- 1Tsinghua-Peking Joint Center for Life Sciences, and MOE Key Laboratory of Bioinformatics, School of Life Sciences, Tsinghua University, Beijing, China
- 2Beijing Key Laboratory of Plant Gene Resources and Biotechnology for Carbon Reduction and Environmental Improvement, College of Life Sciences, Capital Normal University, Beijing, China
- 3The Institute of Plant Quarantine, Chinese Academy of Inspection and Quarantine, Beijing, China
RNA intereferencing (RNAi) pathway regulates antiviral immunity and mediates plant growth and development. Despite considerable research efforts, a few components in RNAi pathway have been revealed, including ARGONAUTEs (AGOs), DICER-LIKEs (DCLs), RNA-dependent RNA polymerase 1 and 6 (RDR1/6), and ALTERED MERISTEM PROGRAM 1 (AMP1). In this study, we performed a forward genetic screening for enhancers of rdr6 via inoculation of CMV2aTΔ2b, a 2b-deficient Cucumber Mosaic Virus that is unable to suppress RNAi-mediated antiviral immunity. We uncover that the membrane-localized flippase Aminophospholipid ATPase 1 (ALA1) cooperates with RDR6 and RDR1 to promote antiviral immunity and regulate fertility in Arabidopsis. Moreover, we find that ALA2, a homolog of ALA1, also participates in antiviral immunity. Our findings suggest that ALA1 and ALA2 act as novel components in the RNAi pathway and function additively with RDR1 and RDR6 to mediate RNAi-based antiviral immunity and plant development.
Introduction
RNA interference (RNAi) mediates plant defense against virus infections (Ding et al., 2004; Incarbone and Dunoyer, 2013; Martinez de Alba et al., 2013). DICER-LIKE ribonucleases (DCLs), such as DCL4, generate viral short interferencing RNAs (siRNAs) (Blevins et al., 2006; Parent et al., 2015), which direct the loading of viral RNAs into ARGONAUTE (AGO) proteins (e.g., AGO1) of the RNA-induced silencing complex (RISC) for the cleavage of viral RNAs (Morel, 2002; Adenot et al., 2006; Arribas-Hernandez et al., 2016), resulting in RNAi-mediated antiviral immunity. RNA-dependent RNA polymerases (RDRs) (Xie et al., 2001; Talmor-Neiman et al., 2006; Cao et al., 2014), including RDR1 and RDR6, promote synthesis of siRNAs by synthesizing long double-strand RNAs (dsRNAs), contributing to the antiviral immunity (Qu et al., 2008; Garcia-Ruiz et al., 2010).
Viruses in turn evolve viral suppressor of RNAi (VSR) to suppress host antiviral immunity. For example, Cucumber Mosaic Virus (CMV) utilizes the VSR protein 2b (Zhang et al., 2006; Diaz-Pendon et al., 2007) to suppress host RNAi-based antiviral immunity and causes severe pathogenic responses in wild-type Arabidopsis, while CMV2aTΔ2b, a CMV mutant without expression of 2b protein, is unable to cause any obvious viral symptoms in wild-type and the single mutants of RDR1 or RDR6, but is able to cause severe pathogenic responses in the RNAi-deficient double mutant rdr1 rdr6 (Wang et al., 2010).
Aminophospholipid transporting ATPases (ALAs) are membrane-localized flippases that are responsible for transporting different lipids, which is essential for asymmetry of membrane lipid bilayers (Lopez-Marques et al., 2010, 2012; Botella et al., 2016). There are 12 Arabidopsis thaliana ALAs in the IV subfamily of ATPases that control plant development or tolerance to temperature stresses (Lopez-Marques et al., 2014). ALA1 is required for plant tolerance to chilling (Gomes et al., 2000). ALA3 regulates pollen germination and pollen tube growth, and adaptability to chilling (Poulsen et al., 2008; McDowell et al., 2013). ALA6 and ALA7 control temperate-regulated pollen tube elongation (McDowell et al., 2015). ALA10 affects lipid uptake to regulate root growth and stomatal control (Botella et al., 2016).
In this study, we performed a forward genetic screening for enhancers of rdr6 with CMV2aTΔ2b infection on M2 population of ethyl methanesulfonate (EMS)-mutagenized rdr6. We show that ALA1 and ALA2 act additively with RDR1 and RDR6 to mediate RNAi-mediated antiviral immunity and development. Our findings discover novel roles of ALA1 and ALA2.
Materials and Methods
Materials and Growth Conditions
The Arabidopsis thaliana mutants ala1-2 (Salk_056947), ala3 (GK-317H04), ala7 (Salk_125598) and ala10 (Salk_024877) were obtained from Arabidopsis Biological Resource Center. The Arabidopsis mutants rdr1-1 (SAIL_672_F11), rdr6-15 (SAIL_617H07), rdr1 rdr6, the L1 line transgenic for GUS, and the 2b-deficient CMV mutant CMV2aTΔ2b were described as previously (Boutet et al., 2003; Wang et al., 2010). The ala1-2 rdr1, ala1-2 rdr6 and ala1-2 rdr1 rdr6 were generated via genetic crossing. Nicotiana benthamiana was grown under a 16-h (28°C)/8-h (22°C) light/dark condition.
For observation of growth defects in Figure 5C and fertility and siliques development in Figure 3, Arabidopsis seeds were sterilized with 20% bleach, plated on Murashige and Skoog (MS) medium, chilled at 4°C for 3 days, and transferred to a growth room under a 16-h (23–25°C)/8-h (18–20°C) light/dark photoperiod for 9 days. The 9-day-old seedlings were transplanted into soil and grew in the same growth room for another ∼3 or ∼6 weeks.
Viral Infection
For viral infection assays, Arabidopsis seedlings were sterilized, plated on MS medium, chilled at 4°C for 3 days, and transferred to a growth room under a 16-h (23–25°C)/8-h (18–20°C) light/dark photoperiod for 9 days. The 9-day-old seedlings were transplanted into soil for growth of another 14 days in a growth room under an 8-h (22–24°C)/16-h (16–19°C) light/dark photoperiod. The 23-day-old plants were inoculated with CMV2aTΔ2b as described previously (Wang et al., 2010), and the disease symptoms were recorded at 21 or 45 days after infection.
EMS Mutagenesis
About 20,000 seeds (M1) of the Arabidopsis mutant rdr6-15 (SAIL_617H07) were soaked with 100 mM phosphate buffer (pH 7.5) overnight at 4°C, washed with sterilized water for five times, and mutagenized with 0.6% ethyl methanesulfonate (EMS) dissolved in phosphate buffer for 8 h at room temperature. The mutagenized seeds were washed with sterilized water for 20 times, and were grown in soil for collection of M2 seeds.
