- Key Laboratory of Bamboo and Rattan Science and Technology of the State Forestry Administration, International Centre for Bamboo and Rattan, Beijing, China
Mini chromosome maintenance 1, agamous, deficiens, and serum response factor (MADS)-box genes are transcription factors which play fundamental roles in flower development and regulation of floral organ identity. However, till date, identification and functions of MADS-box genes remain largely unclear in Phyllostachys edulis. In view of this, we performed a whole-genome survey and identified 34 MADS-box genes in P. edulis, and based on phylogeny, they were classified as MIKCC, MIKC∗, Mα, and Mβ. The detailed analysis about gene structure and motifs, phylogenetic classification, comparison of gene divergence and duplication are provided. Interestingly, expression patterns for most genes were found similar to those of Arabidopsis and rice, indicating that the well-established ABCDE model can be applied to P. edulis. Moreover, we overexpressed PheMADS15, an AP1-like gene, in Arabidopsis, and found that the transgenic plants have early flowering phenotype, suggesting that PheMADS15 might be a regulator of flowering transition in P. edulis. Taken together, this study provides not only insightful comprehension but also useful information for understanding the functions of MADS-box genes in P. edulis.
Introduction
Phyllostachys edulis is one of the most important non-timber forest products in the world. They flower at the end of very long vegetative growth phases, often followed by the death of large areas of P. edulis. They show a cyclic recurrence of flowering, the intervals of which are basically definite varying from a few years to 120 years or longer. In this case, studying the mechanism of P. edulis flowering time is very challenging, and it is quite difficult to determine the key regulatory genes involved in floral formation and transition in P. edulis.
The mini chromosome maintenance 1, agamous, deficiens, and serum response factor (MADS)-box family members, identified originally as floral homeotic genes, are important transcription factors for plant development (Purugganan, 1997; Theissen et al., 2000; Jack, 2001; Nam et al., 2003; Pařenicová et al., 2003). In plants, the type I comprises M-type genes and type II group includes most well-known MIKC-type genes (Arora et al., 2007), named after the four basic components of the MADS (M) domain: the Intervening (I) domain, the Keratin (K) domain, and the C-terminal (C) domain (Kramer et al., 1998). MIKC-type genes have been further divided into two subgroups, MIKCC- and MIKC∗-types, due to different exon/intron structures (Henschel et al., 2002; Kofuji et al., 2003). Type I MADS-box genes have been categorized into Mα, Mβ, Mγ, and Mδ clades based on the phylogenetic relationships of conserved MADS-box domain (Pařenicová et al., 2003). In most plants, type I genes experience a higher number of births and deaths than type II genes, due to more frequent segmental gene duplications and weaker purifying selection (Nam et al., 2004).
The genetic ABCDE model of floral organ development can be applied to dicot plants, mainly in Arabidopsis, snapdragon and petunia (Coen and Meyerowitz, 1991; Angenent and Colombo, 1996; Theißen and Saedler, 2001). Generally, A and B class genes together are required for petal development, B and C class genes cooperate to control stamen development. A and C class genes are, respectively, involved in sepal and carpel development. D class genes function in ovule development, while E class proteins are expressed in all four whorls of floral organs by forming MADS-box protein complexes with proteins of other classes (Pelaz et al., 2000; Favaro et al., 2003; Pinyopich et al., 2003). In Arabidopsis, APETALA1 (AP1) and APETALA2 (AP2) belong to the A-function genes; B-function genes include APETALA3 (AP3), PISTILLATA (PI); the C-function gene is AGAMOUS (AG); SEPALLATA1, 2, 3 and 4 (SEP1, 2, 3, 4/AGL2, 4, 9, 3) form the E-function genes (Yanofsky et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994; Jofuku et al., 1994; Huang et al., 1995; Savidge et al., 1995; Mandel and Yanofsky, 1998; Pelaz et al., 2000; Ditta et al., 2004).
Poaceae family is generally known for monocot crops such as rice (Oryza sativa), maize (Zea mays), wheat (Triticum spp.) and barley (Hordeum vulgare) (Grass Phylogeny Working Group et al., 2001). However, Bambusoideae is quite distinct from other members of Poaceae and is known for its unique floral organization and morphology (Grass Phylogeny Working Group et al., 2001; Rudall et al., 2005; Whipple et al., 2007). In rice and bamboo, each grass spikelet is the structural unit of grass flowers, which consists of a number of florets. In addition, the floret contains four whorls, such as lemma and palea (whorl 1), two lodicules (whorl 2), six stamens (whorl 3), and gynoecium (whorl 4) (Nagasawa et al., 2003). Like eudicots, MADS-box genes in rice and maize are divided into ABCDE gene classes. (Ambrose et al., 2000; Nagasawa et al., 2003; Kater et al., 2006; Yamaguchi et al., 2006; Preston and Kellogg, 2007; Yao et al., 2008; Cui et al., 2010; Li et al., 2011). Few of the MADS-box genes are functionally characterized in different plants. For instance, three AP1/FUL-like genes (OsMADS14, OsMADS15, and OsMADS18) coordinately act in the shoot apical meristem in rice (Kobayashi et al., 2012). Maize Silky1 and rice SPW1 or OsMADS16, orthologs of the Arabidopsis AP3, cause homeotic transformations of stamens to carpels and lodicules to lemma- or palea-like structures (Ambrose et al., 2000; Nagasawa et al., 2003; Whipple et al., 2004). Rice OsMADS3, belonging to the AG homolog gene, is, respectively, involved in stamen, ovule and late anther development (Kramer et al., 2004; Yamaguchi et al., 2006; Hu et al., 2011; Li et al., 2011). In rice, OsMADS13 and OsMADS21, two D class genes, are involved in ovule identity specification and floral meristem termination (Dreni et al., 2007; Li et al., 2011). Simultaneous four rice SEP-like genes, LHS1, OsMADS5, OsMADS7, and OsMADS8, play a key role in all floral organs development (Cui et al., 2010). However, for bamboo, especially P. edulis, fewer genes were reported to play roles in specifying flower development, their genetic interactions remained to be answered (Tian et al., 2005, 2006; Li et al., 2009; Shih et al., 2014). Therefore, it is necessary to systematically study MADS-box genes related to flower development in bamboo.
In Arabidopsis, class A genes are represented by AP1 (Mandel et al., 1992) and AP2 (Jofuku et al., 1994). AP1 and LEAFY (LFY) are floral meristem-identity genes that confer identity on developing floral primordia (Weigel et al., 1992). The LFY, AP1, CAULFLOWER (CAL) and AP2 genes appear to mutually reinforce each other, leading to a sharp transition from vegetative to reproductive development (Ferrándiz et al., 2000). In addition, AP1, AGAMOUS LIKE24 (AGL24) and SHORT VEGETATIVE PHASE (SVP) act redundantly to control the identity of the floral meristem and to repress expression of class B, C, and E genes in Arabidopsis (Gregis et al., 2009). Genetic evidence suggests that SUPPRESSOR OF CONSTANS 1 (SOC1) and FLOWERING LOCUS T (FT) function are closely associated with the activation of AP1 (Teper-Bamnolker and Samach, 2005; Lee and Lee, 2010). Some results strongly show that not only SOC1, but also AP1 can activate LFY (Liljegren et al., 1999; Lee and Lee, 2010). TERMINAL FLOWER 1 (TFL1) is involved in the maintenance of the inflorescence meristem by preventing the expression of floral meristem identity genes such as AP1 and LFY in the shoot apical meristem, which in turn is negatively regulated by LFY (Irish and Sussex, 1990; Schultz and Haughn, 1991; Weigel et al., 1992; Bowman et al., 1993). Moreover, TFL1 function is compromised by constitutive AP1 activity (Liljegren et al., 1999).
Extensive duplications in Poaceae resulted in the expansion and diversification of gene families. Duplications of MADS-box genes have contributed to understanding of the origin and evolution of developmental mechanisms in plant (Alvarez-Buylla et al., 2000a; Shan et al., 2009). Variance in gene family sizes occurred in a number of families in bamboo. P. edulis underwent a whole-genome duplication (WGD) event, which resulted in 5,370 gene losses (28% of the total genes in the collinear regions) in comparison to rice (Peng et al., 2013). In addition, some genes displayed expression subfunctionalization; for example, the genes in flowering promotion pathways (the photoperiod, gibberellin, ambient-temperature pathways) and floral pathway integrator (FPI) genes (Ehrenreich et al., 2009; Fornara et al., 2010) were not highly expressed in bamboo floral tissues. Low expression of FPI genes, which are involved in floral meristem identity, could signify that the flowering promotion pathways in bamboo may be different.
In this study, we performed a comprehensive identification and phylogenetic analysis of the MADS-box gene family in P. edulis. A total of 34 MADS-box genes were identified and subjected to phylogenetic, gene structure, and domain analyses. We also studied the expression patterns of P. edulis MADS-box genes under normal and abiotic stress conditions. Furthermore, expression profiles and anatomical expression were generated to screen candidate genes involved in flower development and the floral transition. The function of one of these genes, PheMADS15, an AP1-like gene, was also characterized in transgenic Arabidopsis. This work provides useful information on the function of this important family of transcription factors in P. edulis, and with both a genome sequence and a transcriptome, future systematic studies can evaluate structure-function relationships.
Materials and Methods
Database Searches for the Identification of MADS-Box Family Members in P. edulis
MADS-box protein sequences of Arabidopsis and O. sativa have been obtained from TAIR (The Arabidopsis Information Resource1) and Rice Genome Annotation Project2, respectively. P. edulis MADS-box protein sequences were collected from Bamboo Genome Database3 and the accession numbers are shown in Supplementary Table S1.
The MADS-box domains were predicted through Hidden Markov Model (HMM) and redundant sequences were removed using the protein alignments with ClustalX 1.83 (Thompson et al., 1997). Information of ID accession numbers, ORF length, amino acids number, molecular weight, and isoelectric point of each protein is provided in Supplementary Table S1. For all MADS-box genes, ExPASY4 was employed to find the molecular weight and PI of each protein, as they were not available in the Bamboo Genome Database.
Phylogenetic Analysis
Multiple sequence alignments of MADS-box full-length proteins were performed using ClustalX 1.835 (Thompson et al., 1997). The un-rooted neighbor-joining method (Saitou and Nei, 1987) was used to construct the phylogenetic tree in MEGA 6.06 (Tamura et al., 2013) software with 1000 bootstrap replicates.
Conserved Motif and Gene Structure Analysis
Multiple EM for Motif Elicitation (MEME) version 4.9.17 (Bailey and Elkan, 1995) was used to identify conserved motifs in candidate sequences with following parameters: number of repetitions = any, maximum number of motifs = 20, minimum width ≥ 6, and maximum width ≤ 200.
The MADS-box full-length cDNA sequences and the corresponding genomic DNA were collected from Bamboo Genome Database8. The Gene Structure Display Server (GSDS9) (Guo et al., 2007) was employed to identify information on intron/exon structure of the MADS-box genes.
Calculation of Ka/Ks Values and Divergence Times Estimation
Alignment of nucleotide sequences of P. edulis MADS-box gene pairs were aligned with ClustalX 1.83, respectively. The DNAsp5 software was used to calculate the synonymous substitution (Ks) and non-synonymous substitution (Ka) rates. Ka/Ks values were used to estimate the two types of substitutions events. The Ks value was calculated for each of the MADS-box gene pairs and then used to calculate the divergence time of the duplication event (T = Ks/2λ) using the formula: T = Ks/2λ (Lynch and Conery, 2000), with the divergence rate λ = 6.5 × 10-9 (Peng et al., 2013).
