- 1Departamento de Genética Molecular de Plantas, Centro Nacional de Biotecnología- Consejo Superior de Investigaciones Científicas, Madrid, Spain
- 2The Botany Department, University of British Columbia, Vancouver, BC, Canada
- 3Centre for Plant Integrative Biology, University of Nottingham, Nottingham, UK
Plant adaptation, growth and development rely on the integration of many environmental and endogenous signals that collectively determine the overall plant phenotypic plasticity. Plant signaling molecules, also known as phytohormones, are fundamental to this process. These molecules act at low concentrations and regulate multiple aspects of plant fitness and development via complex signaling networks. By its nature, phytohormone research lies at the interface between chemistry and biology. Classically, the scientific community has always used synthetic phytohormones and analogs to study hormone functions and responses. However, recent advances in synthetic and combinational chemistry, have allowed a new field, plant chemical biology, to emerge and this has provided a powerful tool with which to study phytohormone function. Plant chemical biology is helping to address some of the most enduring questions in phytohormone research such as: Are there still undiscovered plant hormones? How can we identify novel signaling molecules? How can plants activate specific hormone responses in a tissue-specific manner? How can we modulate hormone responses in one developmental context without inducing detrimental effects on other processes? The chemical genomics approaches rely on the identification of small molecules modulating different biological processes and have recently identified active forms of plant hormones and molecules regulating many aspects of hormone synthesis, transport and response. We envision that the field of chemical genomics will continue to provide novel molecules able to elucidate specific aspects of hormone-mediated mechanisms. In addition, compounds blocking specific responses could uncover how complex biological responses are regulated. As we gain information about such compounds we can design small alterations to the chemical structure to further alter specificity, enhance affinity or modulate the activity of these compounds.
Introduction
From Phenotypes to Molecules: Early Chemical Genomics Approaches
Plant growth, development and adaptation to the environment require the integration of many environmental and endogenous signals that, together with the intrinsic genetic program, determine overall plant responses. In this context, signaling molecules and growth regulators, collectively known as phytohormones, act as central hubs for the integration of complex environmental and cellular signals. Phytohormones such as auxins, cytokinins (CK), gibberellins (GAs), abscisic acid (ABA), ethylene (ET), brassinosteroids (BRs) salicylic acid (SA), jasmonates (JAs), and strigolactones act at low concentrations and, either alone or in combination with other hormones, regulate multiple aspects of plant development, defense and adaptation. The search for both synthetic plant hormones and hormone mimics with increased stability/activity has been central to the development of the agrochemical industry and the “green revolution” in the past century (Brown et al., 2014). Initially, organic chemists used chemically synthesized hormonal derivatives to identify novel compounds mimicking or reversing the phenotypes induced by endogenous phytohormones. For example, the discovery of the structure of the naturally occurring auxin phytohormone indole-3-acetic acid (IAA) allowed chemical synthesis of a wide array of analogs and derivatives, and phenotypic screens. These approaches identified molecules such as 1-naphthaleneacetic acid (1-NAA), 2,4-dichlorophenoxyacetic acid (2,4-D), and 2-methyl-4-chlorophenoxyacetic acid (MCPA) that are still widely used today as growth promoters or herbicides (Overbeek and Vélez, 1946; Grossmann, 2010) (Supplemental Table 1). Similarly, phenotypic screens of functional analogs of the endogenous salicylic acid signals identified compounds such as benzothiadiazole (BTH) and 2,6-dichloroisonicotinic acid (INA) that were employed in the field to enhance plant disease resistance (Conrath et al., 1995; Görlach et al., 1996; Lawton et al., 1996) (Table 1 and Supplemental Table 1).
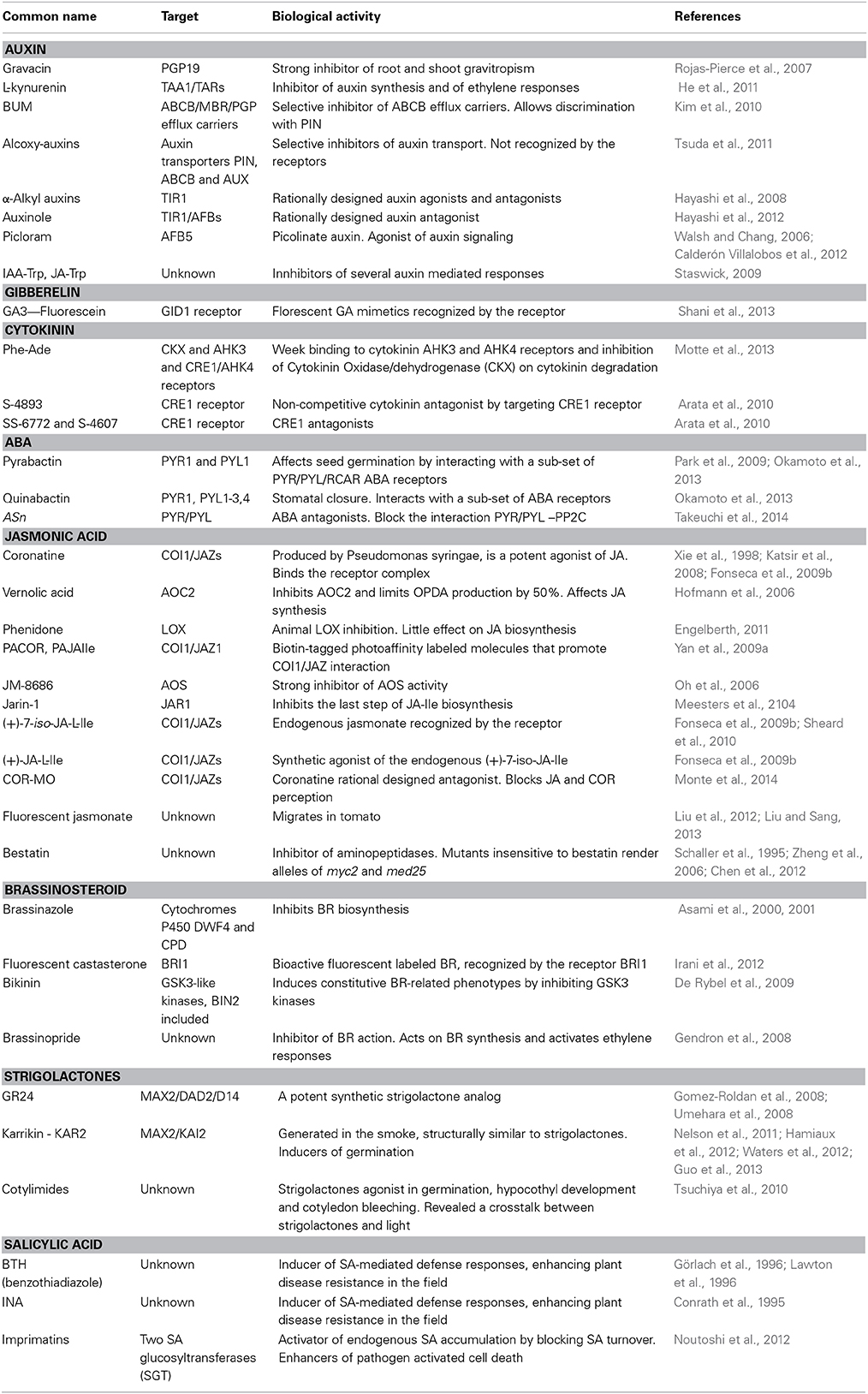
Table 1. List of molecules described in this review including molecular targets, biological activity and references.
A complementary chemical approach for the identification of bioactive molecules mimicking the activity of endogenous hormones can be based on the analysis of plant-interacting organisms. This approach revolves around organisms that have evolved the capability to produce phytohormones or phytohormone mimics to enhance disease susceptibility and counteract plant defenses. For example, characterization of the fungal pathogen Gibberella fujikuroi [responsible for the bakanae disease in rice (Kurosawa, 1926)] allowed the identification of gibberellic acid derived phytohormones (Shimada et al., 2008; Robert-Seilaniantz et al., 2011), and analysis of the bacterium Pseudomonas syringae pv. Tomato was instrumental for the identification of the phytotoxin coronatine (COR) (Feys et al., 1994). This is a jasmonate functional analog that the bacteria use to hijack the plant defense signaling network (Kloek et al., 2001; Brooks et al., 2004; Gimenez-Ibanez and Solano, 2013; Xin and He, 2013) (Table 1 and Supplemental Table 1).
Despite the profound contribution of those early chemical approaches in phytohormone research, these methodologies had two important limitations. Firstly, the success of these approaches relies on the serendipity of identifying a structurally amenable product from a relatively small number of natural sources. Secondly, the large collections of hormonal derivatives frequently lack chirality and their structural diversity is limited to variations in attachments within a restricted number of common skeletons (Brown et al., 2014). Therefore, these approaches only cover a small fraction of the structural possibilities present within the chemical space, and therefore reduce their potential versatility.
From Molecules to Function: Plant Chemical Biology in the Genetic Era
Recent decades have seen the development of a whole host of molecular and genetic tools as well as the release of complete genome sequences. Therefore, genetic strategies such as the isolation of mutations that confer altered hormonal responses and the identification of the downstream target genes have substituted the early chemical approaches and quickly became the preferred methods to elucidate the molecular mechanisms underlying phytohormone action. These genetic approaches have significantly enhanced our understanding of the molecular basis of phytohormone action (for review see Browse, 2009). In spite of its success, the use of well-established genetic tools (such as large collections of knockout and activation tagged mutants) for the identification of components in plant hormonal networks has now reached such a stage that it is becoming increasingly challenging to identify the remaining components. This recalcitrant to genetic approaches is largely due to a combination of gene redundancy, where multiple genes regulate the same process and individual knockouts have no discernable phenotype, and gene lethality, which prevents the identification of loss-of-function mutations in essential genes (Robert et al., 2009; Fernández-Calvo et al., 2011; Acosta et al., 2013).
Fortunately, the development of genetic tools has gone hand in hand with advances in combinatorial synthesis. These advances have enabled access to highly diverse chemical libraries containing both wider spectra of molecular shapes and range of biological targets than traditional combinatorial libraries (Schreiber, 2000; Hicks and Raikhel, 2012). These chemical libraries are being used to overcome many of the limitations of purely genetic approaches. They can be used to address genetic redundancy, as small molecules are capable of modulating the active sites of whole classes of protein targets. They can also address gene lethality, as small molecules can enable the temporal and spatial blockage of specific hormonal responses in a reversible manner (McCourt and Desveaux, 2010; Tóth and van der Hoorn, 2010; Hicks and Raikhel, 2012). Hence, in the last two decades agrochemical biased libraries have been widely used in combined genetic and chemical screens aimed at the dissection of multiple physiological processes in plants. These screens have yielded valuable bioactive compounds such as gravacin (Rojas-Pierce et al., 2007), morlin (DeBolt et al., 2007), sortins (Zouhar et al., 2004; Rosado et al., 2011), hypostatin (Zhao et al., 2007), and endosidins (Robert et al., 2008; Drakakaki et al., 2011) (Figure 1 and Table 1). All these compounds are currently used to modify the activity of individual proteins or protein families in a tuneable, reversible and spatial-temporal controlled manner.
