
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 August 2017
Sec. Extreme Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01535
This article is part of the Research Topic Actinobacteria in Special and Extreme Habitats: Diversity, Function Roles and Environmental Adaptations, Second Edition View all 17 articles
 Bao-Zhu Fang1
Bao-Zhu Fang1 Nimaichand Salam1
Nimaichand Salam1 Ming-Xian Han1,2
Ming-Xian Han1,2 Jian-Yu Jiao1
Jian-Yu Jiao1 Juan Cheng3
Juan Cheng3 Da-Qiao Wei2
Da-Qiao Wei2 Min Xiao1*
Min Xiao1* Wen-Jun Li1,3*
Wen-Jun Li1,3*The phylum Actinobacteria is one of the most ubiquitously present bacterial lineages on Earth. In the present study, we try to explore the diversity of cultivable rare Actinobacteria in Sigangli Cave, Yunnan, China by utilizing a combination of different sample pretreatments and under different culture conditions. Pretreating the samples under different conditions of heat, setting the isolation condition at different pHs, and supplementation of media with different calcium salts were found to be effective for isolation of diverse rare Actinobacteria. During our study, a total of 204 isolates affiliated to 30 genera of phylum Actinobacteria were cultured. Besides the dominant Streptomyces, rare Actinobacteria of the genera Actinocorallia, Actinomadura, Agromyces, Alloactinosynnema, Amycolatopsis, Beutenbergia, Cellulosimicrobium, Gordonia, Isoptericola, Jiangella, Knoellia, Kocuria, Krasilnikoviella, Kribbella, Microbacterium, Micromonospora, Mumia, Mycobacterium, Nocardia, Nocardioides, Nocardiopsis, Nonomuraea, Oerskovia, Pseudokineococcus, Pseudonocardia, Rhodococcus, Saccharothrix, Streptosporangium, and Tsukamurella were isolated from these cave samples.
Caves provide a quasi-extreme environment for living organisms owing to relatively low organic nutrient input and lack of light (Pedersen, 2000). Some ‘sojourners’ such as crickets, spiders, olms and bats are, however, adapted to these unassumingly harsh environments through modifications in their body morphology and other physiological changes (Culver and Pipan, 2009). Unlike these organisms, the microscopic counterparts are abundantly present within the cave environment (Wu et al., 2015; Tomczyk-Żak and Zielenkiewicz, 2016), and they are involved in the dissolution and precipitation of karst minerals (Castanier et al., 2000). Despite many studies on the potential function and diversity of these microbes in the oligotrophic environments, our knowledge on cave microbial diversity and related bioactivities are still limited (Engel et al., 2004).
Earlier studies on caves indicated that bacteria and archaea constitute the majority of the microbial diversity (Barton and Jurado, 2007). Pyrosequencing analyses had determined phyla Proteobacteria, Acidobacteria, and Actinobacteria to be among the dominant taxa on cave environments (Pasić et al., 2010; Wu et al., 2015; Tomczyk-Żak and Zielenkiewicz, 2016). Among the major bacterial lineages, the phylum Actinobacteria are of special interest due to their versatile metabolic activities (Genilloud, 2014; Remenár et al., 2014). They are found ubiquitously in nature. Besides their role in biodegradation and production of ecologically important bioactive metabolites, they are also involved in biomineralization (Gillieson, 1996; Dhami et al., 2013). Based on the biotechnological significance of the phylum Actinobacteria, the basic aim of this work is to study the diversity of cultivable rare Actinobacteria in a karst cave located in Yunnan, China.
Though bacterial richness and diversity within specific environmental samples and their possible physiological role in nature can be established with NextGen sequencing techniques and other bioinformatics tools (Green and Keller, 2006), physiologies can only be verified with pure cultures (Leadbetter, 2003). It is, however, estimated that 99% of the existing microbes have not been cultivated yet (Whitman et al., 1998). It is therefore necessary to utilize various enrichment techniques or media to bring these uncultivated cells into cultures. Some of these techniques of culturing previously uncultivable soil bacteria have been reviewed by Pham and Kim (2012). In their review, major emphases are given upon the modification of growth conditions and use of new culturing methods. In the current, we try to explore the option of using a combination of enrichment techniques including heat-pretreatments of the samples, adjusting the isolation media into a pH gradient and supplementing the media with different calcium salts at different concentration. These techniques have already been established as an effective measure for isolation of diverse rare Actinobacteria (Alferova and Terekhova, 1988; Hayakawa, 2008; Lauber et al., 2009), but have not been exploited to determine the cultivable actinobacterial diversity in caves.
The Sigangli Caves, located in Cangyuan County, Yunnan Province, China, are part of a series of karst caves of the Yunnan–Guizhou Plateau formed from the dissolution of limestone and other calcareous rocks. The plateau, covering an area of over 1.3 × 105 sq. km, was formed during tectonic shifts of Eurasian plate and is the center of karst area in East Asia (Figure 1).
Samples for isolation of Actinobacteria were collected from different part of the caves (23°32′ N, 99°33′ E). The samples (Table 1) include the hard rock forms (sedimentary rocks and cave coral, referred to here as Type 1 sample) and the weathered rock forms (saprolites, sand, debri and arene, referred to here as Type 2 sample). The samples were collected using sterile scalpels or spades and were transferred immediately in falcon tubes or Ziploc bags. These samples were then stored under a low temperature environment until processing for isolation.
