- 1Department of Life Science and Technology, Heilongjiang Bayi Agricultural University, Daqing, China
- 2Key Laboratory of Earth and Planetary Physics, Institute of Geology and Geophysics, Chinese Academy of Sciences, Beijing, China
- 3Laboratory for Marine Geology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 4France-China Biomineralization and Nano-structures Laboratory, Chinese Academy of Sciences, Beijing, China
- 5IMPMC, Centre National de la Recherche Scientifique, UMR 7590, Sorbonne Universités, MNHN, UPMC, IRD UMR 206, Paris, France
- 6France Chimie Me′tallurgique des Terres Rares, ICMPE, UMR 7182, Centre National de la Recherche Scientifique, Thiais, France
Magnetotactic bacteria (MTB) are morphologically and phylogenetically diverse prokaryotes. They can form intracellular chain-assembled magnetite (Fe3O4) or greigite (Fe3S4) nanocrystals each enveloped by a lipid bilayer membrane called a magnetosome. Magnetotactic cocci have been found to be the most abundant morphotypes of MTB in various aquatic environments. However, knowledge on magnetosome biomineralization within magnetotactic cocci remains elusive due to small number of strains that have been cultured. By using a coordinated fluorescence and scanning electron microscopy method, we discovered a unique magnetotactic coccus strain (tentatively named SHHC-1) in brackish sediments collected from the estuary of Shihe River in Qinhuangdao city, eastern China. It phylogenetically belongs to the Alphaproteobacteria class. Transmission electron microscopy analyses reveal that SHHC-1 cells formed many magnetite-type magnetosomes organized as two bundles in each cell. Each bundle contains two parallel chains with smaller magnetosomes generally located at the ends of each chain. Unlike most magnetotactic alphaproteobacteria that generally form magnetosomes with uniform crystal morphologies, SHHC-1 magnetosomes display a more diverse variety of crystal morphology even within a single cell. Most particles have rectangular and rhomboidal projections, whilst others are triangular, or irregular. High resolution transmission electron microscopy observations coupled with morphological modeling indicate an idealized model—elongated octahedral crystals, a form composed of eight {111} faces. Furthermore, twins, multiple twins and stack dislocations are frequently observed in the SHHC-1 magnetosomes. This suggests that biomineralization of strain SHHC-1 magnetosome might be less biologically controlled than other magnetotactic alphaproteobacteria. Alternatively, SHHC-1 is more sensitive to the unfavorable environments under which it lives, or a combination of both factors may have controlled the magnetosome biomineralization process within this unique MTB.
Introduction
Magnetotactic bacteria (MTB) are a group of geographically widespread prokaryotes sharing the common ability to form magnetite (Fe3O4) or greigite (Fe3S4) nanocrystals intracellularly, each surrounded by a lipid bilayer membrane (Bazylinski and Frankel, 2004). These intracellular magnetic particles are called magnetosomes that generally arrange into chain structures (Balkwill et al., 1980). They serve as a cellular biocompass, enabling the MTB cells to passively align along the geomagnetic field lines. Using this compass, MTB can actively swim, to their favorite niches at or just below the oxic-anoxic interface (OAI) in aquatic environments (Frankel et al., 1997; Bazylinski and Frankel, 2004). This process is termed as magneto-aerotaxis (Frankel et al., 1997; Lefèvre et al., 2014). In order to better understand magneto-aerotaxis, the study of the diversity and biomineralization of MTB is required. Moreover, a more comprehensive understanding of MTB is required for the potential application of magnetosomes in biotechnological and biomedical fields (Faivre and Schüler, 2008; Li et al., 2013; Pósfai et al., 2013a). Fossilized remains of MTB (i.e., magnetofossils) have also been tentatively used to retrieve paleomagnetic and paleoenvironmental information from ancient sediments (Hesse, 1994; Yamazaki and Kawahata, 1998; Snowball et al., 2002; Roberts et al., 2011; Larrasoana et al., 2014; Liu et al., 2015), and to trace the origin and evolution of life on Earth and even perhaps Mars (Chang and Kirschvink, 1989; McKay et al., 1996; Thomas-Keprta et al., 2002).
MTB that have been detected so far have many morphotypes, including spirillum, coccus, vibrio, ovoid, rod-shaped, and even multicellular bacteria. These known cultured and uncultured MTB have been assigned to the Alpha-, Gamma-, and Delta-proteobacteria classes of the Proteobacteria phylum (Lefèvre and Bazylinski, 2013), the Nitrospirae phylum (Spring et al., 1993), and even the “Candidatus Omnitrophica” phylum (Kolinko et al., 2012). Despite the morphological and phylogenetic diversity of MTB, the most commonly observed types present in natural environments are coccoid-to-ovoid cells, the so-called magnetococci (Moench and Konetzka, 1978; Spring et al., 1992, 1995; Thornhill et al., 1995; Cox et al., 2002; Flies et al., 2005; Pan et al., 2008, 2009; Lin and Pan, 2009; Zhang et al., 2012; Chen et al., 2015; Abreu et al., 2016). Previous 16S rDNA phylogenetic analyses have shown that magnetococci are also highly diverse. Most magnetococci are not closely related to other Alphaproteobacteria and form an independent clade within the Alphaproteobacteria (i.e., the Magnetococcales order) that is basal to the rest of the group (Bazylinski et al., 2013). This phylogenetic diversity is consistent with a broad morphological diversity of magnetosomes among different species or strains of magnetococci. In particular, the chain assemblages of magnetosomes are diverse, e.g., a single chain (Lefèvre et al., 2009; Pan et al., 2009; Bazylinski et al., 2013), double chains (Pan et al., 2008), multiple chains (Lin et al., 2009), and dispersed aggregates or clusters (Moench and Konetzka, 1978; Flies et al., 2005; Lin and Pan, 2009; Zhang et al., 2012) can occur. Many magnetococci contain sulfur globules and/or calcium polyphosphate inclusions in addition to magnetosomes within a single cell, suggesting their great potential for iron, sulfur, and phosphorus cycling in natural environments (Cox et al., 2002; Araujo et al., 2016). Despite the wide distribution and high diversity of MTB, only three magnetococci strains have been axenically cultured so far: the Magnetococcus marinus strain MC-1 (Bazylinski et al., 2013), the MO-1 strain (Lefèvre et al., 2009), and the Magnetofaba australis stain IT-1 (Morillo et al., 2014). They all form magnetosomes organized as a single chain. In addition, the spatial arrangements and crystal morphologies of magnetosomes remain poorly documented for many uncultured and yet phylogenetically constrained magnetococci.
In order to expand our knowledge of the diversity of magnetococci and the crystallographic properties of their magnetosomes, we studied a unique magnetotactic coccus tentatively named strain SHHC-1 from brackish sediments collected in the estuary of the Shihe River in Qinhuangdao City, eastern China. In order to characterize it, we used a recently developed method called coupled FISH-SEM (Li et al., 2017). This method enabled the rapid phylogenetic and biomineralogical characterization of uncultured MTB at the single-cell level by coordinating fluorescence with scanning electron microscopy (SEM) (Li et al., 2017). Transmission electron microscopy (TEM) analyses were further performed on this magnetococcus to reveal the crystallographic features of its magnetosomes in high detail.
Materials and Methods
Sediment Sampling and Sample Preparation
The MTB-bearing sediment samples were collected from the estuary of the Shihe River in Qinhuangdao city, eastern China. The sampling site was located inside the estuary of Shihe River into the Bohai Sea (39°57′55.6″N, 119°47′9.4″E). The sediment comes from a brackish lacustrine environment with a salinity of ~23.9 ppt, a pH of 7.5 and a temperature of 19°C measured in July 2015. Surface sediments were collected near the shore at water depths of ~1–2 meters and transferred to 1-l plastic bottles with a sediments: water ratio of ~2:1. The bottles were shipped back to the laboratory and stored at room temperature (~20°C) in dim light to set up microcosms. Magnetotacic bacteria in the microcosms were routinely checked with the hanging-drop technique (Schüler, 2002) using an Olympus microscope BX51 equipped with phase-contrast, fluorescence and a DP70 digital camera system (Olympus Corp., Tokyo, Japan). Although they were collected from the same sample site, MTB populations differed among individual microcosms after several weeks of incubation in the laboratory, and even varied within the same microcosm with time (Li et al., 2017). Here, we focused on one microcosm dominated by one group of magnetococci (i.e., SHHC-1) that swam toward the south pole of a bar magnet (north-seeking MTB) (Figure 1A).