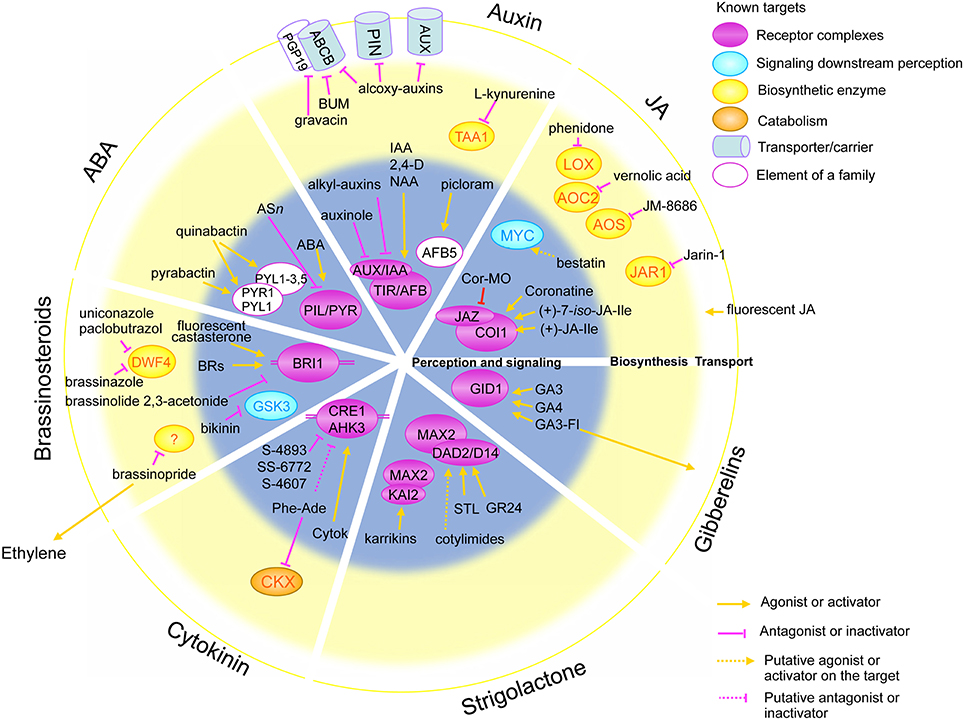
Figure 1. Schematic representation of the molecular targets of small molecules acting in different hormonal pathways. Concentric circles in the background represent the distinct biological processes in hormonal pathways: perception and signaling (gray inner circle), biosynthesis (yellow middle circle) and transport (white outer circle). Circles are divided in quadrants for distinct hormones, from the top clockwise: auxin, jasmonic acid (JA), gibberellins, strigolactones, cytokinins, brassinosteroids and abscisic acid (ABA). Ovals represent the molecular targets: receptor complexes (violet), signaling components (blue), biosynthetic enzyme (yellow) and catabolic enzymes (orange). Cylindrical shapes represent transporters and carriers. Molecules acting as activators are represented with an orange arrow toward their targets, whereas pink blocked arrows highlight antagonists and inhibitor molecules.
We now know that in some cases the mechanisms for perceiving individual hormones are conserved, and the same recognition systems are able to mediate response to several hormones, while in other cases unique perception strategies have evolved for individual molecules (Tan et al., 2007; Murase et al., 2008; Shimada et al., 2008; Park et al., 2009; Lumba et al., 2010; Sheard et al., 2010; Kumari and van der Hoorn, 2011). A paradigmatic example of a conserved recognition system is the “molecular glue” mechanism, first described for the auxin receptor complex, in which the auxin molecule promotes the formation of its receptor complex (Tan et al., 2007; Mockaitis and Estelle, 2008). The F-box TIR1 (TRANSPORT INHIBITOR RESPONSE 1) or AFBs (AUXIN SIGNALING F-BOX) cannot bind, or bind at very low affinity, auxin without the interaction of the co-receptors Aux/IAA (AUXIN RESISTANT/INDOLE-3-ACETIC ACID INDUCIBLE) and the inositol hexakisphosphate (IP6) cofactor. Only the structural modifications produced by the formation of the tetrameter stabilize the hormone perception. The same mechanism also occurs in jasmonate perception, since the hormone induces the formation of the receptor tetramer complex formed by JA-Ile, the F-box COI1 (CORONATINE INSENSITIVE1), the co-receptor JAZ (JASMONATE ZIM DOMAIN PROTEIN) and the inositol pentakisphosphate (IP5) cofactor (Chini et al., 2007; Thines et al., 2007; Sheard et al., 2010). Gibberellins are also sensed by a similar perception system: active GAs promote the establishment of the complex formed by GID1 (GIBBERELLIN INSENSITIVE DWARF1) receptor and the F-box SLY1 (SLEEPY1) (Murase et al., 2008; Shimada et al., 2008). In contrast, other phytohormones are perceived by specific protein complex based on different recognition systems. For example, the PYR1 (PYRABACTIN RESISTANT 1) and PYL (PYRABACTIN RESISTANT-LIKE) receptors bind ABA directly in cooperation with the co-receptors type 2C protein phosphatases, such as ABI1 (ABA INSENSITIVE 1) and ABI2 (ABA INSENSITIVE 2). The subsequent inactivation of the phosphatases induces the SNF1-type kinase activity, which in turn regulates ABA-dependent gene expression and downstream signaling cascades (Weiner et al., 2010). CK perception and signal transduction pathway occur through a phosphorelay similar to bacterial two-component response systems. Briefly, CK binds directly to the membrane-located HISTIDINE KINASE (AHK) receptors. This initiates a phosphorelay cascade where a phosphoryl group is translocated via the HISTIDINE-CONTAINING PHOSPHOTRANSFER PROTEINS (AHPs) and then to the RESPONSE REGULATOR (ARRs) transcription factors. Type-B ARRs regulate the transcription of cytokinin responsive genes and type-A ARRs acting as negative feedback regulators to desensitize plants to excess cytokinin (Kieber and Schaller, 2014).
The discovery of each of these hormonal response mechanisms has enabled the implementation of innovative chemical genomics approaches, and the rational design of chemical tools for phytohormone studies. These will be described in details within this review.
From Function to Targets: Screening for Novel Proteins/Complex Using Screens and Tagged-Molecules
Bioactive chemicals identified from forward or reverse chemical screens are very useful for the dissection of complex biological processes. One advantage of this technique is that it can either target specific proteins or multiple members of redundant gene families. However, the identification of the cognate biochemical target/s remains a very complex process that depends on the type and affinity of the chemical-target interaction, as well as the abundance of the target sites (Robert et al., 2009). Throughout the years, researchers have performed diverse genetic screens in Arabidopsis thaliana for resistance to specific chemicals. These have allowed the subsequent biochemical and genetic identification of cognate targets. For example, chemical genetic screens for resistance to bikinin (an activator of the brassinosteroids responses) showed that it could bind directly and inhibit a subset of the GSK3 (GLYCOGEN SYNTHASE KINASE 3) kinase family (De Rybel et al., 2009) (Table 1 and Supplemental Table 1). Similarly, a screen for resistance to gravacin (a strong inhibitor of root and shoot gravitropism) identified the auxin efflux transporter PGP19 (P-GLYCOPROTEIN 19) as its molecular target (Rojas-Pierce et al., 2007) (Table 1 and Supplemental Table 1).
These genetic-based approaches require further validation of the target since mutations can prevent the drug from reaching the site of action due to either metabolic alterations or uptake/translocation defects. As an alternative, different biochemical tools have been developed for target identification (Kolb and Sharpless, 2003). These include collections of “tagged” chemical libraries that possess reactive moieties permitting the immobilization of active compounds through “click chemistry.” Although there are several potential click reactions, the Copper (I) catalyzed synthesis of triazoles from azides and acetylenes has become the standard for the generation of “click libraries” and the chemical species in those libraries possess an amine handle that enables affinity resin synthesis via reaction with activated carboxylic acid affinity resins (Kolb et al., 2001). For example, a library of tagged molecules was used in a high throughput approach to detect active proteins in the whole proteome of Arabidopsis thaliana (Van der Hoorn et al., 2011). Additionally these compounds can also contain a fluorophore to enable visualization of hits in living cells or other contexts.
In the following sections of this review we will describe recent landmark chemical genomics approaches and place special emphasis on their roles in the elucidation of the molecular mechanisms underlying hormonal regulation, considering all stages from the biosynthesis to the perception of the signal.
Phytohormone Homeostasis
Different endogenous and environmental stimuli regulate the tissue-specific biosynthesis of phytohormones. The synthesis and catabolism of these molecules are tightly regulated as they are very bio-active. For example at least three, partially redundant, biosynthetic pathways have been identified so far for synthesizing auxin (Stepanova et al., 2008; Zhao, 2008). The complete biosynthetic network is not yet fully understood. However, the use of auxin analogs played an important role in identifying many of the mutants impaired in auxin biosynthesis. For example, the tir2 (transport inhibitor response 2) mutant was isolated as an NPA (1-N-naphthylphthalamic acid)-resistant mutant and subsequently shown to encode TAA1 (TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1), one of the key enzymes in the indole-3-pyruvic acid (IPA) auxin biosynthetic pathway (Yamada et al., 2009). However, mutants with modestly perturbed levels in auxin show strong pleiotropic effects and this restrict their usefulness in investigating specific aspects of auxin action. In addition, the enzymes that mediate key biosynthetic steps are often redundant and require the generation of higher order combinations of mutants to detect observable phenotypes. Therefore, the identification of compounds enabling the manipulation or blockage of specific biosynthetic pathways is invaluable. An example of such compound is the recently isolated inhibitor of the auxin biosynthesis L-kynurenine (He et al., 2011) (Figure 1, Table 1 and Supplemental Table 1).
L-kynurenine (Kyn) was originally identified as an inhibitor of the ethylene-induced auxin biosynthesis in roots (He et al., 2011) (Figure 1). Subsequently, He and colleagues demonstrated that Kyn is an alternate substrate for auxin biosynthetic enzymes TAA1/TAR (TRYPTOPHAN AMINOTRANSFERASE RELATED) and that it competitively inhibits TAA1/TAR activity (Stepanova et al., 2008). Strikingly, Kyn binds to the substrate pocket of TAA1/TAR proteins in a highly effective and selective manner, but does not bind to other aminotransferases. The use of Kyn has overcome some genetic limitations of traditional approaches, such as the sterility and lethality of double and triple mutants in the redundant TAA1/TAR gene family, and has enabled the blockage of auxin biosynthesis in specific tissues or plant stages (Stepanova et al., 2008). Kyn has added value to classic genetic studies. For example, by combining Kyn treatments with mutants impaired in auxin biosynthesis, it was recently shown that root-based auxin biosynthesis is required in addition to polar auxin transport to correctly pattern the root xylem axis (Ursache et al., 2014). Most enzymes within the auxin biosynthetic network are well conserved between plant species, including mosses and lichens (Finet and Jaillais, 2012). Consequently, molecules such as kynurenine that inhibit auxin biosynthesis could easily be used on other species, providing a wide range of possible applications.
Many molecules regulate the complex signaling networks responsible for plant defense, however salicylic acid plays a central role in restricting the activity of biotrophic pathogens. Genetic screens for mutants with enhanced disease resistance have mainly uncovered dwarf mutants with elevated SA levels (Murray et al., 2002; Shirano et al., 2002; Grant et al., 2003; Zhang et al., 2003; Vlot et al., 2009). To avoid the pleiotropic effects of plants with altered SA levels, researchers have long sought-after compounds enabling the manipulation of SA in a tuneable and reversible manner. Recently, a high-throughput chemical genomic screen identified the imprimatin family of molecules as enhancers of pathogen-activated cell death (Noutoshi et al., 2012). Imprimatin treatments induce the accumulation of SA, reduce its inactive metabolite SA-glucoside, and enhance plant disease resistance (Table 1 and Supplemental Table 1). Further analyses have shown that imprimatins block SA turnover through specific inhibition of two SA GLUCOSYLTRANSFERASES (SAGTs). Double knockout mutants of these SAGTs are semi-dwarf plants that consistently showed the same SA-accumulation and enhanced disease resistance as imprimatin-treated plants (Noutoshi et al., 2012). Imprimatins offer an exciting way in which synthetic compounds that can be applied to different plant species to trigger accumulation of the active endogenous SA and overcome the pleiotropic effects associated with constitutive high levels of SA. Besides the biotechnological applications, these molecules can also be used in phytohormone research to induce the accumulation of endogenous SA transiently at specific plant developmental stages, avoiding the need to use of semi-dwarf mutant lines in the redundant SAG genes.
