Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics
- 1Animal Nutrition and Health Group, AgResearch Ltd., Palmerston North, New Zealand
- 2Centre of Research Excellence, Riddet Institute, Massey University, Palmerston North, New Zealand
A commentary on
Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics
by Emerson, D., Field, E. K., Chertkov, O., Davenport, K. W., Goodwin, L., Munk, C., et al. Front. Microbiol. 4:254. doi: 10.3389/fmicb.2013.00254
Iron is the fourth most abundant mineral in the Earth's lithosphere (Weber et al., 2006; Emerson et al., 2013), where it is present at a mean concentration of 5% (Hedrich et al., 2011). Iron can exist in two oxidation stages as ferrous (Fe(II)) and ferric (Fe(III)) iron and some bacteria and archaea have evolved to use iron as an obligate or facultative energy source, giving them the name “Iron Oxidizing Bacteria” (FeOB) and “Iron Oxidizing Archaea” (FeOA), respectively (Figure 1). The oxidation from Fe(II) to Fe(III) can occur under both oxic and anoxic conditions within a pH range between 0.5 to 8.4 (Edwards et al., 2000; Weber et al., 2006; Hedrich et al., 2011). Here, the microbially mediated oxidation of iron under (micro-)aerobic circum-neutral conditions will be discussed.
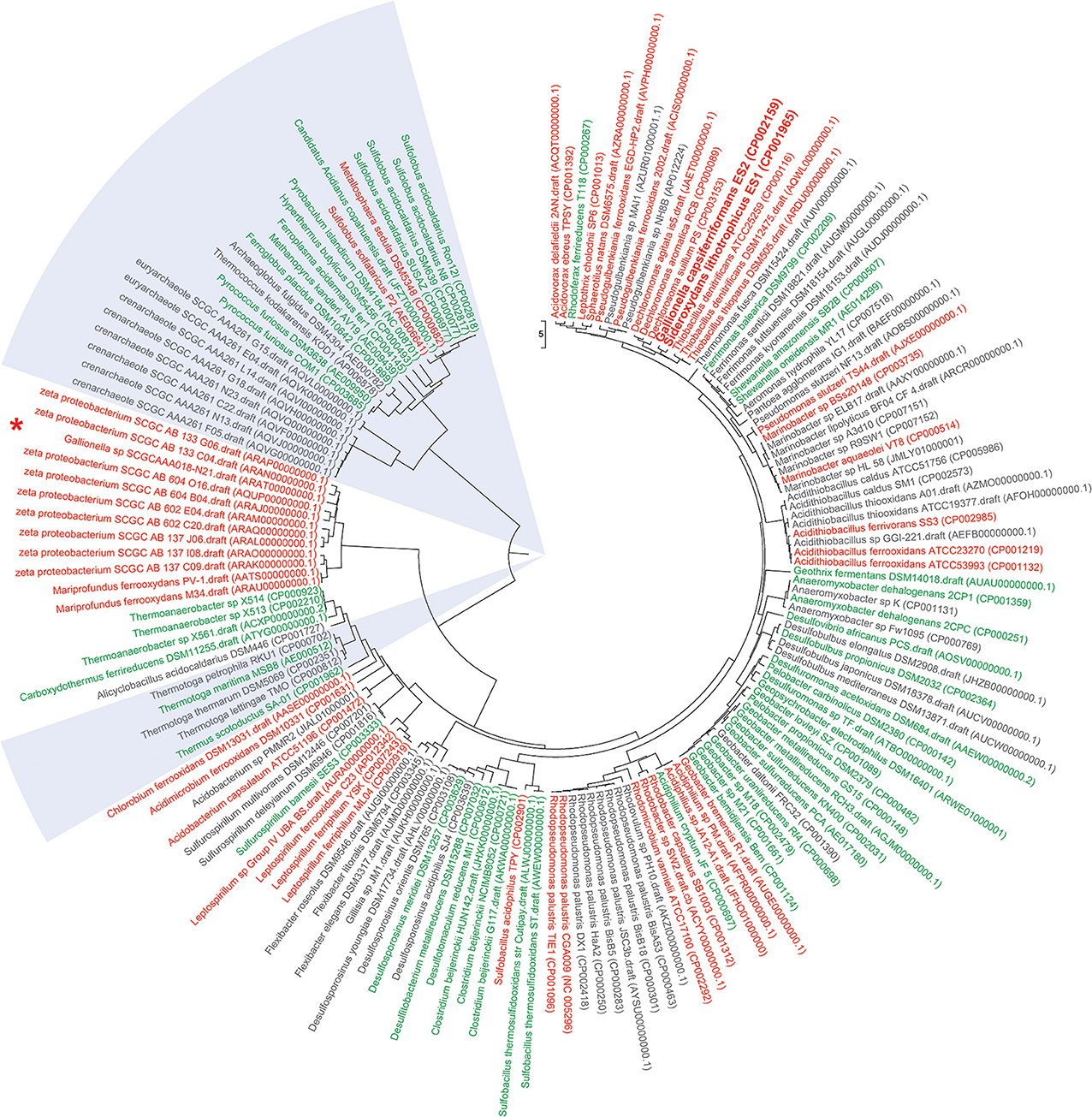
Figure 1. Functional Genome Distribution Tree of iron oxidizing and iron reducing microbes. 158 genomes of publicly available known (and predicted) iron oxidizing and iron reducing bacteria and archaea were included into a Functional Genome Distribution (FGD) analysis (Altermann, 2012). Briefly, draft genomes were downloaded in FASTA format, concatenated and their respective gene models predicted through an updated version of the GAMOLA annotation pipeline (Altermann and Klaenhammer, 2003). Complete genomes were downloaded in Genbank format. All 158 genomes were then subjected to an FGD analysis using the compACTor software. Briefly, within an FGD analysis complete ORFeomes are compared to each other, providing a snapshot of genome-to-genome