Generation of Mutants and Transgenic Plants
Mutations at 698th (-), 1120th (+) and 2216th (+) bp of coding sequence (CDS) of ALA1 (Supplementary Figure 1B), and at the 951th (+) bp of CDS of ALA2 (Supplementary Figure 5B) were introduced into the rdr6 mutant through CRISPR/Cas9 method (Mao et al., 2013). The guide RNA of the CRISPR target was driven by U6 promoter, and Cas9 was under control of a CaMV35S promoter in a modified pCAMBIA1300 vector (Mao et al., 2013). Primers used for construction of vectors are listed in Supplementary Table 1. The constructs were introduced into rdr6 mutants through agrobacterium-mediated flower dip method. The transgenic seeds were selected on MS containing 20 mg/L hygromycin, T2 plants were inoculated with CMV2aTΔ2b. Mutations of ALA1 or ALA2 were confirmed by sequencing.
The CDS of ALA1 was cloned into the pCAMBIA1300 vector through SmaI and XbaI sites for fusion with three FLAG tags under the control of CaMV35S promoter, and introduced into the ala1-2 using agrobacterium-mediated flower dip method.
Whole-Genome Sequencing and Gene Cloning of ENOR Loci
The F2 population generated by crossing enor1 rdr6 or enor2 rdr6 with rdr6 were inoculated with CMV2aTΔ2b. One hundred susceptible plants from F2 population were harvested at 21 days after inoculation to generate a bulked pool for DNA extraction with DNeasy Plant Maxi Kit (QIAGEN, Cat. 68163) and construction of DNA library. Whole genome sequencing was performed with the illumina HiSeq2000 platform. The softwares Skewer, Bowtie2 and SHOREmap were used to analyze the data and isolate mutations (Schneeberger et al., 2009; Sun and Schneeberger, 2015). The SNP-based Cleaved Amplified Polymorphic Sequences (CAPSs) markers generated from comparison of genome sequences of enor1 rdr6 or enor2 rdr6 with rdr6 were used to assist mapping and cloning of ENOR1 and ENOR2.
Immunoblotting Analysis
The total proteins were extracted from plants at 21 days after inoculation with mock or CMV2aTΔ2b. Fifty microgram of total protein for each sample was quantified and loaded for detection of coat protein (CP) of CMV2aTΔ2b. The antibody against coat protein (anti-CP) of CMV2aTΔ2b was produced by Abmart company (Abmart1) with the recombinant protein of the 1st to 154th AA of CP. The anti-CP was used as first antibody (1:6000), and anti-rabbit immunoglobulin antibody was used as the secondary antibody respectively (1:3000). All of the experiments were repeated at least three biological times.
GUS Staining
The L1 line transgenic for the β-glucuronidase (GUS) gene driven by 35S promoter (35S::GUS), in which the GUS activity is very low in all the expanded rosette leaves due to the post transcriptional gene silencing (Boutet et al., 2003), was crossed with ala1-2 to generate ala1-2 with L1 transgene (35S::GUS), named ala1-2 35S::GUS. Eighteen ala1-2 35S::GUS plants were used for histochemical staining of GUS using the method described previously (Shan et al., 2011).
Quantitative Real-Time PCR Analysis
For Figure 6A, the expression of ALA family members was analyzed in Col-0 and ala1-2 at 21 days after inoculation with mock or CMV2aTΔ2b. For Supplementary Figure 4, the accumulation of genomic RNA of CMV2aTΔ2b was analyzed in Col-0 and ala1-2 at 21 days after CMV2aTΔ2b inoculation. The primers used for RNA detection of CMV2aTΔ2b were designed based on the conserved sequences from genomic RNA1 to RNA3 in the 3 prime end. The materials were harvested for RNA extraction using trizol (TRANSGENE, Cat.ET101-01), and reverse transcription was performed according to the kit (TRANSGENE, Cat. AT311-03). Quantitative real-time PCR was performed with EvaGreen 2∗qPCR MasterMix-Low ROX reagents (ABM, Cat. Mastermix-LR) using the ABI7500 real-time PCR system. ACTIN8 was used as the internal control. All of the experiments were repeated at least three biological times. Primers used for quantitative real-time PCR analysis are listed in Supplementary Table 1.
Phylogenetic Analysis
For the phylogenetic analysis shown in Supplementary Figure 6, the evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length (4.45679805) is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (Felsenstein, 1985). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The analysis involved all 12 amino acid sequences of ALA family. All positions containing gaps and missing data were eliminated. There were a total of 794 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). The transcripts, including ALA1 (AT5G04930.1), ALA2 (AT5G44240.1), ALA3 (AT1G59820.1), ALA4 (AT1G17500.1), ALA5 (AT1G72700.1), ALA6 (AT1G54280.1), ALA7 (AT3G13900.1), ALA8 (AT3G27870.1), ALA9 (AT1G68710.1), ALA10 (AT3G25610.1), ALA11 (AT1G13210.1), and ALA12 (AT1G26130.2), were used for phylogenetic analysis.
Subcellular Localization
Coding sequence of ALA1 was cloned into the pJG054 vector for fusion with YFP under control of CaMV35S promoter (YFP-ALA1). The agrobacterium containing YFP-ALA1 or the mCherry-ER-marker were resuspended in the infiltration buffer (10 mM MgCl2, 10 mM MES, 0.2 mM acetosyringone) for 3-5 h, and co-infiltrated into leaves of N. benthamiana. The fluorescence signals were collected with a Zeiss microscope (LSM710) at ∼50 h after co-infiltration. All of the experiments were repeated at least three biological times.
Accession Numbers
The Arabidopsis Genome Initiative numbers for genes mentioned in this letter are as follows: ALA1 (AT5G04930), ALA2 (AT5G44240), ALA3 (AT1G59820), ALA4 (AT1G17500), ALA5 (AT1G72700), ALA6 (AT1G54280), ALA7 (AT3G13900), ALA8 (AT3G27870), ALA9 (AT1G68710), ALA10 (AT3G25610), ALA11 (AT1G13210), ALA12 (AT1G26130), RDR1 (AT1G14790), RDR6 (AT3G49500), and ACTIN8 (AT1G49240).
Results
Identification and Mapping of the enor1 Mutant
We generated M2 population of EMS-mutagenized rdr6, and inoculated M2 with CMV2aTΔ2b to identify mutants that enhanced the susceptibility to CMV2aTΔ2b in rdr6 (referred to as enhancer of rdr6 [enor]), and utilized whole genome sequencing to assist mapping and cloning of ENOR loci (Figure 1A).
