Alkane biosynthesis genes in cyanobacteria and their transcriptional organization
- Genetics and Experimental Bioinformatics, Institute of Biology 3, Faculty of Biology, University of Freiburg, Freiburg, Germany
In cyanobacteria, alkanes are synthesized from a fatty acyl-ACP by two enzymes, acyl–acyl carrier protein reductase and aldehyde deformylating oxygenase. Despite the great interest in the exploitation for biofuel production, nothing is known about the transcriptional organization of their genes or the physiological function of alkane synthesis. The comparison of 115 microarray datasets indicates the relatively constitutive expression of aar and ado genes. The analysis of 181 available genomes showed that in 90% of the genomes both genes are present, likely indicating their physiological relevance. In 61% of them they cluster together with genes encoding acetyl-CoA carboxyl transferase and a short-chain dehydrogenase, strengthening the link to fatty acid metabolism and in 76% of the genomes they are located in tandem, suggesting constraints on the gene arrangement. However, contrary to the expectations for an operon, we found in Synechocystis sp. PCC 6803 specific promoters for the two genes, sll0208 (ado) and sll0209 (aar), which give rise to monocistronic transcripts. Moreover, the upstream located ado gene is driven by a proximal as well as a second, distal, promoter, from which a third transcript, the ~160 nt sRNA SyR9 is transcribed. Thus, the transcriptional organization of the alkane biosynthesis genes in Synechocystis sp. PCC 6803 is of substantial complexity. We verified all three promoters to function independently from each other and show a similar promoter arrangement also in the more distant Nodularia spumigena, Trichodesmium erythraeum, Anabaena sp. PCC 7120, Prochlorococcus MIT9313, and MED4. The presence of separate regulatory elements and the dominance of monocistronic mRNAs suggest the possible autonomous regulation of ado and aar. The complex transcriptional organization of the alkane synthesis gene cluster has possible metabolic implications and should be considered when manipulating the expression of these genes in cyanobacteria.
Introduction
The production of liquid fuels and a variety of chemicals indispensable for daily life depends on fossil resources. Due to their limited availability and with respect to environmental concerns the exploitation of alternative, renewable, and sustainable energy sources is gaining in importance. Thus, the conversion of solar energy and carbon dioxide into biofuels and suitable chemicals, e.g., hydrogen, ethanol, ethylene, isobutyraldehyde, or isoprene by genetically modified and improved phototrophic microorganisms such as cyanobacteria is of high interest (Deng and Coleman, 1999; Takahama et al., 2003; Atsumi et al., 2009; Lindberg et al., 2010; McKinlay and Harwood, 2010; Georgianna and Mayfield, 2012; Peralta-Yahya et al., 2012).
Interestingly, many cyanobacteria are naturally able to produce alkanes (Winters et al., 1969), which are the major constituents of gasoline, diesel, and jet fuels. However, there are also strains such as Synechococcus sp. PCC 7002 in which no alkanes were detectable, indicating that the responsible synthesis pathway is not present in these strains. Considering this information and using a comparative genomics approach, the according genes were identified (Schirmer et al., 2010). In cyanobacteria, alkanes are synthesized from intermediates of the fatty acid metabolism (Figure S1 in Supplementary Material) by two enzymes: acyl–acyl carrier protein reductase (AAR) and aldehyde deformylating oxygenase (ADO) (Schirmer et al., 2010; Li et al., 2011, 2012; Zhang et al., 2013). These enzymes are encoded by the two adjacent genes sll0208 (ado) and sll0209 (aar) in the model strain Synechocystis sp. PCC 6803 (from here: Synechocystis 6803). Interestingly, orthologs of these genes have been found so far only in cyanobacteria, suggesting the possible existence of a link to photoautotrophic life style i.e., oxygenic photosynthesis, but the functional relevance of cyanobacterial alkane biosynthesis in vivo has remained enigmatic thus far.
Cyanobacteria are morphologically very complex and are represented by unicellular (e.g., Synechocystis 6803, Prochlorococcus sp. MIT9313) as well as multicellular strains with differentiated cells (e.g., Anabaena sp. PCC 7120, Nodularia spumigena sp. CCY9414). Moreover, cyanobacteria are found in nearly all light-exposed habitats on earth including extreme environments such as deserts (Cameron, 1962; Garcia-Pichel et al., 2001), hot springs (Miller and Castenholz, 2000), hypersaline water (Reed et al., 1984) as well as Antarctic meltwater ponds (Nadeau and Castenholz, 2000), which underlines also their physiological diversity. Based on morphological properties five subsections were defined. However, due to the insufficient coverage of the cyanobacterial phylum by full genome sequences, truly comprehensive genome analyses have remained impossible for a long time. Recently, 54 additional genomes have been published (Shih et al., 2013), overcoming the lack of genomic data and enabling a comprehensive view on the occurrence and organization of genes important for a particular environmental situation.
For many cyanobacterial strains the expression of ado and aar orthologs is evident since long-to-medium-chain alkanes were detected (Winters et al., 1969; Schirmer et al., 2010). Though, their physiological function as well as the regulation of expression remains elusive. In Synechocystis 6803, alkane synthesis could be abolished by deletion of ado/aar and thus appears not essential, at least under standard growth conditions (Schirmer et al., 2010). To make an impact on cell physiology, genes need to be expressed. Furthermore, if their function is only needed under certain conditions, their expression should be regulated. The presence of upstream genetic regulatory elements can to a great extent serve as evidence for the functional significance of a gene. Thus, a detailed expression analysis might point to certain environmental conditions under which alkane synthesis is physiologically more relevant or even essential. Global transcriptomic analyses using microarrays are powerful approaches to investigate gene regulation and comparative data for manifold environmental conditions are available also for cyanobacteria (Hernandez-Prieto and Futschik, 2012). Moreover, RNA-seq, especially the differential RNA sequencing approach [dRNAseq, (Sharma et al., 2010)] is often used for the analysis of the primary transcriptome, which provides insight into gene expression changes together with detailed information about transcriptional start sites (TSS) and all promoters active at a given moment. By now, the primary transcriptomes of several cyanobacteria are available, supporting the analysis of the transcriptional organization of alkane synthesis (Mitschke et al., 2011a,b; Voß et al., 2013; Voigt et al., 2014; Pfreundt et al., submitted).
In this work, we present a comprehensive analysis of the genomic arrangement of genes encoding ADO and AAR throughout the cyanobacterial phylum. Moreover, we investigated the transcriptional organization of these genes for Synechocystis 6803 but also for Anabaena PCC 7120, N. spumigena CCY9414, Prochlorococcus MIT9313 & MED4, and Trichodesmium erythraeum IMS101. Although a dicistronic or polycistronic arrangement appears conserved at the genomic level among almost all cyanobacteria, we found solid evidence for the independent transcription of ado and aar in all tested strains. Since these strains are rather distantly related, the data indicate that an independent transcription of both genes might be common also for most other cyanobacteria. Additionally, for Synechocystis 6803 we compiled available expression data extracted from 115 microarray datasets, which comprise more than 25 environmental stimuli but reveal only modest conditional changes in gene expression.
Materials and Methods
Cluster Analysis
By using the JGI database and blastP algorithm (threshold E-value = 1e−5), 181 cyanobacterial genomes were screened for orthologs of ADO and acyl-ACP reductase genes. The full list of all genomes included in the study is shown in Table S1 in Supplementary Material. The corresponding protein sequences from Synechocystis 6803 were used as reference. For acetyl-CoA carboxylase, short-chain dehydrogenase, and GTP cyclohydrolase I, the respective sequences from Anabaena sp. PCC 7120 were used. The phylogenetic tree was generated with MEGA V6.0 (Tamura et al., 2013) by using the neighbor joining algorithm based on 16S rRNA sequences that were extracted from the SILVA database (Quast et al., 2013).
Strains and Growth Conditions
The following strains were used: Synechocystis 6803, substrain “PCC-M” (Trautmann et al., 2012), T. erythraeum IMS101 (obtained from Ilana Berman-Frank, Bar-Ilan University, Tel-Aviv, Israel; originally isolated by Prufert-Bebout et al., 1993) and Prochlorococcus MIT9313 & MED4 (courtesy of Sallie W. Chisholm, Massachusetts Institute of Technology, Cambridge, USA). Synechocystis was grown in TES-buffered (20 mM, pH 8.0) BG11 medium (Rippka et al., 1979) at 30°C under continuous white light illumination of 50–80 μmol quanta m−2 s−1 and gentle agitation. To determine the stability of transcripts, cultures were additionally aerated with ambient air through a glass tube and a sterile filter for constant and fast growth. Rifampicin was added as an inhibitor of transcription at a final concentration of 300 μg/ml. Samples were taken before and 3, 5, 10, 15 min after the treatment. T. erythraeum cultures were grown in YBCII medium (Chen et al., 1996) at 25°C and 12/12 h light/dark cycles at ~80 μmol photons m−2 s−1 white light. Prochlorococcus cells were grown at 22°C in AMP1 medium (Moore et al., 2007) under 10–30 μmol quanta m−2 s−1 continuous white cool light. For DNA cloning the E. coli strains Top10F′ and DH5α were used and cultivated in LB medium at 37°C.