Plant Material
Arabidopsis plants were grown under long daylight exposure (16 h light/8 h dark) in light growth incubator maintained at 23°C with 40 to 50% humidity, and an irradiance of approximately 118 μmol m-2 s-1.
The flower buds and flower of P. edulis at different flowering developmental stages were collected in Dajing County, Guilin (E 110°17′-110°47′; N 25°04′-25°48′) in Guangxi Zhuang Autonomous Region from April to August, 2012. Flower development was distinguished by four phases: the floral bud formation stage, the inflorescence growing stage, the bloom stage, the embryo formation stage (Gao et al., 2014).
Expression Profile Analysis
Reads per kilobase of exon model per million mapped reads (RPKM) values of flowering tissues at floral organ development and shoot growth were imported into Cluster 3.0 (de Hoon et al., 2004) for windows and Java TreeView (Saldanha, 2004) to generate the heat maps. RPKM values were shown in Supplementary Table S4.
Quantitative Real-Time PCR (QPCR)
Total RNA was extracted using the Trizol reagent (Invitrogen, USA). The quality and purified RNA was initially assessed on an agarose gel and NanoDrop 8000 spectrophotometer (NanoDrop, Thermo Scientific, Germany), and then the integrity of RNA samples was further evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). For qPCR, Primer 3 Input (version 4.0) was used to design the specific primers according to the MADS-box gene sequences. Detailed descriptions were provided in Supplementary Table S5. Data acquisition and analyses were performed by the Roche Light Cycler software.
Subcellular Localization and Transcriptional Activation
The subcellular localization of PheMADS15 was performed by transfecting GFP-tagged PheMADS15 into rice stem and sheath protoplasts (Zhang et al., 2011). The full-length cDNA of PheMADS15 was fused in frame with the GFP cDNA and ligated between the CaMV 35S promoter and the nopaline synthase terminator. The fluorescence signals in transfected protoplasts were examined using a confocal laser scanning microscope (Leica Microsystems).
The transcriptional activation activity of PheMADS15 was tested by transforming the pGBKT7 construct containing a fusion of PheMADS15 and the GAL4 DNA-binding domain into the yeast strain PJ69-4a. The yeast strain contains the His-3 and LacZ reporter genes. The transformed yeast cells were grown on synthetic defined plates with or without His and assayed for β-galactosidase activity.
In Situ Hybridization
RNA hybridization and immunological detection of the hybridized probes were performed based on the protocol described previously (de Almeida Engler et al., 2001). The specific probes of PheMADS15, PheMADS4-1, PheMADS3, PheMADS21, and PheMADS5 were designed and synthesized by GENEWIZ (Supplementary Table S6). Images were obtained using the Olympus Nikon E600 microscope.
Overexpression
The 35S:PheMADS15 sequence was amplified using specific primers (forward, 5′-GGTACCATGGGGCGCGGGAAGGTG-3′; reverse, 5′-CCCAAGCTTTCATGAAGGACGAGGAAGAGTCTG-3′) by RT-PCR with 2 μl cDNA from leaves of P. edulis. The product was initially cloned into pGEM-T Easy vector and then 35S:pCAMBIA2300 vector. The 35S:PheMADS15 construct was introduced into wild-type Arabidopsis plants (Columbia-0) through Agrobacterium-mediated transformation (Feldmann and Marks, 1987).
Results
Identification and Phylogenetic Analysis of P. edulis MADS-Box Genes
A total of 34 non-redundant MADS-box genes were identified in the P. edulis genome using rice MADS-box domain sequences as queries. To determine the evolutionary relationship of these genes in P. edulis and other species, we constructed a Neighbor-Joining phylogenetic tree of MADS-box proteins from P. edulis, rice and Arabidopsis. According to the previously reports in rice and Arabidopsis, the proteins can be classified into five functional groups (Pařenicová et al., 2003; Arora et al., 2007).
Of the 34 identified P. edulis MADS-box genes, 31 grouped into the type II clade subdividing into 25 MIKCC-type genes and six MIKC∗-type genes (Figure 1). MIKCC-type genes were further divided into nine classic clades: SOC1-like (four genes), E (three genes), SVP-like (three genes), Bs (two genes), C/D (two genes), B (four genes), A (six genes), and OsMADS32-like (one gene). In this study, genes belonging to FLC-clade were absent in P. edulis and rice, which may be specific to Arabidopsis. Interestingly, PheMADS64 was grouped with OsMADS32-like which is a novel monocot MADS-box gene (Sang et al., 2012). However, in contrast to previous research, the OsMADS64 was found to cluster with OsMADS32 to form a OsMADS32-like group instead of the MIKC∗ group (Arora et al., 2007). In the case of type I genes, including Mα and Mβ, PheMADS90 and PheMADS91 were identified as Mβ and PheMADS72 grouped with Mα, but nothing grouped with the Mγ clade. Mγ, FLC-like and ANR1-like MADS-box genes were absent from P. edulis, indicating that these genes might have been lost after the divergence of monocots and dicots. In addition, a phylogenetic tree with bootstrap values was constructed to identify putative orthologs in Arabidopsis, rice and P. edulis using the complete protein sequences (Supplementary Table S1).
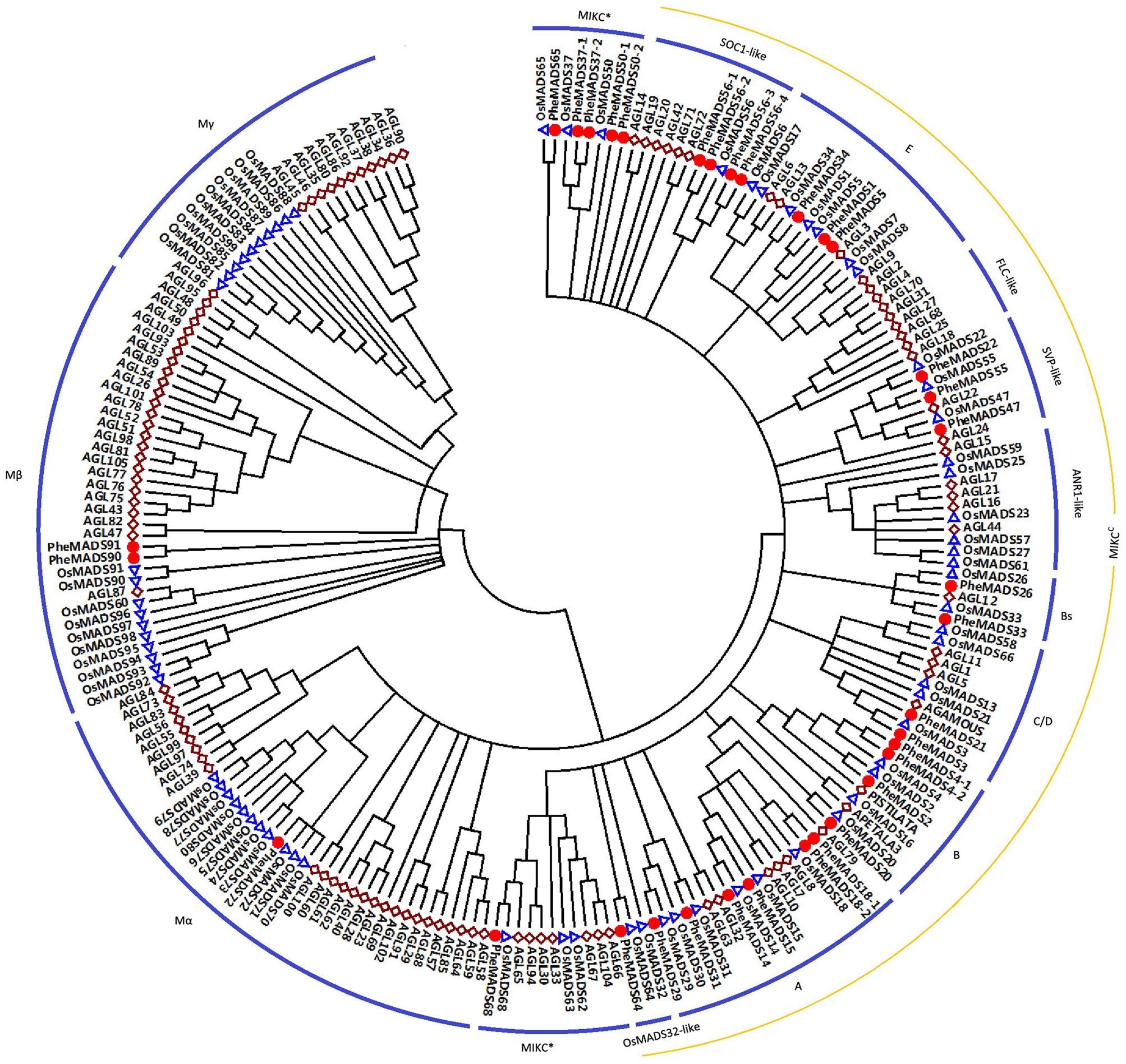
FIGURE 1. Phylogenetic analysis of MADS-box proteins in Phyllostachys edulis, rice and Arabidopsis. A total of 34 MADS-box proteins in P. edulis, 75 in rice and 98 in Arabidopsis were used to construct the NJ tree. The MADS-box proteins in P. edulis were marked by red dots. Branches with less than 50% bootstrap support were collapsed.
Gene Structure and Conserved Motif Distribution Analysis
To better understand the structural diversity of P. edulis MADS-box genes, intron/exon arrangements and conserved motifs were compared with phylogenetics. The MEME motif search tool and GSDS were employed to identify conserved motifs and gene structures in MADS-box genes. Intron/exon arrangements in P. edulis MADS-box genes were different among MIKCC and MIKC∗ genes (Figure 2), similar to reports in Arabidopsis and rice. Nearly half of MIKCC genes lacked introns, but only one MIKC∗ gene lacked an intron (PheMADS50-2). The number of introns in remaining MADS-box genes ranged from 1 to 8. The length of MADS-box proteins varied from 62 to 376 amino acids (Supplementary Table S1).
