Exceptional longevity and muscle and fitness related genotypes: a functional in vitro analysis and case-control association replication study with SNPs THRH rs7832552, IL6 rs1800795, and ACSL1 rs6552828
- 1Graduate School of Health and Sports Science, Juntendo University, Chiba, Japan
- 2Biology Centre, China Institute of Sport Science, Beijing, China
- 3Research Institute of Hospital 12 de Octubre (“i+12”), Madrid, Spain
- 4School of Doctorate Studies and Research, European University of Madrid, Madrid, Spain
- 5Center for Supercentenarian Study, Keio University School of Medicine, Tokyo, Japan
- 6Department of Health Promotion and Exercise, National Institute of Health and Nutrition, Tokyo, Japan
- 7Department of Health Sciences, University of Pavia, Pavia, Italy
- 8INEF, Universidad Politécnica, Madrid, Spain
- 9Department of Physiotherapy and Nursing, Faculty of Health and Sport Science, University of Zaragoza, Huesca, Spain
There are several gene variants that are candidates to influence functional capacity in long-lived individuals. As such, their potential association with exceptional longevity (EL, i.e., reaching 100+ years) deserves analysis. Among them are rs7832552 in the thyrotropin-releasing hormone receptor (TRHR) gene, rs1800795 in the interleukin-6 (IL6) gene and rs6552828 in the coenzyme A synthetase long-chain 1 (ACSL1) gene. To gain insight into their functionality (which is yet unknown), here we determined for the first time luciferase gene reporter activity at the muscle tissue level in rs7832552 and rs6552828. We then compared allele/genotype frequencies of the 3 abovementioned variants among centenarians [n = 138, age range 100–111 years (114 women)] and healthy controls [n = 334, 20–50 years (141 women)] of the same ethnic and geographic origin (Spain). We also studied healthy centenarians [n = 79, 100–104 years (40 women)] and controls [n = 316, 27–81 years (156 women)] from Italy, and centenarians [n = 742, 100–116 years (623 women)] and healthy controls [n = 499, 23–59 years (356 women)] from Japan. The THRH rs7832552 T-allele and ACSL1 rs6552828 A-allele up-regulated luciferase activity compared to the C and G-allele, respectively (P = 0.001). Yet we found no significant association of EL with rs7832552, rs1800795 or rs6552828 in any of the 3 cohorts. Further research is needed with larger cohorts of centenarians of different origin as well as with younger old people.
Introduction
The oldest old population (≥85 years) is rapidly expanding among westerners (Waite, 2004; Robine and Paccaud, 2005). However, aging is associated with an increased risk of loss of functional independence (Christensen et al., 2008). In this regard, centenarians (people aged 100+ years) are the paradigm of healthy aging, as they have usually postponed (or even avoided, in some cases) major age-related diseases, as well as the onset of disability, until they were well into their nineties (Terry et al., 2008). Thus, the search for the gene variants that might influence the likelihood of reaching exceptional longevity (EL, i.e., becoming a centenarian) might help identify targets of “anti-aging” interventions.
In old people, functional independence is dependent on physical fitness, which in turn is determined by several phenotypes such as mainly cardiorespiratory fitness and muscle performance (Garber et al., 2011). Concerning the latter, aging is inevitably associated with a decline in muscle mass and function, i.e., sarcopenia, with an acceleration of this process increasing the risk of mortality (Metter et al., 2004; Ruiz et al., 2008). Thus, those gene variations that can be associated with preservation of muscle mass/function at advanced ages, e.g., the K153R polymorphism in the myostatin (MSTN) gene (Garatachea et al., 2013a) or the I/D polymorphism in the angiotensin converting enzyme (ACE) (Garatachea et al., 2013b) as well as other genetic variations linked with muscle aerobic capacity (e.g., the R577X mutation in the α-actinin-3 (ACTN3) gene), might also influence the likelihood of reaching EL (Fiuza-Luces et al., 2011).
There are other potential candidates to influence muscle phenotypes in long-lived individuals and as such their potential association with EL deserves analysis. In this regard, a recent genome wide scan (GWAS) analysis of 379,319 SNPs in US Caucasians of both genders (aged 50 years on average) revealed an association of lean body mass with 2 SNPs in tight linkage disequilibrium within the thyrotropin-releasing hormone receptor (TRHR) gene, rs16892496 and rs7832552 (Liu et al., 2009). These results were further corroborated in independent cohorts of older Caucasians of both genders, aged 63 (men) and 61 years (women). The functional significance of these 2 variants remains however to be determined. Another candidate is the −174C/G polymorphism (rs1800795) in the interleukin-6 (IL6) gene, where the G-allele is associated with higher transcription in vitro (Fishman et al., 1998) and in vivo conditions (Bennermo et al., 2004). IL6 is a multifunctional cytokine that might be also involved in muscle regeneration (Serrano et al., 2008). Pereira et al recently found that the GG genotype, which is associated with lower IL6 levels and thus with “anti-inflammatory” profile, was associated with better physical performance in community-dweller elderly women (≥65 years) (Pereira et al., 2013).
As for cardiorespiratory fitness [which is usually determined with peak oxygen uptake (VO2peak)], a recent GWAS study in sedentary Caucasians found that, among 324,611 SNPs, the strongest association with the VO2peak response to exercise was found to acyl coenzyme A synthetase long-chain 1 (ACSL1) gene polymorphism rs6552828 (Bouchard et al., 2011). The ACSL1 gene is a candidate to explain individual variability in VO2peak, as well as in some health-related phenotypes, owing to its potential role in aerobic metabolism at the adypocyte, cardiomyocyte, liver and skeletal muscle fiber level (Martin et al., 1997; Coleman et al., 2000; Hall et al., 2003; Mashek et al., 2006; Ellis et al., 2010), yet its functional significance has not been assessed.
