- 1Department of Microbial Pathogenicity Mechanisms, Leibniz Institute for Natural Product Research and Infection Biology – Hans Knoell Institute, Jena, Germany
- 2Department of Microbial Pathogenicity Mechanisms, Friedrich Schiller University, Jena, Germany
- 3Center for Sepsis Control and Care, University Hospital, Jena, Germany
Iron acquisition is a crucial virulence determinant for many bacteria and fungi, including the opportunistic fungal pathogens Candida albicans and C. glabrata. While the diverse strategies used by C. albicans for obtaining iron from the host are well-described, much less is known about the acquisition of this micronutrient from host sources by C. glabrata – a distant relative of C. albicans with closer evolutionary ties to Saccharomyces cerevisiae, which nonetheless causes severe clinical symptoms in humans. Here we show that C. glabrata is much more restricted than C. albicans in using host iron sources, lacking, for example, the ability to grow on transferrin and hemin/hemoglobin. Instead, C. glabrata is able to use ferritin and non-protein-bound iron (FeCl3) as iron sources in a pH-dependent manner. As in other fungal pathogens, iron-dependent growth requires the reductive high affinity (HA) iron uptake system. Typically highly conserved, this uptake mechanism normally relies on initial ferric reduction by cell-surface ferric reductases. The C. glabrata genome contains only three such putative ferric reductases, which were found to be dispensable for iron-dependent growth. In addition and in contrast to C. albicans and S. cerevisiae, we also detected no surface ferric reductase activity in C. glabrata. Instead, extracellular ferric reduction was found in this and the two other fungal species, which was largely dependent on an excreted low-molecular weight, non-protein ferric reductant. We therefore propose an iron acquisition strategy of C. glabrata which differs from other pathogenic fungi, such as C. albicans, in that it depends on a limited set of host iron sources and that it lacks the need for surface ferric reductases. Extracellular ferric reduction by a secreted molecule possibly compensates for the loss of surface ferric reductase activity in the HA iron uptake system.
Introduction
Iron is an essential micronutrient for almost all living organisms (Posey and Gherardini, 2000; Troxell et al., 2012), as it is indispensable for numerous cellular processes such as respiration, synthesis of iron-sulfur-clusters (Fe-S clusters), the tricarboxylic acid (TCA) cycle, and the synthesis of DNA, amino acids, lipids, and sterols (Schaible and Kaufmann, 2004). Although iron is highly abundant in the environment, its bioavailability is low due to the low solubility of its most common form, ferric iron (Fe3+), under aerobic conditions (Haas et al., 2008). For pathogenic microbes, iron acquisition is especially demanding, since they are dependent on the host’s iron supply. However, host iron is typically bound to carrier proteins such as hemoglobin, the transport compound transferrin, or the storage molecule ferritin (Ratledge, 2007). Additionally, iron is also actively withheld from infection sites to restrict the proliferation of invading pathogens. This host strategy is known as nutritional immunity (Hood and Skaar, 2012).
Successful pathogens, including many fungal species, have therefore evolved sophisticated strategies to use iron sources in the host, and these strategies are considered important virulence attributes (Timmerman and Woods, 1999; Almeida et al., 2008; Jung et al., 2008; Newman and Smulian, 2013; Kuznets et al., 2014). The opportunistic fungal pathogen Candida albicans and its distant relative C. glabrata, for example, are members of the normal gastrointestinal and oral flora, but can also cause diseases ranging from superficial mucosal infections to life-threatening systemic infections (e.g., candidemia), especially in immuno-compromised hosts (Perlroth et al., 2007). C. albicans is the most frequently encountered species causing candidemia, while C. glabrata ranks second in Europe and the United States (Guinea, 2014). However, despite colonizing the same host niche and causing similar clinical symptoms, C. glabrata is evolutionarily more closely related to the baker’s yeast Saccharomyces cerevisiae than to C. albicans. In fact, C. glabrata and C. albicans differ remarkably from each other regarding their infection strategy, morphological flexibility, and genome structure, as C. glabrata possesses a haploid genome, and lacks certain virulence-associated genes and metabolic pathways known in other yeasts (Brunke and Hube, 2013). As recently shown, this species difference also extends to the regulatory networks for iron homeostasis (Blankenship and Mitchell, 2011; Chen et al., 2011; Gerwien et al., 2016).
Most pathogenic fungi have three main iron acquisition strategies in common: (i) the receptor-mediated uptake of siderophores (excreted low-molecular weight, high-affinity iron scavengers); (ii) the receptor-mediated heme uptake; and (iii) the reductive high-affinity (HA) iron uptake. Although Candida and Saccharomyces species do not produce their own siderophores, they are generally able to use siderophores produced by other microorganisms (xenosiderophores) (Haas et al., 2008; Nevitt and Thiele, 2011). In C. albicans, heme/hemoglobin is additionally bound by receptors of the Rbt family (predominantly Rbt5 and Rbt51), followed by endocytosis and intracellular degradation by a heme oxygenase (Weissman and Kornitzer, 2004; Weissman et al., 2008).
The reductive HA iron uptake is strictly required for virulence in C. albicans (Ramanan and Wang, 2000; Almeida et al., 2008; Cheng et al., 2013) and is not only crucial for uptake of free iron, but also for acquisition of iron associated with transferrin (Knight et al., 2005) or ferritin (Almeida et al., 2008). Typically, the reductive HA iron uptake consists of three steps: (i) initial extracellular Fe3+ reduction to Fe2+, typically mediated by surface-bound ferric reductases followed by (ii) re-oxidation to Fe3+ by multicopper ferroxidases, and (iii) Fe3+ import by the permease Ftr1. Ferric reduction is therefore important for releasing complexed ferric iron (bound, e.g., to siderophores, transferrin, or ferritin) into the soluble ferrous state (Schröder et al., 2003). In addition to iron uptake, ferric reductases play a central role in intracellular iron transmembrane transport and storage, when present in the vacuolar membrane (Urbanowski and Piper, 1999; Haas et al., 2008), and in some cases they serve as surface cupric reductases (Martins et al., 1998; Woodacre et al., 2008; Jeeves et al., 2011). Consequently, many fungi have large families of ferric reductases with diverse functions encoded in their genome, among them S. cerevisiae (nine characterized ferric reductases) (Dancis et al., 1992; Martins et al., 1998; Yun et al., 2001) and C. albicans (17 putative ferric reductases) (Jeeves et al., 2011; Xu et al., 2014). In contrast, the ferric reductases of C. glabrata are not well characterized yet, and our knowledge of this fungus’ mechanisms for exploiting host iron sources is still incomplete. The aim of this study was therefore to investigate the iron acquisition strategies of C. glabrata, and in particular its spectrum of host iron sources and the nature of the ferric reduction in its high-affinity iron uptake system.
We report here that C. glabrata can use non-protein-bound iron (FeCl3) and ferritin-associated iron, but not iron bound to transferrin, hemin, or hemoglobin, in a pH-dependent manner via the reductive HA iron uptake system. Surprisingly, the two analyzed putative ferric reductases encoded in the C. glabrata genome proved to be dispensable for growth, and C. glabrata lacks surface ferric reductase activity even under iron starvation. Instead, we found that this fungus produces an extracellular low-molecular, non-protein ferric reductant.
Results
Comparative Analysis of Iron Acquisition and Trafficking Systems in C. glabrata, S. cerevisiae, and C. albicans
Fungi use various, partially redundant iron acquisition systems, depending on their current host niche. To find common key components of iron acquisition and trafficking in C. glabrata, S. cerevisiae, and C. albicans, we searched their genome databases (Candida Genome Database, CGD, and Saccharomyces Genome Database, SGD) for orthologs with known or postulated iron-related functions based on previously published data or iron-related mutant phenotypes (Table 1 and Supplementary Table 1). Genes involved in intracellular iron trafficking were found highly conserved between the species (Supplementary Table 1). These code for transporters in membranes of the vacuole, a major iron storage site (Ccc1, Smf1, Smf3, and Fth1), and of the mitochondria, where a constant supply of iron is needed to maintain respiration and iron-sulfur cluster biosynthesis (Mmt1, Atm1, Mrs3, and Mrs4). Similarly, all three species possess the main components of the reductive HA iron uptake (the permease Ftr1, the associated ferroxidases Fet3/Fet34, and ferric reductases; Table 1). Notably, the ferric reductase family is considerably smaller in C. glabrata (3 Fre orthologs) than in C. albicans (17 Fre orthologs) and S. cerevisiae (9 Fre orthologs). With regard to host iron source-related acquisition systems, however, major differences between the species were observed (Table 1). While all three species encode orthologs of transporters for xenosiderophore uptake (Sit1/Arn1-4), both C. glabrata and S. cerevisiae lack orthologs of C. albicans proteins associated with iron acquisition from host ferritin (Als3) or heme and hemoglobin (Rbt5, Rbt51, Csa1, Csa2, and Pga7).
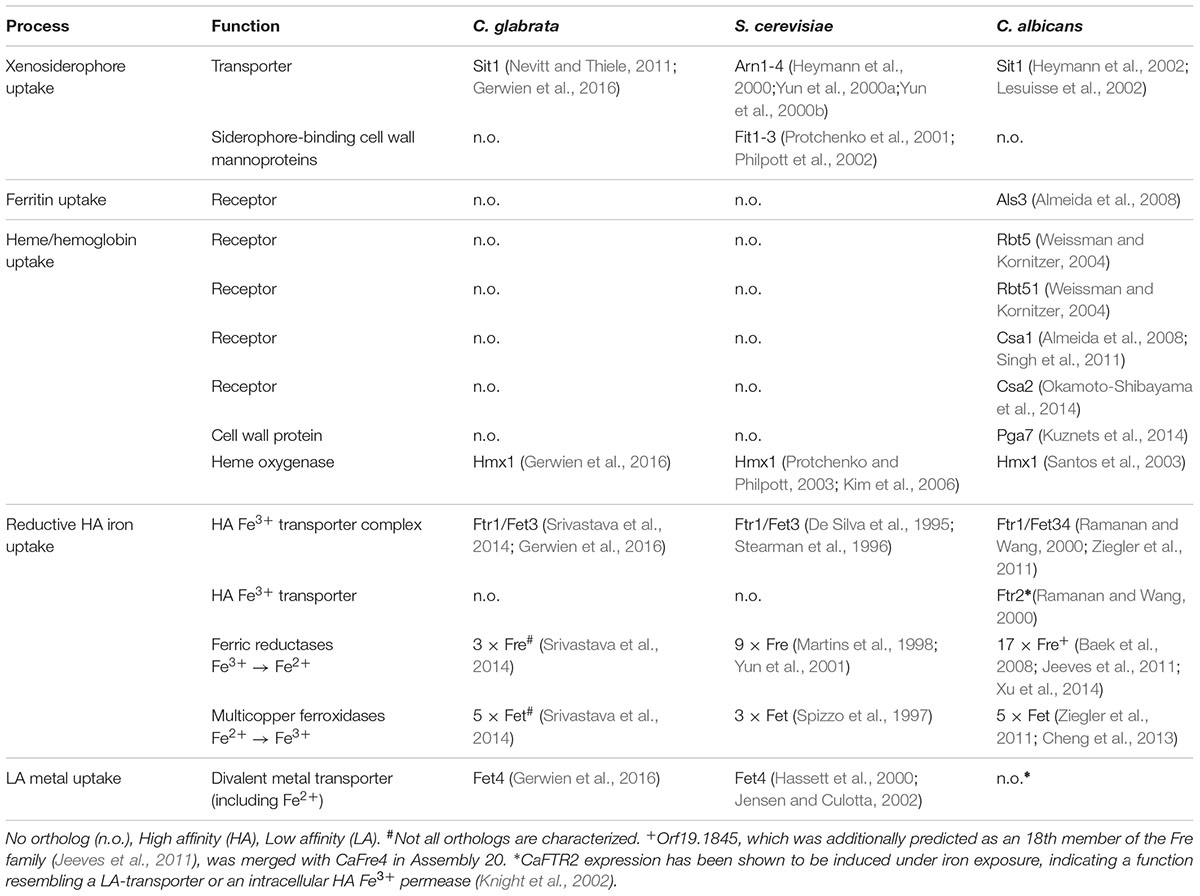
TABLE 1. Iron acquisition systems in Candida glabrata, Saccharomyces cerevisiae and Candida albicans.
