- Department of Microbiology, Immunology, and Pathology, Colorado State University, Fort Collins, CO, United States
The opportunistic pathogen Burkholderia pseudomallei is a saprophytic bacterium and the causative agent of melioidosis, an emerging infectious disease associated with high morbidity and mortality. Although melioidosis is most prevalent during the rainy season in endemic areas, domestic gardens and farms can also serve as a reservoir for B. pseudomallei during the dry season, in part due to irrigation and fertilizer use. In the environment, B. pseudomallei forms biofilms and persists in soil near plant root zones. Biofilms are dynamic bacterial communities whose formation is regulated by extracellular cues and corresponding changes in the nearly universal secondary messenger cyclic dimeric GMP. Recent studies suggest B. pseudomallei loads are increased by irrigation and the addition of nitrate-rich fertilizers, whereby such nutrient imbalances may be linked to the transmission epidemiology of this important pathogen. We hypothesized that exogenous nitrate inhibits B. pseudomallei biofilms by reducing the intracellular concentration of c-di-GMP. Bioinformatics analyses revealed B. pseudomallei 1026b has the coding capacity for nitrate sensing, metabolism, and transport distributed on both chromosomes. Using a sequence-defined library of B. pseudomallei 1026b transposon insertion mutants, we characterized the role of denitrification genes in biofilm formation in response to nitrate. Our results indicate that the denitrification pathway is implicated in B. pseudomallei biofilm growth dynamics and biofilm formation is inhibited by exogenous addition of sodium nitrate. Genomics analysis identified transposon insertional mutants in a predicted two-component system (narX/narL), a nitrate reductase (narGH), and a nitrate transporter (narK-1) required to sense nitrate and alter biofilm formation. Additionally, the results presented here show that exogenous nitrate reduces intracellular levels of the bacterial second messenger c-di-GMP. These results implicate the role of nitrate sensing in the regulation of a c-di-GMP phosphodiesterase and the corresponding effects on c-di-GMP levels and biofilm formation in B. pseudomallei 1026b.
Introduction
Biofilms are dynamic natural communities of adherent bacteria encased in an extracellular polymeric matrix with signal-sensing systems primed to detect and respond to a variety of cues and environmental stimuli (O’Toole et al., 2000; McDougald et al., 2011). Biofilm formation in many pathogenic species is coordinated and positively regulated by the universal bacterial second messenger cyclic dimeric GMP (c-di-GMP), which facilitates the transition from free-swimming planktonic bacteria to sessile adherent communities (Tamayo et al., 2007). Burkholderia pseudomallei is a Gram-negative motile bacillus and saprophyte inhabiting tropical and subtropical soils and surface waters (Dance, 2000). This bacterium has been shown to persist in the environment as a pellicle (a biofilm that forms at the air-liquid interface) and as surface-associated biofilms on plants and in the rhizosphere of rice in endemic areas (Inglis and Sagripanti, 2006; Ramli et al., 2012; Prasertsincharoen et al., 2015). As an opportunistic pathogen, B. pseudomallei is acquired directly from the environment and can cause multiple types of infections in humans and a range of animal hosts (Dance, 2000). For a sapronotic disease agent such as B. pseudomallei (Kuris et al., 2014), it is important to understand the unique epidemiology related to its transition from the environment to establish persistent infections within susceptible hosts.
Burkholderia pseudomallei is found primarily within Southeast Asia and Northern Australia, where it is a prominent cause of bacterial pneumonia and sepsis, respectively (Hatcher et al., 2015). However, a recent report estimates the global distribution of B. pseudomallei is much larger than previously recognized and predicts environmental suitability across the tropics worldwide (Limmathurotsakul et al., 2016). B. pseudomallei infection results in melioidosis, a febrile illness whose clinical manifestations reflect the route of inoculation. Infection most commonly results from cutaneous exposure to contaminated soil or water, but also ingestion and respiratory inhalation (Wiersinga et al., 2012). Occupational exposure while working in rice fields is a primary source for acquisition of infection, but recreational activities such as gardening also expose individuals to B. pseudomallei in endemic areas (Limmathurotsakul et al., 2013; Kaestli et al., 2015). Anthropogenic manipulation of garden soils through irrigation and application of urea- and nitrate-rich fertilizers increases B. pseudomallei populations across different soil types (Kaestli et al., 2015). Nitrates are naturally occurring salts and byproducts of animal wastes, as well as artificial components of fertilizers, the use of which can lead to an imbalance in nutrient-limited soils (Kaestli et al., 2015). B. pseudomallei is associated with soil that contains elevated levels of nitrates and total nitrogen (Palasatien et al., 2008), anthrosol soil (Limmathurotsakul et al., 2016), as well as livestock and animal housing (Palasatien et al., 2008). This association with nitrogen levels is an apparent exception to the general negative association between B. pseudomallei and soil nutrient levels (Hantrakun et al., 2016).
Given the association of increased B. pseudomallei abundance in disturbed soils with elevated nitrate and organic materials, there is a critical need to understand the effects of exogenous nitrogen derivatives on B. pseudomallei physiology and epidemiology. Members of the Burkholderia genus, including the clinical isolate B. pseudomallei K96243, can grow in anaerobic conditions using nitrate as an alternative terminal electron acceptor, much like other facultative anaerobic bacteria (Yabuuchi et al., 1992; Hamad et al., 2011). The effects of anaerobic growth using nitrate on the physiology of B. pseudomallei biofilms remains largely undetermined. Interestingly, genes associated with nitrate reduction are implicated in biofilm formation in B. thailandensis, yet a mechanism of action has not been determined (Andreae et al., 2014). Nitrate sensing and metabolism have also been shown to influence biofilm formation and virulence in Pseudomonas aeruginosa (Van Alst et al., 2007; Zemke et al., 2013) and nitrite is known to inhibit biofilm formation in Staphylococcus aureus (Schlag et al., 2007). Additionally, nitrite metabolism is implicated in modulating antibiotic susceptibility in P. aeruginosa (Zemke et al., 2014, 2015).
To address the effects of exogenous nitrate on B. pseudomallei biofilms, transposon insertional mutants in predicted nitrate metabolism, regulatory, and sensory genes were identified and assessed for their ability to form biofilms in the presence of sodium nitrate. Inhibition of biofilm formation by sodium nitrate was not observed in five of the 21 transposon mutants tested: Bp1026b_I1013::T24 (narL), Bp1026b_I1014::T24 (narX), Bp1026b_I1017::T24 (narH-1), Bp1026b_I1018::T24 (narG-1), and Bp1026b_I1020::T24 (nark-1). The dissimilatory nitrate reductase encoded by the narGHJI operon, which is conserved among both facultative and obligate anaerobes, is a membrane-bound enzyme complex comprised of four subunits responsible for nitrate reduction in the absence of oxygen (Payne, 1981; Sodergren et al., 1988). The narX/narL system is instrumental in nitrate metabolism and regulation as it has been shown to activate nitrate-specific enzymes and repress unnecessary anaerobic respiration genes in response to exogenous nitrate (Egan and Stewart, 1991; Goh et al., 2005). In this study, we show that the alpha and beta subunits of the major nitrate reductase narGHJI-1 along with the narX/narL two-component regulatory system and the nitrate/nitrite transporter narK-1 are responsible for biofilm inhibition based on nitrate availability.
These data demonstrate that nitrate sensory cascades and metabolism are linked to biofilm growth dynamics in B. pseudomallei 1026b. Moreover, we show a significant reduction in c-di-GMP levels in B. pseudomallei grown in the presence of nitrate. We hypothesize that nitrate sensing and nitrate-dependent anaerobic respiration promote a planktonic lifestyle transition in B. pseudomallei that is ultimately a result of depletion of intracellular levels of c-di-GMP and a corresponding decrease in biofilm formation.
Materials and Methods
Bacterial Strains and Growth Conditions
All experiments were performed in biosafety level 3 (BSL-3) facilities within the Regional Biocontainment Laboratory at Colorado State University with approvals from the Center for Disease Control and Prevention and the Colorado State University Institutional Biosafety Committee. B. pseudomallei strain 1026b, a clinical isolate from a human case of septicemic melioidosis in Thailand (Deshazer et al., 1997), was used in this study. Transposon (T24) mutant strains used in this study were generated during the production of a comprehensive two-allele sequence defined transposon mutant library (manuscript in preparation). T24 is a Tn5-derived transposon containing a kanamycin resistance selection marker approved for use in B. pseudomallei that was constructed in Colin Manoil’s laboratory at the University of Washington (manuscript in preparation). Briefly, the two-allele library was assembled via random transposon insertion into gene loci spanning both chromosomes of B. pseudomallei 1026b, resulting in insertions into 4,931 of 6,070 (81.2%) total predicted genes. In total, the library contains two copies of 4,415 genetic mutants that were re-sequenced. All transposon insertion mutants were verified by additional sequencing for these studies. Bacterial strains were grown in Lysogeny Broth (LB) media consisting of 10 g/L tryptone (Fisher Scientific), 5 g/L yeast extract (Becton, Dickinson and Company), and 5 g/L NaCl (Fisher Scientific) as previously described (Plumley et al., 2017). For experiments involving media with sodium nitrate or sodium nitrite, LB was supplemented with a final concentration of 10 mM sodium nitrate (Sigma) or 10 mM sodium nitrite (Matheson, Coleman & Bell) unless otherwise stated. Transposon mutants were grown in LB with 300 μg/mL kanamycin (Gold Biotechnology).
Identification of Putative Nitrogen Metabolism Genes in B. pseudomallei 1026b
A combination of open-access genomic databases were used to assess the comparative identity of genetic orthologs in closely related Burkholderia spp. as well as P. aeruginosa strain PAO1. Predicted nucleotide sequences in the B. pseudomallei 1026b (tax id: 884204) genome for chromosomes I and II (accession numbers NC_017831.1 and NC_017832.1, respectively), were acquired from the NCBI GenBank database using the BLASTN program under default parameters to identify sequences similar to the PAO1 reference genome (GenBank assembly accession: GCA_000414275.1). Functional nitrogen metabolism pathways were evaluated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Ogata et al., 1999). Further comparative genomic analyses, as well as gene annotations and locus positions were deduced using the Burkholderia Genome Database (Winsor et al., 2008) in conjunction with the Pseudomonas Genome Database (Winsor et al., 2015). Sequence comparison among nitrogen metabolism genes was analyzed with the following: B. pseudomallei K96243 (tax id: 272560), B. thailandensis E264 (tax id: 271848), and P. aeruginosa PAO1 (tax id: 208964). Percent identity between genetic orthologs was calculated using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) tool provided by the European Bioinformatics Institute (EMBL-EBI), which calculates a percent identity matrix using Clustal 2.1 (Edgar, 2004). Gene names and functional descriptions were not entirely available for B. pseudomallei 1026b, and in such cases, were assigned based on consensus among the better annotated species detailed above.