A set of pretreatments and inoculation procedures adopted for the isolation of culturable Actinobacteria is listed in Table 2. Samples (2 g) were suspended in 20 ml sterile distilled water and kept in a rotary shaker (180 rpm, 28°C) for 1 h. The suspensions were serially diluted and aliquots of 100 μl of the diluted suspension was plated on freshly prepared agar media (in triplicates). The following isolation media were used: Humic acid-vitamin medium (HV) (Hayakawa and Nonomura, 1987); International Streptomyces Project (ISP) 5 medium (Shirling and Gottlieb, 1966); Cellulose-casamino acid (CC) medium (Yuan et al., 2017); Trehalose-proline agar (HP) (Li et al., 2014); Starch-Casien (SC) medium (Küster and Williams, 1964); B-4 medium (Boquet et al., 1973), and Water agar containing 11 g of agar per liter of water. Each of these media was supplemented with nystatin (50 mg L-1) and nalidixic acid (20 mg L-1) to prevent the growth of fungi and fastidious bacteria. Following incubation for 2 weeks at 28°C, all the colonies developed on the isolation media were counted. Depending on the isolation conditions and the media used, total colony forming units (CFUs) from each treatment were determined. Apart from the spore-forming strains, viabilility of the other vegetative cells were determined by subculturing on YIM 38 medium (Zhao et al., 2010). Heatmaps representing the distribution of these CFUs across the different physiological parameters were generated with R software (R Core Team, Vienna, Austria). The heatmaps are generated by applying ‘heatmap.2’ function of ‘gplots’ package. Further, morphologically distinct colonies were selected and purified on YIM 38 medium. The purified cultures were preserved as lyophilized cultures in skim milk and as glycerol suspensions (20%, v/v) at -80°C.
Genomic DNA was extracted using TIANGENTM Genomic DNA purification kit according to manufacturer’s instructions. The DNA preparations were used as template for PCR amplification using the universal primers 27F: 5′-CAGAGTTTGATCCTGGCT-3′ and 1492F: 5′-AGGAGGTGATCCAGCCGCA-3′. PCR reactions were conducted using iCycler Thermal Cycler (Bio-Rad, USA Laboratories, Inc.) by applying the following conditions: initial denaturation at 94°C for 4 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. Amplified PCR products were verified on 0.8% agarose gel with 2 Kb DNA ladder (Fermentas) as a molecular size reference and sent for sequencing (Sangon Biotech, Shanghai, China).
16S rRNA gene sequences of the isolates were compared with the published 16S rRNA gene sequence database in EzBioCloud server (Yoon et al., 2017) on the basis of pairwise alignment. The strains were identified based on the sequence similarity to their closest homolog. Strains showing identity to bacterial phyla, other than Actinobacteria, were not reflected in the current study. Relative abundance of the Actinobacteria based on the number of strains from each genera were plotted into a scatter-plot using Microsoft Excel 2013.
The partial 16S rRNA gene sequences of all the cultivated actinobacterial strains isolated during the study were deposited in GenBank with the following accession numbers: KX274728–KX274786, MF431270–MF431414 (Supplementary Table S1).
Figure 2 represents the effects of the different enrichment methods on the isolation of Actinobacteria. When heat pretreatments was used as the enrichment techniques (Figure 2A), more CFUs was determined in samples incubated at 40°C for 2 days prior to isolation (Treatment c) than in samples incubated at higher temperatures (Treatments d, e, and f). It is, however, interesting to note that samples kept at room temperature (Treatment b) yield lower CFUs than the one incubated at 40°C. Among the five media during this process, HV agar seems to be a better isolation medium (Figure 2A). When pH was used as the isolation criteria, it was determined that more CFUs were obtained in media adjusted to neutral pH, fewer in alkaline pH and least in circumneutral pH (Figure 2B). If the sample pH is taken into consideration, there is a gradual decrease in the number of CFUs with increase in sample pH, irrespective of the sample types. Lastly, considering the use of calcium salts in isolation, it was determined that CaCO3 yielded more Actinobacteria than (CH3COO)2Ca and CaCl2 (Figure 2C). However, it was not just the salt that is important, the concentration of each salt in the selection media also played an important role. In our case, it was determined that higher CFUs was determined when salt concentration was proportionately at lower concentration (i.e., 0.1 and 0.01%, w/v) than in its absence or at high concentration.
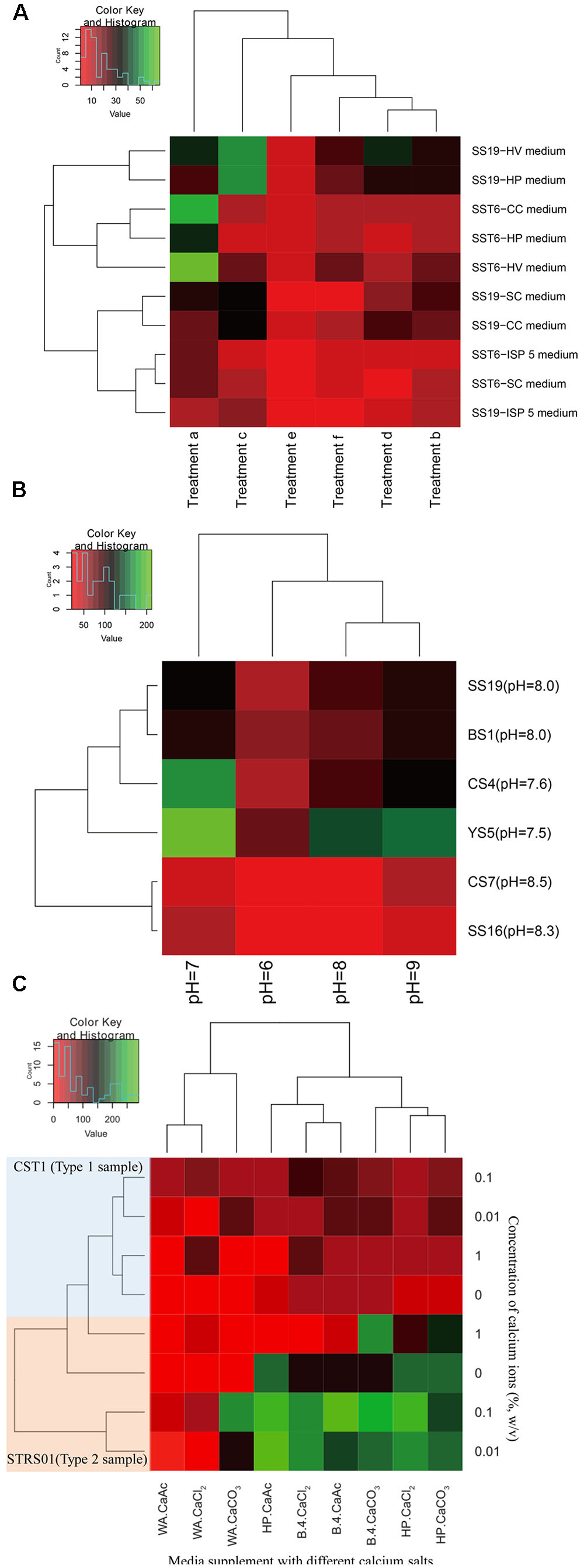
FIGURE 2. Heatmaps indicating the number of CFUs obtained after enrichments of samples collected from a karstic cave in Sigangli, China. The labels (A–C) represent the effects of temperature, pH, and calcium salts on the isolation of rare Actinobacteria, respectively. The heatmaps are generated in R software by applying ‘heatmap.2’ function in ‘gplots’ package.