Thus, although C. glabrata is a successful pathogen with a large overlap in host niches to C. albicans, it seems to lack the components needed for the utilization of the most abundant host iron sources.
Use of Host Iron Sources Is Restricted to Non-protein-bound Iron and Ferritin in C. glabrata
The in silico data indicated a limited set of host iron sources available for utilization by C. glabrata. We thus tested different iron sources commonly found in the host. To this end we grew C. glabrata, S. cerevisiae, and C. albicans without free iron, but in presence of ferritin, transferrin, hemin, or hemoglobin. Addition of 100 μM or 1 mM FeCl3 served as a control for moderate or high iron levels. The uptake of xenosiderophores via C. glabrata Sit1 has been described elsewhere (Nevitt and Thiele, 2011). To account for possible effects of the pH on host iron source stability and solubility (Almeida et al., 2008), we performed these tests under conditions of different pH (Figure 1). Moderate FeCl3 levels and ferritin supported robust growth by all three species under acidic pH conditions (pH 4.5 and 5.8). However, under slightly alkaline conditions (pH 7.3) C. glabrata and S. cerevisiae, but not C. albicans, were unable to grow (Figures 1B,D). An increase in iron concentration to high levels, however, allowed growth of all species (Figure 1C). In agreement with our in silico analysis, C. glabrata and S. cerevisiae were not able to use hemoglobin, hemin, or transferrin over the whole tested range of media pH. In contrast, hemoglobin, hemin, and transferrin were all exploited as iron sources by C. albicans in a pH-dependent manner, in which alkaline pH conditions allowed the most robust growth (Figures 1E–G).
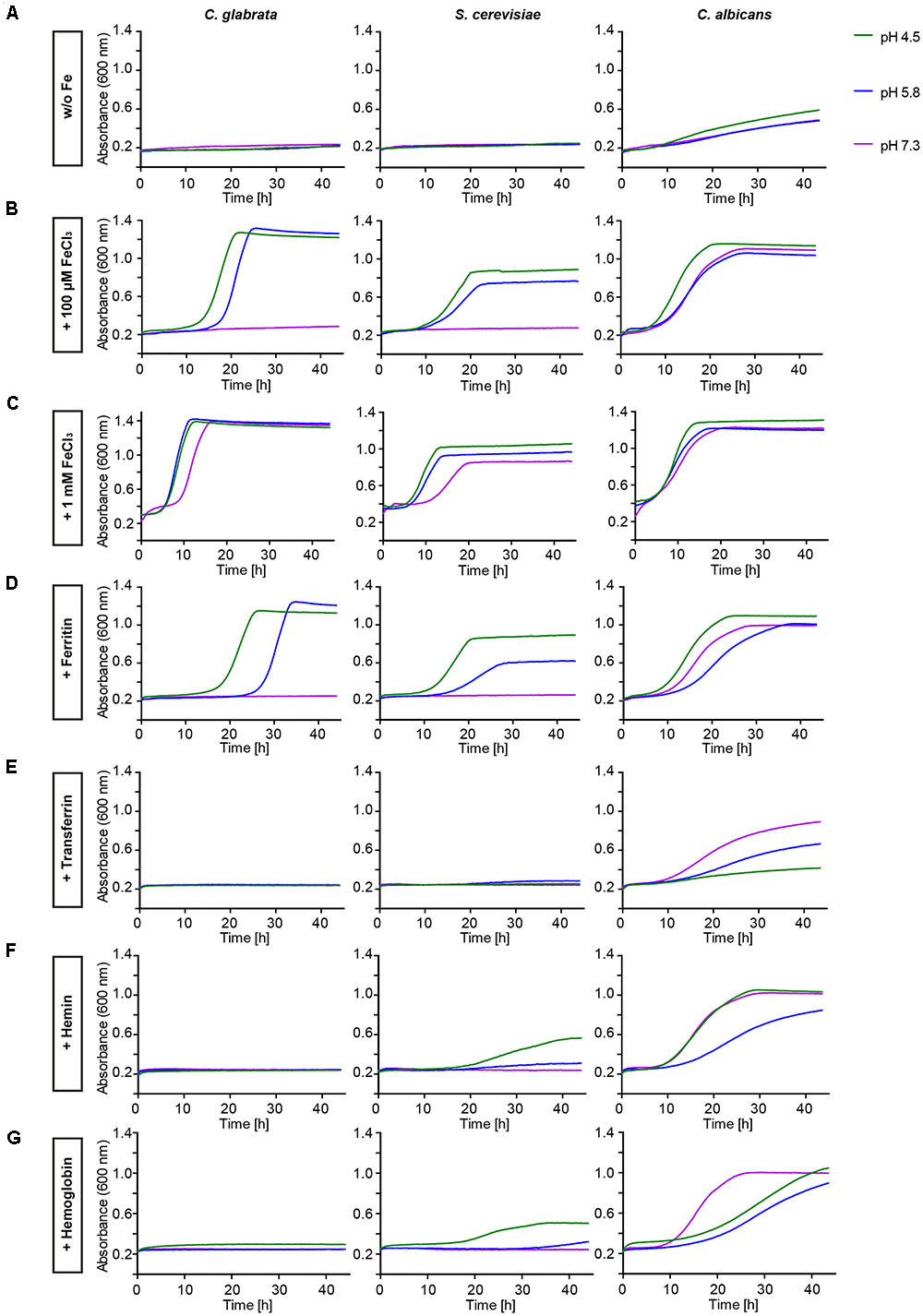
FIGURE 1. Comparative pH-dependent growth of Candida glabrata, Saccharomyces cerevisiae, and Candida albicans on host iron sources. Growth of iron-prestarved [preculture treatment with the iron chelator bathophenanthrolinedisulfonic acid (BPS)] wild-type (WT) strains in liquid SD buffered to pH 4.5, pH 5.8, and pH 7.3 containing: (A) 200 μM BPS (S. cerevisiae, C. glabrata) or 500 μM BPS (C. albicans) (B) BPS + 100 μM FeCl3 (C) BPS + 1 mM FeCl3 (D) BPS + 100 μg/ml ferritin (E) BPS + 100 μg/ml transferrin (F) BPS + 1 μM hemin (G) BPS + 0.1 mg/ml hemoglobin. n = 3, one representative example is shown.
Taken together, C. glabrata is much less versatile than C. albicans in the utilization of host iron sources, and more similar to S. cerevisiae in that respect. C. glabrata thus seems to rely on ferritin and non-protein-bound ferric iron, in combination with acidic pH, for growth under iron-limiting conditions in the host.
Ferritin and FeCl3 Utilization by C. glabrata Depends on the Reductive Iron Uptake System
To determine which pathways allow C. glabrata iron acquisition and growth with ferritin or FeCl3 as iron sources, we investigated deletion mutants lacking genes for two major components of the HA iron uptake system – the HA permease Ftr1 and the associated ferroxidase Fet3. Both mutants displayed severe growth defects with either ferritin as sole iron source or in presence of only moderate FeCl3 levels. Only high-level iron supplementation (1 mM FeCl3) partially restored growth of these mutants, predominantly at acidic pH (Figure 2). Hence, the reductive HA iron uptake, particularly the permease-ferroxidase complex, is needed for ferritin- and FeCl3-dependent growth of C. glabrata.
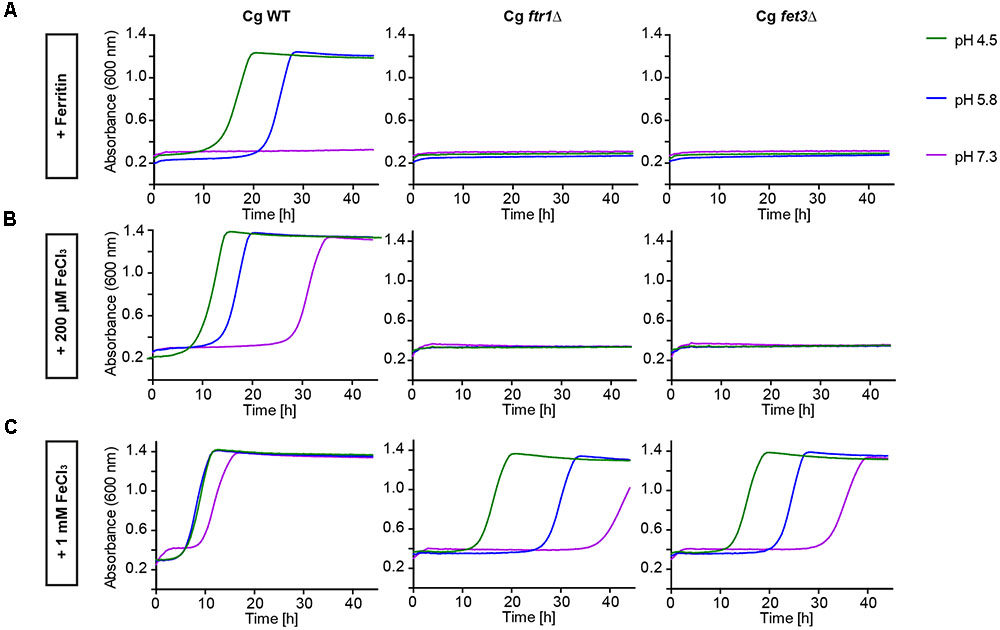
FIGURE 2. Comparative pH-dependent growth of C. glabrata mutants defective in HA reductive iron uptake on host iron sources. Growth of iron-prestarved (preculture treatment with 200 μM BPS) with C. glabrata WT, and mutants defective in HA reductive iron uptake (Cgfrt1Δ and Cgfet3Δ) in liquid SD buffered to pH 4.5, pH 5.8, and pH 7.3 at 37°C containing: (A) 200 μM BPS + 100 μg/ml ferritin. (B) 200 μM BPS + 200 μM FeCl3. (C) 200 μM BPS + 1 mM FeCl3. n = 3, A representative example is shown.