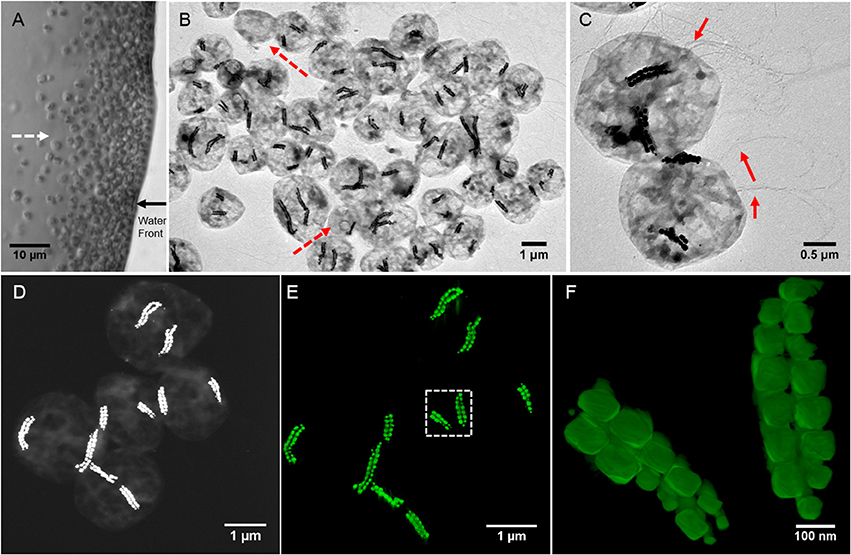
Figure 1. Morphological features of SHHC-1 cells. (A) Optical microscopy image of living SHHC-1 cells swimming out from one small drop of sediment on the left (photograph not shown) along the applied magnetic field lines (indicated by the white dashed line arrow), and accumulating at the edge of the water droplets (indicated by the black solid line arrow). (B) Low-magnification TEM image of SHHC-1 cells collected from sediments. The red dash line arrows indicate other types of MTB or non-MTB cells. (C) TEM image of two SHHC-1 cells showing their magnetosome chains and flagella (indicated by the red line arrows). (D) HAADF-STEM image of five SHHC-1 cells for HAADF-STEM tomography observations. (E) HAADF-STEM tomographic image (three-dimensional visualization) showing the spatial arrangement of double bundles of magnetosome chains within the five SHHC-1 cells shown in (D). (F) HAADF-STEM tomographic image of magnetosome chains within the SHHC-1 cell indicated by the white dashed box in (E).
Approximately 200 ml of slurry from this microcosm was used to magnetically concentrate the SHHC-1 cells by using a modified magnetic separation apparatus previously described in Jogler et al. (2009). The collected MTB cells were washed three times with Milli-Q water and then re-suspended into ~100 μl of Milli-Q water for additional experiments: ~5 μl of MTB cells were used for TEM analyses, ~20 μl were boiled for 10 min and stored at −20°C for PCR amplification of 16S rRNA genes, and the remaining ~75 μl were prepared for coupled FISH-SEM analyses as previously described in Li et al. (2017). TEM samples were stored in a pure N2 atmosphere prior to TEM observations.
Phylogenetic Analyses and Bacterial Identification
SixteenS rRNA genes of the MTB cells were amplified using the universal bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Lane, 1991), and sequenced using protocol described in Li et al. (2017). Briefly, each 50 μl PCR mixture contained 1 μl of template, 25 μl of DreamTaq PCR Master Mix (MBI Fermentas), 2 μl of each primer (10 μM), and 20 μl of Milli-Q water. The PCR conditions were 95°C for 3 min, 30 cycles at 95°C for 1 min, 55°C for 1.5 min, 72°C for 1.5 min, and a final 10 min extension at 72°C. To avoid potential amplification biases, triplicate PCR products from each sample were pooled and purified using 0.8% (w/v) agarose gel electrophoresis (described below). All PCR controls without added template were negative. PCR products were purified using an E.Z.N.A.® Gel Extraction Kit (Omega Bio-tek, Inc. USA). They were ligated with the pMD19-T vector (TaKaRa, Japan), then cloned in Escherichia coli (strain DH5α) competent cells (Tiangen, Beijing, China) according to the manufacturer instructions. Thirty clones were randomly picked and sequenced using the vector primers M13-47 (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and RV-M (5′-GAGCGGATAACAATTTCACACAGG-3′) at the Huada Genome Center (Beijing, China). After discarding sequences of insufficient length (<1,000 bp), the remaining sequences were then aligned with their close relatives using the ClustalW algorithm for manual correction, and a phylogenetic tree was subsequently constructed using the neighbor-joining method (Saitou and Nei, 1987) in the MEGA software package (v. 7.0) (Kumar et al., 2016). Bootstrap values were calculated with 1,000 replicates.
To phylogenetically and morphologically identify the SHHC-1 cells, the probe SHHC1228 (5′-CTCCAGGTCACCCATTCGCCGCTCT-3′, positions 1,228–1,252) was designed to target specifically all 16S rRNA gene sequences identified in this study. Probe specificity was evaluated by using the online probe evaluation tools probeCheck and probeBase (Loy et al., 2008; Greuter et al., 2016). No other sequence in the SILVA111 database has a matching complementary sequence (the minimum number of mismatches is 2). The universal bacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) was used as a positive control probe of bacteria for FISH (Amann et al., 1990). Probe EUB338 was synthesized and fluorescently labeled with fluorescein phosphoramidite FAM at the 5′ end, while the SHHC1228 probe was synthesized and fluorescently labeled with the hydrophilic sulfoindocyanine dye Cy3 at the 5′ end. E. coli cells were added in approximately same amount to the SHHC-1 cells and were used as inner control cells of non-magnetotactic bacteria.
Coordinated FISH-SEM analysis was carried out using the procedure described in Li et al. (2017). Briefly, ~1 μl of cell mixtures (E. coli + SHHC-1) were placed directly on a high-precision cover glass (Paul Marienfeld GmbH & Co. KG, Germany) and dried in air at ambient temperature, followed by gradual dehydration in 50, 80, and 100% ethanol baths (3 min in each bath). In situ hybridization was performed at 46°C for 3 h in 9 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.5], 0.02% [wt/vol] sodium dodecyl sulfate [SDS], 35% [vol/vol] formamide). One microliter of the EUB338 probe (50 ng/μl) and 1 μl of the SHHC1228 probe of interest (50 ng/μl) was then added to the mixture. After 3 h of hybridization the samples were incubated in a washing buffer (0.08 M NaCl, 20 mM Tris-HCl [pH 7.5], 5 mM EDTA [pH 8.0], 0.01% [wt/vol] SDS) at 48°C for 30 min, then washed in Milli-Q water three times, dried in air at ambient temperature and finally observed using an Olympus epifluorescent microscope BX51. After fluorescence microscopy observations, the cells mounted on cover glasses were coated with carbon using a Leica ACE200 Low Vacuum Sputter Coater (Leica Microsystems, Wetzlar, Germany), and observed using a Zeiss Ultra-55 field-emission gun scanning electron microscope (Carl Zeiss, Germany) operating at 5 kV.