In order to analyze their functionality at the muscle level, we measured for the first time luciferase gene reporter activity in TRHR rs16892496 and rs7832552, and also in ACSL1 rs6552828. Only rs7832552 in the TRHR gene was genotyped in this study because it is in tight linkage disequilibrium with rs16892496 (Liu et al., 2009). Based on the hypothesis that common genetic polymorphisms influencing physical fitness may also have an impact on the ability to reach EL, we then compared allele/genotype frequencies of the abovementioned SNPs—together with the IL6 rs1800795 SNP (whose functional significance is already known, as mentioned above)—among Spanish centenarians (cases) and healthy controls matched by ethnic and geographic origin and also in 2 other geographically and ethnically-independent replication cohorts (from Italy and Japan).
Methods
Functional Analysis: Luciferase Reporter Gene
The fragment, including the allele, was directly inserted into the pGL3-promoter at the restriction recognition sites MluI/NheI in the 5′ and XhoI in the 3′ (see below -in bold) of the sequences obtained from the genomes of:
(i) one individual homozygous for the rs16892496 A-allele and one individual homozygous for the rs16892496 C-allele (see below -underlined)
rs16892496 AA
GCTAGCTATGAAAGATCTACGTTAAAACATAAGGTT
AAGCTGTGCAGTGTACAGAAGAGACAAGAAAGTGG
TACTTACTGTGCATAAGGTTGAAGAGCAAGCCCCCA
GTGGGATACAAGTCACTCTCAGGCTTGAAAAATGAG
TAGGCATTCACTAGGCCAACATAAAATACAAGAAGA
CCCTCCAGTCTGCAGAAGTAGTCAATGACTCGAG
rs16892496 CC
GCTAGCTATGAAAGATCTACGTTAAAACATAAGGTT
AAGCTGTGCAGTGTACAGAAGAGACAAGAAAGTGG
TACTTACTGTGCATAAGGTTGAAGAGCAAGCCCCCC
GTGGGATACAAGTCACTCTCAGGCTTGAAAAATGAG
TAGGCATTCACTAGGCCAACATAAAATACAAGAAGA
CCCTCCAGTCTGCAGAAGTAGTCAATGACTCGAG
(ii) one individual homozygous for the rs7832552 C-allele and one homozygous for the rs7832552 T-allele (see below -underlined)
rs7832552-CC
GAGCTCATTAGCCTTGTGACAAAAGCAACGCACTCC
ATTTTGCACACAGTACTTGACTTTATTTTGCTACTGC
CTTGACCTCAAAGGAATGTGATAGTGTGAGGTACGA
ATGCTCTTAATAAACAGGATCGATCAAGGGTGCTTG
ACTCTTGTTGTTCATGTGCAAGTATAGTGGCTTTTT
TGTGCCTCAACAAAACCATCAAGAGTCTCGAG
rs7832552-TT
GAGCTCATTAGCCTTGTGACAAAAGCAACGCACTCC
ATTTTGCACACAGTACTTGACTTTATTTTGCTACTGC
CTTGACCTCAAAGGAATGTGATAGTGTGAGGTATGA
ATGCTCTTAATAAACAGGATCGATCAAGGGTGCTTGA
CTCTTGTTGTTCATGTGCAAGTATAGTGGCTTTTTTG
TGCCTCAACAAAACCATCAAGAGTCTCGAG
(iii) one individual homozygous for the rs6552828 A-allele and one individual homozygous for the rs6552828 G-allele (see below -underlined)
rs6552828-AA
GAGCTCCAAGACATTATAGCCAAAAGAAACAAACAG
ATAAATTGGTGTGCATAAACTTTAAACCAACCACCAG
ATATCTAAAGAGGGAATACAGCACAGTGTTGGAAAG
AAAGTACAGAATAGTATTTGAGATCCTAGATGCAGC
CGGACGCGGTGGCTCATGCCTGTAATCCCAGCACTT
TGGGAAGCCGAGGCGGGTGGATCACCCTCGAG
rs6552828-GG
GAGCTCCAAGACATTATAGCCAAAAGAAACAAACAG
ATAAATTGGTGTGCATAAACTTTAAACCAACCACCAG
ATATCTAAAGAGGGAATACAGCACAGTGTTGGAGAG
AAAGTACAGAATAGTATTTGAGATCCTAGATGCAGC
CGGACGCGGTGGCTCATGCCTGTAATCCCAGCACTT
TGGGAAGCCGAGGCGGGTGGATCACCCTCGAG
We used mice skeletal muscle C2C12 cell lines to study muscle-specific expression. We performed cell cultures, transfections and dual-luciferase reporter assays following the procedures previously reported by our group (He et al., 2011). We used the pRL-SV40 vector as an internal control for variations in transfection efficiency, and the pGL3-promoter vector without an insert as a negative control. The transfected cells were harvested after 48 h, and assayed for firefly and renilla luciferase activity with the dual-luciferase reporter assay system (Promega Biotech, Beijing, China) using a luminometer following the manufacturer's recommendations (TecanGenios Pro, Männedorf, Switzerland). From each measurement, we divided firefly by renilla luciferase activity reading to calculate relative luciferase activity. We performed the experiments in triplicates and expressed relative luciferase activity values as the means±SD of the 3 different measurements.
Subjects
We obtained approval from the local ethics committees [European University of Madrid (Spain), University of Pavia (Italy), and National Institute of Health and Nutrition, (Japan) Medical Research Institute and Keio University (Japan)] and the study followed the tenets of the Declaration of Helsinki for Human Research. Written consent was also obtained from each participant.
Spanish Cohort
Two groups of Spanish subjects were assessed: (i) 138 cases (centenarians, aged 100–111 years, 114 women); and (ii) 334 healthy controls (aged 20–50 years, 141 women). All the subjects were of the same Caucasian (Spanish) descent for ≥3 generations. The major diseases among the centenarians were osteoarthritis (66%), hypertension (57%), dementia (51%) and cardiovascular disease (CVD, 29%). The DNA of a convenience sample of 355 younger disease-free controls with no reported family history of high longevity (>90 years) was collected during 2008-2012 in the European University of Madrid.
