- 1School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW, Australia
- 2Clinical Research Center, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China
- 3Gastrointestinal and Liver Unit, Prince of Wales Hospital, University of New South Wales, Sydney, NSW, Australia
- 4St George and Sutherland Clinical School, University of New South Wales, Sydney, NSW, Australia
Campylobacter concisus was previously shown to be associated with inflammatory bowel disease including Crohn's disease (CD) and ulcerative colitis (UC). C. concisus has two genomospecies (GS). This study systematically examined the colonization of GS1 and GS2 C. concisus in the human gastrointestinal tract. GS1 and GS2 specific polymorphisms in 23S rRNA gene were identified by comparison of the 23S rRNA genes of 49 C. concisus strains. Two newly designed PCR methods, based on the polymorphisms of 23S rRNA gene, were developed and validated. These PCR methods were used to detect and quantify GS1 and GS2 C. concisus in 56 oral and enteric samples collected from the gastrointestinal tract of patients with IBD and healthy controls. Meta-analysis of the composition of the isolated GS1 and GS2 C. concisus strains in previous studies was also conducted. The quantitative PCR methods revealed that there was more GS2 than GS1 C. concisus in samples collected from the upper and lower gastrointestinal tract of both patients with IBD and healthy controls, showing that GS2 C. concisus is better adapted to the human gastrointestinal tract. Analysis of GS1 and GS2 composition of isolated C. concisus strains in previous studies showed similar findings except that in healthy individuals a significantly lower GS2 than GS1 C. concisus strains were isolated from fecal samples, suggesting a potential difference in the C. concisus strains or the enteric environment between patients with gastrointestinal diseases and healthy controls. This study provides novel information regarding the adaptation of different genomospecies of C. concisus in the human gastrointestinal tract.
Introduction
Campylobacter concisus is a Gram-negative motile bacterium, growing under anaerobic, and microaerophilic conditions in the presence of hydrogen gas (Lastovica et al., 2014; Lee et al., 2014). A number of studies reported an association between C. concisus and inflammatory bowel disease (IBD); these studies found that the prevalence of C. concisus in the intestinal tissues of patients with IBD was significantly higher than that in the controls (Zhang et al., 2009; Mahendran et al., 2011; Mukhopadhya et al., 2011; Kirk et al., 2016). IBD is a chronic inflammatory condition of the gastrointestinal tract, presenting as two major clinical forms including Crohn's disease (CD) or ulcerative colitis (UC; Sartor and Mazmanian, 2012). In addition to its association with IBD, some studies suggested that C. concisus may be involved in diarrheal disease (Lindblom et al., 1995; Kalischuk and Inglis, 2011; Nielsen et al., 2013).
C. concisus is part of the oral microbiota, with it being detected in saliva samples of nearly every individual (Zhang et al., 2010). C. concisus strains have different abilities in resistance to the enteric environmental factors such as the bile, some were able to colonize the intestinal tract (Ma et al., 2015). Some oral C. concisus strains may cause enteric diseases; they were found to induce intestinal epithelial death and production of proinflammatory cytokines (Nielsen et al., 2011; Ismail et al., 2012, 2013). Enteric virulent C. concisus strains translocated from the oral cavity to the intestinal tract were suggested to be initiators for a subgroup of IBD (Zhang et al., 2014; Zhang, 2015). C. concisus zonula occludens toxin was found to damage intestinal barrier function and enhance the responses of macrophage to other bacterial species (Mahendran et al., 2016), and phospholipase A was shown to damage human cell membrane (Istivan et al., 2004).
C. concisus has two genomospecies (GS), which can be consistently differentiated by the sequences of C. concisus core-genome, housekeeping genes and a polymerase chain reaction (PCR) targeting the polymorphisms of 23S rRNA gene (Aabenhus et al., 2005; Miller et al., 2012; On et al., 2013; Mahendran et al., 2015; Chung et al., 2016). The C. concisus GS1 and GS2 strains are morphologically similar but have genomospecies-specific genes, with some of the genes suggested to play a role in environmental adaptation (Chung et al., 2016). Previous studies suggested that C. concisus strains of different genomospecies may vary in their pathogenicity. For example, Engberg et al. examined C. concisus strains isolated from diarrheal stool samples and found that bloody diarrhea was present only in individuals infected with GS2 C. concisus (Engberg et al., 2005). A study from Kalischuk et al. found that GS2 C. concisus strains had a greater mean epithelial invasion and translocation than GS1 strains (Kalischuk and Inglis, 2011). Furthermore, oral C. concisus strains that were invasive to intestinal epithelial cells reported by Ismail et al. were GS2 strains (Ismail et al., 2012; Mahendran et al., 2015).
C. concisus was previously detected in clinical samples collected from both the upper and lower human gastrointestinal tract such as saliva, intestinal biopsies, and fecal samples (Zhang et al., 2009, 2010; Mahendran et al., 2011; Mukhopadhya et al., 2011; Kirk et al., 2016). The PCR methods used for the detection of C. concisus in these studies targeted the 16S rRNA gene, which were unable to differentiate C. concisus GS1 and GS2 strains. Therefore, it is not clear whether C. concisus previously detected in these clinical samples were GS1 or GS2.
This study aimed to examine the colonization of GS1 and GS2 C. concisus in the human gastrointestinal tract. We identified C. concisus GS1 and GS2 specific polymorphisms in the 23S rRNA gene. Based on these polymorphisms, we developed C. concisus GS1 and GS2 specific PCR methods and quantified GS1 and GS2 C. concisus in saliva, intestinal biopsies and fecal samples from patients with IBD and healthy controls. This study found that in the gastrointestinal tract of both patients with IBD and healthy controls, there were more GS2 than GS1 C. concisus, showing that GS2 C. concisus is better adapted to the environment of the human gastrointestinal tract. The potential clinical relevance was discussed.
Methods
Identification of C. concisus GS1 and GS2 Specific Polymorphisms in 23S rRNA Gene
In order to design specific PCR methods that can be used for detection and quantification of C. concisus GS1 and GS2, we identified GS1 and GS2 specific polymorphisms in 23S rRNA gene. The 23S rRNA gene of 49 C. concisus strains was examined, including 13 C. concisus strains which had their genomes sequenced in this study and 36 publicly available C. concisus genomes with known GS1 and GS2 identities (Tanner et al., 1981; Deshpande et al., 2013; Chung et al., 2016). The 13 C. concisus genomes were sequenced using the MiSeq method and their GS1 and GS2 identities were determined using six housekeeping genes and 23rRNA genes as described previously (Chung et al., 2016). The sequences of the 23S rRNA gene and the housekeeping genes of these 13 C. concisus strains were submitted to National Center for Biotechnology Information (NCBI) Genbank. The accession numbers for the 23S rRNA gene and the six housekeeping genes were MF351708-MF351720 and MF358606-MF358683, respectively.