Fre Families Differ between C. glabrata, C. albicans, and S. cerevisiae
The initial step of HA iron uptake, the extracellular reduction of ferric iron, is typically mediated by membrane-integral NAD(P)H-dependent ferric reductases (Fre). The C. glabrata genome, in contrast to C. albicans and S. cerevisiae contains only few (three) potential FRE gene orthologs (Table 1), none of them with proven Fre function. We scanned the genomes of C. glabrata, C. albicans, and S. cerevisiae for additional putative ferric reductases. This in silico analysis included a BLAST search for the ferric reductase transmembrane component-like domain (Pfam family PF01794). We identified 17 of the 18 known C. albicans ferric reductases (Jeeves et al., 2011), and the nine ferric reductases already described in S. cerevisiae. Both families show only marginal overlap, which points to different evolutionary origins in both species. Notably, CaFre10 and ScFre1/ScFre2 (green dot, Figure 3A) are known to account for 75% and 90–98% of the whole cell surface reductase activity, respectively (Dancis et al., 1990; Anderson et al., 1992; Georgatsou and Alexandraki, 1994; Knight et al., 2002, 2005), but are non-orthologous. Overall, the gene families of the two species contain ferric as well as cupric reductases, with different cellular localizations depicted as intracellular (mitochondria, ER, vacuole), or extracellular/surface-associated (Figure 3A and Supplementary Table 2). Our analysis furthermore revealed that most (potential) Fres additionally contained a FAD-binding domain, a NAD-binding domain, multiple transmembrane domains and a signal peptide (Figure 3A).
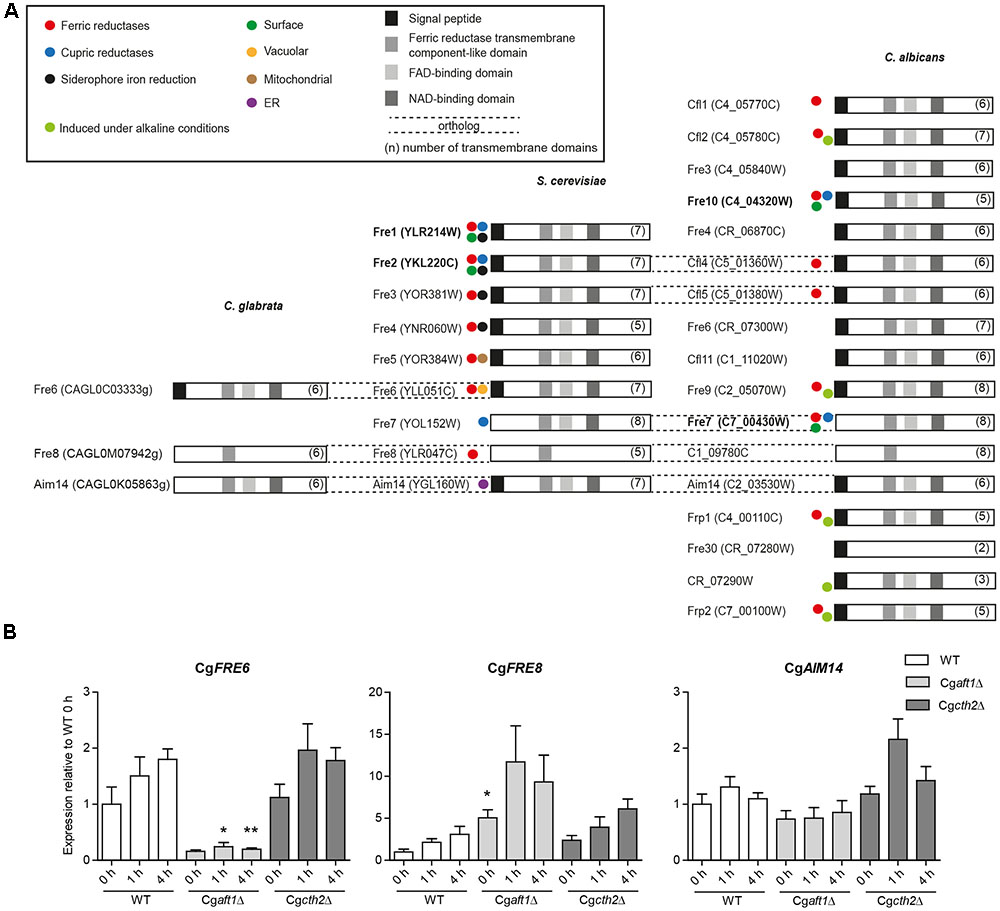
FIGURE 3. Ferric reductase genes in C. glabrata, S. cereviae and C. albicans and expression under iron deprivation in C. glabrata. (A) Species comparative in silico analysis of the ferric reductase families in C. glabrata (3 putative family members), S. cerevisiae (9 family members) and C. albicans (17 putative family members), Orf19.1845 which had additionally been predicted as a 18th member of the Fre family (Jeeves et al., 2011) was merged with CaFre4 in Assembly 20. Dashed lines = gene orthologs in the respective species. Displayed domains (squares) include the signal peptide, a ferric reductase transmembrane component-like domain (PF01794), a FAD-binding domain (PF08022) and a NAD-binding domain (PF08030). The number of transmembrane domains is indicated in parentheses. Colored circles indicate known activity as ferric, cupric or siderophore iron reductases and the postulated localization on the cell surface, in the vacuole, mitochondria or ER according to previous publications (Supplementary Table 2). Major surface ferric reductases are depicted in bold. (B) qRT-PCR expression analysis of the putative C. glabrata ferric reductase genes: CgFRE6, CgFRE8, and CgAIM14 under iron starvation conditions in WT, Cgaft1Δ and Cgcth2Δ. Data is shown in biological triplicates as means ± SEM, for statistical analysis unpaired Student’s t-test was performed in comparison to WT/equal time point (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
A BLAST search in the predicted protein sequences of C. glabrata using the ferric reductase transmembrane component-like domain of major C. albicans and S. cerevisiae surface ferric reductases (CaFre10 and ScFre1) as the query revealed only the three aforementioned putative C. glabrata ferric reductases CgFre6, CgFre8, and CgAim14. No additional, thus far unknown ferric reductases were found. CgFre6 showed 37% protein identity to ScFre6 and contained the characteristic ferric reductase domains – an N-terminal signal peptide, the ferric reductase transmembrane component-like domain, a FAD- and NAD-binding domain, and a transmembrane domain (Figure 3). CgFre8 showed 41% protein identity to ScFre8 and 29.9% identity to its presumed ortholog in C. albicans, C1_09780C. Like its S. cerevisiae and C. albicans orthologs, CgFre8 lacks both, the FAD-/NAD-binding domains and a signal peptide. CgAim14 showed 38 and 28.3% protein identity to its S. cerevisiae and C. albicans orthologs, respectively. In contrast to its cross-species counterparts, however, CgAim14 does not contain any signal peptide. Importantly, the closest S. cerevisiae and C. albicans orthologs of CgFre6, CgFre8, and CgAim14 have all not been described as surface-localized ferric reductases. In addition, and in contrast to CgFre6, the lack of a predicted signal peptide in CgFre8 and CgAim14 suggests that these proteins may not enter the secretory pathway and thus do not have access to the cell surface or the extracellular space.
In conclusion, our in silico analysis confirmed the comparatively small number of three ferric reductase family members and the lack of orthologs of known surface ferric reductases of S. cerevisiae and C. albicans in C. glabrata. The presence of ferric reductase protein domains and a signal peptide in CgFre6, which is absent in the other putative C. glabrata ferric reductases, suggests CgFre6 to be the potential major surface reductase of this species.
Expression of CgFRE6 and CgFRE8 Is Dependent on the Iron Master Regulator Aft1
Expression of uptake-associated ferric reductases is typically induced under iron deprivation (Georgatsou and Alexandraki, 1999; Jeeves et al., 2011). Similarly to S. cerevisiae (Puig et al., 2005), iron acquisition in C. glabrata is largely activated by the transcription factor Aft1, whereas iron consuming processes are inhibited by Cth2-dependent degradation of their mRNA transcripts (Gerwien et al., 2016).
To analyze the iron-dependent regulation of the three potential C. glabrata ferric reductases, we tested for Aft1 and Cth2 dependency of CgFRE6, CgFRE8, and CgAIM14 transcription under iron limitation. We found a moderate induction of CgFRE6 and CgFRE8 transcript levels in the wild-type (WT), in line with their presumed roles as ferric reductases (Figure 3B). CgAFT1 deletion almost completely abolished CgFRE6 expression, but led to an overexpression of CgFRE8. In contrast, both CgFRE6 and CgFRE8 were not significantly affected by CTH2 deletion. Finally, transcript levels of CgAIM14 were largely unaffected by iron deprivation and by deletion of CgAFT1 or CgCTH2.
Taken together, the CgAft1-dependent suppression indicates that there is no apparent role of CgFre8 in iron uptake, while the CgAft1-mediated up-regulation of CgFRE6 transcription supports our in silico prediction of CgFre6 as the main C. glabrata uptake-associated surface ferric reductase.
C. glabrata Fre6 and Fre8 Do Not Confer Ferric Reductase Activity In Vitro
We wanted to measure the contribution of these proteins to the surface ferric reductase activity of C. glabrata by a 2,3,5-triphenyltetrazolium chloride (TTC)- and ferrozine-based assay (Figure 4; see Materials and Methods section). Surprisingly, while C. albicans and S. cerevisiae exhibited surface ferric reductase activity, indicated by red coloration in the TTC assay (supplemented with antimycin A) (Figure 4A) or the formation of a purple halo around colonies in the ferrozine assay (Figure 4B), C. glabrata showed no such activity. This was the case for our standard WT strain and also for a set of clinical strains isolated from different anatomical sites. Iron and copper starvation is known to increase surface reductase activity (Georgatsou and Alexandraki, 1999; Santos et al., 2003). Enhanced activity was indeed observed for the C. albicans WT with the TTC assay, but not using ferrozine (Figures 4C,D and Supplementary Figure 1). In contrast C. glabrata lacked any detectable surface ferric reductase activity even under these inducing conditions (Supplementary Figure 1). We continued to elucidate the function of the C. glabrata ferric reductases by creating Cgfre6Δ and Cgfre8Δ single and double deletion mutants; we were, however, not able to delete CgAIM14. In accordance with the lack of surface ferric reductase activity and in contrast to mutants lacking other major components of the HA iron uptake system (Figure 2), C. glabrata ferric reductase mutants showed no defects in growth with ferritin, under moderate FeCl3 levels (Figures 5A,B) or under copper and iron starvation (Supplementary Figure 1). Thus, neither CgFre6 nor CgFre8 are required for HA iron uptake of C. glabrata.
