Proteomics insights: proteins related to larval attachment and metamorphosis of marine invertebrates
- 1Biological and Environmental Sciences and Engineering, Division of Applied Mathematics and Computer Sciences, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
- 2KAUST Global Collaborative Research, Division of Life Science, The Hong Kong University of Science and Technology, Kowloon, Hong Kong
The transition in an animal from a pelagic larval stage to a sessile benthic juvenile typically requires major morphological and behavioral changes. Larval competency, attachment and initiation of metamorphosis are thought to be regulated by intrinsic chemical signals and specific sets of proteins. However, the molecular mechanisms that regulate larval attachment and metamorphosis in marine invertebrates have yet to be fully elucidated. Despite the many challenges associated with analysis of the larvae proteome, recent proteomic technologies have been used to address specific questions in larval developmental biology. These and other molecular studies have generated substantial amount of information of the proteins and molecular pathways involved in larval attachment and metamorphosis. Furthermore, the results of these studies have shown that systematic changes in protein expression patterns and post-translational modifications (PTMs) are crucial for the transition from larva to juvenile. The degeneration of larval tissues is mediated by protein degradation, while the development of juvenile organs may require PTM. In terms of application, the identified proteins may serve as targets for antifouling compounds, and biomarkers for environmental stressors. In this review we highlight the strengths and limitations of proteomic tools in the context of the study of marine invertebrate larval biology.
Importance of Marine Invertebrates: Barnacles, Bryozoans and Polychaetes
Marine invertebrates are important players in various marine ecosystems, including in biofouling, intertidal and sediment ecosystems. The barnacle Balanus amphitrite, the bryozoan Bugula neritina and the serpulid polychaete Hydroides elegans are the most dominant fouling species, are distributed worldwide, and are commonly used in larval settlement (including both attachment and metamorphosis) studies and antifouling research (Ten Hove, 1974; Khandeparker and Anil, 2007; Aguirre et al., 2008; Cohen, 2011). Marine polychaete worms, including Capitella teleta, Pseudopolydora vexillosa, and Neanthes arenaceodentata, occur in habitats ranging from subtropical to inshore waters, and are used as indicators of marine pollution and as toxicological test animals (Reish and Gerlinger, 1997; Greg and Kristian, 1998; Rouse and Fauchald, 1998; Glasby and Timm, 2008; Blake et al., 2009). Because of high sensitivity to marine contamination mussels have been used as indicators of marine pollution (Romeo et al., 2003). For instance, mussels and barnacles were used as biomonitors in coastal waters (Phillips and Rainbow, 1988) Abalone Haliotis diversicolor supertexta was used for investigating contamination of marine ecosystems by toxic chemicals (Zhou et al., 2010). Calcifying marine invertebrates including polychaetes and barnacles, are capable of physiological and behavioral adaptation to various environmental stressors and thus serve as model organisms in climate change research (Dupont and Thorndyke, 2009; Taylor et al., 2010). Abalone, gastropod mollusk, is used for food and their shells are often used for ornaments. Several invertebrates provide habitat to other species. For example, the mussel beds provide living space to the species living on them. Peracaridan species utilize B. neritina as living space (Conradi et al., 2000).
Larval Development: Competency, Attachment, and Metamorphosis
The population dynamics of marine invertebrates are controlled by recruitment and post-settlement survival. The choice of attachment site is crucial because attachment to an unfavorable substrate can be fatal (Bishop et al., 2006). Factors that influence attachment and metamorphosis, including settlement cues, competency, and habitat, are selective for the larvae of a particular species. For instance, larval metamorphosis in barnacles is a dynamic and rapid process that involves substantial tissue remodeling and differentiation (Thiyagarajan et al., 2007; Maruzzo et al., 2012). Barnacle species have six nauplius stages and a cyprid larval stage, and larva–juvenile transitions may or may not require a metamorphosis inducer (Thiyagarajan et al., 2005). In the absence of cues, the larvae of B. neritina commonly settle within several minutes (Woollacott and Zimmer, 2005). In H. elegans, the fertilized eggs hatch into young trochophore larvae, which develop into competent larvae and metamorphose into young juveniles. The larvae will not settle without suitable settlement cues, and the larval–juvenile transition requires substantial tissue reorganization and major morphological changes (Hadfield et al., 2001). In contrast, the young larvae of P. vexillosa can take 7–10 days to develop competency, and larval metamorphosis is a gradual process that does not require substantial development of juvenile organs (Mok et al., 2008). This type of metamorphosis in the absence of specific inducers is referred to as “spontaneous metamorphosis.” Although past studies have contributed to our understanding of the basic underpinnings of the morphological, ecological and behavioral patterns of larval settlement, the molecular mechanisms that regulate larval attachment and metamorphosis are poorly understood. In recent years, it has been hypothesized that attachment and metamorphic events are regulated by changes in protein expression patterns and intrinsic signaling pathways that influence larval morphology and behavior. To test this hypothesis, proteomics tools are being used increasingly to identify proteins and pathways involved in larval development, despite technological limitations associated with the advancement of larval biology research in this area. Hence, the goal of this review is to describe the progress in marine larval proteomic research in relation to changes in protein expression patterns in different larval stages of marine invertebrates. Specifically, this review highlights: (i) technical challenges in larval proteomics research; (ii) proteome and phosphoproteome insights during larval attachment and metamorphosis; (iii) the involvement of signaling pathways during larval metamorphosis; and (iv) the usefulness of commonly expressed proteins in anti-fouling applications.
Protein Expression Patterns During Larval Development: Two-Dimensional Gel Electrophoresis (2-DE) Proteomics
Proteomics tools and pipelines used in the study of marine larval biology are outlined in Figure 1. A list of studies on larval proteomics is shown in Table 1. In the majority of these studies, the larval proteome was characterized by gel-based (2-DE) or gel-free proteomics, followed by mass spectrometry (MS) analysis. These approaches enabled documentation of changes in protein expression patterns and protein modification status at various stages in the larval lifespan. López et al. (2005) was the first group to use 2-DE proteomics approach to identify larval-specific proteins in the mussel Mytilus galloprovincialis. DeBoer et al. (2007) then conducted comparative proteome and transcriptome analyses to reveal symbiosis-related differences in coral larvae and dinoflagellates and found no significant changes in the protein profiles of the larvae. Thiyagarajan and Qian (2008) used 2-DE proteomics to document the global proteome expression pattern in the larvae of B. amphitrite. They optimized protein extraction and iso-electric focusing (IEF) conditions for a better resolution of protein spots on 2-DE gels and showed that the proteome pattern of cyprid larvae was very distinct from that of attached larvae and newly-metamorphosed juveniles. Another study (Mok et al., 2009) found that there are proteomic changes in the polychaete worm P. vexillosa during larval metamorphosis. They found that the proteome of competent larvae of P. vexillosa was similar to that of juveniles, whereas the proteomes of precompetent and competent larvae were more distinct. Availability of genome sequencing allowed a better characterization of the proteins involved in different biological processes during the development in ascidians (as reviewed in Inaba et al., 2007). Nomura et al. (2009) found the involvement of vitellogenin, elongation factors, actin-binding proteins and chaperones during embryonic development of Ciana intestinalis. In the abalone Haliotis asinina, the first proteomic study characterized protein domains from calcified shell, which were rich in glutamine, glycine, alanine, and aspartate (Marie et al., 2010). All the studies mentioned above have relied on low-resolution immobilized pH gradient (IPG) strips (13 cm in length; pH 3–10) for the first-dimension separation and Coomassie-brilliant blue (CBB) staining of the gels. Aggregation or overlapping of proteins spots limited the resolution of many spots on the 2-DE gels and could not really quantify actual changes in protein abundance. To overcome these shortcomings, Qian et al. (2010) and Zhang et al. (2010a) used combination of different methods to probe the larval proteomes of B. neritina and B. amphitrite, including pre-fractionation of the larval proteomes prior to 2-DE to reduce the sample complexity, sequential fluorescent dye gel staining to increase the detection limit (~0.5–5 ng) and dynamic range (Panfoli et al., 2012) and implementation of narrow-range IPG strips (pH 4–7) to enhance resolution of the protein spots on 2-DE gels. In these studies, they used used a Zoom IEF fractionator (Invitrogen, Carlsbad, CA, USA) to fractionate the total proteome into two subproteomes of pH 3.0–5.4 and pH 5.4–7.0 and high-resolution narrow-range IPG (pH 4–7) strips followed by gel staining with SYPRO® Ruby (Molecular Probes, Eugene, OR, USA) to achieve better protein separation. The number of protein spots detected was more than that detected for unfractionated samples.
