
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 May 2017
Sec. Plant Proteomics and Protein Structural Biology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00810
 Sijia Wei1†
Sijia Wei1† Yangyang Bian2†
Yangyang Bian2† Qi Zhao3
Qi Zhao3 Sixue Chen4
Sixue Chen4 Jiawei Mao2
Jiawei Mao2 Chunxia Song2
Chunxia Song2 Kai Cheng2
Kai Cheng2 Zhen Xiao3
Zhen Xiao3 Chuanfang Zhang3
Chuanfang Zhang3 Weimin Ma3
Weimin Ma3 Hanfa Zou2‡
Hanfa Zou2‡ Mingliang Ye2*
Mingliang Ye2* Shaojun Dai1,3*
Shaojun Dai1,3*Palmella stage is critical for some unicellular algae to survive in extreme environments. The halotolerant algae Dunaliella salina is a good single-cell model for studying plant adaptation to high salinity. To investigate the molecular adaptation mechanism in salinity shock-induced palmella formation, we performed a comprehensive physiological, proteomics and phosphoproteomics study upon palmella formation of D. salina using dimethyl labeling and Ti4+-immobilized metal ion affinity chromatography (IMAC) proteomic approaches. We found that 151 salinity-responsive proteins and 35 salinity-responsive phosphoproteins were involved in multiple signaling and metabolic pathways upon palmella formation. Taken together with photosynthetic parameters and enzyme activity analyses, the patterns of protein accumulation and phosphorylation level exhibited the mechanisms upon palmella formation, including dynamics of cytoskeleton and cell membrane curvature, accumulation and transport of exopolysaccharides, photosynthesis and energy supplying (i.e., photosystem II stability and activity, cyclic electron transport, and C4 pathway), nuclear/chloroplastic gene expression regulation and protein processing, reactive oxygen species homeostasis, and salt signaling transduction. The salinity-responsive protein–protein interaction (PPI) networks implied that signaling and protein synthesis and fate are crucial for modulation of these processes. Importantly, the 3D structure of phosphoprotein clearly indicated that the phosphorylation sites of eight proteins were localized in the region of function domain.
The unicellular algae can develop a vegetative palmella in their life cycle, when exposed to various extreme environment conditions, such as salinity (Takouridis et al., 2015), copper (Sztrum et al., 2012), organic acids (Iwasa and Murakami, 1968), herbicide (Franqueira et al., 2000), oxidative stress (Wang et al., 2004a), and predators (Lurling and Beekman, 2006). The palmella formation in Chlamydomonas reinhardtii (Iwasa and Murakami, 1968; Lurling and Beekman, 2006; Sztrum et al., 2012) and Pediastrum tetras (Ellis, 1972) have been found to be regulated by free calcium concentration, starch accumulation, carotenoid/chlorophyll ratio, as well as the activities of catalase (CAT) and ascorbate peroxidase (APX). Similarly, for green motile flagellates of Haematococcus pluvialis, oxidative stress could induce its green motile cells transforming into enlarged red resting cysts (aplanospores) (Boussiba et al., 1999; Wang et al., 2004a; Han et al., 2012). In H. pluvialis, proteomics study revealed that the abundances of several proteins were induced, which were involved in reactive oxygen species (ROS) scavenging [e.g., superoxide dismutase (SOD), CAT and peroxidase (POD)], photosynthesis [e.g., ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (Rubisco LSU), phosphoglycerate kinase (PGK)], nitrogen assimilation [e.g., glutamine synthetase (GS)], and mitochondrial respiratory (e.g., mitochondrial ATPase β subunit) (Wang et al., 2004a). Moreover, the reduction of chlorophyll content, moderate declines in the maximal photosynthetic rate and the maximum quantum yield of photosystem (PS) II, as well as the significant increase in PS I activity were detected during the transformation of green vegetative cells to red aplanospores in H. pluvialis (Han et al., 2012). These indicates that the early stress response involves multiple enzymatic defense processes, which plays a critical role during the transition of green vegetative cells to red cysts (Wang et al., 2004a).
The halotolerant unicellular green algae Dunaliella salina is a model photosynthetic organism for studying plant adaptation to high salinity (Katz et al., 2007), which can adapt to a wide range of salinities ranging from about 0.05 M NaCl to saturation (around 5.5 M NaCl) (Liska et al., 2004). During their life cycle, D. salina may have a dominant palmella stage formed by a colony-like group of round non-motile cells to cope with extreme conditions (Borowitzka and Siva, 2007). When entering palmella stage, the cells usually lose their flagella and eyespot, become round, and excrete a slime exopolysaccharides (EPSs) layer outside the accumulation of green cells, but when returning to fresh medium at a “normal” salinity, algae cells usually reform their flagella and return to the motile and free-swimming state (Borowitzka and Siva, 2007).
It is reported that high salinity triggers palmella formation in D. salina (Montoya and Olivera, 1993), but the molecular mechanism of palmella formation is still unknown. Previous studies mainly focus on the mechanism of remarkable salinity and osmotic adaptability of D. salina. It is found that the de novo synthesis of compatible solute (i.e., glycerol and β-carotene) (Ben-Amotz and Avron, 1973), higher activity of Na+ extrusion systems (Pick, 2002), as well as active photosynthetic and energy metabolism (Liska et al., 2004; Katz et al., 2007; Alkayal et al., 2010) are crucial for salinity-response in D. salina. Importantly, proteomic research has provided new insights into the high salinity-responsive strategies in D. salina. The abundances of several proteins were salinity-regulated, which were involved in Calvin cycle, starch mobilization, energy production, protein synthesis/degradation, membrane structure stabilization, and signal transduction in D. salina (Liska et al., 2004; Katz et al., 2007). However, whether these fine-tuned molecular mechanism also happened in the process of salinity-induced palmella formation is poorly understood.
In this study, the salinity shock-induced palmella formation was analyzed in D. salina. By integrative analysis of morphology changes, photosynthesis, antioxidant enzyme activities, as well as protein abundance and phosphorylation level, we highlighted several important mechanisms upon palmella formation, such as photosynthetic modulation, ROS scavenging, gene expression regulation, and protein post-translational modification.
D. salina cells were cultivated in a modified medium containing 1 M NaCl under a 16/8 h light/dark cycle (light intensity 100 μmol photons × m−2 × s−1) at 26°C with shaking at 96 rpm (Katz and Avron, 1985). Cells in the stationary phase were transferred to fresh medium containing 1 M NaCl (control) and 3 M NaCl (salinity shock), respectively. After treatment, algae cells were used freshly or stored at −80°C for experiments.
The cell growth was evaluated by the absorbance of cultures at 630 nm in a spectrophotometer (Chen et al., 2009). The cell number was counted using a haemacytometer under light microscope. Three independent biological replicates for each sample were conducted for all the experiments. The morphology of D. salina cells were observed under Olympus BX53 Microscope (Olympus America Inc., Center Valley, PA, USA) equipped with Olympus DP72 digital camera system (Tokyo, Japan).
The cell viability was estimated according to the method of Mendes et al. (2013). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) stock solution (60 μl; 5 mg/ml) was applied to 250 μl cells for 60 min at 26°C with shaking at 96 rpm, and then the samples were centrifuged at 8,000 × g for 5 min. The formazan crystals were dissolved in 150 μl of dimethylsulfoxide and vortexed for 10 min. After centrifugation at 12,000 × g for 5 min, the supernatants were transferred to 96-well cell culture plate. The absorbance at 490 nm was measured using an iMark™ Microplate Reader (Bio-Rad, Richmond, CA, USA).
Carbohydrates were analyzed according to the method of Li et al. (2000). Ten milliliter culture was centrifuged at 12,000 × g for 10 min, and then 2 ml of supernatant was analyzed by the anthrone reagent method (Pistocchi et al., 1997). Glycerol content was determined according to the method of Chen et al. (2011).
The chlorophyll contents were determined using a method described by Lichtenthaler and Wellburn (1983). Photosynthesis oxygen evolution and respiration rate were recorded by Clark-type oxygen electrode (Hansatech, UK) according to the method of Ma et al. (2005). The chlorophyll fluorescence parameters and P700 were detected using a pulse-amplitudemodulated chlorophyll fluorometer (Dual-PAM-100; Walz, Effeltrich, Germany) and an emitter-detector-cuvette assembly with a unit 101ED (ED-101US) according to the method of Ma et al. (2008).
Antioxidant enzyme assays were performed essentially according to Suo et al. (2015). For the sample preparation of SOD, CAT, POD, APX, monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), glutathione reductase (GR), glutathione S-transferase (GST), and glutathione peroxidase (GPX), 0.5 g of cells was homogenized on ice in 3 ml buffer [50 mM phosphate buffer (pH 7.8), 2% PVP-40, and 2 mM ascorbate (AsA)]. After centrifugation at 15,000 × g for 20 min at 4°C, the supernatants were used for enzyme activity assays according to the method of Zhao et al. (2016).
For glycerol-3-phosphate dehydrogenase (GPDH) activity assay, cells were ground to a fine powder in liquid nitrogen and resuspended in extraction buffer (100 mM Tris, 20 mM ascorbic acid, pH 6.9) (Chen et al., 2009). After being centrifuged at 34,000 × g for 20 min at 4°C, the supernatant was collected for GPDH activity assay. GPDH activity was determined using a plant GPDH assay kit according to manufacturer's instructions (IBL Hamburg, Germany).