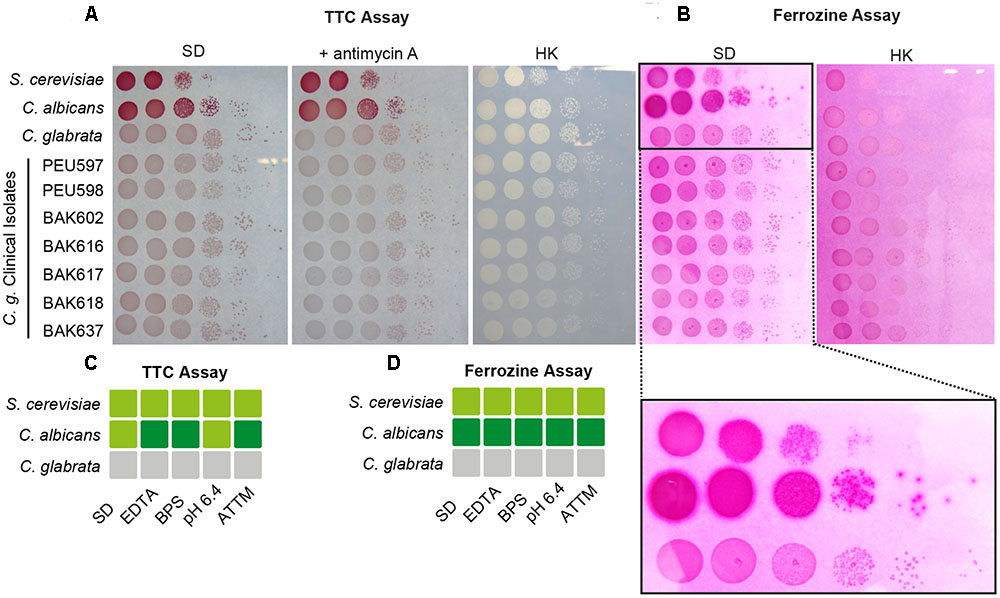
FIGURE 4. Candida glabrata lacks surface ferric reductase activity. Species comparison of surface ferric reductase activity for S. cerevisiae, C. albicans, and C. glabrata WTs as well as C. glabrata clinical isolates derived from different host tissues. Cells were spotted on SD agar and grown over-night (oN) at 37°C. (A) Determination of surface reductase activity with 2,3,5-triphenyltetrazolium chloride (TTC). Red coloration of colonies 1 h after application indicates surface reductase activity. Antimycin A was added to inhibit intracellular respiratory reduction. HK: heat-inactivated plate (70°C, 1 h). (B) Determination of surface ferric reductase activity with ferrozine. Formation of a purple halo (see magnified section) around colonies 5 min after application of ferrozine indicates surface ferric reductase activity. HK: heat-inactivated plate (70°C, 1 h) represents negative control. (C,D) Summary of determined surface reductase activity with TTC (C) and ferrozine (D) under iron- and copper-deprivation inducing conditions (3 μM ethylenediaminetetraacetic acid (EDTA), 10 μM BPS, pH 6.4, 7 μM ammonium tetrathiomolybdate (ATTM), gray box: no surface reductase activity, light-green box: moderate surface reductase activity, dark-green box: strong surface reductase activity.
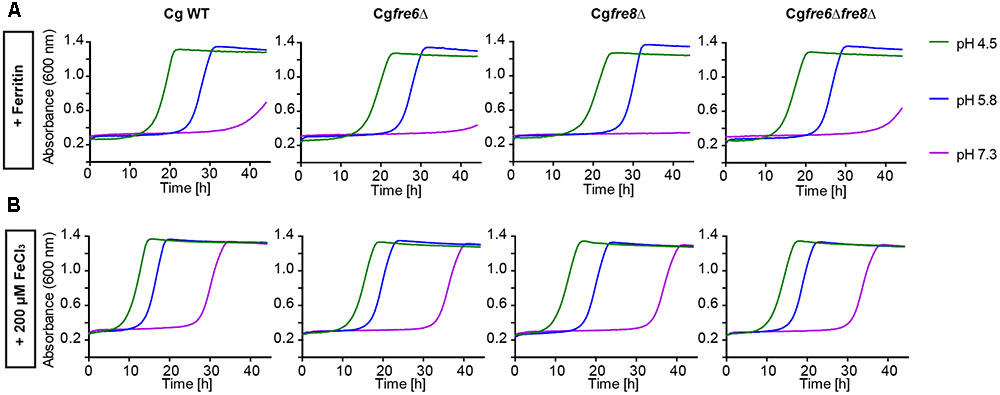
FIGURE 5. Comparative pH-dependent growth of C. glabrata ferric reductase mutants on host iron sources. Growth of iron-prestarved (preculture treatment with 200 μM BPS) with C. glabrata WT, and mutants (Cgfre6Δ, Cgfre8Δ and Cgfre6Δfre8Δ) in liquid SD buffered to pH 4.5, pH 5.8, and pH 7.3 at 37°C containing: (A) 200 μM BPS + 100 μg/ml ferritin. (B) 200 μM BPS + 200 μM FeCl3. n = 3, A representative example is shown.
Taken together, these results point to major differences between the surface ferric reduction properties of C. albicans and S. cerevisiae in comparison to C. glabrata. The absence of measurable ferric reductase activity in C. glabrata argues against surface reductase functions of CgFre6 or CgFre8, and indicates a loss of surface ferric reduction ability in C. glabrata.
C. glabrata, C. albicans, and S. cerevisiae Exhibit Extracellular Ferric Reduction
The importance of surface ferric reduction has been demonstrated in various bacteria and fungi (Schröder et al., 2003). However, some fungi such as Histoplasma capsulatum are also able to secrete either ferric reductases (Zarnowski and Woods, 2005; Zarnowski et al., 2008) or low molecular weight ferric reductants into the environment (Timmerman and Woods, 1999). We therefore postulated that C. glabrata might engage in a similar strategy to compensate for its lack of surface ferric reductase activity. To measure extracellular ferric reduction activity in culture supernatants of C. glabrata, C. albicans, and S. cerevisiae, we monitored conversion of Fe3+ to Fe2+ by spectrophotometric detection of Fe2+-ferrozine complexes, with the reductant 1,4-Dithiothreitol (DTT) as a positive control. For all analyzed species, ferric reduction activities were detected (Figure 6A).
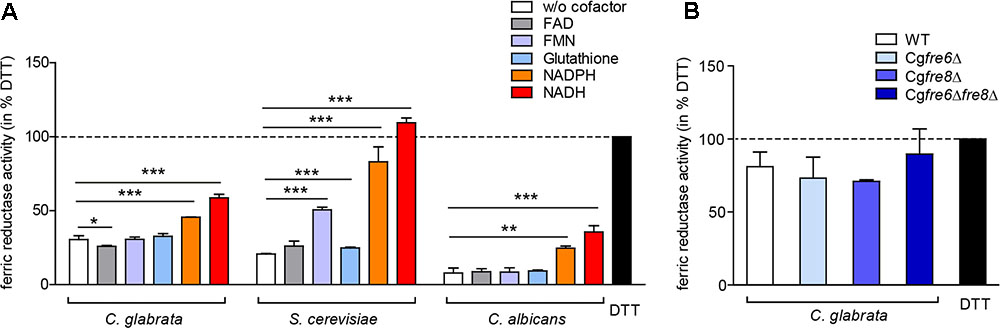
FIGURE 6. Extracellular ferric reduction in the culture supernatants of C. glabrata, C. albicans, and S. cerevisiae. oN cultures (in SD BR, pH 5.8) cells were harvested, the wet cell volume was determined and a cell-free culture supernatant was obtained via filtration. Extracellular ferric reduction in the supernatant was measured continuously for 3 h after addition of the detection mix (containing ferrozine, and ferric ammonium citrate as a ferric iron source) by spectrophotometrical detection of a ferrozine-Fe2+-complex. Data is shown for the 2 h time point after subtraction of the media control, as percentage of the positive control (the reductant DTT). Data is shown in biological triplicates as means ± standard deviation, for statistical analysis unpaired Student’s t-test was performed (∗P < 0.05, ∗∗P < 0.01, or ∗∗∗P < 0.001). (A) Extracellular ferric reduction activity of C. glabrata, S. cerevisiae, and C. albicans WT strains after addition of different cofactors with a final concentration of 250 μg/ml NADH, 250 μg/ml NADPH, 162.5 μM glutathione, 0.5 μM FMN, or 0.5 μM FAD. (B) Extracellular ferric reduction activity of C. glabrata WT, Cgfre6Δ, Cgfre8Δ and Cgfre6Δfre8Δ with NADH as cofactor.
Microbial ferric reductases typically act as oxido-reductases and require a cofactor as electron donor. This cofactor is frequently NAD(P)H (Schröder et al., 2003), and more rarely glutathione (Zarnowski and Woods, 2005; Zarnowski et al., 2008). In addition, certain bacteria additionally require flavins (FAD and FMN) as intermediate electron acceptor (Schröder et al., 2003). Accordingly, we tested the potential cofactor function of glutathione, FAD, FMN, NADH, or NADPH in extracellular ferric reduction (Figure 6A). Only addition of NADH or NADPH led to a significant increase in ferric reduction in all species. A slight increase upon FMN addition was additionally detected in S. cerevisiae supernatants. NADH was therefore chosen as a cofactor for further experiments.
Our data thus show that all three species are able to reduce ferric iron extracellularly.
Extracellular Ferric Reduction Is Independent of Fre6 and Fre8 and Mediated by a Low-Molecular Weight Molecule
To test whether the putative C. glabrata ferric reductases contribute to extracellular ferric reduction, we analyzed culture supernatants of the CgFRE6 and CgFRE8 single and double deletion mutants in presence of NADH. The deletion mutants showed similar extracellular ferric reduction capabilities as the WT (Figure 6B), indicating that CgFre6 and CgFre8 are not only dispensable for surface activity, but also for extracellular ferric reduction by C. glabrata.
To elucidate whether the observed extracellular ferric reduction is dependent on another protein or a smaller molecule, we separated the supernatant into a low molecular weight fraction (<10 kDa) and a high molecular weight fraction (>10 kDa), and optionally treated both with proteinase K (Figure 7A). The majority of extracellular ferric reduction activity was found in the low molecular weight fraction for all species. As this activity was not (C. glabrata and C. albicans) or only slightly (S. cerevisiae) affected by proteinase K treatment, these data point to a small non-protein molecule as the main reducing agent. The high molecular weight fraction contributed only a very small reductase activity in all species, which was almost completely abolished by addition of proteinase K. This points to a minor contribution of a potentially secreted protein. In support of these findings, extracellular ferric reduction was not affected by heat or ultraviolet light (UV) treatment to denature proteins (Figure 7B), or by growth in presence of brefeldin A, an inhibitor of protein secretion in eukaryotes (Kossaczka et al., 1995) (Figure 7C).