similarities, rather than an ancestral outlook. In summary, the 158 genomes comprised 501,624 open reading frames (ORFs) and a total of 79,881,099 individual BLAST analyses were carried out to calculate the level of similarities between all genomes. The resulting dissimilarity matrix was then visualized in Mega6 (Tamura et al., 2013) using the UPGMA algorithm to infer a dissimilarity tree. The early draft sequence of Leptothrix ochracea L12 was found to be too incomplete and was removed from the final tree. Iron oxidizing bacteria and archaea are shown in red; iron reducing bacteria (FeRB) and archaea (FeRA) are depicted in green; likely iron oxidizing/reducing candidate microbes are indicated in gray. Archaeal iron oxidizers and reducers are highlighted in light blue sections. Accession numbers for respective Genbank or assembly entries are listed in brackets behind strain designations. Sideroxydans lithotrophicus ES1 and Gallionella capsiferriformans ES2 are shown in bold. Interestingly, Sideroxydans lithotrophicus ES1 and Gallionella capsiferriformans ES2 form their own sub-cluster and are most similar to Thiobacillus and Dechloromonas genomes. The recently published draft genome of Gallionella sp SCGCAAA018-N21 (marked by “*”) does not fall into the ES1/ES2 cluster, but groups within the larger zeta proteobacterium cluster. This may indicate the need to further investigate whether N21 represents a true Gallionella strain. In most instances, iron oxidizing and iron reducing microbes form their own separate cluster. There are notable exceptions (e.g., Rhodoferax ferrireducens T118 or Geobacter bremensis R1) and it may be possible that those microbes are able to carry out both iron oxidation and iron reduction, depending on their respective environmental circumstances. Most candidate iron oxidizing or iron reducing bacteria and archaea fall within distinct clusters that harbor examples of confirmed phenotypes. This may assist future research in systematically test for iron oxidizing or iron reducing phenotypes.