Phosphoglucomutase (PGM) activity was determined using a PGM colorimetric assay kit according to manufacturer's instructions (BioVision, USA). For the H+-ATPase activity, 0.5 g of cells was used for the preparation of membranes. Plasmalemma vesicles were prepared as described by Kasamo (1988). The H+-ATPase activity was measured according to the method of Liang (1999).
The generation rate of was measured by monitoring the nitrite formation from hydroxylamine in the presence of and the absorbance in the aqueous solution was read at 530 nm (Zhao et al., 2016). The content of H2O2 was measured by monitoring the A410 of titanium-peroxide complex according to the method of Suo et al. (2015).
For the contents of reduced AsA, oxidized AsA (DHA), oxidized glutathione (GSSG), and reduced glutathione (GSH), 0.5 g of cells was homogenized on ice in 3 ml 5% trichloroacetic acid. After centrifugation at 15,000 × g for 15 min at 4°C, the supernatants were used for substrate content assays. Total AsA and reduced AsA were determined by recording the absorbance changes at 525 nm (Kampfenkel et al., 1995). DHA content was estimated from the difference between assays with and without dithiothreitol (DTT). The extracts were also used for GSH/GSSG assay according to the method of Baker et al. (1990). The concentration of GSH was calculated from the differences between total glutathione and GSSG assays (Haghjou et al., 2009).
The proteins for differential stable isotope labeling experiments were extracted in the modified extraction buffer (900 mM sucrose, 100 mM Tris-HCl (pH 8.0), 65 mM DTT, 1 mM MgCl2, 1% Triton X-100 (v/v), 1 mM phenylmethanesulfonyl fluoride, 1% protease inhibitor cocktail (v/v), 1 mM EDTA, 1 mM EDGA, 1 mM NaF, 1 mM Na3VO4, 1 mM Na4O7P2, and 1 mM C3H7Na2O6P) according to the phenol-methanol methods of Suo et al. (2015). Protein concentration was determined using a Quant-kit according to manufacture instructions (GE Healthcare, USA).
The protein was resuspended in the denaturing buffer containing 8 M urea and 100 mM triethylammonium bicarbonate (pH 8.0) (Bian et al., 2012; Song et al., 2014), and then was reduced by DTT and alkylated by iodoacetamide. After that, the solution was diluted with 100 mM triethylammonium bicarbonate and trypsin was added with an enzyme-to-protein ratio of 1/25 (w/w) and incubated at 37°C overnight.
For the light and heavy dimethyl labeling, 500 μl of CH2O (4%, v/v) and CD2O (4%, v/v) was added to the tryptic digests from D. salina cells cultured in 1 M NaCl (control) or 3 M NaCl (salinity shock for 6 h) medium, respectively, then 500 μl of freshly prepared NaBH3CN (0.6 M) was subsequently added. The detailed labeling and desalted procedure were the same as described in the method of Song et al. (2014).
The phosphopeptides from 2.0 mg of labeled protein digestion mixture were enriched by Ti4+-immobilized metal ion affinity chromatography (IMAC) microspheres (Zhou et al., 2008, 2013) and then were resuspended in 30 μl 5% formic acid.
For the reversed-phase (RP) liquid chromatography (LC)-MS/MS analysis, a 12 cm × 75 μm i.d. capillary column packed with C18 AQ particles (3 μm, 120 Å) was used as the separation column, and 0.1% formic acid in water and in acetonitrile was used as mobile phases A and B, respectively (Song et al., 2012).
For the quantitative proteomic analysis, 20 μg tryptic digests were loaded onto the strong cation exchange (SCX) monolithic column and then were eluted by 11 salt stepwise elutions to perform the 2-D RPLC-MS/MS analysis (Wang et al., 2007). Each RPLC-MS/MS elution step was performed in a 72 min gradient from 5 to 35% acetonitrile. For the two-dimensional RPLC-MS/MS phosphoproteomic analysis, a RP-strong cation exchange chromatography (SCX) biphasic column combined with C18 analytical column was used (Bian et al., 2012, 2016). Briefly, the phosphopeptides were manually loaded onto the RP-SCX column and then eluted from the RP segment to SCX segment by a 160 min RP gradient LC-MS/MS (0 mM). Then, a series of stepwise elutions with salt concentrations of 24, 40, 56, 72, 100, 200, and 1,000 mM NH4AC were used to elute phosphopeptides from the SCX column to the second dimensional C18 separation column. Finally, the RPLC-MS/MS separation was performed with a 117 min gradient from 5 to 25% acetonitrile.
The LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) was operated in data-dependent MS/MS acquisition mode. Full mass spectra were acquired in the Orbitrap at a resolution of 60,000 (m/z 400). The 20 most intense precursors were selected for fragmentation via collision induced dissociation in the LTQ. For the phosphopeptide analysis, the multistage activation was enabled. The dynamic exclusion function was set as follows: repeat count 2, repeat duration 30 s, and an exclusion duration of 60 s.
Protein identification and quantification are based on the MaxQuant software (version 1.1.1.36) according to the standard workflow (Cox and Mann, 2008). Because the m/z of the light and heavy labeled peptides were different at MS1 level, this mass difference can be distinguished by the mass spectrometer with resolution (R > ~10,000). Quantification is achieved by comparing the intensities of the labeled peptides at MS1 level (Boersema et al., 2009). At least 1.5 fold change is consider as differential proteins in response to salinity.
The database search was performed on the Andromeda search engine against the Chlorophyta protein database (from NCBI, containing 156,988 entries) concatenated with reversed sequences for evaluating of FDR (Cox et al., 2011). Carbamidomethylation on cysteine was set as a fixed modification, whereas oxidation on methionine was set as a variable modification. For phosphopeptides, Phospho (STY) was also set as variable modifications. FDR were set to 1% at phosphorylation site, peptide, and protein group level. For protein quantitation, only the proteins with a BenjaminiHochberg corrected p-value < 0.05 based on significance B were reported by the MaxQuant software (Cox and Mann, 2008). A more detailed parameters setting for database search was illustrated in our previous studies (Song et al., 2012). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2014) partner repository with the dataset identifier PXD005443 and PXD005501.
Only the phosphorylation sites with a reported localization probability of at least 0.75 were used for further analysis (Sharma et al., 2014). For quantification analysis, we used the intensity-weighted ratio significance values reported by MaxQuant to determine significantly changed phosphorylation sites.
Protein functional domains were predicted using the position-specific iterated BLAST and pattern-hit initiated BLAST programs (http://www.ncbi.nlm.nih.gov/BLAST/). Combined BLAST alignments with Gene Ontology and knowledge from literatures, proteins were classified into different categories.
The subcellular locations of proteins were determined using five internet tools: Yloc (http://abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi), LocTree3 (https://rostlab.org/services/loctree3/), Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/.), ngLOC (http://genome.unmc.edu/ngLOC/index.html), and TargetP (http://www.cbs.dtu.dk/services/TargetP/). The confident results have the consistent predictions from at least two tools. For the inconsistent prediction results among five tools, subcellular localizations for corresponding proteins were predicted based on literatures.
The protein–protein interactions (PPI) were predicted using the web-tool STRING 10 (http://string-db.org). The proteins homologs in Arabidopsis were analyzed by sequence BLASTing in TAIR database (http://www.arabidopsis.org/Blast/index.jsp), and then the homologs were subjected to the molecular interaction tool of STRING 10 for creating the proteome-scale interaction network (Suo et al., 2015).
The protein three-dimensional (3D) structure homology model was pre-calculated in SWISS-MODEL workspace (https://swissmodel.expasy.org) (Arnold et al., 2006). The 3D structures and the phosphorylation site were displayed using the SPDBV (version 4.1) software (Arnold et al., 2006). Multiple sequence alignment was performed with sequences from multiple species using Clustal Omega software (http://www.ebi.ac.uk/Tools/msa/clustalo/).
All physiological results were presented as means ± standard deviation of at least three replicates. Salinity-responsive proteins (SRPs) were quantified in at least two of the three replicates. The data were analyzed by Student's t-test using the statistical software SPSS 18.0 (SPSS Inc., Chicago, IL). A p-value less than 0.05 was considered statistically significant.
The D. salina cells were cultured under medium containing 1 M NaCl (Katz and Avron, 1985). The cell number was increased gradually, and cell growth entered stationary phase after 10 days of culturing (Figure 1A), and then the cells were directly transferred to medium containing 3 M NaCl (Figure 1B and Supplemental Figure S1). The number of palmella cells was about 0.63 ± 0.16 × 106 per milliliter (about 5% of the total cell number) at 2 h after salinity shock (HAS), and reached to 2.08 ± 0.25 × 106 per milliliter (17%) at 6 HAS. The maximum of palmella cell number was detected as 6.09 ± 0.38 × 106 per milliliter (49%) at 1 days after salinity shock (DAS), indicating the D. salina cells formed a palmella stage under the salinity induction at 1 DAS. The palmella stage continued for 3 days and reverted to free-swimming cell stage at 4 DAS (Figure 1B). The change of free-swimming cell number was also reflected from the medium color during this course (Supplemental Figure S1). After salinity shock, the medium color turned from green at 0 HAS to light green gradually at 1 DAS, and then reverted to dark green at 6 DAS (Supplemental Figure S1).
