High Prevalence of Extended-Spectrum β-Lactamase Producing Enterobacteriaceae Among Clinical Isolates From Cats and Dogs Admitted to a Veterinary Hospital in Switzerland
- 1National Centre for Enteropathogenic Bacteria and Listeria, Vetsuisse Faculty, Institute for Food Safety and Hygiene, University of Zürich, Zürich, Switzerland
- 2Vetsuisse Faculty, Institute of Veterinary Bacteriology, University of Zürich, Zürich, Switzerland
Objectives: This study aimed to identify and characterize extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae among clinical samples of companion animals.
Methods: A total of 346 non-duplicate Enterobacteriaceae isolates were collected between 2012 and 2016 from diseased cats (n = 115) and dogs (n = 231). The presence of blaESBL, PMQR genes, and the azithromycin resistance gene mph(A) was confirmed by PCR and sequencing of bla genes. Isolates were further characterized by antimicrobial resistance profiling, multilocus sequence typing, phylogenetic grouping, identification of mutations in the QRDR of gyrA and parC, and screening for virulence-associated genes.
Results: Among the 346 isolates, 72 (20.8%) were confirmed ESBL producers [58 Escherichia coli (E. coli), 11 Klebsiella pneumoniae (K. pneumoniae), and 3 Enterobacter cloacae]. The strains were cultured from urine (n = 45), skin and skin wounds (n = 8), abscesses (n = 6), surgical sites (n = 6), bile (n = 4), and other sites (n = 3). ESBL genes included blaCTX-M-1, 14, 15, 27, 55, and blaSHV-12, predominantly blaCTX-M-15 (54.8%, 40/73), and blaCTX-M-1 (24.7%, 18/73). Further genes included qnrB (4.2%, 3/72), qnrS (9.7%, 7/72), aac(6’)-Ib-cr (47.2%, 34/72), and mph(A) (38.9%, 28/72). Seventeen (23.6%) isolates belonged to the major lineages of human pathogenic K. pneumoniae ST11, ST15, and ST147 and E. coli ST131. The most prevalent ST was E. coli ST410 belonging to phylogenetic group C.
Conclusion: The high prevalence of ESBL producing clinical Enterobacteriaceae from cats and dogs in Switzerland and the presence of highly virulent human-related K. pneumoniae and E. coli clones raises concern about transmission prevention as well as infection management and prevention in veterinary medicine.
Introduction
Members of the family of the Enterobacteriaceae, although natural inhabitants of the intestinal tracts of mammals, may cause urinary tract, skin, ear, soft tissue, and respiratory infections in cats and dogs (1). For uncomplicated infections, first-line therapeutic options are ampicillin, amoxicillin-clavulanate or first- and second-generation cephalosporins, while amikacin, third-generation cephalosporins or fluoroquinolones (enrofloxacin or ciprofloxacin) remain appropriate for severe infections (1, 2). One of the most important mechanisms of antimicrobial resistance in Enterobacteriaceae is the enzymatic inactivation of penicillins and cephalosporins by means of plasmid-mediated extended-spectrum β-lactamases (ESBLs), such as the TEM-, SHV-, or cefotaxime (CTX)-M-group enzymes (3). The emergence of ESBL producing Enterobacteriaceae in healthy and in diseased companion animals constitutes an increasing challenge to infection management in veterinary therapy. Moreover, resistance caused by ESBLs is often associated with resistance to other classes of antibiotics like aminoglycosides, fluoroquinolones, and sulfamethoxazole/trimethoprim (SXT), which are antimicrobials that are critically important in human medicine (4, 5). Additionally, previous studies have shown that multidrug resistant, highly virulent human-related clonal lineages of Enterobacteriaceae, such as Escherichia coli (E. coli), belonging to sequence type (ST)131 and ST648, or Klebsiella pneumoniae (K. pneumoniae) ST11, ST15, and ST147 may be isolated from companion animals (6, 7). Consequently, there is growing concern that ESBL producers in companion animals pose a potential health hazard to humans, either through direct transmission of resistant pathogens from animals to humans, or indirectly through transmission of resistance genes (8, 9). Recent data on the prevalence of ESBL producers in clinical isolates of cats and dogs and the phenotypes and genotypes of such isolates are scarce for Switzerland, and it remains unclear to what extent clinically relevant phylogenetic or clonal lineages occur.
Here, we analyze a collection of clinical feline and canine Enterobacteriaceae obtained during 2012–2016 by (i) identifying ESBL producers within the strain collection, (ii) assessing their antimicrobial resistance profiles, (iii) determining their blaESBL genes and screening for plasmid-mediated fluoroquinolone and azithromycin resistance genes, and by (iii) characterizing E. coli and K. pneumoniae strains by multilocus sequence typing (MLST), and E. coli strains by phylogenetic grouping and virulence gene profiling.
Materials and Methods
Bacterial Isolates
Between 2012 and 2016, 346 clinical Enterobacteriaceae were isolated from diseased cats (n = 115) and dogs (n = 231) admitted to the veterinary clinic of the University of Zürich. The isolates were cultured from urinary samples (n = 273), samples obtained from surgical sites (n = 26), abscess samples (n = 16), skin and skin wound samples (n = 14), bile samples (n = 7), and samples from other sites (n = 10). Strain identification and routine antimicrobial susceptibility profiling was performed using the VITEK® two compact system with AST GN38 cards (Biomérieux, Nürtingen, Germany) according to the manufacturer’s instructions. The identity of Enterobacter cloacae (E. cloacae) was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF–MS, Bruker Daltronics, Bremen, Germany). ESBL producers were screened by using the chromogenic medium Brilliance™ ESBL Agar (Oxoid, Hampshire, UK), according to the manufacturer’s recommendations. All non-duplicate isolates growing on ESBL agar were further analyzed. In accordance with local legislation, ethics approval was not required for this study.
