Microbial Communities in North American Ixodid Ticks of Veterinary and Medical Importance
- 1Department of Basic Sciences, College of Veterinary Medicine, Mississippi State University, Starkville, MS, United States
- 2Department of Food Science, Center for Food Safety, University of Arkansas, Fayetteville, AR, United States
Interest in microbial communities, or microbiota, of blood-feeding arthropods such as ticks (order Parasitiformes, suborder Ixodida) is increasing. Studies on tick microorganisms historically emphasized pathogens of high medical or veterinary importance. Current techniques allow for simultaneous detection of pathogens of interest, non-pathogenic symbionts, like Coxiella-LE and Francisella-LE, and microorganisms of unknown pathogenic potential. While each generation of ticks begins with a maternally acquired repertoire of microorganisms, microhabitats off and on vertebrate hosts can alter the microbiome during the life cycle. Further, blood-feeding may allow for horizontal exchange of various pathogenic microbiota that may or may not also be capable of vertical transmission. Thus, the tick microbiome may be in constant flux. The geographical spread of tick vector populations has resulted in a broader appreciation of tick-borne diseases and tick-associated microorganisms. Over the last decade, next-generation sequencing technology targeting the 16S rRNA gene led to documented snapshots of bacterial communities among life stages of laboratory and field-collected ticks, ticks in various feeding states, and tick tissues. Characterizing tick bacterial communities at population and individual tissue levels may lead to identification of markers for pathogen maintenance, and thus, indicators of disease “potential” rather than disease state. Defining the role of microbiota within the tick may lead to novel control measures targeting tick-bacterial interactions. Here, we review our current understanding of microbial communities for some vectors in the family Ixodidae (hard ticks) in North America, and interpret published findings for audiences in veterinary and medical fields with an appreciation of tick-borne disease.
Ticks as Obligate Parasites: Implications for the Tick Microbiome
Ticks represent a unique group of hematophagous ectoparasites capable of transmitting the greatest variety of microorganisms to vertebrate hosts (1). With few exceptions, motile life stages for all three tick families (Ixodidae, Argasidae, and Nuttaliellidae) require a blood meal. The significance of ticks as vectors is not new. Ticks were the first arthropod to be associated with transmission of a disease agent. In the late 1800s, Smith and Kilbourne established Rhipicephalus (Boophilus) ticks as the vector for (Texas) cattle fever, caused by Babesia bigemina (2). After that discovery, the ability of ticks to acquire, maintain and transmit pathogens became a significant and productive area of scientific research. Historically, tick-borne diseases were described first and then the etiologic agent pursued, as with Texas cattle fever. However, in some cases, tick-associated microorganisms of unknown pathogenicity were identified first, and an association with human or animal disease found later. Better diagnostic assays and heightened recognition of tick-borne diseases contributed to re-descriptions of tick-associated microorganisms. The spotted fever group rickettsia, Rickettsia parkeri, was recovered from Amblyomma maculatum in Texas approximately 60 years prior to the index case, reported in 2004 (3). Additionally, the spirochete, Borrelia miyamotoi, first reported from Ixodes persucatus in Japan, was considered non-pathogenic until the first human cases 15 years later (4, 5). New reports of emerging pathogenic tick-borne bacteria, viruses, and protozoa are not rare (6–9). However, most tick-associated microorganisms are not likely pathogens.
There is increasing interest in the tick microbial community, how it may impact transmission and maintenance of pathogens, and how manipulation of the microbial community may serve as an avenue for tick or pathogen control (10). The microhabitat at the surface of a vertebrate host, such as a human, presents a complex source of organisms for ticks to potentially acquire (11). In addition, microorganisms may be exchanged through co-feeding as well as from the external environment, considering ticks spend approximately 90% of their life off the host. Further, the tick itself, through vertical transmission, contributes to the microbiome of its next generation. There are at least ten bacterial genera that are transovarially (vertically) transmitted, including some found only in ticks (e.g., Coxiella-LE, Francisella-LE, and Midichloria), and some common among various arthropods (e.g., Wolbachia and Arsenophonus) (12). Hawlena et al. found that the microbiomes of the ticks, Dermacentor variabilis and I. scapularis, were most affected by arthropod-related factors, rather than the host or environment (13). Not surprisingly, the preponderance of Francisella-LE and Arsenophonus genera in D. variabilis, and Rickettsia in I. scapularis, in their study likely depended more on vertical transmission. A combination of factors is likely, depending on the tick-associated microorganism.