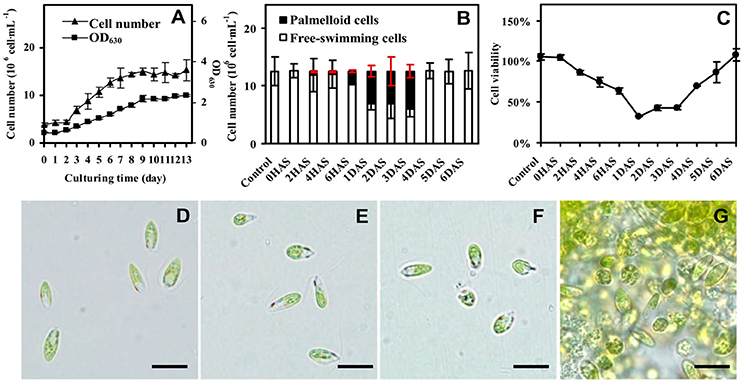
Figure 1. Growth and salinity-induced palmella formation of Dunaliella salina. (A) Cell growth curves of D. salina grown in medium containing 1 M NaCl. (B) The number of palmella cells and free-swimming cells in medium containing 3 M NaCl from 0 h after salinity shock (HAS) to 6 days after salinity shock (DAS). (C) Cell viability in 3 M NaCl medium. (D) Morphology of cells in 1 M NaCl medium. (E) Morphology of cells at 0 HAS. (F) Morphology of free-swimming cells at 2 HAS. (G) Morphology of palmella cells at 6 HAS. The group of accumulated palmella cells were surrounded by exopolysaccharides. Values of cell number and viability are means ± standard deviation (n = 3), bar = 10 μm.
Besides, the change of cell viability evaluated by MTT assay was consistent with the number of free-swimming cells (Figure 1C). The cellular viability was gradually reduced at 6 HAS, reached to the lowest value at 1 DAS, and gradually increased from 4 DAS. In addition, the cell morphology has obvious changes during salinity shock-induced palmella formation. The free-swimming cells under 1 M NaCl were green, biflagellate, and rod to ovoid shaped, as well the cell length was about 12 μm (Figure 1D). The cell morphology didn't show obvious changes at 0 HAS (Figure 1E), but some cells started to lose their flagella and changed to round and immotile at 2 HAS (Figure 1F). More than 17% of free-swimming cells formed immotile palmella at 6 HAS, and the accumulated palmella cells lost their flagella and eyespot, appearing round (Figure 1G). All these indicate that 3 M NaCl shock induces the formation of a short palmella stage, but the palmella cells reverted to free-swimming cells at 4 DAS. During this course, there were three morphotypes at 6 HAS, which were predominantly motile cells, motile form with a prominent palmelloid cells (non-motile, mucilage rich), and palmelloid form with a weakly motile cells (Buchheim et al., 2010). Thus, 6 HAS is a critical time point for the transformation from free-swimming cells to palmella cells.
The glycerol content was significantly increased, and the activity of GPDH for glycerol synthesis was also obviously increased at 6 HAS (Figure 2A). Besides, the content of EPSs was significantly induced, but the cellular carbohydrate content was slightly decreased in cells at 6 HAS (Figure 2B). In addition, the activity of a carbohydrate metabolic enzyme PGM, which catalyzes the bidirectional interconversion of glucose-1-phosphate and glucose-6-phosphate, was significantly reduced at 6 HAS (Figure 2C). Importantly, the activity of plasma membrane (PM) H+-ATPase was significantly induced at 6 HAS, indicating a proton-motive force generator for sucrose transporter was enhanced (Figure 2D).
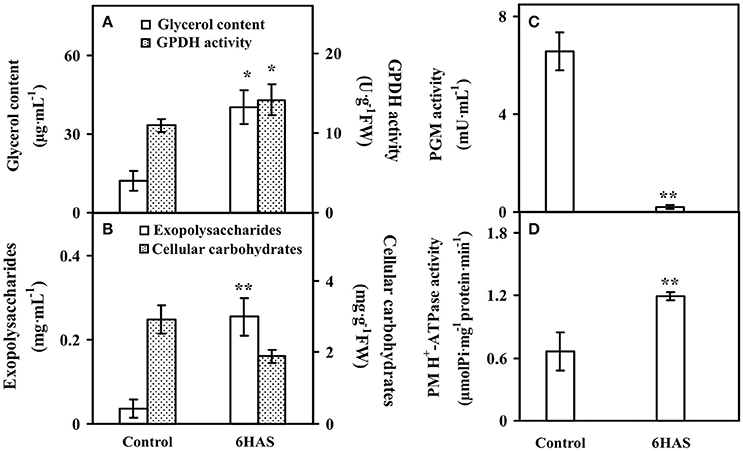
Figure 2. The contents of (A) glycerol, (B) exopolysaccharides (EPSs) and cellular carbohydrates, and activities of (A) glycerol-3-phosphate dehydrogenase (GPDH), (C) phosphoglucomutase (PGM), and (D) plasma membrane (PM) H+-ATPase upon palmella formation of Dunaliella salina. Values are means ± standard deviation based on three independent determinations for control and cells for 6 h after salinity shock (HAS), and bars indicate standard deviations. * and ** indicate values that differ significantly from controls at p < 0.05 and p < 0.01, respectively, according to Student's t-test.
The contents of chlorophylls in control cells and salinity-shocked cells at 6 HAS were detected (Figure 3). The contents of chlorophyll a (Figure 3A), chlorophyll b (Figure 3A), and total chlorophyll (Figure 3B) were all declined, but the chlorophyll a/b (Figure 3B) didn't show obvious changes at 6 HAS.
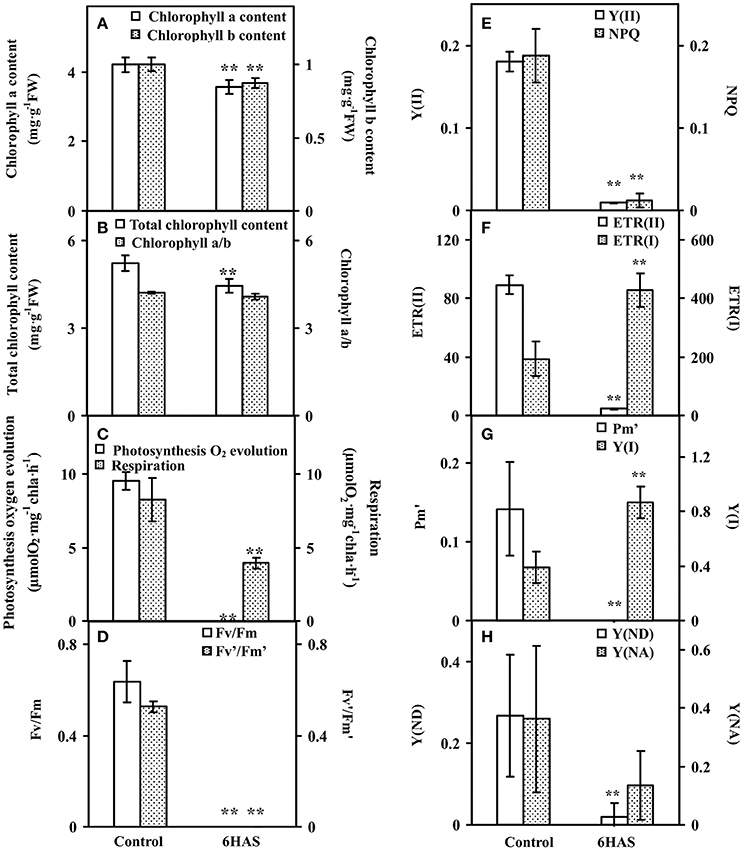
Figure 3. Chlorophyll contents and photosynthetic parameters upon palmella formation of Dunaliella salina. (A) The contents of chlorophyll a and chlorophyll b. (B) Total chlorophyll content and chlorophyll a/b. (C) Photosynthesis oxygen evolution and respiration rate. (D) PSII maximum quantum yield (Fv/Fm) and the excitation capture efficiency of open centers (Fv'/Fm'). (E) Effective PSII quantum yield (Y(II)) and coefficient of non-photochemical quenching (NPQ). (F) Electron transport rates of PSII (ETR(II)) and PSI (ETR(I)). (G) Maximum P700 change (Pm') and photochemical quantum yield of PSI (Y(I)). (H) Non-photochemical quantum yields of PSI caused by donor-side limitation (Y(ND)) and by acceptor-side limitation (Y(NA)). The values are presented as means ± standard deviation (n = 3). ** indicates significant differences among control and 6 h after salinity shock (HAS) is p < 0.01.
Photosynthetic oxygen evolution and respiration rates were all decreased in the salinity- shocked cells when compared with those in the control cells (Figure 3C). In addition, chlorophyll fluorescence parameters were monitored to evaluate the photosynthetic performance. The PSII related parameters, including PSII maximum quantum yield (Fv/Fm) (Figure 3D), the excitation capture efficiency of open centers (Fv'/Fm') (Figure 3D), the effective PSII quantum yield (Y(II)) (Figure 3E), non-photochemical quenching (NPQ) (Figure 3E), and electron transport rate (ETR(II)) (Figure 3F), were declined in salinity-shocked cells. Furthermore, PSI related parameters, such as maximum fluorescence yield (Pm') (Figure 3G), non-photochemical quantum yields of PSI caused by donor-side limitation (Y(ND)) (Figure 3H) and by acceptor-side limitation (Y(NA)) (Figure 3H), were declined, whereas photochemical quantum yield of PSI (Y(I)) (Figure 3G) and electron transport rate (ETR(I)) were increased in salinity-shocked cells (Figure 3F).