Transmission Electron Microscopy Analyses
Conventional TEM observations were performed on a JEM2100 microscope (JEOL Ltd., Tokyo, Japan) operating at 200 kV at the IGG-CAS (Beijing, China). High resolution TEM (HRTEM) observations were carried out on a JEM-2100F microscope (JEOL Ltd., Tokyo, Japan) operating at 200 kV at the IMPMC (Paris, France). This microscope was equipped with a field emission gun, an ultra-high resolution (UHR) pole piece, a JEOL detector with an ultrathin window, and a scanning TEM (STEM) device. STEM Z-contrast images were acquired in high angle annular dark field (HAADF) mode. Chemical composition analyses were performed by energy dispersive X-ray spectrometry (EDXS) elemental mapping in HAADF-STEM mode. HAADF-STEM tomography was performed on a FEI Tecnai F20 microscope at 200 kV at the ICMPE (Thiais, France). Acquisition was made using the Digital MicrographTM (Gatan) STEM tomography module, and 3-D reconstruction was performed using the DigiECT software (available at http://www.digisens3d.com/fr/logiciel-tomographie/soft/23D_Electron_Tomography_Software.html).
The crystal length (along the long axis) and width (perpendicular to the long axis) of the magnetosomes were measured from the TEM images. The shape factor was defined as width/length. Crystal habits of magnetosomes were determined by a combination of Fourier analyses of HRTEM images of individual particles and crystallographic investigations, as previously described in Faivre et al. (2008) and Li et al. (2015). For each particle, the zone axis and in plane crystallographic directions were determined from the 2-D Fast Fourier transform (FFT) of the HRTEM image, enabling the the stereographic projection to be determined. Idealized shapes modeled using the KrystalShaper software package (available at http://www.jcrystal.com/) were then compared to the observed ones.
Results
Morphology and Ultrastructure of SHHC-1 Cells
TEM observations showed that the magnetically concentrated cells contained predominantly one group of magnetococci, called SHHC-1, with two bundles of magnetosome chains, and a few (< 5%) other magnetotactic cocci forming single magnetosome chain or non-MTB cells (Figure 1B). The SHHC-1 cells were coccoid in shape with an average diameter of 2.1 ± 0.4 μm (n = 150). They contained two bundles of flagella, and with an average magnetosome number of 38 ± 17 per cell (n = 150) (Figure 1C). HAADF-STEM tomography observations clearly evidenced that each bundle was composed of two parallel closely assembled magnetosome chains (Figures 1D–F and Supplementary movie). Smaller magnetosomes were often observed at the ends of the chains. Within the same SHHC-1 cell, the two bundles of magnetosome chains were commonly distributed on opposite sides of the cell body in close proximity to the cell membrane. Such an arrangement of the magnetosomes in multiple chains supports the previous assumption that repulsion forces between parallel magnetic dipoles may drive the chains or bundle of chains apart from one another. Thus, the magnetic torque acting on them by the external applied or the geomagnetic field can be effectively transferred to the entire cell body (Hanzlik et al., 1996). In the case of SHHC-1, each bundle of chains may behave as one enlarged SD particle because all the individual magnetosomes may have their magnetic moments uniformly aligned along the chain axis, as previously shown by electron holography observations performed on another uncultured, phylogenetically unknown, magnetococcus strain having two bundles of magnetosome chains (Simpson et al., 2005).
STEM-EDXS mapping (in HAADF mode) clearly shows that the SHHC-1 magnetosomes are rich in Fe and O, consistently with magnetite. Sulfur-rich globules with various sizes, ranging from several to hundreds of nanometers were also often observed within the SHHC-1 cells (Figure 2). These features were consistent with previous observations of many uncultured magnetococci that can form sulfur globules as well as magnetite-type magnetosomes, even when sulfide is absent in the sediment sample (Moench, 1988; Cox et al., 2002; Araujo et al., 2016). This confirmed that most of magnetococci cells may have an autotrophic or mixotrophic metabolism based on the oxidization of reduced sulfur compounds (Araujo et al., 2016).
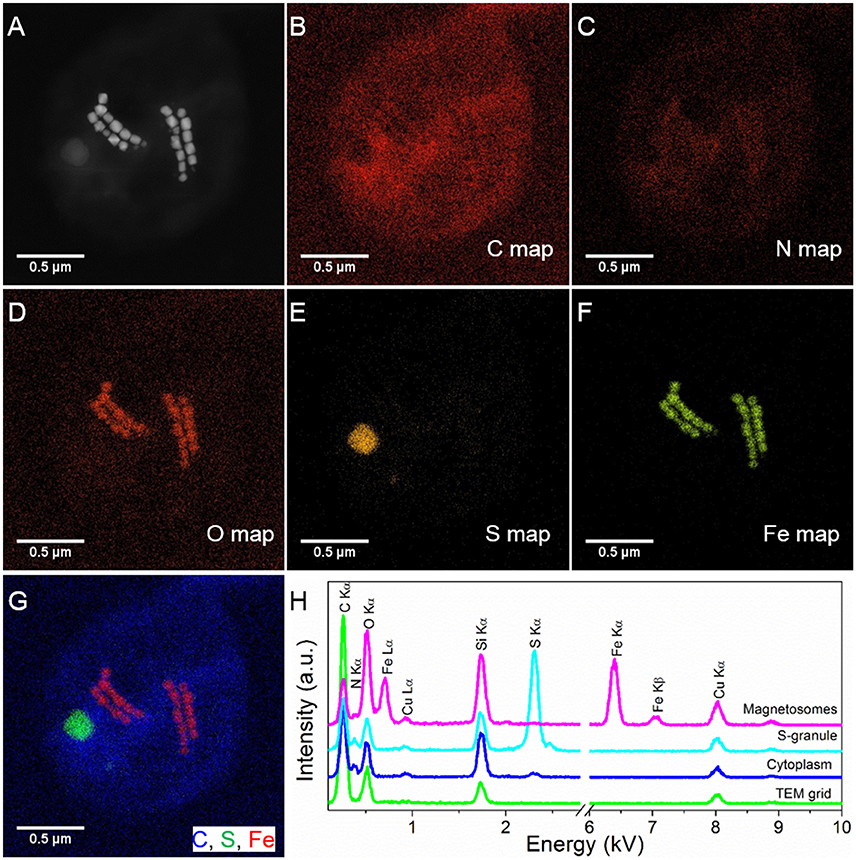
Figure 2. STEM-EDXS mapping (HAADF mode) of SHHC-1 cells. (A–F) HAADF-STEM image of a single SHHC-1 cell (A), and the corresponding chemical maps of C (C Kα) (B), N (N Kα) (C), O (O Kα) (D), S (S Kα) (E), and Fe (Fe Kα) (F). (G) Overlapped map of Fe (red), C (blue), and S (green). (H) EDXS spectra extracted from the carbon film covering TEM grid (green line), cell wall (blue line), sulfur-rich particle (light blue line), and magnetosomes (pink line).
Phylogeny of the SHHC-1 Strain
To phylogenetically and structurally identify the strain SHHC-1, 16S rRNA genes of the magnetically enriched cells were amplified with a total of 30 clones being sequenced. Twenty-three 16S rRNA gene sequences with full-length had more than 99% of sequence identity. Coordinated FISH-SEM analysis using the SHHC1228 FISH probe designed to match the 23 identical 16S rRNA gene sequences was conducted on the magnetically enriched cells. As shown in Figures 3A–D, all bacterial cells, including the inner control E. coli cells, were positively labeled (green) by the 5′-FAM-labeled universal bacterial probe EUB338, while only the group of magnetococci with two bundle of magnetosome chains were positively labeled (red) by the 5′-Cy3-labeled SHHC-1-specific probe SHHC1228. This confirmed that the twenty-three 16S rRNA gene sequences corresponded to strain SHHC-1. Phylogenetic analyses based on the 16S rRNA gene shows that the strain of SHHC-1 affiliates with the Alphaproteobacteria class of the Proteobacteria phylum (Figure 3E). Moreover, it was phylogenetically close to (1) the uncultured magnetotactic coccus sp. clone WHI-8 (97% sequence identity), detected in intertidal sediments of the Xiaoshi Island in the North Yellow Sea (Weihai, China) (Chen et al., 2015), and (2) the uncultured magnetotactic coccus sp. CF24 (AJ863157) (95% sequence identity), detected in marine sediments in Germany (Flies et al., 2005). However, the bacterial and magnetosome morphologies of both WHI-8 and CF24 clones have not been described due to that both studies did not link their 16S rRNA gene sequences to the bacterial cells (Flies et al., 2005; Chen et al., 2015). Furthermore, to our knowledge, the morphological features and spatial arrangement of SHHC-1 magnetosomes have not been reported in any cultured and uncultured magnetococci. Therefore, SHHC-1 represents a novel magnetotactic coccus morphotype which is detected from the estuary of the Shihe River in Qinhuangdao City, eastern China.