Italian Cohort
Two groups of subjects from Northern Italy (mainly from Lombardy and Piedmont) were studied: (i) 79 cases (healthy centenarians, aged 100–104 years, 40 women); and (ii) 316 healthy controls (aged 27–81 years, 156 women). All patients and controls were Caucasian whites of Italian descent for ≥3 generations The Italian centenarians were free of major age-related diseases, i.e., severe cognitive impairment, clinically evident cancer, CVD, renal insufficiency or severe physical impairment (Emanuele et al., 2010). Controls were free of CVD or cerebrovascular disease, cancer, dementia, chronic autoimmune/ inflammatory disorders, renal or hepatic failure, and major psychiatric conditions.
Japanese Cohort
Two groups of subjects of the same Asian (Japanese) descent were assessed: (i) 742 cases (centenarians, aged 100–116 years, 623 women); and (ii) 499 healthy controls (aged 23–59 years, 356 women). The group of cases was gathered from 2 cohorts, which are described in detail elsewhere (Gondo et al., 2006): the Tokyo Centenarians Study (TCS) and the Semi-Supercentenarians Study in Japan (SSC-J). The prevalence of hypertension, CVD and dementia among the Japanese centenarians was of 63.6, 28.8, and 59.4%, respectively. Controls from both genders (aged <60 years, and free of diagnosed CVD and chronic renal failure) were recruited during years 2008–2012.
Genotyping
As mentioned above, only one THRH SNP, rs7832552, was genotyped in the 3 cohorts (and not rs16892496) because the genotype distributions of both SNPs are completely linked according to available HapMap for both European and Asian populations (sorted as a Supplementary file 1) and previous research has shown that both SNPs are in strong linkage disequilibrium (r2 = 0.98). All genotyping was performed only for research purposes with the researchers who performed the genotyping being blinded to the participants' identities. For quality control, a random ~20% of the samples of each cohort were genotyped again, with no differences in the results compared with the initial genotyping.
Spanish Cohort
DNA was extracted from the participants' buccal cells (saliva samples) using a standard phenol chloroform protocol and the genotype analyses were performed in the Biomedicine laboratory at the European University, Madrid (Spain). The DNA samples were diluted with sterile water and stored at −20°C until analysis. Genotyping was performed by Real-Time PCR and using the TaqMan® rs7832552, rs6552828, and rs1800795 SNP genotyping assays with a Step One Real-Time PCR System (Applied Biosystems, Foster City, CA).
Italian Cohort
Genomic DNA was purified from blood leukocytes using the QiaAmp DNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Genotyping was performed at the Cellular Pathophysiology and Clinical Immunology Laboratory (University of Pavia) using the TaqMan® rs7832552, rs6552828, and rs1800795 SNP genotyping assays (Applied Biosystems, Foster City, CA, USA).
Japanese Cohort
Total genomic DNA was extracted from blood leukocytes with a QIAamp DNA Blood Mini or Maxi Kit (Qiagen, Tokyo, Japan). Genotyping of rs7832552, rs6552828, and rs1800795 was performed at the Institute of Health and Sports Science and Medicine (Juntendo University) with Real Time Thermocycler (LightCycler 480, Roche Applied Science, Mannheim, Germany) using TaqMan SNP genotyping assay method. PCR 384-well plates were read on LightCycler 480 using the end-point analysis mode. Allelic discrimination analysis was performed with a LightCycler 480 SW software version 1.5.1.62 (Roche Applied Science, Mannheim, Germany).
Statistical Analysis
One-way analysis of variance was used to compare the relative luciferase activity in the different plasmids of each SNP. Allele frequencies were calculated by gene-counting. We tested Hardy-Weinberg equilibrium (HWE) using χ2-test. Genotype/allele frequencies of cases vs. controls within each cohort (Spanish, Italian, and Japanese) were compared using the χ2-test with Yates' correction and the association between genotypes/alleles and EL within each of the 3 cohorts was analyzed with logistic regression analysis after adjusting for sex. All statistical analyses were performed using the PASW (v. 18.0 for WINDOWS, Chicago) and corrected for multiple comparisons using the Bonferroni's method -that is, the threshold P-value was obtained by dividing 0.05 by the number of studied polymorphisms (P = 0.05/3 = 0.017).
Results
Functional Analysis
The results of luciferase report analyses are presented in Figure 1. All the SNPs we studied showed functional significance, as reflected by differences in luciferase activity between the 3 SNP constructs (all P ≤ 0.001); the THRH rs16892496 A-allele up-regulated luciferase activity compared to the C-allele (upper panel), the THRH rs7832552 T-allele up-regulated luciferase activity compared to the C-allele (middle panel), and the ACSL1 rs6552828 A-allele up-regulated luciferase activity compared to the G-allele (lower panel).
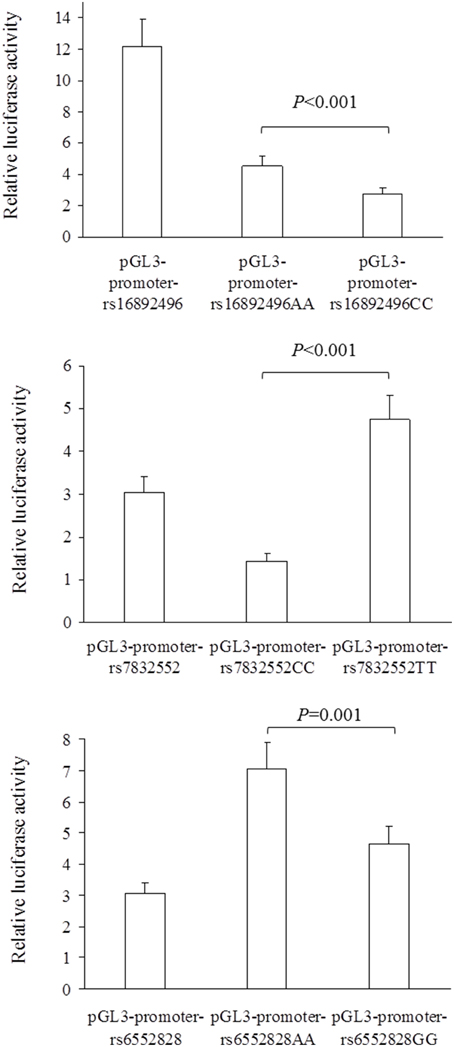
Figure 1. Comparison of relative luciferase activity (i.e., firefly luciferase activity divided by renilla luciferase activity) between plasmids for the THRH rs16892496 (upper panel), THRH rs7832552 (middle panel), and ACSL1 rs6552828 (lower panel). Values are mean ± SD of three different experiments, each performed in triplicate. For the 3 SNPs, statistical significance was reached for all the comparisons between plasmids (all P ≤ 0.001).