RNA Extraction, Northern Blots, and Mapping of RNA 5′ and 3′ Ends
Cyanobacterial cells were harvested in exponential growth phase by vacuum filtration on hydrophilic polyethersulfone filters (Pall Supor-800, 0.8 μm or Supor-450, 0.45 μm), immediately immersed in 1 ml of PGTX solution (Pinto et al., 2009) and frozen in liquid nitrogen. Total RNA was extracted as described (Hein et al., 2013); T. erythraeum and Prochlorococcus cells were subjected to bead beating (bead diameter 0.1–0.5 mm) for 3 × 20 s at 6500 rpm (Precellys, Peqlab, Germany) immediately prior to extraction. For northern blot analysis, 3 (Synechocystis) to 5 μg (T. erythraeum and Prochlorococcus) of total RNA were separated on 1.5% agarose gels, transferred to Hybond-N+ nylon membranes (GE Healthcare) by capillary blotting and cross-linked by UV-illumination (125 mJ). Generation of single-stranded radioactively labeled RNA probes and hybridization with the blotted RNA were performed as described before (Steglich et al., 2008). The sequences of oligonucleotides used to amplify the respective probe templates are shown in Table S6 in Supplementary Material. Signals were visualized by using a Personal Molecular Imager FX system and Quantity One software (Bio-Rad). The half-life of transcripts was calculated after densitometric quantification of the corresponding signals. Mapping of RNA 5′ and 3′ ends was performed by rapid amplification of cDNA ends as described (Argaman et al., 2001). To define precise lengths, RNA molecules were self-ligated, reverse transcribed using gene-specific primers, and amplified by circular PCR as described (Vogel and Hess, 2001). All used RNA and DNA oligonucleotides are listed in Table S6 in Supplementary Material. Reverse transcription was performed at 42°C for 2 h using the Omniscript™ RT system (Qiagen). Prior to sequencing the PCR products were cloned into the pGEM®-T vector (Promega).
Generation of Reporter Strains
The putative promoter elements were fused to a reporter gene by PCR amplification (for oligonucleotides see Table S6 in Supplementary Material), followed by restriction digest with FseI/AgeI and cloning into the promoter probe vector pILA (Kunert et al., 2000). The pILA plasmid contains a promoterless luxAB operon encoding subunits of the luciferase enzyme and sequences for the homologous recombination of the entire promoter probe cassette into the Synechocystis 6803 gene slr0168 on the chromosome. The vector used in this work was modified prior to promoter insertion by introducing the restriction recognition sites FseI and AgeI upstream of the luxAB genes. The sites were introduced by PCR amplification of the original pILA plasmid with the primers “Plux AgeI fw”/“Plux-bla AgeI rev” and with the primers “Plux-bla FseI fw”/“Plux FseI rev”, resulting in two blunt ended products, which were subsequently ligated. The plasmid derivatives were used for transformation of a Synechocystis 6803 strain carrying the luxCDE operon in turn encoding enzymes for the synthesis of decanal, the substrate for the luciferase reaction. The expression cassette harboring the genes luxCDE under control of the strong promoter of the ncRNA Yfr2a (Voß et al., 2009) and a cat gene mediating resistance to chloramphenicol, was integrated into the intergenic region of sll1691 and slr1819 (both hypothetical proteins on the chromosome), which is regarded as a neutral site. Transformation was performed as described (Kunert et al., 2000). Genetically modified cells were initially selected on agar-solidified BG11 medium (0.9% KobeI agar, Roth, Germany) containing 10 μg ml−1 kanamycin (Km, selection of luxAB constructs) and 2 μg ml−1 chloramphenicol (Cm, marker for luxCDE cassette), but the segregation of clones and subsequent cultivation of mutants was performed in presence of 50 μg ml−1 Km and 10 μg ml−1 Cm.
Luciferase Assays
Bioluminescence was measured in vivo as total light counts per second by using a VICTOR3 multiplate reader (PerkinElmer). The cells were grown in the presence of 10 mM glucose. Prior to the measurement, cells were diluted to an OD750 = 0.4 and 200 μl of the suspension were filled into a white 96-well plate (CulturePlate™-96, PerkinElmer). In the multiplate reader the cell suspensions were shaken for 10 s and subsequently total light emission was measured for 1 s. A strain carrying the promotorless luxAB genes served as a negative control.
Results
Occurrence and Genomic Organization of Alkane Synthesis Genes in Cyanobacteria
Today, many cyanobacterial genomes are available in the databases enabling the possibility of comparing genomes with regard to physiological properties such as the production of alkanes. In this study, 181 genomes (Table S1 in Supplementary Material) were screened for orthologs of AAR and ADO (Schirmer et al., 2010; Li et al., 2011, 2012; Zhang et al., 2013). By using the blastP algorithm at an E-value cut off 1e−5, orthologs of both enzymes could be identified in 90% (163/181) of the genomes (Tables S2 and S3 in Supplementary Material), which underlines their importance. Notably, their functional connection is highlighted by the fact that we found not a single genome in which one of the two genes, aar and ado, would have been retained in the absence of the other. Moreover, the genomes were also analyzed for possible synteny at the locus encoding ADO. The different types of arrangements are shown in Figure 1. Interestingly, in 76% (138/181) of the genomes tested, the genes encoding AAR and ADO were found adjacent to each other, indicating a possible operon-like organization. Moreover, in 61% (111/181) of the strains both genes clustered together with additional genes encoding the alpha subunit of acetyl-CoA carboxyl transferase (EC 6.4.1.2) and a short-chain dehydrogenase, strengthening the functional connection to fatty acid metabolism. In most genomes, additionally a gene encoding a GTP cyclohydrolase I (EC 3.5.4.16), which is involved in folate biosynthesis, was found downstream of the other four. However, in a few strains aar and ado are located at different loci as single genes (Figure 1). Additionally, in 18 of the tested genomes AAR and ADO genes were not found (for the full list see Table S4 in Supplementary Material), including the previously studied Synechococcus sp. PCC 7002 and Cyanothece sp. PCC 7424 and in consent with those reports (Schirmer et al., 2010).
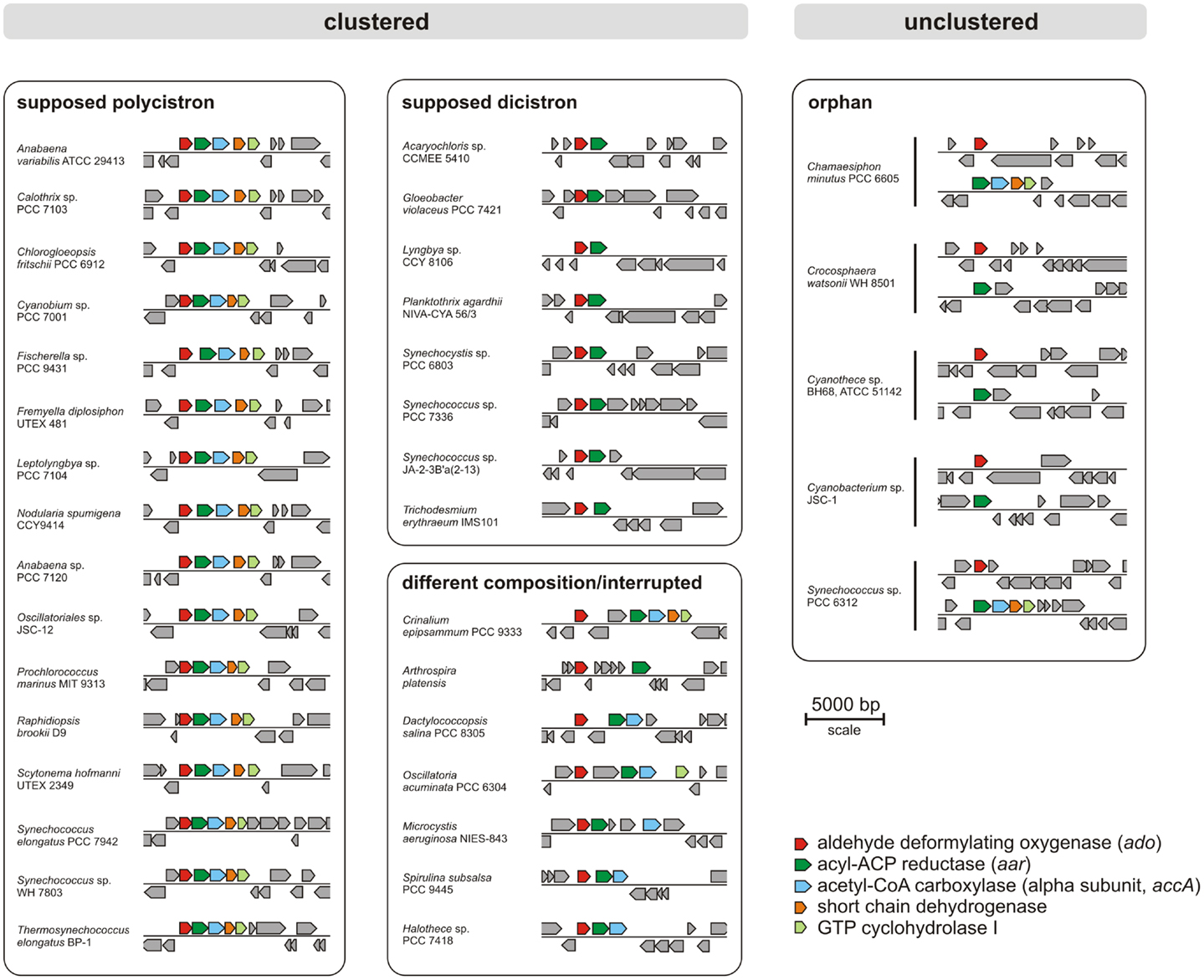
Figure 1. Examples for genomic arrangements of aldehyde deformylating oxygenase orthologs in cyanobacteria. By using JGI database and blastP algorithm (threshold E-value = 1e−5) 181 cyanobacterial genomes were screened for the presence of alkane biosynthesis genes. The corresponding protein sequences from Synechocystis sp. PCC 6803 were used as reference.