Of these total colonies observed, morphologically distinct colonies were further selected, subcultured, and preserved. These include 87 isolates from Type 1 samples and 117 from the Type 2 samples. Sequence analysis of 16S rRNA gene indicated that the strains from the Type 1 samples were distributed to 20 genera in 14 families of the phylum Actinobacteria, while the strains isolated from Type 2 samples were distributed to 21 genera and 16 families. The relative abundance of the strains is represented in Figure 3, and the 16S rRNA gene sequence profile listed in Supplementary Table S1. Besides the most abundant genus Streptomyces, the rare actinobacterial genera Nocardia and Rhodococcus were relatively abundant in both the sample types (18 and 7 strains respectively in Type 1 samples, and 17 and 7 strains in Type 2 samples). While the genus Micromonospora was relatively more abundant in Type 2 samples (19 strains, as compared to 2 in Type 1 samples), the genus Mycobacterium was more in hard rock (7) than in the weathered rock samples (3). Other rare genera that were common to both the sample types include Jiangella, Kribbella, Nocardioides, Nocardiopsis, Nonomuraea, and Streptosporangium. Apart from these common genera, few rare actinobacterial genera were restricted to only one particular sample type. The genera Amycolatopsis, Beutenbergia, Cellulosimicrobium, Gordonia, Isoptericola, Microbacterium, Mumia, Oerskovia, and Pseudokineococcus were isolated only from Type 1 samples while the rare genera Actinocorallia, Actinomadura, Agromyces, Alloactinosynnema, Knoellia, Kocuria, Krasilnikoviella, Pseudonocardia, Saccharothrix, and Tsukamurella were isolated from Type 2 samples.
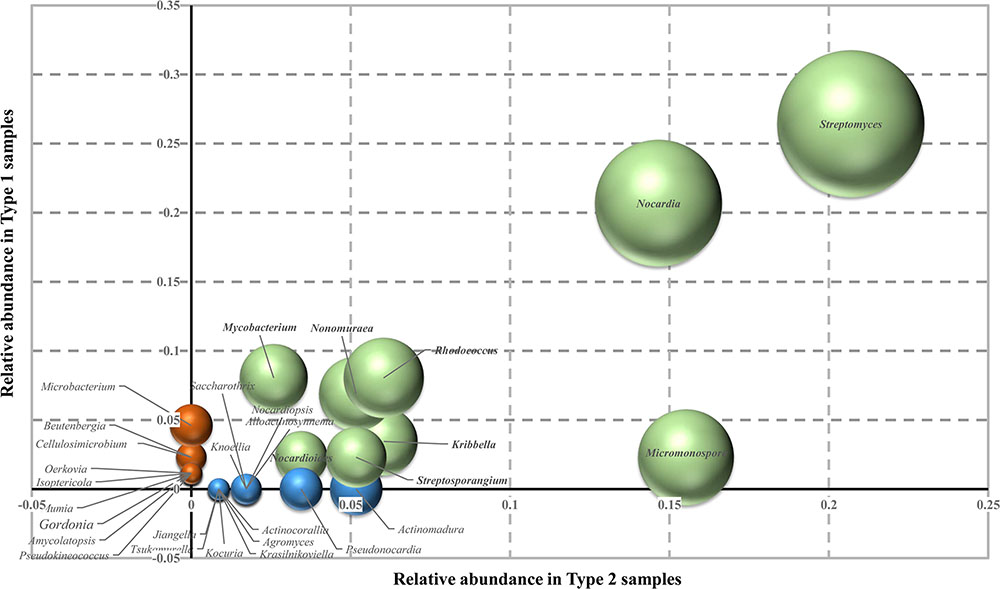
FIGURE 3. Relative abundance of rare Actinobacteria on the two types of cave samples used on this study. Values on the axes represented the relative abundance of each genus on the different samples. Green spheres indicate actinobacterial genera present in both the sample types; red, genera isolated from Type 1 samples only; and blue, genera found only in Type 2 samples. Sizes of the sphere quantify the relative number of strains of each genera. The scatter-plot is generated using Microsoft Excel 2013.