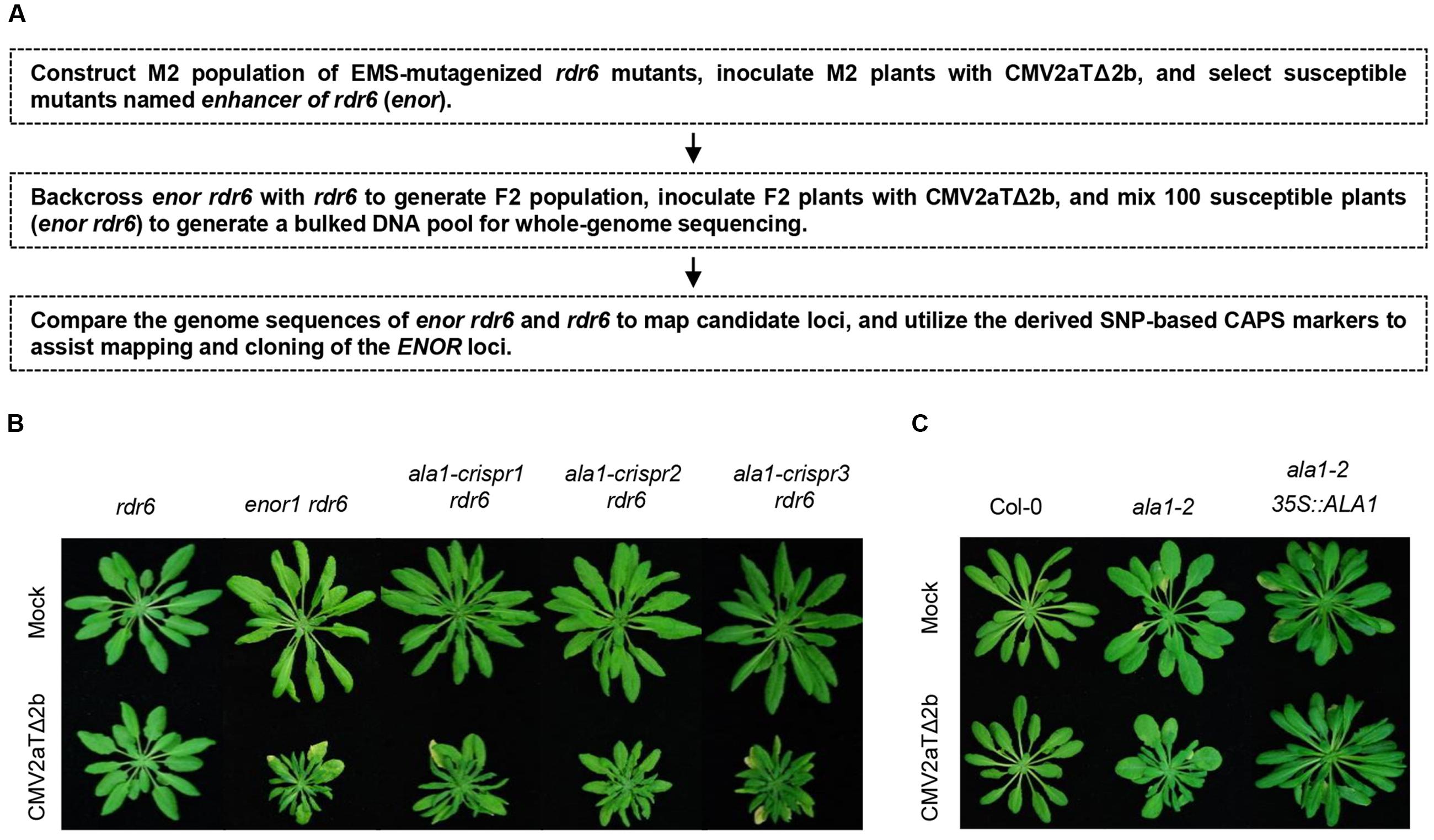
FIGURE 1. ENOR1 encodes ALA1 and mediates antiviral immunity. (A) Schematic diagram of screening of enor (enhancer of rdr6) mutants and mapping of ENOR loci. (B) Phenotypes of rdr6, enor1 rdr6 and the ala1 mutants generated by CRISPR/Cas9 in rdr6 (ala1-crispr1, ala1-crispr2, ala1-crispr3) at 21 days after infection with mock or CMV2aTΔ2b. (C) Phenotypes of Col-0, ala1-2 and the ala1-2 plant transgenic for the ALA1 gene driven by 35S promoter (ala1-2 35S::ALA1) at 21 days after infection with mock or CMV2aTΔ2b.
As shown in Figure 1B, the newly identified mutant enor1 in the rdr6 background, named enor1 rdr6, exhibited severely stunted and clustered leaves after infection with CMV2aTΔ2b. One fourth of F2 population from the cross between enor1 rdr6 and rdr6 were susceptible to CMV2aTΔ2b, demonstrating that enor1 is a recessive mutation. In order to map the ENOR1 locus, we generated a bulked pool of susceptible plants from the F2 population for whole-genome sequencing, screened mutations by comparing the sequences with SHOREmap methods (Schneeberger et al., 2009; Sun and Schneeberger, 2015), and mapped the ENOR1 locus using CAPS markers (Figure 1A). We finally found that only a C to T mutation at the 2965th bp of CDS of AT5G04930, which causes a premature stop codon and generates a HaeIII-based CAPS marker, co-segregated with enor1 (Supplementary Figures 1A,B).
ENOR1 Corresponds to ALA1 and Is Essential for Antiviral Immunity
AT5G04930 encodes ALA1 (Lopez-Marques et al., 2014) that co-localizes with the mCherry-ER-marker (Supplementary Figure 2) (Lopez-Marques et al., 2012). To further genetically verify whether AT5G04930 (ALA1) corresponds to ENOR1 and mediates antiviral immunity, we generated ala1 mutants by the CRISPR/Cas9 genome editing method (Feng et al., 2014; Jia et al., 2016) in the rdr6 background, and examined whether these ala1-crispr rdr6 double mutants exhibit the viral symptoms similar to that of enor1 rdr6 when inoculated with CMV2aTΔ2b. As shown in Figure 1B, all the ala1-crispr rdr6 double mutants were severely susceptible to CMV2aTΔ2b, demonstrating that ALA1 corresponds to ENOR1 and is required for antiviral immunity.
We also obtained a T-DNA insertional mutant (Salk_056947, named ala1-2) of ALA1 (Supplementary Figure 1B), and found that the ala1-2 single mutant was mildly susceptible to CMV2aTΔ2b, less severe than ala1-crispr rdr6 (Figures 1B,C), which also supports of the ALA1 function in antiviral immunity. Moreover, we found that transgenic expression of ALA1 under the control of CaMV 35S promoter fully restored the mutant phenotypes of ala1-2 (Figure 1C).
ALA1 Acts Additively With RDR1 and RDR6 to Regulate Antiviral Immunity
Further analyses of various double mutants and the triple mutant ala1-2 rdr1 rdr6 showed that all the double mutants including ala1-2 rdr6, ala1-2 rdr1, enor1 rdr6, ala1-crispr rdr6 and rdr1 rdr6 exhibited similar symptoms after inoculation with CMV2aTΔ2b, which were much more severe than the single mutant ala1-2, while the triple mutant ala1-2 rdr1 rdr6 showed the most severe symptoms with over-stunted newly born leaves and yellow old chlorotic leaves (Figures 1B,C, 2A,B and Supplementary Figure 3). These results suggest that ALA1 functions additively with RDR1 and RDR6 to mediate plant immunity.