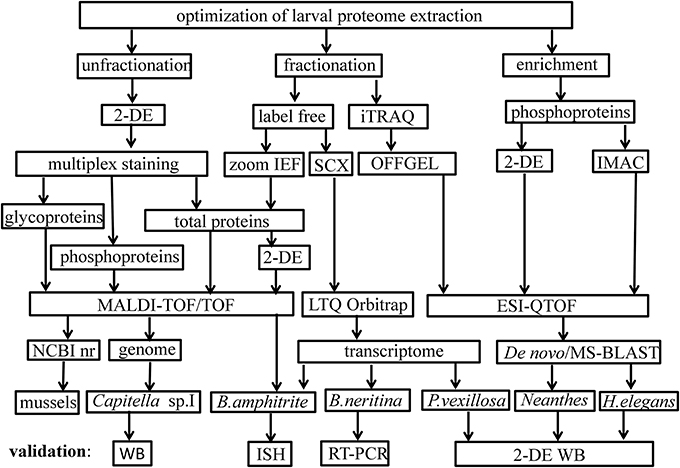
Figure 1. Proteomics tools used to study marine larvae. Abbreviations: 2-DE, two-dimensional-gel electrophoresis; iTRAQ, isobaric tags for relative and absolute quantitation; IEF, isoelectric focusing; SCX, strong cation exchange; IMAC, immobilized metal ion affinity chromatography; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; ESI, electrospray ionization; NCBI nr, national center for biotechnological information non-redundant; WB, Western blot; ISH, in situ hybridization; RT-PCR, real-time polymerase chain reaction.
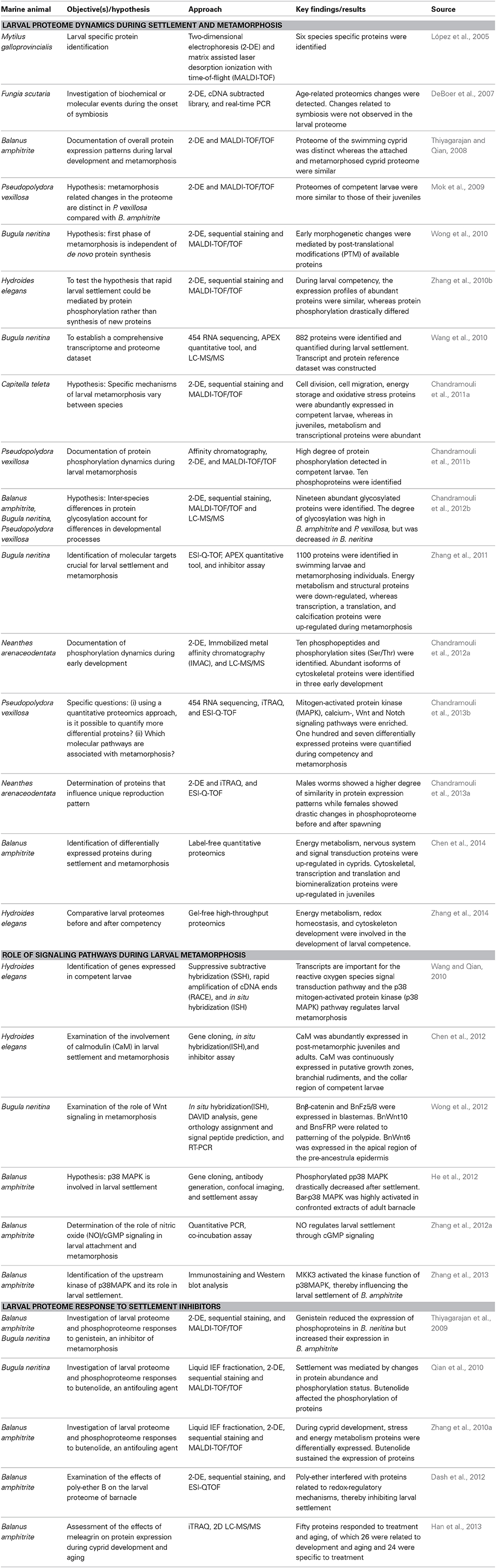
Table 1. List of proteomics investigations focused on larval settlement and metamorphosis of marine invertebrates.
Post-Translational Modifications (PTMs) of Proteins During Larval Metamorphosis
The identification and characterization of protein modifications among marine invertebrates have received substantial attention in recent years. It has been hypothesized that the early phase of metamorphic events depends on PTMs of proteins. For example, in B. neritina, the rapid initial phase of metamorphosis was facilitated by PTMs of existing proteins (Wong et al., 2010). In fact, protein modification often plays a crucial role in many biological processes (Zhou et al., 2007). In particular, protein phosphorylation is a rapid process that regulates many cellular processes and signal transduction pathways (Hunter, 1995). Thiyagarajan et al. (2009) demonstrated the usefulness of multiplex staining and 2-DE to probe the larval phosphoproteome of B. amphitrite and B. neritina. In their study, specific fluorescent dyes including Pro-Q Diamond and Sypro Ruby, were used to detect phosphoproteins and total proteins on a single 2-DE gel. This method enabled parallel analyses of total protein and phosphoprotein expression. The similar tools were used to study phosphoproteome patterns in other marine species. In H. elegans, the larval phosphoproteome pattern changed significantly during competency, whereas the proteome pattern was not affected (Zhang et al., 2010b). In Capitella teleta (this refers to Capitella sp. I in early publication) phosphorylated proteins were essential for the elongation of body segments, production of hooded hooks, and preparation of juvenile tissues during larval metamorphosis (Chandramouli et al., 2011a). Protein enrichment methods have enabled the separation of proteins according to their PTM (e.g., phosphorylation) (Gilchrist et al., 2006; Puente et al., 2006; Cantin et al., 2008). This approach was used successfully by Chandramouli et al. (2011b) to improve the detection and identification of phosphoproteins during larval metamorphosis of P. vexillosa, revealing that competent larvae exhibit a higher degree of phosphorylation than precompetent larvae and juveniles. However, MS analysis of phosphopeptides remains challenging (Leitner et al., 2011). Using immobilized metal affinity chromatography (IMAC) and liquid chromatography (LC)-MS/MS to enrich phosphorylated peptides, Chandramouli et al. (2012a) identified nine phosphopeptides and phosphorylation sites (Thr/Ser) that are crucial for the early development (ova and larval stages) of N. arenaceodentata. This enrichment strategy illustrated an effective method for the identification of phosphoproteins and phosphopeptides in a larval proteome. On the other hand, glycoproteomics is an emerging proteomic technology and can be used to detect specific glycosylation patterns and to identify glycoproteins in complex proteomes (Pan et al., 2011). This method has the advantage of sequential staining, whereby ProQ Emerald fluorescent dye is used to selectively detect glycoproteins on 2-D gels. Using thie method, Chandramouli et al. (2012b) identified 19 abundant glycoproteins involved in the stress response and metabolic processes during larval metamorphosis of three marine invertebrate species. They found that the degree of protein glycosylation in B. amphitrite and P. vexillosa was greater prior to metamorphosis, whereas glycosylation in B. neritina increased right after metamorphosis. Overall, these studies clearly demonstrated emerging proteomics technologies can be used to document both glycosylation and phosphorylation dynamics during larval metamorphosis.