Karstic caves are characterized by low stable temperature (10–15°C), relatively high humidity (90–100%) and total darkness or low level of light, and are often mystical and inaccessible for study. Above that, caves usually constitute a oligotrophic ecosystem with total organic carbon of less than 2 mg/L (Tomczyk-Żak and Zielenkiewicz, 2016). Despite the oligotrophic condition, the average number of microorganisms in this ecosystem have been estimated to be in the range of 106 cells/g of rock (Barton and Jurado, 2007). Despite the estimated large number of bacterial cells in cave rocks, they could isolate around 400 bacteria belonging to phyla Proteobacteria, Firmicutes, and Actinobacteria from a sample of Lechuguilla Cave, New Mexico. Of these strains, nearly 40 strains are assumed to be previously uncultivated species indicating that the diversity of microbes within the caves is impressive (Barton and Jurado, 2007). In another study in Kartchner Caverns, Arizona, 90 unique isolates belonging to Proteobacteria, Firmicutes, and Actinobacteria were isolated, but these bacteria have 16S rRNA gene sequence similarity profiles to known bacteria (Ikner et al., 2007).
Among these group of bacteria, class Actinobacteria, because of its different versatile morphology, were detected and isolated from even the most extreme of environments (Mohammadipanah and Wink, 2016). The main mechanism for the survival of these actinobacteria in these environments is through the formation of different types of spores. Most actinobacterial spores are developed either endogenously (e.g., Dactylosporangium, Thermoactinomyces) or exogenously (e.g., Streptomyces) in response to environmental stress (Kalakoutskii and Agre, 1976). While in karst environments, the presence of minerals, particularly different forms of calcium rocks, trigger sporulation in many Actinobacteria (Kalakoutskii and Agre, 1976). These spores usually remain in dormant state with minimum respiration, but could be made to germinate in defined media by providing an energy source (Salas et al., 1983). In most case of germination, a mild stimuli is required either in the form of heating or supplying germinants that stimulate the disruption of spore cortex (Warth and Strominger, 1972; Ensign, 1978). In our effort to select and isolate diverse Actinobacteria, we considered three sample enrichments methods involving chemical and physical treatments as discussed below.
Among the physical enrichments method for actinobacterial isolation, air drying, dry heating, moist incubation, and desiccation have been found to be an effective for selection of spore-forming rare Actinobacteria (Hayakawa, 2008). Air-drying of soil at 120°C for an hour is usually preferred for isolation of genera Dactylosporangium, Microbispora and Streptosporangium, while limiting the growth of streptomycetes (Jiang et al., 2016). Similarly, air-drying at 100°C for 15 min have been used effectively for isolation of Actinomadura (Jiang et al., 2016). Air-drying the sample at an ambient temperature for a week preferentially select Herbidospora among other bacteria (Jiang et al., 2016). Genus Micromonospora were selectively isolated by pretreatment of samples at 55–65°C for 30 min (Jiang et al., 2016). An effective method proposed for isolation of rare Actinobacteria involved air drying at 80°C for 2 h (Goodfellow, 2010). Preferential selection of several rare Actinobacteria on heat treatments might be related to the spore-forming capability of several groups of Actinobacteria (Jiang et al., 2016). In the current study, we considered to pretreat our samples by air-drying at room temperature, 40, 65, and 110°C for different time intervals for effective isolation of diverse rare actinobacteria (Table 2). An advantages of air-drying is that many Actinobacteria produces spores, and that dry spores have low respiration rate and can survive for longer period of time. During this period of low level of endogenous respiration, the spores did not germinate, but can be germinated readily when a defined medium with organic energy source is provided (Ensign, 1978). During our study, four cells in the heatmap representation indicated CFUs’ count in the range of 50 or above (Figure 2A). These highest OTUs were represented in samples treated at low ambient temperatures. The levels of CFUs was found to decrease with increased pretreatment’s conditions. Least CFUs were obtained in the samples pretreated at 110°C for 1 h. However, if the pretreatment at 110°C for 1 h is accompanied by incubation at 40°C for 2 days, the numbers of sporulating cells that survived the heat treatment increased as indicated in the Figure 2A. This finding could be related to the study of Lapteva et al. (1976) whereby the authors suggested that the germination inhibitors produced in the spores due to heating were neutralized during subsequent incubation at lower temperature. If a comparative analyses were made between the number of CFUs obtained and the strains cultured during our study, positive correlation could be established between the number of spore-forming Actinobacteria obtained and the temperature used for pretreatment. While no strict thermophilic Actinobacteria such as members of the genera Thermomonospora, Dactylosporangium, etc. were isolated during our study, a fair number of rare Actinobacteria with aleurispores (Micromonospora), arthrospores (Actinomadura), sporangiospores (Streptosporangium), or other spore-bearing structures (Amycolatopsis etc.) were obtained. However, in addition to these Actinobacteria, endospore-forming non-Actinobacteria such as Bacillus strains were also obtained during the isolation process. The co-occurrence of these bacteria are, however, found to be very less as confirmed by our preliminary sequencing analysis of the 16S rRNA gene (data not shown).
Actinobacteria are capable of growing under selective conditions of pH or salinity (Mohammadipanah and Wink, 2016). It is because pH of soil strongly influence the biomass, activity and composition of the microbial community, and therefore pH in the isolation media provide a selective pressure for the growth of bacteria (Bååth, 1996; Matthies et al., 1997; Rousk et al., 2009). Unlike fungi which grow preferentially in acidic and moist condition, most Actinobacteria showed optimum growth on slightly alkaline condition (Kontro et al., 2005; Lewin et al., 2016). The isolation of strictly acidophilic Actinobacteria like the genus Streptacidiphilus (Kim et al., 2003; Cho et al., 2008; Golinska et al., 2016) from diverse ecosystems have provided a platform for isolation of Actinobacteria under acidic environments, in addition to the normally preferred slightly alkaline condition. Considering the wide range of pH on which Actinobacteria can dwell with, we considered to compare the CFU’s count under a gradient of pH range of isolation, despite all the sample pHs being in the range of 7.5 to 8.5 (Table 2). During the current study, more actinobacterial CFUs were detected in neutral pH (Figure 2B). This may be because of the easy maintanence of cell’s cytoplasmic pH at close to neutrality (Kontro et al., 2005). While the actinobacterial CFUs did not dwindle much in alkaline pH, it had the least count in acidic isolation media. This finding could also be related with the findings of Rousk et al. (2010) whereby the relative abundance of Actinobacteria was not affected by soil pH, but rather depended on the isolation condition (Lauber et al., 2009).