Transcriptional Organization of Alkane Synthesis Genes in Synechocystis 6803
Data so far available suggest that ado and aar orthologs form an operon, but a genome-wide mapping of TSS in Synechocystis 6803 by using differential RNA sequencing (dRNAseq) revealed that both genes possess their own, specific, TSS (Mitschke et al., 2011a). Thus, they do not seem to be part of an operon since transcription is driven by independent promoters (Figure 2A). This unexpected transcriptional organization was further substantiated by northern blots (Figure 2B). Hybridization of an RNA probe specific for the ado (sll0208) ORF, which has a length of 696 bp, yielded a signal of about 900–1000 nt. This size is too short for a dicistronic mRNA, for which a minimal length of >1700 nt would be expected. Moreover, with an aar-specific probe, again, a transcript too short for a dicistron was detected but which approximately matches with the length of the aar (sll0209) open reading frame (1023 bp). Thus, these hybridization signals are consistent with the generation of independent monocistronic mRNAs from two independent promoters. Interestingly, our data uncovered not only two independent TSS for ado and aar but also a third TSS, which might belong to a putative non-coding RNA, called SyR9, upstream of the two genes (Mitschke et al., 2011a). This finding is supported by the identification of suitable −10 elements within the putative promoter sequences associated with these three TSS (Figure 2C). Hybridization using a SyR9-specific RNA probe yielded two signals – the lower band corresponds to the small transcript of SyR9, the upper band, however, was of approximately the same size as found for the ado transcript (Figure 2B). In this case, the upper signal could represent a combined RNA that included SyR9 and the ado mRNA. To test this possibility, PCR was carried out on cDNA samples prepared from total RNA circularized by RNA ligase and using primers in outbound orientation. One primer was located within the SyR9 segment and the other at the end of the coding region of ado. The obtained fragments were sequenced and revealed the 5′ end mapped for SyR9 to be connected to a sequence finishing 75 nt behind the last nucleotide of the ado stop codon [at genomic pos. 2511439 (complement)]. Thus, this analysis confirmed the SyR9-ado cotranscript and yielded its precise length of 985 nt, consistent with the sizes estimated by northern analysis. We conclude that in Synechocystis expression of ado is driven by two promoters, P1 and P2, and that the smaller accumulating transcript SyR9 results from processing of the SyR9–mRNA cotranscript or alternative termination of transcription.
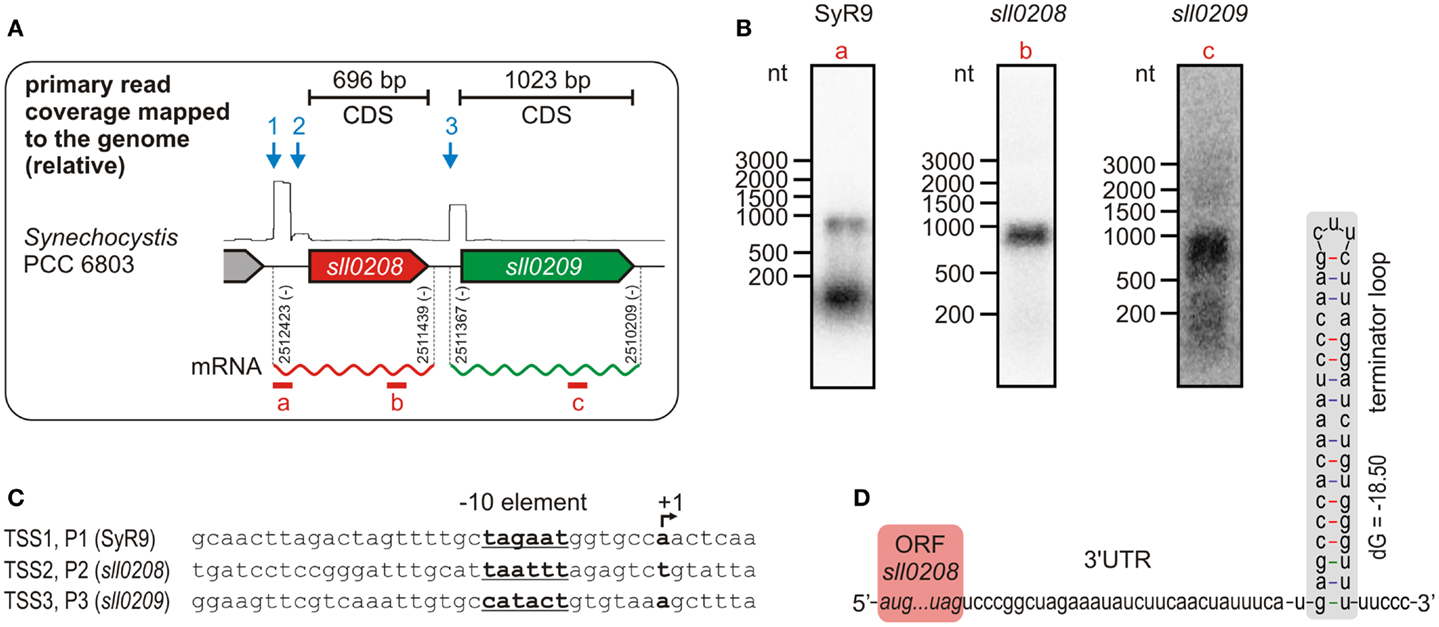
Figure 2. Transcriptional organization of the gene cluster encoding acyl-ACP reductase (aar, sll0209) and aldehyde deformylating oxygenase (ado, sll0208) in Synechocystis sp. PCC 6803. (A) Differential RNA sequencing (dRNAseq, data shown as primary read coverage mapped to the corresponding genomic region) of cells grown under standard conditions revealed independent transcriptional start sites (TSS) for both genes indicating for monocistronic mRNAs. The rectangular shape within the graph results from the enrichment of primary 5′-PPP carrying transcripts representing a TSS and the read length (101 nt) of the sequence analysis. Data were extracted from Kopf et al., 2014. Each TSS is represented by an arrow. The experimentally confirmed 5′ and 3′ ends of the main mRNAs are given by the respective genomic positions. (B) Northern verification for the monocistronic mRNAs and the small transcript SyR9, which accumulates independently from the sll0208 mRNA in Synechocystis sp. PCC 6803. The part covered by the labeled probe is shown by a red bar in (A). Consistent with the TSS mapping all transcripts detected appeared too short for a dicistronic mRNA. (C) Putative promoter elements upstream of the mapped TSS which is designated as +1. (D) The mapped 3′ end of the sll0208 mRNA can be folded into a stem loop secondary structure that is typical for a Rho-independent terminator of transcription.