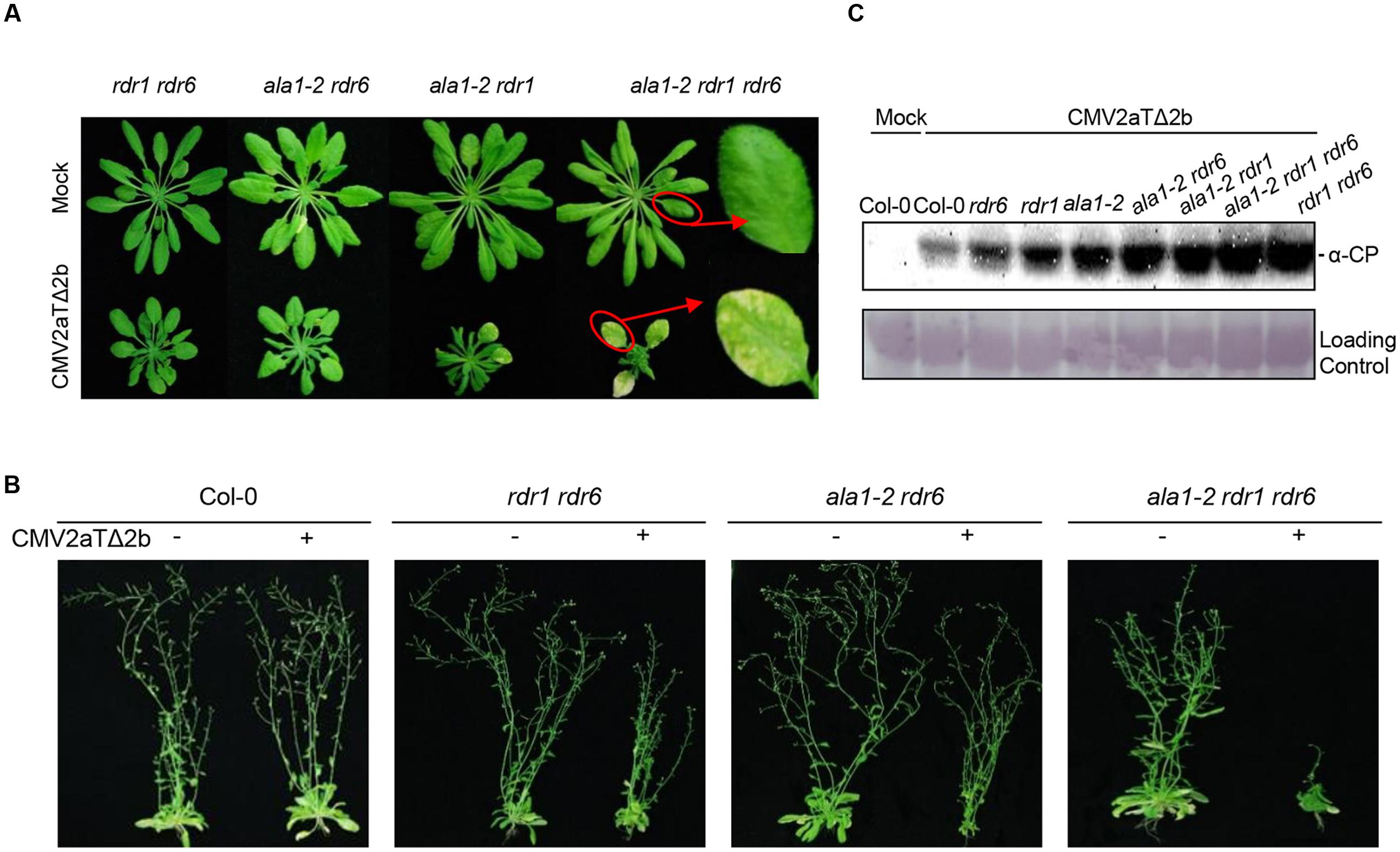
FIGURE 2. ALA1 acts through both RDR1/6-related and -unrelated pathways to mediate antiviral immunity. (A) Phenotypes of rdr1 rdr6, ala1-2 rdr6, ala1-2 rdr1 and ala1-2 rdr1 rdr6 at 21 days after infection with mock or CMV2aTΔ2b. (B) Phenotypes of Col-0, rdr1 rdr6, ala1-2 rdr6 and ala1-2 rdr1 rdr6 at 45 days after infection with mock or CMV2aTΔ2b. (C) Immunoblotting analysis to detect the accumulations of CMV2aTΔ2b from leaves of Col-0, rdr6, rdr1, ala1-2, ala1-2 rdr6, ala1-2 rdr1, ala1-2 rdr1 rdr6 and rdr1 rdr6 at 21 days after infection with mock or CMV2aTΔ2b. The coat protein (CP) of CMV2aTΔ2b was detected by anti-CP (α-CP) antibody. The large subunit of ribulose-1,5-bisphosphate was used as the loading control.
The immunoblot analysis with antibody against the CP of CMV2aTΔ2b showed that CMV2aTΔ2b accumulated much more in ala1-2 than in wild-type, and that the double mutants (ala1-2 rdr6, ala1-2 rdr1 and rdr1 rdr6) accumulated much more CP than the corresponding single mutants (Figure 2C). These results further demonstrate that ALA1 acts additively with RDR1 and RDR6 to mediate RNAi-based antiviral immunity. Interestingly, the triple mutant ala1-2 rdr1 rdr6 showed enhanced susceptibility compared with the double mutant rdr1 rdr6 when inoculated with CMV2aTΔ2b, however, the accumulation of CMV2aTΔ2b was indistinguishable between the triple mutant ala1-2 rdr1 rdr6 and the double mutant rdr1 rdr6, implying that ALA1 mediates plant immunity through both a RDR1/6-related RNAi pathway and RDR1/6–unrelated pathways.
Further phenotypic analysis showed that the ala1-2 rdr1 rdr6 triple mutant also displays developmental defects, including shorter siliques and less fertile siliques (Figures 3A–C). These results imply that ALA1 may function additively with RDR1 and RDR6 to mediate RNAi-regulated plant development, consistent with the previous observations that RNAi, in addition to the RNAi-mediated plant immunity, also mediates plant developmental processes (Yoshikawa et al., 2005).