Gel-Free High-Throughput Proteomics and Comprehensive Larval Proteome Analysis
Although gel-based proteomics provided valuable information about larval protein expression patterns, limitations associated with the identification of low-abundance larval proteins prompted the development of high throughput gel-free approaches (López, 2007; Tomanek, 2011; Slattery et al., 2012). In addition, transcriptome sequence datasets for several marine invertebrates, including B. neritina (Wang et al., 2010), B. amphitrite (Chen et al., 2011; De Gregoris et al., 2011), the coral Acropora millepora (Moya et al., 2012) and P. vexillosa (Chandramouli et al., 2013b), are now available in the public domain, which allows larval biologists to conduct more gel-free proteome analysis. Label-free quantitative methods involving individual sample preparation steps followed by LC-MS/MS analysis. Quantification of the amount of proteins is based on mass spectrum counting of the identified peptides (Wong et al., 2010; Zhang et al., 2011; Chen et al., 2014). Wang et al. (2010) applied next generation RNA sequencing and high-throughput LC-MS-based proteomics to profile larval transcripts and proteins of B. neritina. They generated 131,450 high-quality transcripts which were used as a reference transcriptome for proteome analysis. About 882 proteins were identified and quantified for the swimming and attached larval stages of B. neritina. Zhang et al. (2011) identified >1000 proteins (among which 61 were differentially expressed) in swimming and in two post-attachment stages of B. neritina. These differentially expressed proteins were grouped in the several categories: energy metabolism, cytoskeleton, transcription and translation, and calcification. Chen et al. (2014) identified about 700 proteins in each stage of B. amphitrite (nauplius II, nauplius VI, cyprids and juveniles) using label-free quantitative proteomics and found that proteins related to energy metabolism, nervous system and signal transduction were up-regulated in cyprids, whereas those related to cytoskeleton, transcription and translation, cell differentiation, and biomineralization proteins were up-regulated in juveniles. Zhang et al. (2014) identified about 1500 proteins that are associated with energy metabolism, redox homeostasis, and cytoskeleton development during larval development of H. elegans.
Although label-free proteomic studies provide a robust platform for direct quantitative analysis of the proteome, it is very challenging and complicated to analyze many mass spectra derived from multiple MS runs (Haqqani et al., 2008). To address this issue, peptide labeling methods have been used for relative quantification of protein abundance in marine invertebrates (Chandramouli et al., 2013a,b; Han et al., 2013; Sun et al., 2013). In fact, isobaric tags for relative and absolute quantification (iTRAQ) have been established (Putz et al., 2005) and used to multiplex 4–8 samples (Evans et al., 2012; Martyniuk et al., 2012). Using this method, the peptides are labeled and then the samples are pooled and desalted prior to LC-MS/MS. The protein or peptide abundance is calculated based on the chromatographic peptide peak intensity, or the height of the various labeled peptide samples. In addition, an improved fractionation technique involving the separation of proteins or peptides in a multi-well device according to their isoelectric points (pI) has been developed to reduce the proteome complexity (Agilent Technologies, Böblingen, Germany; Chenau et al., 2008). Chandramouli et al. (2013b) combined peptide OFFGEL fractionation with iTRAQ labeling to identify and quantify proteins involved in key molecular processes during larval metamorphosis of P. vexillosa and identified 107 differentially expressed proteins in precompetent, competent, and juvenile stages. Among these 107 proteins, 14 and 53 proteins were differentially expressed during competency and metamorphosis, respectively. Using the same approach, Chandramouli et al. (2013a) identified a set of proteins that influence the unique pattern of reproduction in the polychaete N. arenaceodentata and demonstrated that male worms have consistent protein expression patterns, whereas female worms (particularly spent females) show marked changes in the phosphoproteome before and after spawning. These studies showed that labeling methods permit the relative and absolute quantification of proteins expressed during a specific developmental stage.
Molecular Pathways Underlying Larval Metamorphosis in Marine Invertebrates
Signaling pathways that are possibly regulate larval metamorphosis of marine invertebratespecies have been a hot topic in recent years. It was reported that cyclic adenosine-monophosphate (cAMP) and calcium signaling in barnacles (Clare et al., 1995; Clare, 1996), inositol-1,4,5-tris-phosphate/diacylglycerol (IP3/DAG) signaling in cnidarians (Fleck, 1997), nitric oxide/cyclic guanosine monophosphate (NO/cGMP), protein kinase C (PKC) in Capitella (Biggers and Laufer, 1999) and sea urchins (Bishop et al., 2001; Amador-Cano et al., 2006), and p38 mediated mitogen-activated protein kinase (MAPK) recently has been implicated in the regulation of larval attachment and metamorphosis in H. elegans and B. amphitrite (Wang and Qian, 2010; He et al., 2012) are possibly involved in larval settlement and metamorphosis of these animals. Furthermore, Zhang et al. (2013) demonstrated that MKK3 (the upstream kinase of p38MAPK) activates the function of p38MAPK, thereby influencing the attachment of B. amphitrite larvae. Nitric oxide (NO) and cyclic GMP (cGMP) signaling molecules have been shown to act as regulators of the initiation of metamorphosis in ascidians and in an echinoid (Bishop et al., 2001; Leise et al., 2001). Bishop and Brandhorst (2001) demonstrated that the NO/cGMP system acts as a negative regulator during metamorphosis of the sea urchin Lytechinus pictus. Zhang et al. (2012a) documented the role of NO and cGMP during larval attachment of B. amphitrite. Recently Ueda and Degnan (2014) reported that NO facilitates the induction of metamorphosis while inhibition of nitric oxide synthase (NOS) reduces rates of metamorphosis in the abalone Haliotis asinine. It appears that nitric oxide signaling can contribute to stress-related responses (up-regulation of HSP-90) and regulate the metamorphic transitions. The role of calcium in signal transduction pathways during larval metamorphosis has been well documented in the polychaete worm Phragmatopoma californica (Ilan et al., 1993). Chen et al. (2012) examined the role of calmodulin (CaM) in larval attachment and metamorphosis of H. elegans Wong et al. (2012) found that the Wnt signaling pathway appears to be important for morphogenesis in B. neritina. Based on the findings above, we anticipate that MAPK signaling is important for larval attachment in B. amphitrite and H. elegans. Wnt signaling typically influences larval metamorphosis of B. neritina. The importance of Ca2+/CaM signaling in the development of polychaetes is of significant interest.