The same approach was taken to determine the precise 5′ and 3′ ends of the aar mRNA. Whereas the 5′ end 72 nt upstream of the start codon resulting from initiation of transcription at promoter P3 was confirmed, its 3′ end was mapped 64 nt downstream of the stop codon [at genomic pos. 2510209 (complement)]. Thus, the total length of the aar mRNA is 1159 nt, consistent with the major signal obtained in the northern hybridization (Figure 2B) and the accumulation of this mRNA as a monocistronic transcript species. This finding receives further strong support from the identification of a putative Rho-independent terminator mapped here at the 3′ end of the ado mRNA (Figure 2D). We conclude that the ado and aar orthologs in Synechocystis 6803 are not transcriptionally organized as an operon. These results clearly impact approaches to manipulate the expression of these genes, as the activity of a strong promoter upstream of a ado–aar two-gene-cassette will lead to a high expression of the first, but not the second gene.
Transcriptional Organization of Alkane Synthesis Genes in Other Cyanobacteria
Synechocystis 6803 has been established as a representative model strain. Nevertheless, cyanobacteria constitute a physiologically and genomically very diverse taxon, hence the independent transcription of ado and aar found in Synechocystis might be different in other cyanobacteria. To test this possibility, we checked the primary transcriptomes that are available for several additional cyanobacteria (Mitschke et al., 2011b; Voß et al., 2013; Voigt et al., 2014; Pfreundt et al., submitted). Interestingly, independent TSS for the two genes exist also in Anabaena PCC 7120, N. spumigena CCY9414, Prochlorococcus MIT9313 & MED4, and T. erythraeum IMS101 (Figure 3A). The respective transcripts were exemplarily verified for three of these strains. In all cases, the main accumulating RNAs that derive from the TSS upstream of ado were shorter than the minimum length of a dicistronic transcript (Figures 3B–D). Moreover, the signal patterns that were observed after hybridization with aar-specific probes were different from those for the ado transcripts. These findings indicate that both genes might be transcriptionally independent from each other also in most other cyanobacteria, which is contrary to their seemingly dicistronic/polycistronic arrangement that appears largely conserved at the genomic level. Interestingly, similar to the arrangement in Synechocystis 6803, a second TSS upstream of ado is active in Trichodesmium, Nodularia, and Prochlorococcus MIT9313, which suggests that ado might be transcribed from two independent promoters also in several other species. For Trichodesmium, this arrangement was verified further by using a labeled probe specific for the sequence around the distal TSS (probe a, locus shown in Figure 3A). A transcript of ~1600 nt was detected, which demonstrates the existence of a long transcript that encompasses also the coding region of the ado gene (Figure 3B). Additionally, the two bands that were obtained when using a probe specific for a sequence downstream of the proximal ado-TSS (probe b) demonstrated the accumulation of two different mRNAs originating from the two TSS (indicated by black arrows).
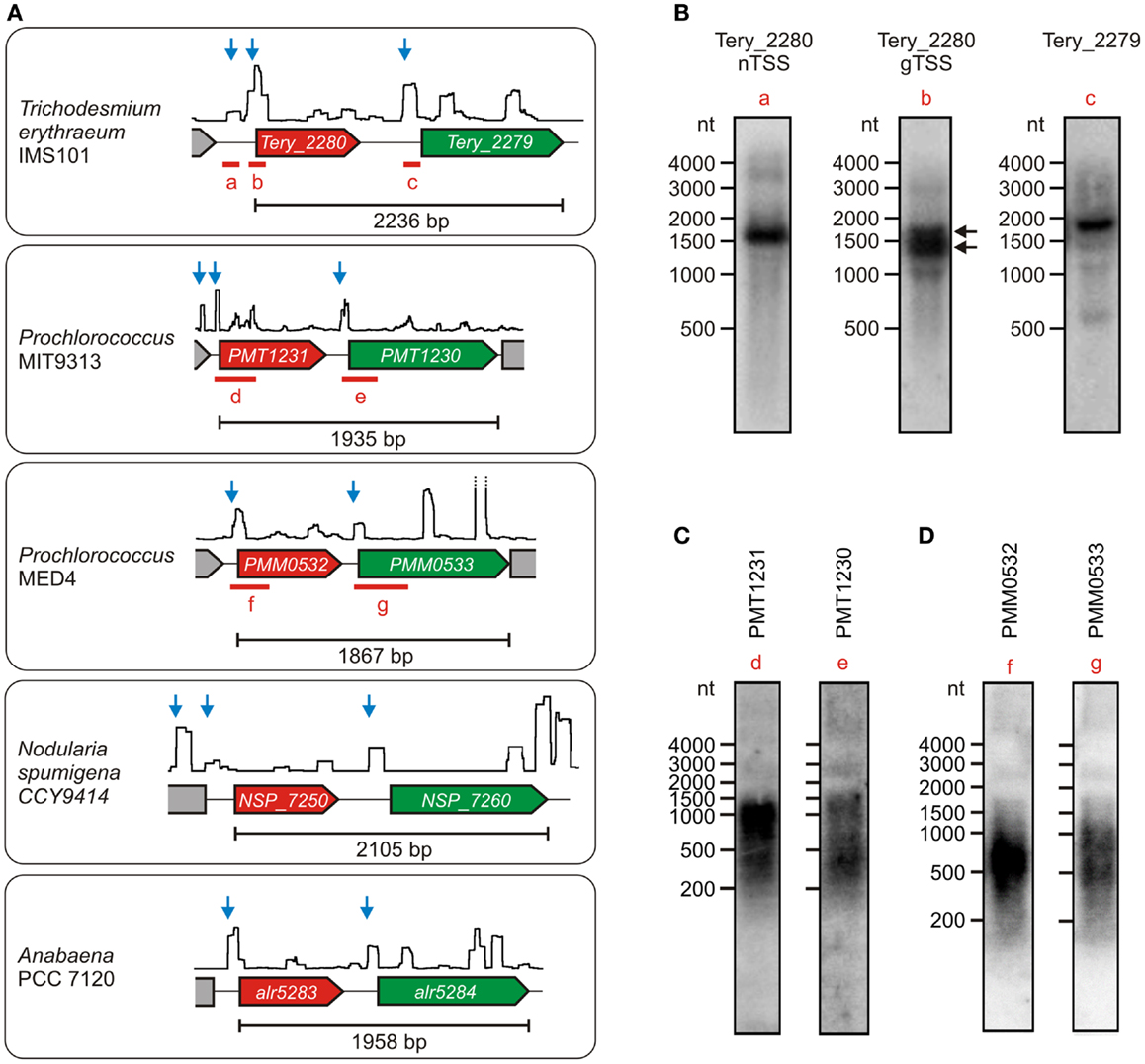
Figure 3. Transcriptional organization of the gene cluster encoding ado (red) and aar (green) in various cyanobacteria under the respective standard conditions. (A) The data are presented analogous to Figure 2. Each TSS potentially belonging to the respective aar/ado mRNA is represented by a blue arrow. Data were extracted from Mitschke et al. (2011b), Voß et al. (2013), Voigt et al. (2014), (Pfreundt et al., submitted). In each case, the minimum length of the dicistronic arrangement is given for the sequence between the start codon of ado and the stop codon of aar. (B) Northern verification of ado (Tery_2280) and aar (Tery_2279) mRNAs in Trichodesmium erythraeum IMS101. The black arrows indicate the two mRNAs originating from the independent TSS upstream of Tery_2280. (C,D) Respective northern blots for both mRNAs in Prochlorococcus sp. MIT9313 and MED4. The corresponding probe loci are given in (A).
Experimental Verification of the Mapped ado and aar Promoters in Synechocystis 6803
In order to experimentally verify the independent TSS for ado and aar, 5′ RACE experiments were performed with RNA extracted from Synechocystis 6803. All three start sites derived from the global mapping of TSS (Mitschke et al., 2011a), could be confirmed [TSS1 at pos. 2512423, TSS2 at pos. 2512315, TSS3 at pos. 2511367; positions are given for the complementary (fwd) strand]. Sequences upstream of the TSS contain putative −10 elements and were assumed as true promoters and therefore designated as P1 (distal ado promoter, SyR9 promoter), P2 (proximal ado promoter), and P3 (aar promoter, see Figure 2). In order to verify the suggested promoter activities, the sequences upstream and around the corresponding TSS were fused to luxAB luciferase genes. With respect to the first transcribed nucleotide (+1) we considered sequence ranges from −116 to +2 for P1, −108 to +106 nt for P2, and −401 to +71 for P3, respectively. Bioluminescence measurements revealed that these sequences indeed contain elements also driving transcription of the reporter gene (Figure 4). P1 and P2 showed about one third of the strength measured for the well-characterized promoter of the petE gene, which is highly active in the presence of copper ions and has been used for heterologous expression in Anabaena as well as Synechocystis (Buikema and Haselkorn, 2001; Tan et al., 2011). The data verified that the expression of ado and aar is regulated independently and that two promoters are found upstream of ado. Moreover, luminescence measured for P2 was close to background levels indicating that luxAB transcription initiated by this promoter is rather weak, consistent with the much lower read number observed for TSS2 in the dRNAseq analysis (Figure 2A).
