1
Department of Psychology, Stanford University, Stanford, CA, USA
2
Department of Radiology, Stanford University School of Medicine, Stanford, CA, USA
A specific polymorphism of the brain-derived neurotrophic factor (BDNF) gene is associated with alterations in brain anatomy and memory; its relevance to the functional connectivity of brain networks, however, is unclear. Given that altered hippocampal function and structure has been found in adults who carry the methionine (met) allele of the BDNF gene and the molecular studies elucidating the role of BDNF in neurogenesis and synapse formation, we examined the association between BDNF gene variants and neural resting connectivity in children and adolescents. We observed a reduction in hippocampal and parahippocampal to cortical connectivity in met-allele carriers within both default-mode and executive networks. In contrast, we observed increased connectivity to amygdala, insula and striatal regions in met-carriers, within the paralimbic network. Because of the known association between the BDNF gene and neuropsychiatric disorder, this latter finding of greater connectivity in circuits important for emotion processing may indicate a new neural mechanism through which these gene-related psychiatric differences are manifest. Here we show that the BDNF gene, known to regulate synaptic plasticity and connectivity in the brain, affects functional connectivity at the neural systems level. In addition, we demonstrate that the spatial topography of multiple high-level resting state networks in healthy children and adolescents is similar to that observed in adults.
Brain-derived neurotrophic factor (BDNF) plays an essential role in the early growth of neural networks in the human brain (McAllister et al., 1999
; Huang and Reichardt, 2001
). BDNF is a polypeptide growth factor in a family of signaling molecules known as neurotrophins. These proteins regulate axonal and dendritic growth (Huang and Reichardt, 2001
), synaptic structure and plasticity (McAllister et al., 1999
; Lu and Gottschalk, 2000
), neurotransmitter release, and long-term potentiation (LTP)-associated-learning (Lu and Gottschalk, 2000
; Chao, 2003
). Considerable progress has been made in understanding the ways in which neurotrophins exert their effects on neuronal health and synaptic plasticity (e.g., cellular signaling and ligand-receptor binding) (Lewin and Barde, 1996
; Stoop and Poo, 1996
); how neurotrophin-related differences in neuronal health and synaptic plasticity may lead to selective impairments in nervous system function, however, is not well understood.
BDNF is a unique neurotrophin because activation of its cognate receptor, tropomyosin-related kinase B (TrkB), results in a wider set of functional consequences than is the case with other Trk receptors (Bath and Lee, 2006
). BDNF is expressed predominantly in the central nervous system, including cortical regions, the hippocampus, limbic structures, the cerebellum, and the olfactory bulb (Huang and Reichardt, 2001
). A common single-nucleotide polymorphism (SNP) in the BDNF gene produces an amino acid substitution (valine to methionine) at codon 66 (val66met) that alters secretion and intracellular trafficking of the mature peptide (Egan et al., 2003
; Chen et al., 2004
). This SNP has been useful in linking BDNF to human cognitive processes (Egan et al., 2003
; Gabrieli and Preston, 2003
; Hariri et al., 2003
; Dempster et al., 2005
; Ho et al., 2006
) and, as a result, interest in BDNF has come from researchers spanning fields from developmental neurobiology to neurodegenerative and psychiatric disorders (Chao, 2003
).
Compared with val-allele homozygotes, individuals with the altered methionine (met) allele of the BDNF gene have been found to exhibit reduced hippocampal volume (Pezawas et al., 2004
; Szeszko et al., 2005
; Bueller et al., 2006
; Frodl et al., 2007
; Chepenik et al., 2009
), memory impairment (Egan et al., 2003
; Hariri et al., 2003
; Dempster et al., 2005
; Ho et al., 2006
), increased susceptibility to schizophrenia and other psychotic disorders (Gratacos et al., 2007
), and abnormal hippocampal activity during memory processing (Egan et al., 2003
; Hariri et al., 2003
). Given the established role of BDNF in mediating processes related to neural excitability, learning, and memory (Korte et al., 1995
; Patterson et al., 1996
; Desai et al., 1999
), this gene is likely implicated in the formation and maintenance of major functional circuits in the human brain. To date, however, the effects of this BDNF polymorphism on the functioning of large-scale neural networks have not been examined.