Spanish Cohort
Rate of genotyping success was as follows: THRH rs7832552, 97.2% in cases and 100% in controls; ACSL rs6552828, 97.2% in cases and 99.1% in controls; IL6 rs1800795, 100% in cases and 94.9% in controls. The distribution of all genotypes was consistent with the HWE in both groups (P > 0.05), except for IL6 rs1800795 in the control group (P < 0.01).
The results of genotype/allele frequency distributions as well as of binary logistic regression adjusted by sex are shown in Table 1 and summarized below. The allele (χ2 = 1.21, P = 0.27) or genotype frequency distributions of THRH rs7832552 did not differ between cases and controls (χ2 = 2.74, P = 0.25). Using logistic regression analysis, no significant associations were found between EL and rs7832552, including when analyzing both sexes separately (data not shown). No differences were found for IL6 rs1800795 in allele (χ2 = 1.01, P = 0.32) or genotype distributions (χ2 = 3.89, P = 0.14), with no significant association with EL after adjusting for sex or when analyzing both sexes separately -data not shown) or for ACSL1 rs6552828 (χ2 = 2.28, P = 0.13 for allele distribution and χ2 = 3.30, P = 0.19 for genotype distribution, with no significant association with EL after adjusting for sex or when analyzing both sexes separately -data not shown).
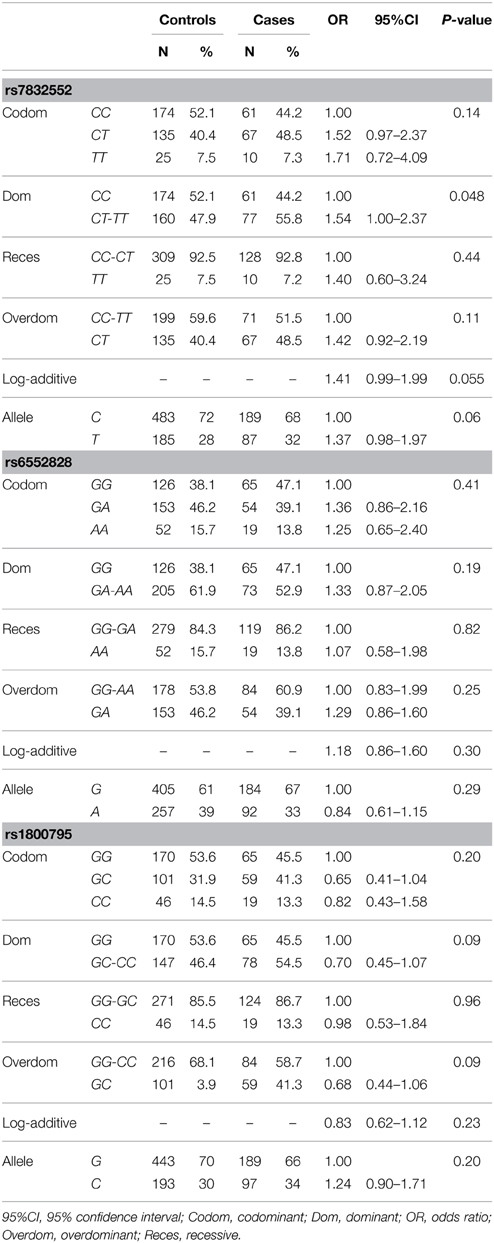
Table 1. Genotype/allele frequencies of THRH rs7832552, ACSL1 rs6552828 and IL6 rs1800795 and results of logistic regression analysis, in the Spanish cohort.
Italian Cohort
Rate of genotyping success was 100% for all gene variants. The distribution of all genotypes was consistent with the HWE in both groups (P > 0.05), except for THRH rs7832552 in the control group (P = 0.04).
The results of genotype/allele frequency distributions as well as of binary logistic regression adjusted by sex are shown in Table 2 and summarized below. The allele (χ2 = 0.003, P = 0.95) and genotype frequency distributions of THRH rs7832552 did not differ between groups (χ2 = 0.26, P = 0.88) and no significant association was found between this polymorphism and EL using logistic regression adjusted by sex, or when analyzing both sexes separately (data not shown). Similar results were found for IL6 rs1800795 (allele frequency: χ2 = 0.82, P = 0.36; genotype frequency: χ2 = 1.054, P = 0.59) and ACSL1 rs6552828 (allele frequency: χ2 = 0.56, P = 0.46; genotype frequency: χ2 = 0.67, P = 0.72), with no significant association between these two polymorphisms and EL using logistic regression adjusted by sex, or when analyzing both sexes separately (data not shown).