To evaluate the ROS level and the dynamics of ROS scavenging system in cells during palmella formation, the generation rate, H2O2 content, the activities of nine antioxidant enzymes, as well as the contents of metabolites in ROS scavenging system were analyzed (Figure 4). The generation rate was declined whereas the H2O2 content and GPX activity were increased at 6 HAS (Figures 4A,E). The activity of SOD was initially increased to catalyze the dismutation of into oxygen and H2O2 at 6 HAS (Figure 4B). However, the activities of POD and CAT in charge of the conversion of H2O2 to H2O were decreased at 6 HAS (Figures 4B,C). Moreover, the activities of enzymes (i.e., APX, MDHAR, DHAR, and GR) as well as the contents of AsA and DHA in GSH-AsA cycle were also decreased at 6 HAS (Figures 4C–E,G), so did the contents of GSSG and GSH (Figure 4H) and the activity of GST (Figure 4F).
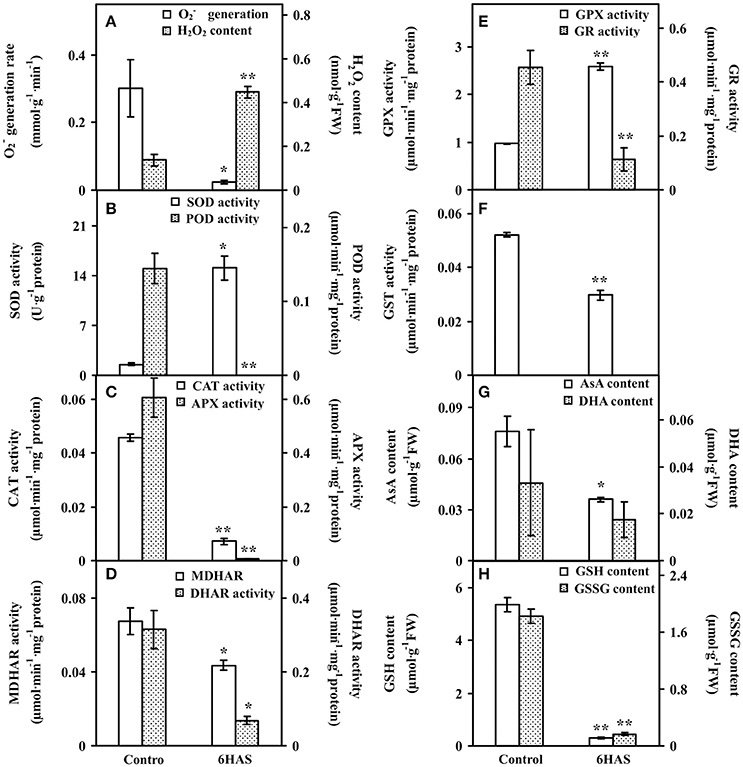
Figure 4. The ROS production, antioxidant enzyme activities, and contents of metabolites in Dunaliella salina. (A) generation rate and H2O2 content. (B) Activities of superoxide dismutase (SOD) and peroxidase (POD). (C) Activities of catalase (CAT) and ascorbate peroxidase (APX). (D) Activities of monodehydroascorbate reductase (MDHAR) and dehydroascorbate reductase (DHAR). (E) Activities of glutathione peroxidase (GPX) and glutathione reductase (GR). (F) Glutathione S-transferase (GST) activity. (G) Contents of reduced ascorbate (AsA) and oxidized ascorbate (DHA). (H) Contents of reduced glutathione (GSH) and oxidized glutathione (GSSG). The values are presented as means ± standard deviation (n = 3). * and ** indicate significant differences among control and 6 h after salinity-shock (HAS) are p < 0.05 and p < 0.01, respectively.
By using differential stable isotope labeling coupled with mass spectrometry approaches, 809 proteins were identified in D. salina cells (Figures 5A,B, Supplemental Table S1, and Supplemental Figure S2). This composes the largest protein database of D. salina so far (Supplemental Table S1). Among them, 509 proteins were quantified in three biological replicates (Figures 5A,B). On the basis of the Gene Ontology, BLAST alignment, KEGG database and information from the literature, the 809 proteins were classified into 14 functional categories (Figure 5C and Supplemental Table S1). Among them, other metabolism (20%), protein synthesis (13%), photosynthesis (13%), carbohydrate and energy metabolism (11%) were over-represented (Figure 5C).
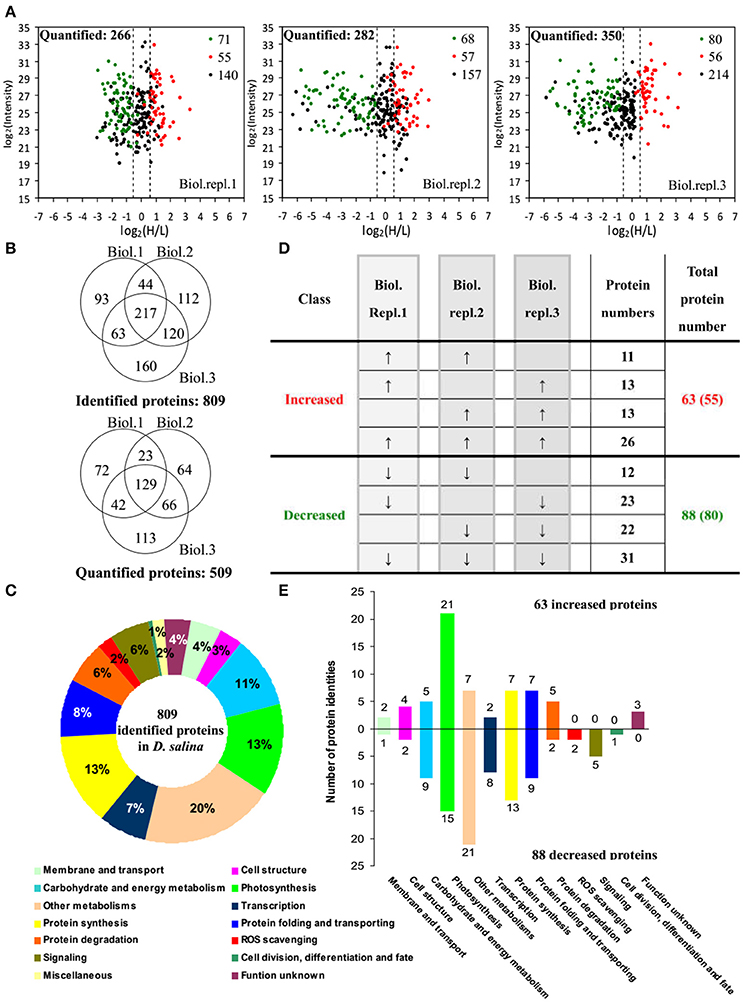
Figure 5. Comparison of proteome in free-swimming cells and cells upon palmella formation of Dunaliella salina. (A) The diagrams showing for each quantified protein the change in abundance between heavy (H; salinity shock) and light (L; control) label (H/L; x axis) as a function of the signal intensity (y axis) recorded in the mass spectrometer. The dashed lines mark the border for a decrease or increase in protein by more than a factor of 1.2 [log 2(H/L) < −0.6 or >0.6]. Most proteins (small black points) did not change in abundance between the two culture conditions. Significantly regulated proteins are depicted as red/green points, and red/green points indicate significantly increased/decreased proteins. (B) Overlap among the three biological replicate experiments for all proteins. 809 proteins were identified and 509 proteins were quantified. Three biological replicate experiments for each sample were performed by LTQ-Orbitrap Velos MS/MS analysis. The obtained results were given in Supplemental Table S1. (C) Functional category of 809 identified proteins in D. salina. (D) Classifications of quantitative proteins significantly change in three biological experiments. A down arrow (↓) indicates significant decrease and an up arrow (↑) significant increase upon palmelloid formation. The protein changes with at least two biological replicates is considered as significant changed protein. There are 63 increased protein species (representing 55 unique proteins) and 88 decreased protein species (80 unique proteins). (E) Functional category of salinity increased and decreased proteins. The columns above and under the x-axis represent the amounts of increased and decreased proteins, respectively. The number of increased/decreased proteins are marked on the column.
Among these proteins, 151 SRPs were quantified in at least two of the three replicates, including 63 salinity-increased proteins, and 88 salinity-decreased proteins (Figure 5D, Tables 1, 2, and Supplemental Table S2). They were classified into 13 functional categories: membrane and transport, cell structure, carbohydrate and energy metabolism, other metabolisms, photosynthesis, transcription, protein synthesis, protein folding and transporting, protein degradation, ROS scavenging, signaling, cell division, differentiation and fate, and function unknown (Figure 5E and Supplemental Table S2). Among them, photosynthesis (24%), other metabolism (19%) and protein synthesis (13%) were predominant (Figure 5E). Most proteins involved in other metabolism, ROS scavenging, transcription, protein synthesis, and signaling were salinity-decreased (Figure 5E and Tables 1, 2). Besides, we found three SRPs involved in membrane and transport, and six involved in cell structure were changed upon palmella formation (Tables 1, 2).