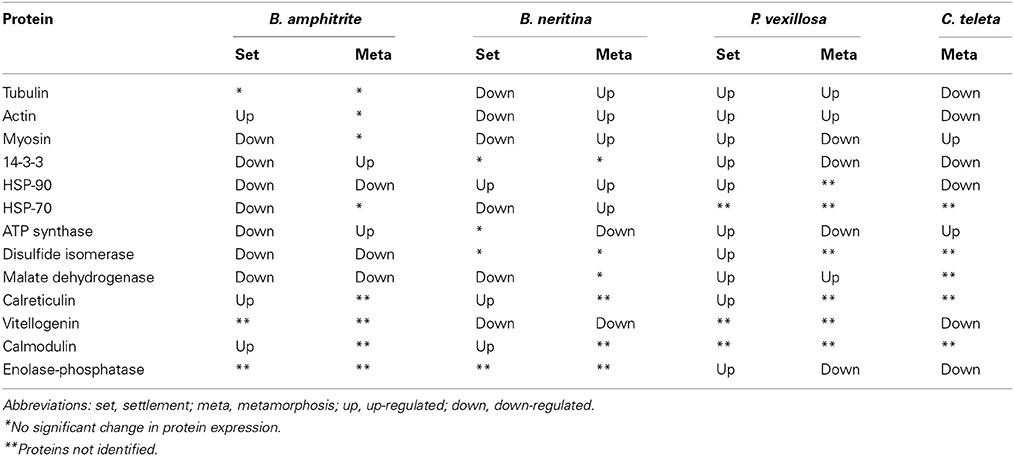
Common Protein Expression
Heyland and Moroz (2006) showed that larval attachment and metamorphosis in marine invertebrates is mediated by common metamorphic events. If this is true, one may expect that larval attachment and metamorphosis are regulated by a common set of proteins in multiple marine invertebrates. To identify common protein expression signatures (PESs) during larval attachment and metamorphosis of B. amphitrite, P. vexillosa, and B. neritina, Chandramouli et al. (2012b) found 11 commonly expressed proteins involved in cytoskeleton, metabolism, and stress responses. A similar set of common proteins were differentially regulated in multiple marine invertebrates. Table 2 lists the commonly regulated proteins and their expression patterns. In general, changes in the expression patterns of cytoskeletal proteins (tubulin, actin, and myosin) were observed in all of the species studied so far, indicating the importance of cytoskeletal dynamics in maintaining the architecture and contractility of larvae. Similarly, proteins related to energy metabolism (ATP synthase, disulfide isomerase, vitellogenin, enolase-phosphatase, and malate dehydrogenase) and the stress response (HSP-90, HSP-70, 14-3-3 protein, and calreticulin) were often differentially expressed during larval attachment and metamorphosis, indicating that these proteins may be synergistically regulated and may control events in the metamorphosis of larvae to juveniles. To identify possible common proteins that are the molecular targets for antifouling compounds, Thiyagarajan et al. (2009) showed that genistein, which is an inhibitor of metamorphosis (Zhou et al., 2009), down-regulated the expression of phosphoproteins in settling larvae of B. neritina. In contrast, this compound induced the expression of phosphoproteins in B. amphitrite. Another potential antifoulant, butenolide (Xu et al., 2010) sustained the expression of stress and energy metabolism proteins in settling larvae of B. neritina and B. amphitrite (Qian et al., 2010; Zhang et al., 2010a). Dash et al. (2012) showed that polyethers affect proteins related to energy metabolism, oxidative stress and chaperones. Han et al. (2013) also found that meleagrin, a potential non-toxic antifoulant, affect the expression of cyprid major protein (CMP) and adhesive plaque matrix proteins (APMPs) and thereby interfere with the larval endocrine system and molting processes (Billinghurst et al., 2000). These studies indicated that proteins involved in oxidative stress, cytoskeletal dynamics, chaperones, apoptosis, and energy homeostasis are possible molecular targets of those antifouling compounds. In contrast, several other types of antifoulants act on ion channels, quorum sensing systems, neurotransmitters, genotoxic damage, and adhesive production (as reviewed in Qian et al., 2013), which indicates voltage dependent anion channels, acetylcholinesterase, phenoloxidase, chitinase, Ran GTPase activating protein, and NADH ubiquinone oxidoreductase are proposed as molecular targets. Because of housekeeping function and responsive to environmental changes, several PESs identified by proteomics studies may not be the direct targets but their differential expression may be an indirect physiological and biochemical response to various antifoulants.
Challenges and Future Directions
Sample Preparation
The complexity of protein samples has always been a major obstacle of proteomic study (Hondermarck, 2003; Speicher, 2007; Shen et al., 2008). Larvae and adults of many marine invertebrates are protected by shell (e.g., B. amphitrite), calcareous tubes (H. elegans), and mucilaginous polysaccharide substances (P. vexillosa and C. teleta). Cell culture and tissue isolation methods have not yet been established for these species. The direct use of whole larvae or juveniles for proteomic studies results in an enormous increase in sample complexity, complicating downstream sample preparation for mass spectrometry (MS) analysis. Highly abundant proteins can mask those of a low abundance, potentially preventing the detection of signaling and receptor proteins that may be crucial for larval attachment and metamorphic transitions. To overcome these problems, we need to improve protein extraction protocols. In addition, larval proteins of polychaetes can rapidly degraded shortly after tissue lysis. To provent degradation, protease inhibitors are required during multiple sample preparation steps (Zhang et al., 2010b; Chandramouli et al., 2011b). In addition, to minimize the possible protein degradation, it is better to freeze the samples in liquid nitrogen or be used immediately. The combined use of strong detergents (8M urea), homogenization, and sonication methods can also significantly improve protein recovery (Sun et al., 2013). It is also highly recommended that prior to 2-DE, larval protein samples shall be purified from other contaminants through the use of acetone precipitation and 2-DE clean-up kits. Multistep strategies in sample preparation, fractionation, and MS separation of peptides may facilitate the detection of these proteins (Millioni et al., 2011). Among these methods, immunoaffinity columns seems to be more suitable for the depletion of high-abundant cytoskeletal proteins (Molnár et al., 2011), after which low-abundant proteins may bind to the column, be eluted with specific buffers, and identified by 2-DE and/or MS. This procedure facilitates the identification of potential protein candidates or biomarkers. Alternately, the enrichment of specific subset of phosphoproteins or peptides from a complex protein sample can be an effective approach (Batalha et al., 2012). Cost-effective specific immune-affinity reagents, such as antibodies and metal, are the subjects for targeted proteomic studies, particularly for elucidating the protein phosphorylation dynamics in signaling pathways. However, the development of protocols for protein enrichment is always challenged by the functional and structural diversity of phosphorylated proteins. Furthermore, we need to maximize the amount of proteins to begin with in order to have sufficient protein for down-stream analysis.
De novo Sequencing and MS BLAST
In general, mass spectra are searched against databases using dedicated softwares, such as Mascot and SEQUEST, and X! Tandem that compare peaks in MS spectra with precomputed peptide fragments (Liska and Shevchenko, 2003). However, this approach is not efficient for identifying proteins that are not included in the database. In de novo sequencing, peptide sequences are obtained from mass spectra without the assistance of databases either manually or via computer programs (Ma and Johnson, 2012). Sequence homology search engines, such as MS driven BLAST (MS BLAST) have been successfully applied in marine proteomics studies. For example, Shevchenko et al. (2005) identified 48 proteins in larval venom of the Brazilian moth Cerodirphia speciosa. Waridel et al. (2007) employed multiple layered integrated search strategy that included MASCOT, de novo sequencing and MS BLAST sequence-similarity search to identify proteins in green alga Dunaliella salina. Sun et al. (2010) identified 65 proteins from golden apple snail P. canaliculata using de novo cross-species method. We combined conventional and customized transcriptome databases search with MS-BLAST sequence similarity search and identified 21 proteins from N. arenaceodentata (Chandramouli et al., 2012a). The de novo sequencing and MS BLAST have shortcomings because they use partial peptide sequences for database search that are very short sequence tags that can be used to search a standard database, such as NCBI nr, SwissProt, Uniprot etc. to identify the homologous peptide.