Identification of blaESBL Genes and Antibiotic Susceptibility Testing
The presence of blaESBL genes was established by PCR, and amplicons were sequenced as described previously using primers listed in Table S1 in Supplementary Material (10–12). For the detection of the CTX-M-25 enzyme group, the newly designed primers Gr. 25 CTX-M fw CCTGTGTTTCGCTGCTGTTGG and Gr. 25 CTX-M rv GGCTCTCTGCCTTCGGCTCC, were used.
Antimicrobial susceptibility testing was performed according to Clinical and Laboratory Standards Institute (CLSI) performance standards (13), using the disk-diffusion method and the antibiotics ampicillin (AM), amoxicillin with clavulanic acid (AMC), azithromycin (AZM), cefazolin, cefepime, CTX, chloramphenicol (C), ciprofloxacin (CIP), fosfomycin (FOS), gentamicin (G), kanamycin (K), nalidixic acid (NA), nitrofurantoin (F/M), streptomycin (S), SXT, and tetracycline (TE) (Becton Dickinson, Allschwil, Switzerland). Results were interpreted according to CLSI standards (13). For azithromycin, an inhibition zone of ≤12 mm was interpreted as resistant. Isolates displaying resistance to three or more classes of antimicrobials (counting β-lactams as one class) were defined as multidrug-resistant (MDR).
Identification of Additional Antimicrobial Resistance Genes
The plasmid-mediated fluoroquinolone resistance genes aac(6')-Ib-cr, qnrA, qnrB, qnrC, qnrD, qnrS, and qepA, and the plasmid-mediated azithromycin resistance gene mph(A) were detected by PCR as described elsewhere using primers listed in Table S1 in Supplementary Material (14, 15).
Quinolone-resistant E. coli strains were examined for mutations in the quinolone resistance-determining regions (QRDRs) of gyrA and parC, using PCR amplification and sequencing primers as described previously using primers listed in Table S1 in Supplementary Material (14).
Synthesis of primers and DNA custom sequencing was carried out by Microsynth (Balgach, Switzerland) and nucleotide sequences were analyzed with CLC Main Workbench 6.6.1. For database searches, the BLASTN program of NCBI1 was used.
Phylogenetic Characterization and MLST
Phylogenetic classification of the E. coli isolates into one of the eight groups, including A, B1, B2, C, D, E, F, (E. coli sensu stricto), or Escherichia clade I, was performed as described by Clermont et al. (16).
Sequence type determination of the E. coli isolates was carried out as described by Wirth et al. (17). Sequences were imported into the E. coli MLST database website2 to determine MLST types. Alleles and STs that had not been previously described were termed new ST, but not assigned new numerical designations by the database.
Sequence type determination of the K. pneumoniae isolates was performed according to previously described methods (18). STs were determined according to the Klebsiella MLST database.3
Virulence Factor (VF) Determination in Uropathogenic E. coli Isolates
Escherichia coli isolated from urinary samples were tested by conventional PCR for the presence of virulence-associated genes that mediate adhesion (p-fimbrial adhesion genes papAH and papEF, and the chaperone-usher fimbria yfcv), toxins (α-hemolysin hlyA), siderophores (the ferric yersiniabactin uptake protein fyuA), serum resistance (traT), and the right-hand terminus of pathogenicity island (PAI) from E. coli strain CFT073, using primers listed in Table S1 in Supplementary Material and conditions described previously (19, 20). The aggregate VF score was defined as the number of unique VF detected for each isolate, counting the PAI marker as one.
Results
During 2012–2016, 20.8% (72/346) of clinical Enterobacteriaceae isolated from cats and dogs were ESBL producers. The isolates originated from 7 cats and 65 dogs, amounting to 6% (7/115) of the feline and 28.1% (65/231) of the canine isolates, respectively. The prevalence of ESBL producers was remarkably higher among isolates from dogs than from cats. Overall, ESBL producers (58 E. coli, 11 K. pneumoniae, and 3 E. cloacae) were cultured from 16.5% (45/273) of the urinary samples, 57.1% (8/14) of the skin and skin wound samples, 37.5% (6/16) of abscess samples, 23% (6/26) of the samples obtained from surgical sites, 57.1% (4/7) of bile samples, and 30% (3/10) of the samples from other sites (Table 1). Among the E. coli from urinary samples, 17% (35/205) were ESBL producers (Table 1).