The sequences of the 23S rRNA gene of these 49 C. concisus strains were compared using MEGA7 (Kumar et al., 2016). From the alignments performed, GS1-specific and GS2-specific polymorphic nucleotides of the 23S rRNA gene were identified. GS1-specific polymorphic nucleotides refer to the nucleotides that are present in the 23S rRNA gene of all GS1 C. concisus strains but absent in all GS2 C. concisus strains, and vice versa.
Development and Validation of Two PCR Methods for Detection and Quantification of GS1 and GS2 C. concisus
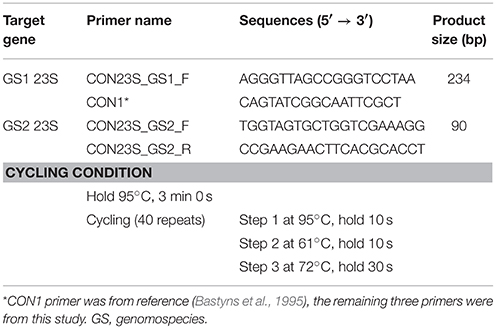
PCR primers were designed by NCBI Primer-BLAST using the sequence of 23S rRNA gene of C. concisus strain 13826 (Ye et al., 2012). Four primers were selected. The sequences of these primers were compared to all species in the NCBI non-redundant database using Primer-BLAST, which showed 100% match to C. concisus and low similarities to other species. One of the primers had a sequence that was identical to the CON1 primer previously designed by Bastyns et al. (1995). Primers CON23S_GS1_F and CON1 were used to amplify GS1 C. concisus (GS1-PCR) with the product size being 234 bp, primers CON23S_GS2_F and CON23S_GS2_R were used to amplify GS2 C. concisus (GS2-PCR) with the product size being 90 bp. The sequences of these primers and the PCR thermocycling conditions are listed in Table 1.
The specificities of GS1-PCR and GS2-PCR were confirmed using 41 C. concisus strains with known GS1 and GS2 identities that were available to us, including the 27 C. concisus strains that we previously performed genome sequencing, the 13 genomes that were sequenced in this study and C. concisus strain 13826 (Chung et al., 2016). These C. concisus strains were cultured on horse blood agar plates and DNA was extracted as previously described (Mahendran et al., 2011; Ismail et al., 2012). An aliquot (5 ng) of the extracted bacterial DNA was subjected to GS1-PCR and GS2-PCR.
The use of the C. concisus GS1 and GS2 specific primers designed in this study in quantitative GS1-real time PCR (GS1-rtPCR) and GS2-real time PCR (GS2-rtPCR) was also validated. DNA samples extracted from GS1 strain P14UCO-S2 and GS2 strain 13826, which were chosen randomly, were used to construct the standard curves and the PCR efficiencies were determined by Rotor gene 6000 software (Dhanasekaran et al., 2010). The SYBR Green I method was used for rtPCR and the reagents were purchased from Bioline (NSW, Australia). Each reaction was prepared with 10 μl of 1 × SensiFAST™ SYBR® No-ROX mix, 0.8 μl of 400 nM forward primer, 0.8 μl of 400 nM reverse primer, mixed with 2 μl of serial diluted bacterial DNA template of concentrations ranged between 0.0015 and 15 ng/μl, the volume was topped up to 20 μl with nuclease-free water. Amplifications were performed in a three step rtPCR using the following cycling conditions: 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, 61°C for 10 s, and 72°C for 30 s (Fite et al., 2004). No primer dimers were observed in the melting curve.
The PCR methods developed in this study were then used to detect and quantify GS1 and GS2 C. concisus in oral and enteric samples collected from patients with IBD and healthy controls.
Detection and Quantification of GS1 and GS2 C. concisus in DNA Samples Extracted from Saliva, Intestinal Biopsies, and Fecal Samples
A total of 56 C. concisus positive samples collected from the human gastrointestinal tract were used for GS1 and GS2 C. concisus quantification, including 27 saliva samples and 29 enteric samples (Tables 3, 4). These samples were collected in a previous study (Mahendran et al., 2011). In that study, multiple intestinal biopsies (each at ileum, caecum, colon, and rectum), were collected from each individual and used for C. concisus detection by the genus PCR. The C. concisus positive intestinal biopsies were used in this study for the quantification of GS1 and GS2 C. concisus. Fecal fluids refer to the liquids containing fecal materials in the draining tubes during the colonoscopy procedure. DNA samples of intestinal biopsies were extracted previously (Mahendran et al., 2011). DNA samples of the saliva and fecal fluids were extracted in this study using the ISOLATE fecal DNA kit (Bioline) following the manufacturer's instructions. Azathioprine, mercaptopurine, and 5-aminosalicylic acid were shown to affect C. concisus growth, however the samples used in this study were collected from the patients who have not received any treatment (Liu et al., 2017).
GS1-PCR and GS2-PCR methods were used to detect the presence of GS1 and GS2 C. concisus in DNA samples extracted from saliva and fecal fluids. PCR reactions had 25 μl in volume, containing 2.5 μl of 10 × DNA polymerase buffer, 2.5 μl of 10 × dNTP, 1.5 μl of 25 mM MgCl2, 1 μl of 10 pmol forward primer, 1 μl of 10 pmol reverse primer, 12.5 μl of nuclease-free water, 2 μl of DNA template and 2 μl of Taq polymerase. DNA templates used were 5 ng DNA extracted from saliva, 100 ng DNA extracted from intestinal biopsies and 50 ng DNA extracted from fecal fluids. A PCR reaction without bacterial DNA was used as the negative control. Positive PCR products were sequenced as previously described using BigDyeTM terminator chemistry (Applied Biosystems, Foster City, USA) (Mahendran et al., 2011). The sequences of the PCR products were compared to the 23S rRNA sequences of the corresponding C. concisus strains (Chung et al., 2016).