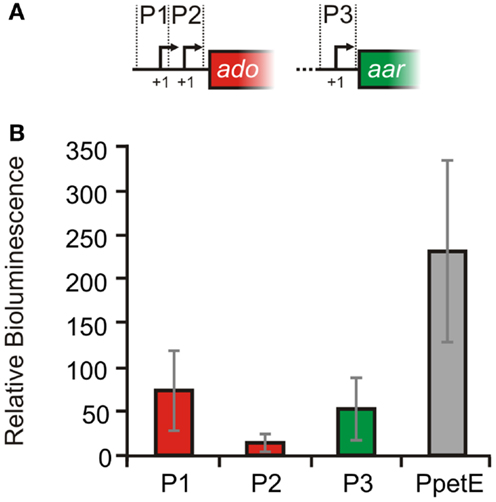
Figure 4. Verification of promoter activities in Synechocystis sp. PCC 6803. (A) Schematic illustration of the putative promoter regions. For the reporter assays luxAB genes were fused with the upstream sequences of either: SyR9 ( =P1, −116 to +2), ado ( =P2, −108 to +106 nt) or aar ( =P3, −401 to +71 nt), and also petE ( =PpetE, −273 to +81, positive control) while +1 represents the first transcribed nucleotide. (B) Bioluminescence was measured in vivo as total light counts per second. A strain harboring a promoterless luxAB operon was used as negative control. The cultures were grown in standard BG11. For the PpetE activity measurements, however, 2 μM CuSO4 was added. Relative bioluminescence was calculated by subtraction of the values obtained from negative control cultures. Data are the mean ± SD of 60 single measurements obtained with two biological replicates ( =independent transformants) in several independent experiments.
Expression of Alkane Synthesis Genes in Synechocystis 6803
The described data suggest the autonomous regulation and possibly independent function of ado and aar genes under certain growth conditions. Thus, a detailed expression analysis could reveal if both are differentially expressed or even contrary regulated. Moreover, it might imply a functional involvement of the genes and possibly alkanes in adapting cyanobacterial physiology according to environmental changes since the function of alkanes in cyanobacteria is still ambiguous. To investigate if ado and aar are differentially expressed we compiled data extracted from CyanoEXpress, a database for microarray experiments performed with samples from Synechocystis (Hernandez-Prieto and Futschik, 2012). Interestingly, there are many conditions for which contrary fold changes for both genes were observed, further verifying their independent transcription. For the full list of fold changes see Table S5 in Supplementary Material. However, for the environmental conditions that are included in the CyanoEXpress database no clear and convincing stimulation of expression was evident. For ado the top log2 fold change (1.52) was observed after 4 h of zinc excess, whereas for aar it was 12 h of iron depletion (log2 fold change = 1.08). Nevertheless, diminished mRNA levels were observed for both, ado and aar, when cells were subjected to oxidative stress by H2O2 treatment. Furthermore, expression of both genes appear dependent on DNA gyrase since a novobiocin treatment in combination with heat stress, low temperature, or salt stress led to significantly reduced mRNA abundances (Table 1).
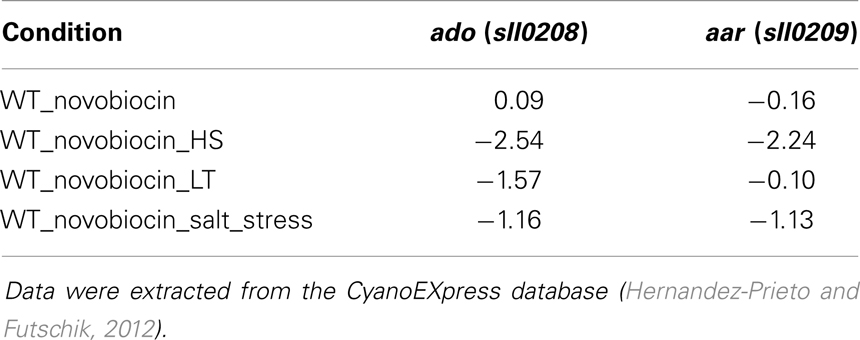
Table 1. Log2 fold changes for ado and aar in Synechocystis 6803 after treatment with novobiocin in addition to heat stress, low temperature, and salt stress.
The actual concentration of an mRNA is the result of two processes, transcription and turnover. Therefore, the stability of an mRNA is of similar importance for gene expression as is the control of transcription. To measure the relative transcript stability, we added the inhibitor of transcription initiation, rifampicin, to our cultures and then followed the disappearance of SyR9 and the mRNAs for ado and aar in a time course experiment (Figure 5). The calculated half-lives of all these transcripts were with <3 min quite short. We observed a transcript stability decreasing from SyR9 with a half-life of 2.2 min over 1.8 min for ado (sll0208), to only 1.6 min for aar (sll0209) at the end of this gene cluster in Synechocystis 6803.
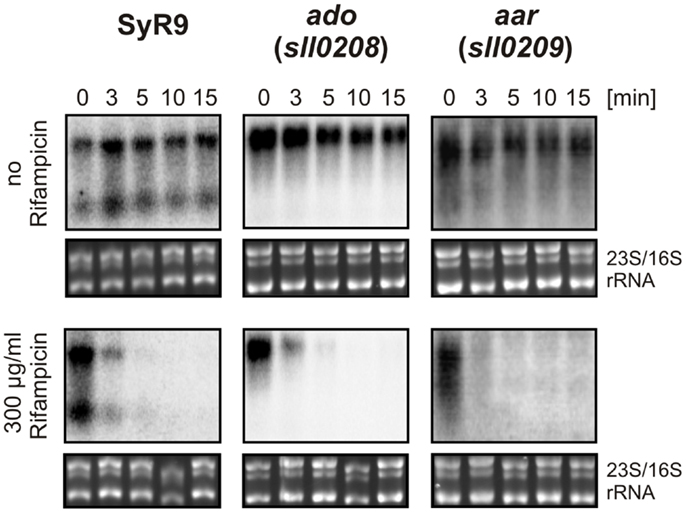
Figure 5. RNA stability measurements. Half-lives of the indicated transcripts were determined in the presence of rifampicin in cultures for the times indicated. For comparison, samples were taken at the identical time points but in the absence of rifampicin. For the control of equal loading, the large rRNA bands stained with ethidium bromide are shown underneath each hybridization.
Discussion
Since the discovery of the cyanobacterial alkane biosynthesis pathway, the major scientific focus has been on the enzymatic properties of ADO and AAR, as well as their potential for biofuel production (Schirmer et al., 2010; Li et al., 2011, 2012; Zhang et al., 2013). Less attention was paid to the expression of the corresponding genes as well as their regulation and possible function in the host organisms. Obviously, in cyanobacteria no other pathways (at least for medium-chain alkanes) exist since a knockout of ado and aar genes abolished heptadecane synthesis in Synechocystis 6803 and no alkanes were detected in Synechococcus sp. PCC 7002, a strain that lacks these two genes naturally (Schirmer et al., 2010). Due to the wealth of new genomic information that has become available within the last 2 years, comparative analyses linking phylogeny and the presence of alkane biosynthesis genes has become straightforward and is yielding new insight. Consistently, orthologs of ADO and AAR were found in all strains for which alkane detection has been reported earlier (see Figure 6). Additionally, alkanes were also detected in many other cyanobacterial isolates for which no genome sequence is available, including the genera Oscillatoria, Microcoleus, Lyngbya, Nostoc, Plectonema, Chlorogloeopsis, and Phormidium (Han et al., 1968; Winters et al., 1969), indicating that ado and aar genes are also present in those species. Surprisingly, orthologs of ADO and AAR are not present in other prokaryotes, associating this pathway to cyanobacteria or certain aspects of their phototrophic life style.
