitivity to the drugs and detergents used in the study. It is not clear whether similar functionalities exist with surA in Salmonella but it does appear to have the same association with virulence in the host. Sydenham et al. (146) found a mutation of surA in S. Typhimurium that exhibited extensive attenuation when introduced to mice orally or intravenously. It has also been demonstrated in several studies that surA is a critical factor for OMP transport and associated with virulence of uro-pathogenic E. coli and Salmonella (64, 147). Using a high-throughput Tn-seq technique to screen the entire genome, Khatiwara et al. (148) identified numerous genes in S. Typhimurium associated with high temperatures with surA identified as a gene associated with growth at 42°C.
Numerous studies have associated virulence factors with thermal changes that mediate DNA topology. These modifications include overall DNA helical conformation “supercoiling,” the degree of helical twists and coiling (25, 149–152), or alterations in the specific-sequence curvature of chromosomal or plasmid DNA (153–156). Some studies have demonstrated that DNA topology plays a role in Salmonella pathogenicity (157, 158). Positive DNA supercoiling after heat exposure causes DNA to be twisted in a right-handed fashion until it generates a knot, as seen mainly in plasmid DNA, and is controlled by DNA gyrase and topoisomerase I. Changes at the level of DNA supercoiling trigger SPI-1 gene expression levels and initiate the subsequent intestinal invasion. Once inside the host cells, the DNA changes its form and as a result, SPI-2 genes are induced (159, 160). For a more detailed discussion of SPI-1 and SPI-2 regulation, please see Ref. (160).
The second mechanism is through a recognized bending DNA sequence “promoter-curvature.” Commonly, this bending DNA region is an AT-rich sequence that has been primarily identified in the 5′-end upstream of the promoter region influencing RNA polymerase binding as a silencing factor. Initially, thermal stress induces some alterations in the DNA topology as bends in the AT-rich sequence regions on the transcriptional level. This can influence the interaction between RNA polymerase and the promoter region, altering gene expression (155, 156).
Cross Protection
The microorganisms’ responses to temperature changes (inflammation, fever) vary from one microorganism to another with cell metabolic changes occurring when sensing external environmental shifts resulting in protection from certain stresses and/or cross protection for other additional stresses (130, 161, 162). This can be a major concern within the host by increasing potential for overall pathogenesis. Prior exposure to prevention strategies utilized within industry before human consumption occurs could increase survivability of Salmonella and their ability to establish infection once ingested (163). When Nielsen et al. (164) compared two different growth forms of S. Typhimurium, immobilized versus planktonic cells, diverse responses were elicited in response to heat shock at 45°C for 30 min. The results revealed that 538 genes were expressed differently with flagellar and virulence genes upregulated in the immobilized heat stressed cells compared to the non-stressed cultures. Greater invasiveness was observed in immobilized HeLa cells after this sublethal treatment compared to decreased invasiveness in the planktonic cells. Based on this study, it would appear that inadequate cooking and heat treatments during food processing could actually increase survival and thermal resistance of Salmonella and other foodborne pathogens through cross protection by increasing virulence capability (164–166). Gruzdev et al. (21) found that desiccated Salmonella cells in sterile deionized water showed high tolerance to dry heat at 60°C with no significant population change within 1 h, in comparison to a 3-log reduction in the number of non-desiccated cells under identical conditions. A previous study also found that Salmonella cells that had previously adapted to desiccation conditions survived substantially longer in aged chicken litter than non-adapted control cells exposed to the same treatment (13).
However, as environmental conditions change, Salmonella must be able to rapidly adapt through alterations in gene expression in order to overcome stress efficiently. For example, this can be accomplished through attachment, which results in a phenotypic change allowing Salmonella to become more resistant to thermal stress than cells in planktonic form (167–170). Multiple studies have concluded that modifications of the membrane fatty acid composition of Salmonella strains were directly associated with their ability to resist thermal treatment where those cells with less membrane fluidity possessed greater thermal resistance (171–174). Similarly, in E. coli, the increase in membrane fluidity also leads to increased synthesis of HSPs, thus suggesting that membrane composition is directly related to thermal resistance (175). Under low temperatures, the physiological state of the cell can switch to a reversible, less fluid like lipid bilayer, whereas under high temperatures, the state of the cell switches to a membrane with higher fluidity. This is regulated by thermosensors (175). A review by O’Bryan et al. (12) noted that foodborne pathogens in contaminated food products possessing a high fat content demonstrated increased pathogenic potential.
A well-known gene encodes for nucleotide-associated protein (H-NS), a histone-like nucleotide-structuring protein, which has been associated with virulence factors as a temperature-dependent phenotype (65–70). This protein is considered a common transcriptional regulator that can be induced by thermal changes in Salmonella. At low temperature, H-NS binds to an AT-rich sequence and forms a complex. When temperature rises to 37°C (host body temperature), the binding capacity is reduced until dissociation occurs, leading to virulence gene expression. This mechanism was demonstrated in E. coli K-12 to control over 60% of the genes regulated by temperature including virulence factors (176). The association of H-NS with virulence has been verified in other pathogens such as Salmonella (69, 177–179), Shigella, Yersinia enterocolitica, and Yersinia pseudotuberculosis (180–182). Two studies were conducted to identify the mechanism of H-NS in Salmonella. The first study was performed on S. Typhimurium LT2, and it was noted that H-NS negatively regulated approximately 254 genes (69). The second study was carried out on S. Typhimurium 14028 (183), and it was discovered that 265 unique Salmonella genes were negatively associated with H-NS which contained low G + C content (183). In both studies, among the identified genes were those present in SPI-1, 2, 3, and 5 (184–186).