DNA samples extracted from saliva, fecal samples and intestinal biopsies were then subjected to GS1-rtPCR and GS2-rtPCR for quantification. The results were presented as copy numbers, amplified using 5 ng DNA extracted from saliva, 100 ng DNA extracted from intestinal biopsies and 50 ng DNA extracted from fecal fluids. The copy numbers of amplified C. concisus GS1 and GS2 in clinical samples were determined using standard curves that were generated using DNA extracted from GS1 strain P14UCO-S2 and GS2 strain 13826, respectively. Non-template control group was included in each run. For each run, the melting curves of individual samples were checked.
Meta-Analysis of the Prevalence of GS1 and GS2 C. concisus in Isolated Oral and Enteric C. concisus Strains in Publicly Available Databases
A literature search was performed in January 2017 and relevant publications were initially identified using the searching terms “Campylobacter concisus” in PubMed and Web of Science. The identified publications were then refined using searching terms “Campylobacter concisus isolation,” “Campylobacter concisus genomospecies,” and “Campylobacter concisus genotypic.” The identified publications following the refined search were manually inspected and studies containing defined C. concisus genomospecies were included in the analysis. In these studies, amplified fragment length polymorphism (AFLP), PCR targeting 23S rRNA gene, sequences of housekeeping genes and sequences of C. concisus core genomes were used for C. concisus genomospecies identification (Aabenhus et al., 2005; Engberg et al., 2005; Kalischuk and Inglis, 2011; On et al., 2013; Mahendran et al., 2015; Chung et al., 2016; Nielsen et al., 2016). These methods were consistent in defining GS1 and GS2 C. concisus (Aabenhus et al., 2005; Engberg et al., 2005; Kalischuk and Inglis, 2011; On et al., 2013; Mahendran et al., 2015; Chung et al., 2016; Nielsen et al., 2016). Given this, GS1 and GS2 C. concisus strains defined by these methods were combined and used for analysis of the prevalence of GS1 and GS2 C. concisus strains.
Statistical Analysis
Fisher's exact test (two tailed) was used to compare the prevalence of GS1 and GS2 C. concisus. Unpaired t-test was used to compare the copy numbers of GS1 and GS2 C. concisus DNA. P < 0.05 was considered statistically significant.
Results
C. concisus GS1 and GS2 Specific Polymorphisms in 23S rRNA Gene
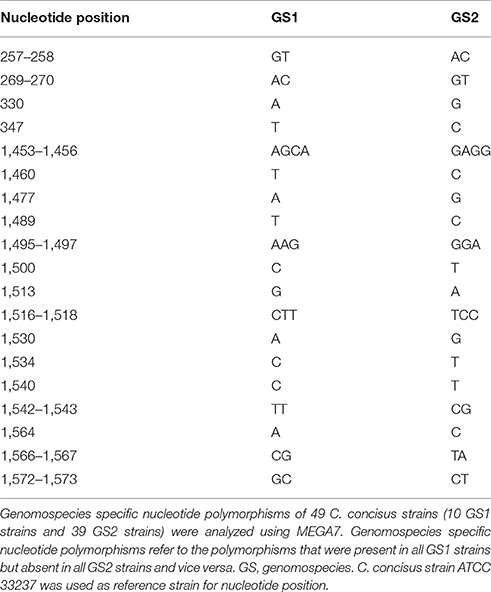
In this study, the genomes of 13 C. concisus strains were sequenced, of which 11 strains were GS2 and two strains were GS1 based on the phylogenetic trees generated using the 23S rRNA genes and housekeeping genes (Supplementary Figure S1). Comparison of the sequences of the 23S rRNA gene of 49 C. concisus strains including those sequenced in this study and publicly available genomes (10 GS1 strains and 39 GS2 strains) revealed 31 GS-specific polymorphisms, at positions 257–258, 269–270, 330, 347, 1,453–1,456, 1,460, 1,477, 1,489, 1,495–1,497, 1,500, 1,513, 1,516–1,518, 1,530, 1,534, 1,540, 1,542–1,543, 1,564, 1,566–1,567, and 1,572–1,573 bp (Table 2, Supplementary Figure S2). Most of the genomospecies-specific polymorphic nucleotides (81%, 25/31) were in the region between 1,453 and 1,575 bp. In addition to the 25 GS-specific nucleotides, this region also contained 22 polymorphic nucleotides that were either strain-specific or occurred in a number of strains. In total, this region contained 47 (38%, 47/123) polymorphic nucleotides (Table 2, Supplementary Figure S2).
The Specificities of GS1-PCR and GS2-PCR and the Amplification Efficiencies of GS1-rtPCR and GS2-rtPCR
The specificities of GS1-PCR and GS2-PCR were confirmed using DNA extracted from 41 C. concisus strains with known GS1 and GS2 identity. GS1-PCR was positive for all 8 GS1 strains and negative for all 33 GS2 strains. GS2-PCR was positive for all 33 GS2 strains and negative for the 8 GS1 strains.
The positive PCR products revealed a single band on agarose gel with the expected sizes (Supplementary Figure S3A). Sequencing the positive PCR products confirmed the identity of C. concisus 23S rRNA gene.
The amplification efficiencies of GS1-rtPCR and GS2-rtPCR were 93.4 and 95.4%, respectively. No primer dimer peaks were observed.
Detection and Quantification of GS1 and GS2 C. concisus in Saliva, Fecal Samples, and Intestinal Biopsies from Patients with IBD and Healthy Controls that were Positive for both GS1 and GS2 C. concisus
The DNA samples extracted from 27 saliva samples, nine fecal samples and 20 intestinal biopsies were all positive for both GS1-PCR and GS2-PCR. These DNA samples were then subjected to GS1-rtPCR and GS2-rtPCR respectively for quantitative measurement of GS1 and GS2 C. concisus.
The majority of the saliva DNA samples (25/27, 92.6%) contained significantly higher copy numbers of GS2 than GS1 C. concisus, which were seen in both patients with IBD (92.3%, 12/13) and healthy controls (92.9%, 13/14; Table 3). The mean GS2/GS1 ratios in saliva samples from patients with IBD were 16 ± 24 and 22 ± 28, respectively, which was not significantly different (P > 0.05).