With the development of next-generation sequencing technology, the number of taxa detected in ticks has risen sharply. Enthusiasm over tick microbiome studies has generated a fresh wave of hypotheses to test and has led to a variety of methodologies to be considered in future studies. Microbiome sequencing based on the 16S rRNA gene continues to be widely utilized, but reliable sequencing data requires high quality DNA of sufficient quantity. Microbial genomic DNA extraction methods may vary based on the sample matrix and target DNA (prokaryotic or eukaryotic). Different bacterial cell wall structures may result in bacterial species bias in an extract, depending on genomic DNA recovery method. Yuan et al. tested six DNA extraction methods for human associated sample microbiome sequencing (14). Each method exhibited different bacterial genomic DNA recovery, with bead beating combined with mutanolysin exhibiting high cell lysis efficiency. Kit contaminants must also be considered (15). As tick microbiome analyses continue, the utility of archived extracts, samples that were not sufficiently cleaned prior to extraction, or extracts not enriched for microorganisms may improve if environmental contaminants can be distinguished from true symbionts, and samples are processed accordingly to minimize loss of these symbionts. In recognizing the limitations of published data, the scientific community and readership can better understand the biological significance of the tick microbiome.
Microbiota Associated with Common Tick Vectors in North America
Microorganisms living in close relationship with ticks are symbionts and can be categorized as obligate or facultative based on the potential for vertical or horizontal transmission and requirement for survival and reproduction, though these characteristics can be difficult to measure (16). Metabolic contributions from arthropod symbionts were recently reviewed (17). Co-infections with tick-borne pathogens, non-pathogens or uncharacterized bacteria are also documented, as well as evidence that the community of symbionts may influence maintenance and transmission of known pathogens (18). The abundance of microorganisms identified and novel associations uncovered continue to generate questions. Here, we highlight bacteria consistently dominating microbiome catalogs of ticks, focusing on some tick species of vector importance in North America. Obligate symbionts are summarized in Figure 1. For further reading, we suggest Narasimhan and Fikrig (10), Bonnet et al. (12), and Duron et al. (19).
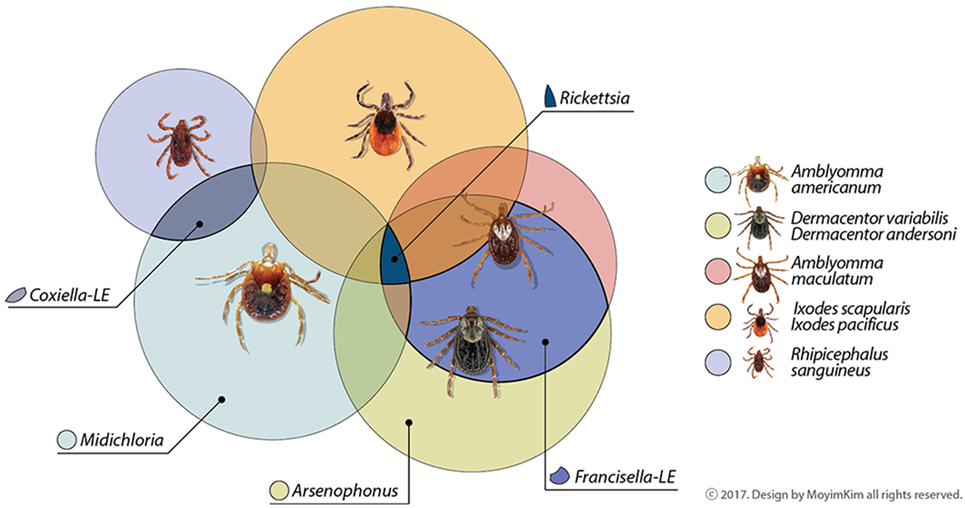
Figure 1. Common maternally inherited symbionts detected in North American tick vectors belong to the Proteobacteria. Genera are in families Rickettsiacea (Rickettsia), Coxiellaceae (Coxiella-LE), Francisellaceae (Francisella-LE), Midichloriacea (Midichloria), and Enterobacteriaceae (Arsenophonus). Some symbionts not depicted here as shared among tick vectors have been occasionally reported in the literature or may be reported as common symbionts in the future as data increase. Other areas of overlap may also occur among tick symbionts; these are suggested by unlabeled areas.
The phylum Proteobacteria comprises the majority of bacterial species detected in ticks (20). Obligate symbionts that predominate include the genera Coxiella-LE in Amblyomma americanum and several Rhipicephalus spp., Francisella-LE in D. variabilis, and A. maculatum, Rickettsia in I. scapularis, and Midichloria in A. americanum (21–23). Wolbachia endosymbionts, which are commonly found in other arthropods and some nematodes, are generally rare in ticks (24–26). Pathogens, when detected, seem to constitute a lower percentage of the bacterial population. Thus, one might predict that their presence does not impact the remaining microbial community. In comparing the community structure of bacteria from pathogen-infected (Anaplasma or Ehrlichia) and uninfected A. americanum, bacterial community structure did not significantly differ (27). Three phyla, Proteobacteria, Bacteriodetes, and Firmicutes, and genera Coxiella-LE and Rickettsia, Flavobacterium, and Bacillus, respectively, were commonly encountered, though there were discrepancies between Illumina sequencing and PCR for Anaplasma and Ehrlichia (27). In addition, while Coxiella-LE was more abundant in female compared to male ticks, the genus exhibited low relative abundance overall, and in comparison to Rickettsia. “Candidatus Rickettsia amblyommii” (now Rickettsia amblyommatis sp. nov.) (28) and “Candidatus Midichloria mitochondrii” dominated in A. americanum from North Carolina (23). Conversely, bacterial communities in A. americanum from Georgia were dominated by Rickettsia and Coxiella-LE, and exhibited frequent contributions from pathogenic (Ehrlichia) and uncharacterized genera (e.g., Midichloria) (29). While Coxiella-LE may be frequently found in A. americanum populations (30), the contribution of this genus to microbial abundance may be minimal. Microbial populations in both colony and field-collected A. americanum appear to increase in diversity during tick feeding at the expense of contributions from Coxiella-LE (31).