A more recent study by Pesingl et al. (187) demonstrated that protein-l-isoaspartyl methyltransferase (PIMT) is required by Salmonella to survive at 42°C and it in turn contributes to virulence capability in poultry (body temperature of 42°C). Proteins were susceptible to damage induced by thermal stress and thus PIMT assisted in prevention and repair of proteins. Under stress, aspartate is converted to iso-aspartate, which can lead to unfolded proteins and modified amino acids residues (188). Pesingl et al. (187) found that PIMT contributes to survival under both thermal and oxidative stress during stationary phase due to its direct role in protection of proteins at elevated temperatures. Therefore, further research is needed in the correlation between the heat shock responses and virulence gene expression and how their respective regulation patterns influence the pathogenic potential of Salmonella.
Conclusions
Salmonella typically encounters various thermal stresses that can be host-specific and can represent a component of the overall immune and physiological response to infection. However, Salmonella spp. have developed thermal resistance mechanisms to overcome these changes in host temperature through the induction of stress response mechanisms. In particular, sigma factors play a leading role in thermal stress response. Preexposure to thermal stress can lead to an increase in pathogenic potential through activation and regulation of genes associated with thermal stress. This thermal stress response can influence the activation of genes associated with virulence and the general stress response allowing for Salmonella to overcome host defenses and establish infection. The type of host can also play a role on the ability to establish infection. A host with a higher body temperature than humans could activate thermal stress resistance mechanisms allowing for easier colonization and establishment of infection compared to a host with a body temperature at 37°C in which these thermal stress resistance mechanisms are not expressed. An understanding of the Salmonella thermal resistance is essential for elucidating survival and infection mechanisms. It could be useful to identify specific targets for prevention and treatment of Salmonella infections. Therefore, it is imperative that the proteins involved in regulation and activation of these genes be thoroughly studied in order develop novel strategies to reduce outbreak cases and infection in all types of hosts.
Author Contributions
The contribution was equally distributed between all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
TD was supported by a scholarship from King Saud University Riyadh, Saudi Arabia. MD is supported by the Food Science Department at the University of Arkansas. SK was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2015R1A6A3A03016811). NJ was supported by a Distinguished Doctoral Fellowship from the University of Arkansas.
References
1. Beier RC, Pillai SD, Phillips TD, Ziprin RL. Preharvest and Postharvest Food Safety: Contemporary Issues and Future Directions. Ames, IA: Wiley-Blackwell (2008).
2. Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. Foodborne illness acquired in the United States – unspecified agents. Emerg Infect Dis (2011) 17:16–22. doi:10.3201/eid1701.P21101
3. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, et al. Foodborne illness acquired in the United States – major pathogens. Emerg Infect Dis (2011) 17:7–15. doi:10.3201/eid1701.P11101
4. Braden CR, Tauxe RV. Emerging trends in foodborne diseases. Infect Dis Clin North Am (2013) 27:517–33. doi:10.1016/j.idc.2013.06.001
5. Pires SM. Assessing the applicability of currently available methods for attributing foodborne disease to sources, including food and food commodities. Foodborne Pathog Dis (2013) 10:206–13. doi:10.1089/fpd.2012.1134
6. Centers for Disease Control and Prevention. Reports of Selected Salmonella Outbreak Investigations. (2015). Available at: http://www.cdc.gov/salmonella/outbreaks.html
7. Linam WM, Gerber MA. Changing epidemiology and prevention of Salmonella infection. Pediatr Infect Dis J (2007) 26:747–8. doi:10.1097/INF.0b013e3181376abc
8. Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol (2010) 2:a000414. doi:10.1101/cshperspect.a000414
9. Droffner ML, Yamamoto N. Prolonged environmental stress via a two step process selects mutants of Escherichia, Salmonella and Pseudomonas that grow at 54°C. Arch Microbiol (1991) 156:307–11. doi:10.1007/BF00263003
10. Park SY, Woodward CL, Kubena LF, Nisbet DJ, Birkhold SG, Ricke SC. Environmental dissemination of foodborne Salmonella in preharvest poultry production: reservoirs, critical factors, and research strategies. Crit Rev Environ Sci Technol (2008) 38:73–111. doi:10.1080/10643380701598227
11. Lynch M, Tauxe R, Hedberg C. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol Infect (2009) 137:307–15. doi:10.1017/S0950268808001969
12. O’Bryan CA, Crandall PG, Martin EM, Griffis CL, Johnson MG. Heat resistance of Salmonella spp., Listeria monocytogenes, Escherichia coli O157:H7, and Listeria innocua M1, a potential surrogate for Listeria monocytogenes, in meat and poultry: a review. J Food Sci (2006) 71:R23–30. doi:10.1111/j.1365-2621.2006.tb15639.x
13. Chen Z, Diao J, Dharmasena M, Ionita C, Jiang X, Rieck J. Thermal inactivation of desiccation-adapted Salmonella spp. in aged chicken litter. Appl Environ Microbiol (2013) 79:7013–20. doi:10.1128/AEM.01969-13
14. Mackey B, Derrick C. Heat shock protein synthesis and thermotolerance in Salmonella typhimurium. J Appl Bacteriol (1990) 69:373–83. doi:10.1111/j.1365-2672.1990.tb01527.x
15. Humphrey TJ, Slater E, McAlpine K, Rowbury RJ, Gilbert RJ. Salmonella enteritidis phage type 4 isolates more tolerant of heat, acid, or hydrogen peroxide also survive longer on surfaces. Appl Environ Microbiol (1995) 61:3161–4.
16. Wilde S, Jørgensen F, Campbell A, Rowbury R, Humphrey T. Growth of Salmonella enterica serovar Enteritidis PT4 in media containing glucose results in enhanced RpoS-independent heat and acid tolerance but does not affect the ability to survive air-drying on surfaces. Food Microbiol (2000) 17:679–86. doi:10.1006/fmic.2000.0363
17. Ng H, Bayne HG, Garibaldi JA. Heat resistance of Salmonella: the uniqueness of Salmonella senftenberg 775W. Appl Microbiol (1969) 17:78–82.