In addition, mitochondria ATP synthase, NADH:ubiquinone oxidoreductase, and glycine cleavage system T protein were accumulated in cells at 6 HAS (Table 1). However, nine chloroplast- and/or cytoplasm- located proteins involved in pentose phosphate pathway, glyco-metabolism, pyruvate metabolism, and starch/sucrose metabolism were decreased at 6 HAS (Table 2). Among 36 photosynthetic SRPs, those in charge of light harvesting, PS I stability, electron transport, and ATP synthesis were increased, but the majority of proteins in carbon fixation and chlorophyll synthesis were decreased (Tables 1, 2), which were in line with the changes of photosynthesis and chlorophyll content upon palmella formation. Furthermore, 28 other metabolism-related SRPs were mainly involved in fatty acid biosynthesis, amino acid metabolism, terpenoid backbone biosynthesis, vitamins metabolism, purine and pyrimidine metabolism, and sulfur metabolism (Tables 1, 2).
Additionally, the protein profiles revealed the transcription, translation, as well as protein processing and fate were altered upon palmella formation. Four decreased histones and four RNA processing-related proteins indicate the transcription was inhibited upon palmella formation (Table 2). Besides, 20 SRPs involved in chloroplastic or cytoplastic protein synthesis (Tables 1, 2). In addition, the changes of 16 protein processing-related proteins imply that the protein folding, processing and transport were altered by salinity shock (Tables 1, 2). Furthermore, five protein degradation-related enzymes were increased, which were localized in cytoplasm, chloroplast, or mitochondria, respectively (Tables 1, 2). Besides, five signaling proteins and one cell division-related protein were decreased upon palmella formation (Table 2).
To investigate the changes of phosphoproteins upon palmella formation, stable isotope dimethyl labeling-based MS approaches were applied to analyze phosphoproteins, 137 phosphorylation sites were identified (Figure 6A), representing 100 phosphoproteins in 12 functional categories (Figure 6C). Among them, 54 phosphorylation sites were quantified (Figure 6B), representing 40 unique phosphoproteins (Figure 6D and Supplemental Tables S3, S4). The amino acid sequences of the 137 identified and 54 quantified phosphorylation sites were shown in Supplemental Table S4. The number of phosphopeptides or phosphorylation sites significantly changed at least a 1.5-fold in three replicates were shown in Supplemental Table S5. Among quantified phosphoproteins, photosynthesis (13%), signaling (13%) and cell structure (13%) were over-represented (Figure 6D). Finally, 35 salinity-responsive phosphoproteins (SRPPs) including 14 salinity-increased phosphoproteins and 22 salinity-decreased phosphoproteins were examined in D. salina during palmella formation (Figure 6E, Table 3, Supplemental Table S6, and Supplemental Figure S3). They were involved in photosynthesis, carbohydrate and energy metabolism, other metabolisms, transcription, protein synthesis, protein folding, protein degradation, signaling, and cell structure (Table 3 and Supplemental Table S6).
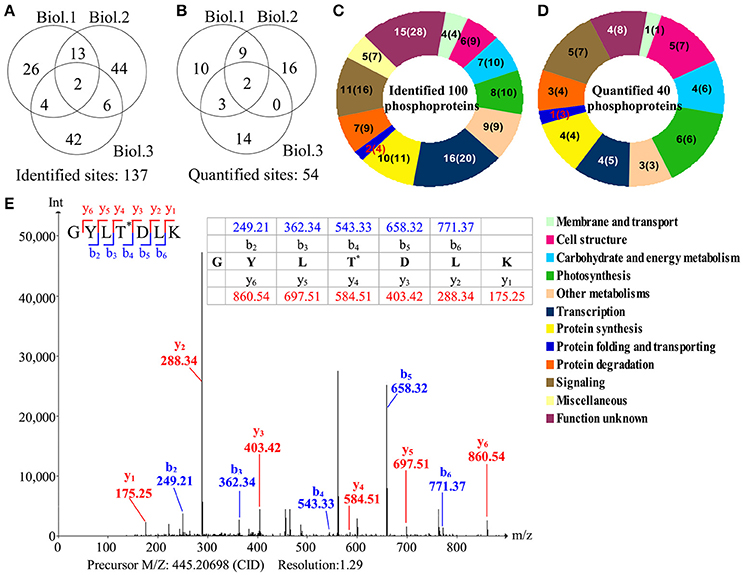
Figure 6. Overview of phosphoproteome upon palmella formation of Dunaliella salina. (A) Overlap of phosphorylation sites among three biological replicates. All phosphorylated sites having a reported localization probability of at least 0.75 were considered to be assigned to a specific residue, and we refer to these as phosphorylated sites. 137 phosphorylated sites were identified. (B) Among the 137 identified sites, 54 phosphorylated sites were quantified. Three biological replicates for each sample were performed by LTQ-Orbitrap Velos MS/MS analysis, and the obtained results were given in Supplemental Table S3. We compared the overlap of identified phosphorylated sites and quantified phosphorylated sites among the three biological replicates. Biol.1/2/3 represent the first/second/third biological replicate. (C) Functional category of 100 identified phosphoproteins (containing 137 identified phosphorylated sites) in D. salina. The numbers inside the parentheses shows how many phosphorylated sites identified in phosphoproteins. (D) Functional category of 40 quantified phosphoproteins (containing 54 quantified phosphorylated sites). The numbers inside the parentheses show how many phosphorylated sites quantified in phosphoproteins. (E) A representative MS/MS spectra of 35 salinity-responsive phosphoproteins. The peptide GYLTDLK from PRP1 splicing factor (gi|307110542).
In this study, all the 35 SRPPs were subjected to SWISS-MODEL database (https://swissmodel.expasy.org) for building the 3D structure homology model. Among them, nine homologs of SRPPs were aligned to their 3D structure homology models (Figure 7 and Supplemental Table S7).
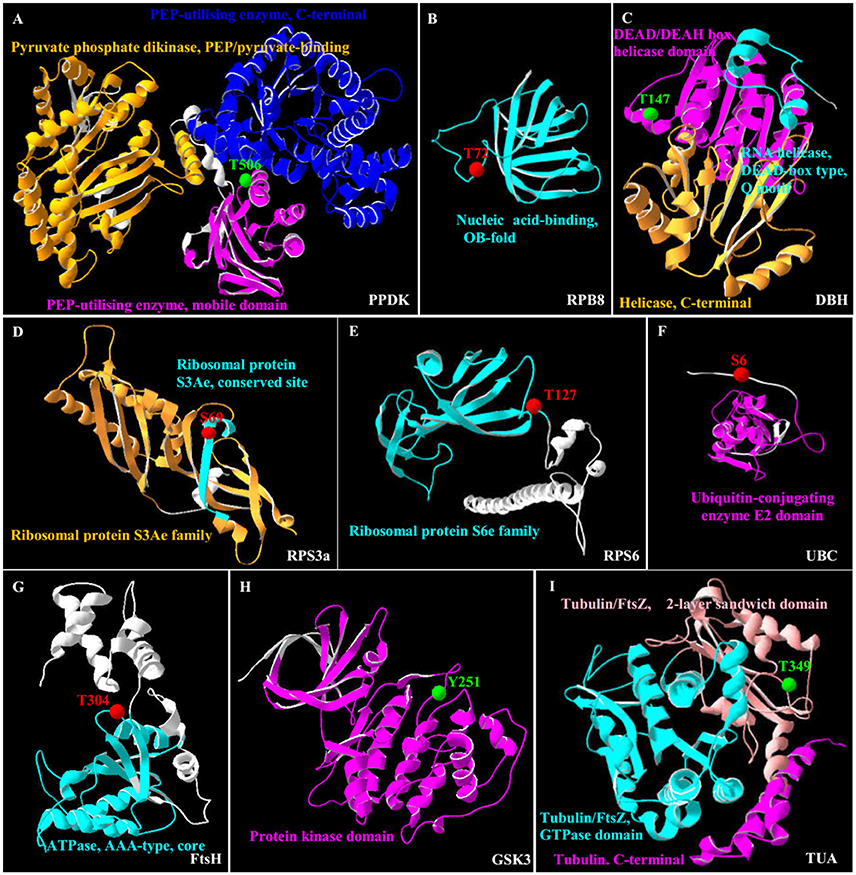
Figure 7. 3D structure of nine salinity-responsive phosphoproteins. The protein 3D structure model were pre-calculated in SWISS-MODEL workspace (https://swissmodel.expasy.org/) (Arnold et al., 2006) and more detail see in Supplemental Table S7. For 35 salinity-responsive phosphoproteins, nine phosphoproteins were built the most suitable protein structure models. (A) Pyruvate phosphate dikinase (PPDK). (B) RNA polymerase subunit 8 domain (RPB8). (C) DEAD-box helicases (DBH). (D) Ribosomal protein S3a (RPS3a). (E) Ribosomal protein S6 (RPS6). (F) Ubiquitin-conjugating enzyme E2 (UBC). (G) Membrane AAA-metalloprotease (FtsH). (H) Glycogen synthase kinase 3 (GSK3). (I) Tubulin α chain (TUA). The red/green balls showed the increased/decreased phosphorylation sites, respectively, and different colorful ribbon showed the different domain.