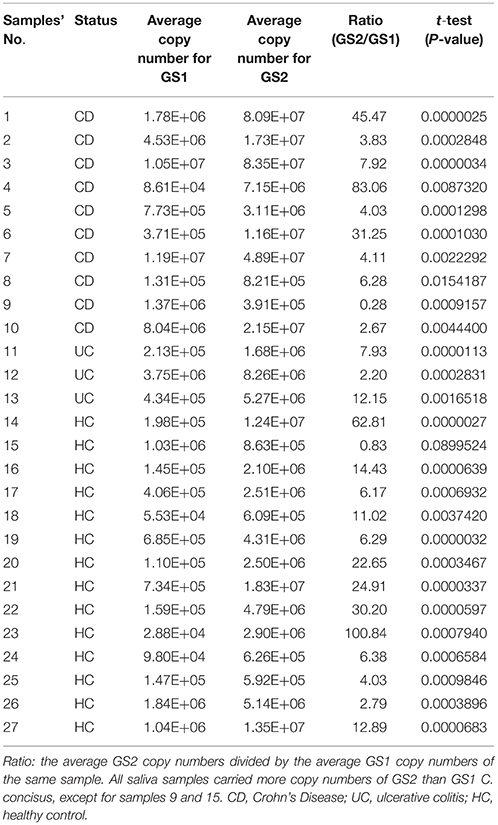
Table 3. Quantification of GS1 and GS2 C. concisus in saliva samples from patients with IBD and healthy controls.
All 29 enteric DNA samples (20 intestinal biopsies and nine fecal samples) contained significantly higher copy numbers of GS2 than GS1 C. concisus (Table 4). The average GS2/GS1 ratios of both intestinal biopsies and fecal samples between patients with IBD and healthy controls were not significantly different (129 ± 71 vs. 131 ± 119 and 62 ± 44 vs. 52 ± 22, respectively, P > 0.05).
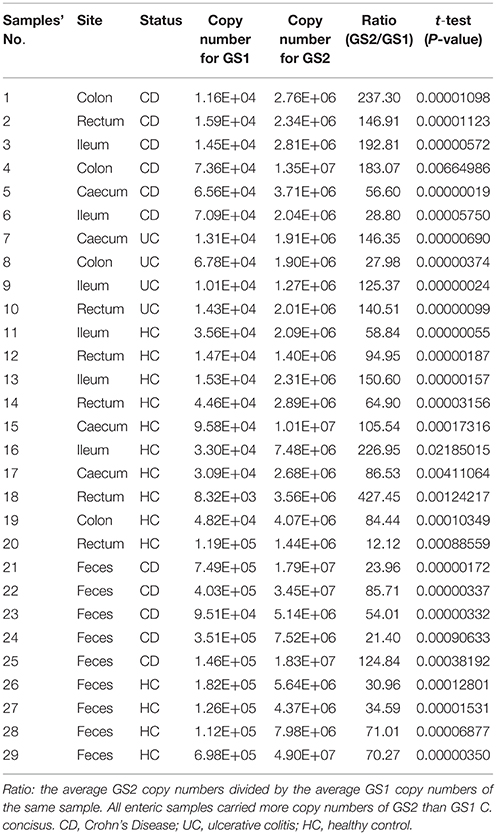
Table 4. Quantification of GS1 and GS2 C. concisus in enteric samples from patients with IBD and healthy controls.
Given that the average GS2/GS1 ratios in saliva, intestinal biopsies and fecal samples between patients with IBD and healthy controls were not significantly different, samples from patients with IBD and controls were combined to compare the GS2/GS1 ratios between samples collected from the upper and lower human gastrointestinal tract. The average GS2/GS1 ratio in the 20 intestinal biopsies was 130 ± 95, which was significantly higher than that of the 27 saliva samples (19 ± 26, P < 0.001) and that of the nine fecal samples (57 ± 34, P < 0.05). The average GS2/GS1 ratio in the fecal samples was significantly higher than that of the saliva samples (P < 0.01; Figure 1).
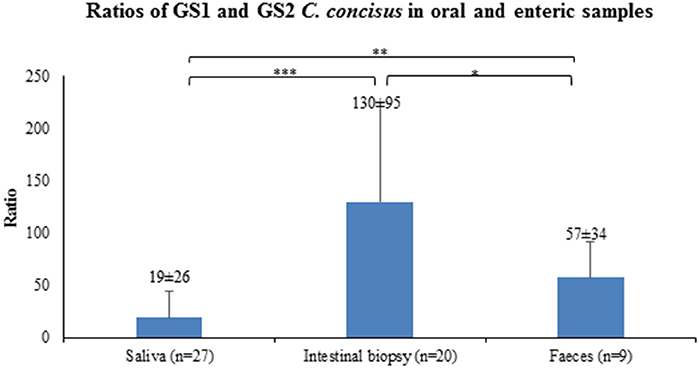
Figure 1. GS2/GS1 C. concisus ratios in oral and enteric samples. The copy numbers of GS1 and GS2 C. concisus determined by quantitative PCR methods were used to calculate the GS2/GS1 ratios in the oral and enteric samples. The average GS2/GS1 ratio in intestinal biopsy samples was 130 ± 95, which was significantly higher than that of the saliva samples (19 ± 26) and fecal samples (57 ± 34). The GS2/GS1 ratio in fecal samples was 57 ± 34, which was significantly higher than that of the saliva samples (19 ± 26). *Indicates statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
Meta-Analysis of the Prevalence of GS1 and GS2 C. concisus in Isolated Oral and Enteric C. concisus Strains
Initial literature search using term “Campylobacter concisus” revealed a total of 149 publications in PubMed and 189 publications in Web of Science. Searching terms “Campylobacter concisus isolation” Campylobacter concisus genomospecies” and “Campylobacter concisus genotypic” were used to further refine the results. According to the criteria described in Methods Section, a total of 260 C. concisus strains obtained from seven original research publications were included in the analysis of the prevalence of GS1 and GS2 C. concisus, of which 194 strains were isolated from fecal samples, nine strains from intestinal biopsies and 57 strains from the oral cavity (Table 5).