Currently, one of the most informative methods for interrogating the integrity of neural networks is to measure activation across distinct brain regions by conducting functional connectivity (FC) analysis on data obtained through whole-brain functional magnetic resonance imaging (fMRI). FC can be operationally defined to refer to temporal correlations across cortical regions representing an index of function (Friston et al., 1993
; Horwitz, 2003
) and anatomical connectivity (Koch et al., 2002
; Quigley et al., 2003
; Hagmann et al., 2008
; Greicius et al., 2009
). In the present study we used fMRI to probe resting FC as a function of BDNF gene variants across the three most frequently examined high-order cognitive- and emotion-processing networks of the brain: the default-mode network (DMN), the executive network (EN), and the salience network (SN). Because neurotrophins profoundly affect the development of the nervous system (Lewin and Barde, 1996
), we examined BDNF and FC in a sample of children and adolescents, in whom neural development is actively progressing and for whom BDNF may therefore be expected to exert significant effects (for detailed discussion, see Casey et al., 2009
).
We hypothesize that because BDNF secretion is decreased in met-allele carriers compared to val-allele homozygotes (Chen et al., 2004
), participants who carry a BDNF met allele will be broadly characterized by reduced FC, although this may differ across the three resting-state networks. Moreover, because BDNF is widely expressed in the hippocampus (Conner et al., 1997
) and because, compared to val-allele homozygotes, met-allele carriers have been found to have poorer memory (Egan et al., 2003
; Dempster et al., 2005
), diminished hippocampal function (Egan et al., 2003
) and smaller hippocampal volume (Pezawas et al., 2004
; Szeszko et al., 2005
; Bueller et al., 2006
; Frodl et al., 2007
; Chepenik et al., 2009
), we hypothesize that differences between genetic groups in resting FC will be particularly pronounced between the hippocampal formation and regions that support memory processing (e.g., parietal association cortices, prefrontal regions, cingulate).
Participants
Participants were 38 children and adolescents (25 females) between the ages of 9 and 16 years (M = 12.2, SD = 2.1). They were recruited through their mothers via Craigslist and other online advertisements and parent networks, and each mother-child pair was compensated $25/hour. All participants had no reported history of brain injury, no behavioral indications of possible mental impairment, no past or present Axis I disorder, were right-handed, fluent in English, and had no learning disorder. Parents and children gave informed consent and assent, respectively, as approved by the Stanford Institutional Review Board.
Procedure
Participants were assessed in two sessions. In the first session, participants were administered the Schedule for Affective Disorders and Schizophrenia for School-Aged Children-Present and Lifetime version (K-SADS) (Geller et al., 1996
, 2001
) to assess current and lifetime psychopathology in order to ensure the absence of any current or past diagnosable DSM-IV psychiatric disorder. During this session, children and parents also provided saliva samples for genetic testing and viewed a video to prepare them for the MRI scan session. In the second session, brain-imaging data were acquired using a whole-brain MRI scanner.
Genetic Data
DNA was extracted from saliva using the Oragene DNA saliva kit. The target 300 bp BDNF gene fragment was amplified using the G196A primer 5′-ATC CGA GGA CAA GGT GGC-3′ (forward) and 5′-CCT CAT GGA CAT GTT TGC AG-3′ (reverse). The PCR amplification was carried out in a final volume of 20 μl consisting of 50 ng of genomic DNA, 200 nM each of sense and antisense primers, 200 μM of dNTP mix, 2 units of Expand High Fidelity PCR System (Roche, Cat# 11-795-078-001). Annealing was carried out at 60 C for 45 s, extension at 72 C for 1 min, and denaturation at 95 C for 30 s for a total of 35 cycles. 5 μl of the PCR products were digested by 10 units of Pml I (New England Biolabs) in a 15 μl reaction at 37 C for 3 h. The digestion mixture was electrophoresed through 7% Polyacrylamide gel (Acrylamide/bis-Acrylamide ratio 19:1) at 150 V for 40 min. 100 bp and 10 bp marker was used to measure the digestion and PCR product size. Allele A (methionine) was not digested showing one 300 bp band, but the G allele (valine) was digested showing two bands, at 180 and 120 bp. As we describe below, this genotyping yielded two groups of children: val-allele homozygotes (n = 23) and met-allele carriers (n = 15).