Cytokinins have been long known to regulate cell division, differentiation as well as many aspects of plant development—including root growth, root/shoot branching architecture and vascular development (Werner and Schmülling, 2009; Hwang et al., 2012). Cytokinins are adenine derivatives, and the incorporation of specific side chains at the N6-position triggers their recognition as ligands for specific receptors or substrates for enzymes regulating their homeostasis. One key group of enzymes catalyzing the oxidative removal of the side chain and thereby degrading cytokinins are the CYTOKININ OXIDASE/DEHYDROGENASE (CKX) family. In Arabidopsis there are 7 members of the CKX family, and each has subtly different substrate specificity (Kowalska et al., 2010). A recent high-throughput chemical screen based on in-vitro cytokinin-dependent shoot regeneration (Motte et al., 2013) identified one novel compound, Phe-Ade (N-phenyl-9H-purin-6-amine) (Figure 1, Table 1 and Supplemental Table 1). Further biochemical studies showed that Phe-Ade induces the accumulation of endogenous cytokinin by acting as a competitive inhibitor of the cytokinin-degrading CKX enzymes and preventing cytokinin catabolism.
Brassinosteroid biosynthesis is regulated by a complex network of three redundant pathways that convert the common precursor campesterol into the active BRs. The BR biosynthetic pathways requires the activity of the cytochrome P450 DWARF 4 (DWF4), a key rate limiting P450 monooxygenase that acts on multiple intermediates in the BR biosynthesis pathways (Asami et al., 2000, 2001; Chung and Choe, 2013) and represents an ideal target to bypass the redundancy of the BR biosynthesis pathways. Uniconazole and paclobutrazol are both inhibitors of P450 monooxygenases that act as weak BR inhibitors and are able to induce accumulation of the precursor campesterol (Asami and Yoshida, 1999) (Figure 1). Subsequent analysis into the structure-activity relationship identified brassinazole (BRZ) as strong inhibitor of BR biosynthesis blocking the cytochrome P450 monooxygenase DWF4 and therefore preventing the hydroxylation of BR precursors (Asami and Yoshida, 1999; Asami et al., 2000) (Table 1 and Supplemental Table 1). BRZ was subsequently used for a genetic screen to isolate BRZ insensitive mutants. bzr1-1D (brassinazole resistant 1) and bes1 (bri1-ems-suppressor 1) mutants respectively showed insensitivity to BRZ and enhanced constitutive BR responses. The phenotypes of these mutants were later shown to be caused by the stabilization of the transcription factors BZR1 (BRASSINAZOLE RESISTANT 1) and BES1/BZR2 (BRI1-EMS-SUPPRESSOR 1/BRASSINAZOLE RESISTANT 1) (Wang et al., 2002; Yin et al., 2005a). BZR1 and BES1/BZR2 are the fundamental activators of the BR signaling pathway, which regulate the expression of most BR responsive genes (Vert and Chory, 2006). The use of BZR is exemplary of the potential of integrating chemical genomics with classical genetics to identify key regulators of a hormone signaling pathway.
Jasmonic acid-isoleucine (JA-Ile) is an end product of the oxylipin biosynthetic pathway and, together with additional oxylipin molecules, it mediates several developmental processes and stress responses (Fonseca et al., 2009a; Wasternack and Hause, 2013). The oxylipin biosynthetic pathway is well understood and several inhibitors of key steps in this pathway have been reported (Wasternack and Hause, 2013). The JA-Ile biosynthesis is believed to start with the conversion of free α-linolenic acid by 13-lipoxygenases. Therefore, several general inhibitors of animal lipoxygenases (such as phenidone, aspirin, ibuprofen and ursolic acid) were tested in plants; however, they show only with limited inhibitory effects on oxylipin biosynthesis in plants, possibly due to functional redundancy or differences between animal and plant lipoxygenases (Wasternack, 1993; Farmer, 1995; Engelberth, 2011).
The subsequent biosynthetic step is catalyzed by the ALLENE OXYDE SYNTHASE (AOS) and ALLENE OXYDE CYCLASE 2 (AOC2), that mediate a non-redundant, coupled reaction producing the first cyclic oxylipin 12-oxo-phytodienoic acid (OPDA). The complete loss of cyclic oxylipins in aos1 mutant generates sterile plants and confirmed the essential role of AOS (Park et al., 2002; Wasternack and Hause, 2013). Therefore, AOS and AOC2 represent ideal targets to inhibit the whole cyclic oxylipin pathway. Vernolic acid is a naturally occurring oxylipin first described as competitive inhibitor of the AOC of maize by Hamberg and Fahlstadius (1990). More recently, the crystal structure of AOC2 determined the direct binding of the competitive JA inhibitor vernolic acid within the AOC2 hydrophobic barrel cavity (Hofmann et al., 2006) (Table 1 and Supplemental Table 1). Biochemical assays also demonstrated that vernolic acid inhibited approximately 50% of the AOC2-mediated production of OPDA in vitro. In addition, the imidazole derivative JM-8686 was designed to inhibit the activity of AOS, most likely by direct binding of the imidazole group of JM-8686 to the heme iron of AOS (Oh et al., 2006). However, the subsequent use of vernolic acid and JM-8686 was very limited because the residual activity of AOS/AOC2 coupled reaction can produce enough cyclic oxylipin to mediate most plant responses.
The final step of the biosynthetic pathway is performed by JASMONOYL-L-ISOLEUCINE SYNTHETASE (JAR1), that synthesizes the bioactive hormone (+)-7-iso-JA-Ile by conjugating JA with the amino acid isoleucine (Staswick and Tiryaki, 2004; Fonseca et al., 2009a). Very recently, Meesters et al. reported jarin-1 as the first small molecule inhibitor of jasmonate responses identified in a chemical screen (Meesters et al., 2104). Jarin-1 inhibits many JA-mediated responses in planta, but did not affect reactions induced by JA-Ile, suggesting an inhibitory activity on the JA-Ile producing enzyme JAR1. Further biochemical data confirmed that jarin-1 impairs the JA-Ile synthesis and inhibits the activity of JAR1, whereas closely related enzymes are not affected. Molecular modeling suggests a direct jarin-1 binding to the active site of JAR1. Overall, jarin-1 is the first direct, specific inhibitor of JAR1 (Meesters et al., 2104) (Figure 1, Table 1 and Supplemental Table 1).
Phytohormone Transport
In plants, most hormones are mobile molecules whose inter or intra-cellular transport is required for function and control of physiological responses. With the textbook exception of auxin polar transport, the molecular mechanisms and components of hormone transport are still relatively unknown. In the case of auxins, genetics and chemistry both played essential roles in identifying and characterizing the three families of auxin transporters, AUX1/LAX (AUXIN RESISTANT 1/LIKE AUXIN RESISTANT), ABCB/MDR/PGP (ATP-BINDING CASSETTE subfamily B/ MULTIDRUG RESISTANCE/ P-GLYCOPROTEIN) and PINs (PIN-FORMED). For example, the aux1 mutant was isolated through exploring the permeability differences between the membrane-permeable auxin 1-NAA and the membrane-impermeable auxin analog 2,4-D (Figure 1, Table 1 and Supplemental Table 1). The aux1 mutant was discovered through its agravitropic phenotype that could only be rescued by 1-NAA (Bennett et al., 1996). AUX1 was subsequently characterized as the first IAA influx carrier (Marchant et al., 1999; Swarup et al., 2001; Yang et al., 2006). Some members of the proteins ABCB/MDR/PGP transporters have been identified as proteins with binding affinity to the auxin transport inhibitor 1- naphthylphthalamic acid (NPA) (Noh et al., 2001; Robert and Friml, 2009; Ma and Robert, 2014) (Figure 1). The initial identification of the PIN family of auxin efflux carriers occurred through the genetic isolation of the pin1 mutant, which shows a phenotype resembling that caused by the pharmacological inhibition of polar auxin transport (Okada et al., 1991; Gälweiler, 1998).
Recently, a chemical genomic screen based on phenotyping a suite of morphological traits such as growth rate and flowering time identified a novel and potent inhibitor of ABCB efflux carriers, BUM (2-[4-(diethylamino)-2-hydroxybenzoyl]benzoic acid). BUM directly binds and inhibits ABCBs, although ABCB1 appears to be the primary target. This binding occurs without directly affecting PIN transporters, and therefore allows the specific analysis discriminating between PIN and ABCB efflux systems (Kim et al., 2010).
Auxin perception allows regulation of the intracellular accumulation of endogenous auxins by modifying the localization of several transporters. As a consequence, it is often difficult to uncouple auxin perception from auxin transport. To overcome this limitation, rationally designed molecules such as alkoxy-IAA derivates (alkoxy-auxins) were developed that specifically target auxin transporters (Tsuda et al., 2011) (Figure 1, Table 1 and Supplemental Table 1). Structural modeling testing the docking of alkoxy-auxins to the TIR1-Aux/IAA receptor suggested that these molecules could not fit into the auxin-binding pocket of the TIR1. It has been shown experimentally that these molecules fail to interfere with auxin perception, Aux/IAA degradation, and the downstream auxin signaling pathway (Tsuda et al., 2011). In contrast, alkoxy-auxins block the auxin transport activity of the PIN, ABCB, and AUX1 transporters in both yeast assays and in planta. Therefore, alkoxy-auxins are meant to become important tools to uncouple perception and transport in complex auxin mediated processes (Tsuda et al., 2011; Ma and Robert, 2014).
Long distance transport has also been reported for several hormones, but the molecular mechanisms are just emerging. ABA, cytokinin, strigolactones and jasmonates were detected in phloem or xylem, suggesting that these molecules could either be actively extruded from the cell or simply cross membranes by diffusion into the vascular tissue (Thorpe et al., 2007; Kudo et al., 2010; Kohlen et al., 2011). As in the case of auxins, small molecules provide useful tools to analyse the transport of other hormones. For example, specific ABC transporters inhibitors such as glibenclamide, verapamil and vanadate have been used to confirm role of the proteins AtABCG25 and AtABCG40 as ABA transporters (Kuromori et al., 2010; Kang et al., 2010).
New evidence exists that gibberellins too are actively transported; Shani and colleagues (2013) synthesized fluorescein labeled GA molecules (GA4- and GA3-fluorescein) that could be visualized in root cells and preferentially accumulate in the endodermal cells (Figure 1, Table 1 and Supplemental Table 1). By using mitochondrial ATP synthesis inhibitors such as antimycin A, oligomycin A and myxothiazol, the researchers demonstrated the specific GA accumulation in the endodermis relies on active, energy dependent mechanisms, suggesting an active GA transport (Shani et al., 2013).
The idea of cytokinin-specific transporters is still an open question (Bishopp et al., 2011a). Podlešáková et al. (2012) generated a series of novel analogs of cytokinin and observed that some of these compounds had different transport affinities, hinting at the possibility of identifying immobile CK analogs. The structure-activity analysis of these immobile CK as well as the identification of their targets might help to define components of the CK transport system.