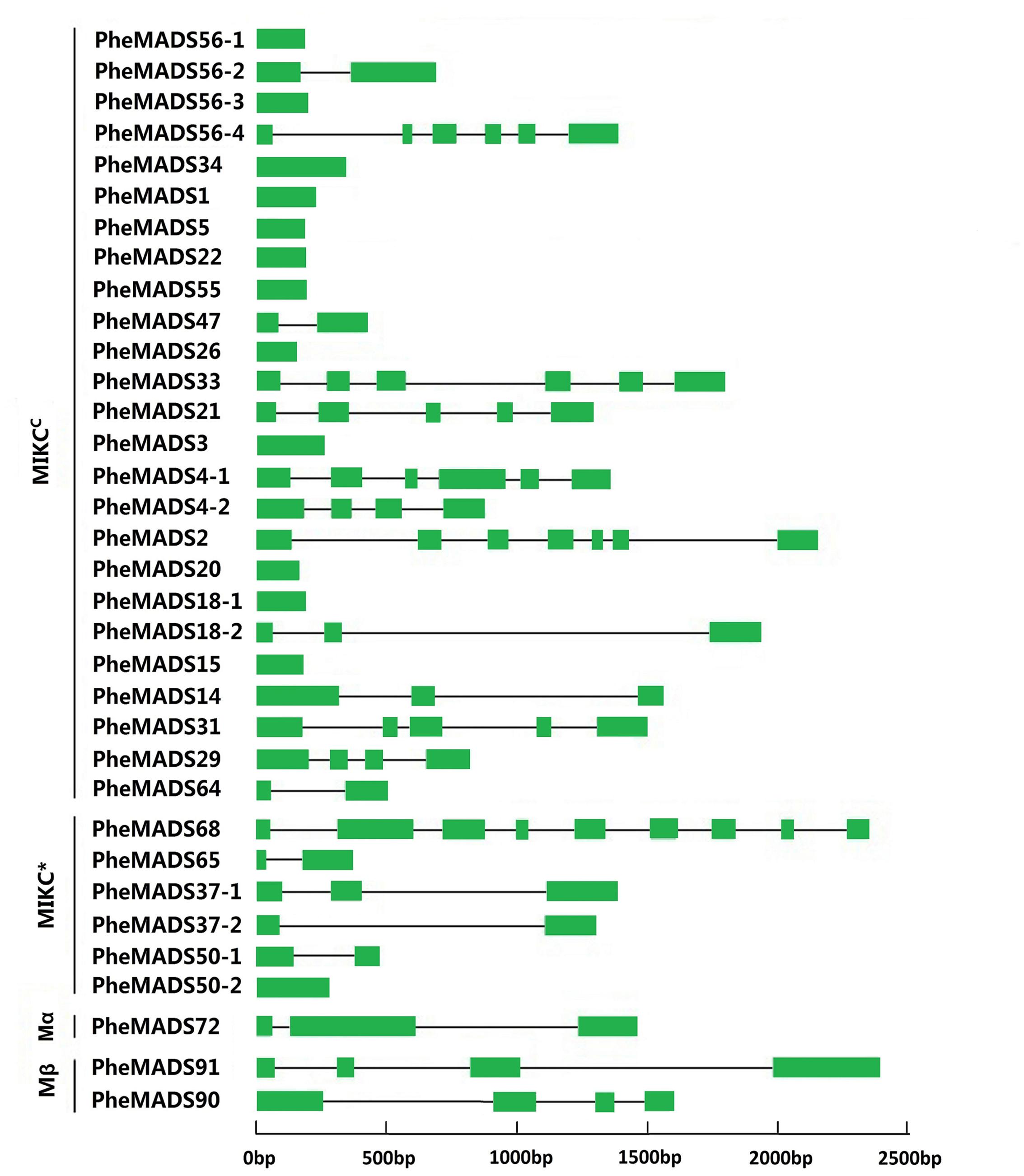
FIGURE 2. Phylogenetic relationship and gene structure analysis of MADS-box genes in P. edulis. Un-rooted neighbor-joining tree was constructed from the alignment of full-length amino acid sequences using the MEGA5 package. Branches with less than 50% bootstrap values were collapsed. Lengths of exons and introns of each MADS-box gene were displayed proportionally. The green solid boxes represent exons; black lines represent introns.
The MEME program was used to analyze conserved motifs in MADS-box proteins followed by SMART annotation, resulting in the identification of 20 conserved motifs (Figure 3 and Supplementary Table S3). In all 34 P. edulis MADS-box proteins, excluding Mγ, most of MIKCC and MIKC∗ groups had motif 1-type MADS domain. Motifs 2, 8, 9, and 10 were localized in the K domain. Motif 4 was also conserved across many of the MADS-box proteins, excluding Mβ, which was found in the I domain. Most of the unconserved motifs (3, 5–7, 11–20) were located in C-terminus, which is typically the most diverse region in MADS-box proteins (Kramer et al., 1998). The sequences and lengths of all the motifs were given in Supplementary Table S3.
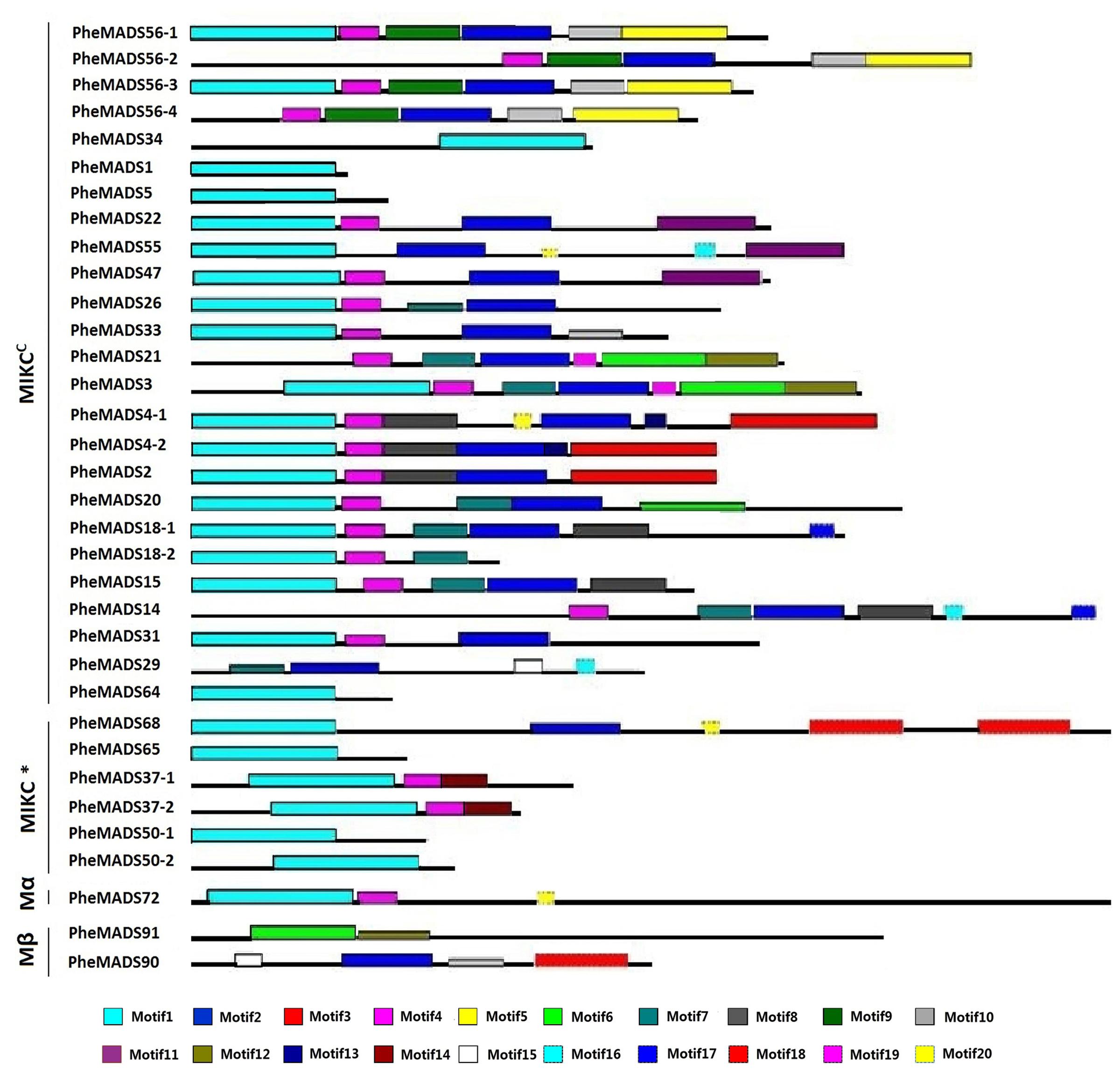
FIGURE 3. Distribution of conserved motifs in P. edulis MADS-box proteins identified using MEME search tool. Schematic representation of motifs identified in P. edulis MADS-box proteins using MEME motif search tool for each group given separately. Each motif is represented by a number in colored box. Length of box does not correspond to length of motif. Order of the motifs corresponds to position of motifs in individual protein sequence. For detail of motifs refer to Supplementary Material.
The Analysis of Expression Patterns of PheMADS-Box Genes during Floral Organ Development
MADS-box gene expression was tested at five broad categories in flowers described by Gao et al. (2014). The expression profiles were expanded by including transcriptomes from the Transcriptome Sequencing Bamboo Genome Database, including: leaves from non-flowering (CK) and four flowering developmental stages (F) of P. edulis (Supplementary Table S4). MADS-box genes were classified into 11 groups based on phylogenetic analysis during flowering developmental stages (Figure 4). The expression levels of the A and B class PheMADS genes were high in F1 and F2 and decreased through floral maturity. In contrast, the expressions of C, D, and E class PheMADS genes were reduced at the floral bud formation stage, increased at the third flowering developmental stage and embryo formation stage. Besides, PheMADS26 (Bs-class), PheMADS68 (MIKC∗-type) and PheMADS72 (Mβ-class) were expressed predominantly at the floral formation stage.
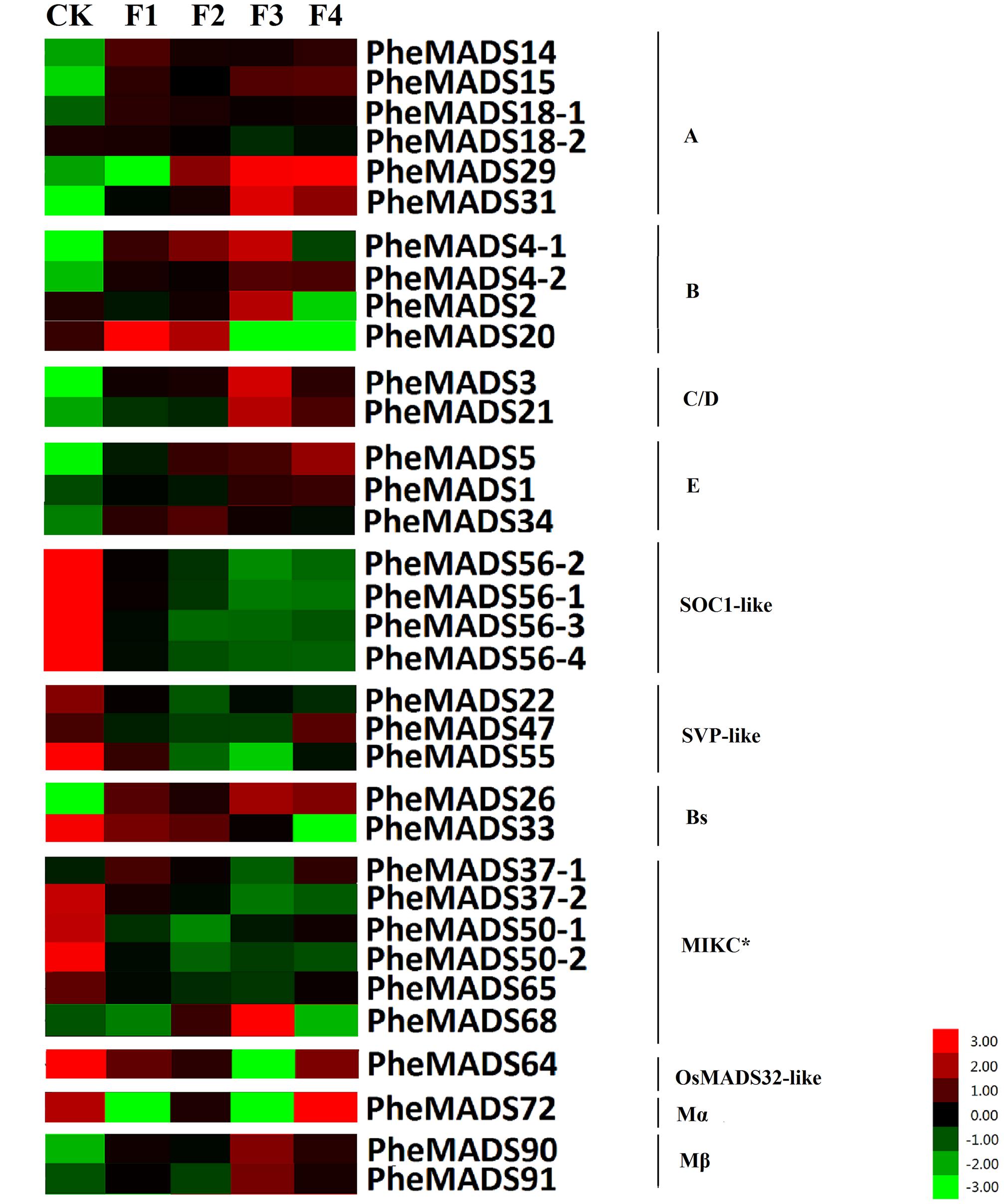
FIGURE 4. Expression Analysis of MADS-box genes in P. edulis. Hierarchical cluster display of expression profile for 34 MADS-box genes showing different expression levels in during floral organ development (CK, F1, F2, F3, and F4). Color bar at the base represents RPKM expression values, thereby green color representing low level expression, black shows medium level expression and red signifies high level expression.