Importantly, the 3D structure clearly indicated that the phosphorylation sites of eight proteins were localized in the region of function domain (Figure 7), presenting useful information for underlying their specific functions. For pyruvate phosphate dikinase (PPDK), T506 was localized in beta turn, and was part of the phosphoenolpyruvate (PEP)-utilizers enzyme mobile domain (IPR008279), which was a “swiveling” beta/beta/alpha domain for catalyzing the transfer of a phosphoryl group from PEP to a histidine residue (Figure 7A). In RNA polymerase subunit 8 (RPB8), T72 was localized in OB-fold nucleic acid binding domain (IPR012340), which was a part of the five-stranded beta-barrel structure for nucleic acid recognition (Figure 7B). Besides, salinity-decreased T147 was located in DEAD-box helicases (DBH), which was part of DEAD/DEAH box helicase domain involved in various aspects of RNA metabolism (Figure 7C). For ribosomal proteins, the salinity-induced S69 of ribosomal protein S3a (RPS3a) was localized in beta sheet (Figure 7D), while T127 of ribosomal protein S6 (RPS6) was located in beta turn, which were all the conserved site of ribosomal protein S3Ae/S6e family (IPR018281/IPR001377) (Figure 7E). The salinity-induced T304 of membrane AAA-metalloprotease (FtsH) was located in beta turn of AAA+ domain (IPR003959), which has conserved alpha-beta-alpha core structure and walker A and B motifs of P-loop NTPase (Figure 7G). The salinity-induced Y251 of glycogen synthase kinase 3 (GSK3) was localized in protein kinase domain (IPR003959), which catalyzes the transfer of the gamma phosphate from ATP to one or more amino acid residues in a protein substrate, resulting in a conformational change affecting protein function (Figure 7H). In addition, salinity-decreased T349 phosphorylation of α-tubulin localized in the carboxy-terminal region which was the alpha domain interface region for motor protein binding (Figure 7I and Supplemental Figure S4). However, the salinity-induced S6 phosphorylation of ubiquitin-conjugating enzyme E2 (UBC) localized outside of its conserved function domain (Figure 7F). Whether the increase of S6 phosphorylation of UBC has an effect on conjugation of ubiquitin to the target protein needs to be confirmed.
To discover the relationship of the 151 SRPs, the PPI networks were generated using the web-tool STRING10 (http://www.string-db.org). After BLASTing in TAIR database (http://www.arabidopsis.org/Blast/index.jsp), 135 homologs in Arabidopsis of the 151 SRPs were analyzed, and then subjected to the molecular interaction tool of STRING 10 for creation of proteome-scale interaction network (Supplemental Table S8). Among them, 118 proteins were depicted in the STRING database, and illuminated in nine functional modules with tightly-connected clusters (stronger associations represented by thicker lines) in the network (Figure 8A). Besides, 23 SRPPs were depicted in the STRING database (Figure 8B and Supplemental Table S9). The relationship of proteins in various modules indicates that signaling and protein synthesis/processing/turnover are crucial for the modulation of light harvesting, carbon assimilation and energy apply, as well as the cytoskeleton upon palmella formation. Importantly, the reverse phosphorylation of proteins plays important roles in regulation of proteins interactions.
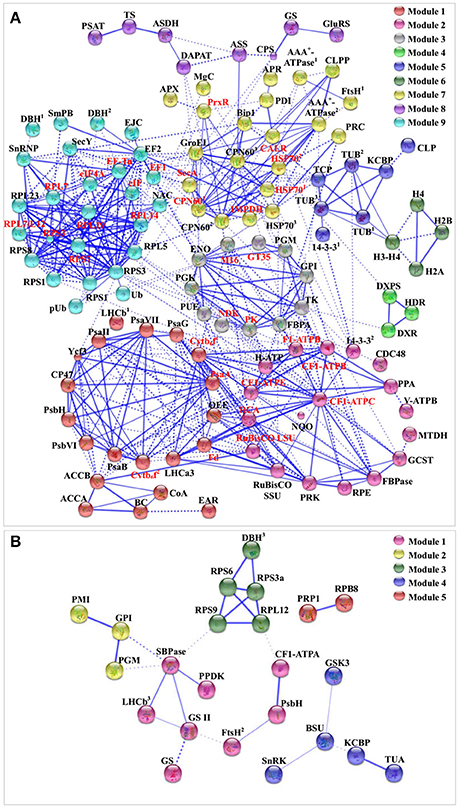
Figure 8. The protein–protein interaction (PPI) network in D. salina revealed by STRING analysis. (A) A total of 151 salinity-responsive proteins represented by 118 unique homologous proteins from Arabidopsis are shown in PPI network. Nine main groups are indicated in different colors. (B) A total of 35 salinity-responsive phosphoproteins represented by 23 unique homologous proteins from Arabidopsis are shown in PPI network. Five main groups are indicated in different colors. The PPI network is shown in the confidence view generated by STRING database. Stronger associations are represented by thicker lines. The abbreviations are referred to Tables 1–3.
D. salina has the ability to grow over an extremely wide range of salinity from less than sea water to NaCl saturation (Montoya and Olivera, 1993). The normal cells of D. salina are ellipsoid to fusiform, without cell wall, but covered with mucilaginous glycocalyx. The biflagellate motile cells contain a single large cup-shaped posterior chloroplast (Borowitzka and Siva, 2007). The osmotic shock, such as rapid decrease/increase of salinity, would induce palmella formation of D. salina (Preetha et al., 2012). The cells in palmella stage lose their flagella and eyespot, become round, and excrete a slime layer to format a multicellular aggregated colonies within a common mucilage (Montoya and Olivera, 1993).
Upon palmella formation, a salt-induced Bin/Amphiphysin/Rvs domain-containing protein would facilitate to bind membrane for membrane curvature modulation in D. salina (Figure 9A and Table 1; Rao and Haucke, 2011). Osmotic and salt stresses cause microtubule disassembly and reorganization (Komis et al., 2002; Shoji et al., 2006). In this study, the salt-induced β-tubulin implies that the tubulin accumulation in cytoplasmic pool of palmella cells may prepare for the flagella reformation of free-swimming cells (Figure 9A), while the salinity-decreased phosphorylation of T349 would facilitate the binding of α-tubulin with motor proteins to regulate the microtubule stability upon palmella formation (Figures 7I, 9A, and Supplemental Figure S4; Ban et al., 2013; Wang et al., 2013).
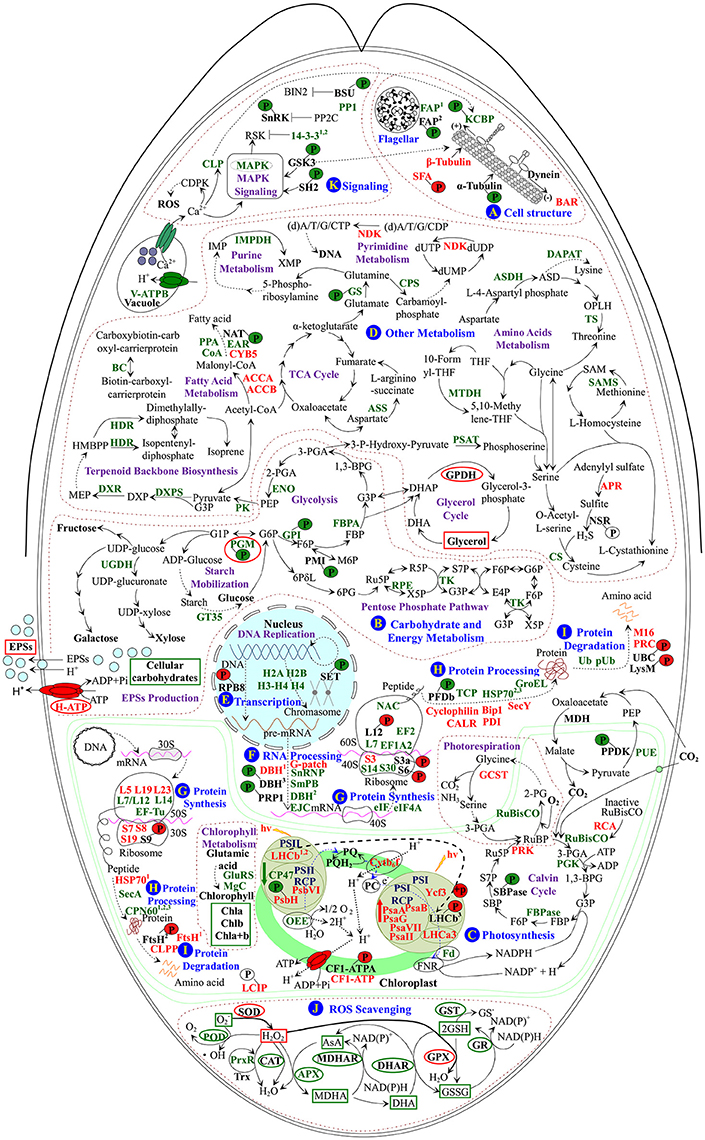
Figure 9. Schematic presentation of the D. salina palmella formation mechanisms. The identified proteins were integrated into subcellular pathways. (A) Cell structure. (B) Carbohydrate and energy metabolism. (C) Photosynthesis. (D) Other metabolisms. (E) Transcription. (F) RNA Processing. (G) Protein synthesis. (H) Protein processing. (I) Protein degradation. (J) ROS scavenging. (K) Signaling. Protein expression patterns, phosphoprotein expression patterns, enzyme activities, and substrate contents are marked with words, P with circles, ellipse, and squares in black (unchanged), red (increased), and green (decreased), respectively. The solid line indicates single-step reaction, and the dashed line indicates multistep reaction. The abbreviations of protein names are referred to Tables 1–3. Other abbreviations used are: 1,3-BPG, 1,3-bisphosphoglycerate; 2-PG, 2-phosphoglycolate; 2-PGA, 2-phosphoglycerate; 30S, chloroplast small ribosomal subunit; 3-PGA, 3-phosphoglycerate; 40S, eukaryotic small ribosomal subunit; 50S, chloroplast large ribosomal subunit; 60S, eukaryotic large ribosomal subunit; 6PG, 6-phosphoguconate; 6PδL, 6-phosphoglucono-δ-lactone; ADP, adenosine diphosphate; AsA, ascorbate; ASD, aspartate-4-semialdehyde; ATP, adenosine triphosphate; BIN2, brassinosteroid insensitive 2; CAT, catalase; CDPK, calcium-dependent protein kinase; CF1-ATPA, ATP synthase CF1 α subunit; Chla/b/a+b, chlorophyll a/b/a+b; (d)A/T/G/C/UDP, (deoxy)adenosine/thymidine/guanosine/deoxycytidine/uridine 5′-diphosphate; (d)A/T/G/C/UTP, (deoxy)adenosine/thymidine/guanosine/deoxycytidine/uridine 5'-triphosphate; DHA, hydroascorbate; DHAP, dihydroxyacetone phosphate; DHAR, dehydroascorbate reductase; DNA, deoxyribonucleic acid; dUMP, deoxy-uridine monophosphate; DXP, 1-deoxy-D-xylulose 5-phosphate; E4P, erythrose 4-phosphate; EPSs, exopolysaccharides; F6P, fructose 6-phosphate; FBP, fructose-1,6-bisphosphate; FNR, ferredoxin-NADP+ reductase; G1P, glucose-1-phosphate; G3P, glyceraldehyde 3-phosphate; G6P, glucose-6-phosphate; GPDH, glycerol-3-phosphate dehydrogenase; GPX, glutathione