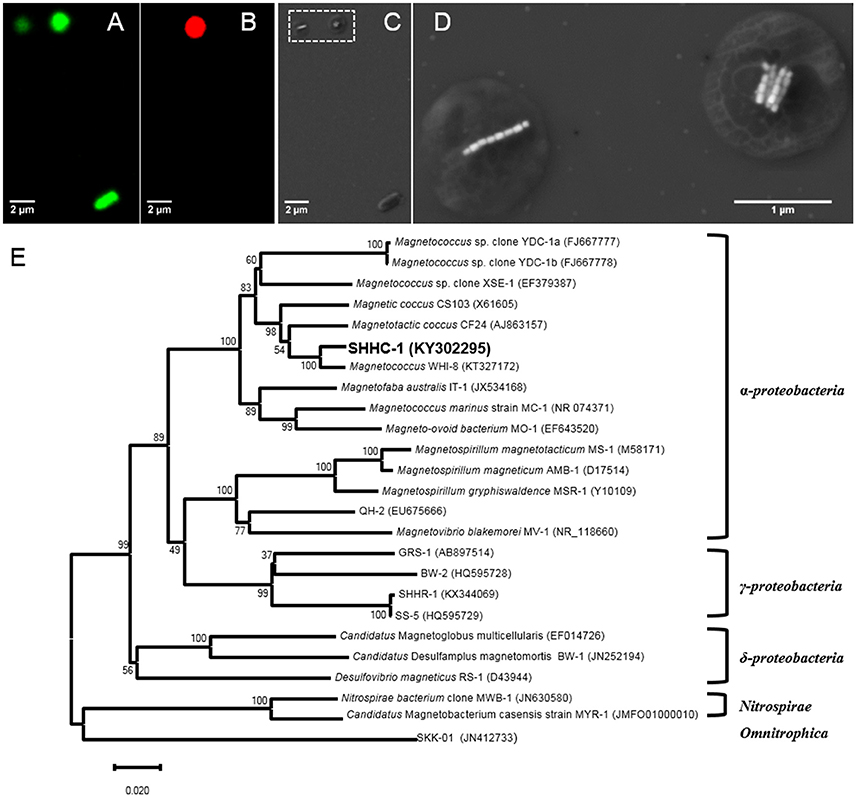
Figure 3. (A–D) Phylogenetic and structural identification of SHHC-1 cells by coupled FISH-SEM. (A) Fluorescence microscopy image of SHHC-1 cells in situ hybridized with the 5′-FAM-labeled universal bacterial probe EUB338. (B) Fluorescence microscopy image of SHHC-1 cells hybridized in situ with the 5′-Cy3-labeled SHHC-1-specific probe SHHC1228. (C) Low-magnification SEM image of the same field of view as in (A). (D) High-magnification SEM images of two magnetococci cells outlined in image (C) by a white dashed square (E) Phylogenetic tree, based on 16S rRNA gene sequences, showing the position of strain SHHC-1 in the Chromatiales order of the Alphaproteobacteria class. Bootstrap values (higher than 50) at nodes are percentages of 1,000 replicates. Bar represents 2% sequence divergence.
Crystal Morphology of SHHC-1 Magnetosomes
Magnetite crystals of SHHC-1 had an average length of 83.3 ± 19.2 nm and an average width of 69.9 ± 17.3 nm with a shape factor of 0.84 ± 0.07 (n = 416) (Figures 4A–C). Both the crystal length and width distributions were negatively skewed and the width/length ratio was nearly constant. These are typical features for prismatic and cuboctahedral magnetite magnetosomes (Devouard et al., 1998; Isambert et al., 2007). This indicates that magnetosomes within SHHC-1 grew homothetically within magnetosome membranes, a mechanism similar to that identified in known magnetotactic Alphaproteobacteria (Li et al., 2009; Jandacka et al., 2013). However, unlike what is generally observed in other cultured and uncultured magnetotactic Alphaproteobacteria where prismatic and cuboctahedral magnetosomes have a relatively consistent morphology (Pósfai et al., 2013a), there is a large diversity of magnetosome shapes in SHHC-1 even within the same cell (Figures 4D,E). Most particles had rectangular (~53%, n = 286) and rhomboidal (~26%) projections, whilst others displayed triangular (~6%) or irregular (~15%) shapes (Figure 4E). HRTEM observations performed on dozens of individual magnetosomes coupled with morphological modeling demonstrates that most particles with larger sizes (i.e., mature magnetosomes) are consistent with elongated octahedrons composed of only {111} faces (Figures 5E–L). Variations in the elongation ratios occur among individual magnetosomes (Supplementary HRTEM images). A few particles generally with smaller sizes (i.e., immature magnetosomes) have a cuboctahedral-like shape based on {111}, {110}, and {100} faces (Figures 5A–D). In addition, crystal defects including twining, multiply twining, and stack dislocations were very often observed within SHHC-1 magnetosomes (~15% of defect frequency, n = 286) (Figures 4E, 6).
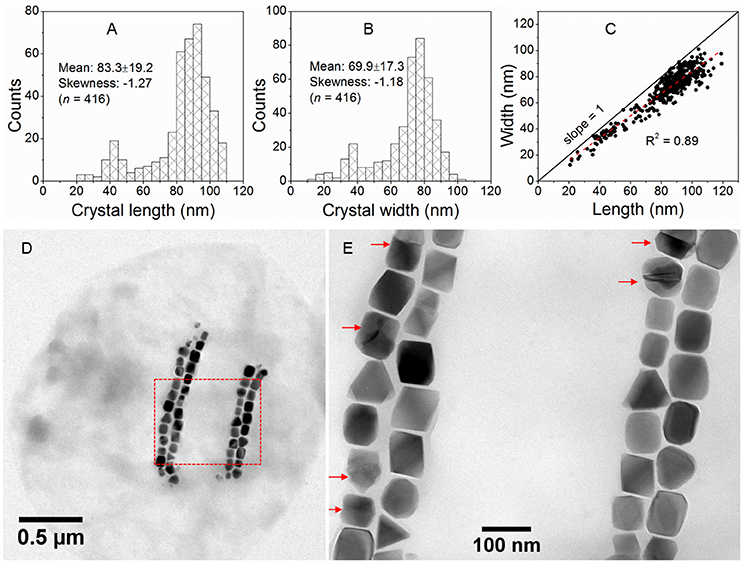
Figure 4. Morphological features of SHHC-1 magnetosomes. (A,B) Histograms of magnetosome length (A) and magnetosome width (B). (C) Plot of crystal length vs. width of magnetosomes showing a linear relationship between crystal length and width of SHHC-1 magnetosomes. (D) Bright-field TEM image of one of the single SHHC-1 cells. (E) Close-up on the area outlined in image (D) by a red dashed rectangle. The red arrows indicate twin crystals of magnetosomes.