From the outer to the inner whorl within the floral organ, the P. edulis flower consisted of four concentric whorls comprising lemma (whorl 1), palea (whorl 1), three lodicules (whorl 2), three stamens (whorl 3) and in the center, pistil (whorl 4) in which the ovule develops (Figures 5F,L,R and Supplementary Figure S3). These organs together formed a floret. Our results indicated that A-class genes, PheMADS14, PheMADS15, and PheMADS18-1 were expressed throughout, and higher at the floral bud formation stage, while PheMADS29 and PheMADS30 were preferentially expressed from F2 to F4 (Figure 4). Based on in situ hybridization analysis, PheMADS15 was expressed in the early spikelet meristem, the primordia of flower organs, and the reproductive organs (Figures 5A,G,M). Based on the phylogenetic tree analysis, the C and D class contain PheMADS3 and PheMADS21 (Figure 1). PheMADS3 and PheMADS21 were mainly expressed in stamen and pistils formation stage (Figure 4). In addition, the in situ hybridization data showed that PheMADS3 and PheMADS21 mRNA were highly expressed in the stamen and developing embryo (Figures 5C,D,I,J,O,P). These data were consistent with those of PheMADS3 and PheMADS21 from RNA-seq. The E class genes in the SEP lineage in P. edulis were PheMADS1, PheMADS5, and PheMADS34. PheMADS1 and PheMADS5 were highly expressed in the third flowering developmental stage and embryo formation stage (Figure 4). The spatial and temporal expression patterns PheMADS5 were detected from the early floral bud to the maturely floral organ by in situ hybridization in P. edulis (Figures 5E,K,Q). However, PheMADS34 was expressed predominantly at the floral bud formation stage and declined during floral development. In addition, the functionally characterized MADS-box genes of rice and Arabidopsis are listed in Supplementary Table S2 which provided support toward ABCDE model. Sense controls for five MADS-box genes are in Figure 6.
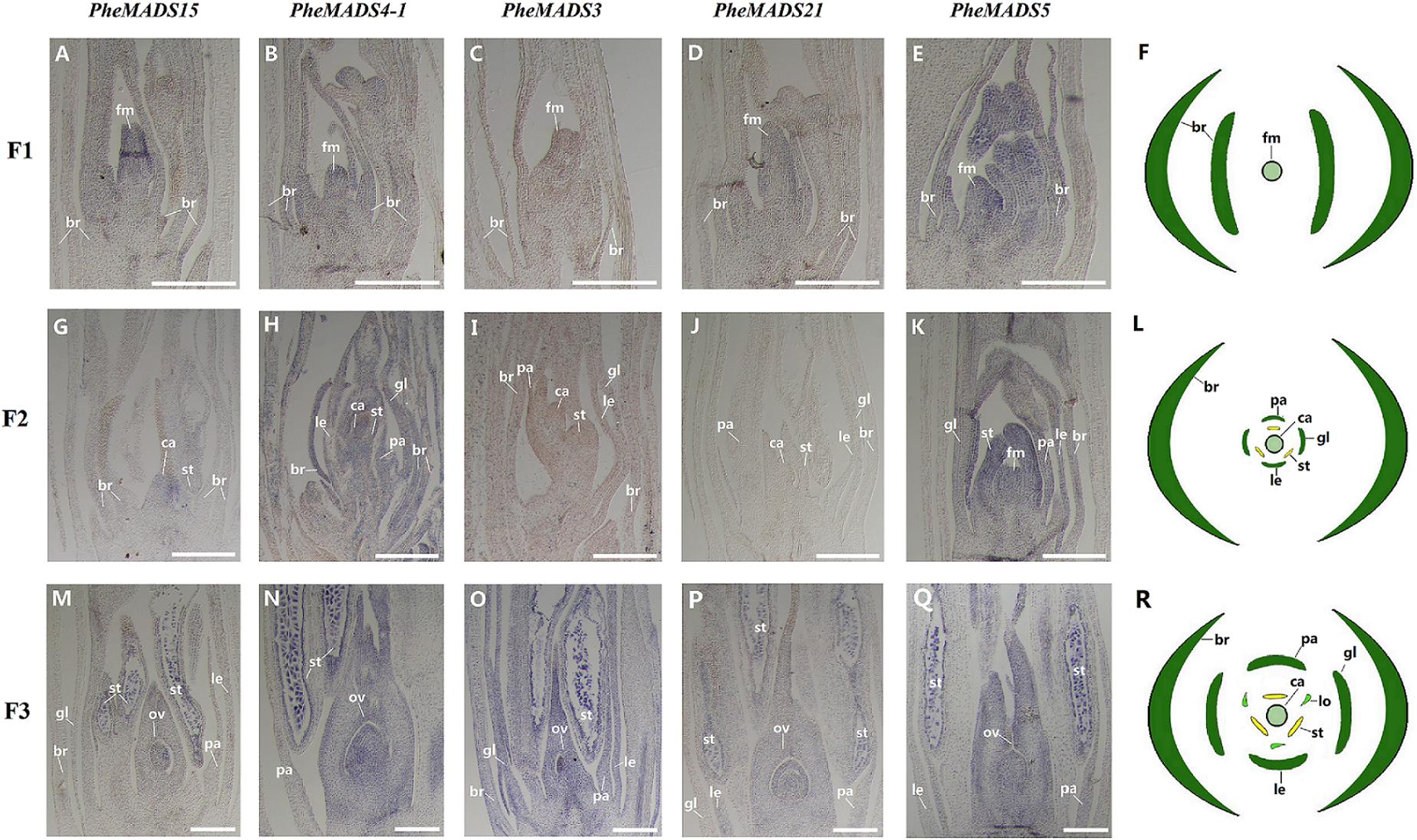
FIGURE 5. In situ analysis of PheMADS15, PheMADS4-1, PheMADS3, PheMADS21, and PheMADS5. F1: The floral bud formation stage. F2: The inflorescence growing stage. F3: The flowers with both pistils and stamens emerging from glumes at bloom stage. A-S: In situ localization of PheMADS15 (A,G,M), PheMADS4-1 (B,H,N), PheMADS3 (C,I,O), PheMADS21 (D,J,P) and PheMADS5 (E,K,Q) transcripts from F1 to F3; floral diagrams of F1 (F), F2 (L), and F3 (R). fm, floral meristem; br, bract; gl, glume; le, lemma; pa, palea; st, stamen; ca, capel; ov, ovule. Bars = 100 μm.
FIGURE 6. Sense controls of PheMADS15, PheMADS4-1, PheMADS3, PheMADS21, and PheMADS5. F1: The floral bud formation stage. F2: The inflorescence growing stage. F3: The flowers with both pistils and stamens emerging from glumes at bloom stage. (A–O) Sense controls for PheMADS15 (A,F,K), PheMADS4-1 (B,G,L), PheMADS3 (C,H,M), PheMADS21 (D,I,N), and PheMADS5 (E,J,O) transcripts from F1 to F3. Bars = 100 μm.
On the contrary, the expression of the remaining genes of PheMADSs was lower than that of ABCDE PheMADS genes in P. edulis floral development. However, PheMADS26 (Bs-class), PheMADS68 (MIKC∗-type) and PheMADS72 (Mβ-class) were expressed predominantly at the floral formation stage (Figure 4). Perhaps these genes might be also involved in the development of flower organs.
To confirm that PheMADS genes from RNA-seq are expressed, eight genes were selected for validation by qPCR (Figure 7). The expression of two PheMADS genes (PheMADS3 and 15) were up-regulated in floral tissues 10-fold more than non-flowering leaves. However, the expression of PheMADS56-1 decreased significantly during flower development. According to the qPCR results, the expression patterns for all eight genes from qPCR were similar to that obtained from the Illumina analysis, thus strengthening the reliability of the RNA-seq data.
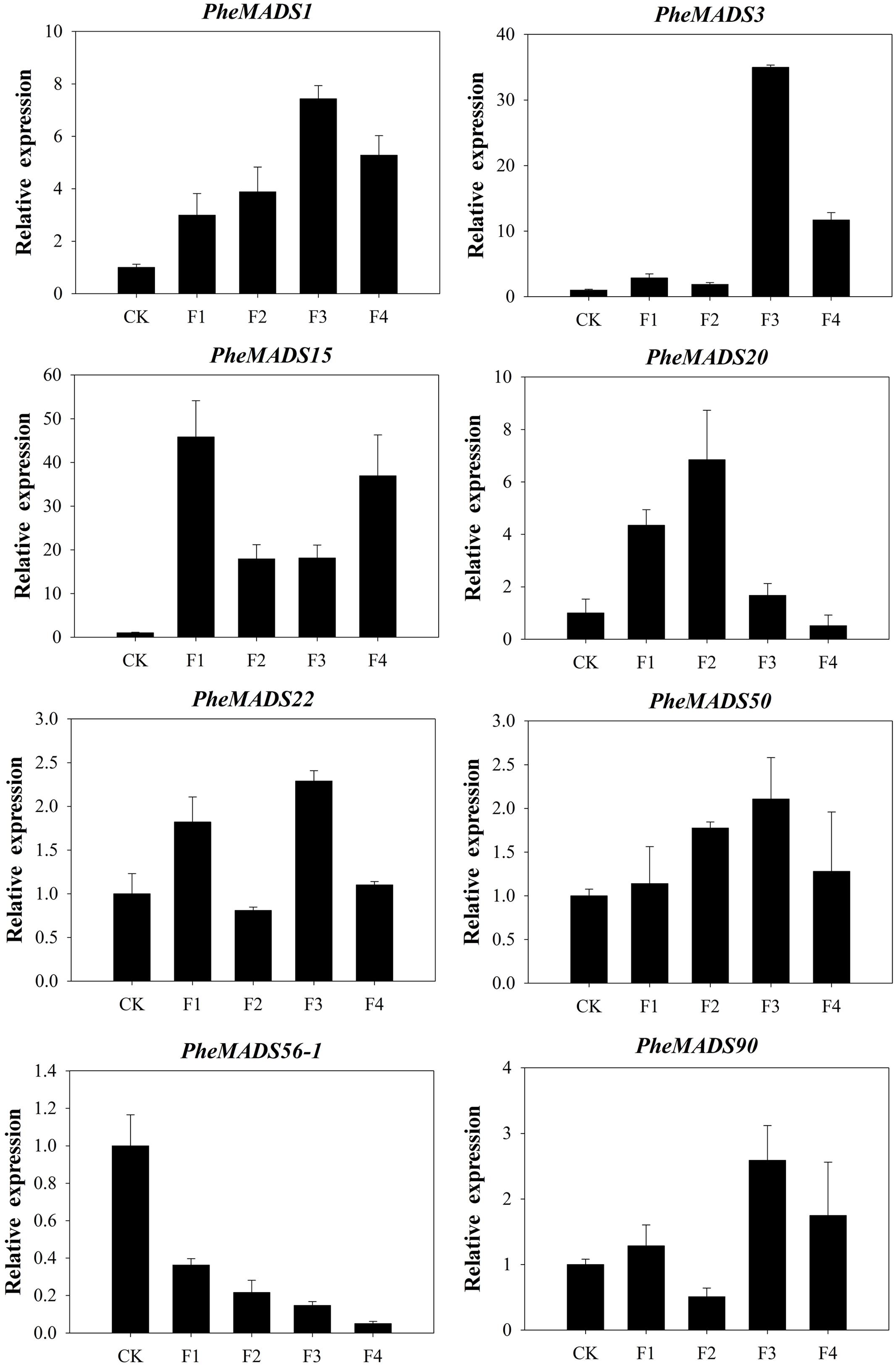
FIGURE 7. The expression profiles of eight selected genes from flowering tissues in different flower developmental stages and leaves of non-flowering plants (CK). The transcript levels were normalized to that of TIP41 (Fan et al., 2013), and the level of each gene in the control was set at 1.0. Error bars represented the SD for three independent experiments.