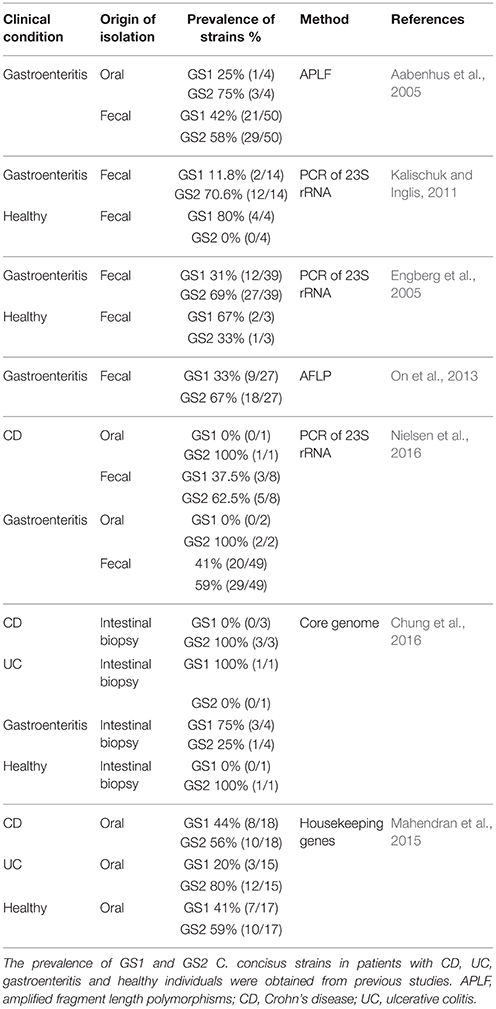
Table 5. Prevalence of GS1 and GS2 C. concisus strains in isolated oral and enteric C. concisus strains from previous studies.
Of the C. concisus strains isolated from saliva samples, there were more GS2 than GS1 strains and the prevalence of GS2 strains was significantly higher than GS1 strains is strains isolated from patients with IBD (68 vs. 32%, P < 0.01; Figure 2A). The prevalence of GS2 and GS1 strains in oral strains isolated from patients with IBD, healthy controls, and gastroenteritis was not significantly different.
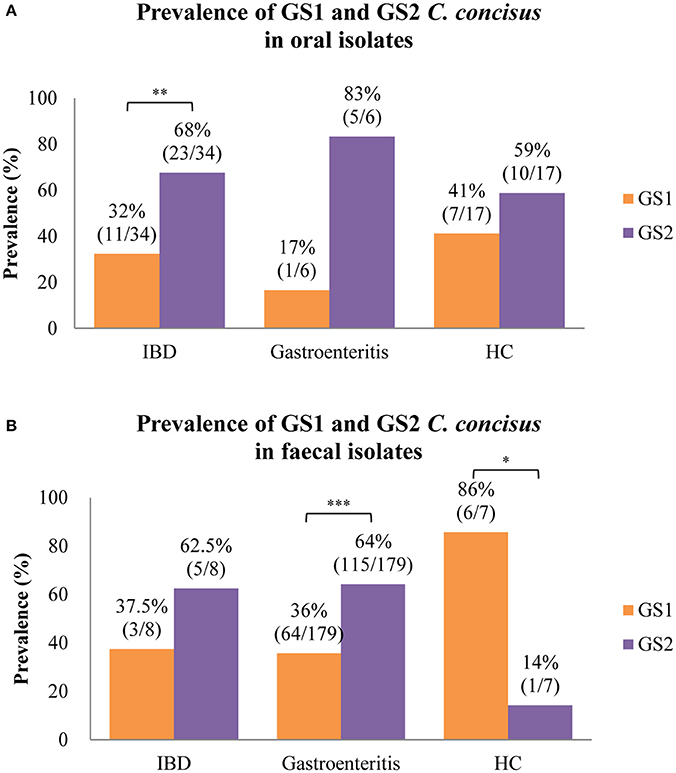
Figure 2. Meta-analysis of the prevalence of GS1 and GS2 C. concisus strains isolated from oral and fecal samples in patients with IBD, gastroenteritis, and healthy controls. (A) GS2 C. concisus has a higher prevalence in the oral cavity of both patients with enteric diseases and healthy controls as compared to GS1 C. concisus. (B) Most of the strains isolated from fecal samples of patients with enteric diseases were GS2 C. concisus, while most of those from healthy controls were GS1 C. concisus. IBD, inflammatory bowel disease. *Indicates statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001). These C. concisus strains were reported in previous studies.
The C. concisus strains isolated from fecal samples from patients with IBD and gastroenteritis also had more GS2 than GS1 C. concisus strains and the prevalence of GS2 in strains isolated from patients with gastroenteritis was significantly higher than the GS1 strains (64 vs. 36%, P < 0.001; Figure 2B). There was less GS2 than GS1 in C. concisus strains isolated from fecal samples from healthy individuals, and the difference was statistically significant (14 vs. 86%, P < 0.05; Figure 2B).
Up until now, only small numbers C. concisus strains isolated from intestinal biopsies had known GS1 and GS2 identities, including one strain from a patient with UC, three strains from patients with CD, four strains from patients with gastroenteritis and one strain from a healthy individual (Table 4). Given the small strain numbers, we did not analyse the prevalence of GS1 and GS2 in C. concisus strains isolated from intestinal biopsies.
Discussion
In this study, we systematically examined the colonization of GS1 and GS2 C. concisus in the human gastrointestinal tract using both quantitative PCR methods and a meta-analysis of isolated C. concisus strains in publicly available publications.
This study firstly identified C. concisus GS1 and GS2 specific polymorphic nucleotides in the 23S rRNA gene. In a recent study, we showed that the sequences of C. concisus core-genome, six housekeeping genes and 23S rRNA gene consistently divided 36 C. concisus strains into two genomospecies (Chung et al., 2016). The finding that the sequences of 23S rRNA gene were able to differentiate GS1 and GS2 C. concisus strains suggests that the C. concisus 23S rRNA gene contains C. concisus genomospecies specific polymorphisms. Indeed, comparison of the sequences of 23S rRNA gene of 49 GS1 and GS2 C. concisus strains conducted in this study identified multiple GS1 and GS2 specific polymorphic nucleotides in the 23S rRNA gene, most of which were in the highly polymorphic region 1,453–1,575 bp (Table 2, Supplementary Figure S2). The GS1 and GS2 multiple specific polymorphic nucleotides provide the molecular base of using this gene to assign GS1 and GS2 C. concisus strains (Chung et al., 2016).