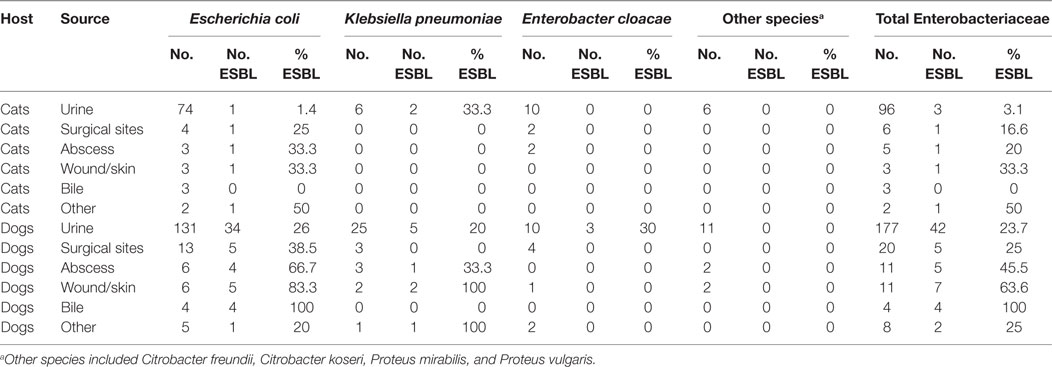
Table 1. Percent and distribution of extended-spectrum β-lactamases (ESBL) producers among clinical Enterobacteriaceae from cats and dogs in Switzerland, 2012–2016.
In addition to their resistance to penicillins and extended-spectrum cephalosporins, the isolates were frequently resistant to quinolones and fluoroquinolones, with 88.9% (64/72) resistant to NA and 83.3% (60/72) resistant to ciprofloxacin. They were also resistant to SXT (76.4%, 55/72), TE (72.2%, 52/72), aminoglycosides streptomycin (45.8%, 33/72), gentamycin (37.5%, 27/72), kanamycin (19.4%, 14/72), chloramphenicol (25%, 18/72), as well as to azithromycin (22.2%, 16/72), and to nitrofurantoin (12.5%, 9/72). One K. pneumoniae isolate (1.4%) was resistant to fosfomycin. Overall, 73.6% (53/72) were MDR and none was pansusceptible (Table S2 in Supplementary Material).
In total, 73 ESBL genes were detected among the 72 isolates, including in 1 K. pneumoniae isolate co-harboring blaCTX-M-15 and blaSHV-12 (Table 2). Among the ESBL genes, blaCTX-M-15 predominated (54.8%, 40/73), followed by blaCTX-M-1 (24.7%, 18/73). Other ESBL genes included blaCTX-M-55 (6.8%, 5/73), blaCTX-M-14 and blaSHV-12 (each 5.5%, 4/73), and blaCTX-M-27 (2.7%, 2/73).
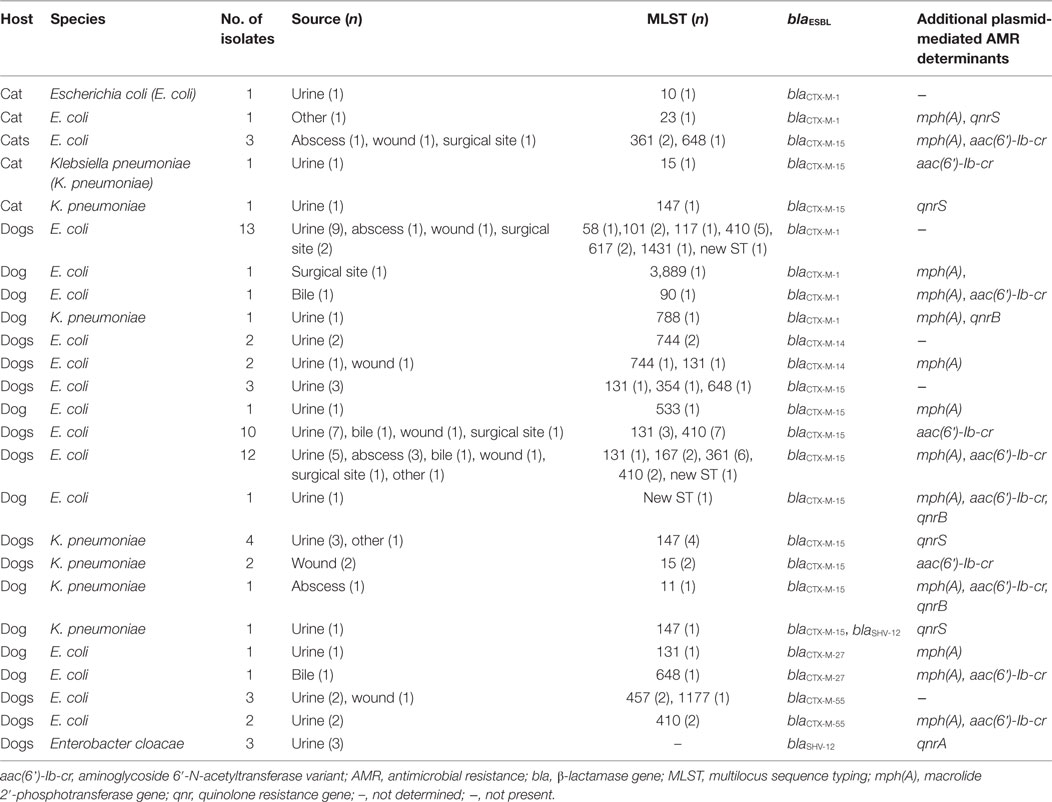
Table 2. Type and distribution of extended-spectrum β-lactamases (ESBL) genes and other plasmid-mediated resistance genes among 72 clinical Enterobacteriaceae isolated from cats and dogs in Switzerland, 2012–2016.