Species-Specific Genome Databases
The major limitation in conducting molecular studies at either the genome or proteome level is the lack of genome sequences (López, 2007; Tomanek, 2011; Slattery et al., 2012). For example, in our study we only managed to identify about 200 proteins in N. arenaceodentata (Chandramouli et al., 2013a). Several studies have addressed this problem by constructing transcriptome databases of marine species under investigation, which has remarkably improved the quality of proteomics dataset. As more genome information becomes available, proteomics will provide a wealth of information about molecular processes that are associated with larval behavior and morphology.
Validation of Proteomic Data
The current proteomic tools were established mainly for model organisms, they need to be selectively optimized for investigating larval proteome. The lack of commercially available species-specific antibodies makes it challenging to experimentally validate proteomics data in functional studies. To overcome this problem, He et al. (2012) generated B. amphitrite species-specific p38 antibodies and overexpressed them in E. coli. The authors demonstrated activation of Bar-p38 MAPK during larval settlement through co-immunoprecipitating p38 antibodies with interacting partner proteins. He et al. (2013) used the same method to generate antibodies for cement proteins, cp20k, to localize those proteins in barnacle cyprids. Zhang et al. (2012b) also used co-immunoprecipitation and SDS-PAGE-LC-MS/MS to identify the binding proteins of icocyanide in three marine invertebrates. Furthermore, there has been no matured method for bridging the gap between gene/protein expression and larval development and behavior in marine species. RNAi or gene knockouts methods may offer the solution but require substantial effort to develop these methods for marine species.
Conclusion
Over the last 7 years, gel-based and gel-free proteomics have been used to study molecular mechanisms of larval attachment and metamorphosis in several species of marine invertebrates. Notably, proteomics studies have provided new tools to systematically document the plasticity and variability of the proteome in larval morphogenesis and behavior as well as the response of larvae to antifoulants and environmental stressors. These larval proteomic investigations have also provided substantial information of protein expression patterns, PTMs, and signaling pathways during larval attachment and metamorphosis, which improved our understanding of possible molecular mechanisms of larval attachment and metamorphosis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region (662413) and an award from King Abdullah University of Science and Technology (SA-C0040/ UK-C0016).
References
Aguirre, J., Martin, J., Braga, J., Betzler, C., Berning, B., and Buckeridge, J. (2008). Densely packed concentrations of sessile barnacles (Cirripedia: Sessilia) from the Early Pliocene of SE Spain. Facies 54, 193–206. doi: 10.1007/s10347-007-0132-2
Amador-Cano, G., Carpizo-Ituarte, E., and Cristino-Jorge, D. (2006). Role of protein kinase C, G-protein coupled receptors, and calcium flux during metamorphosis of the sea urchin Strongylocentrotus purpuratus. Biol. Bull. 210, 121–131. doi: 10.2307/4134601
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Batalha, I. L., Lowe, C. R., and Roque, A. C. (2012). Platforms for enrichment of phosphorylated proteins and peptides in proteomics. Trends Biotechnol. 30, 100–110. doi: 10.1016/j.tibtech.2011.07.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Biggers, W. J., and Laufer, H. (1999). Settlement and metamorphosis of Capitella larvae induced by juvenile hormone-active compounds is mediated by protein kinase C and ion channels. Biol. Bull. 196, 187–198. doi: 10.2307/1542564
Billinghurst, Z., Clare, A. S., Matsumura, K., and Depledge, M. H. (2000). Induction of cypris major protein in barnacle larvae by exposure to 4-n-nonylphenol and 17 beta-oestradiol. Aquat. Toxicol. 47, 203–212. doi: 10.1016/S0166-445X(99)00018-1
Bishop, C. D., Bates, W. R., and Brandhorst, B. P. (2001). Regulation of metamorphosis in ascidians involves NO/cGMP signaling and HSP90. J. Exp. Zool. 289, 374–384. doi: 10.1002/jez.1019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bishop, C. D., and Brandhorst, B. P. (2001). NO/cGMP signaling and HSP90 activity represses metamorphosis in the sea urchin Lytechinus pictus. Biol. Bull. 201, 394–404. doi: 10.2307/1543617
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bishop, C. D., Huggett, M. J., Heyland, A., Hodin, J., and Brandhorst, B. P. (2006). Interspecific variation in metamorphic competence in marine invertebrates: The significance for comparative investigations into the timing of metamorphosis. Integr. Comp. Biol. 46, 662-682. doi: 10.1093/icb/icl043
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Blake, J. A., Grassle, J. P., and Eckelbarger, K. J. (2009). Capitella teleta, a new species designation for the opportunistic and experimental Capitella sp. I, with a review of the literature for confirmed records. Zoosymposia 2, 25–53.
Cantin, G. T., Yi, W., Lu, B., Park, S. K., Xu, T., Lee, J. D., et al. (2008). Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J. Proteome Res. 7, 1346–1351. doi: 10.1021/pr0705441
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chandramouli, K. H., Mok, F. S., Wang, H., and Qian, P.-Y. (2011b). Phosphoproteome analysis during larval development and metamorphosis in the spionid polychaete Pseudopolydora vexillosa. BMC Dev. Biol. 11:31. doi: 10.1186/1471-213X-11-31
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chandramouli, K. H., Ravasi, T., Reish, D., and Qian, P.-Y. (2013a). Proteomic changes between male and female worms of the polychaetous annelid Neanthes arenaceodentata before and after spawning. PLoS ONE 8:e72990. doi: 10.1371/journal.pone.0072990
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chandramouli, K. H., Reish, D., and Qian, P.-Y. (2012a). Gel-based and gel-free identification of proteins and phosphopeptides during egg-to-larva transition in polychaete Neanthes arenaceodentata. PLoS ONE 7:e38814. doi: 10.1371/journal.pone.0038814
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chandramouli, K. H., Soo, L., and Qian, P.-Y. (2011a). Differential expression of proteins and phosphoproteins during larval metamorphosis of the polychaete Capitella sp. I. Proteome Sci. 9:51. doi: 10.1186/1477-5956-9-51
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chandramouli, K. H., Sun, J., Mok, F. S., Liu, L., Qiu, J. W., Ravasi, T., et al. (2013b). Transcriptome and quantitative proteome analysis reveals molecular processes associated with larval metamorphosis in the polychaete Pseudopolydora vexillosa. J. Proteome Res. 12, 1344–1358. doi: 10.1021/pr3010088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chandramouli, K. H., Zhang, Y., Wong, Y. H., and Qian, P.-Y. (2012b). Comparative glycoproteome analysis: dynamics of protein glycosylation during metamorphic transition from pelagic to benthic life stages in three invertebrates. J. Proteome Res. 11, 1330–1340. doi: 10.1021/pr200982k
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Z., Zhang, H., Wang, H., Matsumura, K., Wong, Y. H., Ravasi, T., et al. (2014). Quantitative proteomics study of larval settlement in the barnacle Balanus amphitrite. PLoS ONE 9:e88744. doi: 10.1371/journal.pone.0088744
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Z. F., Matsumura, K., Wang, H., Arellano, S. M., Yan, X., Alam, I., et al. (2011). Toward an understanding of the molecular mechanisms of barnacle larval settlement: a comparative transcriptomic approach. PLoS ONE 6:e22913. doi: 10.1371/journal.pone.0022913
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Z. F., Wang, H., and Qian, P. Y. (2012). Characterization and expression of calmodulin gene during larval settlement and metamorphosis of the polychaete Hydroides elegans. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 162, 113–119. doi: 10.1016/j.cbpb.2012.04.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chenau, J., Michelland, S., Sidibe, J., and Seve, M. (2008). Peptides OFFGEL electrophoresis: a suitable pre-analytical step for complex eukaryotic samples fractionation compatible with quantitative iTRAQ labeling. Proteome Sci. 6:9. doi: 10.1186/1477-5956-6-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clare, A., Thomas, R., and Rittschof, D. (1995). Evidence for the involvement of cyclic AMP in the pheromonal modulation of barnacle settlement. J. Exp. Biol. 198, 655–664.