We then designed and validated specific PCR methods for GS1 and GS2 C. concisus detection and quantification. CON1, CON2, and MUC1 are PCR primers designed by Bastyns et al. aiming to detect C. concisus by amplification of 23S rRNA gene (Bastyns et al., 1995). A number of studies used these PCR primers to define C. concisus genomospecies (Bastyns et al., 1995; Engberg et al., 2005; Kalischuk and Inglis, 2011; Nielsen et al., 2016). In our preliminary experiments, we attempted to use CON1, CON2, and MUC1 primers in rtPCR using SYBR Green I method to quantify GS1 and GS2 C. concisus. However, primer dimers were frequently observed when using CON1, CON2, and MUC1 for examination of GS1 and GS2 C. concisus in enteric samples, interfering with the interpretation of the results. We therefore designed and validated two new PCR methods for detection of GS1 and GS2 C. concisus, respectively. The newly designed PCR methods are specific in differentiating GS1 and GS2 strains, as demonstrated using C. concisus strains with known GS1 and GS2 identity. Furthermore, these PCR primers were suitable for rtPCR to quantify GS1 and GS2 C. concisus using SYBR Green I method; no primer dimers were seen and the amplification efficiencies were 93.4 and 95.4%, respectively (Supplementary Figure S3B). Using the newly developed PCR methods, we quantified GS1 and GS2 C. concisus in the upper and lower human gastrointestinal tract using oral and enteric samples previously collected from patients with IBD and controls.
Using quantitative PCR methods, we found that GS2 C. concisus is better adapted the human gastrointestinal tract than GS1 C. concisus. The copy numbers of GS2 C. concisus were significantly higher than GS1 C. concisus in most of the saliva samples and all enteric samples (Tables 3, 4). We previously found that GS2 and GS1 C. concisus strains have genomospecies-specific genes; one such gene specific to GS2 strains is aquaporin-Z (Chung et al., 2016). Aquaporin-Z is a bacterial membrane-bound water channel protein, which has been shown to mediate entry or exit of water in response to large changes in extracellular osmolarity (Delamarche et al., 1999). The osmolarity in the human gastrointestinal tract varies greatly and rapidly due to the uptake of large variety of liquids or solid food. The aquaporin-Z water channel may have contributed to the better adaption of GS2 C. concisus in the human gastrointestinal tract.
It was found that there were significantly higher GS2/GS1 ratios in intestinal biopsies than the oral cavity and intestinal lumen (Figure 1). This may be due to the environment near the intestinal epithelium being less favorable for GS1 C. concisus to grow as compared to the oral cavity and the intestinal lumen. Previously, we found that some C. concisus strains belonging to both GS1 and GS2, acquired virulence genes from bacteriophages or plasmids (Mahendran et al., 2013; Liu et al., 2016). Given that C. concisus detected in intestinal biopsies was predominantly GS2, GS2 C. concisus strains that have virulence genes are more likely to cause enteric diseases involving damaging the intestinal epithelial cells.
We also analyzed the composition of isolated GS1 and GS2 C. concisus strains in previously reported studies (Table 4). C. concisus strains isolated from the human oral cavity, including both individuals with enteric diseases and healthy individuals, all had more GS2, with the difference in patients with IBD being statistically significant (Figure 2A). Thus, the molecular methods and bacterial isolation consistently detected more GS2 than GS1 C. concisus in the human oral cavity. C. concisus strains isolated from the fecal samples of patients with IBD and gastroenteritis also had more GS2 than GS1 C. concisus strains, consistent with GS2/GS1 ratios detected using molecular methods in the fecal samples from patients with IBD in this study (Figure 2B, Table 4). A significantly higher GS1 than GS2 C. concisus was isolated from fecal samples of healthy controls, which was different from the C. concisus strains isolated from the oral samples and the detection by molecular methods (Figure 2, Table 4). One possible explanation is that the GS2 C. concisus strains in the oral cavity of the healthy individuals are less resistant to the enteric environment as compared to that of the patients with enteric diseases. Alternatively, the enteric environment of patients with IBD and gastroenteritis may be different from that of the healthy controls, the latter may be less favorable to the growth of GS2 C. concisus strains which in turn affects the isolation rate of GS2 from fecal samples. Up until now, only three C. concisus strains isolated from intestinal tissues of patient with CD had known GS1 and GS2 identities, all were GS2 strains (Table 4). Kirk et al. isolated larger numbers of C. concisus strains from intestinal tissues of patients with IBD and healthy controls, however the GS1 and GS2 identities of these strains were not reported (Kirk et al., 2016). Given the small numbers of C. concisus strains isolated from intestinal biopsies with known GS1 and GS2 identities, we did not compare their GS 1 and GS2 composition with what was detected by the quantitative PCR methods.
In summary, this study identified C. concisus GS1 and GS2 specific polymorphisms in 23S rRNA gene and C. concisus GS1- and GS2-specific PCR methods were developed. Using these PCR methods, GS1 and GS2 C. concisus in samples collected from the upper and lower human gastrointestinal tract for the first time were quantified. We also analyzed the GS1 and GS2 composition of the isolated C. concisus strains in previous studies. Based on the quantitative PCR methods, there was more GS2 than GS1 C. concisus in samples collected from the upper and lower gastrointestinal tract of both patients with IBD and healthy controls, showing that GS2 C. concisus is better adapted to the human gastrointestinal tract. Analysis of GS1 and GS2 composition of the isolated C. concisus strains in previous studies showed similar findings except that in healthy individuals a significantly lower GS2 than GS1 C. concisus strains were isolated from fecal samples, suggesting a potential difference in the C. concisus strains or the enteric environment between patients with gastrointestinal diseases and healthy controls. This study provides novel information regarding the adaptation of different genomospecies of C. concisus in the human gastrointestinal tract.
Ethics Statement
The ethics approval was granted by the Ethics Committees of the University of New SouthWales and the South East Sydney Area Health Service, Australia [HREC 09237/SESIAHS 09/078 and HREC08335/SESIAHS (CHN)07/48]. Written informed consent was obtained from all subjects.
Author Contributions
Conceived and designed the experiments: LZ, YW, FL, SR, MG. Performed the experiments: YW, FL, HC, RM, SL. Analyzed the data: YW, FL XZ, SZ. Wrote the paper: YW, FL, LZ, XZ, HC, SR, MG, SZ, and RM. All authors have approved the final version of the manuscript.
Funding
This work is supported by a Faculty Research Grant awarded to LZ from the University of New South Wales (Grant No: PS35329).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2017.00543/full#supplementary-material
Availability of Data and Materials
The dataset supporting the conclusions of this article is included within the article.
Supplementary Figure 1. Phylogenetic trees generated based on 23S rRNA gene and housekeeping genes for 13 C. concisus strains sequenced in this study. The phylogenetic trees of 23S rRNA gene (A) and six housekeeping genes (asd, aspA, atpA, glnA, pgi, and tkt) (B) were generated using maximum likelihood method. C. concisus strains were consistently divided into two genomospecies (GS). C. concisus strains isolated from patients with CD, UC, and healthy controls were colored in red, blue, and green, respectively. C. concisus strains ATCC 33237 and 13826 were used as representative strains for GS1 and GS2, respectively. Bootstrap values higher than 70 were shown. Campylobacter jejuni strain NCTC 11168 was used as outgroups.