In addition to blaESBLs, other plasmid-mediated resistance genes detected among the 72 isolates included aac(6’)-Ib-cr (47.2%, 34/72), mph(A) (38.9%, 28/72), qnrS (9.7%, 7/72), qnrA and qnrB (each 4.2%, 3/72) (Table 2).
The majority of the aac(6’)-Ib-cr genes (88.2%, 30/34), the mph(A) genes (62%, 18/29), the qnrB (66.7%, 2/3), and qnrS genes (85.7%, 6/7) was detected in isolates harboring blaCTX-M-15. All qnrA were detected together with blaSHV-12 in E. cloacae (Table 2).
Phylogenetic analysis of the 58 E. coli isolates revealed a predominance of group C (32.8%, 19/58), followed by group A (31%, 18/58), group B2 and group F (each 12%, 7/58), group B1 (8.6%, 5/58), and group D (3.4%, 2/58) (Table S2 in Supplementary Material).
Among the 58 E. coli isolates, 23 different STs and three new STs were identified (Table 2; Table S3 in Supplementary Material). Most frequently, isolates belonged to ST410 (27.6%, 16/58), followed by a collective of STs occurring only once or twice (24.1%, 14/58), ST361 (13.8%, 8/58), ST131 (12%, 7/58), and ST648, ST744, and new STs (each 5.2%, 3/58). E. coli ST410 and human-related pandemic clone E. coli ST131 were detected only in isolates from dogs. E. coli ST410 was isolated from 33.3% of the urine samples from dogs.
Among the 11 K. pneumoniae isolates, 4 different STs were detected (Table 2; Table S2 in Supplementary Material). The majority (54.5%, 6/11) of the isolates belonged to ST147. Other STs included ST15 (27.3%, 3/11), ST11, and ST788 (both 9.1%, 1/11).
Among the 51 E. coli isolates displaying quinolone resistance, all revealed chromosomal mutations that result in amino acid substitutions in GyrA and ParC. Unusual point mutations Asp87→Tyr in GyrA and Glu84→Gly in ParC were noted for two E. coli ST457 isolates harboring blaCTX-M-55 (Table 3; Table S2 in Supplementary Material).
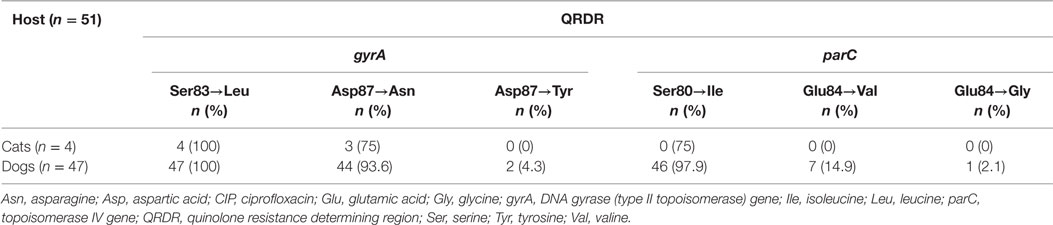
Table 3. Amino acid substitutions in the QRDR of 51 quinolone-resistant extended-spectrum β-lactamases producing Escherichia coli from cats and dogs in Switzerland, 2012–2016.
Virulence factors were distributed unequally among the 35 uropathogenic E. coli isolates (Table 4).
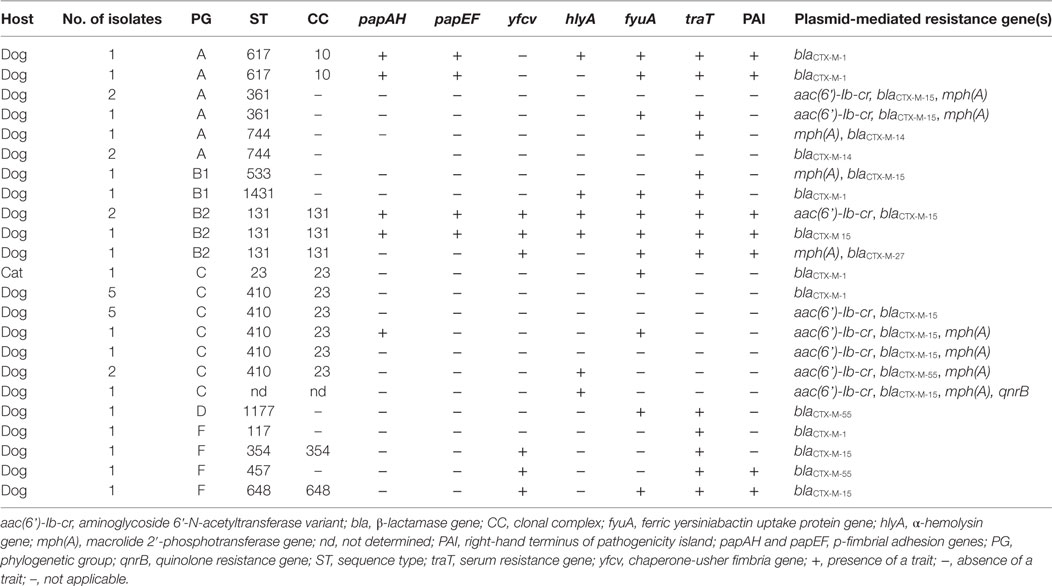
Table 4. Virulence-associated genes detected in 35 uropathogenic extended-spectrum β-lactamases producing Escherichia coli from cats and dogs in Switzerland, 2012–2016.