Comparative Analysis of Predicted Nitrogen Metabolism Clusters
Sequence analyses for the predicted nitrogen metabolism clusters encoded on chromosome I and II from the sequenced genome of B. pseudomallei 1026b (tax id: 884204) were visualized using open-source bioinformatics platforms and genomic sequence databases. We used BLASTN (BLAST, NCBI) under default parameters in conjunction with the Burkholderia Orthologous Groups cataloging system from the Burkholderia Genome Database1 to identify homologous regions. Sequences for B. pseudomallei 1026b chromosome I (accession number: NC_017831.1) and chromosome II (accession number: NC_017832.1) were downloaded from the GenBank database (NCBI, NIH). Sequence fragments were extracted with Geneious version 7.1.72 (Kearse et al., 2012) and visualized with EasyFig version 2.23 (Sullivan et al., 2011) using Python version 2.73. To calculate homology between input sequences, command line parameters for EasyFig included a minimum identity cut-off for BLASTN of 0.60 and a threshold E-value of 1E-3.
Pellicle Biofilm Assays
Overnight cultures were grown in LB medium (kanamycin as indicated for transposon mutants) and sub-cultured 1:50 in 2 mL LB with or without 10 mM sodium nitrate. Pellicles were grown in 17 mm × 100 mm polystyrene tubes (#1485-2810, USA Scientific) at 37°C for 24 h before incubation at room temperature for 14 days without disturbance then photographed.
Static Biofilm Assays
Overnight cultures were grown in LB and diluted to OD600 of 0.1 in LB with or without 10 mM sodium nitrate. Experiments using sodium nitrite were performed in the same manner as those using sodium nitrate. Gradient concentrations {0, 1, 2, 3, 5, 8, 10, 20, 50, and 100 mM} of sodium nitrate, sodium nitrite, or sodium chloride in LB were evaluated as indicated. One hundred microliters was added, in replicates of six, to individual wells of a 96-well polystyrene plate (NuncTM MicrowellTM 96-well microplates #243656, Thermo Scientific) and incubated at 37°C for 24 h. Static biofilms were processed as previously described (Plumley et al., 2017). Briefly, bacterial supernatant was removed from the biofilms and wells were washed once with PBS to remove non-adherent cells. The wells were stained with 0.05% crystal violet (Sigma–Aldrich, St. Louis, MO, United States) and incubated at room temperature for 15 min. Stain was removed and wells were washed again with PBS. Adherent crystal violet was solubilized by addition of 95% EtOH and incubated for 30 min. The solubilized dye was transferred to a new 96-well polystyrene plate and the absorbance was measured at OD600 on a Synergy HT plate reader (BioTek Instruments, Winooski, VT, United States). Significance was calculated with an unpaired Student’s t-test using the Holm-Sidak method to account for multiple comparisons.
Complementation of Bp1026b_I1013::T24 (narL) and Bp1026b_I1014::T24 (narX) Mutants
Complementation constructs were designed for conditional expression of re-integrated genes via IPTG induction from the tac promoter, following select-agent-compliant methods for genetic modification in B. pseudomallei (Choi et al., 2008; Plumley et al., 2017). The full-length sequences of Bp1026b_I1013 (narL) and Bp1026b_I1014 (narX) were amplified using the following PCR primer pairs: (5′-NNNCCCGGGAGGAGGAT ATTCATGACCATACGGGTACTGTT-3′) and (5′-NNNAAGC TTTTATGCCTCGGCCGGATGCG-3′) for Bp1026b_I1013, and (5′-NNNCCCGGGAGGAGGATATTCATGGCTCCCGCC CTCCCCGA-3′) and (5′-NNNAAGCTTCTATGCCGCCTGTC GCGCGT-3′) for Bp1026b_I1014. Cloned genes were ligated into pUC18T-mini-Tn7T-Km-LAC, which uses the tac promoter from Escherichia coli to drive gene expression (Rholl et al., 2011). 5 mM IPTG was used to induce expression from the tac promoter. 5 mM NaNO3 was used to stimulate biofilm inhibition.
Growth Curves
Overnight cultures were grown in LB and diluted to OD600 of 0.1 in LB supplemented with increasing concentrations of sodium nitrate or sodium chloride as indicated. Sodium chloride was supplemented in addition to the original 170 mM NaCl concentration in LB. Each experimental growth condition was repeated in six replicates. One hundred microliters of each culture was added to a 96-well polystyrene plate (NuncTM MicrowellTM 96-well microplates #243656, Thermo Scientific) in triplicate. Plates were incubated at 37°C shaking aerobically at 250 rpm for 48 h. Absorbance (OD600) was measured hourly on a Synergy HT plate reader (BioTek Instruments).
Extraction and Quantification of c-di-GMP
Nucleotide extraction methods using formic acid were adapted from Massie et al. (2012). Overnight cultures of B. pseudomallei 1026b were grown in LB at 37°C shaking (250 rpm). Cultures were diluted 1:50 in 4 mL of M9 Salts minimal medium with or without 10 mM sodium nitrate supplemented and grown statically at 37°C for 18 h in six-well Costar polystyrene plates (#3516, Corning). Cells from 1 mL culture including the pellicle biofilm were collected and resuspended in 100 μL of cold 40% (vol/vol) acetonitrile/40% (vol/vol) methanol/20% (vol/vol) LC-MS-grade water/1% formic acid. An internal control of 2-chloro-adenosine-5′-O-monophosphate (2-Cl-5′-AMP, Axxora, LLC) was used at a final concentration of 100 nM (Irie and Parsek, 2014). The buffer, cells, and matrix were incubated at 65°C for 10 min and then immediately transferred to -20°C for 30 min to ensure complete lysis. Samples were then centrifuged at 16,000 × g at 4°C for 5 min and the supernatant fraction containing c-di-GMP was collected and neutralized with 20 μL 1 M KOH, followed by another spin at 16,000 × g at 4°C for 5 min. A calibration curve using known standards of chemically synthesized c-di-GMP (Axxora, LLC) and 2-Cl-5′-AMP was generated and extracted in identical conditions to experimental samples. Absolute quantification of c-di-GMP was calculated using the linear regression equation generated from known standards and peak areas normalized to the internal standard. Analysis was performed on an Acquity M-Class UPLC (Waters) coupled to a Xevo TQ-S triple quadrupole mass spectrometer (Waters) for LC-MS/MS. A Waters Atlantis dC18 stationary phase column (300 μm × 150 mm, 3.0 μm) was used for chromatographic separations. Protein pellets from the initial cell collection were analyzed for total protein concentration using the Pierce 660 nm Protein Assay (Thermo Scientific) for normalization of the absolute concentration of c-di-GMP. Nucleotide extraction experiments were repeated on two different days using four biological replicate cultures with three technical replicates each. Statistical significance was determined using an unpaired Student’s t-test in GraphPad Prism.
RNA Isolation and Quantitative Real-Time PCR
Burkholderia pseudomallei 1026b was grown statically in M9 salts minimal media with or without 10 mM sodium nitrate supplemented at 37°C in six-well Costar polystyrene plates (Corning). After 18 h of static growth, 1 mL of culture was collected and cells were centrifuged at 12,000 × g for 2 min at 4°C. Bacterial pellets were resuspended in 350 μL RNAprotect Bacteria Reagent (Qiagen) before centrifuging at 5,000 × g for 10 min at 4°C. Bacterial pellets were resuspended in 1 mL QIAzol Lysis Reagent (Qiagen), incubated for 5 min at room temperature, and transferred to screwcap tubes containing 250 μL of 0.1 mm glass disruption beads (RPI Corp.) on ice. Cells were lysed using a TissueLyser II homogenizer (Qiagen) using three rounds of 60 s pulses at 30 Hz with 60 s on ice between each lysis round. After lysis, the cell mixture was incubated at room temperature for 5 min before adding 200 μL chloroform and vortexing for 5 s. Samples were incubated again at room temperature for 5 min before centrifugation at 13,000 × g for 10 min. The aqueous phase was collected and RNA was purified with an RNeasy Mini Kit (Qiagen) using the protocol recommended by the manufacturer. RNA samples were treated with two rounds of TURBO DNase (Ambion) before cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche). qRT-PCR was performed on a LightCycler 480 System with SYBR Green I Master (Roche) using 10 ng total cDNA and the following cycling conditions: pre-incubation at 95°C for 10 min; three-step amplification at 95°C for 10 s, 55°C for 15 s, 72°C for 15 s; melting curve at 95°C for 5 s, 65°C for 1 min, and 97°C for continuous acquisitions per 5°C. Primers were designed using the PrimerQuest tool (IDT DNA Technologies). Previously published 23S rRNA primers were used for normalization (Mima and Schweizer, 2010). All sets of primers were validated for amplification efficiency in a standard curve. The primer set used for cdpA was as follows: forward (5′-AAGCTGGCTGGAGCAAA-3′) and reverse (5′-GCAGATAGTCGCGGTGATAA-3′). The primer set used for Bp1026b_II2523 was as follows: forward (5′-GCAAGATCGAGAGCGTGTT-3′) and reverse (5′-GTGATCGAAGCGGAACAGATAC-3′). The primer set used for Bp1026b_ II0885 was as follows: forward (5′-CTGCACCTACCGCTTCTT-3′) and reverse (5′-AAGCACAGCGAGAAGTAGTC-3′). The primer set used for Bp1026b_I3148 was as follows: forward (5′-CGCTGCATCTGGAGAACTT-3′) and reverse (5′-CCTGAAACGGATCGGTGAG-3′). Transcript abundance was measured using the Pfaffl method (Pfaffl, 2001), taking into account primer amplification efficiency, including three independent biological samples performed in technical triplicates.