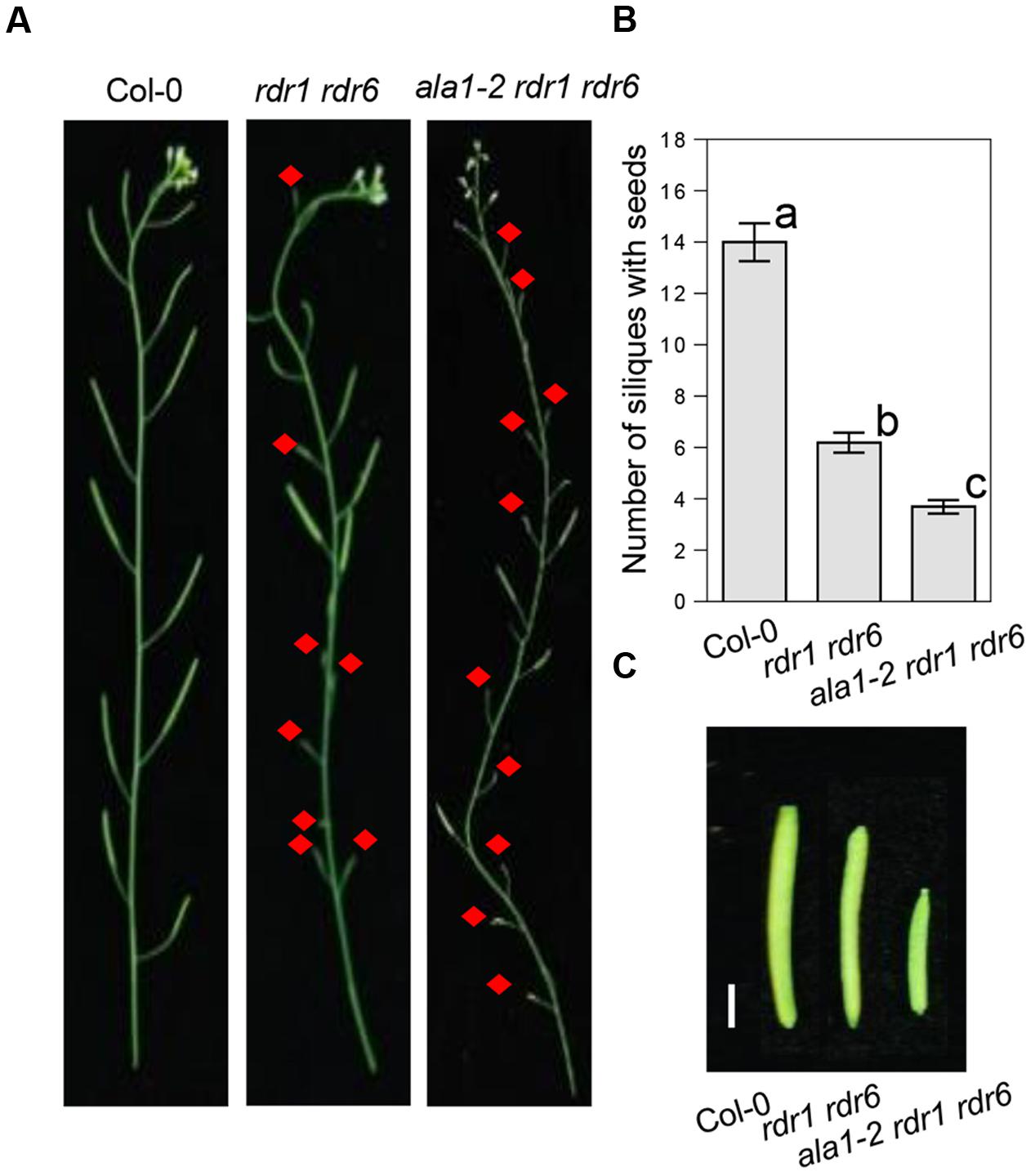
FIGURE 3. ALA1 acts additively with RDR1 and RDR6 to regulate plant development. (A,B) Phenotypes (A) and statistical analysis of numbers of siliques with seeds (B) of the main inflorescences from Col-0, rdr1 rdr6 and ala1-2 rdr1 rdr6. Red arrows indicate sterile siliques. Errors represent ± SE. Lowercase letters indicate significant differences by one-way ANOVA analysis with SPSS software (P < 0.05). (C) Phenotypes of representative fertile siliques from Col-0, rdr1 rdr6 and ala1-2 rdr1 rdr6. The bar represents 2 mm.
ALA1 Is Required for Gene Silencing
Having shown that ALA1 acts additively with RDR1 and RDR6 in RNAi-based antiviral immunity and development, we further verified whether ALA1 affects gene silencing via genetic cross of the ala1-2 mutant with the L1, a transgenic silencing marker line where the GUS transgene driven by the CaMV35S promoter (35S::GUS) was silenced and expressed at low level (Boutet et al., 2003). As shown in Figure 4, the GUS activity was obviously increased in ala1-2 (named ala1-2 35S::GUS). These data demonstrate that mutation in ALA1 abolished the gene silencing on the GUS transgene driven by the 35S promoter, suggesting that ALA1 is indeed required for gene silencing. Consistently, our quantitative real-time PCR analysis showed that the accumulation of CMV2aTΔ2b RNA in ala1-2 was much higher than that in Col-0, further supporting the essential roles of ALA1 in gene silencing and antiviral defense.

FIGURE 4. ALA1 is involved in gene silencing. The effect of ala1-2 on post-transcriptional silencing of 35S::GUS transgene of the L1 line. The 35S::GUS transgene was post-transcriptionally silenced in the L1 line (Boutet et al., 2003); the homozygous ala1-2 mutation prevents silencing of the GUS transgene in all expanded rosette leaves of the L1 line (ala1 35S::GUS), which was identified from F2 population of ala1-2 and L1.
ALA2 Also Participates in Antiviral Immunity
During the screening, we isolated a second enhancer mutant enor2 rdr6 (Figure 5A), in which CP accumulation was similar with that in enor1 rdr6 (Figure 5B). We further found that ENOR2 encodes ALA2 by performing the same mapping and identification procedures as ENOR1 (Supplementary Figure 5A). The ALA2 gene in enor2 rdr6 contained a G to A mutation at the 1995th bp, leading to a premature stop codon (Supplementary Figure 5B), and mutation of ALA2 by CRISPR/Cas9 in rdr6 also resulted in severe susceptibility to CMV2aTΔ2b (Figure 5A and Supplementary Figure 5B), suggesting that ALA2 mediates antiviral immunity. Moreover, we generated the enor1 enor2 rdr6 triple mutant, and found that enor1 enor2 rdr6 displayed severe developmental defects, including stunted leaves, which is similar with CMV2aTΔ2b-infected enor1 rdr6 and enor2 rdr6 (Figure 5C). This results (Figure 5) indicate that both ALA1 and ALA2 act additively with RDR6 to mediate antiviral immunity and plant growth.