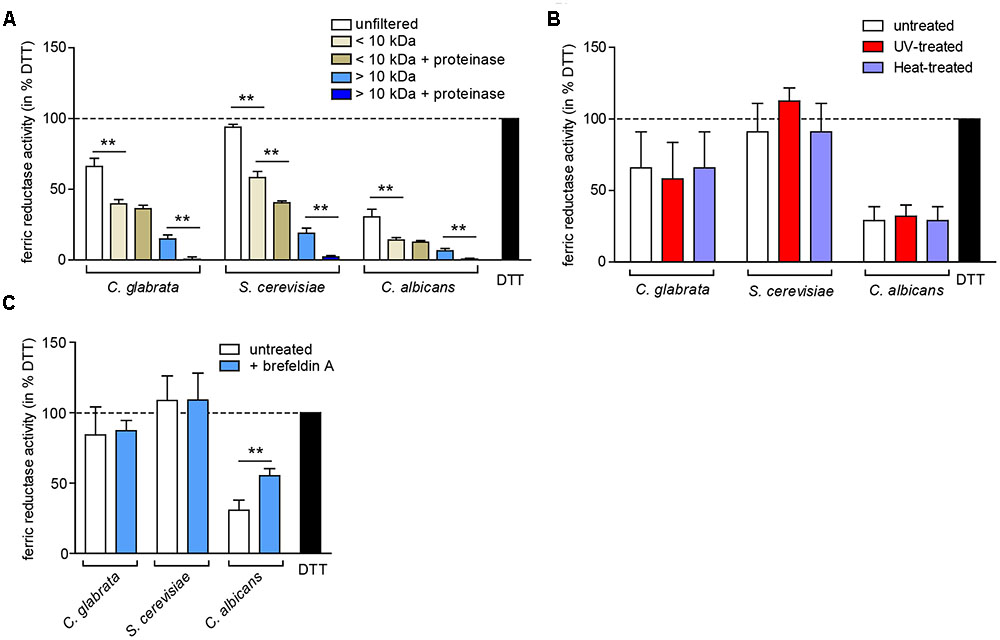
FIGURE 7. The extracellular ferric reduction activity is mediated by a low-molecular non-protein in C. glabrata, C. albicans, and S. cerevisiae. oN cultures (in SD BR, pH 5.8) cells were harvested, the wet cell volume was determined and a cell-free culture supernatant was obtained via filtration. Extracellular ferric reduction in the supernatant was measured continuously for 3 h after addition of the detection mix (containing NADH, ferrozine and ferric ammonium citrate) by spectrophotometrical detection of a ferrozine-Fe2+-complex. Data is shown for the 2 h time point after subtraction of the media control, as percentage of the positive control (the reductant DTT). Data is shown in biological triplicates as means ± standard deviation, for statistical analysis unpaired Student’s t-test was performed (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). (A) Extracellular ferric reduction activity of C. glabrata, S. cerevisiae, and C. albicans WT strains after molecular weight fractioning of the culture supernatants. Analysis has been performed for the high molecular weight fraction (>10 kDa) and the low molecular weight fraction (<10 kDa) with or without addition of proteinase K (100 μg/ml, 30 min, 37°C). (B) Extracellular ferric reduction activity of C. glabrata, S. cerevisiae, and C. albicans WT strains after UV-treatment (120 mJ, 360 s) or heat-treatment (70°C, 20 min). (C) Extracellular ferric reduction activity of C. glabrata, S. cerevisiae, and C. albicans WT strains after addition of the protein secretion inhibitor brefeldin A (10 μg/ml) prior to oN culturing.
In summary, major extracellular ferric reduction activity in the three analyzed species is likely carried out by a low molecular weight non-protein.
Discussion
Exploitation of host iron sources and efficient iron uptake are essential for pathogens to survive and grow in the host. Our experiments confirmed again that C. albicans is able to use a broad spectrum of host iron sources including ferritin (Almeida et al., 2008), transferrin (Knight et al., 2005), hemin (Santos et al., 2003), and hemoglobin (Kuznets et al., 2014). The latter three served as iron sources preferably at alkaline pH, a condition also found in blood, where these iron sources are predominantly encountered by the fungus. Accordingly, our species-comparative analysis of iron acquisition systems detected the known ferritin and heme uptake receptors in C. albicans, while both, S. cerevisiae and C. glabrata seem to lack the necessary genes to exploit these iron sources. We cannot exclude that the current knowledge of iron uptake-related genes is not complete and that there are yet unknown host iron source-specific utilization systems in C. glabrata or S. cerevisiae. However, the fact that both species are unable to grow with transferrin, hemoglobin, or hemin as sole iron source seems to argue against such additional systems. It has been shown previously that hemin is not a suitable iron source for C. glabrata (Nevitt and Thiele, 2011), although earlier studies had reported hemolytic activity by C. glabrata in vitro (Luo et al., 2004; Malcok et al., 2009). This study adds transferrin as another potential iron source which seems inaccessible to C. glabrata.
The inability of S. cerevisiae to use all these host molecules may be considered not too surprising, as this yeast leads a generally non-pathogenic lifestyle. It seems more puzzling why C. glabrata is also not able to exploit transferrin, hemoglobin, or heme: While it underlines its close evolutionary relationship to the baker’s yeast, it raises the question of how this fungus is able to grow in the host environment and cause disease. In fact, the lack of the ability to use iron sources of the blood and the iron-related growth impairment of C. glabrata at alkaline pH suggests that C. glabrata is not well-adapted to typical alkaline host niches. Our data rather implies that C. glabrata depends on a restricted spectrum of host iron sources, especially compared to C. albicans. As non-bound iron is virtually non-existent in the host, these sources seem limited to ferritin and xenosiderophores, produced by other members of the microbial flora (Nevitt and Thiele, 2011).
Which system allows C. glabrata the exploitation of ferritin and the use of non-protein-bound iron? Our in silico analyses show the existence of a low-affinity (LA) iron uptake, a xenosiderophore uptake, and a HA iron uptake system. In previous studies, the disruption of LA iron uptake (Fet4) resulted in no apparent iron-related phenotype (Srivastava et al., 2014; Gerwien et al., 2016), suggesting a minor role for this system, similar maybe to S. cerevisiae (Dix et al., 1994). Iron acquisition mediated by uptake of siderophores produced by other species might play a role during interspecies commensal growth, e.g., in the gut, and it may also enhance resistance to the microbicidal activity of phagocytes (Haas et al., 2008; Nevitt and Thiele, 2011). In agreement with previous studies (Srivastava et al., 2014; Gerwien et al., 2016; Sharma et al., 2016), we found that ferritin and non-protein-bound iron uptake in C. glabrata depends on the third pathway: the reductive HA iron uptake system. However, for ferritin-mediated iron acquisition the detailed mechanism, including the receptors for binding of ferritin and the mode of iron release, remains to be elucidated in C. glabrata.
Ferric reduction is a critical initial step for the reductive HA iron uptake and is usually highly conserved in bacteria and fungi (Schröder et al., 2003). Surprisingly, in contrast to the large gene families in other pathogenic fungi (Dancis et al., 1992; Martins et al., 1998; Yun et al., 2001; Blatzer et al., 2011; Jeeves et al., 2011; Saikia et al., 2014; Xu et al., 2014), we identified in our in silico analysis only three putative ferric reductase orthologs in C. glabrata, none of them orthologous to known surface reductases in C. albicans or S. cerevisiae. Although we cannot formally exclude the presence of other ferric reductases with alternate domains, we were not able to find more proteins with a ferric reductase transmembrane component-like domain in the predicted C. glabrata protein sequences. In line with this, our assays showed, in contrast to C. albicans and S. cerevisiae, no surface ferric reduction ability of C. glabrata in vitro. However, we still observed iron-dependent expression for two of the three putative ferric reductase genes, CgFRE6 and CgFRE8, although deletion of both genes resulted in no apparent iron-related phenotypes. While we failed to delete CgAIM14, the lack of surface reductase activity of the WT as well as the lack of a signal peptide in CgAim14 makes it similarly highly unlikely that it plays a role in surface-based iron acquisition processes.
Our observations for CgFRE6 complement other studies, where a CgFRE6 deletion did not affect iron-dependent growth or intracellular iron accumulation (Srivastava et al., 2014). Deletion also affected virulence of C. glabrata only subtly, with a slightly reduced fungal burden in murine kidneys (Srivastava et al., 2014) and a small increase in survival of a Drosophila melanogaster model (Brunke et al., 2015). Overall, our data thus resemble findings in S. cerevisiae, as ScFRE6 displays a moderate iron-dependent expression (Shakoury-Elizeh et al., 2004), and its deletion resulted in no apparent phenotype under iron limitation (Ahmed Khan et al., 2000). Importantly, ScFre6 is localized at the vacuole (Huh et al., 2003) and not at the cell surface. It seems thus likely that Fre6 in C. glabrata has a similar localization and possibly a similar, thus-far unknown biological function as ScFre6.
In contrast, our observations for CgFre8 (having an iron-dependent gene expression and being dispensable for growth) do not match to what is known for ScFre8. A strain lacking ScFRE8 was unable to grow in low iron and was respiration-deficient (De Freitas et al., 2004), but expression of ScFRE8 was unaffected by changing iron levels (Georgatsou and Alexandraki, 1999). These differences indicate potentially distinct roles for Fre8 in both species, although CgFRE8 deletion was accompanied by lower survival rates in macrophages and diminished intracellular replication (Seider et al., 2014).
In conclusion, we cannot exclude a more subtle biological function of CgFre6 and CgFre8 that depends on additional factors besides iron availability. With these two reductases being dispensable for growth, our data indicates a general loss of surface-associated ferric reduction, which makes C. glabrata – to our knowledge – the first pathogenic fungus to lack this capability.
Consequently, we analyzed whether secreted ferric reductants might be present in the fungus to compensate for the lack of surface ferric reduction. Surprisingly, we found ferric reduction capacity in the supernatants of all three investigated species, and we wondered whether this process was dependent on an enzymatic process. Although excreted ferric reductases have been described previously in H. capsulatum, those are glutathione-dependent (Zarnowski and Woods, 2005; Zarnowski et al., 2008), whereas our data point toward an NAD(P)H-dependent process. In C. albicans it has been suggested that the ferric reductase CaCfl2 is secreted (Sorgo et al., 2011), but no secreted ferric reductases have been described in C. glabrata (or S. cerevisiae), and both species lack direct CaCfl2 orthologs. Furthermore, although CaCfl2 might contribute to C. albicans extracellular ferric reduction, we observed the majority of extracellular ferric reduction in the low-molecular weight fraction in all three species, and protein degradation as well as inhibition of the protein secretory pathway did not influence extracellular ferric reduction. Thus, our observations strongly indicate a secreted non-protein compound to be responsible for the extracellular ferric reduction activity in all three species.
In other fungi, many low-molecular compounds are known which can act as ferric reductants, such as 3-hydroxyanthranilate (3-HAA) and anthranilate in S. cerevisiae (Lesuisse et al., 1992), 3-HAA in C. neoformans (Nyhus et al., 1997; Jacobson et al., 1998; Jung et al., 2008), an unknown compound in H. capsulatum (Timmerman and Woods, 1999), terrein in Aspergillus terreus (Gressler et al., 2015), 2,5-dimethylhydroquinone in the brown-rot fungi such as Gloeophyllum trabeum (De Luca and Wood, 2000; Arantes and Milagres, 2008), as well as secreted oxalic and citric acids in other wood-decaying fungi (Arantes and Milagres, 2008) and possibly in A. niger (Johnson, 2003). However, in most of these species surface-bound ferric reductases are still responsible for the vast majority of iron reduction, and these compounds play only secondary roles. This applies also to C. albicans and S. cerevisiae with their broad array of surface ferric reductases. For these fungi, secreted reductants may benefit the overall reductive capacity, but are not strictly necessary. For C. glabrata, however, the lack of surface ferric reductase activity combined with its limited use of host iron sources makes this ability likely much more important for sustained growth and survival in the host. The nature of C. glabrata’s reductants is still unknown, but 3-HAA [as in S. cerevisiae (Lesuisse et al., 1992)] seems unlikely, as it is a metabolite of the de novo NAD-biosynthetic pathway (Li and Bao, 2007), which, in contrast to S. cerevisiae and C. albicans is not existent in C. glabrata (Kucharczyk et al., 1998; Li and Bao, 2007; Ma et al., 2007). Whether a pathway from host-derived NADH toward 3-HAA could exist is highly speculative, and currently seems a remote possibility at best. The second extracellular ferric reductant described for S. cerevisiae, anthranilate (Lesuisse et al., 1992), is a precursor of tryptophan biosynthesis in fungi (Braus, 1991) and might be a reasonable candidate responsible for the extracellular ferric reduction observed in C. glabrata.