Results
Nitrate Inhibits B. pseudomallei Biofilm Formation
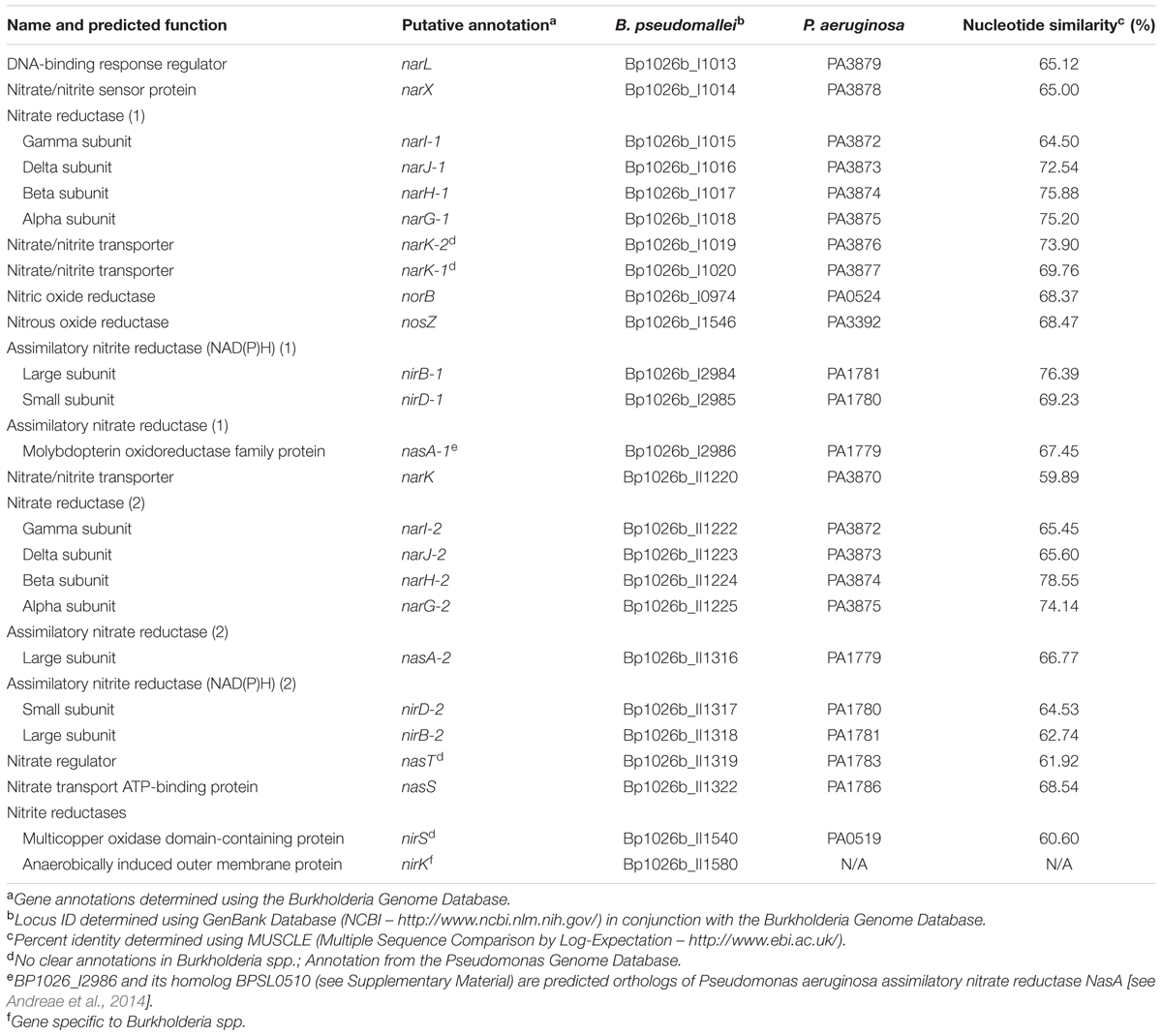
While the effectors of biofilm dispersal and inhibition are extensive and diverse, nitrogenous compounds have been shown to modulate biofilm formation in both Gram-negative and Gram-positive bacteria (Schlag et al., 2007; Van Alst et al., 2007; Zemke et al., 2014). Sodium nitrate (NaNO3), a naturally occurring salt compound and a synthetic agricultural additive, inhibits biofilm formation in a dose-dependent manner (Figure 1A). Likewise, sodium nitrite also inhibits biofilm formation following a similar trend, although the effect is not as robust initially (Figure 1A). To assess the impact of nitrate/nitrite on biofilm formation, wild-type B. pseudomallei 1026b was grown statically for 24 h with increasing concentrations of sodium nitrate or sodium nitrite (Figure 1A). Biofilm inhibition effects were first noted at 1 mM while the most significant biofilm inhibition occurred with the addition of 10 mM sodium nitrate or 10 mM sodium nitrite. There were no additional inhibitory effects on biofilm formation with treatment concentrations greater than 10 mM NaNO3. As such, concentrations of 10 mM NaNO3 were used for all subsequent experiments. To evaluate the effects of sodium concentration on biofilm formation, experiments with only sodium chloride (NaCl) indicated that biofilm inhibition was not significantly altered during biofilm formation under the same conditions (Figure 1A). Viability was also tested by measuring growth kinetics in media supplemented with NaCl (Figure 1B) and NaNO3 (Figure 1C). Growth inhibition was observed only when cells were grown in media supplemented with 100 mM of NaNO3 and 100 mM of NaCl, concentrations that are well beyond those used in this study. No significant differences in growth dynamics were observed at 10 mM NaNO3 suggesting that nitrate dosing at this concentration does not affect cell viability. These results suggest that nitrate sensing and metabolism, and not sodium, is responsible for biofilm inhibition in B. pseudomallei 1026b and the observed inhibition is not the result of decreased cellular viability.
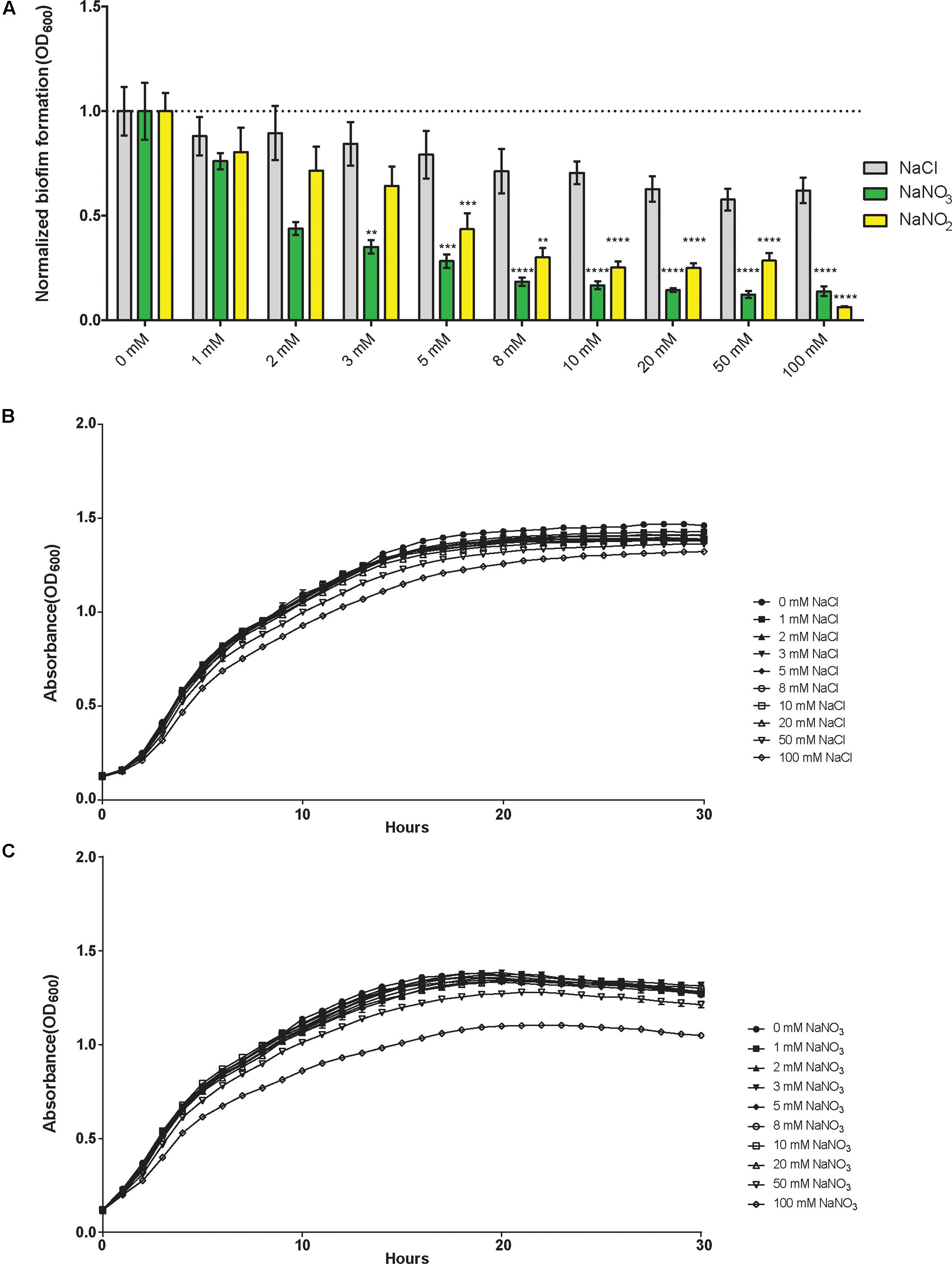
FIGURE 1. Sodium nitrate and sodium nitrite, but not sodium chloride, inhibit biofilm formation in Burkholderia pseudomallei 1026b. (A) Biofilm inhibition was assessed for static biofilm cultures growing at 37°C in the presence of sodium chloride, sodium nitrate, or sodium nitrite at 1, 2, 3, 5, 8, 10, 20, 50, and 100 mM. All experiments were performed with wild-type B. pseudomallei 1026b and inhibition was evaluated relative to wild-type biofilm formation in LB without additional salts added. Asterisks indicate a significant difference (∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001) calculated with an unpaired Student’s t-test using the Holm-Sidak method to account for multiple comparisons (n = 12). Growth curves were generated from cultures grown with shaking at 37°C in the presence of either sodium chloride (B) or sodium nitrate (C) using concentrations as indicated for the biofilm inhibition assay. Absorbance (OD600) measurements were taken hourly for 30 continuous hours.
Bioinformatics Analysis of Predicted Nitrogen Metabolism Genes
Bioinformatics analyses identified 25 B. pseudomallei 1026b genes predicted to be involved in nitrogen metabolism (Table 1), of which 21 were included in this study based on the availability from a two-allele sequence defined transposon library. The genes included in this study were selected based on sequence homology among closely related Burkholderia spp. to P. aeruginosa PAO1, which has an extensively annotated reference genome that has been biochemically validated for the nitrogen metabolism genes. Within the Proteobacteria phylum, the β-proteobacteria, which contains the genus Burkholderia, is closely related to the genus Pseudomonas, which is a γ-proteobacteria (Estrada-De Los Santos et al., 2013), thus inter-genus comparisons described in this study will provide insights into the functions among these conserved nitrogen metabolism proteins. While B. pseudomallei 1026b has many genes predicted to encode enzymes in the denitrification pathway, there is a need for more comprehensive functional annotation in this relatively new clinically isolated strain. Therefore, nucleotide sequence identity was analyzed across three Burkholderia species in relation to P. aeruginosa PAO1 (Table 1 and Supplementary Table 1).
Orthologs of the denitrification enzymes in B. pseudomallei 1026b presented here share 99–100% nucleotide identity to the fully sequenced genome of B. pseudomallei K96243, 90–97% nucleotide identity to B. thailandensis E264, and 60–78% nucleotide identity to the more distant relative P. aeruginosa PAO1 (Supplementary Table 1). This underscores the highly conserved nature of the genes constituting the essential denitrification pathway among these soil-dwelling bacteria. Interestingly, bioinformatics analysis revealed duplications of specific genes implicated in the denitrification pathway in B. pseudomallei 1026b, which is consistent with the predicted genetic supplementation and metabolic redundancy of B. pseudomallei (Holden et al., 2004). The major nitrate reductases are encoded on chromosome I at Bp1026b_I1015 – Bp1026b_I1018 and on chromosome II at Bp1026b_II1222 – Bp1026b_II1225, and share sequence homology of 69–79% (Supplementary Table 2). Additionally, the major nitrite reductase is encoded on chromosome I at Bp1026b_I2984 – Bp1026b_I2985 and on chromosome II at Bp1026b_II1317 – Bp1026b_II1318, sharing only 65% nucleotide identity (Supplementary Table 2).