Duplication and Functional Divergence of MADS-Box Gene Pairs in P. edulis
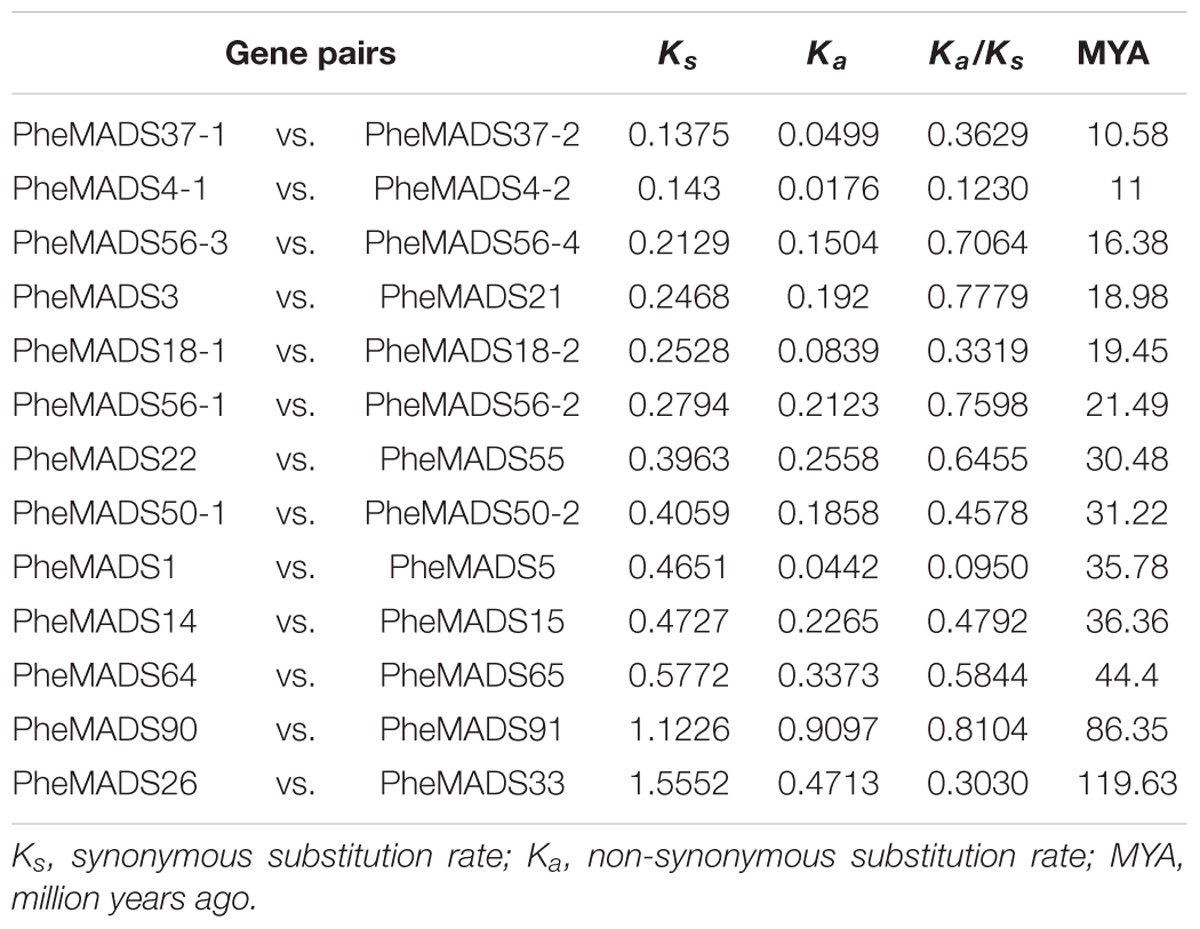
A significant role for gene duplication in the proliferation and evolution of biological complexity of MADS-box genes has been postulated in many divergent plant species (Alvarez-Buylla et al., 2000b; Shan et al., 2009). Most duplicated genes diverge to compartmentalize function (sub-functionalization) or gain novel function (neo-functionalization), and can increase biological complexity (Lynch and Conery, 2000; Ohno, 2013). A rate of 6.5 × 10-9 substitution per synonymous site per year was used to calculate the divergence time between 13 pairs of closely related MADS-box genes in a phylogenetic tree (Gaut et al., 1996). The divergence for most PheMADS-box gene pairs is around 10 to 30 MYA (Million Years Ago) (Table 1), which is a similar time fram as the P. edulis WGD event (Peng et al., 2013), which occurred later than Brachypodium at ∼70 MYA (Wei et al., 2014). In contrast, six gene pairs (PheMADS50-1/50-2, 1/5, 14/15, 64/65, 90/91, and 26/33) diverged 31 to 119 MYA, which does not correlate with the P. edulis WGD.
A Ka/Ks ratio less than 1 is indicative of purifying selection and a ratio greater than 1 is indicative of diversifying selection. With pairwise comparisons we found that for all 13 PheMADS-box gene pairs evolved under purifying selection (Table 1). Interestingly, further analyses indicated that some closely related gene pairs had different expression patterns and subtle functional divergence. Most notably, for MIKC∗-type, PheMADS37-1 expressed predominantly during floral development, whereas PheMADS37-2 expression was detectable in leaves from non-flowering plants. These results indicated that PheMADS-box genes diverged in function whilst also undergoing strong purifying selection.
Identification and Sequence Analysis of the PheMADS15 Gene
To elucidate the role of PheMADS15 in flower formation in P. edulis, we identified the PheMADS15 cDNA encoding a highly conserved MADS domain. PheMADS15 appeared to be a full-length cDNA of 630 bp encoding a protein of 209 amino acids residues (Supplementary Figure S2).
While green fluorescent protein (GFP) alone exhibited a dispersed cytoplasmic distribution, GFP tagged PheMADS15 was indeed located in the nucleus, in accordance with its function as a transcription factor (Figure 8A). In addition, we fused PheMADS15 with the GAL4 DNA-binding domain (GAL4DB) and tested its ability in a yeast reporter construct. PheMADS15 was able to activate the expression of the His-3 and β-Gal reporter gene (Figure 8B). PheMADS15 was highly expressed in early blooming stages (Figure 5A), closely followed by later blooming stages, but just above detection in leaf samples. These results indicated that PheMADS15 might play an important role in flower formation at an early stage, as a transcription factor.
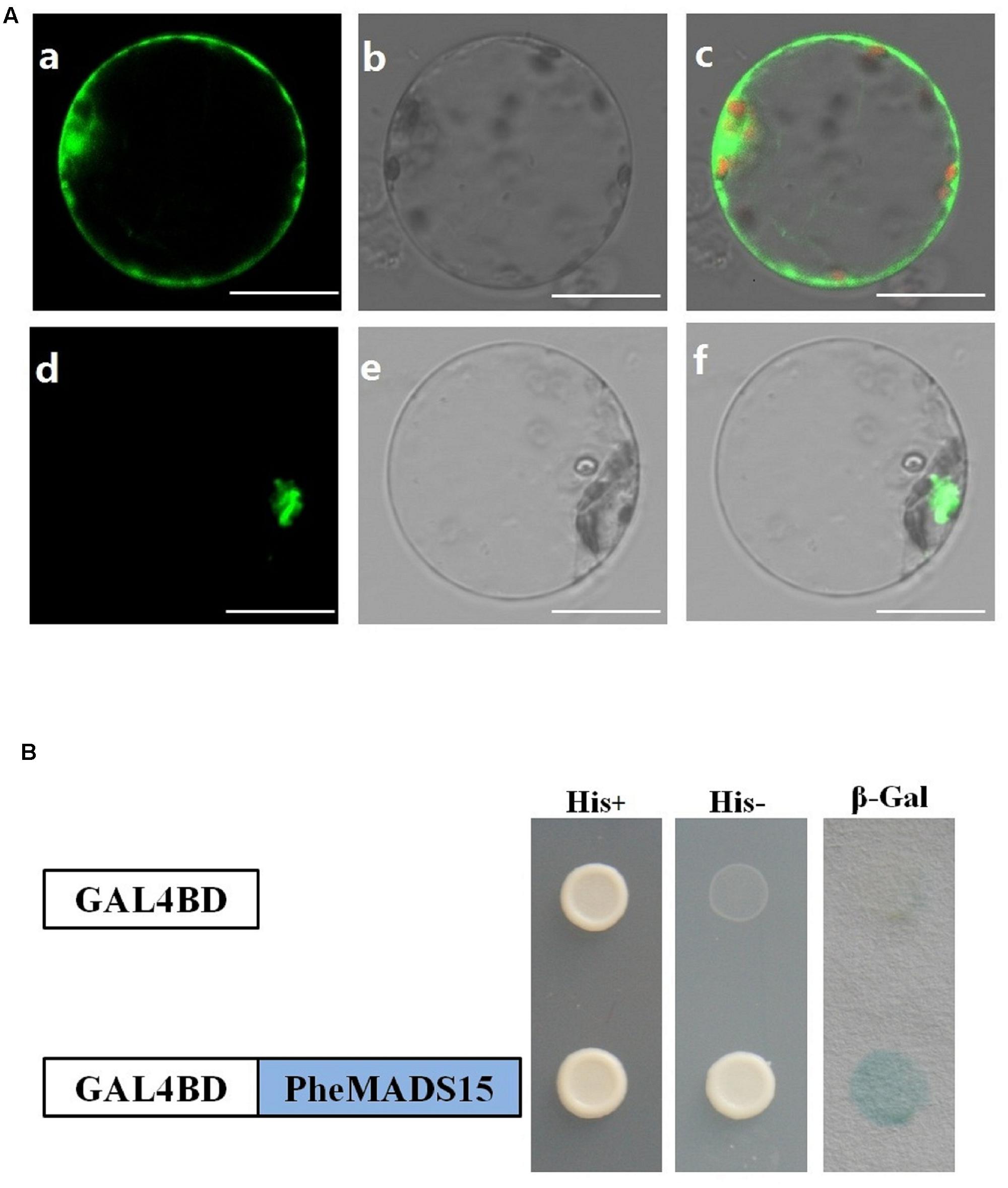
FIGURE 8. Subcellular localization and transcriptional activation analysis of PheMADS15. (A) (a–c), rice protoplasts expressing 35S-GFP, (d–f), rice protoplasts expressing PheMADS15-GFP. Bar = 10 μm. (B) Transcriptional activation analysis of full-length PheMADS15 fused with the GAL4 DNA binding domain in yeast showing its ability to activate the expression of the His-3 and β-Gal reporter genes.