Wounding triggers systemic responses that depend on the de novo synthesis of JA and JA-Ile in distal leaves in Arabidopsis (Koo et al., 2009; Wasternack and Hause, 2013), whereas grafting experiments with mutants excluded systemic formation of JA in tomato (Li et al., 2002; Koo and Howe, 2009). In principle this advocates against the transport of JA or JA-Ile. However, using radioactively labeled molecules, Me-JA, JA and JA-Ile were all found in phloem and/or xylem (Baldwin and Zhang, 1997; Thorpe et al., 2007; Matsuura et al., 2012). In addition, a functional fluorescent-labeled jasmonate probe was reported to migrate in the vascular tissues of tomato plants (Liu et al., 2012; Liu and Sang, 2013). We envision that the development of fluorescent-labeled hormones combined with the use of chemicals inhibiting different transport mechanisms will be essential tools with which to address the transport of jasmonates (Rigal et al., 2014). Many hypotheses have been proposed to explain the nature of systemic wound signals, being electric signals a possibility, and recently glutamate receptor-like genes (GLR), similar to those involved in synaptic activity in animals, have been implicated (Mousavi et al., 2013). In addition, three GLR antagonists were identified through a pharmacological screen for molecules inhibiting the growth of tobacco pollen tubes. Furthermore, the analysis of the GLR agonistic amino acids showed that D-serine is the most active agonist promoting pollen tube growth. D-serine is secreted naturally by the pistil to mediate pollen tube guidance (Michard et al., 2011). As D-serine is a modulator of animal neuronal circuits, this finding shows an astonishing analogy between electrochemical signal transduction in plants and animals.
Phytohormone Perception
Phytohormones are active at very low concentrations due to their high-affinity recognition systems. Since perception is the first step for the activation of downstream signaling cascades, researchers have prioritized the identification hormone receptors and perception components. Although many components of the hormonal perception system were identified by classical genetic approaches, the use of phytohormone analogs and chemical genomics was important for the detailed dissection of the underlying molecular mechanisms through which they function. For example, NPA was instrumental in identifying several components of the auxin pathway. These include TIR1, the founder member of the auxin receptor family TIR1/AFB proteins (Ruegger et al., 1997; Mockaitis and Estelle, 2008).
Coronatine, the bacterial mimic of JA-Ile, was instrumental in the identification of the coi1 (coronatine insensitive 1) mutant. It was subsequently discovered that coi1 was impaired in the F-box component of the JA-Ile receptor (Xie et al., 1998; Sheard et al., 2010). In addition, a small-scale screen of oxylipins, JA precursors and derivatives identified the synthetic isomer (+)-JA-L-Ile as a strong jasmonate agonist (Fonseca et al., 2009b). The structure of coronatine and the synthetic (+)-JA-L-Ile suggests that the stereochemical orientation of the cyclopentanone-ring side chains greatly affects receptor binding. Purification of the two natural epimers demonstrated that pure (−)-JA-L-Ile is inactive and that the active hormone is (+)-7-iso-JA-L-Ile, which is structurally more similar to coronatine (Fonseca et al., 2009b). Besides, the activity of COI1 as the JA-Ile receptor was first demonstrated by using radiolabeled coronatine in competitive binding assays (Katsir et al., 2008). To assess the direct binding of jasmonates to the COI1 receptor, biotin-tagged photoaffinity probes of JAs were designed (Yan et al., 2009a). The coronatine photoaffinity probe (PACOR), which retained weak biological activities, physically binds with the purified COI1 protein, further supporting that COI1 directly binds to COR and serves as a receptor for jasmonate (Yan et al., 2009a). All of these results show clearly the importance of JA-Ile analogs in several of the most important advances in phytohormone research.
The Redundancy/Specificity Paradox of Hormone Receptors
Chemical genomic studies can also be used to address the striking receptor redundancy/specificity paradox. Many components of hormone receptor complexes belong to large gene families and are functionally redundant. For example, the Arabidopsis genome encodes 14 PYR/PYL genes and 12 JAZ genes (Chini et al., 2007; Thines et al., 2007; Park et al., 2009). Although members of these families regulate the same hormone-mediated responses, individual members confer some tissue- and process-specificity.
The auxin perception complex shows the greatest redundancy of all the pathways discussed in this review. It is composed of one F-box member (among the 6 possible TIR1/AFB proteins), one co-receptor (among the 29 possible Aux/IAAs) alongside the single IP6 cofactor. The identification of auxin analogs has helped to address both the redundancy and specificity of various components within the auxin perception machinery. For example, mutations in the auxin receptor, AFB5, were identified in a genetic screen for lines resistant to the picolinate auxin (Walsh and Chang, 2006). afb5 is highly resistant to picolinate auxins (such as picloram or DAS534) but not to other auxin isoforms such as 2,4-D or IAA (Table 1 and Supplemental Table 1). This suggests that picolinate is a highly specific agonist of the auxin pathway (Walsh and Chang, 2006). Interestingly exogenous application of picloram mimics some aspects of auxin responses that application of 2,4-D or IAA application fails to reproduce, such as hypocotyl elongation. Although TIR1 and AFB5 show an almost identical secondary structure, biochemical analyses show that the TIR1–IAA7 and AFB5–IAA7 co-receptor complexes exhibit different auxin-binding affinities (Figure 1). Indeed, a single amino acid substitution has been identified through docking analyses that is responsible for the change in affinities of TIR1 and AFB5 for IAA and picloram (Calderón Villalobos et al., 2012). These data demonstrate that the AFB5-Aux/IAA co-receptor selectively binds picloram, but not IAA, whereas TIR1-Aux/IAA accepts IAA, but not picloram, providing the first mechanistic explanation for specificity in auxin perception.
From Molecules to Functions: the Power of Chemical Genomics
Chemical genomics approaches have also been instrumental in the discovery of the redundant ABA receptors, as different compounds show specificity to certain groups of receptors. Pyrabactin was originally identified as a synthetic inhibitor of only one ABA-mediated response, seed germination (Zhao et al., 2007). A screen was performed for pyrabactin-resistant mutants aiming to identify redundant components of the ABA pathway (Cutler and McCourt, 2005). Indeed, single pyrabactin resistant mutants (pyr) were sensitive to ABA, whereas only multiple mutants in PYR1/PYR1-like (PYL) genes exhibited ABA insensitivity, demonstrating the functional redundancy of family members (Park et al., 2009). Additional studies using small molecules assessed the structural requirements of the binding pocket of the PYR/PYL receptors (Cao et al., 2013; Okamoto et al., 2013). For example, pyrabactin binds and activates two of the PYR/PYL receptors, while quinabactin activates three additional PYR/PYLs (Table 1 and Supplemental Table 1). Since pyrabactin affects ABA-related processes in seeds and quinabactin regulates ABA-dependent stomatal closure, these chemicals are shedding light on the partially redundant functions of the PYR/PYL ABA receptors (Figure 1).
In the case of cytokinin, a chemical genomic approach was employed to identify molecules antagonizing the activity of the cytokinin receptor CRE1 (CYTOKININ RESPONSE 1; Arata et al., 2010). The authors elegantly generated a yeast system based on the Arabidopsis CRE1 gene conferring cytokinin dependent growth. This system allowed a high-throughput screen looking for growth defects in yeast grown in the presence of cytokinin, and identified two compounds (SS-6772 and S-4607) that inhibited the CRE1-dependent yeast growth (Table 1 and Supplemental Table 1). These compounds were chemically quite distinct from previous reported cytokinin receptor antagonists and new variations of these compounds were generated introducing minor modification of the quinazoline ring (Spíchal et al., 2009; Arata et al., 2010; Nisler et al., 2010). A new antagonist, S-4893, was confirmed as a strong inhibitor of cytokinin signaling in both yeast system and in planta. Further biochemical and genetic studies revealed that S-4893 acts as a non-competitive inhibitor of CRE1 not only in Arabidopsis but also in rice, suggesting that this compound operates in a range of plant species to antagonize cytokinin-mediated processes (Figure 1).
Perception of BR occurs at the plasma membrane by the receptor BRASSINOSTEROID INSENSITIVE (BRI1). In order to investigate endocytosis of the receptor-ligand complex, researchers developed a bioactive fluorescent labeled BR, called fluorescent castasterone (AFCS) (Irani et al., 2012) (Figure 1, Table 1 and Supplemental Table 1). They used this tool to show that trafficking and endocytosis of the BRI1-AFCS complex is dependent on clathrin, ARF GTPases and the Rab5 GTPase pathway. However, concanamycin A, a specific inhibitor of the trans-Golgi network/early endosome (TGN/EE) blocked the BRI1-AFCS complex at the TGN/EE without affecting the BR signaling. The integration of these chemical and genetic data showed that retention of active BRI1 at the plasma membrane, rather than in endosomes, is an important factor in activation of BR signaling.
The recent identification of many components of several phytohormone receptor complexes opens the opportunity to generate new molecular tools. Most plant co-receptor complexes are able to perceive their targets in heterologous systems such as yeast. For example, yeast two hybrid (Y2H) systems have been used to induce the formation of TIR1-Aux/IAA complex in an auxin-inducible manner (Calderón Villalobos et al., 2012) and in a similar way JA-Ile or COR promotes COI1-JAZ interaction in yeast (Fonseca et al., 2009a; Chini, 2014). As the hormone co-receptors are the only plant proteins expressed within these heterologous systems, they represent unique tools to identify small molecules directly perturbing the hormone perception. Compounds able to induce the formation of the perception complex can subsequently be used to identify novel active forms of the hormone. In contrast, compounds inhibiting the hormone-dependent co-receptor complex might be direct antagonist molecules.
From Receptor Structures to Molecules: Rational Design of Phytohormone Analogs
In the last decade, the crystal structures of several perception complexes were solved (Tan et al., 2007; Murase et al., 2008; Shimada et al., 2008; Park et al., 2009; Sheard et al., 2010). These structural data open new opportunities for the rational design of antagonist molecules specifically binding to and blocking the active pockets of individual receptors. The methodology of ligand-based rational design has been exploited extensively in medical research, but is just emerging in the agrochemical field (Lamberth et al., 2013). For example, this methodology has permitted the rational design of alfa-alkyl auxin molecules (Figure 1, Table 1 and Supplemental Table 1). These auxin analogs are able to specifically bind and block the formation of the hormone receptor complex was very successful and have allowed systematic structure-activity analysis of the alfa-position of IAA (Hayashi et al., 2008, 2012) (Figure 1). An advanced modification of one of these compounds generated the auxinole molecule (alfa-[2,4-dimethylphenylethyl-2-oxo]-IAA). This binds TIR1 strongly to block the formation of the TIR1-IAA-Aux/IAA receptor complex. Molecular docking studies have provided novel insights of the molecular mechanism of auxinole activity, predicting that auxinole strongly interacts with the Phe82 residue. This residue of TIR1 that is crucial for Aux/IAA recognition and blocks TIR1 activity by interacting with this critical amino acid. Hayashi et al. showed that auxinole and alfa-alkyl auxin molecules retain their antagonistic activity in crop plants such as tomato as well as in distant relatives, such as the moss Physcomitrella patens (Hayashi et al., 2008, 2012).