fMRI Data Acquisition
Magnetic resonance imaging was performed on a 3.0-T GE whole-body scanner. Participants were positioned in a purpose-built single channel T/R head coil and stabilized by clamps and a bite bar formed with dental impression wax (made of Impression Compound Type I, Kerr Corporation, Romulus, MI) to reduce motion-related artifacts during scanning. During the resting-state experiment, participants completed a 6-min scan during which they were instructed to lay still with eyes closed. For this study, 29 axial slices were taken with 4 mm slice thickness. High-resolution T2-weighted fast spin echo structural images (TR = 3000 ms, TE = 68 ms, ETL = 12) were acquired for anatomical reference. A T2*-sensitive gradient echo spiral in/out pulse sequence (Glover and Law, 2001
) was used for all functional imaging (TR = 2000 ms, TE = 30 ms, flip angle = 77°, FOV = 22 cm, 64 × 64). An automated high-order shimming procedure, based on spiral acquisitions, was used to reduce B0 heterogeneity (Kim et al., 2002
). Spiral in/out methods have been shown to increase signal-to-noise ratio and BOLD contrast-to-noise ratio in uniform brain regions, as well as to reduce signal loss in regions compromised by susceptibility-induced field gradients generated near air-tissue interfaces such as PFC (Glover and Law, 2001
). Compared to traditional spiral imaging techniques, spiral in/out methods result in less signal dropout and greater task-related activation in PFC regions (Preston et al., 2004
). A high-resolution volume scan (140 slices, 1 mm slice thickness) was collected for every participant using a spoiled grass gradient recalled (SPGR) sequence for T1 contrast (TR = 3000 ms, TE = 68 ms, TI = 500 ms, flip angle = 11°, FOV = 25 cm, 256 × 256). During the resting-state scan, children’s heart-rate and respiration waveform were recorded.
fMRI Preprocessing
fMRI data were then preprocessed using AFNI
1
(Cox, 1996
). Preprocessing included slice-timing correction, volume registration, smoothing (4 mm), bandpass filtering (0.008 < f < 0.15), and co-registration of functional and anatomical images.
ICA Versus ROI-Based Connectivity Analysis
Previous resting state connectivity analyses have used FSL’s Independent Component Analysis (ICA) software (MELODIC) and an automated MATLAB analysis routine (Greicius et al., 2004
, 2007
). This processing pathway was not suitable for the present study because in a small number of cases (<20%) the MELODIC ICA process separated the executive control network for the participant into two lateralized maps, one right and one left. For this reason, and because in our previous work we demonstrated that seed-based FC and ICA-based network analyses yield comparable DMNs in children (Thomason et al., 2008
), we used ROI-based connectivity analysis to evaluate multiple resting networks in the present study.
Functional Connectivity (FC) Analysis
We conducted ROI-based connectivity analysis on the functional data. Selected seed regions were based on those reported in published studies (Default: 10, −50, 30; Executive: 44, 36, 20; Salience: 38, 26, −10) (Krasnow et al., 2003
; Seeley et al., 2007
; Thomason et al., 2008
), as they have been shown to yield robust connectivity across the major resting-state networks. We began our analysis with image reconstruction using a correction that removes respiratory variations and HR signal extraction. In brief, this correction removes the effect of low-frequency respiratory variations (i.e., the “envelope” of the respiratory belt waveform) and heart rate (average rate in a 6-s sliding window) by first convolving those signals with appropriate filters and then regressing them out of the time series for each voxel (described in Birn et al., 2008
; Chang et al., 2009
).
Following signal extraction, the trace from each participant’s seed region (3D sphere with a radius of 3, centered on the previously presented coordinates) was detrended for three translational and three rotational motion regressors (AFNI 3dDetrend) and was used to calculate the correlation between the seed region and time-course data in all of the other voxels in the brain. Correlation estimates were controlled for estimated translational and rotational motion and a white matter nuisance time-course (3d sphere with radius of 3 mm, centered at (27, −8, 27). In contrast to some of the previous resting-state protocols conducted with adults (e.g., Fox et al., 2005
), in the present analysis rather than removing variance from motion, white matter, cerebral spinal fluid and from global signal, we removed only motion and a timecourse sampled from a white matter region of interest. We made this choice based on recent work indicating that the use of global signal correction may force the presence of anticorrelated networks (Chang et al., 2009
; Murphy et al., 2009
; Weissenbacher et al., 2009
); indeed, this methodological and interpretive topic is currently being actively debated for resting-state studies (Fox et al., 2009
). After correlation coefficients were calculated for each voxel in the brain, we normalized the distribution of these values using Fishers r-to-z transformation. We submitted the resulting correlation maps to main-effects analysis, conducting two-tailed t-tests to identify regions in which whole-brain z-converted regression maps differed between the two genotype groups. We conducted group-level analyses separately for each resting network and report results at p < 0.01.