18. Bunning VK, Crawford RG, Tierney JT, Peeler JT. Thermotolerance of Listeria monocytogenes and Salmonella typhimurium after sublethal heat shock. Appl Environ Microbiol (1990) 56:3216–9.
19. Foster JW, Spector MP. How Salmonella survive against the odds. Annu Rev Microbiol (1995) 49:145–74. doi:10.1146/annurev.mi.49.100195.001045
20. Xavier IJ, Ingham SC. Increased D-values for Salmonella enteritidis following heat shock. J Food Prot (1997) 60:181–4. doi:10.4315/0362-028X-60.2.181
21. Gruzdev N, Pinto R, Sela S. Effect of desiccation on tolerance of Salmonella enterica to multiple stresses. Appl Environ Microbiol (2011) 77(5):1667–73. doi:10.1128/AEM.02156-10
22. Milillo SR, Ricke SC. Synergistic reduction of Salmonella in a model raw chicken media using a combined thermal and acidified organic acid salt intervention treatment. J Food Sci (2010) 75:M121–5. doi:10.1111/j.1750-3841.2009.01510.x
23. Milillo SR, Martin E, Muthaiyan A, Ricke SC. Immediate reduction of Salmonella enterica serotype Typhimurium viability via membrane destabilization following exposure to multiple-hurdle treatments with heated, acidified organic acid salt solutions. Appl Environ Microbiol (2011) 77:3765–72. doi:10.1128/AEM.02839-10
24. Klinkert B, Narberhaus F. Microbial thermosensors. Cell Mol Life Sci (2009) 66(16):2661–76. doi:10.1007/s00018-009-0041-3
25. Hurme R, Rhen M. Temperature sensing in bacterial gene regulation – what it all boils down to. Mol Microbiol (1998) 30:1–6. doi:10.1046/j.1365-2958.1998.01049.x
26. Konkel ME, Tilly K. Temperature-regulated expression of bacterial virulence genes. Microbes Infect (2000) 2:157–66. doi:10.1016/S1286-4579(00)00272-0
27. Yura T, Nakahigashi K. Regulation of the heat-shock response. Curr Opin Microbiol (1999) 2:153–8. doi:10.1016/S1369-5274(99)80027-7
28. Arsène F, Tomoyasu T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol (2000) 55:3–9. doi:10.1016/S0168-1605(00)00206-3
29. Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol (2003) 57:441–66. doi:10.1146/annurev.micro.57.030502.090913
30. Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev (2006) 20:1776–89. doi:10.1101/gad.1428206
31. Guisbert E, Yura T, Rhodius VA, Gross CA. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev (2008) 72:545–54. doi:10.1128/MMBR.00007-08
32. Hayes SA, Dice JF. Roles of molecular chaperones in protein degradation. J Cell Biol (1996) 132:255–8. doi:10.1083/jcb.132.3.255
33. Dougan D, Mogk A, Bukau B. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell Mol Life Sci (2002) 59:1607–16. doi:10.1007/PL00012487
34. Turgay K. Role of proteolysis and chaperones in stress response and regulation. 2nd ed. In: Storz G, Hengge R, editors. Bacterial Stress Responses. Washington, DC: ASM Press (2010). p. 75–90.
35. Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol (2010) 11:777–88. doi:10.1038/nrm2993
36. Lim B, Gross CA. Cellular response to heat shock and cold shock. 2nd ed. In: Storz G, Hengge R, editors. Bacterial Stress Responses. Washington, DC: ASM Press (2011). p. 93–114.
37. Robinson R. Heat shock response regulator is pinned to the membrane. PLoS Biol (2013) 11:e1001736. doi:10.1371/journal.pbio.1001736
38. Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T. Translational induction of heat shock transcription factor σ32: evidence for a built-in RNA thermosensor. Genes Dev (1999) 13:655–65. doi:10.1101/gad.13.6.655
39. Narberhaus F, Waldminghaus T, Chowdhury S. RNA thermometers. FEMS Microbiol Rev (2006) 30:3–16. doi:10.1111/j.1574-6976.2005.004.x
40. Narberhaus F. Translational control of bacterial heat shock and virulence genes by temperature-sensing mRNAs. RNA Biol (2010) 7:84–9. doi:10.4161/rna.7.1.10501
41. Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol (2007) 65:413–24. doi:10.1111/j.1365-2958.2007.05794.x
42. Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res (2010) 38:3834–47. doi:10.1093/nar/gkq124
43. Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res (2011) 39:8258–70. doi:10.1093/nar/gkr314
44. Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol (2012) 10:255–65. doi:10.1038/nrmicro2730
45. Narberhaus F. Control of bacterial heat shock and virulence genes by RNA thermometers. In: Hess WR, Marchfelder A, editors. Regulatory RNAs in Prokaryotes. Vienna: Springer (2012). p. 183–93.