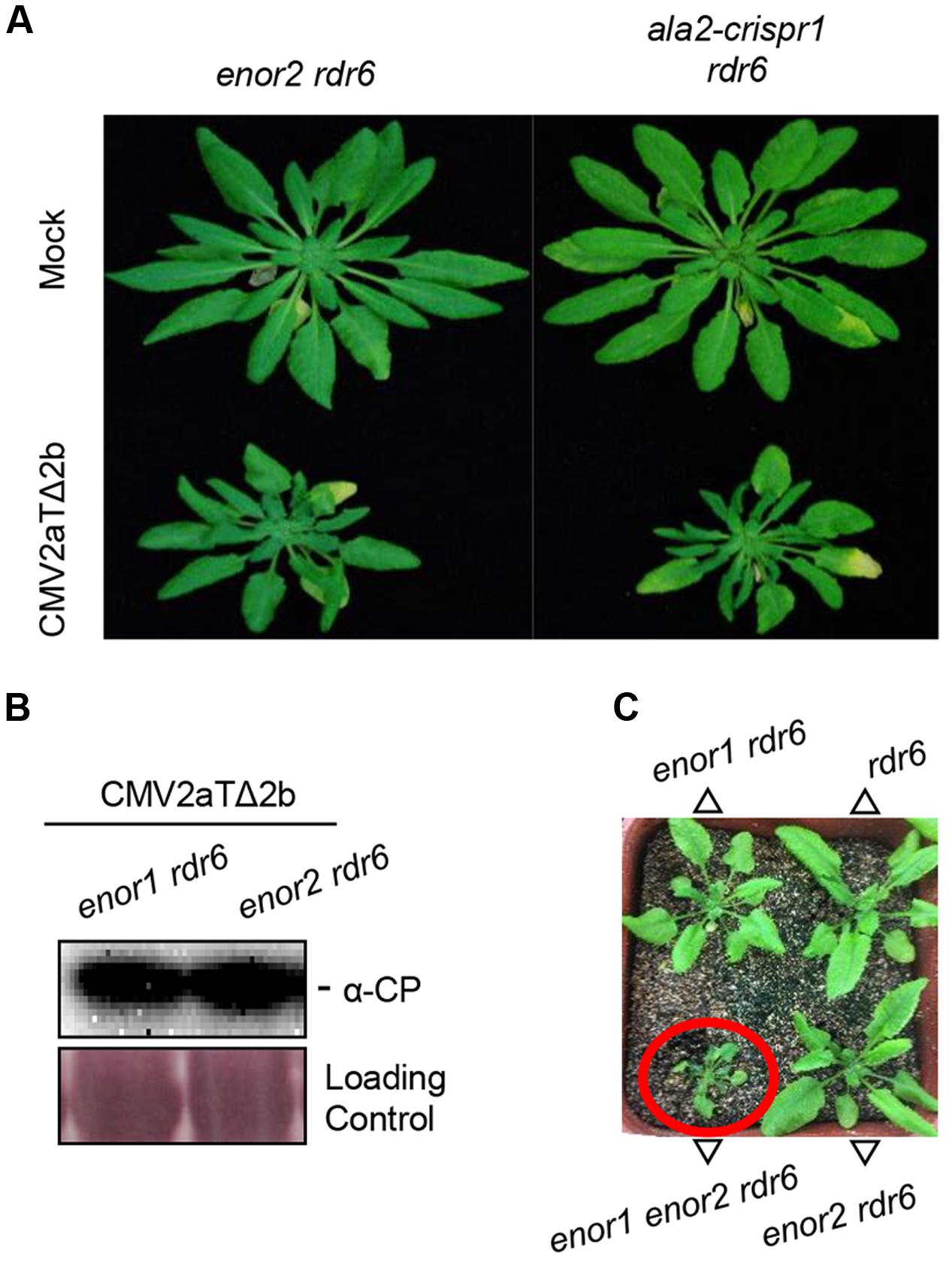
FIGURE 5. ALA2 acts additively with RDR6 and redundantly with ALA1 in antiviral immunity. (A) Phenotypes of enor2 rdr6, and the ala2-crispr1 mutant generated by CRISPR/Cas9 in rdr6 (ala2-crispr1 rdr6) at 21 days after infection with mock or CMV2aTΔ2b. (B) Immunoblotting analysis to detect the accumulations of CMV2aTΔ2b from leaves of enor1 rdr6 and enor2 rdr6 at 21 days after infection with mock or CMV2aTΔ2b. The coat protein (CP) of CMV2aTΔ2b was detected by anti-CP (α-CP) antibody. The large subunit of ribulose-1,5-bisphosphate was used as the loading control. (C) Phenotypes of 30-day-old seedlings of rdr6, enor1 rdr6, enor2 rdr6, and enor1 enor2 rdr6.
Analysis of Other ALAs in Antiviral Immunity
Phylogenetic analysis of the ALA family proteins showed that ALA1 and ALA2 are the closest members, and other members are less related (Supplementary Figure 6). We observed that CMV2aTΔ2b infection dramatically induced the expression of ALA7 and ALA10 in ala1-2, but could not obviously affect the expression of other ALAs in wild-type and ala1-2 (Figure 6A). We next investigated whether other ALA members play a role in antiviral immunity. The T-DNA insertion mutants of ALA3 to ALA12 were inoculated with CMV2aTΔ2b, and the results showed that none of these mutants were susceptible (Figure 6B and data not shown). It remains to be elucidated whether these ALAs function redundantly to mediate RNAi-based antiviral immunity and plant development.