Actinobacteria often colonize the rock walls of caves. In a study on biogeochemical role of Actinobacteria in Altamira Cave (Spain), Actinobacteria-coated spots on the cave walls was found to uptake carbon dioxide gas which is available in abundance in cave (Cuezva et al., 2012). This uptake gas is used by the bacteria to dissolved rock and subsequently generate crystals of calcium carbonate (Cañaveras et al., 2001). While its role in biomineralization is plausible, calcium ions do play specific role in various spore-forming microorganism as well. While measuring the metal ion content in five Streptomyces strains (Salas et al., 1983), the level of calcium was found to be higher in dormant spore than in the vegetative cells. Calcium is mostly found as a complex with dipicolinate and this complex could be acting as secondary stabilizing agent for the spore against environmental stresses (Moir and Smith, 1990). However in the presence of suitable germinants, the spore release the calcium-dipicolinate complex from the core to initiate the process of spore germination (Moir, 2003). One process through which the complex acts is by initiating cortex degradation through structural modification of the peptidoglycan (de Vries, 2004). The use of calcium carbonate in pretreatment for selective isolation of Actinobacteria (El-Nakeeb and Lechevalier, 1963; Alferova and Terekhova, 1988) may be related with the spore formation in Actinobacteria. On the other hand, calcium chloride, when added to isolation media, was found to stimulate the growth of a rare heterotrophic Actinobacteria, Sporichthya (Suzuki et al., 1999). We, therefore, considered to compare the effect of supplementation of three calcium salts in the isolation media including CaCO3 and CaCl2. During our study, all three calcium salts facilitated the isolation of Actinobacteria (Figure 2C). This finding is also in congruence with the finding of Chen et al. (2016) whereby the actinobacterial community structures showed significant correlations with calcium. Warth and Strominger (1972) have determined that germination of bacterial spore required a optimum concentration of approximately 10 mM calcium ions. This observation is similar with our findings where a lower concentration of calcium ions (0.1% or ∼10 mM) provide more CFUs than higher (1%, w/v or ∼0.1 M) or in the absence of calcium salts (Figure 2C). Among the two types of samples, more CFUs were observed in Type 2 than in Type 1 samples. The reason could be implicated on the lower cell concentration on the surface of hard rock (Barton and Jurado, 2007).
In all the above cases, isolation media play the key role for providing the favorable condition for isolation and growth of rare Actinobacteria. It is therefore important to use isolation media that preferentially isolate different group of rare Actinobacteria and select/design set of media with different components to maximize our chance for isolation of unique and other rare Actinobacteria (Tiwari and Gupta, 2013), lest Actinobacteria will be at competitive disadvantages on the solid media against the fastidious bacteria and fungi that usually occupy a larger living space. In the current study, we had selected seven isolation media that have been found effective in isolation of Actinobacteria. Among them, HV agar with/without chemical supplements had been used efficiently by Hayakawa’s group for isolation of many rare Actinobacteria including strains of genera Actinokineospora, Actinomadura, Actinoplanes, Actinosynnema, Catenuloplanes, Cryptosporidium, Dactylosporangium, Geodermatophilus, Herbidospora, Kineosporia, Microbispora, Micromonospora, Microtetraspora, Nonomuraea, Spirilliplanes, Sporichthya, Streptosporangium, and Virgosporangium (Hayakawa, 2008). It may be the wide applicability of this media in isolation of different group of rare Actinobacteria, that we are able to found more CFUs in this medium (Figure 2A). While SC and ISP media were introduced for the isolation of mycelial-producing Actinobacteria particularly genus Streptomyces (Küster and Williams, 1964; Shirling and Gottlieb, 1966), B-4 media was established to be good for isolation of Actinobacteria precipitating calcium carbonate crystals (Boquet et al., 1973). The large amount of Steptomyces among our isolates could be correlated with the use of ISP and SC during our isolation (Supplementary Table S1). The media CC and HP were especially designed in our laboratory to isolate rare Actinobacteria that could utilize complex energy sources (Li et al., 2014; Yuan et al., 2017). Among these two media, HP was more efficient than CC in giving larger CFUs count (Figures 2A,C). The reason behind the larger CFUs in HP could not be ascertained from the current study, however, it is possible that degradation of cellulose required complex enzyme-system and that many Actinobacteria were not able to use cellulose as their energy sources. On the other hand, water agar, which is found to stimulate growth of spore-forming microorganisms, was not effective during our study for the growth of rare Actinobacteria (Figure 2C). Inhibition of certain rare actinobacterial strains by the preferential treatments, however, cannot be completely ruled out.