46. Cimdins A, Roßmanith J, Langklotz S, Bandow JE, Narberhaus F. Differential control of Salmonella heat shock operons by structured mRNAs. Mol Microbiol (2013) 89:715–31. doi:10.1111/mmi.12308
47. Hurme R, Berndt KD, Normark SJ, Rhen M. A proteinaceous gene regulatory thermometer in Salmonella. Cell (1997) 90:55–64. doi:10.1016/S0092-8674(00)80313-X
48. Pallen MJ, Wren BW. The HtrA family of serine proteases. Mol Microbiol (1997) 26:209–21. doi:10.1046/j.1365-2958.1997.5601928.x
49. Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell (2002) 10:443–55. doi:10.1016/S1097-2765(02)00658-5
50. Alba BM, Gross CA. Regulation of the Escherichia coli σE-dependent envelope stress response. Mol Microbiol (2004) 52:613–9. doi:10.1111/j.1365-2958.2003.03982.x
51. Kazmierczak MJ, Wiedmann M, Boor KJ. Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol Rev (2005) 69:527–43. doi:10.1128/MMBR.69.4.527-543.2005
52. Raivio TL. MicroReview: envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol (2005) 56:1119–28. doi:10.111/j.1365-2958.2005.04625.x
53. Schumann W. Thermosensors in eubacteria: role and evolution. J Biosci (2007) 32:549–57. doi:10.1007/s12038-007-0054-8
54. Schumann W. Thermosensory stems in eubacteria. Adv Exp Med Biol (2012) 739:1–16. doi:10.1007/978-1-4614-1704-0_1
55. Dodd CE, Aldsworth TG. The importance of RpoS in the survival of bacterial through food processing. Int J Food Microbiol (2002) 74(3):189–94. doi:10.1016/S0168-1605(01)00679-1
56. Ibanez-Ruiz M, Robbe-Saule V, Hermant D, Labrude S, Norel F. Identification of RpoS (ςS)-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol (2000) 182(20):5749–56. doi:10.1128/JB.182.20.5749-5756.2000
57. Humphreys S, Rowley G, Stevenson A, Kenyon WJ, Spector MP, Roberts M. Role of periplasmic peptidylprolyl isomerases in Salmonella enterica serovar Typhimurium virulence. Infect Immun (2003) 71:5386–8. doi:10.1128/IAI.71.9.5386-5388.2003
58. Henderson B, Allan E, Coates AR. Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect Immun (2006) 74:3693–706. doi:10.1128/IAI.01882-05
59. Lazar SW, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol (1996) 178:1770–3. doi:10.1128/jb.178.6.1770-1773.1996
60. Bos MP, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane. Curr Opin Microbiol (2004) 7:610–6. doi:10.1016/j.mib.2004.10.011
61. Bos MP, Robert V, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane. Annu Rev Microbiol (2007) 61:191–214. doi:10.1146/annurev.micro.61.080706.093245
62. Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev (2007) 21:2473–84. doi:10.1101/gad.1581007
63. Geitner A-J, Varga E, Wehmer M, Schmid FX. Generation of a highly active folding enzyme by combining a parvulin-type prolyl isomerase from SurA with an unrelated chaperone domain. J Mol Biol (2013) 425:4089–98. doi:10.1016/j.jmb.2013.06.038
64. Ünal CM, Steinert M. Microbial peptidyl-prolyl cis/trans isomerases (PPIases): virulence factors and potential alternative drug targets. Microbiol Mol Biol Rev (2014) 78(3):544–71. doi:10.1128/MMBR.00015-14
65. Bolshoy A, Nevo E. Ecologic genomics of DNA: upstream bending in prokaryotic promoters. Genome Res (2000) 10:1185–93. doi:10.1101/gr.10.8.1185
66. Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol (2000) 37:687–95. doi:10.1046/j.1365-2958.2000.01972.x
67. Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J (2003) 84:2467–73. doi:10.1016/S0006-3495(03)75051-6
68. Jáuregui R, Abreu-Goodger C, Moreno-Hagelsieb G, Collado-Vides J, Merino E. Conservation of DNA curvature signals in regulatory regions of prokaryotic genes. Nucleic Acids Res (2003) 31:6770–7. doi:10.1093/nar/gkg882
69. Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog (2007) 3:e38. doi:10.1371/journal.ppat.0020081
70. Dorman CJ, Kane KA. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol Rev (2009) 33:587–92. doi:10.1111/j.1574-6976.2008.00155.x
71. Böhme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, et al. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog (2012) 8:e1002518. doi:10.1371/journal.ppat.1002518
72. Mäkelä PH, Koski P, Riikonen P, Taira S, Holthöfer H, Rhen M. The virulence plasmid of Salmonella encodes a protein resembling eukaryotic tropomyosins. In: Cabello IF, Hormaeche C, Mastroeni P, Bonina L, editors. Biology of Salmonella. New York, NY: Springer (1993). p. 115–20.
74. Clements M, Eriksson S, Tezcan-Merdol D, Hinton JC, Rhen M. Virulence gene regulation in Salmonella enterica. Ann Med (2001) 33:178–85. doi:10.3109/07853890109002075
75. Gal-Mor O, Valdez Y, Finlay BB. The temperature-sensing protein TlpA is repressed by PhoP and dispensable for virulence of Salmonella enterica serovar Typhimurium in mice. Microbes Infect (2006) 8:2154–62. doi:10.1016/j.micinf.2006.04.015
76. Skórko-Glonek J, Wawrzynów A, Krzewski K, Kurpierz K, Lipińiska B. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene (1995) 163:47–52. doi:10.1016/0378-1119(95)00406-V
77. Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell (1999) 97:339–47. doi:10.1016/S0092-8674(00)80743-6
78. Ingmer H, Brøndsted L. Proteases in bacterial pathogenesis. Res Microbiol (2009) 160:704–10. doi:10.1016/j.resmic.2009.08.017
79. Frees D, Brøndsted L, Ingmer H. Bacterial proteases and virulence. In: Dougan EA, editor. Regulated Proteolysis in Microorganisms. Netherlands: Springer (2013). p. 161–92.