Supplementary Figure 2. Comparison of 23S rRNA gene from 49 C. concisus strains. The 23S rRNA gene sequences were compared using MEGA7 and aligned using Clustal Omega (Sievers et al., 2011). GS1 are shaded in gray. Genomospecies specific nucleotide polymorphisms are boxed. PCR primers used in this study are shaded in blue.
Supplementary Figure 3. Validation of GS1-PCR and GS2-PCR specificities and comparison of previously published PCR primers with primers designed in this study in use for GS1-rtPCR and GS2-rtPCR. (A) The positive PCR products obtained from GS1-PCR and GS-2 PCR revealed a single band on agarose gel with expected sizes, which were confirmed to be C. concisus 23S rRNA gene by sequencing. −, Negative control (No DNA template). S, Sample. +, Positive control (DNA template that carried GS1 or GS2 C. concisus DNA) (B) Primer dimers were frequently observed in GS1-rtPCR and GS2-rtPCR when previously published primers were used (right). Primer dimers were absent when primers designed in this study were used (left).
Abbreviations
IBD, inflammatory bowel disease; CD, Crohn's disease; UC, ulcerative colitis; GS, genomospecies; GS1-PCR, Polymerase chain reaction for C. concisus genomospecies 1; GS2-PCR, Polymerase chain reaction for C. concisus genomospecies 2.
References
Aabenhus, R., On, S. L., Siemer, B. L., Permin, H., and Andersen, L. P. (2005). Delineation of Campylobacter concisus genomospecies by amplified fragment length polymorphism analysis and correlation of results with clinical data. J. Clin. Microbiol. 43, 5091–5096. doi: 10.1128/JCM.43.10.5091-5096.2005
Bastyns, K., Chapelle, S., Vandamme, P., Goossens, H., and De Wachter, R. (1995). Specific detection of Campylobacter concisus by PCR amplification of 23S rDNA areas. Mol. Cell. Probes 9, 247–250. doi: 10.1016/S0890-8508(95)90114-0
Chung, H. K., Tay, A., Octavia, S., Chen, J., Liu, F., Ma, R., et al. (2016). Genome analysis of Campylobacter concisus strains from patients with inflammatory bowel disease and gastroenteritis provides new insights into pathogenicity. Sci. Rep 6:38442. doi: 10.1038/srep38442
Delamarche, C., Thomas, D., Rolland, J. P., Froger, A., Gouranton, J., Svelto, M., et al. (1999). Visualization of AqpZ-mediated water permeability in Escherichia coli by cryoelectron microscopy. J. Bacteriol. 181, 4193–4197.
Deshpande, N. P., Kaakoush, N. O., Wilkins, M. R., and Mitchell, H. M. (2013). Comparative genomics of Campylobacter concisus isolates reveals genetic diversity and provides insights into disease association. BMC Genomics 14:585. doi: 10.1186/1471-2164-14-585
Dhanasekaran, S., Doherty, T. M., Kenneth, J., and Group, T. B. T. S. (2010). Comparison of different standards for real-time PCR-based absolute quantification. J. Immunol. Methods 354, 34–39. doi: 10.1016/j.jim.2010.01.004
Engberg, J., Bang, D. D., Aabenhus, R., Aarestrup, F. M., Fussing, V., and Gerner-Smidt, P. (2005). Campylobacter concisus: an evaluation of certain phenotypic and genotypic characteristics. Clin. Microbiol. Infect. 11, 288–295. doi: 10.1111/j.1469-0691.2005.01111.x
Fite, A., MacFarlane, G. T., Cummings, J. H., Hopkins, M. J., Kong, S. C., Furrie, E., et al. (2004). Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut 53, 523–529. doi: 10.1136/gut.2003.031245
Ismail, Y., Lee, H., Riordan, S. M., Grimm, M. C., and Zhang, L. (2013). The effects of oral and enteric Campylobacter concisus strains on expression of TLR4, MD-2, TLR2, TLR5 and COX-2 in HT-29 cells. PLoS ONE 8:e56888. doi: 10.1371/journal.pone.0056888
Ismail, Y., Mahendran, V., Octavia, S., Day, A. S., Riordan, S. M., Grimm, M. C., et al. (2012). Investigation of the enteric pathogenic potential of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease. PLoS ONE 7:e38217. doi: 10.1371/journal.pone.0038217
Istivan, T. S., Coloe, P. J., Fry, B. N., Ward, P., and Smith, S. C. (2004). Characterization of a haemolytic phospholipase A(2) activity in clinical isolates of Campylobacter concisus. J. Med. Microbiol. 53, 483–493. doi: 10.1099/jmm.0.45554-0
Kalischuk, L., and Inglis, G. (2011). Comparative genotypic and pathogenic examination of Campylobacter concisus isolates from diarrheic and non-diarrheic humans. BMC Microbiol. 11:53. doi: 10.1186/1471-2180-11-53
Kirk, K. F., Nielsen, H. L., Thorlacius-Ussing, O., and Nielsen, H. (2016). Optimized cultivation of Campylobacter concisus from gut mucosal biopsies in inflammatory bowel disease. Gut Pathog. 8, 27. doi: 10.1186/s13099-016-0111-7
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lastovica, A. J., On, S. L., and Zhang, L. (2014). “The Family Campylobacteraceae,” in The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer), 307–335.