Clare, A. S. (1996). Signal transduction of barnacle settlement: calcium revisited. Biofouling 10, 141–159. doi: 10.1080/08927019609386276
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cohen, A. N. (2011). The Exotics Guide: Non-native Marine Species of the North American Pacific Coast. Richmond, CA: Center for Research on Aquatic Bioinvasions; Oakland, CA: San Francisco Estuary Institute.
Conradi, M., López-González, P. J., Cervera, J. L., and García-Gómez, J. C. (2000). Seasonality and spatial distribution of peracarids associated with the bryozoan Bugula neritina in Algeciras bay, Spain. J. Crust. Biol. 20, 334–349. doi: 10.1163/20021975-99990045
Dash, S., Chandramouli, K. H., Zhang, Y., and Qian, P.-Y. (2012). Effects of poly-ether B on proteome and phosphoproteome expression in biofouling Balanus amphitrite cyprids. Biofouling 28, 405–415. doi: 10.1080/08927014.2012.679731
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
DeBoer, M. L., Krupp, D. A., and Weis, V. M. (2007). Proteomic and transcriptional analyses of coral larvae newly engaged in symbiosis with dinoflagellates. Comp. Biochem. Physiol. 2, 63–73. doi: 10.1016/j.cbd.2006.11.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Gregoris, T. B., Rupp, O., Klages, S., Knaust, F., Bekel, T., Kube, M., et al. (2011). Deep sequencing of naupliar-, cyprid- and adult-specific normalized Expressed Sequence Tag (EST) libraries of the acorn barnacle Balanus amphitrite. Biofouling 27, 367–374. doi: 10.1080/08927014.2011.577211
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dupont, S., and Thorndyke, M. C. (2009). Impact of CO2-driven ocean acidification on invertebrates early life-history: what we know, what we need to know and what we can do. Biogeosci. Discuss. 6, 3109–3131. doi: 10.5194/bgd-6-3109-2009
Evans, C., Noirel, J., Ow, S. Y., Salim, M., Pereira-Medrano, A. G., Couto, N., et al. (2012). An insight into iTRAQ: where do we standnow? Anal. Bioanal. Chem. 404, 1011–1027. doi: 10.1007/s00216-012-5918-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fleck, J. (1997). “Phosphatidylinositol (PI) signaling and subsequent events in metamorphosis induction of cnidarian larvae,” in Proceedings, 8th International Coral Reef Symposium, Vol. 2 (Panama), 1225–1230.
Gilchrist, A., Au, C. E., Hiding, J., Bell, A. W., Fernandez-Rodriguez, J., Lesimple, S., et al. (2006). Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281. doi: 10.1016/j.cell.2006.10.036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glasby, C. J., and Timm, T. (2008). Global diversity of polychaetes (Polychaeta: Annelida) in freshwater. Hydrobiologia 595, 107–115. doi: 10.1007/s10750-007-9008-2
Greg, W. R., and Kristian, F. (1998). Recent views on the status, delineation, and classification of the Annelida. Am. Zool. 38, 953–964. doi: 10.1093/icb/38.6.953
Hadfield, M. G., Carpizo-Ituarte, E. U., Carmen, K., and Nedved, B. T. (2001). Metamorphic competence, a major adaptive convergence in marine invertebrate larvae. Am. Zool. 41, 1123–1131. doi: 10.1668/0003-1569(2001)041[1123:MCAMAC]2.0.CO;2
Han, Z., Sun, J., Zhang, Y., He, F., Xu, Y., Matsumura, K., et al. (2013). iTRAQ-based proteomic profiling of the barnacle Balanus amphitrite in response to the antifouling compound meleagrin. J. Proteome Res. 12, 2090–2100. doi: 10.1021/pr301083e
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haqqani, A. S., Kelly, J. F., and Stanimirovic, D. B. (2008). Quantitative protein profiling by mass spectrometry using label-free proteomics. Methods Mol. Biol. 439, 241–256. doi: 10.1007/978-1-59745-188-8_17
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
He, L. S., Xu, Y., Matsumura, K., Zhang, Y., Zhang, G., Qi, S. H., et al. (2012). Evidence for the involvement of p38 MAPK activation in barnacle larval settlement. PLoS ONE 7:e47195. doi: 10.1371/journal.pone.0047195
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
He, L. S., Zhang, G., and Qian, P.-Y. (2013). Characterization of two 20kDa-cement protein (cp20k) homologues in Amphibalanus amphitrite. PLoS ONE 8:e64130. doi: 10.1371/journal.pone.0064130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heyland, A., and Moroz, L. L. (2006). Signaling mechanisms underlying metamorphic transitions in animals. Integr. Comp. Biol. 46, 743–759. doi: 10.1093/icb/icl023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hondermarck, H. (2003). Breast cancer: when proteomics challenges biological complexity. Mol. Cell. Proteomics 2, 281–291. doi: 10.1074/mcp.R300003-MCP200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hunter, T. (1995). Protein kinase and phosphatase: the yin and yang of protein phosphorylation and signaling. Cell 80, 225–236. doi: 10.1016/0092-8674(95)90405-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ilan, M., Jensen, R. A., and Morse, D. E. (1993). Calcium control of metamorphosis in polychaete larvae. J. Exp. Zool. 267, 423–430. doi: 10.1002/jez.1402670408
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Inaba, K., Nomura, M., Nakajima, A., and Hozumi, A. (2007). Functional proteomics in Ciona intestinalis: a breakthrough in the exploration of the molecular and cellular mechanism of ascidian development. Dev. Dyn. 236, 1782–1789. doi: 10.1002/dvdy.21121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khandeparker, L., and Anil, A. C. (2007). Underwater adhesion: the barnacle way. Int. J. Adhes. Adhesives 27, 165–172. doi: 10.1016/j.ijadhadh.2006.03.004
Leise, E. M., Thavaradhara, K., Durham, N. R., and Turner, B. E. (2001). Serotonin and nitric oxide regulate metamorphosis in the marine snail Ilyanassa obsoleta. Am. Zool. 41, 258–267. doi: 10.1668/0003-1569(2001)041[0258:SANORM]2.0.CO;2
Leitner, A., Sturm, M., and Lindner, W. (2011). Tools for analyzing the phosphoproteome and other phosphorylated biomolecules: a review. Anal. Chim. Acta 703, 19–30. doi: 10.1016/j.aca.2011.07.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liska, A. J., and Shevchenko, A. (2003). Expanding the organismal scope of proteomics: cross-species protein identification by mass spectrometry and its implications. Proteomics 3, 19–28. doi: 10.1002/pmic.200390004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
López, J. L. (2007). Two-dimensional electrophoresis in proteome expression analysis. J. Chromatogr. 849, 190–202. doi: 10.1016/j.jchromb.2006.11.049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
López, J. L., Abalde, S. L., and Fuentes, J. (2005). Proteomic approach to probe for larval proteins of the mussel Mytilus galloprovincialis. Mar. Biotechnol. (NY) 7, 396–404. doi: 10.1007/s10126-004-4029-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, B., and Johnson, R. (2012). De novo sequencing and homology searching. Mol. Cell. Proteomics 11:O111.014902. doi: 10.1074/mcp.O111.014902