We analyzed motion and found groups differed at p < 0.05. Therefore, before signal was extracted from the seed regions, we detrended the signal for motion. Then, the six motion parameters (three translational, three rotational) were modeled as regressors of non-interest at the individual subject level to adjust the error term appropriately.
Region of Interest (ROI) Analysis
ROI analysis was conducted for hippocampal ROIs created using the AFNI software package and subroutines for creating smoothed (3 mm), resampled (3.75 mm3), binary masks for the left and right hippocampi
2
. Results within the masks were held to a liberal threshold (p < 0.05) for completeness in reporting.
Hippocampal Volume Analysis
To determine whether the connectivity analysis required correction for hippocampal volume, we conducted manual tracing of hippocampal volumes within SPGR images using Insight Toolkit’s SNAP program (Yushkevich et al., 2006
). Tracings were performed in reoriented native space, and hippocampal volumes were divided by total brain volume to control for the potentially confounding factor of head size. The anatomical features used to guide manual tracing of the hippocampus have been described elsewhere (Pruessner et al., 2000
). Briefly, the hippocampal head-body boundary was delineated by the clear appearance of the uncal recess, while the body-tail boundary was delineated by the opening of the crus of the fornix. Final volumes were output using SNAP. Data were analyzed in SPSS16 using a multivariate general linear model framework and including participant age as a covariate. Because there was not a significant gene-group effect in hippocampal volume, F(2, 32) = 0.36, p = 0.69, correction of functional connectivity data for volume differences was not required.
Participants
Participants were 26 Caucasians (68%), 3 Asian Americans (8%), 2 Hispanic Americans (5%), and 7 participants of multi- or bi-racial descent (18%). BDNF genotyping yielded two groups of children: val-homozygotes (n = 23) and met-allele carriers (n = 15). These allelic frequencies were in Hardy-Weinberg equilibrium, χ2 = 0.65, p = 0.42. Demographic data for the two genotype groups are presented in Table 1
. The two groups did not differ significantly with respect to age, F(1,36) = 0.96, p = 0.33, or gender, χ2 = 0.63, p = 0.43. BDNF alleles were distributed similarly in Caucasian and non-Caucasian participants, χ2 = 0.035, p = 0.85.
Network Maps
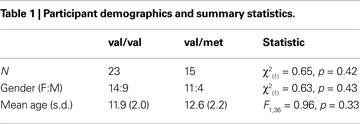
Complete network maps generated using three separate one-sample t-tests (Figure 1
), each collapsing across the full sample, yielded maps similar to those produced in previous studies of adults (Fox et al., 2005
; Fransson, 2005
; Seeley et al., 2007
; Taylor et al., 2008
; Habas et al., 2009
) and children (Thomason et al., 2008
; Kelly et al., 2009
), and regarding reproducibility, see (Meindl et al., 2009
). These maps demonstrate reliable generation of the high-order cognitive and/or affective resting network maps in a sample of children and adolescents that have previously been reported in samples of adults.
Figure 1. Map of neural connectivity for the three major resting-state networks across all subjects (n = 38). p < 0.0001. Red circles denote approximate locations of seed-point ROIs.