For 42.9% (15/35) of E. coli urinary isolates, no VF was detected. Strains with aggregate VF score ≥1 were identified in 34.5% (57.1%/35) of the isolates. VF scores were highest for isolates belonging to ST617 (median 5.5, range 5–6) and ST131 (median 7, range 4–7).
Discussion
This study identified a high prevalence (20.8%) of ESBL-producing Enterobacteriaceae derived from clinical samples of cats and dogs collected during 2012–2016 at the veterinary clinic of the University of Zürich, Switzerland. This is considerably higher than that found in similar studies from pets in the UK (7%) (21), the Netherlands (2%) (22), and France (3.7%) (23), and remarkably higher than the prevalence of 1.6% detected in a European collection of Enterobacteriaceae obtained from diseased companion animals in 2015 (6). In addition, among the uropathogenic E. coli analyzed in this study, the observed prevalence of 16.8% of ESBL producers is considerably higher than that found previously in cats and dogs in Switzerland between 2010 and 2012 (7.5%) (24). Although our data are single-institution based and thus limited, they provide important information on the trends in the burden of infections due to ESBL producers in veterinary medicine in Switzerland.
Overall, a diversity of blaESBL genes was found within three bacterial species. The predominance of blaCTX-M-15, which is highly prevalent in ESBL producers in humans, is comparable to what is found in other studies on isolates from companion animals (21, 23, 25). This gene was the only one that was detected in cats and dogs in Switzerland between 2010 and 2012 (24). Our study shows that in the following years, blaCTX-M-1, blaCTX-M-14, blaCTX-M-27, blaCTX-M-55, and blaSHV-12 harboring Enterobacteriaceae have emerged in cats and dogs in Switzerland.
Second to blaCTX-M-15, blaCTX-M-1 was the most frequent variant identified in this study. The blaCTX-M-1 gene is the most prevalent blaESBL gene among ESBL-producing Enterobacteriaceae isolated food-producing animals and food, in particular chicken and chicken meat (26, 27). Consumption of raw meat represents a risk factor for dogs acquiring pathogenic E. coli, including ESBL producers (28, 29). Moreover, a recent study detected a high prevalence (77.8%) of ESBL producers in raw cat food and demonstrated a strong association of consumption of raw cat food with shedding of ESBL producers by household cats in the Netherlands (30). Further studies are needed to investigate the possibility of raw meat as an origin of the high prevalence of ESBL and the occurrence of CTX-M-1 producers in isolates from companion animals in Switzerland. Similarly, CTX-M-55 has been widely reported in food-producing animals and pets in mainland China (31). This ESBL variant has rarely been detected outside China and its emergence in pets in Switzerland, possibly due to international food and animal trade, warrants attention.
This study identified 17 (23.6%) isolates belonging to major lineages of human pathogenic K. pneumoniae and E. coli. CTX-M-15 producing K. pneumoniae ST11, ST 15, and ST147 represent major international high-risk nosocomial clones (32). K. pneumoniae ST11 and ST15 from companion animals have been involved in nosocomial events in veterinary clinics (7, 33). By contrast, K. pneumoniae ST147 has only very recently been detected in pets in Europe and in Japan (34, 35), and this is to our knowledge the second report on this ST isolated from dogs in Europe.
Pandemic human pathogenic E. coli ST131-producing CTX-M-15 has disseminated globally in hospital and community settings causing a wide spectrum of infections, including urinary tract infection, cystitis, pyelonephritis, and bacteremia, with transmission between humans and their companion animals (cats and dogs in particular) was well documented (36). Since the earlier study period 2010–2012 (24), the prevalence of ESBL-producing uropathogenic E. coli ST131 among feline and canine samples in Switzerland has increased from 0 to 1.5% (4/273), and includes E. coli ST131-CTX-M-15 as well as ST131-CTX-M-27, which is currently emerging in human medicine in Germany, France, and Japan (37, 38).
Other human-related strains detected in this study included E. cloacae harboring blaSHV-12 together with the plasmid-mediated quinolone resistance gene qnrA. The combined presence of blaSHV-12 and qnrA has been described in human clinical E. cloacae isolates in hospitals in France and the UK (39, 40). Although data on ESBL-producing E. cloacae in animals are scarce (22, 41), our results provide evidence that this important pathogen has emerged in companion animals in Switzerland, illustrating their potential for increased dissemination.
In this study, the identification of phylogenetic groups among the E. coli isolates was performed based on the new Clermont scheme (16). Consequently, a number of STs from this study were classified as phylogenetic group F from their original D designation, including E. coli ST117 which is a recognized avian pathogenic lineage (42), E. coli ST354 and ST648, which are frequently detected in humans and animals (9, 43), and the rarely described E. coli ST457. In this study, we detected two isolates belonging to ST457, both harboring the uncommon blaCTX-M-55. E. coli ST457-CTX-M-55 harboring the carbapenemase gene blaKPC-3 was isolated in Italy from a human diagnosed with pneumonia (44), but to our knowledge, this ST has not been associated with disease in companion animals before.