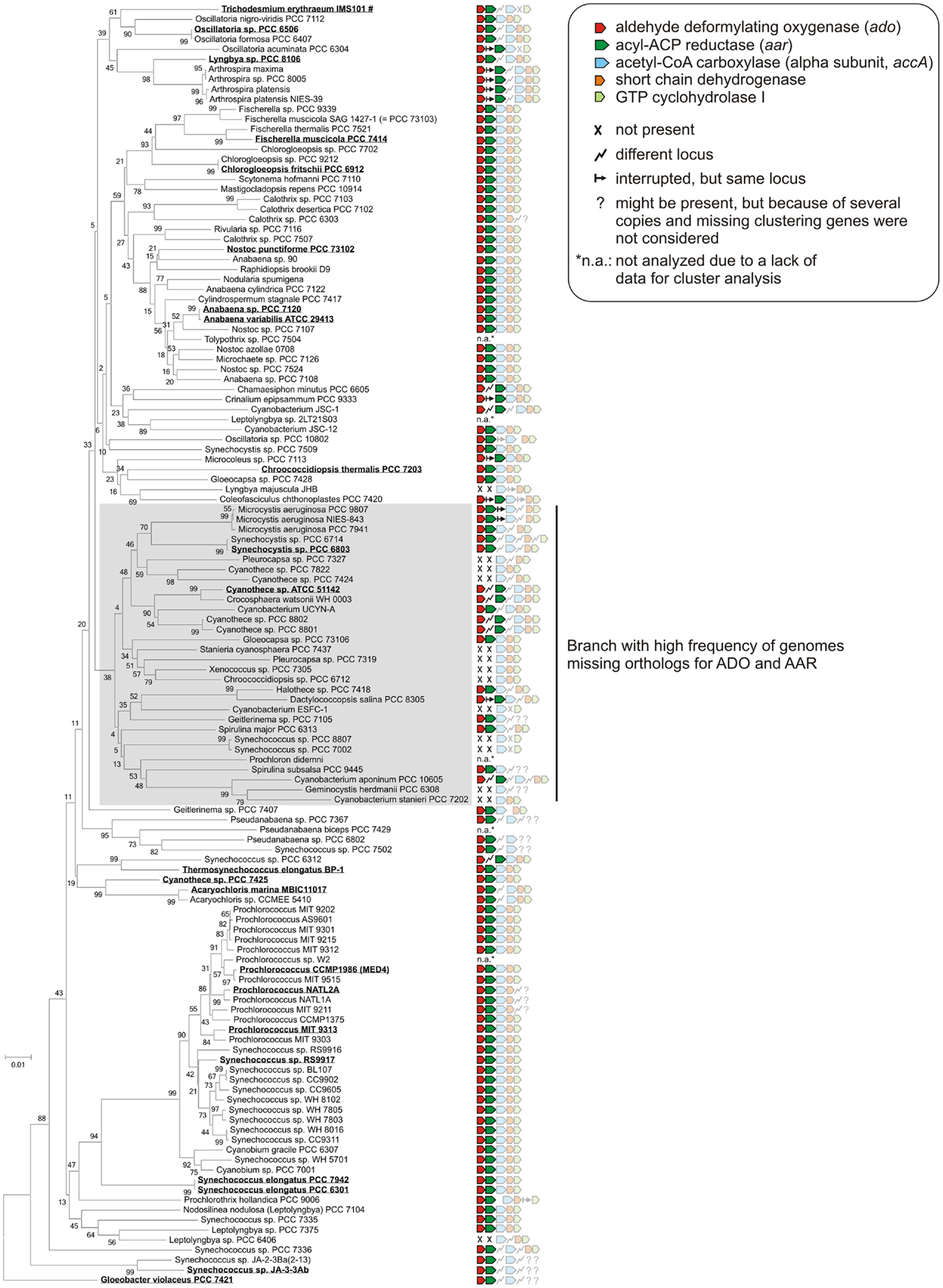
Figure 6. Clustering of alkane biosynthesis genes among the cyanobacterial phylum. The phylogenetic tree was generated by using the neighbor joining algorithm based on 16S rRNA sequences that were extracted from the SILVA database (Quast et al., 2013). Orthologs of aldehyde deformylating oxygenase and acyl-ACP reductase were searched by using blastP (threshold 1e−5) and the protein sequences from Synechocystis sp. PCC 6803 as reference. For the other three genes encoding acetyl-CoA carboxylase, short-chain dehydrogenase, GTP cyclohydrolase I the sequences from Anabaena sp. PCC 7120 were used. Strains for which alkane biosynthesis has been reported (Schirmer et al., 2010; Coates et al., 2014) are highlighted and underlined. Alkanes were measured either directly in cyanobacterial cell extracts (mainly penta- or hepta-decane) or indirectly by alkane detection in recombinant E. coli strains expressing the corresponding ado orthologs in combination with the aar gene from S. elongatus PCC7942. #Alkanes were detected in a natural surface bloom of Trichodesmium erythraeum near Port Aransas in the Gulf of Mexico (Winters et al., 1969).
Multiple Promoters Associated with the Alkane Biosynthesis Genes in Cyanobacteria
The clustering of ado and aar genes is widely conserved throughout the cyanobacterial phylum and frequently three additional genes are found associated with them (Figure 6). Despite their close functional connection and co-appearance in most genomes, our data demonstrate that in all tested strains the ado and aar genes are transcribed from separate, specific promoters. Thus, these two genes can be expressed independently from each other, in turn enabling their different or even divergent regulation. Due to the presence of separate regulatory elements, the dominance of monocistronic mRNAs and additionally the fact that the two genes are split in some species we speculate that physiological situations might exist for which an independent transcription of ado and aar could be advantageous. Additionally, in some strains ado is transcribed from two separate TSS, which increases the transcriptional complexity further. Multiple TSS may indicate different functions. In Synechocystis, for instance, two separate TSS were also reported for the petH gene that encodes two isoforms of ferredoxin:NADP oxidoreductase, while the decision between both isoforms is triggered by the differential use of these two TSS (Omairi-Nasser et al., 2011). Under standard conditions a proximal promoter is active, resulting in a short 5′UTR whereas under nitrogen depletion a more distally located promoter is active that mediates the transcription of petH with a long 5′UTR. Under both conditions only one isoform – a long or a short – is dominant. The different translation initiation sites are dictated by alternative secondary structures only depending on the length of the 5′UTR (Omairi-Nasser et al., 2011). In principle, alternative translation initiation is also imaginable for the ado gene. For the transcription of ado under standard growth conditions the distal promoter P1 appeared as the dominant regulatory element but situations might exist when the main expression is mediated by P2. Two mRNA species hence offer the possibility of alternative RNA foldings, which could then affect translation initiation or the yield of translated protein.
A highly interesting aspect of the observation that ado and aar genes are transcribed from separate and distinct promoters in cyanobacteria are the possible metabolic implications. Their monocistronic layout found in this work allows the regulatory autonomy of these two genes. Thus, they may signify also other, unknown, pathways branching off from aldehyde synthesis that would require their separate and non-stoichiometric expression. Such pathways might, e.g., lead to free fatty acid (from aldehyde oxidation) or even fatty alcohols (from aldehyde reduction) for which separate regulation of these two genes would be required. So, it is quite possible that still other unknown aldehyde-derived pathways remain to be discovered in cyanobacteria.
Genomic Organization of Alkane Synthesis Genes in Cyanobacteria
The initial step of alkane synthesis produces a fatty aldehyde by the conversion of a fatty acyl-ACP thus connecting the pathway to fatty acid metabolism. Interestingly, in many cyanobacteria ado and aar are found adjacent to accA encoding the alpha subunit of acetyl-CoA carboxylase (ACCase, EC 6.4.1.2). ACCase catalyzes the synthesis of malonyl-CoA, which is the first and rate-limiting step in fatty acid synthesis (Figure S1 in Supplementary Material). Accordingly, overexpression of ACCase in Synechocystis leads to an enhancement of hydrocarbon production especially heptadecane (Tan et al., 2011). Moreover, in Anabaena 7120 and also most other strains the gene downstream of accA encodes a protein that possibly belongs to the short-chain dehydrogenase/reductase (SDR) family of NAD- or NADP-dependent oxidoreductases (Joernvall et al., 1995). Interestingly, it shows high similarity to the 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100) that also participates in fatty acid biosynthesis. However, despite the clustering and the functional relation, the accA gene possesses its own TSS in several strains, similar to the transcriptional separation of ado and aar (see Figure 3A, accumulating primary transcripts at the 3′ end of the aar gene), indicating a possibly independent regulation. The functional connection, if any, to the fifth and last gene in this cluster (Figure 6), encoding GTP cyclohydrolase I (EC 3.5.4.16), is less obvious. This enzyme is involved in folate biosynthesis from GTP and during the catalysis formate is generated. There is one parallel as during the conversion of the fatty aldehyde to the final alkane catalyzed by ADO, also formate is generated (Li et al., 2011), but if that has any meaning is currently unknown.