Default-Mode Network (DMN) Comparison
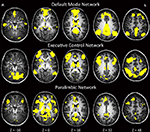
Figure 2
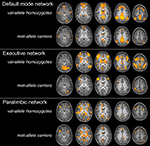
shows val-allele homozygotes have greater and wider extent of suprathreshold functional connectivity within and beyond the DMN than do met-allele carriers. The between-groups whole-brain comparison of DMN statistical maps (Figure 3
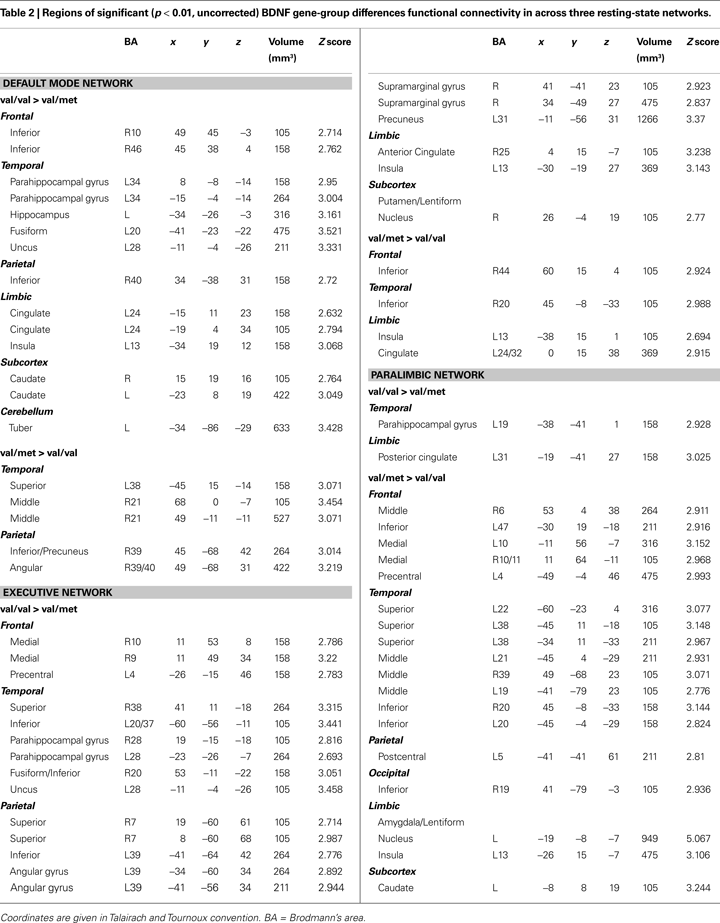
) showed significantly (p < 0.01) increased hippocampal, fusiform, insula, caudate, cingulate, frontal and cerebellar contribution to the DMN in children homozygous for the BDNF val allele. The reverse contrast for areas with significantly greater contribution in met-allele carriers produced fewer significant clusters, and included regions of the parietal and temporal lobes; see Table 2
.
Figure 2. One-sample t-tests within each genetic group depicting group effects for three major resting-state networks at p < 0.0001.
Figure 3. Two-sample t-tests for three major resting-state networks. BDNF gene group differences (val/val > val/met: blue; val/met > val/val: orange) across three resting networks. FG = fusiform gyrus, Ins = insula, Hip = hippocampus, IPL = inferior parietal lobule, PHG = parahippocampal gyrus, SPL = superiorparietal lobe, Pu = putamen, Am = amygdala; p < 0.01.
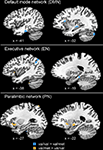
A total of 10 voxels in the between-groups, within-hippocampal, ROI comparison of DMN statistical maps were significant at p < 0.05. The largest significant cluster was k = 5 voxels in the left hippocampus (−34, −19, −7) where resting functional connectivity to the DMN seed region was greater in children homozygous for the BDNF val allele (z = 4.10); there was no significant correlation in children who carried a met allele (z = 0.34). There were no areas in the hippocampal ROI analysis in which the contribution of met-allele carriers to the DMN network were significantly greater than val-allele homozygotes; see Figure 4
.
Figure 4. BDNF gene group differences (val/val > val/met: blue; val/met > val/val: yellow) for each of the three resting networks within hippocampal ROIs.
Executive Network (EN) Comparison
As seen in Figure 2
, val-allele homozygotes have a greater and wider extent of suprathreshold functional connectivity within and beyond the EN than do met-allele carriers. Whole-brain between-groups analysis of the EN (Figure 3
) showed that children homozygous for the BDNF val allele showed significantly increased (p < 0.01) contributions from a number of clusters in temporal and parietal cortices, including parahippocampal and neighboring temporal lobe regions and a large area in the precuneus. In addition, there were frontal areas, insula, anterior cingulate, and striatal regions that showed greater FC in the val-allele homozygotes than in the met-allele carriers within this network. In the reverse contrast, met-allele carriers again had fewer significant clusters, and these were located in inferior frontal and temporal regions, insula, and cingulate cortex.
The between-groups, within-hippocampal ROI comparison of EN resulted in 17 voxels that showed significantly increased contribution to the EN by children homozygous for the BDNF val allele compared to met-allele carriers. Of these, 11 fell within the left hippocampal ROI and 6 fell within the right hippocampus ROI. The largest, most significant cluster was k = 4 voxels in the left hippocampus (−26, −26, −7) where the average correlation to the executive seed was z = 3.87 in val-allele homozygotes compared to z = 1.15 in met-allele carriers. There were no areas in the hippocamal ROI analysis in which met-allele carrier contributions to the EN network were significantly greater than was the case for val-allele homozygotes; see Figure 4
.