In the karstic caves, the primary production usually depends on chemoautotrophic bacteria (Sarbu et al., 1996). Recent studies have, however, revealed that considerable input of organic matter could support the growth of heterotrophic bacteria including Actinobacteria (Arroyo et al., 1997; Groth and Saiz-Jimenez, 1999). These findings instigated the study on diversity of Actinobacteria in several caves located around the world such as Niu Cave (Zhou et al., 2007), Pajsarjeva jama (Pasić et al., 2010), Wind Cave (Chelius and Moore, 2004), Kartchner Caverns (Ikner et al., 2007), Altamira Cave (Cuezva et al., 2009), and Altamira and Tito Bustillo Caves (Schabereiter-Gurtner et al., 2002). Studies of Shabarova and Pernthaler (2010) have resulted in the isolation of Actinobacteria belonging to the genera Arthrobacter, Blastococcus, Curtobacterium, Kribella, Micrococcus, Nocardia, Promicromonspora, Pseudonocardia, Rhodococcus, and Streptomyces. Unlike the above study, significant diversity of rare Actinobacteria were observed in the present study. These Actinobacteria were affiliated to genera Actinocorallia, Actinomadura, Agromyces, Alloactinosynnema, Amycolatopsis, Beutenbergia, Cellulosimicrobium, Gordonia, Isoptericola, Jiangella, Knoellia, Kocuria, Krasilnikoviella, Kribbella, Microbacterium, Micromonospora, Mumia, Mycobacterium, Nocardia, Nocardioides, Nocardiopsis, Nonomuraea, Oerskovia, Pseudokineococcus, Pseudonocardia, Rhodococcus, Saccharothrix, Streptosporangium, and Tsukamurella. The presence of genera Micromonospora, Nocardia, and Rhodococcus as the dominant rare Actinobacteria in our study was consistent with other related studies (Arroyo et al., 1997; Zhou et al., 2007; Valme et al., 2010).
Despite the isolation of varied actinobacterial groups after applying a set of pretreatments and modification of isolation media, our study suffers from few limitations. Firstly, the physicochemical parameters of the sampling site and the co-existence of different minerals/metals were not measured during the study. Lack of these data prevent us from indirect establishment of the interrelationship between the occurrence of different actinobacterial groups and their physiological roles in cave. Secondly, the study was limited to isolation of culturable Actinobacteria. As such, we could not equally verify if the applied methods were effective to deselect non-Actinobacteria. It is also equally possible that certain culturable rare Actinobacteria particularly non-spore formers were deselected due to the stressors provided in our pretreatments. Lastly, the isolation methods have not been replicated in other karst environments or any other habitats. A study on the total microbial composition using NextGen sequencing could provide an idea of the effectiveness of the isolation method. However, it can certainly be stated that the methods provided above proved effective for the isolation of many rare Actinobacteria, comprising of both spore-formers (e.g., Actinocorallia, Alloactinosynnema, Jiangella, Oerskovia etc.) and non-spore formers (such as Agromyces, Beutenbergia, Cellulosimicrobium, Gordonia, Isoptericola, Kocuria, Tsukamurella).
W-JL, B-ZF, and JC designed research and project outline. JC, M-XH, MX, and NS performed isolation, deposition, and identification. B-ZF, J-YJ, and D-QW contructed the heatmap and other related bioinformatic plots. B-ZF, NS, MX, and W-JL drafted the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by National Natural Science Foundation of China (No. 31600015), National Fundamental Fund Project Subsidy Funds of Personnel Training of China (No. J1310025), China Postdoctoral Science Foundation (2016M592567, 2016M602566) and Visiting Scholar Grant of State Key Laboratory of Biocontrol, Sun Yat-sen University (No. SKLBC14F02). W-JL was also supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (2014).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01535/full#supplementary-material
Alferova, I. V., and Terekhova, L. P. (1988). Use of the method of enriching of soil samples with calcium carbonate for isolation of Actinomyces. Antibiot. Khimioter. 33, 888–890.
Arroyo, G., Arroyo, I., and Arroyo, E. (1997). Microbiological analysis of maltravieso cave (Caceres), Spain. Int. Biodeterior. Biodegrad. 40, 131–139. doi: 10.1016/S0964-8305(97)00039-5
Bååth, E. (1996). Adaptation of soil bacterial communities to prevailing pH in different soils. FEMS Microbiol. Ecol. 19, 227–237. doi: 10.1111/j.1574-6941.1996.tb00215.x
Barton, H. A., and Jurado, V. (2007). What’s up down there? Microbial diversity in caves. Microbe 2, 132–138.
Boquet, E., Boronat, A., and Ramos-Cormenzana, A. (1973). Production of calcite (calcium carbonate) crystals by soil bacteria is a general phenomenon. Nature 246, 527–528. doi: 10.1038/246527a0
Cañaveras, J. C., Sanchez-Moral, S., Sloer, V., and Saiz-Jimenez, C. (2001). Microorganisms and microbially induced fabrics in cave walls. Geomicrobiol. J. 18, 223–240. doi: 10.1080/01490450152467769
Castanier, S., Le Métayer-Levrel, G., and Perthuisot, J. P. (2000). “Bacterial roles in the precipitation of carbonate minerals,” in Microbial Sediments, eds R. E. Riding and S. M. Awramik (Heidelberg: Springer-Verlag), 32–39.
Chelius, M. K., and Moore, J. C. (2004). Molecular phylogenetic analysis of archaea and bacteria in Wind Cave, South Dakota. Geomicrobiol. J. 21, 123–134. doi: 10.1080/01490450490266389
Chen, P., Zhang, L., Guo, X., Dai, X., Xi, L., Wang, J., et al. (2016). Diversity, biogeography, and biodegradation potential of Actinobacteria in the deep-sea sediments along the southwest Indian ridge. Front. Microbiol. 7:1340. doi: 10.3389/fmicb.2016.01340
Cho, S. H., Han, J. H., Ko, H. Y., and Kim, S. B. (2008). Streptacidiphilus anmyonensis sp. nov., Streptacidiphilus rugosus sp. nov. and Streptacidiphilus melanogenes sp. nov., acidophilic actinobacteria isolated from Pinus soils. Int. J. Syst. Evol. Microbiol. 58, 1566–1570. doi: 10.1099/ijs.0.65480-0
Cuezva, S., Fernandez-Cortes, A., Porca, E., Pašić, L., Jurado, V., Hernandez-Marin, M., et al. (2012). The biogeochemical role of Actinobacteria in Altamira Cave, Spain. FEMS Microbiol. Ecol. 81, 281–290. doi: 10.1111/j.1574-6941.2012.01391.x
Cuezva, S., Sanchez-Moral, S., Saiz-Jimenez, C., and Cañaveras, J. C. (2009). Microbial communities and associated mineral fabrics in Altamira Cave, Spain. Int. J. Speleol. 38, 83–92. doi: 10.5038/1827-806X.38.1.9
Culver, D. C., and Pipan, T. (2009). The Biology of Caves and Other Subterranean Habitats. Oxford: Oxford University Press, 255.