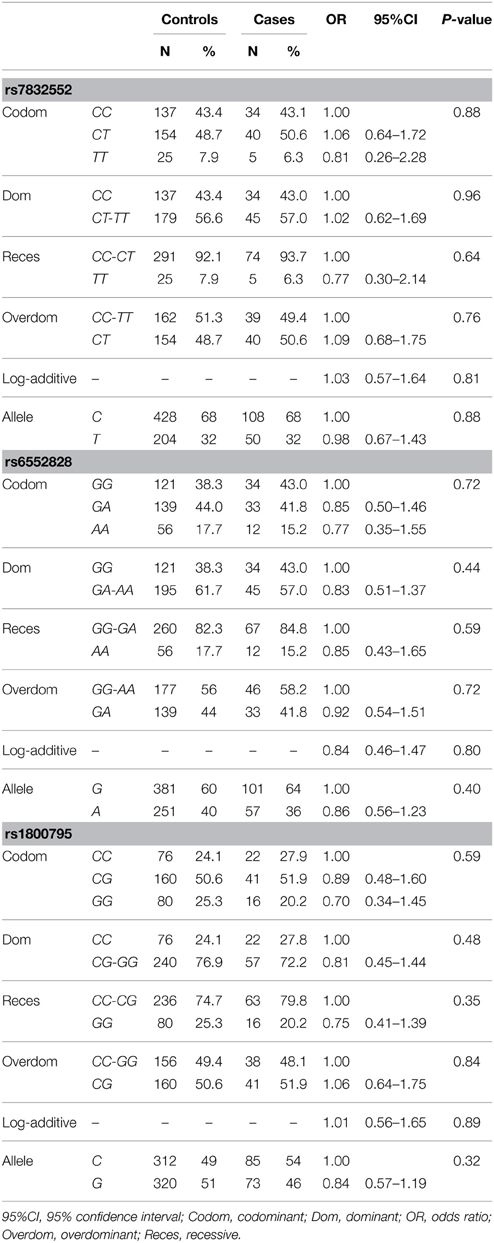
Table 2. Genotype/allele frequencies of THRH rs7832552, ACSL1 rs6552828, and IL6 rs1800795 and results of logistic regression analysis, in the Italian cohort.
Japanese Cohort
Rate of genotyping success was as follows: THRH rs7832552, 97.0% in cases and 100% in controls; IL6 rs1800795, 98.7% in cases and 100% in controls; ACSL1 rs6552828, 95.9% in cases and 99.2% in controls. The distribution of all genotypes was consistent with the HWE in both groups (P > 0.05), except for rs6552828 in centenarians (P = 0.02).
The results of genotype/allele frequency distributions as well as of binary logistic regression adjusted by sex are shown in Table 3 and summarized below. We found no differences between cases and controls for THRH rs7832552 (allele frequency: χ2 = 0.012, P = 0.91; genotype frequency: χ2 = 0.592, P = 0.74), rs1800795 (allele frequency: χ2 = 0.07, P = 0.78; genotype frequency: χ2 = 0.07, P = 0.78), or rs6552828 (allele frequency: χ2 = 0.020, P = 0.89; genotype frequency: χ2 = 3.78, P = 0.15), with no significant association between any of the SNPs and EL using logistic regression adjusted by sex or when analyzing both sexes separately (data not shown).
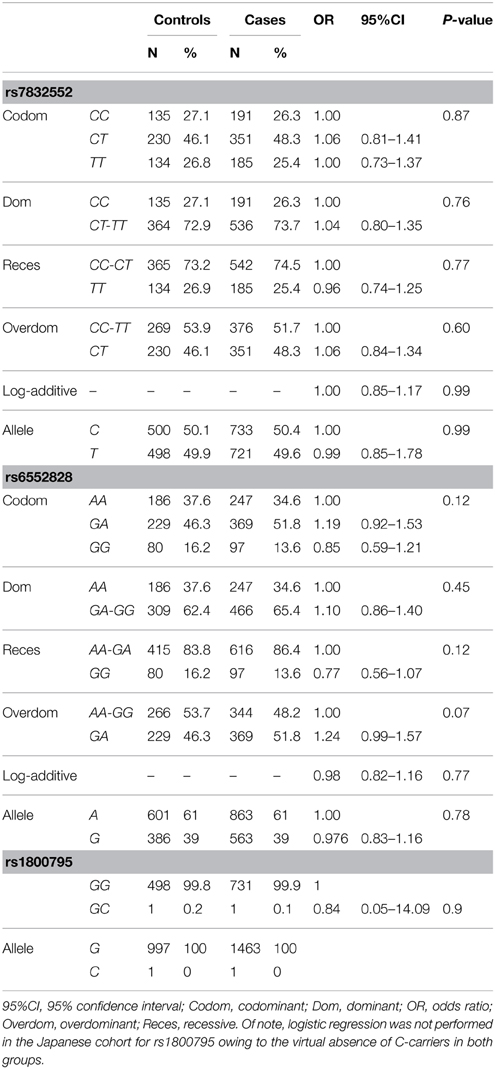
Table 3. Genotype/allele frequencies of THRH rs7832552, ACSL1 rs6552828, and IL6 rs1800795 and results of logistic regression analysis, in the Japanese cohort.
Discussion
The main findings of our study are two-fold. First, all the studied SNPs showed functional significance, as reflected by the results of the luciferase constructs. This is the first attempt to determine (with an in vitro approach) the potential functional consequences of the rs16892496, rs7832552, and rs6552828 SNPs, with the A-allele, T-allele and A-allele up-regulating luciferase activity compared to the other alleles, respectively. The THRH rs7832552 and ACSL1 rs6552828 SNPs are intronic genomic variants and, as such, could potentially alter the stability and/or alternative splicing of mRNA, as well as transcription factor binding (Tabor et al., 2002; Knight, 2005; Mercado et al., 2005; Sasabe et al., 2007). However, we found no association between THRH rs7832552, IL6 rs1800795, and ACSL1 rs6552828 and EL.