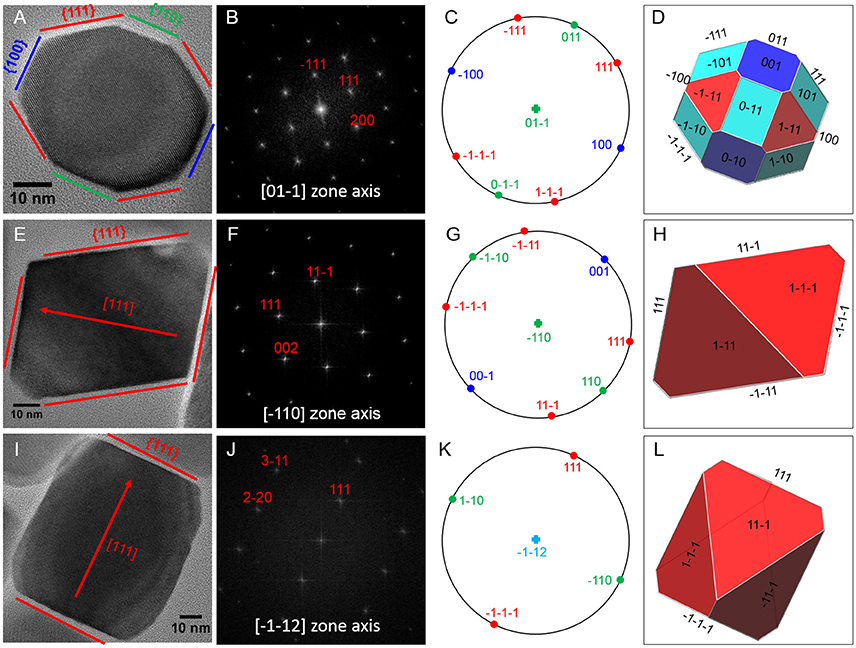
Figure 5. HRTEM analyses of three representative SHHC-1 magnetosomes: (A) an immature magnetosome recorded from a [0–11] zone axis, (E) a mature magnetosome recorded from a [–110] zone axis, and (I) a mature magnetosome recorded from a [–1–12] zone axis. The first column shows HRTEM images of individual magnetosomes (A,E,I); the second column shows their corresponding indexed Fast Fourier Transforms (FFT) patterns (B,F,J); the third columns shows stereographic projections (C,G,K); and the forth columns shows the morphological modes (D,H,L). For mature magnetosomes (E,I), the outlines and lattice fringes of the magnetosomes in their HRTEM images are consistent with the octahedrons based only on {111} faces. The immature one (A) has a cuboctahedral-like shape based on {111}, {110}, and {100} faces.
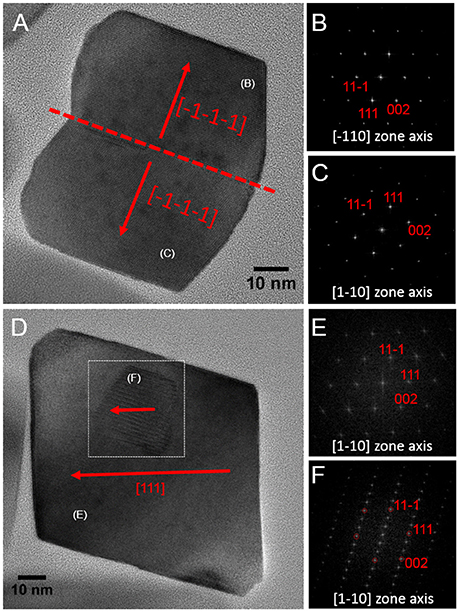
Figure 6. HRTEM images of defected crystals of SHHC-1 magnetosomes. (A) Bright Field HRTEM image of one twin magnetosome crystal with a straight contact (–1–1–1) plane (A); corresponding indexed FFT patterns from the upper (B) and lower (C) parts of the crystal shown in (A). (D) HRTEM of one defected magnetosome containing a stacking fault in it (indicated by white dashed square); corresponding indexed FFT patterns from the normal (E) and the defected (F) areas.
Discussion and Conclusions
FISH with rRNA-targeted oligonucleotide probes has become a widely used method for the phylogenetic identification of MTB cells from various freshwater and marine sediments (Spring et al., 1992, 1995; Flies et al., 2005; Pan et al., 2008; Lin et al., 2011, 2012; Wang et al., 2013; Taoka et al., 2014). However, traditional optical and fluorescence microscopy cannot detect the structural features of magnetosomes due to their limitation in spatial resolution at around 0.2 μm. In the past decades TEM has made significant progress on both spatial and energy resolution, and now provides a powerful platform for obtaining simultaneous structural, compositional, and magnetic characterization of a single MTB cell down to the atomic scale (Pósfai et al., 2013b; Li and Pan, 2015; Li et al., 2015). On the other hand, TEM cannot provide phylogenetic affiliation information on the bacteria. This may explain why only a few uncultured MTB strains have been identified at the single-cell level both phylogenetically and structurally. In an early study, Spring et al. (1998) developed a method for linking the ultrastructure of enriched MTB cells with their 16S rRNA sequence via TEM of ultrathin sections that were hybridized in situ with digoxigenin- and fluorescein-labeled polynucleotide probes. The authors found one ovoid MTB from the Itaipu lagoon in Rio de Janeiro, that has Itaipu I 16S rRNA genotype and forms unusually large magnetosomes (Spring et al., 1998). Based on a targeted phylogenetic and ultrastructural analysis of micromanipulated single cells, Kolinko et al. (2012) detected a large ovoid cell, “SKK-01,” belonging to the proposed “Candidatus Omnitrophica” phylum. Here, the coupled FISH-SEM approach has successfully identified the SHHC-1 cells at the single-cell level from non-magnetotactic bacteria (i.e., E. coli) and from other magnetococci such as those forming single magnetosome chain (Figure 3). This demonstrates the feasibility and effectiveness of this coupled approach on those uncultured MTB cells that are dominant populations within the magnetically enriched cells, providing opportunities for the study of bacterial diversity and magnetosome biomineralization in uncultured magnetococci.
Magnetosomes formed by the cultured M. marinus strain MC-1 were reported to be elongated prismatic magnetites which can be idealized as hexagonal prisms elongated along a <111> direction and composed of six large {110} side faces, two large {111} end faces, and other small {110} and {100} truncated faces (Meldrum et al., 1993; Devouard et al., 1998). Within MC-1, magnetosomes were organized as a single chain with the chain direction parallel to the elongation direction of individual particles, i.e., [111] direction (Meldrum et al., 1993). In contrast, HRTEM observations performed on many individual magnetosomes coupled with morphological modeling demonstrates that most of mature magnetosomes produced by SHHC-1 are slightly elongated octahedrons only composed of {111} faces. This crystal habit is similar to that reported in strain IT-1 by Araujo et al. (2016). However, the spatial arrangement of magnetosomes within strain SHHC-1 are quite different from that in the cultured strains MC-1, MO-1, and IT-1. For instance, there are two bundle chains in SHHC-1 and one single chain in MC-1, MO-1, and IT-1 (Lefèvre et al., 2009; Bazylinski et al., 2013; Araujo et al., 2016; this study). Based on studies of cultured and uncultured MTB strains (Meldrum et al., 1993; Faivre et al., 2008; Lefèvre et al., 2011a; Li and Pan, 2012; Li et al., 2015), a correlation was proposed between the magnetosome mineral habits and the phylogenetic affiliations of MTB (Pósfai et al., 2013a). Here, the formation of octahedral magnetites and the arrangement of bundle magnetosome chains within SHHC-1, supports this correlation and indicates a species- or strain-specific mineral habits and spatial arrangement of magnetosomes.