Paralimbic Network (PN) Comparison
The whole-brain between-groups analysis produced a different pattern of results in the PN. Whereas the DMN and EN comparisons were dominated by regions in which val-allele homozygotes had significantly higher FC than did met-allele carriers, the reverse was true for the PN (see Table 2
); many more regions showed significantly increased connectivity in the PN in met-allele carriers. In a number of cortical areas, amygdala, insula, and caudate, met-allele carriers showed significantly greater FC than did val-allele homozygotes. In contrast, val-allele homozygotes showed significantly greater reliance than did met-allele carriers only in regions of the posterior cingulate and a region of the parahippocampal gyrus.
The hippocampal ROI analysis also yielded different results in the PN. Only three voxels showed significant differences between the gene groups, all of which were located in the left hippocampus. Here, in contrast to what was observed for the other networks, significantly (p < 0.05) greater FC was observed in met-allele carriers than in val-allele homozygotes. In the peak of this difference (−30, −11, −22), which occurred in a region of the hippocampus that was more anterior than were peaks of group differences observed for the other networks, the average correlation to the executive seed was z = 1.95 in val-allele homozygotes and z = 3.84 in met-allele carriers.
Studies of the BDNF gene polymorphism have reliably documented episodic memory deficits associated with the met allele (Egan et al., 2003
; Hariri et al., 2003
). Contemporary models of declarative memory function suggest that strong neural connectivity between neocortical association areas and medial temporal lobe (MTL) regions underlies successful memory encoding, long-term maintenance, and retrieval (Ranganath et al., 2005
). In this study we did not restrict differences in FC to only within-network regions; consequently, we observed reliable effects across networks. More specifically, we observed a reduction in hippocampal and parahippocampal to cortical connectivity at rest in carriers of the met allele within each of the three resting networks we examined: the default-mode, executive, and paralimbic networks (see Table 2
). This work suggests that differences are present between genetic groups both within networks and in the connectivity of these networks to other parts of the brain. Analyses of all three networks provided evidence of higher basal connectivity between MTL structures and neocortical association areas in val-allele homozygotes than in met-allele carriers. These differences in resting FC may result from differences in anatomical connectivity, in which met-allele carriers have less robust cortical-to-MTL projections, particularly in the network important for executive control and in the default-mode network.
Association studies have linked the BDNF gene to substance-related disorders, eating disorders, and schizophrenia; the association of the BDNF gene with major depression disorders and bipolar disorders, however, has been inconclusive (see, for example, meta-analysis by Gratacos et al., 2007
). Contemporary theory suggests that the allele (met vs. val) that confers risk may change across development, differing in trajectory for various disorders (Casey et al., 2009
). Comprised primarily of insular and cingulate cortices that are connected with subcortical limbic structures, the paralimbic network may be the neural network most broadly relevant to psychiatric disorders for the role it is expected to exert in the detection, integration, and filtering of interoceptive, autonomic, and emotional information (Seeley et al., 2007
; Habas et al., 2009
). The insula plays a critical role in the experience of emotion, interoceptive awareness, and bodily homeostasis (Critchley et al., 2004
; Pollatos et al., 2007
). In a complementary fashion, the cingulate cortex is implicated in resolution of emotional conflict, in part through mediation of MTL structures, in particular, the amygdala (Etkin et al., 2006
). Overall, the system is posited to play an important role in response selection and orientation toward action by integrating its numerous inputs. Given the unique role of the paralimbic neural network in integrating sensory stimuli and in generating responses to those stimuli, it is noteworthy that the two genotype groups differ in resting connectivity in this circuit. In the present study we demonstrate that connectivity between this network and neocortical association areas and the amygdala is enhanced in met-allele carriers. It is possible, therefore, that greater connectivity between the cortico-limbic structures underlies both the risks associated with this allele (e.g., disorders, schizophrenia) and the protective effects of this allele (e.g., reduced substance abuse, lower levels of neuroticism).