A large number (26.4%, 19/72) of isolates changed designation from the original phylogenetic group A to group C. Most isolates in this group belonged to ST410 and were of low virulence. However, the panel of VFs selected for this study was limited in number and represents only a subset of known VFs. Other important determinants of virulence may have been missed due to this limitation. Nevertheless, the pathogenic potential of ST410 has been documented previously, together with strong evidence for clonal dissemination of E. coli ST410 between the avian wildlife, humans, and companion animals in Germany (45, 46). CTX-M-15-producing E. coli ST410 was also identified as a veterinary hospital strain in the UK (21). Although currently available reports on blaESBLs in ST410 are limited to blaCTX-M-15, our results demonstrate that this ST can also harbor blaCTX-M-1 and blaCTX-M-55, both variants that occur among food-producing animals (26, 31). Here, we provide further evidence for the pathogenic potential of this ST in companion animals and suggest that, in addition to its potential as an international clone for the dissemination of blaCTX-M-15, it may contribute to the dispersion of other resistance genes, including other blaESBL variants, aac(6’)-Ib-cr, and mph(A). The high prevalence (38.9%) of isolates harboring plasmid-mediated mph(A) which confers reduced susceptibility to azithromycin is of concern, since this macrolide is considered a last-resort antimicrobial agent for shigellosis (47). Furthermore, azithromycin represents an option for the treatment of Gram-negative rods expressing MDR, including carbapenem-resistant isolates of Pseudomonas aeruginosa, K. pneumoniae, and Acinetobacter baumannii (48), and is the only antimicrobial under consideration for the treatment of enterohemorrhagic E. coli in humans (49).
In conclusion, this study provides information on the prevalence, the blaESBL variants and the genotypes of ESBL-producing isolates in cats and dogs in Switzerland. The occurrence of potentially high-risk human-related K. pneumoniae and E. coli clones, as well as E. cloacae harboring blaSHV-12 and qnrA genes, previously described in humans suggests transmission events between companion animals as well as the possibility of the presence of a common source. This collection of ESBL-producing Enterobacteriaceae from cats and dogs identifies E. coli phylogroup C ST410 as a frequent MDR, ESBL-producing clone among clinical isolates from dogs in Switzerland that warrants further attention. The clinical significance of phylogroup C strains as etiological agents of extraintestinal disease and disseminators of antimicrobial resistance in companion animals remains to be investigated. Understanding the epidemiological and molecular features of ESBL-producing Enterobacteriaceae in companion animals can be helpful for infection management and prevention in veterinary as well as in human medicine.
Author Contributions
RS designed the study. AZ, KZ, SNS, and SS carried out the microbiological and molecular biological tests. AZ, KZ, SNS, and MN-I analyzed and interpreted the data. MN-I drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Marina Morach for technical assistance. This work was partly supported by the Swiss Federal Office of Public Health, Division Communicable Diseases.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fvets.2018.00062/full#supplementary-material.
Footnotes
- ^http://www.ncbi.nlm.nih.gov/blast/ (Accessed: May 17, 2017).
- ^http://enterobase.warwick.ac.uk (Accessed: June 6, 2017).
- ^http://bigsdb.pasteur.fr/klebsiella/ (Accessed: June 24, 2017).
References
1. Koenig A. Gram-negative bacterial infection. In: Greene CE, editor. Infectious Diseases of the Dog and Cat. St Louis: Elsevier Saunders (2012). p. 349–59.
2. Weese JS, Blondeau JM, Boothe D, Breitschwerdt EB, Guardabassi L, Hillier A, et al. Antimicrobial use guidelines for treatment of urinary tract disease in dogs and cats: antimicrobial guidelines working group of the international society for companion animal infectious diseases. Vet Med Int (2011) 2011:263768. doi:10.4061/2011/263768
3. Bush K. Proliferation and significance of clinically relevant β-lactamases. Ann N Y Acad Sci (2013) 1277:84–90. doi:10.1111/nyas.12023
4. Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, et al. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis (2008) 14:195–200. doi:10.3201/eid1402.070350
5. World Health Organization. Critically Important Antimicrobials for Human Medicine – 5th Revision, 2016. Geneva: World Health Organization (2017). Available from: http://apps.who.int/iris/bitstream/10665/255027/1/9789241512220-eng.pdf?ua=1 (Accessed: February 01, 2018).
6. Bogaerts P, Huang TD, Bouchahrouf W, Bauraing C, Berhin C, El Garch F, et al. Characterization of ESBL- and AmpC-producing Enterobacteriaceae from diseased companion animals in Europe. Microb Drug Resist (2015) 21:643–50. doi:10.1089/mdr.2014.0284
7. Ewers C, Stamm I, Pfeifer Y, Wieler LH, Kopp PA, Schønning K, et al. Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J Antimicrob Chemother (2014) 69:2676–80. doi:10.1093/jac/dku217
8. Pomba C, Rantala M, Greko C, Baptiste KE, Catry B, van Duijkeren E, et al. Public health risk of antimicrobial resistance transfer from companion animals. J Antimicrob Chemother (2017) 72:957–68. doi:10.1093/jac/dkw481
9. Ewers C, Bethe A, Stamm I, Grobbel M, Kopp PA, Guerra B, et al. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemother (2014) 69:1224–30. doi:10.1093/jac/dkt516
10. Geser N, Stephan R, Korczak BM, Beutin L, Hächler H. Molecular identification of extended-spectrum-β-lactamase genes from Enterobacteriaceae isolated from healthy human carriers in Switzerland. Antimicrob Agents Chemother (2012) 56:1609–12. doi:10.1128/AAC.05539-11
11. Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob Chemother (2006) 57:154–5. doi:10.1093/jac/dki412
12. Zurfluh K, Nüesch-Inderbinen M, Morach M, Berner AZ, Hächler H, Stephan R. Extended-spectrum ß-lactamase-producing-Enterobacteriaceae in vegetables imported from the Dominican Republic, India, Thailand and Vietnam. Appl Environ Microbiol (2015) 81:3115–20. doi:10.1128/AEM.00258-15
13. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. Wayne, PA: Clinical and Laboratory Standards Institute (2016). CLSI SupplementM100S.