The same principle of rational design used around the crystal structure of the COI1/JAZ co-receptor to design a COR-derivative that binds to COI1 but spatially impedes the interaction of the COI-JAZ co-receptors (Figure 1). This compound, COR O-methyloxime (COR-MO), shows a strong activity inhibiting the formation of the COI1-JAZ perception complex and preventing JAZ degradation (Monte et al., 2014). COR-MO reverses the effects induced by exogenous JA-Ile or COR treatments on several JA-mediated responses efficiently, thereby underpinning its usefulness in dissecting the JA-pathway (Table 1 and Supplemental Table 1). Moreover, COR-MO enhances plant defense by preventing the effectiveness of the bacterial effector COR during Pseudomonas syringae infections. As this compounds works in a variety of different plant species, it further highlights the potential of such compounds in biotechnological and agronomical processes (Monte et al., 2014) (Figure 1).
In contrast to JA-Ile and auxins, which act as molecular glues by holding receptor complexes together, ABA binds within a cavity in its receptor where it induces conformational changes that in turn promote the interaction with the active site of group-A PROTEIN PHOSPHATASE 2C (PP2Cs) (Melcher et al., 2010). Following the resolution of the crystal structure of several ABA/PYR/PP2C complexes, Takeuchi and colleagues designed a series of ABA analogs (ASn) with long alkyl chains of the ABA 3′ ring CH, that they predicted would spatially block the PYL-PP2C interaction (Table 1 and Supplemental Table 1). A six-carbon alkyl chain was sufficient to produce a potent ABA antagonist able to block multiple ABA-mediated responses in vivo such as germination, the expression of known downstream response genes and PP2A activity (Takeuchi et al., 2014) (Figure 1).
Brassinolide (BL) is a potent brassinosteroid that binds the BR receptor BRI1 directly and induces the interaction between BRI1 and SERK1 (SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1; Santiago et al., 2013). Based on the crystal structure of the BRI1-BL complex Muto and colleagues generated a alkylated version of BL called brassinolide-2,3-acetonide. This compound was able to bind BRI1 but sterically interferes with the SRRK1 interaction (Muto and Todoroki, 2013) (Figure 1, Table 1). Indeed, brassinolide-2,3-acetonide showed a clear BL antagonistic effect in rice seedlings and opens the opportunity to develop a set of chemical tools modulating BR perception and further dissect the BR response pathway.
Collectively, these examples of antagonist molecules highlight the usefulness of the structure-based design of hormone analogs specifically binding for and blocking the active pocket of the receptors. This approach provides a novel methodology for generating bioactive hormone analogs.
Specificity and Redundancy
Another important contribution of chemical genomic screens is the possibility to assess specificity within signaling pathways or specific developmental processes. Essentially this notion is based on the fact that chemicals can overcome functional redundancy by inhibiting multiple members of a redundant protein family (Cutler and McCourt, 2005). A good example described earlier is pyrabactin, a compound affecting a single ABA-mediated response, germination (Zhao et al., 2007). The analyses of the first pyrabactin resistant (pyr) and further pyr/pyl mutants revealed nicely the functional redundancy of the 14-member PYR/PYL family for multiple ABA responses (Park et al., 2009).
Bikinin was identified in a screen for molecules inducing constitutive BR-related phenotypes such as hypocotyl elongation, petiole elongation and pale, blade shaped leaves (De Rybel et al., 2009). Strikingly, bikinin induces BR responses in mutants deficient in BR biosynthesis, perception and signaling. Bikinin also stimulates BR responses in bin2-1, a gain of function mutation in BIN2 (BRASSINOSTEROID-INSENSITIVE2). BIN2 is a GSK3 (GLYCOGEN SYNTHASE KINASE3) kinase that phosphorylates and inactivates the key transcription factors in the BR pathway, BZR1 and BES1/BZR2 (He et al., 2002). Bikinin acts as a competitive inhibitor of ATP binding and binds BIN2 directly causing the inhibition of seven of the 10 GSK3 kinases (Vert and Chory, 2006; De Rybel et al., 2009; Yan et al., 2009b). One bikinin-inhibited GSK3 kinases, ASK θ, interacts directly with and phosphorylates BEH2 (BES1/BZR1 HOMOLOG 2), a BR responsive transcription factor closely related to BZR1 and BES1/BZR2 (Yin et al., 2005b; Rozhon et al., 2010). Therefore, the discovery of bikinin allowed the identification of new components of the BR pathway and also enabled the conditional blockage of multiple key regulators in BR signaling, providing an essential tool to study the BR regulatory mechanisms.
Bestatin is an inhibitor of aminopeptidase and powerful inducer of JA- and wound-response genes in tomato (Schaller et al., 1995). The root growth inhibitory effect of bestatin depends on the key transcription factor of the JA pathway MYC2 but seems independent of the JA-Ile receptor COI1 (Figure 1). Therefore, Zheng et al. (2006) used bestatin to identify new components of the wounding signaling pathway dependent on JA-Ile and MYC2. Several bestatin resistant mutants (ber) were isolated, some of which allelic to jin1/myc2. In addition, ber6 carries a mutation in MED25/PFT1 (MEDIATOR 25/PHYTOCHROME AND FLOWERING TIME 1). This gene encodes for a subunit of the eukaryotic transcription mediator system (Zheng et al., 2006). MED25/PFT1 was first described as a positive regulator of shade avoidance and has subsequently been shown to also be required for plant defense (Cerdán and Chory, 2003; Kidd et al., 2009). Recent studies showed that MYC2, MYC3 and MYC4 have redundant roles in plant defense; MED25 directly interacts with MYC2 and it is required for MYC2-dependent pathogen defense (Fernández-Calvo et al., 2011; Çevik et al., 2012; Chen et al., 2012; Schweizer et al., 2013). MYC2, MYC3, and MYC4 are regulated by light quality and are involved in shade avoidance responses (Robson et al., 2010; Chico et al., 2014). Therefore, the use of bestatin to isolate mutants in MED25/PFT1 suggested the redundant role of the MYC2, MYC3, and MYC4 in defense and shade avoidance responses. The use of bestatin can potentially identify new regulators of the MYCs and help to assess redundancy.
Strigolactones have long been studied because of their importance in stimulating the growth of the parasitic Striga and Orobanche on several crops. Structure-activity relationship analyses showed that several analogs mimic strigolactone functions (reviewed by Janssen and Snowden, 2012). Different structural requirements regulate strigolactone-mediated processes such as seed germination, hyphal branching of arbuscular mycorrizal fungi and shoot branching inhibition (Kondo et al., 2007; Zwanenburg et al., 2009; Akiyama et al., 2010; Fukui et al., 2011, 2013; Boyer et al., 2012; reviewed by Zwanenburg and Pospísil, 2013). Furthermore, newly synthesized strigolactone competitive analogs suggest that Arabidopsis, Orobanche and arbuscular mychorrial fungi possess variations in the sensitivity to strigolactone analogs, providing additional support to the idea that variations in strigolactone receptors among the different species should exist (Cohen et al., 2013; reviewed by Janssen and Snowden, 2012).
Karrikins are compounds structurally similar to strigolactones. They promote germination, but unlike strigolactones, karrikins are not produced in plants, but instead are found in the smoke of burning plant material. Despite this, in many ways they behave as hormones, as small quantities of the signal is sufficient to trigger a signal transduction pathway. Genetic screenings for karrikin insensitive mutants showed that karrikins perception share a common mechanism with strigolactones. The F-box MAX2 (MORE AXILLARY BRANCHES 2) is required to perceive both kinds of compounds. In both cases an alfa/beta hydrolase fold protein [KAI2 (KARRIKIN INSENSITIVE 2) or DAD2/D14] is part of the receptor complex (Nelson et al., 2011; Hamiaux et al., 2012; Waters et al., 2012; Guo et al., 2013). The strigolactone and karrikin pathways are a good example of how structurally similar molecules rely on similar—or even common—perception mechanisms and confer overlapping physiological responses while maintaining their identity in terms of structure-function (Figure 1, Table 1 and Supplemental Table 1).
Phytohormone Crosstalk
It has been well documented that most biological processes are not regulated by a single hormone but rather by complex signaling networks controlled by multiple hormones or other signaling components. For example, auxin and cytokinin act in consort to control the formation of the embryonic root, root meristem size, root branching, vascular pattern and shoot phylotaxy (Dello Ioio et al., 2008; Bishopp et al., 2011b; Besnard et al., 2014). Chemical genomic approaches not only facilitate the dissection of hormonal pathways but could also shed light on the non-linear networks within which they operate as well as identify new downstream biological functions. For example, screening for compounds that affect gravitropism led to the identification of multiple chemicals that affect membrane trafficking in auxin dependent and independent manners (Surpin et al., 2005). Such important and sometimes unexpected results demonstrate the power of screens for small molecules regulating related biological processes.
Recently, a chemical genomic screen for molecules perturbing germination identified cotylimides, a class of compounds structurally similar to strigolactones that recapitulate cotyledon bleaching promoted by GR24 (Tsuchiya et al., 2010). The subsequent screen for mutants insensitive to cotylimides isolated several suppressor lines showing elongated hypocotyls, a phenotype commonly observed in mutants defective in light signaling. The analysis of strigolactones in light responses showed that these compounds mimic light in seedling growth and increase the accumulation of HY5 (ELONGATED HYPOCOTYL5), a protein directly targeted by COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC 1) for degradation. Tsuchiya and colleagues elegantly revealed the molecular mechanism behind this response. Strigolactones mediate the nuclear exclusion of COP1, this stabilizes HY5, which in turn reduces hypocotyl elongation.
As described earlier in this review, L-kynurenine was identified in a chemical genomic screen. This compound was instrumental in showing that auxin induces the nuclear accumulation of the key activator of the ET pathway, EIN3 (ETHYLENE-INSENSITIVE3). Kyn was used to unravel a positive feedback loop between auxin biosynthesis and ET signaling (He et al., 2011), that was not detectable by conventional genetic analysis.
Brassinopride (BRP) was identified in a screen for inhibitors of brassinosteroids, based on the inhibition of BR-mediated hypocotyl growth in the dark and activation of the BR-responsive reporter gene CPD:GUS (Gendron et al., 2008) (Figure 1). The site of action of BRP has not been defined yet, however, application of brassinolide can reverse BR related effects of BRP. This suggests that BRP could perturb BR biosynthesis. Unexpectedly, BRP also induced exaggerated formation of apical hooks, resembling plant subjected to ET treatments. Additionally, the apical hook phenotype could be blocked by a chemical inhibitor of ethylene perception or by ET insensitive mutants, suggesting that BRP activates ET responses. Phenotypic analysis of ET and BR mutants in combination with analysis of the effects of BRP analogs, revealed a crosstalk between ET and BR in etiolated seedlings. Moreover, variation among BRP analogs suggest that modifying the side groups of BRP can have specific effects on BR or ET functions (Gendron et al., 2008).
Recently, it has also been proposed that hormone derivates can interfere with different signaling pathways (Katsir et al., 2008; Staswick, 2009; Gutierrez et al., 2012). For example, hormone conjugation is a common process in plants to activate, store or deactivate phytohormones, however, some conjugates show unexpected activity. When combined with either auxin or JA, tryptophan (Trp) conjugates with indole-3-acetic acid (IAA-Trp) or with jasmonic acid (JA-Trp). These conjugates act as an inhibitors of auxin responses, preventing several auxin-mediated responses, such as gravitropism, lateral root production and expression of known auxin response genes (Staswick, 2009). The evidence that an endogenous JA derivate inhibits the auxin pathway adds a novel level of regulation between the jasmonate and auxin pathways. It was hypothesized that IAA-Trp and JA-Trp could directly interfere with the auxin receptor TIR1-IAA/Aux as the TIR1 is required for IAA-Trp and JA-Trp inhibition and because these conjugated compounds are structurally similar to the active forms. However, IAA-Trp and JA-Trp do not directly alter the IAA-dependent interaction between TIR1 and Aux/IAA7 and their precise mode of action remains unknown (Staswick, 2009).