Rather than MTB based biomineralization, nanocrystals of magnetite with octahedral, cubotachedral or prismatic shapes can also be produced by processes such as the co-precipitation of ferrous and ferric ions in aqueous solutions (Faivre et al., 2005), thermal decomposition of Mg–Fe carbonates (Golden et al., 2001), and extracellularly, biologically-induced mineralization by iron-reducing bacteria (Zhang et al., 1998; Vali et al., 2004). These non-MTB processes may produce magnetite crystals with some features (e.g., crystal habits, narrow size distribution in the single magnetic domain size range, pure chemistry) similar to those of magnetosomes. This makes the identification of magnetofossils from ancient sediments or rocks more complicated and sometimes misleading (Golden et al., 2004; Benzerara and Menguy, 2009; Jimenez-Lopez et al., 2010). In addition, the crystal shape, grain size and chemical composition of magnetosomes can be modified by growing MTB cells under high oxygen environments or within the culture medium rich in some other transition metals or inorganic chemicals (Devouard et al., 1998; Taylor and Barry, 2004; Faivre et al., 2008; Lefèvre et al., 2011b; Li and Pan, 2012; Li et al., 2016). Moreover, genetic factors may also have an impact on the formation of irregular shaped magnetite magnetosomes (Lohsse et al., 2014). Our study shows that some uncultured MTB produce magnetosome magnetites with octahedral and irregular shapes and with a high frequency of twin crystals. On the one hand, these features do not meet some of the criteria of biogenicity enacted for magnetofossils, such as crystallographic perfection and unusual crystal morphology (Thomas-Keprta et al., 2000). This indicates that besides crystal morphology, other robust criteria such as their chain structure, chemical composition, and the co-presence of reliable magnetofossils (e.g., bullet-shaped SD magnetites) should be carefully tested in order to precisely identify magnetofossils (Li et al., 2013). On the other hand, the observed variety of morphologies and the high frequency of observed crystal defects indicate that the magnetosome formation within the strain SHHC-1 might be less biologically controlled or that they were affected by unfavorable environments in which they live in. Therefore, more coupled fluorescence and electron microscopy studies on natural MTB cells, as well as systematic studies on magnetosome biomineralization within cultured and uncultured MTB are needed to better understand the correlation between magnetosome mineral habits and MTB phylogenies and/or habitats. This has important implications on the magnetosome biomineralization and further understanding magneto-aerotaxis processes.
Nucleotide Sequence Accession Numbers
The sequence obtained in this study was deposited in GenBank under accession number KY302295.
Author Contributions
JL, ZC, and YP designed the research. HZ, FW, PL, WL, and CW prepared samples and carried out microbiological experiments. JL, NM, KB, and EL carried out TEM experiments. JL, HZ, and FW performed FISH-SEM experiments. JL and HZ prepared the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (grants 41522402, 41374004, 41330104, and 41621004) and the Open Project Support from the State Key Laboratory of Lithospheric Evolution (SKLLE, Beijing). The TEM facility at IMPMC was purchased owing to a support by Region Ile-de-France grant SESAME 2000 E 1435. We thank Dr. Hitesh G. Changela for his constructive comments and improving the English. We are also grateful to the TEM engineers at the IGGCAS in Beijing (Mr. Gu Lixin and Mr. Tang Xu) and at the IMPMC in Paris (Mr. Guigner Jean-Michel) in Paris for smoothly running the facilities. J.H.L. benefits from the discussions in Coffice 442 of the Institute of Geology and Geophysics, Chinese Academy of Sciences (IGGCAS).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00969/full#supplementary-material
References
Abreu, F., Carolina, A., Araujo, A. C., Leão, P., Silva, K. T., Carvalho, F. M., de Lima Cunha, O., et al. (2016). Culture-independent characterization of novel psychrophilic magnetotactic cocci from Antarctic marine sediments. Environ. Microbiol. 18, 4426–4441. doi: 10.1111/1462-2920.13388
Amann, R. I., Krumholz, L., and Stahl, D. A. (1990). Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172, 762–770. doi: 10.1128/jb.172.2.762-770.1990
Araujo, A. C. V., Morillo, V., Cypriano, J., Teixeira, L. C. R. S., Leão, P., Lyra, S., et al. (2016). Combined genomic and structural analyses of a cultured magnetotactic bacterium reveals its niche adaptation to a dynamic environment. BMC Genomics 17:726. doi: 10.1186/s12864-016-3064-9
Balkwill, D. L., Maratea, D., and Blakemore, R. P. (1980). Ultrastructure of a magnetic spirillum. J. Bacteriol. 141, 1399–1408.
Bazylinski, D. A., and Frankel, R. B. (2004). Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2, 217–230. doi: 10.1038/nrmicro842
Bazylinski, D. A., Williams, T. J., Lefèvre, C. T., Berg, R. J., Zhang, C. L. L., Bowser, S. S., et al. (2013). Magnetococcus marinus gen. nov., sp nov., a marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. nov., Magnetococcales ord. nov.) at the base of the Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 63, 801–808. doi: 10.1099/ijs.0.038927-0
Benzerara, K., and Menguy, N. (2009). Looking for traces of life in minerals. Comptes Rendus Palevol. 8, 617–628. doi: 10.1016/j.crpv.2009.03.006
Chang, S. B. R., and Kirschvink, J. L. (1989). Magnetofossils, the magnetization of sediments, and the evolution of magnetite biomineralization. Annu. Rev. Earth Planet. Sci. 17, 169–195. doi: 10.1146/annurev.earth.17.1.169
Chen, H. T., Li, J. H., Xing, X., Du, Z. J., and Chen, G. J. (2015). Unexpected Diversity of Magnetococci in Intertidal Sediments of Xiaoshi Island in the North Yellow Sea. J. Nanomaterials 2015:902121. doi: 10.1155/2015/902121
Cox, B. L., Popa, R., Bazylinski, D. A., Lanoil, B., Douglas, S., Belz, A., et al. (2002). Organization and elemental analysis of P-, S-, and Fe-rich inclusions in a population of freshwater magnetococci. Geomicrobiol. J. 19, 387–406. doi: 10.1080/01490450290098504
Devouard, B., Pósfai, M., Hua, X., Bazylinski, D. A., Frankel, R. B., and Buseck, P. R. (1998). Magnetite from magnetotactic bacteria: size distributions and twinning. Am. Mineral. 83, 1387–1398. doi: 10.2138/am-1998-1101
Faivre, D., and Schüler, D. (2008). Magnetotactic bacteria and magnetosomes. Chem. Rev. 108, 4875–4898. doi: 10.1021/cr078258w
Faivre, D., Menguy, N., Guyot, F., Lopez, O., and Zuddas, P. (2005). Morphology of nanomagnetite crystals: implications for formation conditions. Am. Mineral. 90, 1793–1800. doi: 10.2138/am.2005.1853
Faivre, D., Menguy, N., Pósfai, M., and Schüler, D. (2008). Environmental parameters affect the physical properties of fast-growing magnetosomes. Am. Mineral. 93, 463–469. doi: 10.2138/am.2008.2678
Flies, C. B., Peplies, J., and Schüler, D. (2005). Combined approach for characterization of uncultivated magnetotactic bacteria from various aquatic environments. Appl. Environ. Microbiol. 71, 2723–2731. doi: 10.1128/aem.71.5.2723-2731.2005
Frankel, R. B., Bazylinski, D. A., Johnson, M. S., and Taylor, B. L. (1997). Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73, 994–1000. doi: 10.1016/S0006-3495(97)78132-3
Golden, D. C., Ming, D. W., Morris, R. V., Brearley, A., Lauer, H. V., Treiman, A. H., et al. (2004). Evidence for exclusively inorganic formation of magnetite in Martian meteorite ALH84001. Am. Mineral. 89, 681–695. doi: 10.2138/am-2004-5-602
Golden, D. C., Ming, D. W., Schwandt, C. S., Lauer, H. V., Socki, R. A., Morris, R. V., et al. (2001). A simple inorganic process for formation of carbonates, magnetite, and sulfides in Martian meteroite ALH84001. Am. Miner. 86, 956–956. doi: 10.2138/am-2001-2-321
Greuter, D., Loy, A., Horn, M., and Rattei, T. (2016). ProbeBase—an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Res. 44, D586–D589. doi: 10.1093/nar/gkv1232
Hanzlik, M., Winklhofer, M., and Petersen, N. (1996). Spatial arrangement of chains of magnetosomes in magnetotactic bacteria. Earth Planet. Sci. Lett. 145, 125–134. doi: 10.1016/S0012-821X(96)00191-4
Hesse, P. P. (1994). Evidence for bacterial paleoecological origin of mineral magnetic cycles in oxic and sub-oxic Tasman Sea sediments. Mar. Geol. 117, 1–17. doi: 10.1016/0025-3227(94)90003-5
Isambert, A., Menguy, N., Larquet, E., Guyot, F., and Valet, J. P. (2007). Transmission electron microscopy study of magnetites in a freshwater population of magnetotactic bacteria. Am. Mineral. 92, 621–630. doi: 10.2138/am.2007.2278
Jandacka, P., Alexa, P., Pistora, J., Li, J., Vojtkova, H., and Hendrych, A. (2013). Size distributions of nanoparticles from magnetotactic bacteria as signatures of biologically controlled mineralization. Am. Mineral. 98, 2105–2114. doi: 10.2138/am.2013.4429
Jimenez-Lopez, C., Romanek, C. S., and Bazylinski, D. A. (2010). Magnetite as a prokaryotic biomarker: a review. J. Geophys. Res. 115, G00G03. doi: 10.1029/2009jg001152
Jogler, C., Lin, W., Meyerdierks, A., Kube, M., Katzmann, E., Flies, C., et al. (2009). Toward cloning of the magnetotactic metagenome: identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from Different Aquatic Sediments. Appl. Environ. Microbiol. 75, 3972–3979. doi: 10.1128/AEM.02701-08
Kolinko, S., Jogler, C., Katzmann, E., Wanner, G., Peplies, J., and Schüler, D. (2012). Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ. Microbiol. 14, 1709–1721. doi: 10.1111/j.1462-2920.2011.02609.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester:John Wiley & Sons), 115–175.