Although more research is obviously needed, we found no evidence that higher THRH expression (as theoretically associated with the T-allele or CT-TT genotypes vs. CC) might favor EL. Yet a GWAS study reported that the TRHR rs7832552 SNP was associated with lean body mass in US Caucasians (Liu et al., 2009). Subjects carrying the theoretically highest expressing (TT) genotype had 2.55 kg higher lean body mass compared to the other subjects. There is some scientific rationale in postulating that higher TRHR expression might help preservation of muscle mass in long-lived individuals: TRHR stimulates the hypothalamic-pituitary-thyroid axis, thereby leading to the release of thyroxin, a hormone that plays an important role in the development of skeletal muscle as well as in attenuating age-related changes in tissue function (Larsson et al., 1994). Although no association was found here, the IL6 rs1800795 might be also a candidate to influence EL. Carriage of the “low-producing” C-allele has been positively associated with longevity in Turkish population (Kayaalti et al., 2011), whereas the high-producing GG-genotype has been linked with higher survival in elderly females from Sweden (Cederholm et al., 2007). On the other hand, the rs1800795 polymorphism has been linked with longevity in Italian centenarians from Treviso (Albani et al., 2009) although this finding was not replicated in other European cohorts (Di Bona et al., 2009) (including from other parts of Italy, Albani et al., 2009), or in US elders (Walston et al., 2009). As for ACSL1 rs6552828, our data do not show an association of this polymorphism with EL despite the involvement of this gene in aerobic metabolism in the heart, liver, adipose and skeletal muscle tissues (Martin et al., 1997; Coleman et al., 2000; Hall et al., 2003; Mashek et al., 2006; Ellis et al., 2010). Recent research did not report an association between rs6552828 and an important age-related disease condition, the metabolic syndrome (Phillips et al., 2010).
A strength and novelty from our design stems from the use of a luciferase construct study to assess functionality of the 3 SNPs at the specific muscle tissue-level. However, our study has several limitations. First, besides the fact that we did not assess the functionality of the SNPs in vivo in blood samples and especially in muscle biopsies (which is understandable due to ethical reasons), we used convenience samples, which increases the risk of bias induced by population stratification. The SNPs rs1800795 and rs7832552 did not meet HWE in Spanish and Italian controls, respectively. In this regard, deviation from HWE does not necessarily reflect genotyping errors (Leal, 2005; Zou and Donner, 2006), with ~10% of all genotype–phenotype association studies actually showing failure of 1+ genotype distributions to meet HWE (Trikalinos et al., 2006). Second, we selected 3 SNPs based on previous GWAS, i.e., those showing associations of rs16892496 and rs7832552 with lean body mass (Liu et al., 2009) and of rs6552828 with VO2peak (Bouchard et al., 2011). Besides differences in terms of population-specificity (ethic/geographic origin, age) between these 2 GWAS and our cohorts, an additional problem is that GWAS are generally successful to find very penetrant dominant genetic variants, but less useful to discover rarer variants, with a likely modest effect on some phenotypes, such as those that could be potentially associated with EL. Finally, a potential confounder of genetic association studies is differences in date of birth, e.g., the centenarians and controls of our study were born in the early 1900s and after 1930, respectively (Lewis and Brunner, 2004). In this regard, the potential demographic biases of longevity studies like ours performing cross-sectional comparisons of genotype/allele frequencies between controls and long lived individuals could be overcome by adding demographic information to genetic data (Yashin et al., 1999; Passarino et al., 2006; Dato et al., 2007). Thus, genetic–demographic methods should be applied in future studies in the field because they allow the estimation of hazard rates and survival functions in relation to candidate genes (Yashin et al., 1999; Passarino et al., 2006; Dato et al., 2007).
In summary, despite the potential functional consequences of the SNPs we studied (rs16892496, rs7832552, and rs6552828) none of them was associated with EL. Similarly, no association was found for rs1800795. More research is needed in the field with other cohorts, using larger population samples, as well as younger elderly (e.g., aged 65–85 years) to assess the potential link between these genetic variants and the human aging process.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by the Fondo de Investigaciones Sanitarias (FIS, ref. # PI12/00914) from Spain, the programs Grant-in-Aid for Challenging Exploratory Research (24650414 to NF) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, a grant-in-aid for scientific research from the Ministry of Health, Labor, and Welfare of Japan (to MM), the National Natural Science Foundation of China (Grand Code:31100853) and the China Institute of Sport Science (2014-08). Carmen Fiuza-Luces is supported by a Postdoctoral Sara Borrell contract (file number CD14/00005) of the Institute of Health Carlos III (ISCIII).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fnagi.2015.00059/abstract
References
Albani, D., Batelli, S., Polito, L., Prato, F., Pesaresi, M., Gajo, G. B., et al. (2009). Interleukin-6 plasma level increases with age in an Italian elderly population (“The Treviso Longeva”-Trelong-study) with a sex-specific contribution of rs1800795 polymorphism. Age 31, 155–162. doi: 10.1007/s11357-009-9092-5
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bennermo, M., Held, C., Stemme, S., Ericsson, C. G., Silveira, A., Green, F., et al. (2004). Genetic predisposition of the interleukin-6 response to inflammation: implications for a variety of major diseases? Clin. Chem. 50, 2136–2140. doi: 10.1373/clinchem.2004.037531