Overexpression of PheMADS15 in Arabidopsis Plants (Wild-Type) Promotes Flowering Time
To further investigate the role of PheMADS15 in the transcriptional regulation of flowering time, PheMADS15 was overexpressed in Arabidopsis (WT). At least 54 transgenic plants expressing PheMADS15 were generated and examined for their morphology in the T1 generation (Supplementary Figure S1). The overexpressed plants showed an early flowering phenotype (Figures 9A,B). We further investigated the expression of SOC1, LFY, and TFL1 in the T3 generation to ascertain the downstream effects of this construct (Figure 9C). SOC1 and LFY had a dramatic expression increases, while TFL1 expression was rather low in compared to wild type (Figure 9C), which was a similar phenomenon exhibited by overexpression of Arabidopsis AP1 (Liljegren et al., 1999).
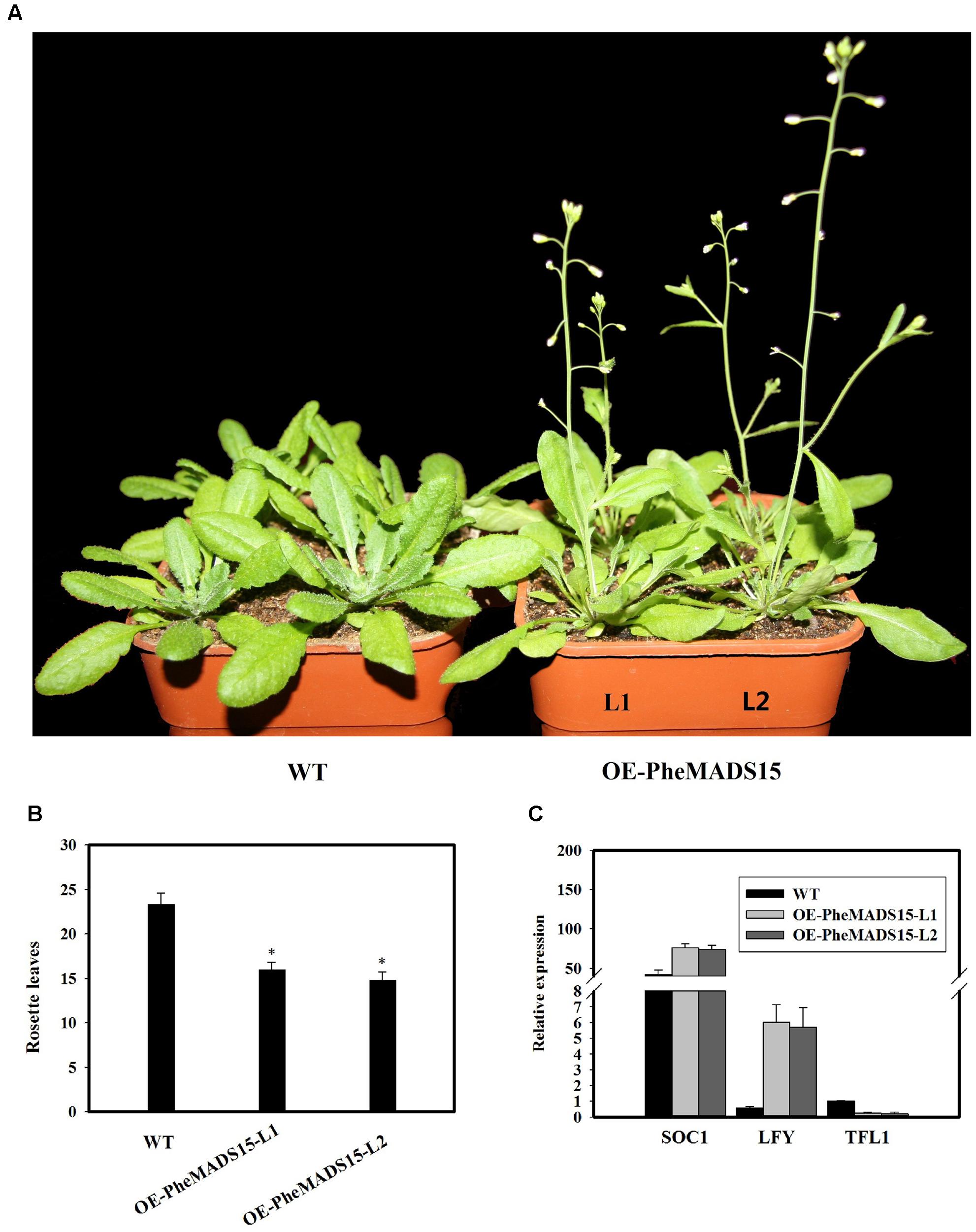
FIGURE 9. Analysis of an early flowering phenotype by overexpression of PheMADS15 in Arabidopsis. (A) The flowering phenotype of wild-type (WT), OEPheMADS15-L1 and OEPheMADS15-L2 grown for 4 weeks at 23°C under long-day conditions. (B) Flowering time scored as the number of rosette leaves at flowering of wild-type and transgenic plants at 23°C under long-day conditions. (C) Transcript levels of SOC1, LFY, and TFL1 in wild-type and transgenic plants (L1 and L2) were evaluated by qPCR. Arabidopsis β-tubulin expression was used as a control. Total RNA from 5-week-old whole- Arabidopsis tissues, including leaves and shoot apex, were used for PheMADS15, SOC1, LFY, and TFL1 examination. Error bars indicate standard deviations. Asterisks indicate a statistically significant difference between wild-type and transgenic plants (P < 0.05 by student’s t-test).
Discussion
The Slow Birth and Death Rate for MADS-Box Genes of P. edulis
The MADS-box gene family in plants has expanded though gene duplication events owing to multiple whole genome duplication events in many plants (Gaut, 2002; Paterson et al., 2004; Yu et al., 2005). Most of the Type II MADS-box genes that mainly control flower development were generally associated with some whole genome duplication events (Causier et al., 2005). On the contrary, the duplications inducing more Type I MADS-box genes can be attributed to smaller scale local duplication events (Nam et al., 2004). We found that P. edulis had a comparable number of MADS-box genes in type II group, but significantly fewer of Mα and Mβ genes than rice and Arabidopsis, indicating that P. edulis genome experienced tandem duplications (Vogel et al., 1992; Guo et al., 2004). Six pairs (PheMADS37-1/37-2, PheMADS4-1/4-2, PheMADS56-3/56-4, PheMADS3/21, PheMADS18-1/18-2, and PheMADS56-1/56-2) in P. edulis had very consistent divergence times, suggesting that these gene pairs followed the WGD event of P. edulis. However, for Mβ, PheMADS90/91 divergence was estimated at about 86 MYA and represented a anciently duplicated gene pair, indicating a smaller scale local duplication event. Thus, for P. edulis, fewer duplication events led to a slower birth and death rate after bamboo diverged from other grasses (Peng et al., 2013).
ABCDE Genes Have Important Functional Conservation and Diversification among P. edulis, Rice and Arabidopsis
MADS-box genes have been found to evolve through neofunctionalization or subfunctionalization after gene duplication events (Irish and Litt, 2005). Moreover, we found that homologous MADS-box genes had different expression profiles, which offered some evidence about functional divergence occurring after the divergence of P. edulis, rice and Arabidopsis (Vogel et al., 1992).
In Arabidopsis, AP1 played an important role in the determination of the identity of sepals and petals and furthermore specifies floral meristem identity (Kater et al., 2006). The AP1 homolog OsMADS14 was highly expressed in inflorescence and caryopses through transcript analysis (Pelucchi et al., 2002). Besides, OsMADS15 and OsMADS18 were activated in the meristem at phase transition in rice (Kobayashi et al., 2012). In Bambusa edulis, as the A class gene, BeMADS14 was expressed throughout, but higher in the lemma and pistil, BeMADS15 was detected in the lemma and palea (Shih et al., 2014). Based on RNA-seq analysis, PheMADS14 showed a similar expression pattern, but very low expression in floral organs differentiation stage (Figure 4). Meanwhile PheMADS15 mRNA obviously accumulated in the meristem at phase transition by in situ hybridization. These data showed that PheMADS15 was involved in flower bud differentiation. The expression pattern of PheMADS18-1 and PheMADS18-2 which were detected in flower bud formation, was different from OsMADS18 with high expression in leaves following germination (Fornara et al., 2004). In P. edulis, PheMADS29 and PheMADS31 were mainly expressed in mature organs and developing caryopses. These data were consistent with that of OsMADS29, which was expressed in seed development of rice (Yang et al., 2012). Our results showed that five AP1-like genes were uniformly expressed in P. edulis floral organs. This similar expression pattern in floral organs was also shown for AP1-like genes in Arabidopsis (Mandel et al., 1992) and rice (Arora et al., 2007).
In Arabidopsis, AP3 and PI, two class B floral organ identity genes, belonged to the DEF-like and GLO-like gene groups, respectively (Jack et al., 1992; Goto and Meyerowitz, 1994). Rice in situ hybridization data showed that two PI homologs, OsMADS2 and OsMADS4 played important roles in lodicule and stamen development (Yao et al., 2008). Whether of PI or AP3 lineage, the mRNA of B class genes (PheMADS2, PheMADS4-1 and PheMADS4-2) showed a similar expression pattern: mainly in stamen development (F3) (Figure 4). Rice OsMADS16/SPW1 and maize SILKY1 (SL) mRNA were detected mainly in the lodicules and stamen primordia during floral development, but not in developing carpels (Ambrose et al., 2000; Nagasawa et al., 2003). In wheat, the expression of TaAP3 was obviously accumulated in mature female organs, but the function of TaAP3 was unknown (Paollacci et al., 2007). To further explore the spatial and temporal expression pattern of B class genes, a stronger expression of PheMADS4-1 was observed in stamen by in situ hybridization (Figures 5B,H,N). For B. edulis, BeMADS2, the PI/GLO-like gene, also displayed similar expression patterns with PheMADS4-1, was highly expressed in anthers (Shih et al., 2014). This result correlated with that of PI and AP3. However, only PheMADS20 was strongly expressed in the spikelet primordia before lemma and palea initiation (F1) (Figure 4).