80. Lindquist S, Craig E. The heat-shock proteins. Annu Rev Genet (1988) 22:631–77. doi:10.1146/annurev.ge.22.120188.003215
81. Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol (2005) 12:842–6. doi:10.1038/nsmb993
82. Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci (2005) 62:2460–76. doi:10.1007/s00018-005-5190-4
83. Nakamoto H, Vigh L. The small heat shock proteins and their clients. Cell Mol Life Sci (2007) 64:294–306. doi:10.1007/s00018-006-6321-2
84. Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev (1998) 12:3788–96. doi:10.1101/gad.12.24.3788
85. Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Annu Rev Microbiol (2001) 55:591–624. doi:10.1146/annurev.micro.55.1.591
86. Heimann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol (2002) 46:47–110. doi:10.1016/S0065-2911(02)46002-X
87. Bashyam MD, Hasnain SE. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect Genet Evol (2004) 4:301–8. doi:10.1016/j.meegid.2004.04.003
88. Ades SE. Regulation by destruction: design of the σE envelope stress response. Curr Opin Microbiol (2008) 11:535–40. doi:10.1016/j.mib.2008.10.004
89. Kumar A, Starke CB, DeZalia M, Moran CP. Surfaces of Spo 0A and RNA polymerase sigma factor A that interact at the spoIIG promoter in Bacillus subtilis. J Bacteriol (2004) 186:200–6. doi:10.1128/JB.186.1.200-206.2004
90. Kumar A, Moran CP. Promoter activation by repositioning of RNA polymerase. J Bacteriol (2008) 190:3110–7. doi:10.1128/JB.00096-08
91. de la Fuente M, Valera S, Martínez-Guitarte JL. ncRNAs and thermoregulation: a view in prokaryotes and eukaryotes. FEBS Lett (2012) 586:4061–9. doi:10.1016/j.febslet.2012.10.018
92. Bang IS, Frye JG, McClelland M, Velayudhan J, Fang FC. Alternative sigma factor interactions in Salmonella: σE and σH promote antioxidant defences by enhancing σS levels. Mol Microbiol (2005) 56:811–23. doi:10.1111/j.1365-2958.2005.04580.x
93. Doucleff M, Pelton JG, Lee PS, Nixon BT, Wemmer DE. Structural basis of DNA recognition by the alternative sigma-factor, σ54. J Mol Biol (2007) 369:1070–8. doi:10.1016/j.jmb.2007.04.019
94. Treviño-Quintanilla LG, Freyre-González JA, Martínez-Flores I. Anti-sigma factors in E. coli: common regulatory mechanisms controlling sigma factors availability. Curr Genomics (2013) 14:378. doi:10.2174/1389202911314060007
95. Merrick M. In a class of its own – the RNA polymerase sigma factor σ;54 (σN). Mol Microbiol (1993) 10:903–9. doi:10.1111/j.1365-2958.1993.tb00961.x
96. Buck M, Gallegos M-T, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent σ54 (σN) transcription factor. J Bacteriol (2000) 182:4129–36. doi:10.1128/JB.182.15.4129-4136.2000
97. Boor KJ. Bacterial stress responses: what doesn’t kill them can make them stronger. PLoS Biol (2006) 4:18. doi:10.1371/journal.pbio.0040023
98. Paget M, Helmann JD. The σ70 family of sigma factors. Genome Biol (2003) 4:203. doi:10.1186/gb-2003-4-1-203
99. Kültz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol (2005) 67:225–57. doi:10.1146/annurev.physiol.67.040403.103635
100. Lund PA. Microbial molecular chaperones. Adv Microb Physiol (2001) 44:93–140. doi:10.1016/S0065-2911(01)44012-4
101. Zhao K, Liu M, Burgess RR. The global transcriptional response of Escherichia coli to induced σ32 protein involves σ32 regulon activation followed by inactivation and degradation of σ32 in vivo. J Biol Chem (2005) 280:17758–68. doi:10.1074/jbc.M500393200
102. Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJ, Struhl K, et al. Extensive functional overlap between σ factors in Escherichia coli. Nat Struct Mol Biol (2006) 13:806–14. doi:10.1038/nsmb1130
103. Österberg S, Peso-Santos TD, Shingler V. Regulation of alternative sigma factor use. Annu Rev Microbiol (2011) 65:37–55. doi:10.1146/annurev.micro.112408.134219
104. Lim B, Miyazaki R, Neher S, Siegele DA, Ito K, Walter P, et al. Heat shock transcription factor σ32 co-opts the signal recognition particle to regulate protein homeostasis in E. coli. PLoS Biol (2013) 11:e1001735. doi:10.1371/journal.pbio.1001735
105. Tomoyasu T, Yuki T, Morimura S, Mori H, Yamanaka K, Niki H, et al. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol (1993) 175:1344–51. doi:10.1128/jb.175.5.1344-1351.1993
106. Akiyama Y, Ogura T, Ito K. Involvement of FtsH in protein assembly into and through the membrane. I. Mutations that reduce retention efficiency of a cytoplasmic reporter. J Biol Chem (1994) 269:5218–24.
107. Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, et al. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol (1999) 31:833–44. doi:10.1046/j.1365-2958.1999.01221.x
108. Katz C, Ron EZ. Dual role of FtsH in regulating lipopolysaccharide biosynthesis in Escherichia coli. J Bacteriol (2008) 190:7117–22. doi:10.1128/JB.00871-08
109. Herman C, Thévenet D, Bouloc P, Walker GC, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev (1998) 12:1348–55. doi:10.1101/gad.12.9.1348
110. Lies M, Maurizi MR. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J Biol Chem (2008) 283:22918–29. doi:10.1074/jbc.M801692200
111. Alix E, Blanc-Potard AB. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J (2008) 27:546–57. doi:10.1038/sj.emboj.7601983
112. Helander IM, Mattila-Sandholm T. Permeability barrier of the Gram-negative bacterial outer membrane with special reference to nisin. Int J Food Microbiol (2000) 60:153–61. doi:10.1016/S0168-1605(00)00307-X
113. Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev (2003) 67:593–656. doi:10.1128/MMBR.67.4.593-656.2003
114. Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta (2009) 1794(5):808–16. doi:10.1016/j.bbapap.2008.11.005
115. Ades SE. Control of the alternative sigma factor σE in Escherichia coli. Curr Opin Microbiol (2004) 7:157–62. doi:10.1016/j.mib.2004.02.010
116. Duguay AR, Silhavy TJ. Quality control in the bacterial periplasm. Biochim Biophys Acta (2004) 1694:121–34. doi:10.1016/j.bbamcr.2004.04.012
117. Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol (2006) 4:383–94. doi:10.1038/nrmicro1394
118. Hayden JD, Ades SE. The extracytoplasmic stress factor, sigmaE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One (2008) 3:e1573. doi:10.1371/journal.pone.0001573
119. Erickson JW, Gross CA. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev (1989) 3:1462–71. doi:10.1101/gad.3.9.1462
120. Wang Q, Kaguni J. A novel sigma factor is involved in expression of the rpoH gene of Escherichia coli. J Bacteriol (1989) 171:4248–53. doi:10.1128/jb.171.8.4248-4253.1989
121. Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K. The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature. J Bacteriol (1995) 177:2918–22. doi:10.1128/jb.177.10.2918-2922.1995
122. De Las Penas A, Connolly L, Gross CA. SigmaE is an essential sigma factor in Escherichia coli. J Bacteriol (1997) 179:6862–4. doi:10.1128/jb.179.21.6862-6864.1997
123. Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol (1998) 28:1059–66. doi:10.1046/j.1365-2958.1998.00865.x
124. Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, et al. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet (2009) 5:e1000651. doi:10.1371/journal.pgen.1000651
125. Ensgraber M, Loos M. A 66-kilodalton heat shock protein of Salmonella typhimurium is responsible for binding of the bacterium to intestinal mucus. Infect Immun (1992) 60:3072–8.