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bouchard, C., Sarzynski, M. A., Rice, T. K., Kraus, W. E., Church, T. S., Sung, Y. J., et al. (2011). Genomic predictors of the maximal O(2) uptake response to standardized exercise training programs. J. Appl. Physiol. 110, 1160–1170. doi: 10.1152/japplphysiol.00973.2010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cederholm, T., Persson, M., Andersson, P., Stenvinkel, P., Nordfors, L., Madden, J., et al. (2007). Polymorphisms in cytokine genes influence long-term survival differently in elderly male and female patients. J. Intern. Med. 262, 215–223. doi: 10.1111/j.1365-2796.2007.01803.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Christensen, K., McGue, M., Petersen, I., Jeune, B., and Vaupel, J. W. (2008). Exceptional longevity does not result in excessive levels of disability. Proc. Natl. Acad. Sci. U.S.A. 105, 13274–13279. doi: 10.1073/pnas.0804931105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Coleman, R. A., Lewin, T. M., and Muoio, D. M. (2000). Physiological and nutritional regulation of enzymes of triacylglycerol synthesis. Annu. Rev. Nutr. 20, 77–103. doi: 10.1146/annurev.nutr.20.1.77
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dato, S., Carotenuto, L., and De Benedictis, G. (2007). Genes and longevity: a genetic-demographic approach reveals sex- and age-specific gene effects not shown by the case-control approach (APOE and HSP70.1 loci). Biogerontology 8, 31–41. doi: 10.1007/s10522-006-9030-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Di Bona, D., Vasto, S., Capurso, C., Christiansen, L., Deiana, L., Franceschi, C., et al. (2009). Effect of interleukin-6 polymorphisms on human longevity: a systematic review and meta-analysis. Ageing Res. Rev. 8, 36–42. doi: 10.1016/j.arr.2008.09.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ellis, J. M., Li, L. O., Wu, P. C., Koves, T. R., Ilkayeva, O., Stevens, R. D., et al. (2010). Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metab. 12, 53–64. doi: 10.1016/j.cmet.2010.05.012
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Emanuele, E., Fontana, J. M., Minoretti, P., and Geroldi, D. (2010). Preliminary evidence of a genetic association between chromosome 9p21.3 and human longevity. Rejuvenation Res. 13, 23–26. doi: 10.1089/rej.2009.0970
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fishman, D., Faulds, G., Jeffery, R., Mohamed-Ali, V., Yudkin, J. S., Humphries, S., et al. (1998). The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J. Clin. Invest. 102, 1369–1376. doi: 10.1172/JCI2629
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fiuza-Luces, C., Ruiz, J. R., Rodriguez-Romo, G., Santiago, C., Gomez-Gallego, F., Yvert, T., et al. (2011). Are ‘endurance’ alleles ‘survival’ alleles? Insights from the ACTN3 R577X polymorphism. PLoS ONE 6:e17558. doi: 10.1371/journal.pone.0017558
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Garatachea, N., Marin, P. J., and Lucia, A. (2013b). The ACE DD genotype and D-allele are associated with exceptional longevity: a meta-analysis. Ageing Res. Rev. 12, 1079–1087. doi: 10.1016/j.arr.2013.04.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Garatachea, N., Pinos, T., Camara, Y., Rodriguez-Romo, G., Emanuele, E., Ricevuti, G., et al. (2013a). Association of the K153R polymorphism in the myostatin gene and extreme longevity. Age 35, 2445–2454. doi: 10.1007/s11357-013-9513-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Garber, C. E., Blissmer, B., Deschenes, M. R., Franklin, B. A., Lamonte, M. J., Lee, I. M., et al. (2011). American college of sports medicine position stand. Quantity and quality of exercise for developing and maintaining cardiorespiratory, musculoskeletal, and neuromotor fitness in apparently healthy adults: guidance for prescribing exercise. Med. Sci. Sports Exerc. 43, 1334–1359. doi: 10.1249/MSS.0b013e318213fefb
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gondo, Y., Hirose, N., Arai, Y., Inagaki, H., Masui, Y., Yamamura, K., et al. (2006). Functional status of centenarians in Tokyo, Japan: developing better phenotypes of exceptional longevity. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 61, 305–310. doi: 10.1093/gerona/61.3.305
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hall, A. M., Smith, A. J., and Bernlohr, D. A. (2003). Characterization of the Acyl-CoA synthetase activity of purified murine fatty acid transport protein 1. J. Biol. Chem. 278, 43008–43013. doi: 10.1074/jbc.M306575200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
He, Z. H., Hu, Y., Li, Y. C., Yvert, T., Santiago, C., Gomez-Gallego, F., et al. (2011). Are calcineurin genes associated with athletic status? A function, replication study. Med. Sci. Sports Exerc. 43, 1433–1440. doi: 10.1249/MSS.0b013e31820e7f38
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kayaalti, Z., Sahiner, L., Durakoglugil, M. E., and Soylemezoglu, T. (2011). Distributions of interleukin-6 (IL-6) promoter and metallothionein 2A (MT2A) core promoter region gene polymorphisms and their associations with aging in Turkish population. Arch. Gerontol. Geriatr. 53, 354–358. doi: 10.1016/j.archger.2011.01.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Knight, J. C. (2005). Regulatory polymorphisms underlying complex disease traits. J. Mol. Med. 83, 97–109. doi: 10.1007/s00109-004-0603-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Larsson, L., Li, X., Teresi, A., and Salviati, G. (1994). Effects of thyroid hormone on fast- and slow-twitch skeletal muscles in young and old rats. J. Physiol. 481(Pt 1), 149–161. doi: 10.1113/jphysiol.1994.sp020426
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Leal, S. M. (2005). Detection of genotyping errors and pseudo-SNPs via deviations from Hardy-Weinberg equilibrium. Genet. Epidemiol. 29, 204–214. doi: 10.1002/gepi.20086
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lewis, S. J., and Brunner, E. J. (2004). Methodological problems in genetic association studies of longevity–the apolipoprotein E gene as an example. Int. J. Epidemiol. 33, 962–970. doi: 10.1093/ije/dyh214
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, X. G., Tan, L. J., Lei, S. F., Liu, Y. J., Shen, H., Wang, L., et al. (2009). Genome-wide association and replication studies identified TRHR as an important gene for lean body mass. Am. J. Hum. Genet. 84, 418–423. doi: 10.1016/j.ajhg.2009.02.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martin, G., Schoonjans, K., Lefebvre, A. M., Staels, B., and Auwerx, J. (1997). Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARalpha and PPARgamma activators. J. Biol. Chem. 272, 28210–28217. doi: 10.1074/jbc.272.45.28210