peroxidase; GR, glutathione reductase; GSH, reduced glutathione; GSSG, oxidized glutathione; GST, glutathione S-transferase; HMBPP, 1-hydroxy-2-methyl-2-butenyl 4-diphosphate; hv, light energy; IMP, inosine-5′-monophosphate; M6P, D-mannose 6-phosphate; MDH, malate dehydrogenase; MDHA, monohydroascorbate; MDHAR, monodehydroascorbate reductase; MEP, 2-C-methyl-D-erythritol 4-phosphate; mRNA, messenger ribonucleic acid; NSR, nitrite/sulfite reductase; OPLH, O-phospho-L-homoserine; P, phosphorylation site; PC, plastocyanin; PEP, phosphoenolpyruvate; Pi, inorganic phosphate; POD, peroxidase; PP2C, protein phosphatase 2C; PQ, plastoquinone; PQH2, reduced plastoquinone; pre-mRNA, precursor messenger ribonucleic acid; PSI, photosystem I; PSII, photosystem II; PSII-RCP, photosystem II reaction center protein; PSI-RCP, photosystem I reaction center protein; R5P, ribose-5-phosphate; ROS, reactive oxygen species; RSK, ribosomal S6 kinase; Ru5P, ribulose 5-phosphate; RuBP, ribulose-1,5-bisphosphate; S7P, sedoheptulose 7-phosphate; SAM, S-adenosylmethionine; SBP, sedoheptulose-1,7-bisphosphate; SOD, superoxide dismutase; TCA, tricarboxylic acid; THF, tetrahydrofolate; Trx, thioredoxin; X5P, xylulose-5-phosphate; XMP, xanthosine-5′-phosphate.
We found the flagella formation was modulated in response to salinity (Figure 9A). Flagellar associated proteins function as a molecular chaperon, and their declined abundance and phosphorylation level would contribute to the flagella lost during the palmella formation. Consistently, the abundance and phosphorylation at S581 of kinesin-like calmodulin binding protein (KCBP) were also decreased, but the normalized phosphorylation was unchanged upon palmella formation (Figure 9A). KCBP was tightly connected with tubulin in PPI network (Figure 8). The highly conserved KCBP is mainly localized near the basal body in D. salina (Shi et al., 2013) and C. reinhardtii (Dymek et al., 2006), which has a myosin tail homology-4 region in the N-terminal tail and a calmodulin-binding region following the motor domain (Dymek et al., 2006). KCBP plays an important role in microtubule-based intracellular motility. Therefore, the decrease of the abundance and phosphorylation of KCBP implies its motor activity may be inhibited upon palmella formation (Figure 9A and Table 3), but we cannot confirm its activity was regulated by phosphorylation. In addition, the increased abundance and phosphorylation level of striated fiber-assemblin would facilitate to regulate the flagellar root microtubule stability upon palmella formation (Figure 9A; Lechtreck et al., 2002).
The Dunaliella cells were enclosed in a thin elastic plasma membrane surrounded by mucus “surface coat”, but lack rigid polysaccharide cell wall (Ben-Amotz and Avron, 1990). It is known that accumulation of glycerol in cells was crucial for osmotic homeostasis in D. salina in response to salinity (Pick, 2002; Liska et al., 2004). In this study, salinity shock-induced accumulation of glycerol and increase of the activity of glycerol metabolic enzyme are important for palmella formation (Figure 2A).
Similarly, total EPS contents were all increased in halotolerant cyanobacterium Microcoleus vaginatus (Chen et al., 2006), halotolerant bacterium Rhodopseudomonas acidophila (Sheng et al., 2006), and medicinal mushroom Phellinus linteus (Zou et al., 2006). In this study, EPS slime layer surrounded D. salina cells was enhanced (Figure 1G), due to the content of EPSs was increased upon palmella formation (Figure 2B). The salinity-induced EPSs would facilitate the accumulation of water and the reduction of ion influx, protecting the membrane system upon palmella formation. The EPSs around D. salina cells were excreted from cells through plasma membrane sugar transporters, which were energy-dependent H+-symporters (Büttner and Sauer, 2000). Its energization was via the proton-motive force generated by the PM H+-ATPase, and the modulation of H+-ATPase activity would immediately affect the sugar transport kinetics (Doidy et al., 2012). In this study, the abundance and activity of PM H+-ATPase were all increased to activate the efflux of sugars upon palmella formation (Figures 2D, 9B, and Table 1; Carpaneto et al., 2010). We also found ten carbohydrate metabolism-related proteins were all salinity-decreased upon palmella formation, which were involved in glycolysis, pentose phosphate pathway, as well as starch mobilization and glucose metabolism, respectively (Figure 9B and Tables 2, 3). The normalized phosphorylation level and activity of PGM were all decreased upon salinity-induced palmella formation (Table 3 and Supplemental Table S1), which implies that the phosphorylation-dependent activity of PGM is inhibited in D. salina under salinity-induced oxidative stress. Moreover, the phosphorylation level of glucose-6-phosphate isomerase and phosphomannose isomerase were also reduced upon palmella formation. It has been reported that glycolytic enzymes were oxidized and inactivated when cells were subjected to oxidative stress, leading to a metabolic reshuffling of glucose equivalents through the pentose phosphate pathway for providing a necessary reducing power (NADPH) for antioxidant defense mechanism in cells (Shanmuganathan et al., 2004). These indicate that carbohydrate metabolism may be reduced in cells upon salinity-induced palmella formation, similar to the salinity-decreased total carbohydrates in halotolerant cyanobacterium M. vaginatus (Chen et al., 2006).
In addition, ROS homeostasis is crucial for rapid metabolism transition upon salinity-shock palmella formation. In this study, the salinity shock-decreased activities of most ROS scavenging enzymes (i.e., POD, APX, MDHAR, DHAR, GR, and GST), the abundances of 2-Cys peroxiredoxin (PrxR) and APX (Figures 4B–F, 9J and Table 2), as well as the contents of GSSG and GSH (Figure 4H) imply that GSH-AsA cycle, PrxR/thioredoxin pathway, and APX pathway might be inhibited under salinity shock. However, the salinity-increased activities of SOD and GPX indicate that the dismutation of superoxide into oxygen and H2O2 and subsequently reduction in GPX pathway are enhanced for ROS scavenging upon palmella formation (Figures 4B,E, 9J). Importantly, the abundance of SOD was oxidative stress-increased during the early transition of green vegetative cells to red cysts in H. pluvialis (Wang et al., 2004a). Thus, the enhanced SOD and GPX pathways may be the key strategy for ROS homeostasis upon salinity-induced palmella formation.
Upon salinity shock-induced palmella formation, the photosynthesis of D. salina was significantly inhibited, being reflected from the obvious decline of chlorophyll content and photosynthesis oxygen evolution. A similar phenomenon (Figures 1D–G) also occurred upon copper-induced palmella formation of C. reinhardtii (Sztrum et al., 2012), and the transformation process of green vegetative cells into red aplanospores of H. pluvialis (Scibilia et al., 2015).
The excess light energy absorbance was reduced. The decline of the abundances of chlorophyll biosynthesis-related enzymes (i.e., Mg-protoporyphyrin IX chelatase, and gln-glu-trna synthetase) indicated that the chlorophyll biosynthesis may be inhibited upon palmella formation (Figure 9C and Table 2), which was also observed in H. pluvialis in response to nitrogen starvation (Scibilia et al., 2015). Besides, the decrease of chloroplast oxygen-evolving protein and PSII 47 kDa protein would attribute to the salt-inhibited photosynthesis oxygen evolution activity, which was supposed to facilitate the reduction of excess light energy absorbance in palmella cells (Figure 9C and Table 2).