126. Humphreys S, Stevenson A, Bacon A, Weinhardt AB, Roberts M. The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect Immun (1999) 67:1560–8.
127. McMeechan A, Roberts M, Cogan TA, Jørgensen F, Stevenson A, Lewis C, et al. Role of the alternative sigma factors σE and σS in survival of Salmonella enterica serovar Typhimurium during starvation, refrigeration and osmotic shock. Microbiology (2007) 153:263–9. doi:10.1099/mic.0.29235-0
128. Lewis C, Skovierova H, Rowley G, Rezuchova B, Homerova D, Stevenson A, et al. Salmonella enterica serovar Typhimurium HtrA: regulation of expression and role of the chaperone and protease activities during infection. Microbiology (2009) 155:873–81. doi:10.1099/mic.0.023754-0
129. Raspoet R, Shearer N, Appia-Ayme C, Haesebrouck F, Ducatelle R, Thompson A, et al. A genome-wide screen identifies Salmonella Enteritidis lipopolysaccharide biosynthesis and the HtrA heat shock protein as crucial factors involved in egg white persistence at chicken body temperature. Poult Sci (2014) 93:1263–9. doi:10.3382/ps.2013-03711
130. Sirsat SA, Burkholder KM, Muthaiyan A, Dowd SE, Bhunia AK, Ricke SC. Effect of sublethal heat stress on Salmonella Typhimurium virulence. J Appl Microbiol (2011) 110:813–22. doi:10.1111/j.1365-2672.2011.04941.x
131. Johnson K, Charles I, Dougan G, Pickard D, O’gaora P, Costa G, et al. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol (1991) 5:401–7. doi:10.1111/j.1365-2958.1991.tb02122.x
132. Li S-R, Dorrell N, Everest PH, Dougan G, Wren BW. Construction and characterization of a Yersinia enterocolitica O: 8 high-temperature requirement (htrA) isogenic mutant. Infect Immun (1996) 64:2088–94.
133. Yamamoto T, Hanawa T, Ogata S, Kamiya S. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect Immun (1996) 64:2980–7.
134. Button JE, Silhavy TJ, Ruiz N. A suppressor of cell death caused by the loss of σE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J Bacteriol (2007) 189:1523–30. doi:10.1128/JB.01534-06
135. Cano DA, Martínez-Moya M, Pucciarelli MG, Groisman EA, Casadesús J, García-Del Portillo F. Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect Immun (2001) 69:6463–74. doi:10.1128/IAI.69.10.6463-6474.2001
136. Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. The alternative sigma factor σE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol Microbiol (2002) 43:771–82. doi:10.1046/j.1365-2958.2002.02787.x
137. Miticka H, Rowley G, Rezuchova B, Homerova D, Humphreys S, Farn J, et al. Transcriptional analysis of the rpoE gene encoding extracytoplasmic stress response sigma factor σE in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett (2003) 226:307–14. doi:10.1016/S0378-1097(03)00600-1
138. Kenyon WJ, Sayers DG, Humphreys S, Roberts M, Spector MP. The starvation-stress response of Salmonella enterica serovar Typhimurium requires σE-, but not CpxR-regulated extracytoplasmic functions. Microbiology (2002) 148:113–22. doi:10.1099/00221287-148-1-113
139. Crouch ML, Becker LA, Bang IS, Tanabe H, Ouellette AJ, Fang FC. The alternative sigma factor σE is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol Microbiol (2005) 56:789–99. doi:10.1111/j.1365-2958.2005.04578.x
140. Arié JP, Sassoon N, Betton JM. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol Microbiol (2001) 39:199–210. doi:10.1046/j.1365-2958.2001.02250.x
141. Horne SM, Kottom TJ, Nolan LK, Young KD. Decreased intracellular survival of an fkpA mutant of Salmonella typhimurium Copenhagen. Infect Immun (1997) 65:806–10.