Bioinformatics analyses also identified putative nitrogen metabolism genes with no clear annotations across Burkholderia spp. Therefore, Bp1026b_I1019, Bp1026b_I1020, Bp1026b_II1319, and Bp1026b_II1540 were assigned genetic annotations based on the P. aeruginosa PAO1 reference genome. Two genes were identified as assimilatory nitrate reductases (Bp1026b_I2986 and Bp1026b_II1316) based on sequence homology to PAO1; however, no annotated homologs were available throughout Burkholderia spp., thus the annotation was assigned to be consistent with P. aeruginosa PAO1. Lastly, Bp1026b_II1580, which is predicted to encode an anaerobically induced outer membrane nitrite reductase, was specific to Burkholderia spp. with no representative homolog in P. aeruginosa.
Based on the information available from the previously described open-source genome databases and the genetic functional pathway predictions provided by the KEGG database, we mapped and compared the genetic arrangement of the predicted nitrogen metabolism enzymes across both chromosomes (Figures 2A,B). The narX/narL predicted two-component signaling system lies downstream and adjacent to the primary nitrate reductase on chromosome I, which is flanked by the cytoplasmic membrane-associated nitrate/nitrite transporters (Figure 2A). Another gene cluster of interest for these studies is the assimilatory nitrite reductase on chromosome I, with the adjacent molybdopterin oxidoreductase nasA, which is predicted to function as an assimilatory nitrate reductase (Ogawa et al., 1995) (Figure 2B). Interestingly, the nitrate metabolism gene clusters found on chromosome I are replicated in similar configurations on chromosome II. The relatively equal distribution of denitrification-associated loci across both B. pseudomallei chromosomes is not surprising considering that chromosome II is not an accessory genome and also encodes for essential and central metabolism genes (Holden et al., 2004); however, the predicted nitrate metabolism loci on chromosome II are not directly homologous to those on chromosome I. This disparity in sequence identity among similarly annotated gene loci with parallel genetic arrangements indicates that the nitrate metabolism clusters are not direct paralogs and may be required for specific metabolic functions based on extracellular cues.
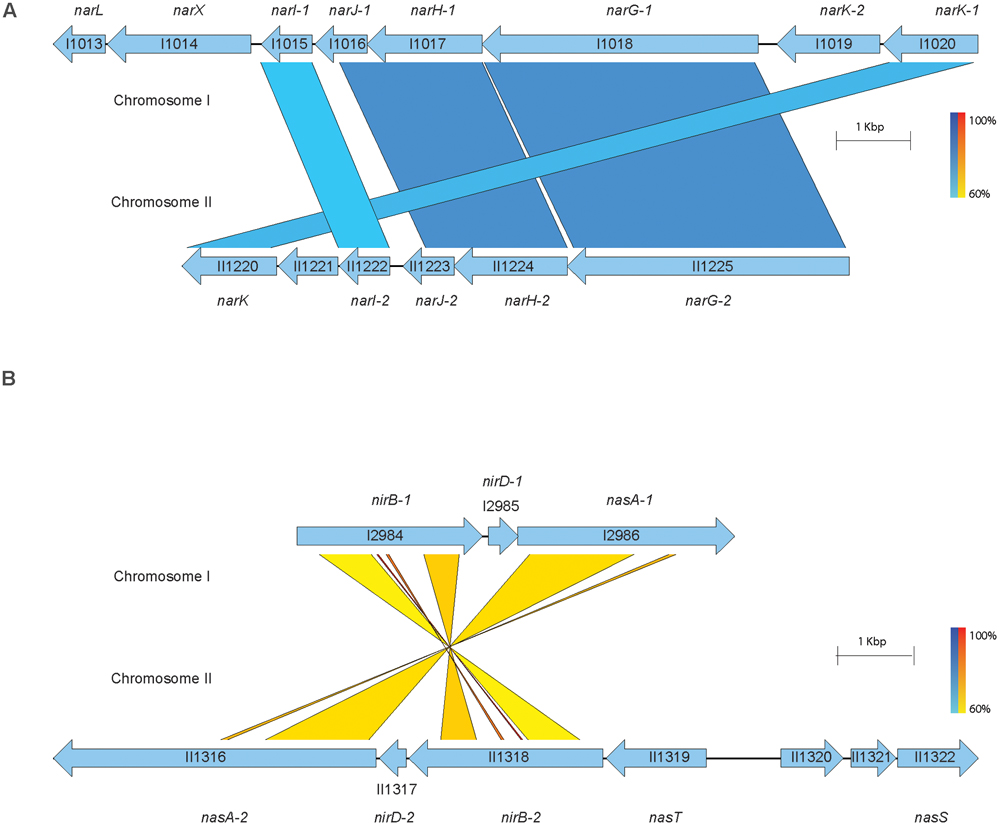
FIGURE 2. Predicted nitrogen metabolism gene clusters in B. pseudomallei 1026b. The putative nitrate and nitrite reductases, transporters, and nitrate-sensing gene clusters from the sequenced genome of B. pseudomallei 1026b are shown. A total of 21 loci spanning B. pseudomallei chromosomes I and II included in this study are illustrated here. Bp1026b_I1546, Bp1026b_II1540, and Bp1026b_II1580 were excluded. Coding sequences for the represented nitrate metabolism loci are depicted by arrows in relation to positive or negative strand orientation with open reading frames and intergenic regions illustrated to scale. The results of BLASTN annotations are depicted by color coded bars spanning gene loci across the two chromosomes with a minimum percent identity of 0.60 and a threshold E-value of 1E-3. Blue bars depict direct sequence homology and yellow-to-red bars depict homology amid sequence inversion, on a color density gradient indicating the percent homology. (A) The major predicted nitrate reductase narGHJI is encoded on chromosome I (Bp1026b_I1015 – 1018) as well as on chromosome II (Bp1026b_II1222 – 1225). Adjacent to the nitrate reductases are predicted nitrate transporters (Bp1026b_I1019, Bp1026b_I1020, Bp1026b_II1220) and the predicted two-component signaling system (Bp1026b_I1013 – I1014). (B) The major nitrite reductase nirBD is encoded on chromosome I (Bp1026b_I2984 – 2985) as well as on chromosome II (Bp1026b_II1317 – 1318). Adjacent to the nitrite reductase is the predicted assimilatory nitrate reductase nasA encoded by Bp1026b_I2986 on chromosome I and Bp1026b_II1316 on chromosome II. The predicted nitrate regulator, nasT, and nitrate transport ATP-binding loci, nasS, are encoded on chromosome II at Bp1026b_II1319 and Bp1026b_II1322, respectively.
Five Genes within the Denitrification Pathway Contribute to Pellicle Biofilm Inhibition in the Presence of Exogenous Nitrate in B. pseudomallei
We hypothesized that pellicle biofilms cannot form in the presence of exogenous nitrate under the conditions tested. Pellicle biofilms have been historically well characterized in the Gram-positive soil-dwelling bacterium Bacillus subtilis (Armitano et al., 2014), as well as pathogenic Gram-negative species such as P. aeruginosa (Cole et al., 1989) and Burkholderia cenocepacia (Fazli et al., 2013). B. pseudomallei forms pellicle biofilms in static cultures in vitro and at the surface-liquid interface near the root zone of plants. Wild-type and transposon insertional mutants of genes in the denitrification pathway were grown statically with or without sodium nitrate to assess the impact of nitrate on pellicle formation. The wild type did not form a pellicle biofilm in the presence of 10 mM NaNO3, while a robust pellicle biofilm was observed in LB without nitrate (Figure 3). Furthermore, pellicle formation was inhibited by sodium nitrate in 16 of 21 transposon insertional mutants (Supplementary Figure 1). Interestingly, five transposon insertional mutants in the denitrification pathway were resistant to inhibition of pellicle formation by exogenous nitrate (Figure 3 and Supplementary Figure 1). Transposon insertions in narL, a DNA-binding response regulator, and narX, a nitrate sensor protein, which comprise a two-component signaling system regulated by nitrate, were resistant to pellicle inhibition by nitrate. Similarly, transposon mutants with insertions in narG-1 and narH-1, which are predicted to be the major nitrate reductase alpha and beta subunits were also insensitive to inhibition of pellicle formation by nitrate. Interestingly, transposon mutations into narG-1 and narH-1 did not confer resistance to biofilm inhibition by sodium nitrite (Supplementary Figure 2), indicating that nitrite may inhibit biofilm formation independently of the major nitrate reductase. It is curious that only the cytoplasmic subunits of the major nitrate reductase were hit during our transposon screen for the biofilm inhibitory phenotype via sodium nitrate, an observation that may provide insights into the respiratory electron transfer pathway in B. pseudomallei. Additionally, it is worth noting that the membrane-anchored narI-1 gamma subunit of the nitrate reductase was not available in our transposon mutant library, and as such, its relation to biofilm dynamics remains uncharacterized. We identified a fifth transposon mutant with an insertion in narK-1, encoding a predicted nitrate/nitrite transporter that was also insensitive to pellicle biofilm inhibition by nitrate. These results suggest that this genetic quintet, Bp1026b_I1013 (narL), Bp1026b_I1014 (narX), Bp1026b_I1017 (narH-1), Bp1026b_I1018 (narG-1), Bp1026b_I1020 (nark-1), contributes to the regulation and formation of B. pseudomallei biofilms when exogenous nitrate is encountered.
FIGURE 3. Five transposon insertions in the predicted nitrate metabolism pathway do not respond to inhibition of pellicle biofilm formation mediated by the addition of sodium nitrate. Wild-type B. pseudomallei 1026b forms a distinct pellicle biofilm when grown statically in liquid culture; however, it is unable to form a pellicle biofilm in the presence of 10 mM NaNO3. Transposon insertion mutants in the following genes {Bp1026b_I1013::T24 (narL), Bp1026b_I1014::T24 (narX), Bp1026b_I1017::T24 (narH-1), Bp1026b_I1018::T24 (narG-1), and Bp1026b_I1020::T24 (nark-1)} were identified that form pellicle biofilms growing statically in LB + 10 mM NaNO3. Pellicles were grown in 2 mL media at room temperature and photographed at 14 days.
Quantification of Biofilm Formation
Biofilm inhibition by nitrate was assessed in the same array of transposon insertion mutants using the quantitative biofilm growth method (O’Toole and Kolter, 1998) to calculate results from static biofilm assays. In conjunction with the direct observation of pellicle formation, the static biofilm assay provides quantitative analysis on the biofilm-forming capabilities for each strain in this study. Biofilm formation was significantly abolished with nitrate treatment in 16 of the 21 transposon mutants; however, biofilm formation of transposon mutants in narL, narX, narG-1, narH-1, and narK-1 was not significantly altered in the presence of nitrate (Figure 4A). These results corroborated the qualitative observations of the pellicle biofilm assay, as the same five mutants were similarly resistant to the biofilm inhibitory effects of nitrate in both assays. Planktonic growth for narL, narX, narG-1, narH-1, and narK-1 was similar when compared to wild-type B. pseudomallei 1026b (Supplementary Figure 3).