The biogeochemistry of iron poses several challenges to iron oxidizing microbes (for a detailed review see Weber et al., 2006). In circum-neutral environments and in the presence of oxygen, rapid abiotic oxidation of Fe(II) occurs, with half-life times of less than 60 s (Emerson et al., 2010). This abiotic oxidation to poorly soluble iron-oxyhydroxides reduces the availability of Fe(II) for biological process significantly. Microbial oxidation of Fe(II) therefore generally occurs in microaerophilic environments, which extend the half-life time of ferrous iron while at the same time increasing the amount of soluble Fe(II) (Bonnefoy and Holmes, 2012). Further, the oxidation from ferrous to ferric iron under low oxygen partial pressure increases the Gibbs free energy yield from 29 to −90 kJ mol−1 Fe(II) (Emerson et al., 2010).
Iron oxidizing bacteria and archaea have contributed on a global scale to shape the lithosphere (Emerson and Moyer, 2002; Weber et al., 2006). Evidence of their impact on the global iron cycle has been dated back to Pre-Cambrian age where they are thought to have potentially aided in the deposition of banded iron formations (Hedrich et al., 2011). Even today they have a major impact on terrestrial and aquatic systems. FeOB and FeOA can be found in habitats ranging from deep sea vents to freshwater systems and the rhizosphere and can form thick mats and deposits from a few millimeters to tens of centimeters thick. More recently, these microbes have made their appearance in man-made environments and industrial processes (Valdes et al., 2008). Most commonly known may be their contribution to biofouling and corrosion where the build-up of ferric iron can lead to a reduction in the water flow rates and water quality (Hedrich et al., 2011; Mcbeth et al., 2011).
Iron oxidizing microbes and their geological significance were first described in the 19th century (Ehrenberg, 1837; Harder, 1919). They are of key importance to global biogeochemical processes and more recently have gained interest for their impact on man-made environments (Hedrich et al., 2011). Yet, progress in understanding the ecology, physiology and genetic analysis of iron oxidizing microbes has been slow (Weber et al., 2006). Although iron oxidizing microbes can be readily observed in nature due to their striking morphology (such as the iconic stalk-forming Gallionella) and ability to form large microbial mats, they have proven elusive to cultivation under laboratory conditions. This elusiveness has caused confusion in the past, where taxonomy was inferred exclusively by morphological and ecological characteristics (Emerson et al., 2010). Recent advances in culturing techniques (Fabisch et al., 2013; Tischler et al., 2013) and culture-independent high-throughput DNA sequencing methods are providing a clearer phylogenetic picture of these bacteria (Emerson et al., 2010).
The recent availability of metagenomic data in particular has greatly accelerated our understanding of iron oxidizing microbes (Wang et al., 2011; Yelton et al., 2013). Only 54 bacterial and 13 archaeal genomes of FeOB are publicly accessible to date (Weber et al., 2006; Cardenas et al., 2010; Emerson et al., 2010; Hedrich et al., 2011), on which the emerging sequence-based taxonomic framework is built upon (Figure 1). New information from cultivated strains still refines this framework and the recently introduced class of marine Zetaproteobacteria (Emerson et al., 2007) is an excellent example of this rapid taxonomic evolution. An overview of publicly available genomes of iron oxidizing (and reducing) microbes and their relation to each other is presented in Figure 1.
While metagenomic datasets are valuable in improving our understanding of the presence and composition of iron oxidizing microbial communities (Ionescu et al., 2012; Kato et al., 2013; Singer et al., 2013), they rely on a robust gene-based taxonomic framework. Such a framework and detailed metabolic reconstruction of individual microbial strains require access to (draft) genome sequences and, consequently, to pure strains of iron oxidizing microbes (Kato et al., 2013). Despite the requirement for new strains of iron oxidizing microbes, isolation and characterization is challenging and time consuming. For example, Gallionella were first described in 1837 (Ehrenberg, 1837) and are amenable to cultivation. However, it was not until the mid-1990s where cultivation of two further strains, Gallionella ferruginea and Gallionella capsiferriformans ES-2, was successful (Hallbeck et al., 1993; Emerson and Moyer, 1997). Similarly, the isolation and characterization of other freshwater Fe(II) oxidizing Betaproteobacteria such as Sideroxydans sp. ES-1, that is metabolically flexible and can utilize reduced sulfur compounds and fix nitrogen, were also achieved only in the 1990s (Emerson and Moyer, 1997).