Larrasoana, J. C., Liu, Q., Hu, P., Roberts, A. P., Mata, P., Civis, J., et al. (2014). Paleomagnetic and paleoenvironmental implications of magnetofossil occurrences in late Miocene marine sediments from the Guadalquivir Basin, SW Spain. Front. Microbiol. 5:71. doi: 10.3389/fmicb.2014.00071
Lefèvre, C. T., and Bazylinski, D. A. (2013). Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol. Mol. Biol. Rev. 77, 497–526. doi: 10.1128/mmbr.00021-13
Lefèvre, C. T., Bennet, M., Landau, L., Vach, P., Pignol, D., Bazylinski, Dennis A., et al. (2014). Diversity of magneto-aerotactic behaviors and oxygen sensing mechanisms in cultured magnetotactic bacteria. Biophys. J. 107, 527–538. doi: 10.1016/j.bpj.2014.05.043
Lefèvre, C. T., Bernadac, A., Yu-Zhang, K., Pradel, N., and Wu, L. F. (2009). Isolation and characterization of a magnetotactic bacterial culture from the Mediterranean Sea. Environ. Microbiol. 11, 1646–1657. doi: 10.1111/j.1462-2920.2009.01887.x
Lefèvre, C. T., Menguy, N., Abreu, F., Lins, U., Pósfai, M., Prozorov, T., et al. (2011b). A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science 334, 1720–1723. doi: 10.1126/science.1212596
Lefèvre, C. T., Pósfai, M., Abreu, F., Lins, U., Frankel, R. B., and Bazylinski, D. A. (2011a). Morphological features of elongated-anisotropic magnetosome crystals in magnetotactic bacteria of the Nitrospirae phylum and the Deltaproteobacteria class. Earth Planet. Sci. Lett. 312, 194–200. doi: 10.1111/j.1462-2920.2011.02505.x
Li, J. H., and Pan, Y. X. (2012). Environmental factors affect magnetite magnetosome synthesis in Magnetospirillum magneticum AMB-1: implications for biologically controlled mineralization. Geomicrobiol. J. 29, 362–373. doi: 10.1080/01490451.2011.565401
Li, J. H., and Pan, Y. X. (2015). Applications of transmission electron microscopy in the Earth science. Sci. Sin. Terrae 45, 1359–1382. (In Chinese) doi: 10.1007/s11430-015-5172-9
Li, J. H., Benzerara, K., Bernard, S., and Beyssac, O. (2013). The link between biomineralization and fossilization of bacteria: insights from field and experimental studies. Chem. Geol. 359, 49–69. doi: 10.1016/j.chemgeo.2013.09.013
Li, J. H., Menguy, N., Arrio, M. A., Sainctavit, P., Juhin, A., Wang, Y. Z., et al. (2016). Controlled cobalt doping in the spinel structure of magnetosome magnetite: new evidences from element- and site-specific X-ray magnetic circular dichroism analyses. J. R. Soc. Interface. 13:20160355. doi: 10.1098/rsif.2016.0355
Li, J., Menguy, N., Gatel, C., Boureau, V., Snoeck, E., Patriarche, G., et al. (2015). Crystal growth of bullet-shaped magnetite in magnetotactic bacteria of the Nitrospirae phylum. J. R. Soc. Interface 12:20141288. doi: 10.1098/rsif.2014.1288
Li, J. H., Pan, Y. X., Chen, G. J., Liu, Q. S., Tian, L. X., and Lin, W. (2009). Magnetite magnetosome and fragmental chain formation of Magnetospirillum magneticum AMB-1: transmission electron microscopy and magnetic observations. Geophys. J. Int. 177, 33–42. doi: 10.1111/j.1365-246X.2009.04043.x
Li, J. H., Zhang, H., Menguy, N., Benzerara, K., Wang, F., Lin, X., et al. (2017). Single-cell resolution study of uncultured magnetotactic bacteria via fluorescence-coupled electron microscopy. Appl. Environ. Microbiol. 83:e00409-17. doi: 10.1128/AEM.00409-17
Lin, W., and Pan, Y. X. (2009). Uncultivated magnetotactic cocci from Yuandadu Park in Beijing, China. Appl. Environ. Microbiol. 75, 4046–4052. doi: 10.1128/AEM.00247-09
Lin, W., Jogler, C., Schüler, D., and Pan, Y. (2011). Metagenomic analysis reveals unexpected subgenomic diversity of magnetotactic bacteria within the Nitrospirae phylum. Appl. Environ. Microbiol. 77, 323–326 doi: 10.1128/AEM.01476-10
Lin, W., Li, J., and Pan, Y. (2012). Newly isolated but uncultivated magnetotactic bacterium of the Phylum Nitrospirae from Beijing, China. Appl. Environ. Microbiol. 78, 668–675. doi: 10.1128/aem.06764-11
Lin, W., Li, J., Schüler, D., Jogler, C., and Pan, Y. X. (2009). Diversity analysis of magnetotactic bacteria in Lake Miyun, northern China, by restriction fragment length polymorphism. Syst. Appl. Microbiol. 32, 342–350. doi: 10.1016/j.syapm.2008.10.005
Liu, S., Deng, C., Xiao, J., Li, J., Paterson, G., Chang, L., et al. (2015). Insolation driven biomagnetic response to the Holocene Warm Period in semi-arid East Asia. Sci. Rep. 5:8001. doi: 10.1038/srep08001
Lohsse, A., Borg, S., Raschdorf, O., Kolinko, I., Tompa, E., Posfai, M., et al. (2014). Genetic bissection of the mamAB and mms6 operons reveals a aene set essential for magnetosome biogenesis in Magnetospirillum gryphiswaldense. J. Bacteriol. 196, 2658–2669. doi: 10.1128/JB.01716-14
Loy, A., Arnold, R., Tischler, P., Rattei, T., Wagner, M., and Horn, M. (2008). probeCheck - a central resource for evaluating oligonucleotide probe coverage and specificity. Environ.Microbiol. 10, 2894–2898. doi: 10.1111/j.1462-2920.2008.01706.x
McKay, D. S., Gibson, E. K., ThomasKeprta, K. L., Vali, H., Romanek, C. S., Clemett, S. J., et al. (1996). Search for past life on Mars: possible relic biogenic activity in Martian meteorite ALH84001. Science 273, 924–930. doi: 10.1126/science.273.5277.924
Meldrum, F. C., Mann, S., Heywood, B. R., Frankel, R. B., and Bazylinski, D. A. (1993). Electron microscopy study of magnetosomes in a cultured Coccoid magnetotactic bacterium. Proc. R. Soc. Lond. B 251, 231–236. doi: 10.1098/rspb.1993.0034
Moench, T. T. (1988). Bilophococcus magnetotacticus gen. nov. sp. nov., a motile, magnetic coccus. Antonie Van Leeuwenhoek 54, 483–496. doi: 10.1007/BF00588385
Moench, T. T., and Konetzka, W. A. (1978). A novel method for the isolation and study of a magnetotactic bacterium. Arch. Microbiol. 119, 203–212. doi: 10.1007/BF00964274
Morillo, V., Abreu, F., Araujo, A. C., Almeida, L. G., Enrich-Prast, A., and Farina, M. (2014). Isolation, cultivation and genomic analysis of magnetosome biomineralization genes of a new genus of South-seeking magnetotactic cocci within the Alphaproteobacteria. Front Microbiol. 5:72. doi: 10.3389/fmicb.2014.00072