Lee, H., Ma, R., Grimm, M. C., Riordan, S. M., Lan, R., Zhong, L., et al. (2014). Examination of the anaerobic growth of Campylobacter concisus strains. Int. J. Microbiol. 2014:476047. doi: 10.1155/2014/476047
Lindblom, G., Sjogren, E., Hansson-Westerberg, J., and Kaijser, B. (1995). Campylobacter upsaliensis, C. sputorum sputorum and C. concisus as common causes of diarrhoea in Swedish children. Scand. J. Infect Dis. 27, 187–188. doi: 10.3109/00365549509019006
Liu, F., Lee, H., Lan, R., and Zhang, L. (2016). Zonula occludens toxins and their prophages in Campylobacter species. Gut. Pathog. 8, 1–11. doi: 10.1186/s13099-016-0125-1
Liu, F., Ma, R., Riordan, S. M., Grimm, M. C., Liu, L., Wang, Y., et al. (2017). Azathioprine, mercaptopurine, and 5-aminosalicylic acid affect the growth of IBD-associated Campylobacter species and other enteric microbes. Front. Microbiol. 8:527. doi: 10.3389/fmicb.2017.00527
Ma, R., Sapwell, N., Chung, H. K., Lee, H., Mahendran, V., Leong, R. W., et al. (2015). Investigation of the effects of pH and bile on the growth of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease and controls. J. Med. Microbiol. 64, 438–445. doi: 10.1099/jmm.0.000013
Mahendran, V., Liu, F., Riordan, S., Grimm, M., Tanaka, M., and Zhang, L. (2016). Examination of the effects of Campylobacter concisus zonula occludens toxin on intestinal epithelial cells and macrophages. Gut. Pathog. 8, 18. doi: 10.1186/s13099-016-0101-9
Mahendran, V., Octavia, S., Demirbas, O. F., Sabrina, S., Ma, R., Lan, R., et al. (2015). Delineation of genetic relatedness and population structure of oral and enteric Campylobacter concisus strains by analysis of housekeeping genes. Microbiology 161, 1600–1612. doi: 10.1099/mic.0.000112
Mahendran, V., Riordan, S. M., Grimm, M. C., Tran, T. A., Major, J., Kaakoush, N. O., et al. (2011). Prevalence of Campylobacter species in adult Crohn's disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS ONE 6:e25417. doi: 10.1371/journal.pone.0025417
Mahendran, V., Tan, Y. S., Riordan, S. M., Grimm, M. C., Day, A. S., Lemberg, D. A., et al. (2013). The prevalence and polymorphisms of zonula occluden toxin gene in multiple Campylobacter concisus strains isolated from saliva of patients with inflammatory bowel disease and controls. PLoS ONE 8:e75525. doi: 10.1371/journal.pone.0075525
Miller, W. G., Chapman, M. H., Yee, E., On, S. L., McNulty, D. K., Lastovica, A. J., et al. (2012). Multilocus sequence typing methods for the emerging Campylobacter Species C. hyointestinalis, C. lanienae, C. sputorum, C. concisus, and C. curvus. Front. Cell Infect. Microbiol. 2:45. doi: 10.3389/fcimb.2012.00045
Mukhopadhya, I., Thomson, J. M., Hansen, R., Berry, S. H., El-Omar, E. M., and Hold, G. L. (2011). Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS ONE 6:e21490. doi: 10.1371/journal.pone.0021490
Nielsen, H., Ejlertsen, T., Engberg, J., and Nielsen, H. (2013). High incidence of Campylobacter concisus in gastroenteritis in North Jutland, Denmark: a population-based study. Clin. Microbiol. Infect. 19, 445–450. doi: 10.1111/j.1469-0691.2012.03852.x
Nielsen, H. L., Nielsen, H., Ejlertsen, T., Engberg, J., Günzel, D., Zeitz, M., et al. (2011). Oral and fecal Campylobacter concisus strains perturb barrier function by apoptosis induction in HT-29/B6 intestinal epithelial cells. PLoS ONE 6, e23858. doi: 10.1371/journal.pone.0023858
Nielsen, H. L., Nielsen, H., and Torpdahl, M. (2016). Multilocus sequence typing of Campylobacter concisus from Danish diarrheic patients. Gut Pathog. 8, 44. doi: 10.1186/s13099-016-0126-0
On, S., Siemer, B., Chung, P., and Lastovica, A. (2013). Characterisation of Campylobacter concisus strains from South Africa using amplified fragment length polymorphism (AFLP) profiling and a genomospecies-specific polymerase chain reaction (PCR) assay: identification of novel genomospecies and correlation with clinical data. Afr. J. Microbiol Res. 7, 1845–1851. doi: 10.5897/AJMR12.2182
Sartor, R. B., and Mazmanian, S. K. (2012). Intestinal microbes in inflammatory bowel diseases. Am. J. Gastroenterol. Suppl. 1, 15–21. doi: 10.1038/ajgsup.2012.4
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W. Z., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Tanner, A. C. R., Badger, S., Lai, C., Listgarten, M. A., Visconti, R. A., and Socransky, S. S. (1981). Wolinella gen. nov., WoZinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and Description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from Humans with Periodontal Disease. Int. J. Syst. Bacteriol. 31, 432–445. doi: 10.1099/00207713-31-4-432
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:1. doi: 10.1186/1471-2105-13-134
Zhang, L. (2015). Oral Campylobacter species: initiators of a subgroup of inflammatory bowel disease? World J. Gastroenterol. 21, 9239–9244. doi: 10.3748/wjg.v21.i31.9239
Zhang, L., Budiman, V., Day, A. S., Mitchell, H., Lemberg, D. A., Riordan, S. M., et al. (2010). Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J. Clin. Microbiol. 48, 2965–2967. doi: 10.1128/JCM.02391-09
Zhang, L., Lee, H., Grimm, M. C., Riordan, S. M., Day, A. S., and Lemberg, D. A. (2014). Campylobacter concisus and inflammatory bowel disease. World J. Gastroenterol. 20, 1259–1267. doi: 10.3748/wjg.v20.i5.1259
Keywords: Campylobacter concisus, Crohn's disease, inflammatory bowel disease, 23S rRNA, genomospecies
Citation: Wang Y, Liu F, Zhang X, Chung HKL, Riordan SM, Grimm MC, Zhang S, Ma R, Lee SA and Zhang L (2017) Campylobacter concisus Genomospecies 2 Is Better Adapted to the Human Gastrointestinal Tract as Compared with Campylobacter concisus Genomospecies 1. Front. Physiol. 8:543. doi: 10.3389/fphys.2017.00543
Received: 17 May 2017; Accepted: 12 July 2017;
Published: 03 August 2017.
Edited by:
Stephen J. Pandol, Cedars-Sinai Medical Center, United StatesReviewed by:
Adonis Sfera, Loma Linda University, United StatesShin Hamada, Tohoku University, Japan
Copyright © 2017 Wang, Liu, Zhang, Chung, Riordan, Grimm, Zhang, Ma, Lee and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Zhang, l.zhang@unsw.edu.au
†These authors have contributed equally to this work.
 Yiming Wang
Yiming Wang Fang Liu
Fang Liu Xiang Zhang2
Xiang Zhang2 Heung Kit Leslie Chung
Heung Kit Leslie Chung Michael C. Grimm
Michael C. Grimm Li Zhang
Li Zhang