Since the initial description of both Gallionella capsiferriformans ES-2 and Sideroxydans lithotrophicus ES-1 a number of publications have investigated their morphological, phenotypical and industrial characteristics (Hallbeck and Pedersen, 1995; De Vet et al., 2012; Krepski et al., 2012), but no information was available on their genetic blueprint. The recent publication by Emerson et al. (2013), presenting the comparative analysis of genomic sequences of both Sideroxydans lithotrophicus ES-1 and Gallionella capsiferriformans ES-2, has filled this gap. By combining morphological characteristics with in silico comparative genomic analyses the authors were able to demonstrate that, although isolated from the same habitat, an iron-mat from ferruginous groundwater in Michigan (Emerson and Moyer, 1997), both strains belong to different genera in the recently proposed order Gallionellales. Importantly, the genomic comparison and metabolic pathway reconstruction highlighted a number of similarities and differences between both strains: 60% of the predicted genes were homologous at the 30% similarity level and both strains share a number of core metabolic capabilities for environmental sensing (chemotaxis), motility (pili), the uptake of nutrients through ABC transporters, energy metabolism, and detoxification of heavy metals, although strain ES-2 harbors a greater array of detoxification systems than ES-1 implying a superior ability to persist in habitats rich in environmental toxins. Notably, ~10% of both genomes is dedicated to environmental sensing and signal transduction which is a significantly higher number of genes than found in most other microbial genomes. Both strains harbor complete sets of chemotaxis genes and in combination with the elaborate environmental sensing array, controlled movement along the opposing gradients of Fe(II) and oxygen may facilitate optimal positioning of individual cells.
One of the problems aerobic Fe(II) oxidizing microbes must overcome is the presence of highly reactive oxygen species (ROS) that are produced via Fenton chemistry in the presence of oxygen and iron. Catalase and superoxide dismutase are the most common defense mechanisms against ROS and surprisingly both strains feature only a minimal set of catalase and peroxidase genes. This may be compensated by a number of truncated hemoglobins that are highly conserved and found in other FeOB such as M. ferrooxydans PV-1. These hemoglobins may represent an intracellular oxygen buffer that reduces the occurrence of ROS by binding oxygen under microaerophilic conditions and high concentrations of Fe(II).
Interestingly, strains ES-1 and ES-2 differ in their strategies to generate ATP. While ES-2 relies on the alternative complex III and a bacterial ATPase, ES-1 employs Cytochrome bc1, and a bacterial/archaeal-type ATPase in addition to the bacterial ATPase. The advantages of a second ATPase are presently unknown and the distribution of such redundant systems throughout the genomes of iron oxidizing microbes may provide clues to their respective niche adaptation in the future.
Both strains harbor prophage that may have been acquired independently and do not share homologous genes. However, the ES-1 prophage shares significant nucleic acid similarity (24%) to a prophage gene cluster identified in Mariprofundus ferrooxydans PV-1 (Singer et al., 2011), possibly indicating shared hosts in the past. Neither of the two strains features CRISPR sequences that have long been associated with phage defense, posing an interesting question on the evolutionary pressure of bacteriophage in iron oxidizing communities. It is generally accepted that phage and their respective hosts are locked into an ongoing arms race that shapes their evolution (Stern and Sorek, 2011). Emerging genomes of iron oxidizing microbes will provide deeper insights into the defense mechanisms these microbes deploy to evade phage infection and killing.
Neither of the two strains features any of the unique extracellular structures such as sheaths or stalks often associated with FeOB. Genome analysis revealed that strain ES-1 feature a single large gene cluster dedicated to exopolymer synthesis, while ES-2 harbors two separate large EPS clusters in addition to two gene clusters possibly involved in cellulose synthesis. The presence of exopolysaccharide (EPS) gene clusters may provide clues how those strains prevent self-encrustation by iron-oxyhydroxides but further in vitro research is required to unravel these survival strategies.