Future Prospectives
Plant chemical biology has enabled significant advances in phytohormone research. In addition to the classical phytohormone analogs widely used by the scientific community, we have recently witnessed the extensive use of chemical genomic approaches. The straightforward availability of large, diverse chemical libraries and the natural chemical resources will surely facilitate a further extension of this methodology and the identification of novel compounds regulating many biological processes of hormone synthesis, transport and response. Complementary, rational design of novel molecules and molecule labeling are emerging but successful strategies.
Therefore, we envision the continued use of plant chemical biology not only to identify novel components or regulation mechanisms of phytohormone pathways but also to better understand their mode of action and molecular networks. The discovery of molecules with certain specificity constrains will certainly contribute toward more rational and sustainable agriculture systems.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to those authors whose excellent contribution in the field could not be cited here due to space limitation. This work was partially funded by the National Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant RGPIN-2014-06468 to Abel Rosado and funding from the Canada Research Chairs program. John Vaughan-Hirsch is funded through the BBSRC Doctoral Training Programme. Anthony Bishopp is supported by the Royal Society through a University Research Fellowship. Andrea Chini is supported by a “Ramon y Cajal” fellowship (RYC-2010-05680) and this work was partially funded the Ministerio de Economía y Competitividad project BIO2013-44407-R, the AECID AP/040886/11 and the CSIC i-COOP060. This project was supported by NSTIP strategic technologies programs, number (11-BIO-2119-02) in the Kingdom of Saudi Arabia to Andrea Chini.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00709/abstract
References
Acosta, I. F., Gasperini, D., Chételat, A., Stolz, S., Santuari, L., and Farmer, E. E. (2013). Role of NINJA in root jasmonate signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 15473–15478. doi: 10.1073/pnas.1307910110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Akiyama, K., Ogasawara, S., Ito, S., and Hayashi, H. (2010). Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 51, 1104–1117. doi: 10.1093/pcp/pcq058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arata, Y., Nagasawa-Iida, A., Uneme, H., Nakajima, H., Kakimoto, T., and Sato, R. (2010). The phenylquinazoline compound S-4893 is a non-competitive cytokinin antagonist that targets Arabidopsis cytokinin receptor CRE1 and promotes root growth in Arabidopsis and rice. Plant Cell Physiol. 51, 2047–2059. doi: 10.1093/pcp/pcq163
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Asami, T., Min, Y. K., Nagata, N., Yamagishi, K., Takatsuto, S., Fujioka, S., et al. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123, 93–100. doi: 10.1104/pp.123.1.93
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Asami, T., Mizutani, M., Fujioka, S., Goda, H., Min, Y. K., Shimada, Y., et al. (2001). Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J. Biol. Chem. 276, 25687–25691. doi: 10.1074/jbc.M103524200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Asami, T., and Yoshida, S. (1999). Brassinosteroid biosynthesis inhibitors. Trends Plant Sci. 4, 348–353.
Baldwin, I. T., and Zhang, Z.-P. (1997). Transport of [2- 14 C]jasmonic acid from leaves to roots mimics wound-induced changes in endogenous jasmonic acid pools in Nicotiana sylvestris. Planta 203, 436–441. doi: 10.1007/s004250050211
Bennett, M. J., Marchant, A., Green, H. G., May, S. T., Ward, S. P., Millner, P. A., et al. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950. doi: 10.1126/science.273.5277.948
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Besnard, F., Refahi, Y., Morin, V., Marteaux, B., Brunoud, G., Chambrier, P., et al. (2014). Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505, 417–421. doi: 10.1038/nature12791
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bishopp, A., Benková, E., and Helariutta, Y. (2011b). Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr. Opin. Plant Biol. 14, 10–16. doi: 10.1016/j.pbi.2010.08.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bishopp, A., Lehesranta, S., Vaten, A., Help, H., El-Showk, S., Scheres, B., et al. (2011a). Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 21, 927–932. doi: 10.1016/j.cub.2011.04.049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boyer, F.-D., de Saint Germain, A., Pillot, J.-P., Pouvreau, J.-B., Chen, V. X., Ramos, S., et al. (2012). Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol. 159, 1524–1544. doi: 10.1104/pp.112.195826
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brooks, D. M., Hernández-Guzmán, G., Kloek, A. P., Alarcón-Chaidez, F., Sreedharan, A., Rangaswamy, V., et al. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant. Microbe. Interact. 17, 162–174. doi: 10.1094/MPMI.2004.17.2.162
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, M. Q., Rosado, A., and Raikhel, N. V. (2014). “From herbal remedies to cutting-edge science, a historical perspective of plant chemical biology,” in Plant Chemical Biology, ed D. Audenaert (Hoboken, NJ: John Wiley & Sons, Inc), 1–17.
Browse, J. (2009). The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 70, 1539–1546. doi: 10.1016/j.phytochem.2009.08.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Calderón Villalobos, L. I. A., Lee, S., De Oliveira, C., Ivetac, A., Brandt, W., Armitage, L., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485. doi: 10.1038/nchembio.926
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cao, M., Liu, X., Zhang, Y., Xue, X., Zhou, X. E., Melcher, K., et al. (2013). An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants. Cell Res. 23, 1043–1054. doi: 10.1038/cr.2013.95
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cerdán, P. D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423, 881–885. doi: 10.1038/nature01636
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Çevik, V., Kidd, B. N., Zhang, P., Hill, C., Kiddle, S., Denby, K. J., et al. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160, 541–555. doi: 10.1104/pp.112.202697
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, R., Jiang, H., Li, L., Zhai, Q., Qi, L., Zhou, W., et al. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24, 2898–2916. doi: 10.1105/tpc.112.098277
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chico, J.-M., Fernández-Barbero, G., Chini, A., Fernández-Calvo, P., Díez-Díaz, M., and Solano, R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26, 1967–1980. doi: 10.1105/tpc.114.125047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chini, A. (2014). Application of yeast-two hybrid assay to chemical genomic screens: a high-throughput system to identify novel molecules modulating plant hormone receptor complexes. Methods Mol. Biol. 1056, 35–43. doi: 10.1007/978-1-62703-592-7_4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chung, Y., and Choe, S. (2013). The Regulation of Brassinosteroid Biosynthesis in Arabidopsis. CRC. Crit. Rev. Plant Sci. 32, 396–410. doi: 10.1080/07352689.2013.797856
Cohen, M., Prandi, C., Occhiato, E. G., Tabasso, S., Wininger, S., Resnick, N., et al. (2013). Structure-function relations of strigolactone analogs: activity as plant hormones and plant interactions. Mol. Plant 6, 141–152. doi: 10.1093/mp/sss134
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conrath, U., Chen, Z., Ricigliano, J. R., and Klessig, D. F. (1995). Two inducers of plant defense responses, 2,6-dichloroisonicotinec acid and salicylic acid, inhibit catalase activity in tobacco. Proc. Natl. Acad. Sci. U.S.A. 92, 7143–7147. doi: 10.1073/pnas.92.16.7143
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cutler, S., and McCourt, P. (2005). Dude, where's my phenotype? Dealing with redundancy in signaling networks. Plant Physiol. 138, 558–559. doi: 10.1104/pp.104.900152
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
DeBolt, S., Gutierrez, R., Ehrhardt, D. W., and Somerville, C. (2007). Nonmotile cellulose synthase subunits repeatedly accumulate within localized regions at the plasma membrane in Arabidopsis hypocotyl cells following 2,6-dichlorobenzonitrile treatment. Plant Physiol. 145, 334–338. doi: 10.1104/pp.107.104703
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dello Ioio, R., Nakamura, K., Moubayidin, L., Perilli, S., Taniguchi, M., Morita, M. T., et al. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384. doi: 10.1126/science.1164147
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Rybel, B., Audenaert, D., Vert, G., Rozhon, W., Mayerhofer, J., Peelman, F., et al. (2009). Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16, 594–604. doi: 10.1016/j.chembiol.2009.04.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Drakakaki, G., Robert, S., Szatmari, A.-M., Brown, M. Q., Nagawa, S., Van Damme, D., et al. (2011). Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 17850–17855. doi: 10.1073/pnas.1108581108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Engelberth, J. (2011). Selective inhibition of jasmonic acid accumulation by a small α, β-unsaturated carbonyl and phenidone reveals different modes of octadecanoid signalling activation in response to insect elicitors and green leaf volatiles in Zea mays. BMC Res. Notes 4:377. doi: 10.1186/1756-0500-4-377
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Farmer, E. E. (1995). Fatty acid signalling in plants and their associated microorganisms. Plant Mol. Biol. 5, 1423–1437. doi: 10.1007/BF00016483
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J.-M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feys, B., Benedetti, C. E., Penfold, C. N., and Turner, J. G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 5, 751–759. doi: 10.1105/tpc.6.5.751
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Finet, C., and Jaillais, Y. (2012). Auxology: when auxin meets plant evo-devo. Dev. Biol. 369, 19–31. doi: 10.1016/j.ydbio.2012.05.039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fonseca, S., Chico, J. M., and Solano, R. (2009a). The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12, 539–547. doi: 10.1016/j.pbi.2009.07.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., et al. (2009b). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fukui, K., Ito, S., and Asami, T. (2013). Selective mimics of strigolactone actions and their potential use for controlling damage caused by root parasitic weeds. Mol. Plant 6, 88–99. doi: 10.1093/mp/sss138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fukui, K., Ito, S., Ueno, K., Yamaguchi, S., Kyozuka, J., and Asami, T. (2011). New branching inhibitors and their potential as strigolactone mimics in rice. Bioorg. Med. Chem. Lett. 21, 4905–4908. doi: 10.1016/j.bmcl.2011.06.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gälweiler, L. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. doi: 10.1126/science.282.5397.2226
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gendron, J. M., Haque, A., Gendron, N., Chang, T., Asami, T., and Wang, Z. Y. (2008). Chemical genetic dissection of brassinosteroid-ethylene interaction. Mol. Plant 1, 368–379. doi: 10.1093/mp/ssn005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gimenez-Ibanez, S., and Solano, R. (2013). Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front. Plant Sci. 4:72. doi: 10.3389/fpls.2013.00072
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pagès, V., Dun, E. A., Pillot, J.-P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189–194. doi: 10.1038/nature07271
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Görlach, J., Volrath, S., Knauf-Beiter, G., Hengy, G., Beckhove, U., Kogel, K. H., et al. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8, 629–643. doi: 10.1105/tpc.8.4.629
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grant, J. J., Chini, A., Basu, D., and Loake, G. J. (2003). Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol. Plant. Microbe. Interact. 16, 669–680. doi: 10.1094/MPMI.2003.16.8.669
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grossmann, K. (2010). Auxin herbicides: current status of mechanism and mode of action. Pest Manag. Sci. 66, 113–120. doi: 10.1002/ps.1860
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guo, Y., Zheng, Z., La Clair, J. J., Chory, J., and Noel, J. P. (2013). Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 8284–8289. doi: 10.1073/pnas.1306265110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gutierrez, L., Mongelard, G., Floková, K., Pacurar, D. I., Novák, O., Staswick, P., et al. (2012). Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24, 2515–2527. doi: 10.1105/tpc.112.099119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hamberg, M., and Fahlstadius, P. (1990). Allene oxide cyclase: a new enzyme in plant lipid metabolism. Arch. Biochem. Biophys. 276, 518–526. doi: 10.1016/0003-9861(90)90753-L