We found lower resting connectivity in widespread brain regions in met-allele carriers than in val-allele homozygotes in both the default-mode and executive networks. We emphasized the differences observed in parahippocampal and hippocampal regions because of the major role these structures play in memory formation and the known association between the met allele and poorer episodic memory (Egan et al., 2003
).
In this paper, we interpret the obtained genotype differences as reflecting greater coherence between the MTL and our seed regions in individuals who are homozygous for the polymorphism of BDNF that confers higher levels of that growth factor in the MTL. Based on our analysis, the relations between our seeds and the MTL peaks range by group from z = 0.3 to 4.1. The finding that val-allele homozygotes have greater FC between hippocampal and both executive and salience networks is surprising given previous reports suggesting that these networks are negatively correlated with the MTL (Fox et al., 2005
). It is possible that by not performing global normalization, our results are more reliable in assessing the nature of the relation between these seeds and MTL, and this may account for the observed difference (Chang et al., 2009
; Murphy et al., 2009
; Weissenbacher et al., 2009
).
As we point out above, effective memory processing relies on distributed cortical and subcortical brain regions. In fact, the resting-state approach has been particularly fruitful in demarcating major memory networks by examining healthy participants and showing that regions of the precuneus, posterior cingulate, and parietal lobule that show greater activity to successfully remembered items are the same regions that are connected at rest to the hippocampal formation (Vincent et al., 2006
), and by drawing conceptual links between memory impairments in Alzheimer patients and reduced resting-state connectivity in the DMN in the hippocampus (Greicius et al., 2004
). Collectively, studies examining resting-state networks have found that levels of connectivity and integrity in these networks are related to the anomalies in the psychological and cognitive operations served by those networks (for reviews of DMN and neuropsychiatric disorders, see Greicius, 2008
; Broyd et al., 2009
). In the present study we found reduced connectivity in default-mode and executive networks in a number of regions known to be implicated in memory processing (hippocampus, precuneus, BA 31, inferior parietal lobule). We suggest, therefore, that reductions in FC in met-allele carriers observed for the DMN and EN may represent anomalous development of these cognitive networks in children and adolescents, a process that may give rise to the kinds of memory impairments previously reported in adult BDNF met-allele carriers (Egan et al., 2003
; Hariri et al., 2003
; Dempster et al., 2005
; Ho et al., 2006
).
There are a number of caveats concerning this work that warrant mention. First, genotype interacts with several factors, including age, pubertal status, and gender, to influence the intermediate phenotype of brain biology. As is typical of most neuroimaging studies, our sample size limited the number of analyses that we could reliably conduct. Moreover, our sample of children is relatively heterogeneous, and other factors will certainly contribute variance to the observed findings. Second, the data are not reported using multiple comparisons correction. Without this correction we have accepted more chance for Type 1 error. Our future work will attempt to replicate these findings in an independent sample. The final caveat concerns the ROI-based approach and the selection of seed regions. We have attempted to differentiate connectivity between each network and the medial temporal lobe, but have done so using seed regions that are not completely independent of one another. Indeed, investigators have documented some degree of overlap among these networks, and between these networks and sensorimotor networks (Seeley et al., 2007
; Habas et al., 2009
). Consequently, resting state networks should not be interpreted as independent networks of the brain, but instead, as having some degree of interplay.
In summary, the reduced mnemonic function, altered MTL function, reduced hippocampal and cortical volume, and disposition to affective illness attributed to the BDNF met allele, combined with the functional connectivity analysis in our study, suggest that reduced BDNF has implications for the formation of large-scale neural networks. Our results extend previous fMRI findings by identifying altered neural system-level functional cooperativity in carriers of the met-allele. It remains for future research to elucidate the nature of the associations among these measures in affecting specific behavioral phenotypes. The role of exogenously applied neurotrophins, for example, is an active area of investigation, and advances in this area will inform our understanding of how these large-scale networks are formed and maintained. In addition, this work will help to generate novel hypotheses about the neural underpinnings of phenotypes associated with specific BDNF alleles. For instance, increased FC in paralimbic networks may also characterize individuals with an eating disorder, given the association of the met allele with that phenotype. The present results highlight the importance of integrating neural and genetic data in elucidating phenotypic behaviors.
The authors do not have a direct or indirect interest, financial or otherwise that is in conflict with or that might reasonably tend to influence the reporting of this study.