de Vries, Y. P. (2004). The role of calcium in bacterial spore germination. Microbes Environ. 19, 199–202. doi: 10.1264/jsme2.19.199
Dhami, N. K., Reddy, M. S., and Mukherjee, A. (2013). Biomineralization of calcium carbonates and their engineered applications: a review. Front. Microbiol. 4:314. doi: 10.3389/fmicb.2013.00314
El-Nakeeb, M. A., and Lechevalier, H. A. (1963). Selective isolation of aerobic actinomycetes. Appl. Microbiol. 11, 75–77.
Engel, A. S., Stern, L. A., and Bennett, P. C. (2004). Microbial contributions to cave formation: new insights into sulfuric acid speleogenesis. Geology 32, 369–372. doi: 10.1130/G20288.1
Ensign, J. C. (1978). Formation, properties, and germination of actinomycete spores. Ann. Rev. Microbiol. 32, 185–219. doi: 10.1146/annurev.mi.32.100178.001153
Genilloud, O. (2014). The re-emerging role of microbial natural products in antibiotic discovery. Antonie Van Leeuwenhoek 106, 173–188. doi: 10.1007/s10482-014-0204-6
Gillieson, D. (1996). Caves: Processes, Development, and Management. Oxford: Blackwell Publishers Ltd, 324.
Golinska, P., Dahm, H., and Goodfellow, M. (2016). Streptacidiphilus toruniensis sp. nov., isolated from a pine forest soil. Antonie Van Leeuwenhoek 109, 1583–1591. doi: 10.1007/s10482-016-0759-5
Goodfellow, M. (2010). “Selective isolation of Actinobacteria,” in Manual of Industrial Microbiology and Biotechnology, eds R. Baltz, A. Demain, J. Davies, A. Bull, B. Junker, L. Katz, et al. (Washington, DC: ASM Press), 13–27.
Green, B. D., and Keller, M. (2006). Capturing the uncultivated majority. Curr. Opin. Biotechnol. 17, 236–240. doi: 10.1016/j.copbio.2006.05.004
Groth, I., and Saiz-Jimenez, C. (1999). Actinomycetes in hypogean environments. Geomicrobiol. J. 16, 1–8. doi: 10.1080/014904599270703
Hayakawa, M. (2008). Studies on the isolation and distribution of rare actinomycetes in soil. Actinomycetologica 22, 12–19. doi: 10.3209/saj.SAJ220103
Hayakawa, M., and Nonomura, H. (1987). Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509. doi: 10.1016/0385-6380(87)90108-7
Ikner, L. A., Toomey, R. S., Nolan, G., Neilson, J. W., Pryor, B. M., and Maier, R. M. (2007). Culturable microbial diversity and the impact of tourism in Kartchner Caverns, Arizona. Microb. Ecol. 53, 30–42. doi: 10.1007/s00248-006-9135-8
Jiang, Y., Li, Q., Chen, X., and Jiang, C. (2016). “Isolation and cultivation methods of Actinobacteria,” in Actinobacteria – Basics and Biotechnological Applications, ed. D. Dhanasekaran (Rijeka: InTech), 39–57.
Kalakoutskii, L. V., and Agre, N. S. (1976). Comparative aspects of development and differentiation in actinomycetes. Bacteriol. Rev. 40, 469–524.
Kim, S. B., Lonsdale, J., Seong, C. N., and Goodfellow, M. (2003). Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici (1943)AL) emend. Rainey et al. 1997. Antonie Van Leeuwenhoek 83, 107–116. doi: 10.1023/A:1023397724023
Kontro, M., Lignell, U., Hirvonen, M. R., and Nevalainen, A. (2005). pH effects on 10 Streptomyces spp. growth and sporulation depend on nutrients. Lett. Appl. Microbiol. 41, 32–38. doi: 10.1111/j.1472-765X.2005.01727.x
Küster, E., and Williams, S. T. (1964). Selection of media for isolation of Streptomycetes. Nature 202, 928–929. doi: 10.1038/202928a0
Lapteva, E. A., Agre, N. S., and Kalakoutskii, L. V. (1976). Restoration of viability in streptomycete spores heated at 100°C. Microbiologiya 45, 3.