142. Weski J, Ehrmann M. Genetic analysis of 15 protein folding factors and proteases of the Escherichia coli cell envelope. J Bacteriol (2012) 194:3225–33. doi:10.1128/JB.00221-12
143. Łasica AM, Jagusztyn-Krynicka EK. The role of Dsb proteins of Gram-negative bacteria in the process of pathogenesis. FEMS Microbiol Rev (2007) 31:626–36. doi:10.1111/j.1574-6976.2007.00081.x
144. Tormo A, Almiron M, Kolter R. surA, an Escherichia coli gene essential for survival in stationary phase. J Bacteriol (1990) 172:4339–47. doi:10.1128/jb.172.8.4339-4347.1990
145. Tamae C, Liu A, Kim K, Sitz D, Hong J, Becket E, et al. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J Bacteriol (2008) 190:5981–8. doi:10.1128/JB.01982-07
146. Sydenham M, Douce G, Bowe F, Ahmed S, Chatfield S, Dougan G. Salmonella enterica serovar Typhimurium surA mutants are attenuated and effective live oral vaccines. Infect Immun (2000) 68:1109–15. doi:10.1128/IAI.68.3.1109-1115.2000
147. Behrens-Kneip S. The role of SurA factor in outer membrane protein transport and virulence. Int J Med Microbiol (2010) 300:421–8. doi:10.1016/j.ijmm.2010.04.012
148. Khatiwara A, Jiang T, Sung S-S, Dawoud T, Kim JN, Bhattacharya D, et al. Genome scanning for conditionally essential genes in Salmonella enterica serotype Typhimurium. Appl Environ Microbiol (2012) 78:3098–107. doi:10.1128/AEM.06865-11
149. Dorman CJ. DNA supercoiling and environmental regulation of gene expression in pathogenic bacteria. Infect Immun (1991) 59(3):745–9.
150. Marko JF, Siggia ED. Fluctuations and supercoiling of DNA. Science (1994) 265:506–8. doi:10.1126/science.8036491
151. Tse-Dinh Y-C, Haiyan Q, Menzel R. DNA supercoiling and bacterial adaptation: thermotolerance and thermoresistance. Trends Microbiol (1997) 5:323–6. doi:10.1016/S0966-842X(97)01080-9
152. Travers A, Muskhelishvili G. DNA supercoiling – a global transcriptional regulator for enterobacterial growth? Nat Rev Microbiol (2005) 3:157–69. doi:10.1038/nrmicro1088
153. Owen-Hughes TA, Pavitt GD, Santos DS, Sidebotham JM, Hulton CS, Hinton JC, et al. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell (1992) 71:255–65. doi:10.1016/0092-8674(92)90354-F
154. Pérez-Martín J, Rojo F, De Lorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev (1994) 58:268–90.
155. Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr Opin Genet Dev (2003) 13:179–84. doi:10.1016/S0959-437X(03)00025-X
156. Browning DF, Grainger DC, Busby SJ. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol (2010) 13:773–80. doi:10.1016/j.mib.2010.09.013
157. López-García P, Forterre P. DNA topology and the thermal stress response, a tale from mesophiles and hyperthermophiles. Bioessays (2000) 22:738–46. doi:10.1002/1521-1878(200008)22:8<738:AID-BIES7>3.0.CO;2-5
158. Adamčík J, Víglaský V, Valle F, Antalík M, Podhradský D, Dietler G. Effect of bacteria growth temperature on the distribution of supercoiled DNA and its thermal stability. Electrophoresis (2002) 23:3300–9. doi:10.1002/1522-2683(200210)23:19<3300:AID-ELPS3300>3.0.CO;2-Q
159. Cameron AD, Stoebel DM, Dorman CJ. DNA supercoiling is differentially regulated by environmental factors and FIS in Escherichia coli and Salmonella enterica. Mol Microbiol (2011) 80:85–101. doi:10.1111/j.1365-2958.2011.07560.x
160. Cameron A, Dorman CJ. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet (2012) 8:e1002615. doi:10.1371/journal.pgen.1002615
161. Dawoud TM, Khatiwara A, Park SH, Davis ML, Baker CA, Ricke SC, et al. Heat survival of phenotype microarray profiling of Salmonella Typhimurium mutants. Curr Microbiol (2017) 74:257–67. doi:10.1007/s00284-016-1170-1
162. You Y, Yousef AE. Resistance of Listeria monocytogenes to heat after adaptation to environmental stresses. J Food Prot (1996) 59:465–71. doi:10.4315/0362-028X-59.5.465
163. Wesche AM, Gurtler JB, Marks BP, Ryser ET. Stress, sublethal injury, resuscitation, and virulence of bacterial foodborne pathogens. J Food Prot (2009) 72(5):1121–38. doi:10.4315/0362-028X-72.5.1121
164. Nielsen M-B, Knudsen GM, Danino-Appleton V, Olsen JE, Thomsen LE. Comparison of heat stress responses of immobilized and planktonic Salmonella enterica serovar Typhimurium. Food Microbiol (2013) 33:221–7. doi:10.1016/j.fm.2012.09.020
165. Jarvis NA, O’Bryan CA, Dawoud TM, Park SH, Kwon YM, Crandall PG, et al. An overview of Salmonella thermal destruction during food processing and preparation. Food Control (2016) 68:280–90. doi:10.1016/j.foodcont.2016.04.006
166. Knudsen GM, Nielsen MB, Grassby T, Danino-Appleton V, Thomsen LE, Colquhoun IJ, et al. A third mode of surface-associated growth: immobilization of Salmonella enterica serovar Typhimurium modulates the RpoS-directed transcriptional programme. Environ Microbiol (2012) 14:1855–75. doi:10.1111/j.1462-2920.2012.02703.x
167. Humphrey T, Wilde S, Rowbury R. Heat tolerance of Salmonella typhimurium DT104 isolates attached to muscle tissue. Lett Appl Microbiol (1997) 25:265–8. doi:10.1046/j.1472-765X.1997.00222.x
168. Orta-Ramirez A, Marks BP, Warsow CR, Booren AM, Ryser ET. Enhanced thermal resistance of Salmonella in whole muscle compared to ground beef. J Food Sci (2005) 70:359–62. doi:10.1111/j.1365-2621.2005.tb11480.x