Microbiomes of laboratory-reared and wild ticks are likely influenced by generations of rearing in a specific environment and on specific hosts. Short-term environmental changes in a population do not appear to significantly impact the microbiome. The Rickettsia genus is rarely detected in colony-reared A. americanum ticks compared to overwhelming infection rates of R. amblyommatis sp. nov. (“Candidatus R. amblyommii”) in wild A. americanum (28, 32–34). In colony-reared A. americanum nymphs, there was an overall loss in microbial diversity regardless of whether nymphal ticks molted and aged outdoors or in the laboratory; Coxiella-LE was present in all tick groups, though Rickettsia was not detected in any group (34). These findings support short-term changes in the environment as having a minimal contribution on microbial communities. A closed related tick, A. maculatum shares a significant portion of its geographical range with A. americanum in the USA but differs in its phenology, microhabitat, host preferences, and obligate symbionts. Microbiome analyses of A. maculatum revealed that salivary glands, but not midgut tissues, of laboratory-reared and field-collected ticks commonly contained an abundance of Francisella-LE, while the genus Rickettsia was predominantly located in midguts. In addition, bacteria from the family Enterobacteriacea, which have not been commonly detected in A. americanum, constituted a greater portion of sequencing reads from both collections of ticks (26).
Ixodes scapularis has public and veterinary health importance as a vector for agents including B. burgdorferi, Anaplasma phagocytophilum, and the protozoan, Babesia. The increasing incidence of Powassan virus II (deer tick virus) in New England recently renewed anxiety over this high profile vector (35). Disease prevalence varies geographically, with states in the northeastern USA contributing the majority of case reports for I. scapularis-associated diseases such as Lyme disease, in comparison to states in the southern USA within the I. scapularis range (36). Geographic variations in microbiota are documented in I. scapularis in the Mid Atlantic USA, South Atlantic USA, and New England. Enterobacteriaceae were dominant in North Carolina I. scapularis populations, whereas Rickettsia (Rickettsiaceae) were dominant from populations in other USA states; interestingly, a population of I. scapularis with high levels of Borrelia, in comparison to known Borrelia-endemic sites in New England, was identified in Virginia (37). Overall, male I. scapularis demonstrated greater diversity compared to females, an observation noted in other tick microbiome studies (23, 37). In an earlier study, 16S rRNA amplicons from I. scapularis were separated by temporal temperature gradient gel electrophoresis and sequenced, revealing the dominance of Rickettsia in New York populations (38). Additional genera included not only Borrelia and Anaplasma, but also Pseudomonas, Ralstonia and Rhodococcus, which encompass species found in the soil, members of Enterobacteriaceae, and Moraxella, that are considered commensal species in mucosal membranes.
Deforestation, increased urbanization, warmer winters and longer transitional autumn, and spring seasons will help expand the dissemination of some tick species into more northern geographical regions as well as to higher elevations (39, 40). With this expansion is an increasing opportunity for ticks to contact different microbial communities outside of the microbial consortia encountered in traditional geographical regions. This is important because the vector microbiome may affect vector competence. Ixodes scapularis larvae reared under primarily sterile conditions and fed on gentamycin-treated mice underwent an altered microbiome (“dysbiosis”). Larvae demonstrated diminished feeding and B. burgdorferi colonization, as well as lower expression of the glycoprotein, peritrophin, suggesting an intimate association between gut microbiome composition and integrity of the peritrophic membrane (41). Whether the overall microbiome, or niche-specific variations in tick tissues are relevant to tick-borne pathogen maintenance and transmission may depend on the pathogen’s natural history, including its ability to be vertically transmitted and its requirement for a vertebrate reservoir. As B. burgdorferi does not utilize transovarial transmission for maintenance, efficient acquisition from a reservoir host is likely critical for vector competency. The presence of certain pathogens may also modify tick microbiome composition. Abraham et al. demonstrated that I. scapularis infected with A. phagocytophilum, are induced to express the I. scapularis anti-freeze glycoprotein, resulting in diminished biofilm formation in the tick gut and allowing for A. phagocytophilum colonization (42). Dysbiosis here acted to enhance pathogen establishment.