Recently, the draft genome sequences of Gallionella sp. SCGC AAA018-N21 have been released (BioSample: SAMN02256456) and a future comparative genomics analyses will undoubtedly reveal further insights in lifestyle adaptation and contribute to a more robust taxonomic framework.
The research presented by Emerson et al. contributes significantly to our understanding of neutrophilic freshwater iron-oxidizing bacteria and their taxonomic relations to each other. However, the intriguing features found in both genomes—either commonly shared or specific to the individual strains—have created a large, new research space waiting to be investigated.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The author thanks Sandra Kittelmann and Adrian Cookson for critical review of the manuscript.
References
Altermann, E. (2012). Tracing lifestyle adaptation in prokaryotic genomes. Front. Microbiol. 3:48. doi: 10.3389/fmicb.2012.00048
Altermann, E., and Klaenhammer, T. R. (2003). GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. OMICS 7, 161–169. doi: 10.1089/153623103322246557
Bonnefoy, V., and Holmes, D. S. (2012). Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environ. Microbiol. 14, 1597–1611. doi: 10.1111/j.1462-2920.2011.02626.x
Cardenas, J. P., Valdes, J., Quatrini, R., Duarte, F., and Holmes, D. S. (2010). Lessons from the genomes of extremely acidophilic bacteria and archaea with special emphasis on bioleaching microorganisms. Appl. Microbiol. Biotechnol. 88, 605–620. doi: 10.1007/s00253-010-2795-9
De Vet, W. W. J. M., Dinkla, I. J. T., Abbas, B. A., Rietveld, L. C., and Van Loosdrecht, M. C. M. (2012). Gallionella spp. in trickling filtration of subsurface aerated and natural groundwater. Biotechnol. Bioeng. 109, 904–912. doi: 10.1002/bit.24378
Edwards, K. J., Bond, P. L., Gihring, T. M., and Banfield, J. F. (2000). An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287, 1796–1799. doi: 10.1126/science.287.5459.1796
Ehrenberg, C. G. (1837). Remarks on the real occurrence of fossil infusoria, and their extenisve diffusion. Taylor's Sci. Memoirs 1, 400–413, plate V.
Emerson, D., Field, E. K., Chertkov, O., Davenport, K. W., Goodwin, L., Munk, C., et al. (2013). Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front. Microbiol. 4:254. doi: 10.3389/fmicb.2013.00254
Emerson, D., Fleming, E. J., and Mcbeth, J. M. (2010). Iron-oxidizing bacteria: an environmental and genomic perspective. Annu. Rev. Microbiol. 64, 561–583. doi: 10.1146/annurev.micro.112408.134208
Emerson, D., and Moyer, C. (1997). Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 63, 4784–4792.
Emerson, D., and Moyer, C. L. (2002). Neutrophilic Fe-oxidizing bacteria are abundant at the Loihi Seamount hydrothermal vents and play a major role in Fe oxide deposition. Appl. Environ. Microbiol. 68, 3085–3093. doi: 10.1128/AEM.68.6.3085-3093.2002
Emerson, D., Rentz, J. A., Lilburn, T. G., Davis, R. E., Aldrich, H., Chan, C., et al. (2007). A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS ONE 2:e667. doi: 10.1371/journal.pone.0000667
Fabisch, M., Beulig, F., Akob, D. M., and Kusel, K. (2013). Surprising abundance of Gallionella-related iron oxidizers in creek sediments at pH 4.4 or at high heavy metal concentrations. Front. Microbiol. 4:390. doi: 10.3389/fmicb.2013.00390
Hallbeck, L., and Pedersen, K. (1995). Benefits associated with the stalk of Gallionella ferruginea, evaluated by comparison of a stalk-forming and a non-stalk-forming strain and biofilm studies in situ. Microb. Ecol. 30, 257–268. doi: 10.1007/BF00171933
Hallbeck, L., Stahl, F., and Pedersen, K. (1993). Phylogeny and phenotypic characterization of the stalk-forming and iron-oxidizing bacterium Gallionella ferruginea. J. Gen. Microbiol. 139, 1531–1535. doi: 10.1099/00221287-139-7-1531
Harder, E. C. (1919). “Iron-depositing bacteria and their geological relations,” in US Geological Survey Professional Paper (Washington, DC), 85.