The reason for the promoting effect of NAD(P)H on extracellular ferric reduction in C. glabrata and C. albicans (this study) as well as S. cerevisiae [this study and (Lesuisse et al., 1992)] still remains to be elucidated. NAD(P)H is a potential cofactor for oxido-reductases, however, our data indicate that non-enzymatic processes are mainly responsible for extracellular ferric reduction. In some fungi anthranilate can react with NAD(P)H in an enzyme-dependent process, producing the iron-chelating compound 2,3-dihydroxybenzoate (Anderson and Dagley, 1981; Kamath and Vaidyanathan, 1990; Chang et al., 2003), which is also a precursor of the iron siderophore enterobactin in Salmonella enterica and Escherichia coli (Graziano et al., 1974; Raymond et al., 2003). Anthranilate production by C. glabrata is likely (see above), however, the enzyme-independent nature of extracellular reduction speaks against a connection between anthranilate and NAD(P)H in C. glabrata.
Taken together, we propose here a novel iron acquisition strategy for C. glabrata, which differs from C. albicans and other pathogenic fungi. This strategy is characterized by the dependence on a restricted host iron source spectrum (ferritin and non-protein-bound iron) and a lack of surface ferric reduction activity while still relying on the HA iron uptake system. Instead C. glabrata possibly employs extracellular ferric reduction, mediated by a non-protein ferric reductant of low molecular weight.
Materials and Methods
Strains
The C. glabrata deletion mutant Cgfre6Δ was generated in an ATCC 2001 background by replacing the CgFRE6 with a TEF1 promoter-driven nourseothricin resistance (NAT1) cassette (Schwarzmüller et al., 2014). The cassette was amplified with 500–1,000 bp gene-specific flanks by PCR (primer: Fre6 na fwd × Fre6 na rev) from a previously existing mutant (Schwarzmüller et al., 2014). The PCR-amplified insert (primer Fre6 fwd × Fre6 rev) was used for C. glabrata transformation using a modified heat shock method (Sanglard et al., 1996) with 45°C heat shock for 15 min. For the double mutant Cgfre6Δfre8Δ, CgFRE6 was deleted in an ATCC 2001 his3Δ background strain (Schwarzmüller et al., 2014) as described above, followed by CgFRE8 deletion using CgHIS3 as an auxotrophy selection marker (including 861 bp promoter sequence and 232 bp terminator sequence): A HIS3 fragment (primer HIS3 gene fwd × HIS3 gene rev) was fused with 1,000 bp FRE8-specific 5′- and 3′-flanks (primer Fre8-fwd-pUC19 × Fre8-HIS3 5′flank-BC, Fre8-HIS3 3′flank-BC × Fre8-rev-pUC19) and integrated into a XbaI-linearized pUC19 vector using the Infusion HD Cloning Kit (Clontech). The PCR-amplified insert (primer Fre8 fwd × Fre8 rev) was used for transformation. The transformants were plated onto YPD agar (2% agar, 1% yeast extract, 2% peptone, 2% glucose) containing 250 μg/ml nourseothricin, or SD agar supplemented with 0.079% complete supplement mix (CSM) without histidine (Formedium), 2.5 μM Cu2O, 2.5 μM CuSO4, 25 μM FeCl3, and 25 μM FeSO4. Knockout strains were verified by PCR (primer gene naCoP1 × gene naCoP4) and Sanger sequencing. Strains used in this study are shown in Table 2 and all primers are listed in Supplementary Table 3.
Growth Curves
Strains were cultivated over-night (oN) at 37°C (C. glabrata) or 30°C (C. albicans and S. cerevisiae) in liquid synthetic defined media (SD: 0.67% YNB, 2% Glucose, 0.079% CSM [Formedium]) with 180°rpm shaking. They were then transferred to citrate-buffered SD (pH 5.8) containing 500 μM (for C. albicans) or 200 μM (for C. glabrata and S. cerevisiae) of the extracellular iron chelator bathophenanthrolinedisulfonic acid (BPS) to induce iron starvation. BPS precultures were incubated for 22 h, washed three times by centrifugation and re-elution in iron-free water, and then resuspended in iron-free water to on OD600 of 0.1. The yeast suspension (20 μl) was added to 180 μl citrate-buffered SD (pH 4.5, 5.8, or 7.3) containing 200 μM BPS and supplemented with either 100 μM FeCl3 (stock in 1% HCl), 100 μg/ml horse ferritin (stock in iron-free 5 mM HEPES, 0.15 M NaCl, 4× filtered through 50 KDa molecular weight cut-off columns [Amicon Ultra 0.5 ml]), 100 μg/ml transferrin (Calbiochem, stock in 0.15 M iron-free Na2CO3, 2× filtered through 50 KDa columns [Amicon Ultra 0.5 ml]), 0.1 mg/ml bovine hemoglobin (Sigma, stock in H2O), or 1 μM hemin (Sigma, stock in DMSO). Growth was recorded over 2 days in biological triplicates by OD600 measurement every 30 min (with intermittent shaking) at 37°C in a Tecan Infinite 200 ELISA reader.
TTC-Based Surface Reductase Activity Assay
SD oN cultures were washed twice with iron-free water, and adjusted to 2 × 107 cells/ml in iron-free water for spotting of 1:10 serial dilutions on unbuffered SD agar containing the metal chelator ethylenediaminetetraacetic acid – (EDTA) (3 μM), the iron chelator BPS (10 μM), or the copper chelator ammonium tetrathiomolybdate – (ATTM) (7 μM). For more alkaline conditions, SD agar was phosphate-buffered to pH 6.4. Plates were incubated oN at 37°C. For the negative control, one plate was inactivated after oN cultivation by heat treatment (1 h, 72°C). Subsequently, 1.5% agarose was melted in 1 × TAE and supplemented with 0.1% TTC (Roth), plus 20 μg/ml antimycin A (Sigma) to inhibit mitochondrial reduction activity (Hsu et al., 2011) where indicated. The plates were overlaid with this TTC agarose, and formation of red formazan, indicating cell surface reduction, was detected in biological triplicates after 1 h incubation.
Ferrozine-Based Surface Ferric Reductase Activity Assay
Serial dilution growth plates were prepared as described before, but with an overlay mixture of 0.5% agarose, 10 mM MgCl2, 0.1 mg/ml NADH, 1 mg/ml ferrozine, 5 mg/ml ferric ammonium citrate instead. Formation of a purple halo around colonies, indicating surface ferric reductase activity, was documented in biological triplicates after 5 min incubation.
Ferric Reductases Assay in Culture Supernatants
Ferric reduction activity was measured for all three species from stationary oN cultures. Preliminary experiments revealed that low pH medium [such as unbuffered rich medium (YPD) and minimal medium (SD)] or a low pH due to secretion of fungal metabolic products lead to increased background signals (not shown). We therefore chose buffered nutrient-limited medium for our assays, which resulted in the lowest background noise and satisfactory growth for all tested species – SD BR medium [0.67% YNB, 2% Glucose, 50% Britton Robinson buffer (40 mM H3BO4, 40 mM Acetic acid, 40 mM H3PO4, 3 g/L KCl, pH 5.8)]. Precultures were prepared in SD BR, and cultures were treated with an inhibitor of Golgi/ER-dependent protein secretion, brefeldin A (10 μg/ml) (Kossaczka et al., 1995) where indicated. The average wet cell volume per 200 μl oN culture was determined by cell volume tubes (TPP) for normalization. The cell-free supernatant was obtained by centrifugation for 4 min at 4,000 ×g and filtering through Minisart syringe filters (Sigma, 0.2 μm). Supernatant and media control were treated identically: Heat-treatment (70°C, 20 min), UV-treatment (120 mJ, 360 s), fractionation on molecular weight (Ultra 0.5 ml Amicon centrifugal protein filters, 10 kDa molecular weight cut-off with restoration of original volume of the high molecular weight fraction in adequate medium), or proteinase K-treatment (100 μg/ml, 30 min, 37°C). Culture supernatant or control (180 μl) were combined with 20 μl detection mix consisting of five volumes Britton-Robinson buffer, 1 volume 1 M MgCl2, 0.5 volumes 15 mg/ml ammonium ferric citrate, 1 volume 20 mM ferrozine, and 2.5 volumes of potential cofactor (stock either 10 mg/ml NADH, 10 mg/ml NADPH, 6.5 mM glutathione, 20 μM FMN, or 20 μM FAD). As positive control, 5 mM of the reductant DTT was added. Iron reduction (as ferrous iron binding to ferrozine) was detected by OD568 measurement every 5 min for 4 h at 37°C in a Tecan Infinite 200 ELISA reader in biological triplicates. Data of the 2 h time point was normalized to the initial wet cell volume and adjusted to a scale from 0% (medium control) to 100% (DTT).
Sample Preparation for RNA Isolation
Sample preparation and RNA isolation was conducted as described previously (Gerwien et al., 2016). Briefly, an SD oN culture was harvested, washed, and 1 × 107 cells/ml were inoculated into iron-free citrate-buffered SD (pH 5.8) supplemented with 5 μM FeCl3 and grown for 4 h at 37°C and 180 rpm. The 0 h sample was immediately frozen in liquid nitrogen and stored at -80°C. The remaining culture was washed four times with iron-free SD (start time set as first contact with the medium) and incubated in the same medium for the 1 and 4 h samples. RNA was isolated with the RNAeasy Mini Kit (Qiagen), and quality was verified using the Agilent 2100 Bioanalyzer Nanochip system according to the manufacturer’s protocol. The concentration was determined using a NanoDrop 1000 instrument.
Quantitative Real-Time PCR (qRT-PCR)
Quantitative real-time PCR was conducted as described previously (Gerwien et al., 2016). Briefly, 600 ng high-quality RNA was treated with DNAse (Epicentre) and reversely transcribed into cDNA using oligo-dT primers and Superscript III (Invitrogen). 1 μl diluted cDNA (1:20) was used for gene expression analysis in a C1000 thermocycler (Bio-Rad, CFX96 Realtime system) with the EvaGreen system (Bio & Sell). The expression rates were determined in biological triplicates and normalized to the house keeping genes EFB1 and EFT2 using Bio-Rad CFX Manager 3.1.
In Silico Analysis, Databases and Statistics
Information about gene orthologs, protein structure, and BLAST results were obtained from the Candida Genome Database (CGD)1 and the Saccharomyces Genome Database (SGD)2. BLAST search for proteins with a predicted ferric reductase domain in C. glabrata (pfam family PF01794, ferric reductase transmembrane component-like domain) was performed using the domain sequence of ScFre1 and CaFre10 as query. Additionally, the presence of additional conserved protein domains were predicted by CGD and SGD-associated prediction tools (SignalP, TMHMM, Pfam) and verified by SMART3. Subcellular localization was predicted by TargetP 1.14. GraphPad Prism 5 (GraphPad Inc.) was used for statistics. All data are reported as the mean ± SEM or standard deviation where appropriate, and two-tailed, unpaired Student’s t-test was performed, if not stated otherwise. Statistically significant results were marked as ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Author Contributions
The authors FG, LK, and BH designed the study. FG, AS, and SW performed and the experiments and acquired the data. FG, AS, and LK evaluated and interpreted the results. FG designed the figures and FG, SB, LK, and BH wrote and revised the manuscript.