Dermacentor andersoni and D. variabilis are two key vectors for Rickettsia rickettsii, the agent of Rocky Mountain spotted fever. Dermacentor spp. are predominantly colonized by Francisella-LE, although other arthropod symbiotic genera are common, including Arsenophonus and Rickettsia (21, 43, 44). In D. andersoni, microbial diversity shifted over three generations and could be manipulated with tetracycline imbibed with the blood meal (43). Interestingly, Acinetobacter was found to increase in salivary glands over three tick generations, and most significantly in ticks fed on tetracycline-treated calves, at the expense of Arsenophonus, Rickettsia, and Francisella-LE abundance. Acinetobacter, a ubiquitous genus found in soil, water, and in normal animal flora, includes species that are now considered pathogenic; further, multidrug-resistant strains of A. baumannii are being increasingly found in hospital settings (45). Francisella-LE and Arsenophonus were common endosymbionts in D. variabilis removed from wild Peromyscus leucopus mice collected in Indiana, whereas Acinetobacter was not detected (44). Ixodes scapularis, also collected from P. leucopus in this study, did not have evidence of Francisella-LE or Arsenophonus but was dominated rather by the Rickettsia endosymbiont (44).
Rhipicephalus sanguineus, a common tick vector found infesting dogs world-wide, will also attach and transmit disease agents to other hosts, including humans. This is significant considering R. sanguineus is responsible for transmission of canine bacterial and protozoan pathogens (e.g., Ehrlichia canis, Babesia vogeli, and Hepatozoon canis) and was recently implicated as the vector in a Rocky Mountain spotted fever outbreak in Arizona, where both dogs and humans were infected (46). Similar to A. americanum, the primary endosymbiont is Coxiella-LE (19). However, there is minimal information using next-generation sequencing about additional symbionts in the R. sanguineus microbiome. Using PCR and sequencing, other genera detected from R. sanguineus outside of North America include Rickettsia, a genus most closely related to Wolbachia, and Midichloria (47–49). An initial assessment of the Rhipicephalus (Boophilus) microplus microbial community provides further insight as it demonstrated a preponderance of Coxiella-LE associated with female ovaries and eggs (50). Borrelia, Wolbachia, and potential environmental contaminants including Staphylococcus spp. and Streptococcus spp. were also detected (50).
Blood Feeding in Driving the Tick Microbiome
The host blood meal and the process of imbibing blood induce physiological changes in ticks that appear to affect the tick microbiome. In fact, the vertebrate host species may itself affect the microbiome. For example, nymphs of the Lyme disease vector in the western USA, I. pacificus, demonstrated reduced microbiome species richness, replaced by a preponderance of Rickettsia, after feeding on the western fence lizard, a host that is refractory to B. burgdorferi. In comparison, nymphs that fed on B. burgdorferi reservoir hosts, mice, exhibited greater diversity at the expense of Rickettsia; they also demonstrated a significant reduction of microbial populations through maturing life stage (51). At a minimum, these data imply a relationship between host blood meal and microbiome diversity. While blood meal causes significant changes within the microenvironment of a tick, the extent to which this affects microbial populations may ultimately depend on microbial adaptability.
As erythrocytes lyse in the tick gut lumen, midgut epithelial cells take up hemoglobin and other proteins through endocytosis, where intracellular digestion occurs. Heme, one of the products of digestion, must be detoxified due to its ability to damage tissues through the production of reactive oxygen species and free radicals (52). Kumar et al. found that experimentally increasing oxidative stress in A. maculatum, using RNA interference to silence the catalase gene (CAT), did not decrease estimated total bacterial loads from salivary glands and midgut tissues (53). In fact, knockdown of Cu/Zn-SOD or Mn-SOD, two enzymes that additionally function to decrease oxidative stress, actually increased bacterial load in the midgut with reciprocal effects on bacterial load in the salivary glands; in contrast, copy numbers of the pathogen, R. parkeri, decreased in these tissues after Cu/Zn-SOD silencing, suggesting a differential effect on different bacterial species (54). The digested blood meal itself may be a direct source of antimicrobial compounds. In midgut extracts from D. variabilis that were capillary-fed B. burgdorferi, α- and β-chain hemoglobin fragments, as well as host ubiquitin in a complex with ribosomal S30, were associated with inhibition of the non-pathogen, Micrococcus luteus. As expected, B. burgdorferi spirochetes in midguts of D. variabilis, which are incompetent vectors for B. burgdorferi, were not viable, though spirochetes remained intact (55). Interestingly, in I. scapularis, glutathione peroxidase (GPx/Salp25D), a homolog to the peroxiredoxin antioxidants, has a protective effect on B. burgdorferi during acquisition by feeding on an infected host, though no effect on spirochete transmission (20). Ixodes scapularis GPx/Salp25D also has an apparent protective effect on the pathogen, Anaplasma marginale (56). In the A. maculatum-Rickettsia system, the function of Salp25D and other selenoprotein antioxidants is not entirely straightforward. Knockdown of the selenocysteine elongation factor resulted in significantly decreased transcription of various selenoprotein antioxidants, with the exception of SalpD25 (GPx), and resulted in a loss of antioxidant capacity in midgut and salivary gland tissues of field-collected A. maculatum. However, while R. parkeri levels expectedly diminished in the midgut, levels doubled in salivary glands (57).