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Leadbetter, J. R. (2003). Cultivation of recalcitrant microbes: cells are alive, well and revealing their secrets in the 21st century laboratory. Curr. Opin. Microbiol. 6, 274–281. doi: 10.1016/S1369-5274(03)00041-9
Lewin, G. R., Carlos, C., Chevrette, M. G., Horn, H. A., McDonald, B. R., Stankey, R. J., et al. (2016). Evolution and ecology of Actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 70, 235–254. doi: 10.1146/annurev-micro-102215-095748
Li, J., Dong, J. D., Yang, J., Luo, X. M., and Zhang, S. (2014). Detection of polyketide synthase and nonribosomal peptide synthetase biosynthetic genes from antimicrobial coral-associated actinomycetes. Antonie Van Leeuwenhoek 106, 623–635. doi: 10.1007/s10482-014-0233-1
Matthies, C., Erhard, H. P., and Drake, H. L. (1997). Effects of pH on the comparative culturability of fungi and bacteria from acidic and less acidic forest soils. J. Basic Microbiol. 37, 335–343. doi: 10.1002/jobm.3620370506
Mohammadipanah, F., and Wink, J. (2016). Actinobacteria from arid and desert habitats: diversity and biological activity. Front. Microbiol. 6:1541. doi: 10.3389/fmicb.2015.01541
Moir, A. (2003). Bacterial spore germination and protein mobility. Trends Microbiol. 11, 452–454. doi: 10.1016/j.tim.2003.08.001
Moir, A., and Smith, D. A. (1990). The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44, 531–553. doi: 10.1146/annurev.mi.44.100190.002531
Pasić, L., Kovce, B., Sket, B., and Herzog-Velikonja, B. (2010). Diversity of microbial communities colonizing the walls of a Karstic cave in Slovenia. FEMS Microbiol. Ecol. 71, 50–60. doi: 10.1111/j.1574-6941.2009.00789.x
Pedersen, K. (2000). Exploration of deep intraterrestrial microbial life: current perspectives. FEMS Microbiol. Lett. 185, 9–16. doi: 10.1111/j.1574-6968.2000.tb09033.x
Pham, V. H. T., and Kim, J. (2012). Cultivation of unculturable soil bacteria. Trends Biotechnol. 30, 475–484. doi: 10.1016/j.tibtech.2012.05.007
Remenár, M., Karelová, E., Harichová, J., Zámocký, M., Krčová, K., and Ferianc, P. (2014). Actinobacteria occurrence and their metabolic characteristics in the nickel-contaminated soil sample. Biologia 69, 1453–1463. doi: 10.2478/s11756-014-0451-z
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Rousk, J., Brookes, P. C., and Bååth, E. (2009). Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl. Environ. Microbiol. 75, 1589–1596. doi: 10.1128/AEM.02775-08
Salas, J. A., Guijarro, J. A., and Hardisson, C. (1983). High calcium content in Streptomyces spores and its release as an early event during spore germination. J. Bacteriol. 155, 1316–1323.
Sarbu, S. M., Kane, T. C., and Kinkle, B. K. (1996). A chemoautotrophically based cave ecosystem. Science 272, 1953–1955. doi: 10.1126/science.272.5270.1953
Schabereiter-Gurtner, C., Saiz-Jimenez, C., Pinar, G., Lubitz, W., and Rölleke, S. (2002). Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in Tito Bustillo cave, Spain, and on its Palaeolithic paintings. Environ. Microbiol. 4, 392–400. doi: 10.1046/j.1462-2920.2002.00303.x
Shabarova, T., and Pernthaler, J. (2010). Karst pools in subsurface environments: collectors of microbial diversity or temporary residence between habitat types. Environ. Microbiol. 12, 1061–1074. doi: 10.1111/j.1462-2920.2009.02151.x
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Suzuki, S., Okuda, T., and Komatsubara, S. (1999). Selective isolation and distribution of Sporichthya strains in soil. Appl. Environ. Microbiol. 65, 1930–1935.
Tiwari, K., and Gupta, R. K. (2013). Diversity and isolation of rare actinomycetes: an overview. Crit. Rev. Microbiol. 39, 256–294. doi: 10.3109/1040841X.2012.709819
Tomczyk-Żak, K., and Zielenkiewicz, U. (2016). Microbial diversity in caves. Geomicrobiol. J. 33, 20–38. doi: 10.1080/01490451.2014.1003341
Valme, J., Leonila, L., Veronica, R. N., and Patrick, B. (2010). Pathogenic and opportunistic microorganisms in caves. Int. J. Speleol. 39, 15–24. doi: 10.5038/1827-806X.39.1.2
Warth, A. D., and Strominger, J. L. (1972). Structure of the peptiodoglycan from spores of Bacillus subtilis. Biochemistry 11, 1389–1396. doi: 10.1021/bi00758a010
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583. doi: 10.1073/pnas.95.12.6578
Wu, Y., Tan, L., Liu, W., Wang, B., Wang, J., Cai, Y., et al. (2015). Profiling bacterial diversity in a limestone cave of the western Loess Plateau of China. Front. Microbiol. 6:244. doi: 10.3389/fmicb.2015.00244
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, J., Kim, Y., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1618. doi: 10.1099/ijsem.0.001755
Yuan, C. G., Chen, X., Jiang, Z., Chen, W., Liu, L., Xian, W. D., et al. (2017). Altererythrobacter lauratis sp. nov., and Altererythrobacter palmitatis sp. nov., isolated from a Tibetan hot spring. Antonie Van Leeuwenhoek 110, 1077–1086. doi: 10.1007/s10482-017-0882-y
Zhao, G. Z., Li, J., Qin, S., Huang, H. Y., Zhu, W. Y., Xu, L. H., et al. (2010). Streptomyces artemisiae sp. nov. isolated from surface-sterilized tissue of Artemisia annua L. Int. J. Syst. Evol. Microbiol. 60, 27–32. doi: 10.1099/ijs.0.011965-0
Keywords: Sigangli Cave, rare Actinobacteria, heat pretreatment, pH, calcium salts
Citation: Fang B-Z, Salam N, Han M-X, Jiao J-Y, Cheng J, Wei D-Q, Xiao M and Li W-J (2017) Insights on the Effects of Heat Pretreatment, pH, and Calcium Salts on Isolation of Rare Actinobacteria from Karstic Caves. Front. Microbiol. 8:1535. doi: 10.3389/fmicb.2017.01535
Received: 26 February 2017; Accepted: 28 July 2017;
Published: 08 August 2017.
Edited by:
Baolei Jia, Chung-Ang University, South KoreaReviewed by:
Isao Yumoto, National Institute of Advanced Industrial Science and Technology, JapanCopyright © 2017 Fang, Salam, Han, Jiao, Cheng, Wei, Xiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Jun Li, liwenjun3@mail.sysu.edu.cn Min Xiao, xiaomin8@mail.sysu.edu.cn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.