Funding
This work has been supported in part by the German Research Foundation (DFG) within the priority program SPP1580 (www.spp1580.uni-bonn.de, HU 528/16-2; LK and BH), and the Centre for Sepsis Control and Care (CSCC, www.cscc.uniklinikum-jena.de/cscc/en/CSCC.html, grant 01EO1002; BH) by the German Federal Ministry of Education and Health (BMBF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
FG and BH are members of International Leibniz Research School for Microbial and Biomolecular Interactions (ILRS) and the graduate school, Jena School for Microbial Communication (JSMC). We thank our student assistants Ali A. Sheikh and Fabrice Hille for their practical contributions to this work, and Markus Gressler for his valuable input.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01055/full#supplementary-material
Footnotes
- ^www.candidagenome.org
- ^www.yeastgenome.org
- ^smart.embl-heidelberg.de
- ^www.cbs.dtu.dk/services/TargetP/
References
Ahmed Khan, S., Zhang, N., Ismail, T., El-Moghazy, A. N., Butt, A., Wu, J., et al. (2000). Functional analysis of eight open reading frames on chromosomes XII and XIV of Saccharomyces cerevisiae. Yeast 16, 1457–1468. doi: 10.1002/1097-0061(200012)16:16<1457::AID-YEA635>3.0.CO;2-3
Almeida, R. S., Brunke, S., Albrecht, A., Thewes, S., Laue, M., Edwards, J. E., et al. (2008). the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 4:e1000217. doi: 10.1371/journal.ppat.1000217
Anderson, G. J., Lesuisse, E., Dancis, A., Roman, D. G., Labbe, P., and Klausner, R. D. (1992). Ferric iron reduction and iron assimilation in Saccharomyces cerevisiae. J. Inorg. Biochem. 47, 249–255. doi: 10.1016/0162-0134(92)84070-4
Anderson, J. J., and Dagley, S. (1981). Catabolism of tryptophan, anthranilate, and 2,3-dihydroxybenzoate in Trichosporon cutaneum. J. Bacteriol. 146, 291–297.
Arantes, V., and Milagres, A. M. (2008). Response of Wolfiporia cocos to iron availability: alterations in growth, expression of cellular proteins, Fe3+-reducing activity and Fe3+-chelators production. J. Appl. Microbiol. 104, 185–193.
Baek, Y. U., Li, M., and Davis, D. A. (2008). Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot. Cell 7, 1168–1179. doi: 10.1128/EC.00108-08
Blankenship, J. R., and Mitchell, A. P. (2011). Candida albicans adds more weight to iron regulation. Cell Host Microbe 10, 93–94. doi: 10.1016/j.chom.2011.08.001
Blatzer, M., Binder, U., and Haas, H. (2011). The metalloreductase FreB is involved in adaptation of Aspergillus fumigatus to iron starvation. Fungal Genet. Biol. 48, 1027–1033. doi: 10.1016/j.fgb.2011.07.009
Braus, G. H. (1991). Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 55, 349–370.
Brunke, S., and Hube, B. (2013). Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell Microbiol. 15, 701–708. doi: 10.1111/cmi.12091
Brunke, S., Quintin, J., Kasper, L., Jacobsen, I. D., Richter, M. E., Hiller, E., et al. (2015). Of mice, flies–and men? Comparing fungal infection models for large-scale screening efforts. Dis. Model. Mech. 8, 473–486. doi: 10.1242/dmm.019901
Chang, H. K., Mohseni, P., and Zylstra, G. J. (2003). Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J. Bacteriol. 185, 5871–5881. doi: 10.1128/JB.185.19.5871-5881.2003
Chen, C., Pande, K., French, S. D., Tuch, B. B., and Noble, S. M. (2011). An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10, 118–135. doi: 10.1016/j.chom.2011.07.005
Cheng, X., Xu, N., Yu, Q., Ding, X., Qian, K., Zhao, Q., et al. (2013). Novel insight into the expression and function of the multicopper oxidases in Candida albicans. Microbiology 159, 1044–1055. doi: 10.1099/mic.0.065268-0
Dancis, A., Klausner, R. D., Hinnebusch, A. G., and Barriocanal, J. G. (1990). Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 2294–2301. doi: 10.1128/MCB.10.5.2294
Dancis, A., Roman, D. G., Anderson, G. J., Hinnebusch, A. G., and Klausner, R. D. (1992). Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. U.S.A. 89, 3869–3873. doi: 10.1073/pnas.89.9.3869
De Freitas, J. M., Kim, J. H., Poynton, H., Su, T., Wintz, H., Fox, T., et al. (2004). Exploratory and confirmatory gene expression profiling of mac1Delta. J. Biol. Chem. 279, 4450–4458. doi: 10.1074/jbc.M212308200
De Luca, N. G., and Wood, P. M. (2000). Iron uptake by fungi: contrasted mechanisms with internal or external reduction. Adv. Microb. Physiol. 43, 39–74. doi: 10.1016/S0065-2911(00)43002-X
De Silva, D. M., Askwith, C. C., Eide, D., and Kaplan, J. (1995). The FET3 gene product required for high affinity iron transport in yeast is a cell surface ferroxidase. J. Biol. Chem. 270, 1098–1101. doi: 10.1074/jbc.270.3.1098
Dix, D. R., Bridgham, J. T., Broderius, M. A., Byersdorfer, C. A., and Eide, D. J. (1994). The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 269, 26092–26099.
Fonzi, W. A., and Irwin, M. Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728.
Georgatsou, E., and Alexandraki, D. (1994). Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 3065–3073. doi: 10.1128/MCB.14.5.3065
Georgatsou, E., and Alexandraki, D. (1999). Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast 15, 573–584. doi: 10.1002/(SICI)1097-0061(199905)15:7<573::AID-YEA404>3.0.CO;2-7
Gerwien, F., Safyan, A., Wisgott, S., Hille, F., Kaemmer, P., Linde, J., et al. (2016). A novel hybrid iron regulation network combines features from pathogenic and nonpathogenic yeasts. MBio 7:e01782-16. doi: 10.1128/mBio.01782-16
Graziano, J. H., Grady, R. W., and Cerami, A. (1974). The identification of 2, 3-dihydroxybenzoic acid as a potentially useful iron-chelating drug. J. Pharmacol. Exp. Ther. 190, 570–575.
Gressler, M., Meyer, F., Heine, D., Hortschansky, P., Hertweck, C., and Brock, M. (2015). Phytotoxin production in Aspergillus terreus is regulated by independent environmental signals. Elife 4:e07861. doi: 10.7554/eLife.07861
Guinea, J. (2014). Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 20(Suppl. 6), 5–10. doi: 10.1111/1469-0691.12539
Haas, H., Eisendle, M., and Turgeon, B. G. (2008). Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46, 149–187. doi: 10.1146/annurev.phyto.45.062806.094338
Hassett, R., Dix, D. R., Eide, D. J., and Kosman, D. J. (2000). The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem. J. 351(Pt 2), 477–484. doi: 10.1042/bj3510477
Heymann, P., Ernst, J. F., and Winkelmann, G. (2000). Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 186, 221–227. doi: 10.1111/j.1574-6968.2000.tb09108.x
Heymann, P., Gerads, M., Schaller, M., Dromer, F., Winkelmann, G., and Ernst, J. F. (2002). The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect. Immun. 70, 5246–5255. doi: 10.1128/IAI.70.9.5246-5255.2002
Hood, M. I., and Skaar, E. P. (2012). Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10, 525–537. doi: 10.1038/nrmicro2836
Hsu, P. C., Yang, C. Y., and Lan, C. Y. (2011). Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot. Cell 10, 207–225. doi: 10.1128/EC.00158-10
Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., et al. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. doi: 10.1038/nature02026
Jacobson, E. S., Goodner, A. P., and Nyhus, K. J. (1998). Ferrous iron uptake in Cryptococcus neoformans. Infect. Immun. 66, 4169–4175.
Jeeves, R. E., Mason, R. P., Woodacre, A., and Cashmore, A. M. (2011). Ferric reductase genes involved in high-affinity iron uptake are differentially regulated in yeast and hyphae of Candida albicans. Yeast 28, 629–644. doi: 10.1002/yea.1892
Jensen, L. T., and Culotta, V. C. (2002). Regulation of Saccharomyces cerevisiae FET4 by oxygen and iron. J. Mol. Biol. 318, 251–260. doi: 10.1016/S0022-2836(02)00093-1
Johnson, S. (2003). Do mitochondria regulate cellular iron homeostasis through citric acid and haem production? Implications for cancer and other diseases. Med. Hypotheses 60, 106–111. doi: 10.1016/S0306-9877(02)00342-0
Jung, W. H., Sham, A., Lian, T., Singh, A., Kosman, D. J., and Kronstad, J. W. (2008). Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog. 4:e45. doi: 10.1371/journal.ppat.0040045
Kamath, A. V., and Vaidyanathan, C. S. (1990). New pathway for the biodegradation of indole in Aspergillus niger. Appl. Environ. Microbiol. 56, 275–280.
Kim, D., Yukl, E. T., Moenne-Loccoz, P., and Montellano, P. R. (2006). Fungal heme oxygenases: functional expression and characterization of Hmx1 from Saccharomyces cerevisiae and CaHmx1 from Candida albicans. Biochemistry 45, 14772–14780. doi: 10.1021/bi061429r
Knight, S. A., Lesuisse, E., Stearman, R., Klausner, R. D., and Dancis, A. (2002). Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology 148, 29–40. doi: 10.1099/00221287-148-1-29
Knight, S. A., Vilaire, G., Lesuisse, E., and Dancis, A. (2005). Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect. Immun. 73, 5482–5492. doi: 10.1128/IAI.73.9.5482-5492.2005
Kossaczka, Z., Drgonova, J., Podobova, B., Betina, V., and Farkas, V. (1995). Accumulation of Golgi-specific mannosyltransferases in Candida albicans cells grown in the presence of brefeldin A. Can. J. Microbiol. 41, 971–977. doi: 10.1139/m95-135
Kucharczyk, R., Zagulski, M., Rytka, J., and Herbert, C. J. (1998). The yeast gene YJR025c encodes a 3-hydroxyanthranilic acid dioxygenase and is involved in nicotinic acid biosynthesis. FEBS Lett. 424, 127–130. doi: 10.1016/S0014-5793(98)00153-7
Kuznets, G., Vigonsky, E., Weissman, Z., Lalli, D., Gildor, T., Kauffman, S. J., et al. (2014). A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog. 10:e1004407. doi: 10.1371/journal.ppat.1004407
Lesuisse, E., Knight, S. A., Camadro, J. M., and Dancis, A. (2002). Siderophore uptake by Candida albicans: effect of serum treatment and comparison with Saccharomyces cerevisiae. Yeast 19, 329–340. doi: 10.1002/yea.840
Lesuisse, E., Simon, M., Klein, R., and Labbe, P. (1992). Excretion of anthranilate and 3-hydroxyanthranilate by Saccharomyces cerevisiae: relationship to iron metabolism. J. Gen. Microbiol. 138, 85–89. doi: 10.1099/00221287-138-1-85
Li, Y. F., and Bao, W. G. (2007). Why do some yeast species require niacin for growth? Different modes of NAD synthesis. FEMS Yeast Res. 7, 657–664. doi: 10.1111/j.1567-1364.2007.00231.x
Luo, G., Samaranayake, L. P., Cheung, B. P., and Tang, G. (2004). Reverse transcriptase polymerase chain reaction (RT-PCR) detection of HLP gene expression in Candida glabrata and its possible role in in vitro haemolysin production. APMIS 112, 283–290. doi: 10.1111/j.1600-0463.2004.apm11204-0509.x
Ma, B., Pan, S. J., Zupancic, M. L., and Cormack, B. P. (2007). Assimilation of NAD(+) precursors in Candida glabrata. Mol. Microbiol. 66, 14–25. doi: 10.1111/j.1365-2958.2007.05886.x
Malcok, H. K., Aktas, E., Ayyildiz, A., Yigit, N., and Yazgi, H. (2009). Hemolytic activities of the Candida species in liquid medium. Eurasian J. Med. 41, 95–98.