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marie, B., Marie, A., Jackson, D. J., Dubost, L., Degnan, B. M., Milet, C., et al. (2010). Proteomic analysis of the organic matrix of the abalone Haliotis asinina calcified shell. Proteome Sci. 8:54. doi: 10.1186/1477-5956-8-54
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martyniuk, C. J., Alvarez, S., Lo, B. P., Elphick, J. R., and Marlatt, V. L. (2012). Hepatic protein expression networks associated with masculinization in the female fathead minnow (Pimephales promelas). J. Proteome Res. 11, 4147–4161. doi: 10.1021/pr3002468
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maruzzo, D., Aldred, N., Clare, A. S., and Høeg, J. T. (2012). Metamorphosis in the cirripede crustacean Balanus amphitrite. PLoS ONE 7:e37408. doi: 10.1371/journal.pone.0037408
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Millioni, R., Tolin, S., Puricelli, L., Sbrignadello, S., Fadini, G. P., Tessari, P., et al. (2011). High abundance proteins depletion vs low abundance proteins enrichment: comparison of methods to reduce the plasma proteome complexity. PLoS ONE 6:e19603. doi: 10.1371/journal.pone.0019603
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mok, F. S., Thiyagarajan, V., and Qian, P.-Y. (2008). Larval development and metamorphic behaviour of subtropical spionid polychaete Pseudopolydora vexillosa. J. Exp. Mar. Biol. Ecol. 357, 99–108. doi: 10.1016/j.jembe.2007.12.029
Mok, F. S., Thiyagarajan, V., and Qian, P.-Y. (2009). Proteomic analysis during larval development and metamorphosis of the spionid polychaete Pseudopolydora vexillosa. Proteome Sci. 7:44. doi: 10.1186/1477-5956-7-44
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Molnár, E., Fábián, G., Klem, J., Darula, Z., Hunyadi-Gulyás, E., Medgyesi, A., et al. (2011). Removal of nonspecific binding proteins from cell and tissue extracts using 2-aminobenzimidazole-tethered affinity resin. Pharmazie 66, 662–665. doi: 10.1691/ph.2011.1027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moya, A., Huisman, L., Ball, E. E., Hayward, D. C., Grasso, L. C., Chua, C. M., et al. (2012). Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2-driven acidification during the initiation of calcification. Mol. Ecol. 21, 2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nomura, M., Nakajima, A., and Inaba, K. (2009). Proteomic profiles of embryonic development in the ascidian Ciona intestinalis. Dev Biol. 325, 468–481. doi: 10.1016/j.ydbio.2008.10.038
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pan, S., Chen, R., Aebersold, R., and Brentnall, T. A. (2011). Mass spectrometry based glycoproteomics–from a proteomics perspective. Mol. Cell. Proteomics 10:R110.003251. doi: 10.1074/mcp.R110.003251
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Panfoli, I., Calzia, D., Santucci, L., Ravera, S., Bruschi, M., and Candiano, G. (2012). A blue dive: from “blue fingers” to “blue silver.” A comparative overview of staining methods for in-gel proteomics. Expert Rev. Proteomics 9, 627–634. doi: 10.1586/epr.12.63
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Phillips, D. J. H., and Rainbow, P. S. (1988). Barnacles and mussels as biomonitors of trace elements: a comparative study. Mar. Ecol. Prog. Ser. 49, 83–93. doi: 10.3354/meps049083
Puente, L. G., Voisin, S., Lee, R. E., and Megeney, L. A. (2006). Reconstructing the regulatory kinase pathways of myogenesis from phosphopeptide data. Mol. Cell. Proteomics 5, 2244–2251. doi: 10.1074/mcp.M600134-MCP200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Putz, S., Reinders, J., Reinders, Y., and Sickmann, A. (2005). Mass spectrometry-based peptide quantification: applications and limitations. Expert Rev Proteomics. 2, 381–392. doi: 10.1586/14789450.2.3.381
Qian, P.-Y., Chen, L., and Xu, Y. (2013). Molecular mechanisms of antifouling compounds. Biofouling 29, 381–400. doi: 10.1080/08927014.2013.776546
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qian, P.-Y., Wong, Y. H., and Zhang, Y. (2010). Changes in the proteome and phosphoproteome expression in the bryozoan Bugula neritina larvae in response to the antifouling agent butenolide. Proteomics 10, 3435–3446. doi: 10.1002/pmic.201000199
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reish, D. J., and Gerlinger, T. V. (1997). A review of the toxicological studies with polychaetous annelids. Bull. Mar. Sci. 60, 584–607.
Romeo, M., Hoarau, P., Garello, G., Gnassia-Barelli, M., and Girard, J. P. (2003). Mussel transplantation and biomarkers as useful tools for assessing water quality in the NW Mediterranean. Environ. Pollut. 122, 369–378. doi: 10.1016/S0269-7491(02)00303-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rouse, G. W., and Fauchald, K. (1998). Recent views on the status, delineation, and classification of the Annelida. Am. Zool. 38, 953–964.
Shen, H., Li, X., Bieberich, C. J., and Frey, D. D. (2008). Reducing sample complexity in proteomics by chromatofocusing with simple buffer mixtures. Methods Mol. Biol. 424, 187–203. doi: 10.1007/978-1-60327-064-9_16
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shevchenko, A., de Sousa, M. M., Waridel, P., Bittencourt, S. T., de Sousa, M. V., and Shevchenko, A. (2005). Sequence similarity-based proteomics in insects: characterization of the larvae venom of the Brazilian moth Cerodirphia speciosa. J. Proteome Res. 4, 862–869. doi: 10.1021/pr0500051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Slattery, M., Ankisetty, S., Corrales, J., Marsh-Hunkin, K. E., Gochfeld, D. J., Willett, K. L., et al. (2012). Marine proteomics: a critical assessment of an emerging technology. J. Nat. Prod. 75, 1833–1877. doi: 10.1021/np300366a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, J., Mu, H., Zhang, H., Chandramouli, K. H., Qian, P.-Y., Wong, C. K., et al. (2013). Understanding the regulation of estivation in a freshwater snail through iTRAQ-based comparative proteomics. J. Proteome Res. 12, 5271–5280. doi: 10.1021/pr400570a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, J., Zhang, Y., Thiyagarajan, V., Qian, P.-Y., and Qiu, J. W. (2010). Protein expression during the embryonic development of a gastropod. Proteomics 10, 2701–2711. doi: 10.1002/pmic.200900846
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Taylor, P. D., Vinn, O., Kudryavtsev, A., and Schopf, J. W. (2010). Raman spectroscopic study of the mineral composition of cirratulid tubes (Annelida, Polychaeta). J. Struct. Biol. 171, 402–405. doi: 10.1016/j.jsb.2010.05.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ten Hove, H. A. (1974). Notes on Hydroideselegans (Haswell 1883) and Mercierellaenigmatica Fauvel 1923, alien serpulid polychaetes introduced into the Netherlands. Amsterdam: Bulletin Zoologisch Museum Universiteit van Amsterdam.