Hedrich, S., Schlomann, M., and Johnson, D. B. (2011). The iron-oxidizing proteobacteria. Microbiology 157, 1551–1564. doi: 10.1099/mic.0.045344-0
Ionescu, D., Siebert, C., Polerecky, L., Munwes, Y. Y., Lott, C., Hausler, S., et al. (2012). Microbial and chemical characterization of underwater fresh water springs in the Dead Sea. PLoS ONE 7:e38319. doi: 10.1371/journal.pone.0038319
Kato, S., Chan, C., Itoh, T., and Ohkuma, M. (2013). Functional gene analysis of freshwater iron-rich flocs at circumneutral pH and isolation of a stalk-forming microaerophilic iron-oxidizing bacterium. Appl. Environ. Microbiol. 79, 5283–5290. doi: 10.1128/AEM.03840-12
Krepski, S. T., Hanson, T. E., and Chan, C. S. (2012). Isolation and characterization of a novel biomineral stalk-forming iron-oxidizing bacterium from a circumneutral groundwater seep. Environ. Microbiol. 14, 1671–1680. doi: 10.1111/j.1462-2920.2011.02652.x
Mcbeth, J. M., Little, B. J., Ray, R. I., Farrar, K. M., and Emerson, D. (2011). Neutrophilic iron-oxidizing “zetaproteobacteria” and mild steel corrosion in nearshore marine environments. Appl. Environ. Microbiol. 77, 1405–1412. doi: 10.1128/AEM.02095-10
Singer, E., Emerson, D., Webb, E. A., Barco, R. A., Kuenen, J. G., Nelson, W. C., et al. (2011). Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS ONE 6:e25386. doi: 10.1371/journal.pone.0025386
Singer, E., Heidelberg, J. F., Dhillon, A., and Edwards, K. J. (2013). Metagenomic insights into the dominant Fe(II) oxidizing Zetaproteobacteria from an iron mat at Lo ihi, Hawai l. Front. Microbiol. 4:52. doi: 10.3389/fmicb.2013.00052
Stern, A., and Sorek, R. (2011). The phage-host arms race: shaping the evolution of microbes. Bioessays 33, 43–51. doi: 10.1002/bies.201000071
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tischler, J. S., Jwair, R. J., Gelhaar, N., Drechsel, A., Skirl, A.-M., Wiacek, C., et al. (2013). New cultivation medium for “Ferrovum” and Gallionella-related strains. J. Microbiol. Methods 95, 138–144. doi: 10.1016/j.mimet.2013.07.027
Valdes, J., Pedroso, I., Quatrini, R., Dodson, R. J., Tettelin, H., Blake, R., et al. (2008). Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597. doi: 10.1186/1471-2164-9-597
Wang, J., Vollrath, S., Behrends, T., Bodelier, P. L., Muyzer, G., Meima-Franke, M., et al. (2011). Distribution and diversity of Gallionella-like neutrophilic iron oxidizers in a tidal freshwater marsh. Appl. Environ. Microbiol. 77, 2337–2344. doi: 10.1128/AEM.02448-10
Weber, K. A., Achenbach, L. A., and Coates, J. D. (2006). Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 4, 752–764. doi: 10.1038/nrmicro1490
Keywords: iron oxidizing bacteria, comparative genomics, taxonomy, life style adaptation, biogeochemistry, iron cycle
Citation: Altermann E (2014) Invited commentary: lubricating the rusty wheel, new insights into iron oxidizing bacteria through comparative genomics. Front. Microbiol. 5:386. doi: 10.3389/fmicb.2014.00386
Received: 25 April 2014; Accepted: 10 July 2014;
Published online: 30 July 2014.
Edited by:
Martin G. Klotz, University of North Carolina at Charlotte, USAReviewed by:
Jennifer Glass, Georgia Institute of Technology, USAArpita Bose, Harvard University, USA
Copyright © 2014 Altermann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: eric.altermann@agresearch.co.nz
 Eric Altermann
Eric Altermann