There may be interesting metabolic implications with the presence of Enterobacteriaceae in ticks, considering normal aerobic respiration by this group is associated with release of reactive oxygen species. Survival mechanisms to withstand the resultant oxidative stress are well-documented in E. coli and are conserved in related bacteria, with homologous regulators present in members of the phylum Proteobacteria (58). In field-collected A. maculatum removed from hosts 8 days post-infestation, Enterobacteriaceae constituted the second most abundant group in the midgut, and was also strongly represented in laboratory-reared A. maculatum; Arsenophonus was not among the Enterobacteriaceae identified (26). Ixodes scapularis from North Carolina were dominated by Enterobacteriaceae, unlike I. scapularis from Virginia, South Carolina, Connecticut, and New York (37). Van Truren et al. also observed an inverse relationship between Enterobacteriaceae and the genus Rickettsia (family Rickettsiaceae) in Ixodes spp. populations; Rickettsia was rare in North Carolina sites dominated by Enterobacteriaceae (37).
Tick feeding is also associated with measurable changes in osmolarity within the gut environment, which in turn may affect microorganisms. In I. scapularis, morphology and motility of B. burgdorferi spirochetes were altered, and expression of key virulence factors stimulated, in association with fluctuations in osmolarity during tick feeding (59). Finally, once present within the tick, ticks may respond to microorganisms though their innate immune system, consisting of both humoral and cellular mechanisms (60). However, the mechanisms by which the tick immune system impacts the composition of the tick microbiome in the midgut and other tissues are poorly understood and beyond the scope of this review.
Conclusion
Over the last few decades, the scientific community has witnessed and driven the development of next generation sequencing platforms for microbiome sequencing of the gastrointestinal tracts in humans, animals, and insects. Because ticks are significant vectors associated with emerging pathogens, understanding the contributions of the tick microbiome in tick physiology and pathogen stability may lead to novel approaches to vector and pathogen control. With the tick genome described (61), the opportunity to apply the corresponding transcriptomics and proteomics, as well as metabolomics, may help to uncover some of the answers to the interaction between the tick host and its microbiome, and in turn how these interactions impact the presence of pathogens.
Author Contributions
Contributed to review of the literature, identification of relevant publications to include, and manuscript preparation; involved in editing drafts prior to submission: AV-S, SP, SK, and SR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Support for preparation of this review was provided internally through the laboratories and institutions of Drs. AV-S and SR. In addition, the AV-S laboratory was supported in part by NIH COBRE P20GM103646 during initial manuscript preparation. This support allowed for a new collaboration to be established with the SR laboratory in a novel area of research for the AV-S laboratory.
References
1. Liu XY, Bonnet SI. Hard tick factors implicated in pathogen transmission. PLoS Negl Trop Dis (2014) 8:e2566. doi:10.1371/journal.pntd.0002566
2. Smith T, Kilbourne FL. Investigations into the nature, causation and prevention of Texas or southern cattle fever. U S Dept Apric Bur Anim Ind Bull (1893) 1:301–24.