169. Tuntivanich V, Orta-Ramirez A, Marks B, Ryser ET, Booren A. Thermal inactivation of Salmonella in whole muscle and ground turkey breast. J Food Prot (2008) 71:2548–51. doi:10.4315/0362-028X-71.12.2548
170. Mogollón MA, Marks BP, Booren AM, Orta-Ramirez A, Ryser ET. Effect of beef product physical structure on Salmonella thermal inactivation. J Food Sci (2009) 74:M347–51. doi:10.1111/j.1750-3841.2009.01253.x
171. Sampathkumar B, Khachatourians GG, Korber DR. Treatment of Salmonella enterica serovar Enteritidis with a sublethal concentration of trisodium phosphate or alkaline pH induces thermotolerance. Appl Environ Microbiol (2004) 70:4613–20. doi:10.1128/AEM.70.8.4613-4620.2004
172. Álvarez-Ordóñez A, Fernández A, López M, Arenas R, Bernardo A. Modifications in membrane fatty acid composition of Salmonella typhimurium in response to growth conditions and their effect on heat resistance. Int J Food Microbiol (2008) 123:212–9. doi:10.1016/j.ijfoodmicro.2008.01.015
173. Álvarez-Ordóñez A, Fernández A, López M, Bernardo A. Relationship between membrane fatty acid composition and heat resistance of acid and cold stressed Salmonella Senftenberg CECT 4384. Food Microbiol (2009) 26:347–53. doi:10.1016/j.fm.2008.11.002
174. Balamurugan S, Dugan ME. Growth temperature associated protein expression and membrane fatty acid composition profiles of Salmonella enterica serovar Typhimurium. J Basic Microbiol (2010) 50:507–18. doi:10.1002/jobm.201000037
175. de Mendoza D, Klages Ulrich A, Cronan JE. Thermal regulation of membrane fluidity in Escherichia coli. Effects of overproduction of beta-ketoacyl-acyl carrier protein synthase I. J Biol Chem (1983) 258(4):2098–101.
176. White-Ziegler CA, Davis TR. Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. J Bacteriol (2009) 191:1106–10. doi:10.1128/JB.00599-08
177. Olekhnovich IN, Kadner RJ. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol (2007) 189:6882–90. doi:10.1128/JB.00905-07
178. Baños RC, Vivero A, Aznar S, García J, Pons M, Madrid C, et al. Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet (2009) 5:e1000513. doi:10.1371/journal.pgen.1000513
179. Martínez LC, Banda MM, Fernández-Mora M, Santana FJ, Bustamante VH. HilD induces expression of Salmonella pathogenicity island 2 genes by displacing the global negative regulator H-NS from ssrAB. J Bacteriol (2014) 196:3746–55. doi:10.1128/JB.01799-14
180. Heroven AK, Nagel G, Tran HJ, Parr S, Dersch P. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol Microbiol (2004) 53:871–88. doi:10.1111/j.1365-2958.2004.04162.x
181. Ellison DW, Miller VL. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J Bacteriol (2006) 188:5101–12. doi:10.1128/JB.00862-05
182. Baños RC, Pons JI, Madrid C, Juárez A. A global modulatory role for the Yersinia enterocolitica H-NS protein. Microbiology (2008) 154:1281–9. doi:10.1099/mic.0.2007/015610-0
183. Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science (2006) 313:236–8. doi:10.1126/science.1128794
184. Duong N, Osborne S, Bustamante VH, Tomljenovic AM, Puente JL, Coombes BK. Thermosensing coordinates a cis-regulatory module for transcriptional activation of the intracellular virulence system in Salmonella enterica serovar Typhimurium. J Biol Chem (2007) 282:34077–84. doi:10.1074/jbc.M707352200
185. Shapiro RS, Cowen LE. Thermal control of microbial development and virulence: molecular mechanisms of microbial temperature sensing. MBio (2012) 3:e212–38. doi:10.1128/mBio.00238-12
186. Steinmann R, Dersch P. Thermosensing to adjust bacterial virulence in a fluctuating environment. Future Microbiol (2013) 8:85–105. doi:10.2217/fmb.12.129
187. Pesingl PK, Kumawat M, Behera P, Dixit SK, Agarwal RK, Goswami TK, et al. Protein-l-isoaspartyl methyltransferase (PIMT) is required for survival of Salmonella Typhimurium at 42°C and contributes to the virulence in poultry. Front Microbiol (2017) 7(8):361. doi:10.3389/fmicb.2017.00361
Keywords: Salmonella, thermal stress response, heat shock proteins, sigma factor, virulence
Citation: Dawoud TM, Davis ML, Park SH, Kim SA, Kwon YM, Jarvis N, O’Bryan CA, Shi Z, Crandall PG and Ricke SC (2017) The Potential Link between Thermal Resistance and Virulence in Salmonella: A Review. Front. Vet. Sci. 4:93. doi: 10.3389/fvets.2017.00093
Received: 20 March 2017; Accepted: 01 June 2017;
Published: 14 June 2017
Edited by:
Michael Kogut, Agricultural Research Service (USDA), United StatesReviewed by:
Wolfgang Ludwig Köster, University of Saskatchewan, CanadaVivek A. Kuttappan, Novus International Inc., United States
Copyright: © 2017 Dawoud, Davis, Park, Kim, Kwon, Jarvis, O’Bryan, Shi, Crandall and Ricke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven C. Ricke, sricke@uark.edu
†Present address: Turki M. Dawoud, Botany and Microbiology Department, Science College, King Saud University, Riyadh, Saudi Arabia
 Turki M. Dawoud1,2†
Turki M. Dawoud1,2†
 Morgan L. Davis
Morgan L. Davis Si Hong Park
Si Hong Park Sun Ae Kim
Sun Ae Kim Young Min Kwon
Young Min Kwon Nathan Jarvis
Nathan Jarvis Corliss A. O’Bryan
Corliss A. O’Bryan Steven C. Ricke
Steven C. Ricke