In Arabidopsis, AG was a typical class C gene (Yanofsky et al., 1990). As proposed by the ABC model, the AG gene was essential to specify stamen and carpel identity and floral determinacy. In rice, analysis of osmads3 osmads58 double mutant revealed the fact that OsMADS3 and OsMADS58 were involved in reproductive organ identity and accumulation of lodicules in the whorl 3 and whorl 4 (Dreni et al., 2011). Besides, in wheat, TaAG-1 and TaAG-2 transcripts were highly expressed in the stamens and pistils (Paollacci et al., 2007). PheMADS3, was also detected in stamens, carpels and ovule primordial by in situ hybridization (Figure 5O). PheMADS21 was also part of the AG-lineage and mainly expressed in anthers and pistils, with especially high levels in anthers by in situ hybridization (Figure 5P). In Arabidopsis, the class D gene, STK, was exclusively expressed in ovules (Pinyopich et al., 2003). In rice, two class D genes have been identified, namely OsMADS13 (Lopez-Dee et al., 1999) and OsMADS21 (Lee et al., 2003) based on phylogenetic reconstruction. The expression pattern of OsMADS13 which was specifically expressed in the ovule was very similar to maize ZAG2 and Arabidopsis STK (Schmidt et al., 1993; Lopez-Dee et al., 1999; Dreni et al., 2007). Moreover, the expression region of OsMADS21 which was highly expressed in the inner cell layers of the ovary and in the ovule integuments, overlapped with that of OsMADS13 (Dreni et al., 2007). The expression pattern of PheMADS21 was slightly different from the Arabidopsis ortholog STK and the rice ortholog OsMADS21. Thus, much deeper investigations are needed to further substantiate the classification and functioning of PheMADS3 and PheMADS21.
In Arabidopsis, the E class genes, such as Arabidopsis SEP genes were involved in the specification of sepal, petal, stamen, carpel, and ovule identity and interact with the class A, B, C, and D genes to form higher order MADS-box protein complexes (Honma and Goto, 2001; Pelaz et al., 2001; Favaro et al., 2003). In P. edulis, three E class genes, such as PheMADS1, PheMADS5, and PheMADS34 belonged to the SEP lineage. On in situ hybridization analysis, PheMADS5 was highly expressed throughout the floral meristem and subsequently detected in palea, lemma, and anthers in the mature flower (Figures 5E,K,Q). In rice, OsMADS5, a SEP-like gene, caused homeotic transformation of all floral organs except the lemma into leaf-like organs (Cui et al., 2010). The expression of PheMADS34 which was high in flower bud formation, was similar to OsMADS34. However, OsMADS34 played a key role in lemma/palea, lodicules, stamens, and carpel (Gao et al., 2010). These findings show many significant differences can be observed between rice and moso bamboo. Future functional studies will have to explore biological function of these PheMADS genes.
Overexpression of PheMADS15 Promotes Flowering Time
In rice, OsMADS14 and OsMADS15 were previously identified as flowering regulators (Kim et al., 2007; Lu et al., 2012). Here, we report the identification and characterization of a MADS-box gene from P. edulis, PheMADS15, which through ectopic overexpression triggered earlier flowering time in Arabidopsis. PheMADS15, an AP1-like gene, is highly expressed during flower bud morphological differentiation (Figure 5A) and it is located in the nucleus (Figure 8B). LFY and AP1 which were expressed in the converted floral meristems primarily control Arabidopsis flower meristems (Shannon and Meeks-Wagner, 1991; Weigel et al., 1992; Ferrándiz et al., 2000). In this study, early flowering time was also observed in 35S: PheMADS15 transgenic Arabidopsis. Meanwhile, the expression level of LFY and SOC1 was up-regulated in 35S:PheMADS15 transgenic Arabidopsis compared with wild type Arabidopsis. SOC1 promoted floral transitions and is considered as one of the core regulators of flowering in Arabidopsis (Moon et al., 2003; Liu et al., 2008; Lee and Lee, 2010; Dorca-Fornell et al., 2011). AP1 is another positive regulator of LFY and is highly expressed in converted floral meristems (Liljegren et al., 1999). In addition, OsMADS14, OsMADS15, and OsMADS18, three AP1/FUL-like genes, were involved in the regulation of flowering time (Kobayashi et al., 2012). This leads us to suspect that PheMADS15 promotes flowering time by regulating LFY and SOC1 directly or indirectly. This is consistent with the previous reports about some AP1-like genes (Weigel et al., 1992), suggesting that PheMADS15 is a functional ortholog of Arabidopsis AP1. Further research on the transcriptional regulatory network mediated by PheMADSs will increase knowledge surrounding the transcriptional regulation of flowering time in P. edulis.
Author Contributions
ZC, WG, and LL performed all the experiments, data analysis and wrote the paper. XL and SM analyzed data. DH, YM, JL, and QB revised the manuscript. JG designed and supervised experiments.
Funding
This work was supported by National High Technology Research and Development Program of China “Moso Bamboo Functional Genomics Research” (Grant No.2013AA102607-4), the National Natural Science Foundation of China (31570673) and Fundamental Research Funds of ICBR [grant No.1632015009].
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank CC (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for providing rice protoplast transformation technology platform and Prof. Xiaoting Qi (Capital Normal University, Beijing, China) for providing the yeast strain PJ69-4a.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00656/full#supplementary-material
Footnotes
- ^http://www.arabidopsis.org/
- ^http://rice.plantbiology.msu.edu/
- ^http://202.127.18.221/bamboo/down.php
- ^http://web.expasy.org/compute_pi/
- ^http://www.clustal.org/
- ^http://www.megasoftware.net/mega.html
- ^http://alternate.meme-suite.org/tools/meme
- ^http://www.bamboogdb.com/
- ^http://gsds.cbi.pku.edu.cn/
References
Alvarez-Buylla, E. R., Liljegren, S. J., Pelaz, S., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000a). MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24, 457–466.
Alvarez-Buylla, E. R., Pelaz, S., Liljegren, S. J., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000b). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. U.S.A. 97, 5328–5333.
Ambrose, B. A., Lerner, D. R., Ciceri, P., Padilla, C. M., Yanofsky, M. F., and Schmidt, R. J. (2000). Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5, 569–579. doi: 10.1016/S1097-2765(00)80450-5
Angenent, G. C., and Colombo, L. (1996). Molecular control of ovule development. Trends Plant Sci. 1, 228–232. doi: 10.1016/S1360-1385(96)86900-0
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242. doi: 10.1186/1471-2164-8-242
Bailey, T. L., and Elkan, C. (1995). “The value of prior knowledge in discovering motifs with MEME,” in Proceedings of the Third International Conference on Intelligent Systems for Molecular Biology, Vol. 3, Menlo Park, CA, 21–29.
Bowman, J. L., Alvarez, J., Weigel, D., Meyerowitz, E. M., and Smyth, D. R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–721.
Causier, B., Castillo, R., Zhou, J., Ingram, R., Xue, Y., Schwarz-Sommer, Z., et al. (2005). Evolution in action: following function in duplicated floral homeotic genes. Curr. Biol. 15, 1508–1512. doi: 10.1016/j.cub.2005.07.063
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Cui, R., Han, J., Zhao, S., Su, K., Wu, F., Du, X., et al. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61, 767–781. doi: 10.1111/j.1365-313X.2009.04101.x
de Almeida Engler, J., De Groodt, R., Van Montagu, M., and Engler, G. (2001). In situ hybridization to mRNA of Arabidopsis tissue sections. Methods 23, 325–334. doi: 10.1006/meth.2000.1144
de Hoon, M. J., Imoto, S., Nolan, J., and Miyano, S. (2004). Open source clustering software. Bioinformatics 20, 1453–1454. doi: 10.1093/bioinformatics/bth078
Ditta, G., Pinyopich, A., and Robles, P. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. doi: 10.1016/j.cub.2004.10.028
Dorca-Fornell, C., Gregis, V., Grandi, V., Coupland, G., Colombo, L., and Kater, M. M. (2011). The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 67, 1006–1017. doi: 10.1111/j.1365-313X.2011.04653.x
Dreni, L., Jacchia, S., Fornara, F., Fornari, M., Ouwerkerk, P. B., An, G., et al. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52, 690–699. doi: 10.1111/j.1365-313X.2007.03272.x
Dreni, L., Pilatone, A., Yun, D., Erreni, S., Pajoro, A., Caporali, E., et al. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23, 2850–2863. doi: 10.1105/tpc.111.087007
Ehrenreich, I. M., Hanzawa, Y., Chou, L., Roe, J. L., Kover, P. X., and Purugganan, M. D. (2009). Candidate gene association mapping of Arabidopsis flowering time. Genetics 183, 325–335. doi: 10.1534/genetics.109.105189
Fan, C., Ma, J., Guo, Q., Li, X., Wang, H., and Lu, M. (2013). Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 8:e56573. doi: 10.1371/journal.pone.0056573
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15, 2603–2611. doi: 10.1105/tpc.015123
Feldmann, K. A., and Marks, M. D. (1987). Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: a non-tissue culture approach. Mol. Genet. Genomics 208, 1–9. doi: 10.1007/BF00330414
Ferrándiz, C., Gu, Q., Martienssen, R., and Yanofsky, M. F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734.
Fornara, F., de Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550. doi: 10.1016/j.cell.2010.04.024
Fornara, F., Paøenicová, L., Falasca, G., Pelucchi, N., Masiero, S., Ciannamea, S., et al. (2004). Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS-box genes. Plant Physiol. 135, 2207–2219. doi: 10.1104/pp.104.045039
Gao, J., Zhang, Y., Zhang, C., Qi, F., Li, X., Mu, S., et al. (2014). Characterization of the floral Transcriptome of moso bamboo (Phyllostachys edulis). at Different flowering developmental stages by transcriptome sequencing and RNA-Seq analysis. PLoS ONE 9:e98910. doi: 10.1371/journal.pone.0098910
Gao, X., Liang, W., Yin, C., Ji, S., Wang, H., Su, X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153, 728–740. doi: 10.1104/pp.110.156711
Gaut, B. S. (2002). Evolutionary dynamics of grass genomes. New Phytol. 154, 15–28. doi: 10.1046/j.1469-8137.2002.00352.x
Gaut, B. S., Morton, B. R., McCaig, B. C., and Clegg, M. T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. U.S.A. 93, 10274–10279. doi: 10.1073/pnas.93.19.10274
Goto, K., and Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Gene. Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Grass Phylogeny Working Group, Barker, N. P., Clark, L. G., Davis, J. I, Duvall, M. R., Guala, G. F., et al. (2001). Phylogeny and subfamilial classification of the grasses (Poaceae). Ann. Mo. Bot. Gard. 88, 373–457. doi: 10.2307/3298585
Gregis, V., Sessa, A., Dorca-Fornell, C., and Kater, M. M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60, 626–637. doi: 10.1111/j.1365-313X.2009.03985.x
Guo, A. Y., Zhu, Q. H., Chen, X., and Luo, J. C. (2007). GSDS: a gene structure display server. Yi Chuan 29, 1023–1026. doi: 10.1360/yc-007-1023
Guo, X. Y., Xu, G. H., Zhang, Y., Hu, W. M., and Fan, L. J. (2004). Small-scale duplications play a significant role in rice genome evolution. Rice Sci. 12, 173–178.