This project was supported by awards from the National Institute of Mental Health [MH081583 to MET; MH074849 to IHG, RR009874 to GHG], and by a NARSAD Young Investigator Award to MET. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health. The authors thank Dr. Michael Greicius for assistance with independent components analysis. The authors thank Emily L. Dennis, Melissa L. Henry, Sarah Victor, and Rebecca Johnson for their assistance in acquiring the scan data, and for data processing, and Yamanda Wright and Lindsey Sherdell for their assistance in participant recruitment, screening, and conducting structured behavioral interviews. Additionally, the authors acknowledge the critical analysis provided by the Reviewers and thank them for their contribution to the work.
- ^ http://afni.nimh.nih.gov/afni
- ^ We should note that AFNI ROIs are based on an adult-derived brain template. Validity of using the same stereotactic space for adults and children in this age range (>9 years old) has been demonstrated empirically (Burgund et al., 2002 ).
Casey, B. J., Glatt, C. E., Tottenham, N., Soliman, F., Bath, K., Amso, D., Altemus, M., Pattwell, S., Jones, R., Levita, L., McEwen, B., Magarinos, A. M., Gunnar, M., Thomas, K. M., Mezey, J., Clark, A. G., Hempstead, B. L., and Lee, F. S. (2009). Brain-derived neurotrophic factor as a model system for examining gene by environment interactions across development. Neuroscience 164, 108–120.
Chen, Z. Y., Patel, P. D., Sant, G., Meng, C. X., Teng, K. K., Hempstead, B. L., and Lee, F. S. (2004). Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J. Neurosci. 24, 4401–4411.
Chepenik, L. G., Fredericks, C., Papademetris, X., Spencer, L., Lacadie, C., Wang, F., Pittman, B., Duncan, J. S., Staib, L. H., Duman, R. S., Gelernter, J., and Blumberg, H. P. (2009). Effects of the brain-derived neurotrophic growth factor val66met variation on hippocampus morphology in bipolar disorder. Neuropsychopharmacology 34, 944–951.
Egan, M. F., Kojima, M., Callicott, J. H., Goldberg, T. E., Kolachana, B. S., Bertolino, A., Zaitsev, E., Gold, B., Goldman, D., Dean, M., Lu, B., and Weinberger, D. R. (2003). The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 112, 257–269.
Frodl, T., Schule, C., Schmitt, G., Born, C., Baghai, T., Zill, P., Bottlender, R., Rupprecht, R., Bondy, B., Reiser, M., Moller, H. J., and Meisenzahl, E. M. (2007). Association of the brain-derived neurotrophic factor Val66Met polymorphism with reduced hippocampal volumes in major depression. Arch. Gen. Psychiatry 64, 410–416.
Geller, B., Zimerman, B., Williams, M., Bolhofner, K., Craney, J. L., DelBello, M. P., and Soutullo, C. (2001). Reliability of the Washington University in St. Louis Kiddie Schedule for Affective Disorders and Schizophrenia (WASH-U-KSADS) mania and rapid cycling sections. J. Am. Acad. Child Adolesc. Psychiatry 40, 450–455.
Gratacos, M., Gonzalez, J. R., Mercader, J. M., de Cid, R., Urretavizcaya, M., and Estivill, X. (2007). Brain-derived neurotrophic factor Val66Met and psychiatric disorders: meta-analysis of case-control studies confirm association to substance-related disorders, eating disorders, and schizophrenia. Biol. Psychiatry 61, 911–922.
Ho, B. C., Milev, P., O’Leary, D. S., Librant, A., Andreasen, N. C., and Wassink, T. H. (2006). Cognitive and magnetic resonance imaging brain morphometric correlates of brain-derived neurotrophic factor Val66Met gene polymorphism in patients with schizophrenia and healthy volunteers. Arch. Gen. Psychiatry 63, 731–740.
Pruessner, J. C., Li, L. M., Serles, W., Pruessner, M., Collins, D. L., Kabani, N., Lupien, S., and Evans, A. C. (2000). Volumetry of hippocampus and amygdala with high-resolution MRI and three-dimensional analysis software: minimizing the discrepancies between laboratories. Cereb. Cortex 10, 433–442.
Szeszko, P. R., Lipsky, R., Mentschel, C., Robinson, D., Gunduz-Bruce, H., Sevy, S., Ashtari, M., Napolitano, B., Bilder, R. M., Kane, J. M., Goldman, D., and Malhotra, A. K. (2005). Brain-derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Mol. Psychiatry 10, 631–636.