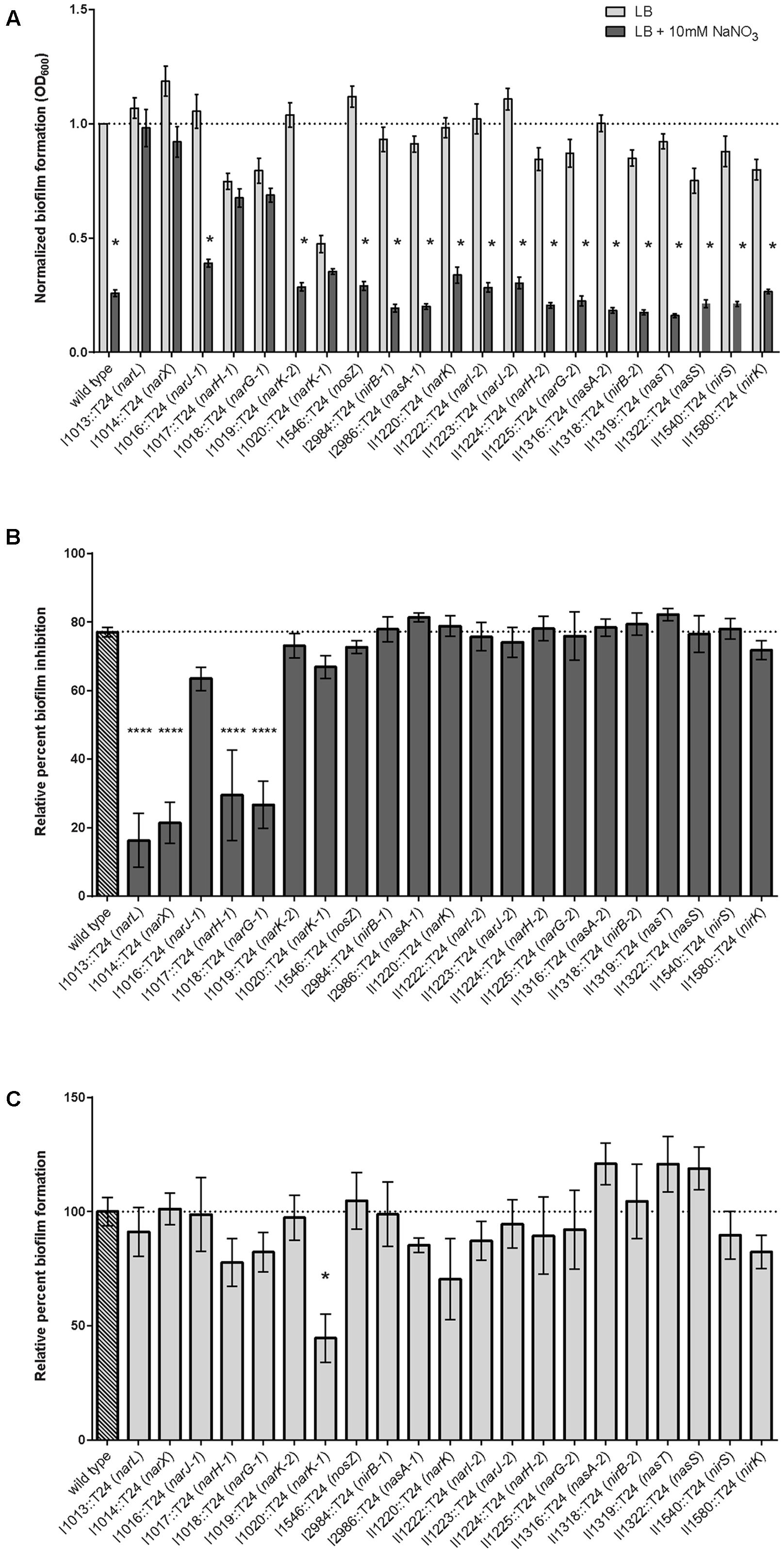
FIGURE 4. Quantitative evaluation of relative biofilm formation and biofilm inhibition in the presence and absence of sodium nitrate. (A) Biofilm formation was evaluated when grown statically in the presence of 10 mM NaNO3. Biofilm formation of the wild-type and 16 of the 21 transposon mutants identified in this study were inhibited by treatment with NaNO3, while five mutants {Bp1026b_I1013::T24 (narL), Bp1026b_I1014::T24 (narX), Bp1026b_I1017::T24 (narH-1), Bp1026b_I1018::T24 (narG-1), and Bp1026b_I1020::T24 (nark-1)} were not inhibited by treatment with NaNO3. Bars are representative of the means for individually normalized values for biofilm formation relative to the wild type. Asterisks indicate a significant difference (p < 0.0001) calculated with an unpaired Student’s t-test using the Holm-Sidak method to account for multiple comparisons (n = 12). (B) Percent biofilm inhibition for all mutants was calculated relative to inhibition of the wild type in the presence of 10 mM NaNO3. Bars are representative of the means for each mutant relative to the mean for the wild type. (C) Biofilm formation for all mutants was calculated relative to wild type biofilm formation in LB medium. Bars are representative of the means for each mutant relative to the mean for the wild type.
To further assess the effects of nitrate metabolism genes on biofilm inhibition, we calculated the percent of relative biofilm inhibition for all mutant strains in this study (Figure 4B). While wild-type biofilm formation was reduced by nearly 80% when sodium nitrate is encountered, transposon insertions in narL, narX, narG-1, and narH-1 did not respond to the inhibitory effects of treatment with sodium nitrate. The decrease in the percentage of relative biofilm formation for the transposon-inactivated narK-1 mutant (Figure 4C) indicates that this mutant is affected by nitrate more so than the narL, narX, narG-1, and narH-1 mutants, and is also reflective of a fundamental biofilm-forming defect in the narK-1 mutant. Taken together, these results provide quantitative measurements of biofilm inhibition and formation in the presence of exogenous nitrate, and support our conclusions regarding the role of these functionally inactivated genetic loci as observed in the pellicle biofilm assay.
To confirm that the transposon mutant insertions identified in the T24-mutagenesis screen were causal to the observed phenotypes, we complemented I1013::T24 (narL) and I1014::T24 (narX) with full-length genes which were conditionally expressed with IPTG induction. The resulting constructs, Bp1026b_I1013::T24 Ptac-I1013 (narL) and Bp1026b_I1014::T24 Ptac-I1014 (narX) produced similar amounts of biofilm as the wild-type empty-vector (EV) control strain in the absence of NaNO3 (Figure 5). Conditional expression of narL in Bp1026b_I1013::T24 Ptac-I1013 and narX in Bp1026b_I1014::T24 Ptac-I1014 resulted in an approximate 50% reduction in biofilm formation with the addition of 5 mM NaNO3 that was equivalent to the wild-type EV control strain (Figure 5). These data further support the hypothesis that the narX/narL two-component signaling system, which is predicted to respond to nitrate sensing, is implicated in biofilm growth dynamics in B. pseudomallei.
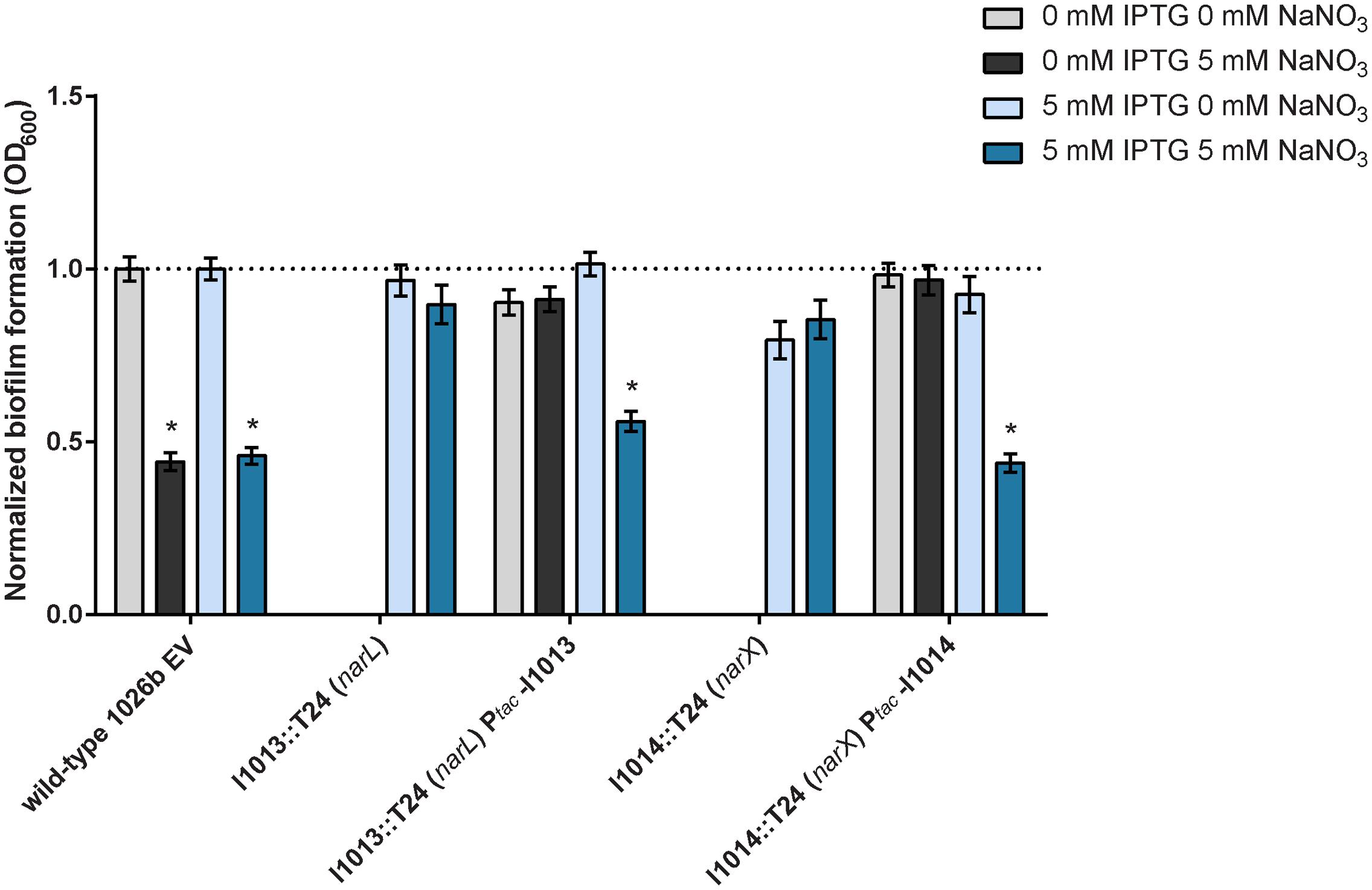
FIGURE 5. Complementation of the I1013::T24 (narL) and I1014::T24 (narX) mutants that are resistant to the inhibitory effects of exogenous nitrate. Mutants of the predicted nitrate-sensing two-component system narX/narL were complemented with full-length Bp1026b_I1013 (narL) and Bp1026b_I1014 (narX). The genes were re-introduced in the T24-mutant background at a neutral site and expression was conditionally induced using 5 mM IPTG. Biofilm formation was assessed in the presence and absence of 5 mM NaNO3. Asterisks indicate a significant difference (p < 0.0001) calculated with an unpaired Student’s t-test.