Martins, L. J., Jensen, L. T., Simon, J. R., Keller, G. L., and Winge, D. R. (1998). Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 273, 23716–23721. doi: 10.1074/jbc.273.37.23716
Nevitt, T., and Thiele, D. J. (2011). Host iron withholding demands siderophore utilization for Candida glabrata to survive macrophage killing. PLoS Pathog. 7:e1001322. doi: 10.1371/journal.ppat.1001322
Newman, S. L., and Smulian, A. G. (2013). Iron uptake and virulence in Histoplasma capsulatum. Curr. Opin. Microbiol. 16, 700–707. doi: 10.1016/j.mib.2013.09.001
Nyhus, K. J., Wilborn, A. T., and Jacobson, E. S. (1997). Ferric iron reduction by Cryptococcus neoformans. Infect. Immun. 65, 434–438.
Okamoto-Shibayama, K., Kikuchi, Y., Kokubu, E., Sato, Y., and Ishihara, K. (2014). Csa2, a member of the Rbt5 protein family, is involved in the utilization of iron from human hemoglobin during Candida albicans hyphal growth. FEMS Yeast Res. 14, 674–677. doi: 10.1111/1567-1364.12160
Perlroth, J., Choi, B., and Spellberg, B. (2007). Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med. Mycol. 45, 321–346. doi: 10.1080/13693780701218689
Philpott, C. C., Protchenko, O., Kim, Y. W., Boretsky, Y., and Shakoury-Elizeh, M. (2002). The response to iron deprivation in Saccharomyces cerevisiae: expression of siderophore-based systems of iron uptake. Biochem. Soc. Trans. 30, 698–702. doi: 10.1042/bst0300698
Posey, J. E., and Gherardini, F. C. (2000). Lack of a role for iron in the Lyme disease pathogen. Science 288, 1651–1653. doi: 10.1126/science.288.5471.1651
Protchenko, O., Ferea, T., Rashford, J., Tiedeman, J., Brown, P. O., Botstein, D., et al. (2001). Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276, 49244–49250. doi: 10.1074/jbc.M109220200
Protchenko, O., and Philpott, C. C. (2003). Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J. Biol. Chem. 278, 36582–36587. doi: 10.1074/jbc.M306584200
Puig, S., Askeland, E., and Thiele, D. J. (2005). Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120, 99–110. doi: 10.1016/j.cell.2004.11.032
Ramanan, N., and Wang, Y. (2000). A high-affinity iron permease essential for Candida albicans virulence. Science 288, 1062–1064. doi: 10.1126/science.288.5468.1062
Ratledge, C. (2007). Iron metabolism and infection. Food Nutr. Bull. 28, S515–S523. doi: 10.1177/15648265070284s405
Raymond, K. N., Dertz, E. A., and Kim, S. S. (2003). Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U.S.A. 100, 3584–3588. doi: 10.1073/pnas.0630018100
Saikia, S., Oliveira, D., Hu, G., and Kronstad, J. (2014). Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect. Immun. 82, 839–850. doi: 10.1128/IAI.01357-13
Sanglard, D., Ischer, F., Monod, M., and Bille, J. (1996). Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40, 2300–2305.
Santos, R., Buisson, N., Knight, S., Dancis, A., Camadro, J. M., and Lesuisse, E. (2003). Haemin uptake and use as an iron source by Candida albicans: role of CaHMX1-encoded haem oxygenase. Microbiology 149, 579–588. doi: 10.1099/mic.0.26108-0
Schaible, U. E., and Kaufmann, S. H. (2004). Iron and microbial infection. Nat. Rev. Microbiol. 2, 946–953. doi: 10.1038/nrmicro1046
Schröder, I., Johnson, E., and De Vries, S. (2003). Microbial ferric iron reductases. FEMS Microbiol. Rev. 27, 427–447. doi: 10.1016/S0168-6445(03)00043-3
Schwarzmüller, T., Ma, B., Hiller, E., Istel, F., Tscherner, M., Brunke, S., et al. (2014). Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog. 10:e1004211. doi: 10.1371/journal.ppat.1004211
Seider, K., Gerwien, F., Kasper, L., Allert, S., Brunke, S., Jablonowski, N., et al. (2014). Immune evasion, stress resistance, and efficient nutrient acquisition are crucial for intracellular survival of Candida glabrata within macrophages. Eukaryot. Cell 13, 170–183. doi: 10.1128/EC.00262-13
Shakoury-Elizeh, M., Tiedeman, J., Rashford, J., Ferea, T., Demeter, J., Garcia, E., et al. (2004). Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 1233–1243. doi: 10.1091/mbc.E03-09-0642
Sharma, V., Purushotham, R., and Kaur, R. (2016). The phosphoinositide 3-kinase regulates retrograde trafficking of the iron permease CgFtr1 and iron homeostasis in Candida glabrata. J. Biol. Chem. 291, 24715–24734. doi: 10.1074/jbc.M116.751529
Singh, R. P., Prasad, H. K., Sinha, I., Agarwal, N., and Natarajan, K. (2011). Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J. Biol. Chem. 286, 25154–25170. doi: 10.1074/jbc.M111.233569
Sorgo, A. G., Heilmann, C. J., Dekker, H. L., Bekker, M., Brul, S., De Koster, C. G., et al. (2011). Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot. Cell 10, 1071–1081. doi: 10.1128/EC.05011-11
Spizzo, T., Byersdorfer, C., Duesterhoeft, S., and Eide, D. (1997). The yeast FET5 gene encodes a FET3-related multicopper oxidase implicated in iron transport. Mol. Gen. Genet. 256, 547–556. doi: 10.1007/pl00008615
Srivastava, V. K., Suneetha, K. J., and Kaur, R. (2014). A systematic analysis reveals an essential role for high-affinity iron uptake system, haemolysin and CFEM domain-containing protein in iron homoeostasis and virulence in Candida glabrata. Biochem. J. 463, 103–114. doi: 10.1042/BJ20140598
Stearman, R., Yuan, D. S., Yamaguchi-Iwai, Y., Klausner, R. D., and Dancis, A. (1996). A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271, 1552–1557. doi: 10.1126/science.271.5255.1552
Timmerman, M. M., and Woods, J. P. (1999). Ferric reduction is a potential iron acquisition mechanism for Histoplasma capsulatum. Infect. Immun. 67, 6403–6408.
Troxell, B., Xu, H., and Yang, X. F. (2012). Borrelia burgdorferi, a pathogen that lacks iron, encodes manganese-dependent superoxide dismutase essential for resistance to streptonigrin. J. Biol. Chem. 287, 19284–19293. doi: 10.1074/jbc.M112.344903
Urbanowski, J. L., and Piper, R. C. (1999). The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 274, 38061–38070. doi: 10.1074/jbc.274.53.38061
Weissman, Z., and Kornitzer, D. (2004). A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 53, 1209–1220. doi: 10.1111/j.1365-2958.2004.04199.x
Weissman, Z., Shemer, R., Conibear, E., and Kornitzer, D. (2008). An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol. 69, 201–217. doi: 10.1111/j.1365-2958.2008.06277.x
Woodacre, A., Mason, R. P., Jeeves, R. E., and Cashmore, A. M. (2008). Copper-dependent transcriptional regulation by Candida albicans Mac1p. Microbiology 154, 1502–1512. doi: 10.1099/mic.0.2007/013441-0
Xu, N., Qian, K., Dong, Y., Chen, Y., Yu, Q., Zhang, B., et al. (2014). Novel role of the Candida albicans ferric reductase gene CFL1 in iron acquisition, oxidative stress tolerance, morphogenesis and virulence. Res. Microbiol. 165, 252–261. doi: 10.1016/j.resmic.2014.03.001
Yun, C. W., Bauler, M., Moore, R. E., Klebba, P. E., and Philpott, C. C. (2001). The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 276, 10218–10223. doi: 10.1074/jbc.M010065200
Yun, C. W., Ferea, T., Rashford, J., Ardon, O., Brown, P. O., Botstein, D., et al. (2000a). Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake. J. Biol. Chem. 275, 10709–10715.
Yun, C. W., Tiedeman, J. S., Moore, R. E., and Philpott, C. C. (2000b). Siderophore-iron uptake in saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters. J. Biol. Chem. 275, 16354–16359.
Zarnowski, R., Cooper, K. G., Brunold, L. S., Calaycay, J., and Woods, J. P. (2008). Histoplasma capsulatum secreted gamma-glutamyltransferase reduces iron by generating an efficient ferric reductant. Mol. Microbiol. 70, 352–368. doi: 10.1111/j.1365-2958.2008.06410.x
Zarnowski, R., and Woods, J. P. (2005). Glutathione-dependent extracellular ferric reductase activities in dimorphic zoopathogenic fungi. Microbiology 151, 2233–2240. doi: 10.1099/mic.0.27918-0
Keywords: Candida glabrata, iron, ferric reductase, fungi, extracellular ferric reduction
Citation: Gerwien F, Safyan A, Wisgott S, Brunke S, Kasper L and Hube B (2017) The Fungal Pathogen Candida glabrata Does Not Depend on Surface Ferric Reductases for Iron Acquisition. Front. Microbiol. 8:1055. doi: 10.3389/fmicb.2017.01055
Received: 19 April 2017; Accepted: 26 May 2017;
Published: 08 June 2017.
Edited by:
Sven Krappmann, University of Erlangen-Nuremberg, GermanyReviewed by:
Christian Perez, University of Würzburg, GermanyDaniel Kornitzer, Technion – Israel Institute of Technology, Israel
Copyright © 2017 Gerwien, Safyan, Wisgott, Brunke, Kasper and Hube. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lydia Kasper, lydia.kasper@leibniz-hki.de
 Franziska Gerwien
Franziska Gerwien Abu Safyan
Abu Safyan Stephanie Wisgott1
Stephanie Wisgott1 Sascha Brunke
Sascha Brunke Lydia Kasper
Lydia Kasper Bernhard Hube
Bernhard Hube