Henschel, K., Kofuji, R., Hasebe, M., Saedler, H., Munster, T., and Theissen, G. (2002). Two ancient classes of MIKC-type MADS-box genes are present in the Moss Physcomitrella patens. Mol. Biol. Evol. 19, 801–814. doi: 10.1093/oxfordjournals.molbev.a004137
Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. doi: 10.1038/35054083
Hu, L. F., Liang, W. Q., Yin, C. S., Cui, X., Zong, J., Wang, X., et al. (2011). Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23, 515–533. doi: 10.1105/tpc.110.074369
Huang, H., Tudor, M., and Weiss, C. A. (1995). The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant. Mol. Biol. 28, 549–567. doi: 10.1007/BF00020401
Irish, V. F., and Litt, A. (2005). Flower development and evolution: gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 15, 454–460. doi: 10.1016/j.gde.2005.06.001
Irish, V. F., and Sussex, I. M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2, 741–753. doi: 10.1105/tpc.2.8.741
Jack, T. (2001). Plant development going MADS. Plant Mol. Biol. 46, 515–520. doi: 10.1023/A:1010689126632
Jack, T., Brockman, L. L., and Meyerowitz, E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS-box and is expressed in petals and stamens. Cell 68, 683–697. doi: 10.1016/0092-8674(92)90144-2
Jofuku, K. D., Den Boer, B., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Kater, M. M., Dreni, L., and Colombo, L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57, 3433–3444. doi: 10.1093/jxb/erl097
Kim, S. L., Lee, S., Kim, H. J., Nam, H. G., and An, G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494. doi: 10.1104/pp.107.103291
Kobayashi, K., Yasuno, N., and Sato, Y. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS-box genes and PAP2, a SEPALLATA MADS-box gene. Plant Cell 24, 1848–1859. doi: 10.1105/tpc.112.097105
Kofuji, R., Sumikawa, N., Yamasaki, M., Kondo, K., Ueda, K., Ito, M., et al. (2003). Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 20, 1963–1977. doi: 10.1093/molbev/msg216
Kramer, E. M., Dorit, R. L., and Irish, V. F. (1998). Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149, 765–783.
Kramer, E. M., Jaramillo, M. A., and Di Stilio, V. S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS-box genes in angiosperms. Genetics 166, 1011–1023. doi: 10.1534/genetics.166.2.1011
Lee, J., and Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Lee, S., Jeon, J. S., An, K., Moon, Y. H., Lee, S., Chung, Y. Y., et al. (2003). Alteration of floral organ identity in rice through ectopic expression of OsMADS16. Planta 217, 904–911. doi: 10.1007/s00425-003-1066-8
Li, D., Yang, C., Li, X., Gan, Q., Zhao, X., and Zhu, L. (2009). Functional characterization of rice OsDof12. Planta 229, 1159–1169. doi: 10.1007/s00425-009-0893-7
Li, H., Liang, W., Hu, Y., Zhu, L., Yin, C., Xu, J., et al. (2011). Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13 and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23, 2536–2552. doi: 10.1105/tpc.111.087262
Liljegren, S. J., Gustafson Brown, C., Pinyopich, A., Ditta, G. S., and Yanofsky, M. F. (1999). Interactions among APETALA1, LEAFY and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11, 1007–1018. doi: 10.1105/tpc.11.6.1007
Liu, C., Chen, H., Er, H. L., Soo, H. M., Kumar, P. P., Han, J. H., et al. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135, 1481–1491. doi: 10.1242/dev.020255
Lopez-Dee, Z. P., Wittich, P., Enrico, P. M., Rigola, D., Del Buono, I., Gorla, M. S., et al. (1999). OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev. Genet. 25, 237–244. doi: 10.1002/(SICI)1520-6408(1999)25:3<237::AID-DVG6>3.0.CO;2-L
Lu, S. J., Wei, H., Wang, Y., Wang, H. M., Yang, R. F., Zhang, X. B., et al. (2012). Overexpression of a transcription factor OsMADS15 modifies plant architecture and flowering time in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 30, 1461–1469. doi: 10.1007/s11105-012-0468-9
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Mandel, M. A., Gustafson Brown, C., Savidge, B., and Yanofsky, M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. doi: 10.1038/360273a0
Mandel, M. A., and Yanofsky, M. F. (1998). The Arabidopsis AGL 9 MADS-box gene is expressed in young flower primordia. Sex. Plant Reprod. 11, 22–28. doi: 10.1007/s004970050116
Moon, J., Suh, S. S., Lee, H., Choi, K. R., Hong, C. B., Paek, N. C., et al. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623. doi: 10.1046/j.1365-313X.2003.01833.x
Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. doi: 10.1242/dev.00294
Nam, J., dePamphilis, C. W., Ma, H., and Nei, M. (2003). Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 20, 1435–1447. doi: 10.1093/molbev/msg152
Nam, J., Kim, J., Lee, S., An, G., Ma, H., and Nei, M. (2004). Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. U.S.A. 101, 1910–1915. doi: 10.1073/pnas.0308430100
Paollacci, A. R., Tanzarella, O. A., Porceddu, E., Varotto, S., and Ciaffi, M. (2007). Molecular and phylogenetic analysis of MADS-box genes of MIKC type and chromosome location of SEP-like genes in wheat (Triticum aestivum L.). Mol. Genet. Genomics 278, 689–708. doi: 10.1007/s00438-007-0285-2
Pařenicová, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis new openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Paterson, A., Bowers, J., and Chapman, B. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. U.S.A. 101, 9903–9908. doi: 10.1073/pnas.0307901101
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pelaz, S., Tapia-López, R., Alvarez-Buylla, E. R., and Yanofsky, M. F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11, 182–184. doi: 10.1016/S0960-9822(01)00024-0
Pelucchi, N., Fornara, F., Favalli, C., Masiero, S., Lago, C., Pè, E. M., et al. (2002). Comparative analysis of rice MADS-box genes expressed during flower development. Sex. Plant Reprod. 15, 113–122. doi: 10.1007/s00497-002-0151-7
Peng, Z., Lu, Y., Li, L., Zhao, Q., Feng, Q., Gao, Z., et al. (2013). The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 45, 456–461. doi: 10.1038/ng.2569
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Preston, J. C., and Kellogg, E. A. (2007). Conservation and divergence of APETALA1/FRUITFULL-like gene function in grasses: evidence from gene expression analyses. Plant J. 52, 69–81. doi: 10.1111/j.1365-313X.2007.03209.x
Purugganan, M. D. (1997). The MADS-box floral homeotic gene lineages predate the origin of seed plants: phylogenetic and molecular clock estimates. J. Mol. Evol. 45, 392–396. doi: 10.1007/PL00006244
Rudall, P. J., Stuppy, W., Cunniff, J., Kellogg, E. A., and Briggs, B. G. (2005). Evolution of reproductive structures in grasses (Poaceae) inferred by sister-group comparison with their putative closest living relatives, Ecdeiocoleaceae. Am. J. Bot. 92, 1432–1443. doi: 10.3732/ajb.92.9.1432
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Saldanha, A. J. (2004). Java Treeview-extensible visualization of microarray data. Bioinformatics 20, 3246–3248. doi: 10.1093/bioinformatics/bth349
Sang, X., Li, Y., and Luo, Z. (2012). CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS-box protein, regulates floral organ identity in rice. Plant Physiol. 160, 788–807. doi: 10.1104/pp.112.200980
Savidge, B., Rounsley, S. D., and Yanofsky, M. F. (1995). Temporal relationship between the transcription of two Arabidopsis MADS-box genes and the floral organ identity genes. Plant Cell 7, 721–733. doi: 10.1105/tpc.7.6.721
Schmidt, R. J., Veit, B., Mandel, M. A., Mena, M., Hake, S., and Yanofsky, M. F. (1993). Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell 5, 729–737. doi: 10.1105/tpc.5.7.729
Schultz, E. A., and Haughn, G. W. (1991). LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3, 771–781. doi: 10.1105/tpc.3.8.771
Shan, H., Zahn, L., Guindon, S., Wall, P. K., Kong, H., Ma, H., et al. (2009). Evolution of plant MADS-box transcription factors: evidence for shifts in selection associated with early angiosperm diversification and concerted gene duplications. Mol. Biol. Evol. 26, 2229–2244. doi: 10.1093/molbev/msp129
Shannon, S., and Meeks-Wagner, D. R. (1991). A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3, 877–892. doi: 10.1105/tpc.3.9.877
Shih, M. C., Chou, M. L., Yue, J. J., Hsu, C. T., Chang, W. J., Ko, S. S., et al. (2014). BeMADS1 is a key to delivery MADSs into nucleus in reproductive tissues-De novo characterization of Bambusa edulis transcriptome and study of MADS genes in bamboo floral development. BMC Plant Biol. 14:179. doi: 10.1186/1471-2229-14-179
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Teper-Bamnolker, P., and Samach, A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17, 2661–2675. doi: 10.1105/tpc.105.035766
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Munster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. doi: 10.1023/A:1006332105728
Theißen, G., and Saedler, H. (2001). Plant biology: floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tian, B., Chen, Y., Li, D., and Yan, Y. (2006). Cloning and characterization of a bamboo Leafy Hull Sterile1 homologous gene: full length research paper. Mitochondr. DNA 17, 143–151.
Tian, B., Chen, Y., Yan, Y., and Li, D. (2005). Isolation and ectopic expression of a bamboo MADS-box gene. Chin. Sci. Bull. 50, 217–224. doi: 10.1007/BF02897530
Vogel, J. P., Garvin, D. F., and Mockler, T. C. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. doi: 10.1016/0092-8674(92)90295-N
Wei, B., Zhang, R.-Z., Guo, J.-J., Liu, D.-M., Li, A.-L., Fan, R.-C., et al. (2014). Genome-wide analysis of the MADS-box gene family in Brachypodium distachyon. PLoS ONE 9:e84781. doi: 10.1371/journal.pone.0084781
Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F., and Meyerowitz, E. M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. doi: 10.1016/0092-8674(92)90295-N
Whipple, C. J., Ciceri, P., and Padilla, C. M. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 2004, 6083–6091. doi: 10.1242/dev.01523
Whipple, C. J., Zanis, M. J., Kellogg, E. A., and Schmidt, R. J. (2007). Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc. Natl. Acad. Sci. U.S.A. 104, 1081–1086. doi: 10.1073/pnas.0606434104
Yamaguchi, T., Lee, D. Y., Miyao, A., Hirochika, H., An, G. H., and Hirano, H.-Y. (2006). Functional diversification of the two C-class MADS box genes OsMADS3 and OsMADS58 in Oryza sativa. Plant Cell 18, 15–28. doi: 10.1105/tpc.105.037200
Yang, X., Wu, F., Lin, X., Du, X., Chong, K., Gramzow, L., et al. (2012). Live and let die - the Bsister MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa). PLoS ONE 7:e51435. doi: 10.1371/journal.pone.0051435
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Yao, S.-G., Ohmori, S., Kimizu, M., and Yoshida, H. (2008). Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol. 49, 853–857. doi: 10.1093/pcp/pcn050
Yu, J., Wang, J., Lin, W., Li, S., Li, H., Zhou, J., et al. (2005). The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:e38. doi: 10.1371/journal.pbio.0030038
Keywords: Phyllostachys edulis, MADS-box, floral organ, ABCDE model, PheMADS15
Citation: Cheng Z, Ge W, Li L, Hou D, Ma Y, Liu J, Bai Q, Li X, Mu S and Gao J (2017) Analysis of MADS-Box Gene Family Reveals Conservation in Floral Organ ABCDE Model of Moso Bamboo (Phyllostachys edulis). Front. Plant Sci. 8:656. doi: 10.3389/fpls.2017.00656
Received: 13 January 2017; Accepted: 10 April 2017;
Published: 03 May 2017.
Edited by:
Borja Cascales-Miñana, University of Liège, BelgiumReviewed by:
Chengcai Chu, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, ChinaVeronica Gregis, Università degli Studi di Milano, Italy
Copyright © 2017 Cheng, Ge, Li, Hou, Ma, Liu, Bai, Li, Mu and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Gao, gaojianicbr@163.com
†These authors have contributed equally to this work.
 Zhanchao Cheng†
Zhanchao Cheng† Jian Gao
Jian Gao