Exogenous Nitrate Reduces Intracellular c-di-GMP in a Static Biofilm Assay
Biofilm formation and dispersal mechanisms are ultimately dependent on the level of intracellular c-di-GMP in many pathogenic bacteria (Tamayo et al., 2007), thus we hypothesized that biofilm inhibition via exogenous nitrate sensing regulates c-di-GMP concentrations in B. pseudomallei 1026b. We analyzed the intracellular concentration of c-di-GMP via liquid chromatography tandem-mass spectrometry (LC-MS/MS) using a triple quadrupole mass spectrometer. The intracellular concentration of c-di-GMP in wild-type bacteria grown statically in minimal media with 10 mM sodium nitrate was reduced by 38% as compared to biofilms cultivated statically in the absence of nitrate (Figure 6A). For static biofilms cultivated without sodium nitrate, we observed an average concentration of 80.46 nmol c-di-GMP per mg of total protein and for biofilms cultivated in the presence of 10 mM nitrate the average concentration was 50.19 nmol c-di-GMP per mg total protein (Figure 6A). These results suggest that nitrate metabolism regulates intracellular c-di-GMP levels in B. pseudomallei 1026b biofilms.
FIGURE 6. Quantification of cyclic dimeric GMP (c-di-GMP) in response to sodium nitrate and relative transcript abundance of cdpA, the major phosphodiesterase in B. pseudomallei. (A) The intracellular concentration of the bacterial second messenger c-di-GMP is significantly reduced in B. pseudomallei 1026b grown statically in nitrate-supplemented media. Error bars indicate standard error for four biological replicates extracted on different days. Statistical significance was determined using the Sidak-Bonferroni method across multiple T-tests (∗∗p < 0.01). (B) cdpA transcript level is upregulated nearly twofold in B. pseudomallei 1026b grown statically in nitrate-supplemented media. Three additional transcripts that encode for a phosphodiesterase (I3148), a diguanylate cyclase (II2523), and a composite diguanylate cyclase/phosphodiesterase (II0885) did not show statistical differences in the levels of transcript. Fold change in transcript level was calculated using the Pfaffl method considering primer amplification efficiency and normalized to the reference transcript for 23S rRNA. Statistical significance was determined using a one-tailed heteroscedastic Student’s t-test (∗p < 0.05).
The Phosphodiesterase cdpA Is Upregulated in Response to Nitrate in a Static Biofilm Assay
In a simplified model of c-di-GMP regulation, a decrease in intracellular c-di-GMP levels suggests either decreased diguanylate cyclase activity or increased phosphodiesterase activity. We chose to evaluate the expression of previously described diguanylate cyclases and phosphodiesterases that are known to alter biofilm dynamics in B. pseudomallei (Lee et al., 2010; Plumley et al., 2017). C-di-GMP metabolism in bacteria, including B. pseudomallei 1026b, is classically mediated by diguanylate cyclases and phosphodiesterases that contain conserved GG(D/E)EF and EAL catalytic domains, respectively (Tamayo et al., 2007; Plumley et al., 2017). B. pseudomallei 1026b contains six predicted proteins with conserved EAL domains and five predicted GGDEF-EAL composite proteins (Plumley et al., 2017); however, only one of these predicted proteins, Bp1026b_I2284, has been shown to potentially function as a classical phosphodiesterase (Plumley et al., 2017). Bp1026b_I2284 (cdpA) is a previously described phosphodiesterase and known regulator of c-di-GMP levels in B. pseudomallei KHW (Lee et al., 2010). Recently, Bp1026b_I2284 (cdpA) has also been shown to contribute to motility and is therefore potentially implicated in reducing c-di-GMP levels in B. pseudomallei 1026b during planktonic growth (Plumley et al., 2017). Thus, we hypothesized that cdpA transcript expression is regulated during nitrate metabolism in B. pseudomallei 1026b, leading to a decrease in intracellular c-di-GMP and inhibition of biofilm formation.
During the transition between biofilm and planktonic states multiple phosphodiesterases and diguanylate cyclases may be expressed in a coordinated manner to alter the levels of c-di-GMP in response to stimulatory signals and conditions. To begin to address the complexity associated with c-di-GMP signaling during nitrate metabolism, we evaluated the transcription of previously identified phosphodiesterase and diguanylate cyclase genes (Plumley et al., 2017). Bp1026b_II0885, a composite protein that is similar to cdpA, was chosen for analysis based on the conservation of the EAL domain and GGDEF catalytic domains (Plumley et al., 2017). Bp1026b_I3148 was also chosen based on the high level of conservation of the EAL domain (Plumley et al., 2017). To evaluate the effects on diguanylate cyclase gene expression, we chose to analyze Bp1026b_II2523 based on its conserved GGEEF domain and its previously observed role in the thermoregulation of biofilm growth dynamics (Plumley et al., 2017).
To assess the impact of exogenous nitrate on the expression of c-di-GMP phosphodiesterases and diguanylate cyclases, wild-type B. pseudomallei 1026b was grown statically under biofilm-inducing conditions in minimal media with or without 10 mM NaNO3. The expression of cdpA was significantly higher, representing a nearly twofold increase in cells grown in 10 mM NaNO3-supplemented minimal media when normalized to 23S (Figure 6B). Interestingly, the expression values for the additional two transcripts that potentially encode c-di-GMP phosphodiesterases showed a similar trend to that of cdpA; however, these analyses lacked statistical significance (Figure 6B). The expression of Bp1026b_II2523, a putative diguanylate cyclase showed no significant response to nitrate, which indicates it may not be involved in the nitrate-dependent biofilm inhibition cascade under the conditions tested. Thus, while nitrate has a clear regulatory effect on cdpA, it is possible that other phosphodiesterases and diguanylate cyclases that remain to be tested also contribute to the intracellular concentration of c-di-GMP. Moreover we showed that cdpA is not required for nitrate- or nitrite-dependent biofilm inhibition (Supplementary Figure 4). Nonetheless, these results indicate that nitrate can potentially increase phosphodiesterase activity in B. pseudomallei 1026b and can explain the significant decrease in c-di-GMP concentration during growth under identical conditions. Taken together with the absolute quantification of c-di-GMP, these data support our hypothesis that exogenous nitrate inhibits B. pseudomallei 1026b biofilms by reducing the intracellular concentration of c-di-GMP.
Discussion
Environmental acquisition of opportunistic pathogens such as B. pseudomallei requires a lifestyle transition that prompts a shift from the natural reservoir to establish infections in susceptible human and animal hosts. Biofilm communities serve as a predominant natural habitat for the saprophytic B. pseudomallei, which colonizes and persists in the rhizosphere of plants in wet soils of endemic areas (Prasertsincharoen et al., 2015), thus efforts to better understand the dissemination from this environment would serve to mitigate public health risks related to the emerging infectious disease melioidosis. The particular effectors that serve as biofilm inhibition signals and dispersal cues for B. pseudomallei are poorly understood, although previous studies link anthropogenic disturbances to increased bacterial presence in endemic areas (Kaestli et al., 2009; Limmathurotsakul et al., 2013, 2016). Notably, and of particular interest to our present study, are the observations by Kaestli et al. (2015) that addition of nitrate-rich fertilizer increased B. pseudomallei bacterial loads across a variety of soils. Since nitrate (NO3-) and nitrite (NO2-) can serve as terminal electron acceptors for anaerobic respiration in denitrifying bacteria (Payne, 1981) and the Burkholderia genera are capable of denitrification (Kaestli et al., 2015), we tested the hypothesis that nitrate inhibits biofilm formation in B. pseudomallei 1026b.
The results presented in this study indicate that exogenous addition of nitrate, and to a lesser extent, nitrite, inhibit biofilm formation. While both nitrate and nitrite have been shown to repress biofilm formation in P. aeruginosa (Van Alst et al., 2007) and S. aureus (Schlag et al., 2007), respectively, this is the first study to address the effects of nitrate on B. pseudomallei biofilm formation. We identified a total of 21 genes, which were predicted to encode nitrate metabolism enzymes in the denitrification pathway, of which five transposon mutants no longer responded to biofilm inhibition by nitrate. This analysis identified the predicted two-component signaling system narX/narL, and implicates this conserved regulatory system in nitrate metabolism as well as control of biofilm growth dynamics in B. pseudomallei. The nitrate-sensing two-component system, which is conserved among aerobes and anaerobes alike, mediates gene expression when narX senses environmental nitrate, thereby stimulating a phosphorylation cascade that causes narL to bind and regulate DNA transcription (Walker and DeMoss, 1993). Thus, the narX/narL system acts as a primary sensor of exogenous nitrate, and our observations implicate this system in the genetic control of biofilm formation. Given the genetic arrangement of this two-component system adjacent to the predicted major nitrate reductase on chromosome I (Figure 2), future studies will address whether the DNA-binding and regulatory effects of NarL are targeted to the upstream narGHJI-1 operon.
Our results also indicate that two subunits in the major dissimilatory nitrate reductase narGHJI-1, which is tasked with reducing nitrate to nitrite inside the bacterial cell, contributes to exogenous nitrate sensing and control of biofilm formation when nitrate is present. Biofilm formation is also inhibited by nitrite (Figure 1A), and although this genetic mechanism has not been investigated in the present study, our results indicate that this phenotype occurs independently of the alpha and beta subunits of the primary nitrate reductase narGH-1 (Supplementary Figure 2). Additionally, biofilm formation of a transposon mutant insertion in one of the four nitrate/nitrite transporter genes, narK-1, was no longer inhibited in the presence of nitrate. These results suggest that the five genes narL, narX, narG-1, narH-1, and narK-1 are implicated in mediating the biofilm growth dynamics of B. pseudomallei when exogenous nitrate is encountered in the environment. Given that the narX/narL two-component system is adjacent to the major nitrate reductase encoded by the narGHJI-1 operon, a prominent hypothesis is that this system is required for transcriptional regulation of the nitrate reductase, similar to the nitrate regulatory network of P. aeruginosa (Schobert and Jahn, 2010). Interestingly, while there is another predicted dissimilatory nitrate reductase encoded on chromosome II by the narGHJI-2 operon, transposon mutant insertions within these loci do not function like narGHJI-1 under the conditions tested. Moreover, we did not find evidence for homologs of the narX/narL two-component signaling system for nitrate sensing on chromosome II in our bioinformatics analyses.
Burkholderia pseudomallei is a facultative anaerobe that is found in soils approximately 30 cm deep (Palasatien et al., 2008), a depth which becomes relatively hypoxic in rice paddy soils (Andreae et al., 2014), and nitrate becomes increasingly more important as a terminal electron acceptor for anaerobic respiration. Similarly, biofilms are intrinsically hypoxic microenvironments as oxygen and nutrient diffusion are physically limited, leading to increased dependence on nitrate anions as an alternative terminal electron acceptor (Andreae et al., 2014). Based on our in vitro results, our working hypothesis is that excessive environmental nitrate triggers dispersal of hypoxic, nutrient-limited sessile cells residing in the biofilm community leading to increased abundance of planktonic bacteria. This working model is illustrated in the context of the denitrification pathway for B. pseudomallei 1026b and its potential connection to c-di-GMP metabolic regulatory cascades (Figure 7). It is important to mention that the information regarding c-di-GMP signaling and metabolism in B. pseudomallei is scarce, given the importance of this second messenger molecule in the regulation of biofilm dynamics (Plumley et al., 2017). Given the well-established connection between c-di-GMP and biofilm formation in many bacterial species, and the important transition from biofilm to planktonic cells in a variety of natural and host environments, it is vital to further identify and characterize the environmental cues that modulate c-di-GMP levels in the Tier 1 select agent B. pseudomallei.