Regulation of Alkane Synthesis Gene Expression
It is interesting to note the short half-lives of <3 min under our standard growth conditions, which we observed for all three major transcripts originating from the syR9/ado/aar locus. Consequently, slight changes in transcript stability could impact the mRNA accumulation and therefore the expression of these genes. Theoretically, a differential expression of ado and aar might point to conditions under which alkanes are physiologically more relevant. However, examining the wealth of expression data available for Synechocystis we found no clear evidence for differences in the abundance of ado or aar mRNA that could be linked to differences in the growth conditions. Expression may well be regulated in a multi-factorial fashion and not exclusively dependent on a single stimuli. Alternatively, the synthesis rate of alkanes could also be regulated by other factors than at the level of transcription or transcript stability, e.g., by modulating enzyme activities. Nevertheless, a regulatory cascade targeting ado and aar might exist but the stimuli remain ambiguous. Interestingly, most of the strains lacking AAR and ADO orthologs, and most probably alkanes, appear on one branch of the phylogenetic tree (Figure 6). Assuming a general importance of alkane synthesis, it is tempting to speculate that their physiological function in some strains of this clade might have been replaced or compensated by other mechanisms or compounds. Indeed, in these strains a likely compensatory polyketide synthase pathway which produces 1-alkenes is present (Coates et al., 2014).
Physiological Functions of Cyanobacterial Alkane Synthesis
Although the presence of ADO and AAR appears to be restricted to cyanobacteria, alkanes were also detected in other prokaryotes (Ladygina et al., 2006), where different types of enzymes must be responsible for their biosynthesis. Indeed, alkanes are widely distributed throughout nature, including plants (Cheesbrough and Kolattukudy, 1984; Bernard et al., 2012), green algae (Dennis and Kolattukudy, 1991), and higher animals (Cheesbrough and Kolattukudy, 1988), where they are mainly a component of surface waxes and function as water barrier or in insects where they serve as pheromones (Blomquist and Jackson, 1979; Howard and Blomquist, 2005). Therefore, the physiological function of alkanes is obviously not restricted to cyanobacteria and a more general role can be assumed also for other prokaryotes. Since fatty acid synthesis is mainly necessary for membrane generation, alkanes might also have an influence on membrane composition and fluidity under particular conditions. However, the functional relevance of alkanes under optimal growth conditions might be low since the pathway can be deleted without any obvious growth phenotype (Schirmer et al., 2010) and due to the fact that 10% of all cyanobacterial genomes lack the ADO–AAR pathway altogether.
Cyanobacteria are equipped with several mechanisms to deal with superfluous electrons generated from excess light energy absorbed. Therefore, it might appear as an attractive additional mechanism if newly synthesized alkanes would serve as electron sinks under certain conditions. In such a scenario, one would expect massive accumulation of alkanes under suitable conditions, e.g., high light stress. To the best of our knowledge this has not been observed in wildtype strains thus far, rendering this possibility highly speculative. For instance, the heptadecane content in various cyanobacteria typically ranges between 0.02 and 0.1% of cell dry weight (Coates et al., 2014). For Synechocystis 6803, amounts of ~200 μg/L (0.13% of cell dry weight) were reported (Wang et al., 2013), which is relatively low compared to dominant molecules such as chlorophyll a (~8 mg/L), glycogen (~200 mg/L), or the osmoprotectant glucosylglycerol [~100 mg/L, representative values for cells of Microcystis firma shocked with 770 mM NaCl, (Erdmann et al., 1992)]. However, for genetically engineered Synechocystis strains it was also shown that redirecting the carbon flux to acyl-ACP and overexpressing alkane biosynthetic genes simultaneously can significantly increase the yield of heptadecane to 26 mg/L (1.1% of cell dry weight; Wang et al., 2013). Indeed, these data indicate that alkane accumulation is basically possible.
Nevertheless, the complex transcriptional organization of the alkane synthesis gene cluster needs to be taken into account when manipulating the expression of these genes in situ. For example, the insertion of strong or controllable promoters upstream of the ado (sll0208) gene in Synechocystis 6803 is very likely to have an effect on the transcription of this but not of the downstream located aar gene due to the presence of the Rho-independent terminator of transcription in between.
Author Contributions
Wolfgang R. Hess designed the study. Stephan Klähn and Wolfgang R. Hess supervised the research. Stephan Klähn performed genomic analyses. Claudia Steglich and Desirée Baumgartner performed verification experiments for the promoter mapping. Desirée Baumgartner executed physiological experiments, Northern blots, and promoter analyses for Synechocystis. Ulrike Pfreundt and Karsten Voigt performed northern verification for Trichodesmium and Prochlorococcus, respectively. Verena Schön generated the decanal producing host strain for luciferase reporter assays in Synechocystis. Stephan Klähn and Wolfgang R. Hess evaluated and interpreted the data. Stephan Klähn, Desirée Baumgartner, and Wolfgang R. Hess wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7-ENERGY-2010-1 DirectFuel) under grant agreement no. 256808 and from the German Research Foundation with grant HE 2544 9-1.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fbioe.2014.00024/abstract
References
Argaman, L., Hershberg, R., Vogel, J., Bejerano, G., Wagner, E. G., Margalit, H., et al. (2001). Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11, 941–950. doi:10.1016/S0960-9822(01)00270-6
Atsumi, S., Higashide, W., and Liao, J. C. (2009). Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27, 1177–1180. doi:10.1038/nbt.1586
Bernard, A., Domergue, F., Pascal, S., Jetter, R., Renne, C., Faure, J.-D., et al. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24, 3106–3118. doi:10.1105/tpc.112.099796
Blomquist, G. J., and Jackson, L. L. (1979). Chemistry and biochemistry of insect waxes. Prog. Lipid Res. 17, 319–345. doi:10.1016/0079-6832(79)90011-9
Buikema, W. J., and Haselkorn, R. (2001). Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. U.S.A. 98, 2729–2734. doi:10.1073/pnas.051624898
Cameron, R. E. (1962). Species of Nostoc Vaucher occurring in the Sonoran desert in Arizona. Trans. Am. Microsc. Soc. 81, 379–384. doi:10.2307/3223790
Cheesbrough, T. M., and Kolattukudy, P. E. (1984). Alkane biosynthesis by decarbonylation of aldehydes catalyzed by a particulate preparation from Pisum sativum. Proc. Natl. Acad. Sci. U.S.A. 81, 6613–6617. doi:10.1073/pnas.81.21.6613
Cheesbrough, T. M., and Kolattukudy, P. E. (1988). Microsomal preparation from an animal tissue catalyzes release of carbon monoxide from a fatty aldehyde to generate an alkane. J. Biol. Chem. 263, 2738–2743.
Chen, Y.-B., Zehr, J. P., and Mellon, M. (1996). Growth and nitrogen fixation of the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. IMS 101 in defined media: evidence for a circadian rhythm. J. Phycol. 32, 916–923. doi:10.1111/j.0022-3646.1996.00916.x
Coates, R. C., Podell, S., Korobeynikov, A., Lapidus, A., Pevzner, P., Sherman, D. H., et al. (2014). Characterization of cyanobacterial hydrocarbon composition and distribution of biosynthetic pathways. PLoS ONE 9:e85140. doi:10.1371/journal.pone.0085140
Deng, M. D., and Coleman, J. R. (1999). Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 65, 523–528.