3. Paddock CD, Sumner JW, Comer JA, Zaki SR, Goldsmith CS, Goddard J, et al. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin Infect Dis (2004) 38:805–11. doi:10.1086/381894
4. Platonov AE, Karan LS, Kolyasnikova NM, Makhneva NA, Toporkova MG, Maleev VV, et al. Humans infected with relapsing fever spirochete Borrelia miyamotoi, Russia. Emerg Infect Dis (2011) 17:1816–23. doi:10.3201/eid1710.101474
5. Gugliotta JL, Goethert HK, Berardi VP, Telford SR III. Meningoencephalitis from Borrelia miyamotoi in an immunocompromised patient. N Engl J Med (2013) 368:240–5. doi:10.1056/NEJMoa1209039
6. Vincent-Johnson NA, Macintire DK, Lindsay DS, Lenz SD, Baneth G, Shkap V, et al. A new Hepatozoon species from dogs: description of the causative agent of canine hepatozoonosis in North America. J Parasitol (1997) 83:1165–72. doi:10.2307/3284379
7. Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev (2013) 26:657–702. doi:10.1128/CMR.00032-13
8. Wormser GP, Pritt B. Update and commentary on four emerging tick-borne infections: Ehrlichia muris-like agent, Borrelia miyamotoi, deer tick virus, heartland virus, and whether ticks play a role in transmission of Bartonella henselae. Infect Dis Clin North Am (2015) 29:371–81. doi:10.1016/j.idc.2015.02.009
9. Vayssier-Taussat M, Kazimirova M, Hubalek Z, Hornok S, Farkas R, Cosson JF, et al. Emerging horizons for tick-borne pathogens: from the ’one pathogen-one disease’ vision to the pathobiome paradigm. Future Microbiol (2015) 10:2033–43. doi:10.2217/fmb.15.114
10. Narasimhan S, Fikrig E. Tick microbiome: the force within. Trends Parasitol (2015) 31:315–23. doi:10.1016/j.pt.2015.03.010
11. Schommer NN, Gallo RL. Structure and function of the human skin microbiome. Trends Microbiol (2013) 21:660–8. doi:10.1016/j.tim.2013.10.001
12. Bonnet SI, Binetruy F, Hernandez-Jarguin AM, Duron O. The tick microbiome: why non-pathogenic microorganisms matter in tick biology and pathogen transmission. Front Cell Infect Microbiol (2017) 7:236. doi:10.3389/fcimb.2017.00236
13. Hawlena H, Rynkiewicz E, Toh E, Alfred A, Durden LA, Hastriter MW, et al. The arthropod, but not the vertebrate host or its environment, dictates bacterial community composition of fleas and ticks. ISME J (2013) 7:221–3. doi:10.1038/ismej.2012.71
14. Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One (2012) 7:e33865. doi:10.1371/journal.pone.0033865
15. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol (2014) 12:87. doi:10.1186/s12915-014-0087-z
16. Haine ER. Symbiont-mediated protection. Proc Biol Sci (2008) 275:353–61. doi:10.1098/rspb.2007.1211
17. Rio RV, Attardo GM, Weiss BL. Grandeur alliances: symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol (2016) 32:739–49. doi:10.1016/j.pt.2016.05.002
18. Clay K, Fuqua C. The tick microbiome: diversity, distribution and influence of the internal microbial community for a blood-feeding disease vector. Critical Needs and Gaps in Understanding Prevention, Amelioration, and Resolution of Lyme and Other Tick-Borne Diseases: The Short-Term and Long-Term Outcomes. Washington, DC: The National Academies Press (2010). p. 435–57.
19. Duron O, Binetruy F, Noel V, Cremaschi J, McCoy KD, Arnathau C, et al. Evolutionary changes in symbiont community structure in ticks. Mol Ecol (2017) 26:2905–21. doi:10.1111/mec.14094
20. Narasimhan S, Sukumaran B, Bozdogan U, Thomas V, Liang X, DePonte K, et al. A tick antioxidant facilitates the Lyme disease agent’s successful migration from the mammalian host to the arthropod vector. Cell Host Microbe (2007) 2:7–18. doi:10.1016/j.chom.2007.06.001
21. Rounds MA, Crowder CD, Matthews HE, Philipson CA, Scoles GA, Ecker DJ, et al. Identification of endosymbionts in ticks by broad-range polymerase chain reaction and electrospray ionization mass spectrometry. J Med Entomol (2012) 49:843–50. doi:10.1603/ME12038
22. Lalzar I, Friedmann Y, Gottlieb Y. Tissue tropism and vertical transmission of Coxiella in Rhipicephalus sanguineus and Rhipicephalus turanicus ticks. Environ Microbiol (2014) 16:3657–68. doi:10.1111/1462-2920.12455
23. Ponnusamy L, Gonzalez A, Van Treuren W, Weiss S, Parobek CM, Juliano JJ, et al. Diversity of Rickettsiales in the microbiome of the lone star tick, Amblyomma americanum. Appl Environ Microbiol (2014) 80:354–9. doi:10.1128/AEM.02987-13
24. Slatko BE, Luck AN, Dobson SL, Foster JM. Wolbachia endosymbionts and human disease control. Mol Biochem Parasitol (2014) 195:88–95. doi:10.1016/j.molbiopara.2014.07.004
25. Zhang X, Norris DE, Rasgon JL. Distribution and molecular characterization of Wolbachia endosymbionts and filarial nematodes in Maryland populations of the lone star tick (Amblyomma americanum). FEMS Microbiol Ecol (2011) 77:50–6. doi:10.1111/j.1574-6941.2011.01089.x