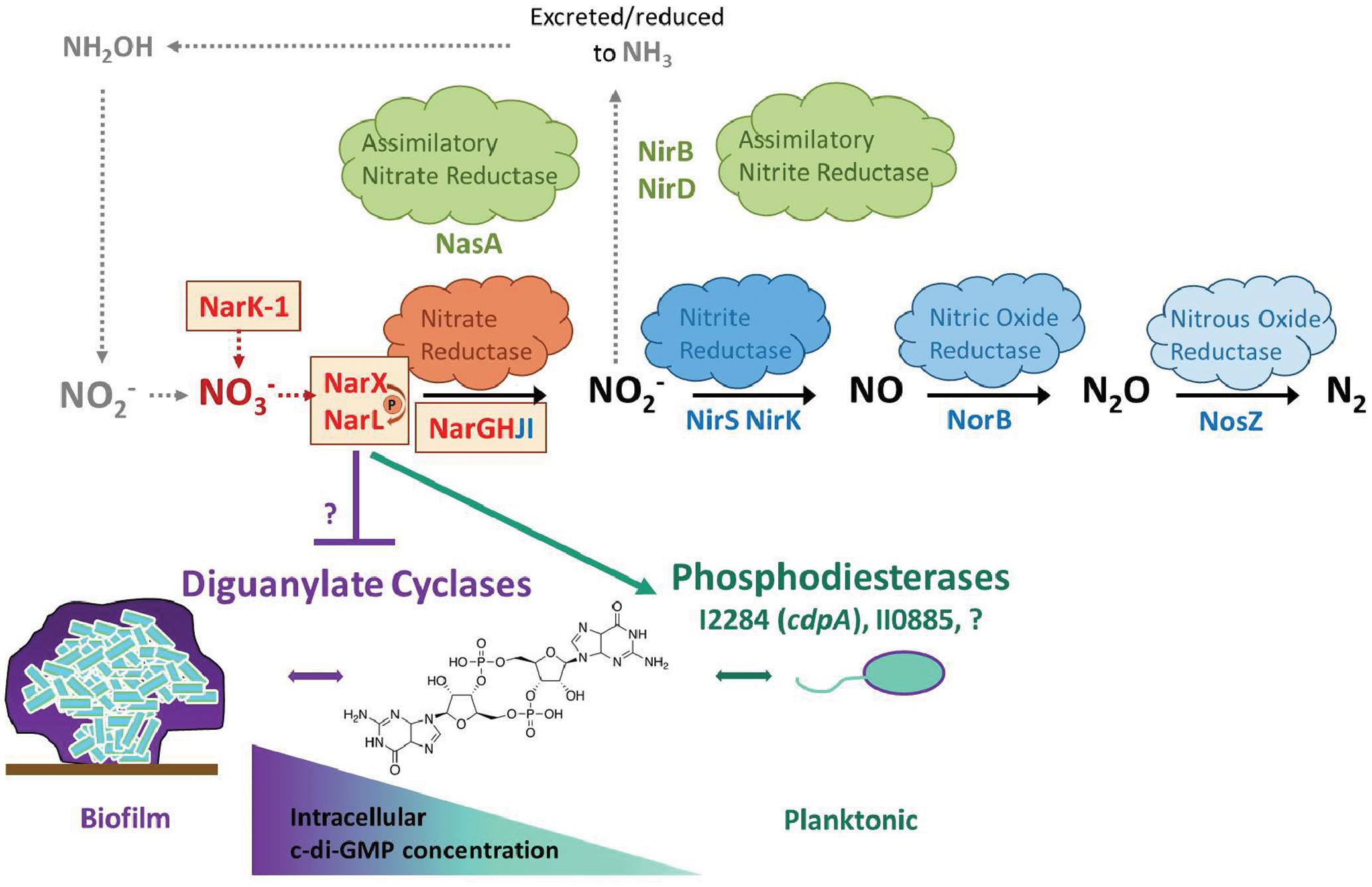
FIGURE 7. Working model for nitrate sensing and metabolism in relation to biofilm dynamics in B. pseudomallei. This model serves to illustrate nitrate metabolism in the context of biofilm dynamics for B. pseudomallei. The process of denitrification is carried out by four individual enzyme complexes: nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase. Additionally, B. pseudomallei encodes assimilatory nitrate and assimilatory nitrite reductases. The process of nitrification, depicted by gray arrows, is not intrinsic to B. pseudomallei; however, it is a relevant source of nitrate in the environment, produced by nitrifying bacteria. The five genes identified in this study are highlighted in bold red with surrounding boxes, in proximity to the major nitrate reductase (also highlighted in red). The biofilm life-cycle is illustrated as a state of constant flux mediated by intracellular c-di-GMP concentration. A proposed mechanism by which the NarX/NarL two-component nitrate-sensing system modulates c-di-GMP is illustrated, although the connection to diguanylate cyclases and additional phosphodiesterases remains to be determined.
The results presented here provide evidence that a primary mechanism for control of B. pseudomallei biofilm formation is through the regulation of the intracellular concentration of the bacterial second messenger, c-di-GMP, via nitrate sensing and metabolism. The role of c-di-GMP in the regulation of bacterial behaviors, which include biofilm formation, is known to have significant implications on the ability of pathogenic bacteria to cause disease (Tamayo et al., 2007). The model of c-di-GMP flux inside a cell generally dictates that more intracellular c-di-GMP molecules corresponds to a biofilm and sessile state, while a reduced intracellular concentration stimulates motility and favors a planktonic lifestyle; however, many of the extracellular cues that activate these signaling cascades remain uncharacterized (Figure 7) (Tamayo et al., 2007). Quantification of intracellular c-di-GMP levels in response to exogenous nitrate revealed that nitrate metabolism can affect the levels of this secondary messenger in B. pseudomallei 1026b. We hypothesize that the signaling transduction cascade initiated by nitrate sensing as controlled by the narX/narL two-component system activates not only dissimilatory nitrate reduction through the narGHJI-1 complex but also regulates phosphodiesterase enzymes tasked with rapidly reducing intracellular concentrations of c-di-GMP. Our results indicate a significant reduction of c-di-GMP levels in static cultures grown in the presence of sodium nitrate, in addition to increased expression of cdpA, which encodes a known phosphodiesterase. These findings collectively indicate that the mechanism of nitrate sensing is linked to control of c-di-GMP levels in the cell.
Modulation of c-di-GMP levels is a primary mechanism bacteria use to respond to extracellular signals and cues as they relate to control of autoaggregation, biofilm formation, and motility during the transition from sessile to planktonic growth states. Therefore, such a mechanism is at the crux of controlling bacterial dissemination from environmental reservoirs to host-associated infections for sapronotic disease agents such as B. pseudomallei. CdpA, which has been characterized as a key phosphodiesterase in B. pseudomallei KHW (Lee et al., 2010) and at the genetic level in B. pseudomallei 1026b (Plumley et al., 2017), has also been implicated in the regulatory control of biofilm formation in B. cepacia J2315 (Ferreira et al., 2013). Our assessment of cdpA expression from B. pseudomallei 1026b revealed significant upregulation of this transcript in response to elevated nitrate availability, with a corresponding decrease of intracellular c-di-GMP levels. These data reveal an important connection between increased phosphodiesterase activity and reduction of this key second messenger molecule in response to the addition of nitrate. We expect global c-di-GMP metabolic cascades to be simultaneously activated by an exogenous nitrate signal, and given that we have previously identified 23 gene loci predicted to encode c-di-GMP metabolic or binding enzymes, there is undoubtedly more complex regulation in addition to cdpA activity (Plumley et al., 2017). Future studies will address nitrate’s global transcriptional regulation of c-di-GMP metabolism in addition to exopolysaccharide, capsule, pili, flagellar, quorum sensing, and antibiotic efflux machinery, among other mechanisms pertinent to the biofilm-to-planktonic transition.
The complexity of the regulatory network for denitrifying growth in hypoxic conditions has been extensively examined across several bacterial species (Campbell, 1977), implicating several transcriptional regulators of nitrate metabolism such as the narX/narL system in addition to quorum sensing and nutrient availability. Our study implicates nitrate metabolism in biofilm growth dynamics in B. pseudomallei, and identifies specific gene loci that may be targeted for regulation by a combination of systems. Furthermore, we provide evidence that the second messenger, c-di-GMP, is involved in biofilm inhibition in response to exogenous addition of nitrate. Future analyses will assess the regulation of the enzymes described in this study and investigate the effects of denitrification on c-di-GMP metabolism as it relates to the pathogenesis of B. pseudomallei.
Author Contributions
MM and BB designed this study. MM and BP performed the experiments. MM analyzed the data and MM, BP, and BB wrote the paper. BB supervised data collection, analysis, and presentation of these results.
Funding
This research was supported by a Super Cluster grant from the Infectious Disease Research Center at Colorado State University and funds from the Boettcher Foundation’s Webb-Waring Biomedical Research Program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge and thank Colin Manoil and Larry Gallagher at the University of Washington for providing preliminary transposon sequence insertion information prior to publication. We also acknowledge Lisa Wolfe and Corey Broeckling at the Proteomics and Metabolomics Facility at Colorado State University for analyzing our c-di-GMP extracts and fine-tuning our extraction methods through thoughtful discussion. Additionally, we thank Eric Bruger and Chris Waters for their advice on c-di-GMP extractions using formic acid. We are also grateful to Grace Borlee, Ankunda Kariisa, and Kevin Martin for their critical review and comments on the manuscript prior to publication.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01353/full#supplementary-material
Footnotes
References
Andreae, C. A., Titball, R. W., and Butler, C. S. (2014). Influence of the molybdenum cofactor biosynthesis on anaerobic respiration, biofilm formation and motility in Burkholderia thailandensis. Res. Microbiol. 165, 41–49. doi: 10.1016/j.resmic.2013.10.009
Armitano, J., Méjean, V., and Jourlin-Castelli, C. (2014). Gram-negative bacteria can also form pellicles. Environ. Microbiol. Rep. 6, 534–544. doi: 10.1111/1758-2229.12171
Campbell, R. (1977). Microbial Ecology: Basic Microbiology, Vol. 5. London: Blackwell Scientific Publications.
Choi, K., Mima, T., Casart, Y., Rholl, D., Kumar, A., Beacham, I. R., et al. (2008). Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl. Environ. Microbiol. 74, 1064–1075. doi: 10.1128/AEM.02430-07
Cole, E. C., Rutala, W. A., Carson, J. L., and Alfano, E. M. (1989). Pseudomonas pellicle in disinfectant testing: electron microscopy, pellicle removal, and effect on test results. Appl. Environ. Microbiol. 55, 511–513.