14. Zurfluh K, Abgottspon H, Hächler H, Nüesch-Inderbinen M, Stephan R. Quinolone resistance mechanisms among extended-spectrum beta-lactamase (ESBL) producing Escherichia coli isolated from rivers and lakes in Switzerland. PLoS One (2014) 9(4):e95864. doi:10.1371/journal.pone.0095864
15. Ojo KK, Ulep C, Van Kirk N, Luis H, Bernardo M, Leitao J, et al. The mef(A) gene predominates among seven macrolide resistance genes identified in gram-negative strains representing 13 genera, isolated from healthy Portuguese children. Antimicrob Agents Chemother (2004) 48:3451–6. doi:10.1128/AAC.48.9.3451-3456.2004
16. Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep (2013) 5:58–65. doi:10.1111/1758-2229.12019
17. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol (2006) 60:1136–51. doi:10.1111/j.1365-2958.2006.05172.x
18. Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol (2005) 43:4178–82. doi:10.1128/JCM.43.8.4178-4182.2005
19. Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis (2000) 181:261–72. doi:10.1086/315217
20. Spurbeck RR, Dinh PC, Walk ST, Stapleton AE, Hooton TM, Nolan LK, et al. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect Immun (2012) 80:4115–22. doi:10.1128/IAI.00752-12
21. Timofte D, Maciuca IE, Williams NJ, Wattret A, Schmidt V. Veterinary hospital dissemination of CTX-M-15 extended-spectrum beta-lactamase-producing Escherichia coli ST410 in the United Kingdom. Microb Drug Resist (2016) 22:609–15. doi:10.1089/mdr.2016.0036
22. Dierikx CM, van Duijkeren E, Schoormans AH, van Essen-Zandbergen A, Veldman K, Kant A, et al. Occurrence and characteristics of extended-spectrum-β-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J Antimicrob Chemother (2012) 67:1368–74. doi:10.1093/jac/dks049
23. Dahmen S, Haenni M, Châtre P, Madec JY. Characterization of blaCTX-M IncFII plasmids and clones of Escherichia coli from pets in France. J Antimicrob Chemother (2013) 68:2797–801. doi:10.1093/jac/dkt29
24. Huber H, Zweifel C, Wittenbrink MM, Stephan R. ESBL-producing uropathogenic Escherichia coli isolated from dogs and cats in Switzerland. Vet Microbiol (2013) 162:992–6. doi:10.1016/j.vetmic.2012.10.029
25. Shaheen BW, Nayak R, Foley SL, Kweon O, Deck J, Park M, et al. Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the United States. Antimicrob Agents Chemother (2011) 55:5666–75. doi:10.1128/AAC.00656-11
26. Zurfluh K, Wang J, Klumpp J, Nüesch-Inderbinen M, Fanning S, Stephan R. Vertical transmission of highly similar blaCTX-M-1-harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front Microbiol (2014) 5:519. doi:10.3389/fmicb.2014.00519
27. Abgottspon H, Stephan R, Bagutti C, Brodmann P, Hächler H, Zurfluh K. Characteristics of extended-spectrum cephalosporin-resistant Escherichia coli isolated from Swiss and imported poultry meat. J Food Prot (2014) 77:112–5. doi:10.4315/0362-028X.JFP-13-120
28. Glaser CA, Powers EL, Greene CE. Zoonotic infections of medical importance in immunocompromised humans. In: Greene CE, editor. Infectious Diseases of the Dog and Cat. St Louis: Elsevier Saunders (2012). p. 1141–62.
29. Weese JS, Rousseau J, Arroyo L. Bacteriological evaluation of commercial canine and feline raw diets. Can Vet J (2005) 46:513–6.