Pan, H., Zhu, K., Song, T., Yu-Zhang, K., Lefevre, C., Xing, S., et al. (2008). Characterization of a homogeneous taxonomic group of marine magnetotactic cocci within a low tide zone in the China Sea. Environ. Microbiol. 10, 1158–1164. doi: 10.1111/j.1462-2920.2007.01532.x
Pan, Y. X., Lin, W., Tian, L. X., Zhu, R. X., and Petersen, N. (2009). Combined approaches for characterization of an uncultivated Magnetotactic coccus from Lake Miyun near Beijing. Geomicrobiol. J. 26, 313–320. doi: 10.1080/01490450902748633
Pósfai, M., Kasama, T., and Dunin-Borkowski, R. E. (2013b). Biominerals at the nanoscale: transmission electron microscopy methods for studying the special properties of biominerals. Miner. Nanoscale 14, 377–435. doi: 10.1180/EMU-notes.14.11
Pósfai, M., Lefèvre, C., Trubitsyn, D., Bazylinski, D. A., and Frankel, R. B. (2013a). Phylogenetic significance of composition and crystal morphology of magnetosome minerals. Front. Microbiol. 4:344. doi: 10.3389/fmicb.2013.00344
Roberts, A. P., Florindo, F., Villa, G., Chang, L., Jovane, L., Bohaty, S. M., et al. (2011). Magnetotactic bacterial abundance in pelagic marine environments is limited by organic carbon flux and availability of dissolved iron. Earth Planet. Sci. Lett. 310, 441–452. doi: 10.1016/j.epsl.2011.08.011
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schüler, D. (2002). The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int. Microbiol. 5, 209–214. doi: 10.1007/s10123-002-0086-8
Simpson, E. T., Kasama, T., Pósfai, M., Buseck, P. R., Harrison, R. J., and Dunin-Borkowski, R. E. (2005). Magnetic induction mapping of magnetite chains in magnetotactic bacteria at room temperature and close to the Verwey transition using electron holography. J. Phys. Conf. Ser. 17, 108–121. doi: 10.1088/1742-6596/17/1/017
Snowball, I., Zillen, L., and Sandgren, P. (2002). Bacterial magnetite in Swedish varved lake-sediments: a potential bio-marker of environmental change. Quat. Int. 88, 13–19. doi: 10.1016/s1040-6182(01)00069-6
Spring, S., Amann, R., Ludwig, W., Schleifer, K. H., and Petersen, N. (1992). Phylogenetic diversity and identification of nonculturable magnetotactic bacteria. Syst. Appl. Microbiol. 15, 116–122. doi: 10.1016/S0723-2020(11)80147-5
Spring, S., Amann, R., Ludwig, W., Schleifer, K. H., Schüler, D., Poralla, K., et al. (1995). Phylogenetic analysis of uncultured magnetotactic bacteria from the alpha-subclass of Proteobacteria. Syst. Appl. Microbiol. 17, 501–508. doi: 10.1016/S0723-2020(11)80068-8
Spring, S., Amann, R., Ludwig, W., Schleifer, K. H., van Gemerden, H., and Petersen, N. (1993). Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Appl. Environ. Microbiol. 59, 2397–2403.
Spring, S., Lins, U., Amann, R., Schleifer, K. H., Ferreira, L., Esquivel, D., et al. (1998). Phylogenetic affiliation and ultrastructure of uncultured magnetic bacteria with unusually large magnetosomes. Arch. Microbiol. 169, 136–147. doi: 10.1007/s002030050553
Taoka, A., Kondo, J., Oestreicher, Z., and Fukumori, Y. (2014). Characterization of uncultured giant rod-shaped magnetotactic Gammaproteobacteria from a fresh water pond in Kanazawa, Japan. Microbiology 160, 2226–2234. doi: 10.1099/mic.0.078717-0
Taylor, A. P., and Barry, J. C. (2004). Magnetosomal matrix: ultrafine structure may template biomineralization of magnetosomes. J. Microsc. 213, 180–197. doi: 10.1111/j.1365-2818.2004.01287.x
Thomas-Keprta, K. L., Bazylinski, D. A., Kirschvink, J. L., Clemett, S. J., McKay, D. S., Wentworth, S. J., et al. (2000). Elongated prismatic magnetite crystals in ALH84001 carbonate globules: potential Martian magnetofossils. Geochim. Cosmochim. Acta 64, 4049–4081. doi: 10.1016/s0016-7037(00)00481-6
Thomas-Keprta, K. L., Clemett, S. J., Bazylinski, D. A., Kirschvink, J. L., McKay, D. S., Wentworth, S. J., et al. (2002). Magnetofossils from ancient Mars: a robust biosignature in the Martian meteorite ALH84001. Appl. Environ. Microbiol. 68, 3663–3672. doi: 10.1128/aem.68.8.3663-3672.2002
Thornhill, R. H., Burgess, J. G., and Matsunaga, T. (1995). PCR for direct detection of indigenous uncultured magnetic cocci in sediment and phylogenetic analysis of amplified 16S ribosomal DNA. Appl. Environ. Microbiol. 61, 495–500.
Vali, H., Weiss, B., Li, Y. L., Sears, S. K., Kim, S. S., Kirschvink, J. L., et al. (2004). Formation of tabular single-domain magnetite induced by Geobacter metallireducens GS-15. Prco. Natl. Acad. Sci. U.S.A. 101, 16121–16126. doi: 10.1073/pnas.0404040101
Wang, Y., Lin, W., Li, J., and Pan, Y. (2013). High diversity of magnetotactic δ-proteobacteria in a freshwater niche. Appl. Environ. Microbiol. 79, 2813–2817. doi: 10.1128/aem.03635-12
Yamazaki, T., and Kawahata, H. (1998). Organic carbon flux controls the morphology of magnetofossils in marine sediments. Geology 26, 1064–1066.
Zhang, C. L., Vali, H., Romanek, C. S., Phelps, T. J., and Liu, S. V. (1998). Formation of single-domain magnetite by a thermophilic bacterium. Am. Mineral. 83, 1409–1418. doi: 10.2138/am-1998-1103
Keywords: magnetotactic cocci, magnetosome, biomineralization, octahedron, coordinated FISH-SEM, TEM
Citation: Zhang H, Menguy N, Wang F, Benzerara K, Leroy E, Liu P, Liu W, Wang C, Pan Y, Chen Z and Li J (2017) Magnetotactic Coccus Strain SHHC-1 Affiliated to Alphaproteobacteria Forms Octahedral Magnetite Magnetosomes. Front. Microbiol. 8:969. doi: 10.3389/fmicb.2017.00969
Received: 09 December 2016; Accepted: 15 May 2017;
Published: 30 May 2017.
Edited by:
Bradley M. Tebo, Oregon Health & Science University, United StatesReviewed by:
Dirk Schüler, University of Bayreuth, GermanySebastian Kolinko, Lawrence Berkeley National Laboratory, United States
Copyright © 2017 Zhang, Menguy, Wang, Benzerara, Leroy, Liu, Liu, Wang, Pan, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhua Li, lijinhua@mail.iggcas.ac.cn
Zhibao Chen, chenzhibao@byau.edu.cn
 Heng Zhang
Heng Zhang Nicolas Menguy4,5
Nicolas Menguy4,5 Fuxian Wang
Fuxian Wang Karim Benzerara
Karim Benzerara Yongxin Pan
Yongxin Pan Jinhua Li
Jinhua Li