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mashek, D. G., Li, L. O., and Coleman, R. A. (2006). Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J. Lipid Res. 47, 2004–2010. doi: 10.1194/jlr.M600150-JLR200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mercado, P. A., Ayala, Y. M., Romano, M., Buratti, E., and Baralle, F. E. (2005). Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 33, 6000–6010. doi: 10.1093/nar/gki897
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Metter, E. J., Talbot, L. A., Schrager, M., and Conwit, R. A. (2004). Arm-cranking muscle power and arm isometric muscle strength are independent predictors of all-cause mortality in men. J. Appl. Physiol. 96, 814–821. doi: 10.1152/japplphysiol.00370.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Passarino, G., Montesanto, A., Dato, S., Giordano, S., Domma, F., Mari, V., et al. (2006). Sex and age specificity of susceptibility genes modulating survival at old age. Hum. Hered. 62, 213–220. doi: 10.1159/000097305
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pereira, D. S., Mateo, E. C., de Queiroz, B. Z., Assumpcao, A. M., Miranda, A. S., Felicio, D. C., et al. (2013). TNF-alpha, IL6, and IL10 polymorphisms and the effect of physical exercise on inflammatory parameters and physical performance in elderly women. Age 35, 2455–2463. doi: 10.1007/s11357-013-9515-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Phillips, C. M., Goumidi, L., Bertrais, S., Field, M. R., Cupples, L. A., Ordovas, J. M., et al. (2010). Gene-nutrient interactions with dietary fat modulate the association between genetic variation of the ACSL1 gene and metabolic syndrome. J. Lipid Res. 51, 1793–1800. doi: 10.1194/jlr.M003046
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Robine, J. M., and Paccaud, F. (2005). Nonagenarians and centenarians in Switzerland, 1860-2001: a demographic analysis. J. Epidemiol. Commun. Health 59, 31–37. doi: 10.1136/jech.2003.018663
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ruiz, J. R., Sui, X., Lobelo, F., Morrow, J. R. Jr., Jackson, A. W., Sjostrom, M., et al. (2008). Association between muscular strength and mortality in men: prospective cohort study. BMJ 337:a439. doi: 10.1136/bmj.a439
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sasabe, T., Furukawa, A., Matsusita, S., Higuchi, S., and Ishiura, S. (2007). Association analysis of the dopamine receptor D2 (DRD2) SNP rs1076560 in alcoholic patients. Neurosci. Lett. 412, 139–142. doi: 10.1016/j.neulet.2006.10.064
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Serrano, A. L., Baeza-Raja, B., Perdiguero, E., Jardi, M., and Munoz-Canoves, P. (2008). Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 7, 33–44. doi: 10.1016/j.cmet.2007.11.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tabor, H. K., Risch, N. J., and Myers, R. M. (2002). Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat. Rev. Genet. 3, 391–397. doi: 10.1038/nrg796
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Terry, D. F., Sebastiani, P., Andersen, S. L., and Perls, T. T. (2008). Disentangling the roles of disability and morbidity in survival to exceptional old age. Arch. Intern. Med. 168, 277–283. doi: 10.1001/archinternmed.2007.75
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Trikalinos, T. A., Salanti, G., Khoury, M. J., and Ioannidis, J. P. (2006). Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am. J. Epidemiol. 163, 300–309. doi: 10.1093/aje/kwj046
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Walston, J. D., Matteini, A. M., Nievergelt, C., Lange, L. A., Fallin, D. M., Barzilai, N., et al. (2009). Inflammation and stress-related candidate genes, plasma interleukin-6 levels, and longevity in older adults. Exp. Gerontol. 44, 350–355. doi: 10.1016/j.exger.2009.02.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yashin, A. I., De Benedictis, G., Vaupel, J. W., Tan, Q., Andreev, K. F., Iachine, I. A., et al. (1999). Genes, demography, and life span: the contribution of demographic data in genetic studies on aging and longevity. Am. J. Hum. Genet. 65, 1178–1193. doi: 10.1086/302572
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zou, G. Y., and Donner, A. (2006). The merits of testing Hardy-Weinberg equilibrium in the analysis of unmatched case-control data: a cautionary note. Ann. Hum. Genet. 70(Pt 6), 923–933. doi: 10.1111/j.1469-1809.2006.00267.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: centenarians, polymorphisms, luciferase reporter, gene association study, muscle and sarcopenia
Citation: Fuku N, He Z, Sanchis-Gomar F, Pareja-Galeano H, Tian Y, Arai Y, Abe Y, Murakami H, Miyachi M, Zempo H, Naito H, Yvert T, Verde Z, Venturini L, Fiuza-Luces C, Santos-Lozano A, Rodriguez-Romo G, Ricevuti G, Hirose N, Emanuele E, Garatachea N and Lucia A (2015) Exceptional longevity and muscle and fitness related genotypes: a functional in vitro analysis and case-control association replication study with SNPs THRH rs7832552, IL6 rs1800795, and ACSL1 rs6552828. Front. Aging Neurosci. 7:59. doi: 10.3389/fnagi.2015.00059
Received: 09 January 2015; Accepted: 07 April 2015;
Published: 06 May 2015.
Edited by:
Adolfo Lopez De Munain, Hospital Universitario Donostia, SpainReviewed by:
Giuseppe Passarino, University of Calabria, ItalySerena Dato, University of Calabria, Italy
Copyright © 2015 Fuku, He, Sanchis-Gomar, Pareja-Galeano, Tian, Arai, Abe, Murakami, Miyachi, Zempo, Naito, Yvert, Verde, Venturini, Fiuza-Luces, Santos-Lozano, Rodriguez-Romo, Ricevuti, Hirose, Emanuele, Garatachea and Lucia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alejandro Lucia, School of Doctorate Studies and Research, European University of Madrid, C/Tajo S/N, Urbanización El Bosque 28670 Villaviciosa de Odón, Madrid, Spain, alejandro.lucia@uem.es
†These authors have contributed equally to this work.
‡Share senior authorship.
 Noriyuki Fuku
Noriyuki Fuku Zi-hong He2†
Zi-hong He2†  Fabian Sanchis-Gomar
Fabian Sanchis-Gomar Haruka Murakami
Haruka Murakami Motohiko Miyachi
Motohiko Miyachi Hisashi Naito
Hisashi Naito Thomas Yvert
Thomas Yvert Letizia Venturini
Letizia Venturini Gabriel Rodriguez-Romo
Gabriel Rodriguez-Romo Giovanni Ricevuti
Giovanni Ricevuti Nobuyoshi Hirose
Nobuyoshi Hirose Nuria Garatachea
Nuria Garatachea Alejandro Lucia
Alejandro Lucia