30. Baede VO, Broens E, Spaninks M, Timmerman A, Graveland H, Wagenaar J, et al. Raw pet food as a risk factor for shedding of extended-spectrum beta-lactamase producing Enterobacteriaceae in household cats. PLoS One (2017) 12(11):e0187239. doi:10.1371/journal.pone.0187239
31. Rao L, Lv L, Zeng Z, Chen S, He D, Chen X, et al. Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003-2012. Vet Microbiol (2014) 172:534–41. doi:10.1016/j.vetmic.2014.06.013
32. Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev (2011) 35:736–55. doi:10.1111/j.1574-6976.2011.00268.x
33. Wohlwend N, Endimiani A, Francey T, Perreten V. Third-generation-cephalosporin-resistant Klebsiella pneumoniae isolates from humans and companion animals in Switzerland: spread of a DHA-producing sequence type 11 clone in a veterinary setting. Antimicrob Agents Chemother (2015) 59:2949–55. doi:10.1128/AAC.04408-14
34. Ovejero CM, Escudero JA, Thomas-Lopez D, Hoefer A, Moyano G, Montero N, et al. Highly tigecycline-resistant Klebsiella pneumoniae sequence type 11 (ST11) and ST147 isolates from companion animals. Antimicrob Agents Chemother (2017) 61(6):e2640–2616. doi:10.1128/AAC.02640-16
35. Sato T, Harada K, Usui M, Tsuyuki Y, Shiraishi T, Tamura Y, et al. Tigecycline susceptibility of Klebsiella pneumoniae complex and Escherichia coli isolates from companion animals: the prevalence of tigecycline-nonsusceptible K. pneumoniae complex, including internationally expanding human pathogenic lineages. Microb Drug Resist (2017). doi:10.1089/mdr.2017.0184
36. Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev (2014) 27:543–74. doi:10.1128/CMR.00125-13
37. Ghosh H, Doijad S, Falgenhauer L, Fritzenwanker M, Imirzalioglu C, Chakraborty T. blaCTX-M-27-encoding Escherichia coli sequence type 131 lineage C1-M27 clone in clinical isolates, Germany. Emerg Infect Dis (2017) 23:1754–6. doi:10.3201/eid2310.170938
38. Birgy A, Bidet P, Levy C, Sobral E, Cohen R, Bonacorsi S. CTX-M-27-producing Escherichia coli of sequence type 131 and clade C1-M27, France. Emerg Infect Dis (2017) 23:885. doi:10.3201/eid2305.161865
39. Cambau E, Lascols C, Sougakoff W, Bébéar C, Bonnet R, Cavallo JD, et al. Occurrence of qnrA-positive clinical isolates in French teaching hospitals during 2002–2005. Clin Microbiol Infect (2006) 12:1013–20. doi:10.1111/j.1469-0691.2006.01529.x
40. Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev (2009) 22:664–89. doi:10.1128/CMR.00016-09
41. Haenni M, Saras E, Ponsin C, Dahmen S, Petitjean M, Hocquet D, et al. High prevalence of international ESBL CTX-M-15-producing Enterobacter cloacae ST114 clone in animals. J Antimicrob Chemother (2016) 71:1497–500. doi:10.1093/jac/dkw006
42. Mora A, López C, Herrera A, Viso S, Mamani R, Dhabi G, et al. Emerging avian pathogenic Escherichia coli strains belonging to clonal groups O111:H4-D-ST2085 and O111:H4-D-ST117 with high virulence-gene content and zoonotic potential. Vet Microbiol (2012) 156:347–52. doi:10.1016/j.vetmic.2011.10.033-
43. Vangchhia B, Abraham S, Bell JM, Collignon P, Gibson JS, Ingram PR, et al. Phylogenetic diversity, antimicrobial susceptibility and virulence characteristics of phylogroup F Escherichia coli in Australia. Microbiology (2016) 162:1904–12. doi:10.1099/mic.0.000367
44. Accogli M, Giani T, Monaco M, Giufrè M, García-Fernández A, Conte V, et al. Emergence of Escherichia coli ST131 sub-clone H30 producing VIM-1 and KPC-3 carbapenemases, Italy. J Antimicrob Chemother (2014) 2014(69):2293–6. doi:10.1093/jac/dku132
45. Schaufler K, Semmler T, Wieler LH, Wöhrmann M, Baddam R, Ahmed N, et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410 – another successful pandemic clone. FEMS Microbiol Ecol (2016) 92(1):fiv155. doi:10.1093/femsec/fiv155
46. Falgenhauer L, Imirzalioglu C, Ghosh H, Gwozdzinski K, Schmiedel J, Gentil K, et al. Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int J Antimicrob Agents (2016) 47:457–65. doi:10.1016/j.ijantimicag.2016.03.019
47. Baker KS, Dallman TJ, Ashton PM, Day M, Hughes G, Crook PD, et al. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect Dis (2015) 15:913–21. doi:10.1016/S1473-3099(15)00002-X
48. Lin L, Nonejuie P, Munguia J, Hollands A, Olson J, Dam Q, et al. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant gram-negative bacterial pathogens. EBioMedicine (2015) 2:690–8. doi:10.1016/j.ebiom.2015.05.021
Keywords: extended-spectrum β-lactamase, clinical, genotypes, cats, dogs
Citation: Zogg AL, Simmen S, Zurfluh K, Stephan R, Schmitt SN and Nüesch-Inderbinen M (2018) High Prevalence of Extended-Spectrum β-Lactamase Producing Enterobacteriaceae Among Clinical Isolates From Cats and Dogs Admitted to a Veterinary Hospital in Switzerland. Front. Vet. Sci. 5:62. doi: 10.3389/fvets.2018.00062
Received: 06 February 2018; Accepted: 12 March 2018;
Published: 27 March 2018
Edited by:
Timothy J. Johnson, University of Minnesota, United StatesReviewed by:
Guadalupe Virginia Nevárez-Moorillón, Autonomous University of Chihuahua, MexicoMaria José Saavedra, Universidade de Trás-os-Montes e Alto Douro, Portugal
Copyright: © 2018 Zogg, Simmen, Zurfluh, Stephan, Schmitt and Nüesch-Inderbinen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Magdalena Nüesch-Inderbinen, magdalena.nueesch-inderbinen@uzh.ch
 Anna Lena Zogg1
Anna Lena Zogg1
 Roger Stephan
Roger Stephan Magdalena Nüesch-Inderbinen
Magdalena Nüesch-Inderbinen