Dance, D. A. B. (2000). Melioidosis as an emerging global problem. Acta Trop. 74, 115–119. doi: 10.1016/S0001-706X(99)00059-5
Deshazer, D., Brett, P. J., Carlyon, R., and Woods, D. E. (1997). Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179, 2116–2125. doi: 10.1128/jb.179.7.2116-2125.1997
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Egan, S. M., and Stewart, V. (1991). Mutational analysis of nitrate regulatory gene narL in Escherichia coli K-12. J. Bacteriol. 173, 4424–4432. doi: 10.1128/jb.173.14.4424-4432.1991
Estrada-De Los Santos, P., Vinuesa, P., Martínez-Aguilar, L., Hirsch, A. M., and Caballero-Mellado, J. (2013). Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr. Microbiol. 67, 51–60. doi: 10.1007/s00284-013-0330-9
Fazli, M., Mccarthy, Y., Givskov, M., Ryan, R. P., and Tolker-Nielsen, T. (2013). The exopolysaccharide gene cluster Bcam1330-Bcam1341 is involved in Burkholderia cenocepacia biofilm formation, and its expression is regulated by c-di-GMP and Bcam1349. Microbiologyopen 2, 105–122. doi: 10.1002/mbo3.61
Ferreira, A. A., Silva, I. I., Oliveira, V. V., Becker, J. J., Givskov, M., Ryan, R. P., et al. (2013). Comparative transcriptomic analysis of the Burkholderia cepacia tyrosine kinase bceF mutant reveals a role in tolerance to stress, biofilm formation, and virulence. Appl. Environ. Microbiol. 79, 3009–3020. doi: 10.1128/AEM.00222-13
Goh, E., Bledsoe, P. J., Chen, L., Gyaneshwar, P., Stewart, V., and Igo, M. M. (2005). Hierarchical control of anaerobic gene expression in Escherichia coli K-12: the nitrate-responsive NarX-NarL regulatory system represses synthesis of the fumarate-responsive DcuS-DcuR regulatory system. J. Bacteriol. 187, 4890–4899. doi: 10.1128/JB.187.14.4890
Hamad, M. A., Austin, C. R., Stewart, A. L., Higgins, M., Vázquez-Torres, A., and Voskuil, M. I. (2011). Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob. Agents Chemother. 55, 3313–3323. doi: 10.1128/AAC.00953-10
Hantrakun, V., Rongkard, P., Oyuchua, M., Amornchai, P., Day, N. P. J., Peacock, S. J., et al. (2016). Soil nutrient depletion is associated with the presence of Burkholderia pseudomallei. Appl. Environ. Microbiol. 82, 7086–7092. doi: 10.1128/AEM.02538-16
Hatcher, C. L., Muruato, L. A., and Torres, A. G. (2015). Recent advances in Burkholderia mallei and B. pseudomallei research. Curr. Trop. Med. Rep. 2, 62–69. doi: 10.1007/s40475-015-0042-2
Holden, M. T. G., Titball, R. W., Peacock, S. J., Cerdeño-Tárraga, A. M., Atkins, T., Crossman, L. C., et al. (2004). Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U.S.A. 101, 14240–14245. doi: 10.1073/pnas.0403302101
Inglis, T. J. J., and Sagripanti, J. L. (2006). Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl. Environ. Microbiol. 72, 6865–6875. doi: 10.1128/AEM.01036-06
Irie, Y., and Parsek, M. R. (2014). LC/MS/MS-based quantitative assay for the secondary messenger molecule, c-di-GMP. Methods Mol. Biol. 1149, 271–279. doi: 10.1007/978-1-4939-0473-0
Kaestli, M., Harrington, G., Mayo, M., Chatfield, M. D., Harrington, I., Hill, A., et al. (2015). What drives the occurrence of the melioidosis bacterium Burkholderia pseudomallei in domestic gardens? PLoS Negl. Trop. Dis. 9:e0003635. doi: 10.1371/journal.pntd.0003635
Kaestli, M., Mayo, M., Harrington, G., Ward, L., Watt, F., Hill, J. V., et al. (2009). Landscape changes influence the occurrence of the melioidosis bacterium Burkholderia pseudomallei in soil in Northern Australia. PLoS Negl. Trop. Dis. 3:e364. doi: 10.1371/journal.pntd.0000364
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kuris, A. M., Lafferty, K. D., and Sokolow, S. H. (2014). Sapronosis: a distinctive type of infectious agent. Trends Parasitol. 30, 386–393. doi: 10.1016/j.pt.2014.06.006
Lee, H. S., Gu, F., Ching, S. M., Lam, Y., and Chua, K. L. (2010). CdpA is a Burkholderia pseudomallei cyclic di-GMP phosphodiesterase involved in autoaggregation, flagellum synthesis, motility, biofilm formation, cell invasion, and cytotoxicity. Infect. Immun. 78, 1832–1840. doi: 10.1128/IAI.00446-09
Limmathurotsakul, D., Golding, N., Dance, D. A. B., Messina, J. P., Pigott, D. M., Moyes, C. L., et al. (2016). Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat. Microbiol. 1:15008. doi: 10.1038/nmicrobiol.2015.8
Limmathurotsakul, D., Kanoksil, M., Wuthiekanun, V., Kitphati, R., deStavola, B., Day, N. P. J., et al. (2013). Activities of daily living associated with acquisition of melioidosis in Northeast Thailand: a matched case-control study. PLoS Negl. Trop. Dis. 7:e2072. doi: 10.1371/journal.pntd.0002072
Massie, J. P., Reynolds, E. L., Koestler, B. J., Cong, J.-P., Agostoni, M., and Waters, C. M. (2012). Quantification of high-specificity cyclic diguanylate signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 12746–12751. doi: 10.1073/pnas.1115663109
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D., and Kjelleberg, S. (2011). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50. doi: 10.1038/nrmicro2695
Mima, T., and Schweizer, H. P. (2010). The BpeAB-OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob. Agents Chemother. 54, 3113–3120. doi: 10.1128/AAC.01803-09
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Ogawa, K., Akagawa, E., Yamane, K., Sun, Z., Celle, M. L. A., Zuber, P., et al. (1995). The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J. Bacteriol. 177, 1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
O’Toole, G. A., and Kolter, R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461. doi: 10.1046/j.1365-2958.1998.00797.x
Palasatien, S., Lertsirivorakul, R., Royros, P., Wongratanacheewin, S., and Sermswan, R. W. (2008). Soil physicochemical properties related to the presence of Burkholderia pseudomallei. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl. 1), S5–S9. doi: 10.1016/S0035-9203(08)70003-8
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Plumley, B. A., Martin, K. H., Borlee, G. I., Marlenee, N. L., Burtnick, M. N., Brett, P. J., et al. (2017). Thermoregulation of biofilm formation in Burkholderia pseudomallei is disrupted by mutation of a putative diguanylate cyclase. J. Bacteriol. 199:e00780-16. doi: 10.1128/JB.00780-16
Prasertsincharoen, N., Constantinoiu, C., Gardiner, C., Warner, J., and Elliman, J. (2015). Effects of colonization of the roots of domestic rice (Oryza sativa L. cv. Amaroo) by Burkholderia pseudomallei. Appl. Environ. Microbiol. 81, 4368–4375. doi: 10.1128/AEM.00317-15
Ramli, N. S. K., Eng Guan, C., Nathan, S., and Vadivelu, J. (2012). The effect of environmental conditions on biofilm formation of Burkholderia pseudomallei clinical isolates. PLoS ONE 7:e44104. doi: 10.1371/journal.pone.0044104
Rholl, D. A., Papp-wallace, K. M., Andrew, P., Vasil, M. L., and Bonomo, R. A. (2011). Molecular investigations of PenA-mediated β-lactam resistance in Burkholderia pseudomallei. Front. Microbiol. 2:139. doi: 10.3389/fmicb.2011.00139
Schlag, S., Nerz, C., Birkenstock, T. A., Altenberend, F., and Götz, F. (2007). Inhibition of staphylococcal biofilm formation by nitrite. J. Bacteriol. 189, 7911–7919. doi: 10.1128/JB.00598-07
Schobert, M., and Jahn, D. (2010). Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int. J. Med. Microbiol. 300, 549–556. doi: 10.1016/j.ijmm.2010.08.007
Sodergren, E. J., Hsu, P. Y., and DeMoss, J. A. (1988). Roles of the narJ and narI gene products in the expression of nitrate reductase in Escherichia coli. J. Biol. Chem. 263, 16156–16162.
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tamayo, R., Pratt, J. T., and Camilli, A. (2007). Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61, 131–148. doi: 10.1146/annurev.micro.61.080706.093426
Van Alst, N. E., Picardo, K. F., Iglewski, B. H., and Haidaris, C. G. (2007). Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect. Immun. 75, 3780–3790. doi: 10.1128/IAI.00201-07
Walker, M. S., and DeMoss, J. A. (1993). Phosphorylation and dephosphorylation catalyzed in vitro by purified components of the nitrate sensing system, NarX and NarL. J. Biol. Chem. 268, 8391–8393.
Wiersinga, W. J., Currie, B. J., and Peacock, S. J. (2012). Melioidosis. N. Engl. J. Med. 367, 1035–1044. doi: 10.1056/NEJMra1204699
Winsor, G. L., Griffiths, E. J., Lo, R., Dhillon, B. K., Shay, J. A., and Brinkman, F. S. L. (2015). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, 646–653. doi: 10.1093/nar/gkv1227
Winsor, G. L., Khaira, B., Van Rossum, T., Lo, R., Whiteside, M. D., and Brinkman, F. S. L. (2008). The Burkholderia genome database: facilitating flexible queries and comparative analyses. Bioinformatics 24, 2803–2804. doi: 10.1093/bioinformatics/btn524
Yabuuchi, E., Kosako, Y., Oyaizu, H., Yano, I., Hotta, H., Hashimoto, Y., et al. (1992). Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 40, 1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x
Zemke, A., Lashua, L., Gladwin, M., and Bomberger, J. (2013). P2: sodium nitrite disrupts biotic biofilm formation by Pseudomonas aeruginosa on polarized human airway epithelial cells. Nitric Oxide 31, S13–S14. doi: 10.1016/j.niox.2013.02.004
Zemke, A. C., Gladwin, M. T., and Bomberger, J. M. (2015). Sodium nitrite blocks the activity of aminoglycosides against Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 59, 3329–3334. doi: 10.1128/AAC.00546-15
Zemke, A. C., Shiva, S., Burns, J. L., Moskowitz, S. M., Pilewski, J. M., Gladwin, M. T., et al. (2014). Nitrite modulates bacterial antibiotic susceptibility and biofilm formation in association with airway epithelial cells. Free Radic. Biol. Med. 77, 307–316. doi: 10.1016/j.freeradbiomed.2014.08.011
Keywords: Burkholderia pseudomallei, biofilm, sodium nitrate, c-di-GMP, melioidosis
Citation: Mangalea MR, Plumley BA and Borlee BR (2017) Nitrate Sensing and Metabolism Inhibit Biofilm Formation in the Opportunistic Pathogen Burkholderia pseudomallei by Reducing the Intracellular Concentration of c-di-GMP. Front. Microbiol. 8:1353. doi: 10.3389/fmicb.2017.01353
Received: 04 May 2017; Accepted: 04 July 2017;
Published: 25 July 2017.
Edited by:
Catherine Ayn Brissette, University of North Dakota, United StatesReviewed by:
Matias Castro, Fundación Ciencia & Vida, ChileDavid Salvador Zamorano Sanchez, University of California, Santa Cruz, United States
Copyright © 2017 Mangalea, Plumley and Borlee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bradley R. Borlee, brad.borlee@colostate.edu
 Mihnea R. Mangalea
Mihnea R. Mangalea Brooke A. Plumley
Brooke A. Plumley Bradley R. Borlee
Bradley R. Borlee