Dennis, M. W., and Kolattukudy, P. E. (1991). Alkane biosynthesis by decarbonylation of aldehyde catalyzed by a microsomal preparation from Botryococcus braunii. Arch. Biochem. Biophys. 287, 268–275. doi:10.1016/0003-9861(91)90478-2
Erdmann, N., Fulda, S., and Hagemann, M. (1992). Glucosylglycerol accumulation during salt acclimation of two unicellular cyanobacteria. J. Gen. Microbiol. 138, 363–368. doi:10.1099/00221287-138-2-363
Garcia-Pichel, F., Lopez-Cortes, A., and Nubel, U. (2001). Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl. Environ. Microbiol. 67, 1902–1910. doi:10.1128/AEM.67.4.1902-1910.2001
Georgianna, D. R., and Mayfield, S. P. (2012). Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488, 329–335. doi:10.1038/nature11479
Han, J., McCarthy, E. D., Calvin, M., and Benn, M. H. (1968). Hydrocarbon constituents of the blue-green algae Nostoc muscorum, Anacystis nidulans, Phormidium luridium and Chlorogloea fritschii. J. Chem. Soc. C 2785–2791. doi:10.1039/J39680002785
Hein, S., Scholz, I., Voß, B., and Hess, W. R. (2013). Adaptation and modification of three CRISPR loci in two closely related cyanobacteria. RNA Biol. 10, 852–864. doi:10.4161/rna.24160
Hernandez-Prieto, M. A., and Futschik, M. E. (2012). CyanoEXpress: a web database for exploration and visualisation of the integrated transcriptome of cyanobacterium Synechocystis sp. PCC6803. Bioinformation 8, 634–638. doi:10.6026/97320630008634
Howard, R. W., and Blomquist, G. J. (2005). Ecological, behavioral, and biochemical aspects of insect hydrocarbons. Annu. Rev. Entomol. 50, 371–393. doi:10.1146/annurev.ento.50.071803.130359
Joernvall, H., Persson, B., Krook, M., Atrian, S., Gonzalez-Duarte, R., Jeffery, J., et al. (1995). Short-chain dehydrogenases/reductases (SDR). Biochemistry 34, 6003–6013. doi:10.1021/bi00018a001
Kopf, M., Klähn, S., Scholz, I., Matthiessen, J. K. F., Hess, W. R., and Voss, B. (2014). Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. doi:10.1093/dnares/dsu018
Kunert, A., Hagemann, M., and Erdmann, N. (2000). Construction of promoter probe vectors for Synechocystis sp. PCC 6803 using the light-emitting reporter systems Gfp and LuxAB. J. Microbiol. Methods 41, 185–194. doi:10.1016/S0167-7012(00)00162-7
Ladygina, N., Dedyukhina, E. G., and Vainshtein, M. B. (2006). A review on microbial synthesis of hydrocarbons. Process Biochem. 41, 1001–1014. doi:10.1016/j.procbio.2005.12.007
Li, N., Chang, W.-C., Warui, D. M., Booker, S. J., Krebs, C., and Bollinger, J. M. Jr. (2012). Evidence for only oxygenative cleavage of aldehydes to alk(a/e)nes and formate by cyanobacterial aldehyde decarbonylases. Biochemistry 51, 7908–7916. doi:10.1021/bi300912n
Li, N., Nørgaard, H., Warui, D. M., Booker, S. J., Krebs, C., and Bollinger, J. M. Jr. (2011). Conversion of fatty aldehydes to alka(e)nes and formate by a cyanobacterial aldehyde decarbonylase: cryptic redox by an unusual dimetal oxygenase. J. Am. Chem. Soc. 133, 6158–6161. doi:10.1021/ja2013517
Lindberg, P., Park, S., and Melis, A. (2010). Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 12, 70–79. doi:10.1016/j.ymben.2009.10.001
McKinlay, J. B., and Harwood, C. S. (2010). Photobiological production of hydrogen gas as a biofuel. Curr. Opin. Biotechnol. 21, 244–251. doi:10.1016/j.copbio.2010.02.012
Miller, S. R., and Castenholz, R. W. (2000). Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 66, 4222–4229. doi:10.1128/AEM.66.10.4222-4229.2000
Mitschke, J., Georg, J., Scholz, I., Sharma, C. M., Dienst, D., Bantscheff, J., et al. (2011a). An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 108, 2124–2129. doi:10.1073/pnas.1015154108
Mitschke, J., Vioque, A., Haas, F., Hess, W. R., and Muro-Pastor, A. M. (2011b). Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. U.S.A. 108, 20130–20135. doi:10.1073/pnas.1112724108
Moore, L. R., Coe, A., Zinser, E. R., Saito, M. A., Sullivan, M. B., Lindell, D., et al. (2007). Culturing the marine cyanobacterium Prochlorococcus. Limnol. Oceanogr. Methods 5, 353–362. doi:10.4319/lom.2007.5.353
Nadeau, T.-L., and Castenholz, R. W. (2000). Characterization of psychrophilic oscillatorians (cyanobacteria) from Antarctic meltwater ponds. J. Phycol. 36, 914–923. doi:10.1046/j.1529-8817.2000.99201.x
Omairi-Nasser, A., de Gracia, A. G., and Ajlani, G. (2011). A larger transcript is required for the synthesis of the smaller isoform of ferredoxin:NADP oxidoreductase. Mol. Microbiol. 81, 1178–1189. doi:10.1111/j.1365-2958.2011.07739.x
Peralta-Yahya, P. P., Zhang, F., del Cardayre, S. B., and Keasling, J. D. (2012). Microbial engineering for the production of advanced biofuels. Nature 488, 320–328. doi:10.1038/nature11478
Pinto, F. L., Thapper, A., Sontheim, W., and Lindblad, P. (2009). Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol. Biol. 10:79. doi:10.1186/1471-2199-10-79
Prufert-Bebout, L., Paerl, H. W., and Lassen, C. (1993). Growth, nitrogen fixation, and spectral attenuation in cultivated Trichodesmium species. Appl. Environ. Microbiol. 59, 1367–1375.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi:10.1093/nar/gks1219
Reed, R. H., Chudek, J. A., Foster, R., and Stewart, W. D. P. (1984). Osmotic adjustment in cyanobacteria from hypersaline environments. Arch. Microbiol. 138, 333–337. doi:10.1007/BF00410900
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61. doi:10.1099/00221287-111-1-1
Schirmer, A., Rude, M. A., Li, X., Popova, E., and del Cardayre, S. B. (2010). Microbial biosynthesis of alkanes. Science 329, 559–562. doi:10.1126/science.1187936
Sharma, C. M., Hoffmann, S., Darfeuille, F., Reignier, J., Findeiss, S., Sittka, A., et al. (2010). The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255. doi:10.1038/nature08756
Shih, P. M., Wu, D., Latifi, A., Axen, S. D., Fewer, D. P., Talla, E., et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 1053–1058. doi:10.1073/pnas.1217107110
Steglich, C., Futschik, M. E., Lindell, D., Voss, B., Chisholm, S. W., and Hess, W. R. (2008). The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 4:e1000173. doi:10.1371/journal.pgen.1000173
Takahama, K., Matsuoka, M., Nagahama, K., and Ogawa, T. (2003). Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus. J. Biosci. Bioeng. 95, 302–305. doi:10.1263/jbb.95.302
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi:10.1093/molbev/mst197
Tan, X., Yao, L., Gao, Q., Wang, W., Qi, F., and Lu, X. (2011). Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab. Eng. 13, 169–176. doi:10.1016/j.ymben.2011.01.001
Trautmann, D., Voss, B., Wilde, A., Al-Babili, S., and Hess, W. R. (2012). Microevolution in cyanobacteria: re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 19, 435–448. doi:10.1093/dnares/dss024
Vogel, J., and Hess, W. R. (2001). Complete 5’ and 3’ end maturation of group II intron-containing tRNA precursors. RNA 7, 285–292. doi:10.1017/S1355838201001960
Voigt, K., Sharma, C. M., Mitschke, J., Lambrecht, S. J., Voß, B., Hess, W. R., et al. (2014). Comparative transcriptomics in two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J. doi:10.1038/ismej.2014.57
Voß, B., Bolhuis, H., Fewer, D. P., Kopf, M., Möke, F., Haas, F., et al. (2013). Insights into the physiology and ecology of the brackish-water-adapted cyanobacterium Nodularia spumigena CCY9414 based on a genome-transcriptome analysis. PLoS ONE 8:e60224. doi:10.1371/journal.pone.0060224
Voß, B., Georg, J., Schön, V., Ude, S., and Hess, W. R. (2009). Biocomputational prediction of non-coding RNAs in model cyanobacteria. BMC Genomics 10:123. doi:10.1186/1471-2164-10-123
Wang, W., Liu, X., and Lu, X. (2013). Engineering cyanobacteria to improve photosynthetic production of alka(e)nes. Biotechnol. Biofuels 6, 69. doi:10.1186/1754-6834-6-69
Winters, K., Parker, P. L., and Baalen, C. V. (1969). Hydrocarbons of blue-green algae: geochemical significance. Science 163, 467–468. doi:10.1126/science.163.3866.467
Keywords: alkane biosynthesis, start sites of transcription, cyanobacteria, operon, promoter, sRNA
Citation: Klähn S, Baumgartner D, Pfreundt U, Voigt K, Schön V, Steglich C and Hess WR (2014) Alkane biosynthesis genes in cyanobacteria and their transcriptional organization. Front. Bioeng. Biotechnol. 2:24. doi: 10.3389/fbioe.2014.00024
Received: 30 April 2014; Paper pending published: 23 May 2014;
Accepted: 26 June 2014; Published online: 14 July 2014.
Edited by:
Toivo Kallas, University of Wisconsin-Oshkosh, USAReviewed by:
M. Kalim Akhtar, University College London, UKXuefeng Lu, Chinese Academy of Sciences, China
Copyright: © 2014 Klähn, Baumgartner, Pfreundt, Voigt, Schön, Steglich and Hess. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wolfgang R. Hess, Genetics and Experimental Bioinformatics, Institute of Biology 3, Faculty of Biology, University of Freiburg, Schänzlestrasse 1, Freiburg D-79104, Germany e-mail: wolfgang.hess@biologie.uni-freiburg.de
 Stephan Klähn
Stephan Klähn Desirée Baumgartner
Desirée Baumgartner Ulrike Pfreundt
Ulrike Pfreundt
 Wolfgang R. Hess
Wolfgang R. Hess