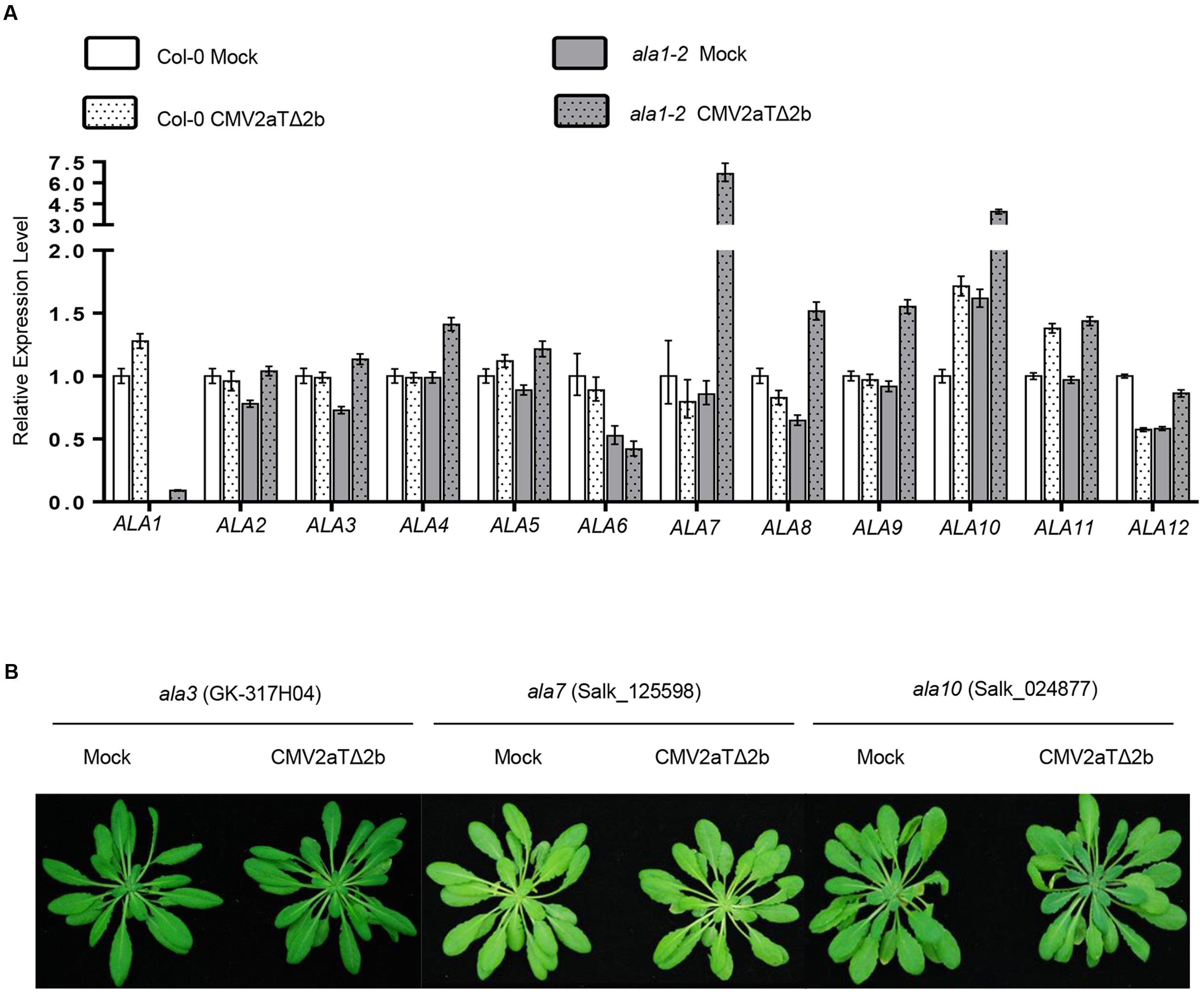
FIGURE 6. Analysis of ALA members in antiviral silencing. (A) Quantitative real-time PCR showed the relative expression levels of ALA family members in Col-0 and ala1-2 after inoculation with mock or CMV2aTΔ2b. The data are means (±SE) from three biological repeats. (B) Phenotypes of the ala3, ala7 and ala10 mutants at 21 days after infection with mock or CMV2aTΔ2b.
Discussion
It is well known that the RNAi pathway regulates plant growth, development and immunity. Previous studies have revealed that AGOs, DCLs, RDR1 and RDR6 are essential components of RNAi pathway (Ding and Voinnet, 2007; Qu et al., 2008; Wang et al., 2010; Cao et al., 2014). In this study, we developed an effective forward genetic screening using 2b-deficient CMV2aTΔ2b, and defined ALA1 and ALA2, membrane-localized proteins (Figures 1, 5 and Supplementary Figure 2) (Lopez-Marques et al., 2010, 2012), as the new components in the RNAi pathway. ALA1 plays an essential role in gene silencing, and acts additively with RDR1/6 to mediate RNAi-based antiviral immunity and plant development (Figures 2–4). ALA2 also participates in antiviral defense and development, and acts redundantly with ALA1 in regulation of plant development in rdr6 background (Figure 5C).
A recent study showed that AMP1, a novel key component in RNAi pathway, associates with AGO1 and mediates miRNA-targeted translational inhibition of mRNA on ER membrane (Li et al., 2013). miRNA-guided cleavage can also occur on ER membrane-bound polysomes (Li et al., 2016). These studies take ER into a central stage of small RNAs-mediated silencing (Ma et al., 2013; Li et al., 2016). On the other hand, viruses recruit ER membrane and manipulate lipid synthesis, transport and metabolism to form a circumstance essential for viral replication and morphogenesis (Fernández de Castro et al., 2016). Our finding that the ER membrane-localized ALA1 and ALA2 are essential players in silencing pathway and antiviral immunity would help to study and understand both the small RNAs machinery on ER membrane and the roles of lipid transport in silencing and antiviral defense. It would be interesting to investigate whether ALA1 and ALA2 associate with AMP1 and/or AGO1 to mediate gene silencing and antiviral immunity.
Author Contributions
DX designed the study; BZ, HG, DW, and TQ performed experiments; DX, TQ, BZ, HG, GX, SS, HJ, and SZ analyzed the data. BZ, HG, TQ, and DX wrote the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant NO. 31230008 and 31630085).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Shouwei Ding (University of California, Berkeley) for sharing the rdr1, rdr6 and rdr1 rdr6 mutants, and Dr. Jiankang Zhu (Center for Plant Stress Biology, Chinese Academy of Science) for the CRISPR/Cas9 vector.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00422/full#supplementary-material
Footnotes
References
Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., et al. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16, 927–932. doi: 10.1016/j.cub.2006.03.035
Arribas-Hernandez, L., Marchais, A., Poulsen, C., Haase, B., Hauptmann, J., Benes, V., et al. (2016). The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell 28, 1563–1580. doi: 10.1105/tpc.16.00121
Blevins, T., Rajeswaran, R., Shivaprasad, P. V., Beknazariants, D., Si-Ammour, A., Park, H. S., et al. (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34, 6233–6246. doi: 10.1093/nar/gkl886
Botella, C., Sautron, E., Boudiere, L., Michaud, M., Dubots, E., Yamaryo-Botte, Y., et al. (2016). ALA10, a phospholipid flippase, controls FAD2/FAD3 desaturation of phosphatidylcholine in the ER and affects chloroplast lipid composition in Arabidopsis thaliana. Plant Physiol. 170, 1300–1314. doi: 10.1104/pp.15.01557
Boutet, S., Vazquez, F., Liu, J., Béclin, C., Fagard, M., Gratias, A., et al. (2003). Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13, 843–848. doi: 10.1016/S0960-9822(03)00293-8
Cao, M., Du, P., Wang, X., Yu, Y. Q., Qiu, Y. H., Li, W., et al. (2014). Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 14613–14618. doi: 10.1073/pnas.1407131111
Diaz-Pendon, J. A., Li, F., Li, W. X., and Ding, S. W. (2007). Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19, 2053–2063. doi: 10.1105/tpc.106.047449
Ding, S. W., Li, H., Lu, R., Li, F., and Li, W. X. (2004). RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 102, 109–115. doi: 10.1016/j.virusres.2004.01.021
Ding, S. W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi: 10.1016/j.cell.2007.07.039
Felsenstein, J. (1985). Confidence-limits on phylogenies - an approach using the Bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Feng, Z. Y., Mao, Y. F., Xu, N. F., Zhang, B. T., Wei, P. L., Yang, D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 4632–4637. doi: 10.1073/pnas.1400822111
Fernández de Castro, I., Tenorio, R., and Risco, C. (2016). Virus assembly factories in a lipid world. Curr. Opin. Virol. 18, 20–26. doi: 10.1016/j.coviro.2016.02.009
Garcia-Ruiz, H., Takeda, A., Chapman, E. J., Sullivan, C. M., Fahlgren, N., Brempelis, K. J., et al. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22, 481–496. doi: 10.1105/tpc.109.073056
Gomes, E., Jakobsen, M. K., Axelsen, K. B., Geisler, M., and Palmgren, M. G. (2000). Chilling tolerance in arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell 12, 2441–2453. doi: 10.1105/tpc.12.12.2441