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hamiaux, C., Drummond, R. S. M., Janssen, B. J., Ledger, S. E., Cooney, J. M., Newcomb, R. D., et al. (2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036. doi: 10.1016/j.cub.2012.08.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hayashi, K.-I., Tan, X., Zheng, N., Hatate, T., Kimura, Y., Kepinski, S., et al. (2008). Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 5632–5637. doi: 10.1073/pnas.0711146105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hayashi, K., Neve, J., Hirose, M., Kuboki, A., Shimada, Y., Kepinski, S., et al. (2012). Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem. Biol. 7, 590–598. doi: 10.1021/cb200404c
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
He, J. X., Gendron, J. M., Yang, Y., Li, J., and Wang, Z. Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 10185–10190. doi: 10.1073/pnas.152342599
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
He, W., Brumos, J., Li, H., Ji, Y., Ke, M., Gong, X., et al. (2011). A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23, 3944–3960. doi: 10.1105/tpc.111.089029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hicks, G. R., and Raikhel, N. V. (2012). Small molecules present large opportunities in plant biology. Annu. Rev. Plant Biol. 63, 261–282. doi: 10.1146/annurev-arplant-042811-105456
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hofmann, E., Zerbe, P., and Schaller, F. (2006). The crystal structure of Arabidopsis thaliana allene oxide cyclase: insights into the oxylipin cyclization reaction. Plant Cell 18, 3201–3217. doi: 10.1105/tpc.106.043984
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hwang, I., Sheen, J., and Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Irani, N. G., Di Rubbo, S., Mylle, E., Van den Begin, J., Schneider-Pizon, J., Hnilikova, J., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8, 583–589. doi: 10.1038/nchembio.958
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Janssen, B. J., and Snowden, K. C. (2012). Strigolactone and karrikin signal perception: receptors, enzymes, or both? Front. Plant Sci. 3:296. doi: 10.3389/fpls.2012.00296
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kang, J., Hwang, J.-U., Lee, M., Kim, Y.-Y., Assmann, S. M., Martinoia, E., et al. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 2355–2360. doi: 10.1073/pnas.0909222107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y., and Howe, G. A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 105, 7100–7105. doi: 10.1073/pnas.0802332105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kidd, B. N., Edgar, C. I., Kumar, K. K., Aitken, E. A., Schenk, P. M., Manners, J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252. doi: 10.1105/tpc.109.066910
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kieber, J. J., and Schaller, G. E. (2014). Cytokinins. Arab. B. 2014:e0168. doi: 10.1199/tab.0168
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J.-Y., Henrichs, S., Bailly, A., Vincenzetti, V., Sovero, V., Mancuso, S., et al. (2010). Identification of an ABCB/P-glycoprotein-specific Inhibitor of Auxin Transport by Chemical Genomics. J. Biol. Chem. 285, 23307–23315. doi: 10.1074/jbc.M110.105981
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kloek, A. P., Verbsky, M. L., Sharma, S. B., Schoelz, J. E., Vogel, J., Klessig, D. F., et al. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26, 509–522. doi: 10.1046/j.1365-313x.2001.01050.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kohlen, W., Charnikhova, T., Liu, Q., Bours, R., Domagalska, M. A., Beguerie, S., et al. (2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 155, 974–987. doi: 10.1104/pp.110.164640
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kolb, H. C., Finn, M. G., and Sharpless, K. B. (2001). Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 40, 2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kolb, H. C., and Sharpless, K. B. (2003). The growing impact of click chemistry on drug discovery. Drug Discov. Today 8, 1128–1137. doi: 10.1016/S1359-6446(03)02933-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kondo, Y., Tadokoro, E., Matsuura, M., Iwasaki, K., Sugimoto, Y., Miyake, H., et al. (2007). Synthesis and seed germination stimulating activity of some imino analogs of strigolactones. Biosci. Biotechnol. Biochem. 71, 2781–2786. doi: 10.1271/bbb.70398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koo, A. J., and Howe, G. A. (2009). The wound hormone jasmonate. Phytochemistry 70, 1571–1580. doi: 10.1016/j.phytochem.2009.07.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koo, A. J. K., Gao, X., Jones, A. D., and Howe, G. A. (2009). A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59, 974–986. doi: 10.1111/j.1365-313X.2009.03924.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kowalska, M., Galuszka, P., Frébortová, J., Šebela, M., Béres, T., Hluska, T., et al. (2010). Vacuolar and cytosolic cytokinin dehydrogenases of Arabidopsis thaliana: heterologous expression, purification and properties. Phytochemistry 71, 1970–1978. doi: 10.1016/j.phytochem.2010.08.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kudo, T., Kiba, T., and Sakakibara, H. (2010). Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52, 53–60. doi: 10.1111/j.1744-7909.2010.00898.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumari, S., and van der Hoorn, R. A. L. (2011). A structural biology perspective on bioactive small molecules and their plant targets. Curr. Opin. Plant Biol. 14, 480–488. doi: 10.1016/j.pbi.2011.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuromori, T., Miyaji, T., Yabuuchi, H., Shimizu, H., Sugimoto, E., Kamiya, A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. U.S.A. 107, 2361–2366. doi: 10.1073/pnas.0912516107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kurosawa, E. (1926). Experimental studies on the nature of the substance secreted by the “bakanae” fungus. Nat. HistSoc. Formosa 16, 213–227.
Lamberth, C., Jeanmart, S., Luksch, T., and Plant, A. (2013). Current challenges and trends in the discovery of agrochemicals. Science 341, 742–746. doi: 10.1126/science.1237227
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lawton, K. A., Friedrich, L., Hunt, M., Weymann, K., Delaney, T., Kessmann, H., et al. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10, 71–82. doi: 10.1046/j.1365-313X.1996.10010071.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, L., Li, C., Lee, G. I., and Howe, G. A. (2002). Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. U.S.A. 99, 6416–6421. doi: 10.1073/pnas.072072599
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, S., and Sang, R. (2013). Bioactive fluorescent jasmonate designed by molecular modeling and its migration in tomato visualized by fluorescent molecular imaging. Tetrahedron 69, 844–848. doi: 10.1016/j.tet.2012.10.105
Liu, S., Wang, W.-H., Dang, Y.-L., Fu, Y., and Sang, R. (2012). Rational design and efficient synthesis of a fluorescent-labeled jasmonate. Tetrahedron Lett. 53, 4235–4239. doi: 10.1016/j.tetlet.2012.06.006
Lumba, S., Cutler, S., and McCourt, P. (2010). Plant nuclear hormone receptors: a role for small molecules in protein-protein interactions. Annu. Rev. Cell Dev. Biol. 26, 445–469. doi: 10.1146/annurev-cellbio-100109-103956
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, Q., and Robert, S. (2014). Auxin biology revealed by small molecules. Physiol. Plant. 151, 25–42. doi: 10.1111/ppl.12128
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marchant, A., Kargul, J., May, S. T., Muller, P., Delbarre, A., Perrot-Rechenmann, C., et al. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18, 2066–2073. doi: 10.1093/emboj/18.8.2066
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matsuura, H., Takeishi, S., Kiatoka, N., Sato, C., Sueda, K., Masuta, C., et al. (2012). Transportation of de novo synthesized jasmonoyl isoleucine in tomato. Phytochemistry 83, 25–33. doi: 10.1016/j.phytochem.2012.06.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCourt, P., and Desveaux, D. (2010). Plant chemical genetics. New Phytol. 185, 15–26. doi: 10.1111/j.1469-8137.2009.03045.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meesters, C., Mönig, T., Oeljeklaus, J., Krahn, D., Westfall, C. S., Hause, B., et al. (2104). A chemical inhibitor of jasmonate signaling targets JAR1 in Arabidopsis thaliana. Nat. Chem. Biol. 10, 830–836. doi: 10.1038/nchembio.1591
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Melcher, K., Zhou, X. E., and Xu, H. E. (2010). Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling. Curr. Opin. Struct. Biol. 20, 722–729. doi: 10.1016/j.sbi.2010.09.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Michard, E., Lima, P. T., Borges, F., Silva, A. C., Portes, M. T., Carvalho, J. E., et al. (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332, 434–437. doi: 10.1126/science.1201101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mockaitis, K., and Estelle, M. (2008). Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24, 55–80. doi: 10.1146/annurev.cellbio.23.090506.123214
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Monte, I., Hamberg, M., Chini, A., Gimenez-Ibanez, S., García-Casado, G., Porzel, A., et al. (2014). Rational design of a ligand-based antagonist of jasmonate perception. Nat. Chem. Biol. 10, 671–676. doi: 10.1038/nchembio.1575
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Motte, H., Galuszka, P., Spíchal, L., Tarkowski, P., Plíhal, O., Šmehilová, M., et al. (2013). Phenyl-adenine, identified in a LIGHT-DEPENDENT SHORT HYPOCOTYLS4-assisted chemical screen, is a potent compound for shoot regeneration through the inhibition of CYTOKININ OXIDASE/DEHYDROGENASE activity. Plant Physiol. 161, 1229–1241. doi: 10.1104/pp.112.210716
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mousavi, S. A. R., Chauvin, A., Pascaud, F., Kellenberger, S., and Farmer, E. E. (2013). GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500, 422–426. doi: 10.1038/nature12478
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murase, K., Hirano, Y., Sun, T. P., and Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. doi: 10.1038/nature07519
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murray, S. L., Thomson, C., Chini, A., Read, N. D., and Loake, G. J. (2002). Characterization of a novel, defense-related Arabidopsis mutant, cir1, isolated by luciferase imaging. Mol. Plant. Microbe. Interact. 15, 557–566. doi: 10.1094/MPMI.2002.15.6.557
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muto, T., and Todoroki, Y. (2013). Brassinolide-2,3-acetonide: a brassinolide-induced rice lamina joint inclination antagonist. Bioorg. Med. Chem. 21, 4413–4419. doi: 10.1016/j.bmc.2013.04.048
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nelson, D. C., Scaffidi, A., Dun, E. A., Waters, M. T., Flematti, G. R., Dixon, K. W., et al. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 8897–8902. doi: 10.1073/pnas.1100987108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nisler, J., Zatloukal, M., Popa, I., Dolezal, K., Strnad, M., Spíchal, L., et al. (2010). Cytokinin receptor antagonists derived from 6-benzylaminopurine. Phytochemistry 71, 823–830. doi: 10.1016/j.phytochem.2010.01.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Noh, B., Murphy, A. S., and Spalding, E. P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13, 2441–2454. doi: 10.1105/tpc.13.11.2441
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Noutoshi, Y., Okazaki, M., Kida, T., Nishina, Y., Morishita, Y., Ogawa, T., et al. (2012). Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 24, 3795–3804. doi: 10.1105/tpc.112.098343
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oh, K., Asami, T., Matsui, K., Howe, G. A., and Murofushi, N. (2006). Characterization of novel imidazole derivative, JM-8686, a potent inhibitor of allene oxide synthase. FEBS Lett. 580, 5791–5796. doi: 10.1016/j.febslet.2006.09.044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okada, K., Ueda, J., Komaki, M. K., Bell, C. J., and Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684. doi: 10.1105/tpc.3.7.677
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okamoto, M., Peterson, F. C., Defries, A., Park, S.-Y., Endo, A., Nambara, E., et al. (2013). Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc. Natl. Acad. Sci. U.S.A. 110, 12132–12137. doi: 10.1073/pnas.1305919110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Overbeek, J. V., and Vélez, I. (1946). Use of 2,4-dichlorophenoxyacetic acid as a selective herbicide in the tropics. Science 103, 472–473. doi: 10.1126/science.103.2677.472