26. Budachetri K, Browning RE, Adamson SW, Dowd SE, Chao CC, Ching WM, et al. An insight into the microbiome of the Amblyomma maculatum (Acari: Ixodidae). J Med Entomol (2014) 51:119–29. doi:10.1603/ME12223
27. Trout Fryxell RT, DeBruyn JM. The microbiome of Ehrlichia-infected and uninfected lone star ticks (Amblyomma americanum). PLoS One (2016) 11:e0146651. doi:10.1371/journal.pone.0155559
28. Karpathy SE, Slater KS, Goldsmith CS, Nicholson WL, Paddock CD. Rickettsia amblyommatis sp. nov.,a spotted fever group rickettsia associated with multiple species of Amblyomma ticks in North and South America. Int J Syst Evol Microbiol (2016) 66(12):5236–43. doi:10.1099/ijsem.0.001502
29. Williams-Newkirk AJ, Rowe LA, Mixson-Hayden TR, Dasch GA. Characterization of the bacterial communities of life stages of free living lone star ticks (Amblyomma americanum). PLoS One (2014) 9:e102130. doi:10.1371/journal.pone.0102130
30. Clay K, Klyachko O, Grindle N, Civitello D, Oleske D, Fuqua C. Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol Ecol (2008) 17:4371–81. doi:10.1111/j.1365-294X.2008.03914.x
31. Heise SR, Elshahed MS, Little SE. Bacterial diversity in Amblyomma americanum (Acari: Ixodidae) with a focus on members of the genus Rickettsia. J Med Entomol (2010) 47:258–68. doi:10.1093/jmedent/47.2.258
32. Zhang X, Ren X, Norris DE, Rasgon JL. Distribution and infection frequency of ’Candidatus Rickettsia amblyommii’ in Maryland populations of the lone star tick (Amblyomma americanum) and culture in an Anopheles gambiae mosquito cell line. Ticks Tick Borne Dis (2012) 3:38–42. doi:10.1016/j.ttbdis.2011.09.010
33. Jiang J, Yarina T, Miller MK, Stromdahl EY, Richards AL. Molecular detection of Rickettsia amblyommii in Amblyomma americanum parasitizing humans. Vector Borne Zoonotic Dis (2010) 10:329–40. doi:10.1089/vbz.2009.0061
34. Menchaca AC, Visi DK, Strey OF, Teel PD, Kalinowski K, Allen MS, et al. Preliminary assessment of microbiome changes following blood-feeding and survivorship in the Amblyomma americanum nymph-to-adult transition using semiconductor sequencing. PLoS One (2013) 8:e67129. doi:10.1371/journal.pone.0067129
35. Piantadosi A, Rubin DB, McQuillen DP, Hsu L, Lederer PA, Ashbaugh CD, et al. Emerging cases of Powassan virus encephalitis in New England: clinical presentation, imaging, and review of the Literature. Clin Infect Dis (2016) 62:707–13. doi:10.1093/cid/civ1005
36. Adams D, Fullerton K, Jajosky R, Sharp P, Onweh D, Schley A, et al. Summary of notifiable infectious diseases and conditions – United States, 2013. MMWR Morb Mortal Wkly Rep (2015) 62:1–122. doi:10.15585/mmwr.mm6253a1
37. Van Treuren W, Ponnusamy L, Brinkerhoff RJ, Gonzalez A, Parobek CM, Juliano JJ, et al. Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl Environ Microbiol (2015) 81:6200–9. doi:10.1128/AEM.01562-15
38. Moreno CX, Moy F, Daniels TJ, Godfrey HP, Cabello FC. Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ Microbiol (2006) 8:761–72. doi:10.1111/j.1462-2920.2005.00955.x
39. Dantas-Torres F. Climate change, biodiversity, ticks and tick-borne diseases: the butterfly effect. Int J Parasitol Parasites Wildl (2015) 4:452–61. doi:10.1016/j.ijppaw.2015.07.001
40. Parola P, Raoult D. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin Infect Dis (2001) 32:897–928. doi:10.1086/319347
41. Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, et al. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe (2014) 15:58–71. doi:10.1016/j.chom.2013.12.001
42. Abraham NM, Liu L, Jutras BL, Yadav AK, Narasimhan S, Gopalakrishnan V, et al. Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc Natl Acad Sci U S A (2017) 114:E781–90. doi:10.1073/pnas.1613422114
43. Clayton KA, Gall CA, Mason KL, Scoles GA, Brayton KA. The characterization and manipulation of the bacterial microbiome of the Rocky Mountain wood tick, Dermacentor andersoni. Parasit Vectors (2015) 8:632. doi:10.1186/s13071-015-1245-z
44. Rynkiewicz EC, Hemmerich C, Rusch DB, Fuqua C, Clay K. Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol Ecol (2015) 24:2566–79. doi:10.1111/mec.13187
45. Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol (2007) 5:939–51. doi:10.1038/nrmicro1789
46. Demma LJ, Traeger MS, Nicholson WL, Paddock CD, Blau DM, Eremeeva ME, et al. Rocky Mountain spotted fever from an unexpected tick vector in Arizona. N Engl J Med (2005) 353:587–94. doi:10.1056/NEJMoa050043
47. Satta G, Chisu V, Cabras P, Fois F, Masala G. Pathogens and symbionts in ticks: a survey on tick species distribution and presence of tick-transmitted micro-organisms in Sardinia, Italy. J Med Microbiol (2011) 60:63–8. doi:10.1099/jmm.0.021543-0
48. Inokuma H, Raoult D, Brouqui P. Detection of Ehrlichia platys DNA in brown dog ticks (Rhipicephalus sanguineus) in Okinawa Island, Japan. J Clin Microbiol (2000) 38:4219–21.