Incarbone, M., and Dunoyer, P. (2013). RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 18, 382–392. doi: 10.1016/j.tplants.2013.04.001
Jia, Y. X., Ding, Y. L., Shi, Y. T., Zhang, X. Y., Gong, Z. Z., and Yang, S. H. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212, 345–353. doi: 10.1111/nph.14088
Li, S., Le, B., Ma, X., Li, S., You, C., Yu, Y., et al. (2016). Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. Elife 5:e22750. doi: 10.7554/eLife.22750
Li, S., Liu, L., Zhuang, X., Yu, Y., Liu, X., Cui, X., et al. (2013). MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153, 562–574. doi: 10.1016/j.cell.2013.04.005
Lopez-Marques, R. L., Poulsen, L. R., Hanisch, S., Meffert, K., Buch-Pedersen, M. J., Jakobsen, M. K., et al. (2010). Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA alpha-subunit. Mol. Biol. Cell 21, 791–801. doi: 10.1091/mbc.E09-08-0656
Lopez-Marques, R. L., Poulsen, L. R., and Palmgren, M. G. (2012). A putative plant aminophospholipid flippase, the Arabidopsis P4 ATPase ALA1, localizes to the plasma membrane following association with a beta-subunit. PLoS ONE 7:e33042. doi: 10.1371/journal.pone.0033042
Lopez-Marques, R. L., Theorin, L., Palmgren, M. G., and Pomorski, T. G. (2014). P4-ATPases: lipid flippases in cell membranes. Pflugers. Arch. 466, 1227–1240. doi: 10.1007/s00424-013-1363-4
Ma, X., Cao, X. F., Mo, B. X., and Chen, X. M. (2013). Trip to ER MicroRNA-mediated translational repression in plants. RNA Biol. 10, 1586–1592. doi: 10.4161/rna.26313
Mao, Y., Zhang, H., Xu, N., Zhang, B., Gou, F., and Zhu, J. K. (2013). Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6, 2008–2011. doi: 10.1093/mp/sst121
Martinez de Alba, A. E., Elvira-Matelot, E., and Vaucheret, H. (2013). Gene silencing in plants: a diversity of pathways. Biochim. Biophys. Acta 1829, 1300–1308. doi: 10.1016/j.bbagrm.2013.10.005
McDowell, S. C., Lopez-Marques, R. L., Cohen, T., Brown, E., Rosenberg, A., Palmgren, M. G., et al. (2015). Loss of the Arabidopsis thaliana P4-ATPases ALA6 and ALA7 impairs pollen fitness and alters the pollen tube plasma membrane. Front. Plant Sci. 6:197. doi: 10.3389/fpls.2015.00197
McDowell, S. C., Lopez-Marques, R. L., Poulsen, L. R., Palmgren, M. G., and Harper, J. F. (2013). Loss of the Arabidopsis thaliana P(4)-ATPase ALA3 reduces adaptability to temperature stresses and impairs vegetative, pollen, and ovule development. PLoS ONE 8:e62577. doi: 10.1371/journal.pone.0062577
Morel, J. B. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell Online 14, 629–639. doi: 10.1105/tpc.010358
Parent, J. S., Bouteiller, N., Elmayan, T., and Vaucheret, H. (2015). Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 81, 223–232. doi: 10.1111/tpj.12720
Poulsen, L. R., Lopez-Marques, R. L., Mcdowell, S. C., Okkeri, J., Licht, D., Schulz, A., et al. (2008). The Arabidopsis P-4-ATPase ALA3 localizes to the Golgi and requires a beta-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 20, 658–676. doi: 10.1105/tpc.107.054767
Qu, F., Ye, X., and Morris, T. J. (2008). Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 14732–14737. doi: 10.1073/pnas.0805760105
Saitou, N., and Nei, M. (1987). The Neighbor-Joining Method - a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schneeberger, K., Ossowski, S., Lanz, C., Juul, T., Petersen, A. H., Nielsen, K. L., et al. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6, 550–551. doi: 10.1038/nmeth0809-550
Shan, X., Wang, J., Chua, L., Jiang, D., Peng, W., and Xie, D. (2011). The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 155, 751–764. doi: 10.1104/pp.110.166595
Sun, H., and Schneeberger, K. (2015). SHOREmap v3.0: fast and accurate identification of causal mutations from forward genetic screens. Methods Mol. Biol. 1284, 381–395. doi: 10.1007/978-1-4939-2444-8_19
Talmor-Neiman, M., Stav, R., Klipcan, L., Buxdorf, K., Baulcombe, D. C., and Arazi, T. (2006). Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 48, 511–521. doi: 10.1111/j.1365-313X.2006.02895.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Wang, X. B., Wu, Q., Ito, T., Cillo, F., Li, W. X., Chen, X., et al. (2010). RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 484–489. doi: 10.1073/pnas.0904086107
Xie, Z., Fan, B., Chen, C., and Chen, Z. (2001). An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 98, 6516–6521. doi: 10.1073/pnas.111440998
Yoshikawa, M., Peragine, A., Park, M. Y., and Poethig, R. S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175. doi: 10.1101/gad.1352605
Zhang, X., Yuan, Y. R., Pei, Y., Lin, S. S., Tuschl, T., Patel, D. J., et al. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. doi: 10.1101/gad.1495506
Keywords: ALA, Arabidopsis, 2b, CMV, RNA interference (RNAi), virus
Citation: Zhu B, Gao H, Xu G, Wu D, Song S, Jiang H, Zhu S, Qi T and Xie D (2017) Arabidopsis ALA1 and ALA2 Mediate RNAi-Based Antiviral Immunity. Front. Plant Sci. 8:422. doi: 10.3389/fpls.2017.00422
Received: 28 December 2016; Accepted: 13 March 2017;
Published: 07 April 2017.
Edited by:
Jari Valkonen, University of Helsinki, FinlandReviewed by:
Zonghua Wang, Fujian Agriculture and Forestry University, ChinaHong-Gu Kang, Texas State University, USA
Copyright © 2017 Zhu, Gao, Xu, Wu, Song, Jiang, Zhu, Qi and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daoxin Xie, daoxinlab@tsinghua.edu.cn Tiancong Qi, qitiancong@163.com
†These authors have contributed equally to this work.
 Biyun Zhu
Biyun Zhu Hua Gao1†
Hua Gao1† Daoxin Xie
Daoxin Xie