Park, J.-H., Halitschke, R., Kim, H. B., Baldwin, I. T., Feldmann, K. A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12. doi: 10.1046/j.1365-313X.2002.01328.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, S.-Y. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Podlešáková, K., Zalabák, D., Cudejková, M., Plíhal, O., Szüèová, L., Doležal, K., et al. (2012). Novel cytokinin derivatives do not show negative effects on root growth and proliferation in submicromolar range. PLoS ONE 7:e39293. doi: 10.1371/journal.pone.0039293
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rigal, A., Ma, Q., and Robert, S. (2014). Unraveling plant hormone signaling through the use of small molecules. Front. Plant Sci. 5:373. doi: 10.3389/fpls.2014.00373
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Robert, H. S., and Friml, J. (2009). Auxin and other signals on the move in plants. Nat. Chem. Biol. 5, 325–332. doi: 10.1038/nchembio.170
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Robert, S., Chary, S. N., Drakakaki, G., Li, S., Yang, Z., Raikhel, N. V., et al. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. U.S.A. 105, 8464–8469. doi: 10.1073/pnas.0711650105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Robert, S., Raikhel, N. V., and Hicks, G. R. (2009). Powerful partners: Arabidopsis and chemical genomics. Arabidopsis Book 7:e0109. doi: 10.1199/tab.0109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. G. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Robson, F., Okamoto, H., Patrick, E., Harris, S.-R., Wasternack, C., Brearley, C., et al. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22, 1143–1160. doi: 10.1105/tpc.109.067728
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rojas-Pierce, M., Titapiwatanakun, B., Sohn, E. J., Fang, F., Larive, C. K., Blakeslee, J., et al. (2007). Arabidopsis P-glycoprotein19 participates in the inhibition of gravitropism by gravacin. Chem. Biol. 14, 1366–1376. doi: 10.1016/j.chembiol.2007.10.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rosado, A., Hicks, G. R., Norambuena, L., Rogachev, I., Meir, S., Pourcel, L., et al. (2011). Sortin1-hypersensitive mutants link vacuolar-trafficking defects and flavonoid metabolism in Arabidopsis vegetative tissues. Chem. Biol. 18, 187–197. doi: 10.1016/j.chembiol.2010.11.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rozhon, W., Mayerhofer, J., Petutschnig, E., Fujioka, S., and Jonak, C. (2010). ASKtheta, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 62, 215–223. doi: 10.1111/j.1365-313X.2010.04145.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ruegger, M., Dewey, E., Hobbie, L., Brown, D., Bernasconi, P., Turner, J., et al. (1997). Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9, 745–757. doi: 10.1105/tpc.9.5.745
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santiago, J., Henzler, C., and Hothorn, M. (2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341, 889–892. doi: 10.1126/science.1242468
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schaller, A., Bergey, D. R., and Ryan, C. A. (1995). Induction of wound response genes in tomato leaves by bestatin, an inhibitor of aminopeptidases. Plant Cell 7, 1893–1898. doi: 10.1105/tpc.7.11.1893
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schreiber, S. L. (2000). Target-oriented and diversity-oriented organic synthesis in drug discovery. Science 287, 1964–1969. doi: 10.1126/science.287.5460.1964
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schweizer, F., Fernández-Calvo, P., Zander, M., Diez-Diaz, M., Fonseca, S., Glauser, G., et al. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25, 3117–3132. doi: 10.1105/tpc.113.115139
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shani, E., Weinstain, R., Zhang, Y., Castillejo, C., Kaiserli, E., Chory, J., et al. (2013). Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 110, 4834–4839. doi: 10.1073/pnas.1300436110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shimada, A., Ueguchi-Tanaka, M., Nakatsu, T., Nakajima, M., Naoe, Y., Ohmiya, H., et al. (2008). Structural basis for gibberellin recognition by its receptor GID1. Nature 456, 520–523. doi: 10.1038/nature07546
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shirano, Y., Kachroo, P., Shah, J., and Klessig, D. F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14, 3149–3162. doi: 10.1105/tpc.005348
Spíchal, L., Werner, T., Popa, I., Riefler, M., Schmülling, T., and Strnad, M. (2009). The purine derivative PI-55 blocks cytokinin action via receptor inhibition. FEBS J. 276, 244–253. doi: 10.1111/j.1742-4658.2008.06777.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Staswick, P. E. (2009). The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 150, 1310–1321. doi: 10.1104/pp.109.138529
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Staswick, P. E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 8, 2117–2127. doi: 10.1105/tpc.104.023549
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L. M., Xie, D.-Y., Dolezal, K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. doi: 10.1016/j.cell.2008.01.047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Surpin, M., Rojas-Pierce, M., Carter, C., Hicks, G. R., Vasquez, J., and Raikhel, N. V. (2005). The power of chemical genomics to study the link between endomembrane system components and the gravitropic response. Proc. Natl. Acad. Sci. U.S.A. 102, 4902–4907. doi: 10.1073/pnas.0500222102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Swarup, R., Friml, J., Marchant, A., Ljung, K., Sandberg, G., Palme, K., et al. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15, 2648–2653. doi: 10.1101/gad.210501
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takeuchi, J., Okamoto, M., Akiyama, T., Muto, T., Yajima, S., Sue, M., et al. (2014). Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions. Nat. Chem. Biol. 10, 477–482. doi: 10.1038/nchembio.1524
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tan, X., Calderon-Villalobos, L. I. A., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. doi: 10.1038/nature05731
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thorpe, M. R., Ferrieri, A. P., Herth, M. M., and Ferrieri, R. A. (2007). 11C-imaging: methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta 226, 541–551. doi: 10.1007/s00425-007-0503-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tóth, R., and van der Hoorn, R. A. L. (2010). Emerging principles in plant chemical genetics. Trends Plant Sci. 15, 81–88. doi: 10.1016/j.tplants.2009.11.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsuchiya, Y., Vidaurre, D., Toh, S., Hanada, A., Nambara, E., Kamiya, Y., et al. (2010). A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat. Chem. Biol. 6, 741–749. doi: 10.1038/nchembio.435
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsuda, E., Yang, H., Nishimura, T., Uehara, Y., Sakai, T., Furutani, M., et al. (2011). Alkoxy-auxins are selective inhibitors of auxin transport mediated by PIN, ABCB, and AUX1 transporters. J. Biol. Chem. 286, 2354–2364. doi: 10.1074/jbc.M110.171165
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. doi: 10.1038/nature07272
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ursache, R., Miyashima, S., Chen, Q., Vatén, A., Nakajima, K., Carlsbecker, A., et al. (2014). Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development 141, 1250–1259. doi: 10.1242/dev.103473
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van der Hoorn, R. A. L., Colby, T., Nickel, S., Richau, K. H., Schmidt, J., and Kaiser, M. (2011). Mining the Active Proteome of Arabidopsis thaliana. Front. Plant Sci. 2:89. doi: 10.3389/fpls.2011.00089
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vert, G., and Chory, J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441, 96–100. doi: 10.1038/nature04681
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Walsh, D. P., and Chang, Y.-T. (2006). Chemical genetics. Chem. Rev. 106, 2476–2530. doi: 10.1021/cr0404141
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, Z.-Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. doi: 10.1016/S1534-5807(02)00153-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wasternack, C. (1993). Ursolic acid inhibits synthesis of jasmonate-induced proteins in barley leaves. Phytochemistry 35, 49–54. doi: 10.1016/S0031-9422(00)90507-7
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Waters, M. T., Nelson, D. C., Scaffidi, A., Flematti, G. R., Sun, Y. K., Dixon, K. W., et al. (2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295. doi: 10.1242/dev.074567
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weiner, J. J., Peterson, F. C., Volkman, B. F., and Cutler, S. R. (2010). Structural and functional insights into core ABA signaling. Curr. Opin. Plant Biol. 13, 495–502. doi: 10.1016/j.pbi.2010.09.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Werner, T., and Schmülling, T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12, 527–538. doi: 10.1016/j.pbi.2009.07.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xin, X.-F., and He, S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51, 473–498. doi: 10.1146/annurev-phyto-082712-102321
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamada, M., Greenham, K., Prigge, M. J., Jensen, P. J., and Estelle, M. (2009). The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 151, 168–179. doi: 10.1104/pp.109.138859
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yan, J., Zhang, C., Gu, M., Bai, Z., Zhang, W., Qi, T., et al. (2009a). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. doi: 10.1105/tpc.109.065730
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yan, Z., Zhao, J., Peng, P., Chihara, R. K., and Li, J. (2009b). BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150, 710–721. doi: 10.1104/pp.109.138099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, Y., Hammes, U. Z., Taylor, C. G., Schachtman, D. P., and Nielsen, E. (2006). High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16, 1123–1127. doi: 10.1016/j.cub.2006.04.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005a). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. doi: 10.1016/j.cell.2004.11.044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005b). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. doi: 10.1016/j.cell.2004.11.044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Goritschnig, S., Dong, X., and Li, X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15, 2636–2646. doi: 10.1105/tpc.015842
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, Y. (2008). The role of local biosynthesis of auxin and cytokinin in plant development. Curr. Opin. Plant Biol. 11, 16–22. doi: 10.1016/j.pbi.2007.10.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, Y., Chow, T. F., Puckrin, R. S., Alfred, S. E., Korir, A. K., Larive, C. K., et al. (2007). Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nat. Chem. Biol. 3, 716–721. doi: 10.1038/nchembio.2007.32
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zheng, W., Zhai, Q., Sun, J., Li, C.-B., Zhang, L., Li, H., et al. (2006). Bestatin, an inhibitor of aminopeptidases, provides a chemical genetics approach to dissect jasmonate signaling in Arabidopsis. Plant Physiol. 141, 1400–1413. doi: 10.1104/pp.106.080390
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zouhar, J., Hicks, G. R., and Raikhel, N. V. (2004). Sorting inhibitors (Sortins): chemical compounds to study vacuolar sorting in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 9497–9501. doi: 10.1073/pnas.0402121101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zwanenburg, B., Mwakaboko, A. S., Reizelman, A., Anilkumar, G., and Sethumadhavan, D. (2009). Structure and function of natural and synthetic signalling molecules in parasitic weed germination. Pest Manag. Sci. 65, 478–491. doi: 10.1002/ps.1706
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zwanenburg, B., and Pospísil, T. (2013). Structure and activity of strigolactones: new plant hormones with a rich future. Mol. Plant 6, 38–62. doi: 10.1093/mp/sss141
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: phytohormones, chemical genomics, hormone perception and signaling, hormone crosstalk, plant chemical biology, jasmonates, agonist and antagonist, small molecules
Citation: Fonseca S, Rosado A, Vaughan-Hirsch J, Bishopp A and Chini A (2014) Molecular locks and keys: the role of small molecules in phytohormone research. Front. Plant Sci. 5:709. doi: 10.3389/fpls.2014.00709
Received: 28 September 2014; Accepted: 26 November 2014;
Published online: 17 December 2014.
Edited by:
Erich Kombrink, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
Joseph M. Jez, Washington University in St. Louis, USAClaus Wasternack, Leibniz Insitute of Plant Biochemistry, Germany
Copyright © 2014 Fonseca, Rosado, Vaughan-Hirsch, Bishopp and Chini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Chini, Departamento de Genética Molecular de Plantas, Centro Nacional de Biotecnología- Consejo Superior de Investigaciones Científicas, Campus Universidad Autónoma, C/ Darwin 3, 28049 Madrid, Spain e-mail: achini@cnb.csic.es
 Sandra Fonseca
Sandra Fonseca Abel Rosado2
Abel Rosado2 Anthony Bishopp
Anthony Bishopp Andrea Chini
Andrea Chini