Thiyagarajan, V., Hung, O. S., Chiu, M. Y., Wu, R. S. S., and Qian, P.-Y. (2005). Growth and survival of juvenile barnacle Balanus amphitrite: interactive effects of cyprid energy reserve and habitat. Mar. Ecol. Prog. Ser. 299, 229-237. doi: 10.3354/meps299229
Thiyagarajan, V., Pechenik, J. A., Gosselin, L., and Qian, P.-Y. (2007). Juvenile growth in barnacles: combined effect of delayed metamorphosis and sub-lethal exposure of cyprids to low salinity stress. Mar. Ecol. Prog. Ser. 344, 173-184. doi: 10.3354/meps06931
Thiyagarajan, V., and Qian, P.-Y. (2008). Proteomic analysis of larvae during development, attachment, and metamorphosis in the fouling barnacle, Balanus amphitrite. Proteomics 8, 3164–3172. doi: 10.1002/pmic.200700904
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thiyagarajan, V., Wong, T., and Qian, P.-Y. (2009). 2D gel-based proteome and phosphoproteome analysis during larval metamorphosis in two major marine biofouling invertebrates. J. Proteome Res. 8, 2708–2719. doi: 10.1021/pr800976u
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tomanek, L. (2011). Environmental proteomics: changes in the proteome of marine organisms in response to environmental stress, pollutants, infection, symbiosis, and development. Ann. Rev. Mar. Sci. 3, 373–399. doi: 10.1146/annurev-marine-120709-142729
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ueda, N., and Degnan, S. M. (2014). Nitric oxide is not a negative regulator of metamorphic induction in the abalone Haliotis asinine. Front. Mar. Sci. 1:21. doi: 10.3389/fmars.2014.00021
Wang, H., and Qian, P.-Y. (2010). Involvement of a novel p38 mitogen activated protein kinase in larval metamorphosis of the polychaete Hydroides elegans (Haswell). J. Exp. Zool. B Mol. Dev. Evol. 314, 390–402. doi: 10.1002/jez.b.21344
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H., Zhang, H., Wong, Y. H., Voolstra, C., Ravasi, T., Bajic, V., et al. (2010). Rapid transcriptome and proteome profiling of a non-model marine invertebrate, Bugula neritina. Proteomics 10, 2972–2981. doi: 10.1002/pmic.201000056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Waridel, P., Frank, A., Thomas, H., Surendranath, V., Sunyaev, S., Pevzner, P., et al. (2007). Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics 7, 2318–2329. doi: 10.1002/pmic.200700003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wong, Y. H., Arellano, S. M., Zhang, H., Ravasi, T., and Qian, P.-Y. (2010). Dependency on de novo protein synthesis and proteomic changes during metamorphosis of the marine bryozoan Bugula neritina. Proteome Sci. 8:25. doi: 10.1186/1477-5956-8-25
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wong, Y. H., Wang, H., Ravasi, T., and Qian, P.-Y. (2012). Involvement of Wnt signaling pathways in the metamorphosis of the bryozoan Bugula neritina. PLoS ONE 7:e33323. doi: 10.1371/journal.pone.0033323
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Woollacott, R. M., and Zimmer, R. L. (2005). Attachment and metamorphosis of the cheilo- ctenostome bryozoan Bugula neritina (Linne). J. Morphol. 134, 351–382. doi: 10.1002/jmor.1051340307
Xu, Y., He, H. P., Schulz, S., Liu, X., Fusetani, N., Xiong, H. R., et al. (2010). Potent antifouling compounds produced by marine Streptomyces. Bioresour. Technol. 101, 1331–1336. doi: 10.1016/j.biortech.2009.09.046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, G., He, L. S., Wong, Y. H., and Qian, P.-Y. (2013). MKK3 was Involved in larval settlement of the barnacle Amphibalanus amphitrite through activating the kinase activity of p38MAPK. PLoS ONE 8:e69510. doi: 10.1371/journal.pone.0069510
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, H., Wong, Y. H., Wang, H., Chen, Z., Arellano, S. M., Ravasi, T., et al. (2011). Quantitative proteomics identify molecular targets that are crucial in larval settlement and metamorphosis of Bugula neritina. J. Proteome Res. 10, 349–360. doi: 10.1021/pr100817v
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., He, L. S., Zhang, G., Xu, Y., Lee, O. O., Matsumura, K., et al. (2012a). The regulatory role of the NO/cGMP signal transduction cascade during larval attachment and metamorphosis of the barnacle Balanus (=Amphibalanus) amphitrite. J. Exp. Biol. 215, 3813–3822. doi: 10.1242/jeb.070235
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Sun, J., Xiao, K., Arellano, S. M., Thiyagarajan, V., and Qian, P.-Y. (2010b). 2D gel-based multiplexed proteomic analysis during larval development and metamorphosis of the biofouling polychaete tubeworm Hydroides elegans. J. Proteome Res. 9, 4851–4860. doi: 10.1021/pr100645z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Sun, J., Zhang, H., Chandramouli, K. H., Xu, Y., He, L. S., et al. (2014). Proteomic profiling during pre-competent to competent transition of the biofouling polychaete Hydroides elegans. Biofouling 30, 921–928. doi: 10.1080/08927014.2014.951341
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Xu, Y., Arellano, S. M., Xiao, K., and Qian, P.-Y. (2010a). Comparative proteome and phosphoproteome analyses during cyprid development of the barnacle Balanus (=Amphibalanus) amphitrite. J. Proteome Res. 9, 3146–3157. doi: 10.1021/pr1000384
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y. F., Kitano, Y., Nogata, Y., Zhang, Y., and Qian, P.-Y. (2012b). The mode of action of isocyanide in three aquatic organisms, Balanus amphitrite, Bugula neritina and Danio rerio. PLoS ONE 7:e45442. doi: 10.1371/journal.pone.0045442
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, H., Liu, Y., Chui, J., Guo, K., Shun, Q., Lu, W., et al. (2007). Investigation on glycosylation patterns of proteins from human liver cancer cell lines based on the multiplexed proteomics technology. Arch. Biochem. Biophys. 459, 70–78. doi: 10.1016/j.abb.2006.10.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, J., Cai, Z. H., Li, L., Gao, Y. F., and Hutchinson, T. H. (2010). A proteomics based approach to assessing the toxicity of bisphenol A and diallyl phthalate to the abalone (Haliotis diversicolor supertexta). Chemosphere 79, 595–604. doi: 10.1016/j.chemosphere.2010.01.052
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, X. J., Zhang, Z., Xu, Y., Jin, C. L., He, H. P., Hao, X. J., et al. (2009). Flavone and isoflavone derivatives of terrestrial plants as larval settlement inhibitor of the barnacle Balanus amphitrite. Biofouling 25, 69–76. doi: 10.1080/08927010802455941
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: marine invertebrates, larvae metamorphosis, biofouling, proteomics, post-translational modifications
Citation: Chandramouli KH, Qian P-Y and Ravasi T (2014) Proteomics insights: proteins related to larval attachment and metamorphosis of marine invertebrates. Front. Mar. Sci. 1:52. doi: 10.3389/fmars.2014.00052
Received: 11 April 2014; Accepted: 22 September 2014;
Published online: 31 October 2014.
Edited by:
Andrew Stanley Mount, Clemson University, USAReviewed by:
Ying Zhang, University of Rhode Island, USAJian-Wen Qiu, Hong Kong Baptist University, Hong Kong
Copyright © 2014 Chandramouli, Qian and Ravasi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei-Yuan Qian, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong e-mail: boqianpy@ust.hk;
Timothy Ravasi, Integrative Systems Biology Lab, Division of Biological and Environmental Sciences and Engineering, Division of Applied Mathematics and Computer Sciences, King Abdullah University of Science and Technology, Building 2, Thuwal 23955-6900, Saudi Arabia e-mail: timothy.ravasi@kaust.edu.sa
 Kondethimmanahalli H. Chandramouli
Kondethimmanahalli H. Chandramouli Pei-Yuan Qian
Pei-Yuan Qian Timothy Ravasi
Timothy Ravasi