49. Harrus S, Perlman-Avrahami A, Mumcuoglu KY, Morick D, Eyal O, Baneth G. Molecular detection of Ehrlichia canis, Anaplasma bovis, Anaplasma platys, Candidatus Midichloria mitochondrii and Babesia canis vogeli in ticks from Israel. Clin Microbiol Infect (2011) 17:459–63. doi:10.1111/j.1469-0691.2010.03316.x
50. Andreotti R, Perez de Leon AA, Dowd SE, Guerrero FD, Bendele KG, Scoles GA. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol (2011) 11:6. doi:10.1186/1471-2180-11-6
51. Swei A, Kwan JY. Tick microbiome and pathogen acquisition altered by host blood meal. ISME J (2017) 11:813–6. doi:10.1038/ismej.2016.152
52. Graca-Souza AV, Maya-Monteiro C, Paiva-Silva GO, Braz GR, Paes MC, Sorgine MH, et al. Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem Mol Biol (2006) 36:322–35. doi:10.1016/j.ibmb.2006.01.009
53. Kumar D, Budachetri K, Meyers VC, Karim S. Assessment of tick antioxidant responses to exogenous oxidative stressors and insight into the role of catalase in the reproductive fitness of the Gulf Coast tick, Amblyomma maculatum. Insect Mol Biol (2016) 25:283–94. doi:10.1111/imb.12218
54. Crispell G, Budachetri K, Karim S. Rickettsia parkeri colonization in Amblyomma maculatum: the role of superoxide dismutases. Parasit Vectors (2016) 9:291. doi:10.1186/s13071-016-1579-1
55. Sonenshine DE, Hynes WL, Ceraul SM, Mitchell R, Benzine T. Host blood proteins and peptides in the midgut of the tick Dermacentor variabilis contribute to bacterial control. Exp Appl Acarol (2005) 36:207–23. doi:10.1007/s10493-005-2564-0
56. Kocan KM, Zivkovic Z, Blouin EF, Naranjo V, Almazan C, Mitra R, et al. Silencing of genes involved in Anaplasma marginale-tick interactions affects the pathogen developmental cycle in Dermacentor variabilis. BMC Dev Biol (2009) 9:42. doi:10.1186/1471-213X-9-42
57. Adamson SW, Browning RE, Budachetri K, Ribeiro JM, Karim S. Knockdown of selenocysteine-specific elongation factor in Amblyomma maculatum alters the pathogen burden of Rickettsia parkeri with epigenetic control by the Sin3 histone deacetylase corepressor complex. PLoS One (2013) 8:e82012. doi:10.1371/journal.pone.0082012
58. Chiang SM, Schellhorn HE. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys (2012) 525:161–9. doi:10.1016/j.abb.2012.02.007
59. Bontemps-Gallo S, Lawrence K, Gherardini FC. Two different virulence-related regulatory pathways in Borrelia burgdorferi are directly affected by osmotic fluxes in the blood meal of feeding Ixodes ticks. PLoS Pathog (2016) 12:e1005791. doi:10.1371/journal.ppat.1005791
60. Hajdusek O, Sima R, Ayllon N, Jalovecka M, Perner J, de la Fuente J, et al. Interaction of the tick immune system with transmitted pathogens. Front Cell Infect Microbiol (2013) 3:26. doi:10.3389/fcimb.2013.00026
Keywords: tick vectors, microbiome, next-generation sequencing, pathogens, endosymbionts
Citation: Varela-Stokes AS, Park SH, Kim SA and Ricke SC (2017) Microbial Communities in North American Ixodid Ticks of Veterinary and Medical Importance. Front. Vet. Sci. 4:179. doi: 10.3389/fvets.2017.00179
Received: 20 June 2017; Accepted: 04 October 2017;
Published: 20 October 2017
Edited by:
Edwin Claerebout, Ghent University, BelgiumReviewed by:
Andrei Daniel Mihalca, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaAnja Joachim, Veterinärmedizinische Universität Wien, Austria
Copyright: © 2017 Varela-Stokes, Park, Kim and Ricke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea S. Varela-Stokes, stokes@cvm.msstate.edu
†Present address: Si Hong Park, Department of Food Science and Technology, Oregon State University, Corvallis, OR, United States
 Andrea S. Varela-Stokes
Andrea S. Varela-Stokes Si Hong Park
Si Hong Park Sun Ae Kim2
Sun Ae Kim2  Steven C. Ricke
Steven C. Ricke