The stability and activity of PSII were modulated by salinity shock. The PSII photochemistry activity [e.g., Fv/Fm, Fv'/Fm', and Y(II)] and linear electron flux were all obviously reduced upon palmella formation, which was similar with what happened in high light-stressed D. salina (Gu et al., 2014). However, the abundances of two PSII assembly-related proteins [PSII protein H (PsbH) and PSII protein VI] were increased upon palmella formation (Figure 9C and Table 1). PsbH is a 10-kDa phosphoprotein associated with the inner antenna PSII 47 kDa protein, which is conserved and essential for assembling PSII in algae and higher plants (Summer et al., 1997). The psbH mutants appeared a PSII deficient phenotype and lack of a functional PSII complex (Summer et al., 1997). Interestingly, we also found T8 phosphorylation of PsbH was decreased upon palmella formation (Figure 9C and Table 3). However, a T2 knockdown mutant of PsbH in C. reinhardtii has similar phonotype of wild-type strains, indicating that T2 phosphorylation of PsbH probably doesn't affect its function (O'Connor et al., 1998). Besides, PSII protein VI is the beta subunit of cytochrome b559 essential for PSII assembly, whose increase would enhance the cyclic electron transport (CET) or in a side path of electron flow for protecting PSII from photoinhibition (Figure 9C and Table 1; Burda et al., 2003).
Enhancement of CET would facilitate ATP synthesis. The salinity-suppressed ETR(II) and salinity-increased ETR(I) abundance implied that CET was tend to enhanced upon palmella formation in D. salina. The increased abundances of electron carrier protein cytochrome b6/f complex and various PSI complex proteins (i.e., P700 chlorophyll a apoprotein A1, P700 chlorophyll a apoprotein A2, PsaG, PSI reaction center subunit II, PSI subunit VII, assembly protein Ycf3, and light-harvesting chlorophyll a/b protein 3) would contribute to the induced CET (Figure 9C and Table 1). This would facilitate funneling excess electrons to generate ATP without increasing oxygen evolution (Zhang et al., 2010; Gu et al., 2014). Interestingly, the normalized phosphorylation level of light harvesting chlorophyll a/b binding proteins (LHCII) of PSII (LHCb) was enhanced upon palmella formation (Figure 9C and Table 3). The reversible phosphorylation of LHCII is generally considered as an adaptation mechanism to balance energy distribution between PSII and PSI for regulating redox homeostasis in chloroplasts (Grieco et al., 2012). Our proteomic results revealed that the abundances of several enzymes in Calvin cycle were decreased upon salinity shock-induced palmella formation, such as ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), phosphoglycerate kinase, and fructose-1, 6-bisphosphatase (Figure 9C and Table 2). The photosynthetic CO2 fixation was inhibited due to the down-regulation of transcript levels of Calvin cycle enzymes in D. salina under high salt stress (Liska et al., 2004). Similarly, the decreased RuBisCO was also examined in H. pluvialis during the early transition of green vegetative cells to red cysts under oxidative stress (Wang et al., 2004a). Additionally, the abundances of chloroplastic ribulose phosphate-3-epimerase and transketolase also imply that the Calvin cycle may be inhibited in D. salina under salinity shock (Figure 9B and Table 2).
We also found the phosphorylation level of chloroplast-localized PPDK was decreased upon palmella formation (Figure 9C and Table 3), and PPDK was tightly connected with sedoheptulose-1,7-bisphosphatase in PPI network (Figure 8B). PPDK catalyzes the formation of PEP that is the initial acceptor of CO2 in C4 pathway. In U. linza, the elevated PPDK transcription and enzyme activity enhanced C4 carbon metabolism under high salinity stress (Xu et al., 2013). In this study, the phosphorylation site T506 of PPDK is localized in beta turn of PEP-utilizers enzyme mobile domain (IPR008279) region (Figure 7A). It is reported that the reverse phosphorylation of PPDK (T456 in maize) would regulate the switch of its active (dephosphorylation) and inactive (phosphorylation) states (Chastain et al., 2002). Thus, the decreased phosphorylation at T506 of PPDK (homologous site with maize) would induce its activity, resulting in enhanced C4 pathway activity upon the salinity-induced palmella formation.
The photosynthetic oxygen evolution, PSII activity, and photosynthetic CO2 fixation were all reduced after free-swimming cells lost their flagella to be immotile (Figures 1, 3). Additionally, the abundances of several enzymes involved in fatty acid metabolism, amino acids metabolism, terpenoid backbone biosynthesis, and purine/pyrimidine metabolism were decreased (Figure 9D and Tables 2, 3), implying these pathways may be inhibited upon palmella formation. Similarly, proteins involved in hydrophobic fatty acid biosynthesis and amino acid synthesis were decreased in C. reinhardtii in response to salt stress for less than 24 h (Mastrobuoni et al., 2012) and nitrogen starvation for 6 h (Longworth et al., 2012). Besides, amino acid metabolism-related genes were down-regulated in brown alga Ectocarpus siliculosus under hypersaline conditions (Dittami et al., 2011). In addition, several nitrogen assimilation and pyruvate kinase metabolism-related proteins were decreased during the oxidative stress-induced early transition of green vegetative cells to red cysts in H. pluvialis (Wang et al., 2004a). All these indicate that the palmelloid cells become dormant under salinity shock condition. However, the energy production was induced upon palmella formation. This is consistent with what happened during oxidative stress-induced early transition of green vegetative cells to red cysts in H. pluvialis (Wang et al., 2004a), as well as in hypersaline-stressed brown alga E. siliculosus (Dittami et al., 2011), which is crucial for energy supply in dormant or stressed algae cells.
The metabolic changes were regulated by gene expression pattern upon palmella formation. We found the abundances of ten transcription-related proteins and phosphorylation level of RPB8 were changed (Figure 9E and Tables 1–3). Three decreased RNA processing-related proteins (i.e., Sm protein B, nucleolar protein snoRNP, and exon junction complex) indicates the pre-mRNA splicing and localization are probably reduced (Figure 9F and Table 2; Tange et al., 2004). However, G-patch domain-containing protein, which functions in RNA recognition, RNA binding or splicing, was increased upon palmella formation (Figure 9F and Table 1). In addition, DBH is involved in various aspects of RNA metabolism (e.g., nuclear transcription, pre-mRNA splicing, and RNA decay) was salinity-altered, In our results, the salinity-reduced DBH is tightly connected with other protein synthesis-related proteins (Figure 8), and its phosphorylation site T147 located in helices (Figure 7C), implying the phosphorylation is perhaps involved in the regulation of DBH function (Figure 9F).
Besides, the phosphorylation at T72 of RPB8 was salinity-induced upon palmella formation, and T72 is localized in OB-fold nucleic acid binding domain (IPR012340) for nucleic acid recognition (Figures 7B, 9E and Table 3). Similarly, the S2 phosphorylation of RNA polymerases (RNA pol) II was increased in wild-type fission yeast under nitrogen starvation (Sukegawa et al., 2011), and RNA pol α subunit were also stress-increased in other algae, such as copper-stressed marine alga Scytosiphon gracilis (Contreras et al., 2010), salt-treated Bifidobacterium longum NCIMB 8809 (Sánchez et al., 2005), and acid (low pH)-stressed Streptococcus mutans (Len et al., 2004). Although the function of RPB8 phosphorylation remains unknown, its increase would affect RNA pol III assembly and catalytic function upon palmella formation of D. salina (Voutsina et al., 1999). Additionally, RPB8 was tightly connected with PRP1 splicing factor (PRP1) in PPI network (Figure 8B). Interestingly, the phosphorylation at T255 of PRP1 was decreased upon palmella formation (Figure 9F and Table 3). The N-terminus highly-conserved site of PRP1 can be phosphorylated by PRP4 during the spliceosome activation (Lützelberger et al., 2010). The decline of PRP1 phosphorylation implies that spliceosome activity is probably salinity reduced upon palmella formation.
De novo protein synthesis plays an important role in abiotic stress adaptation in plants. We found that eukaryotic initiation factor (eIF), eIF 4A-like protein, elongation factor (EF) 1 alpha 2 and EF2 were all reduced (Figure 9G and Table 2), indicating that translation initiation and peptide elongation are decreased upon palmella formation. Importantly, protein phosphorylation is involved in the initiation of protein translation (Jackson et al., 2010). We found that the phosphorylation levels of RPS6 (T127), RPS3a (S69), and ribosomal protein L12 (RPL12) (S27), and other ribosomal proteins (RPS3 and RPL23) were increased, but the abundances of ribosomal proteins S14 and ribosomal proteins L7 were decreased during palmella formation (Figure 9G and Tables 1–3). Among them, RPS6 phosphorylation in plants leads to the selective recruitment of ribosomal mRNAs to polysomes, regulating in the growth pattern of plants in response to environment changes (Reinbothe et al., 2010). In addition, the phosphorylation sites of RPS6 and RPS3a were all the conserved site of ribosomal protein family (Figures 7D,E). Therefore, the induced phosphorylation of T127 in RPS6 could be responsible for the elevated level of protein synthesis upon palmella formation. Besides, RPS3 is crucial in translation initiation via participation in rearrangements of the 40S subunit structure and promotion of subsequent recognition of the start codon and interaction with mRNA (Valášek, 2012). In our studies, the increased RPS3 abundance and the phosphorylation at S69 of RPS3a would contribute to translation initiation upon palmella formation. In addition, the RPL12 is a member of ribosomal L7/12 stalk, which functions in restoring the biological activity of “core” ribosomal particles (Ilag et al., 2005). Although the phosphorylation of RPL12 was reported, its exact function is still not clear.
Under stress conditions, the misfolded proteins accumulated in algae cells should be refolded or removed. We found that 23 proteins and four phosphoproteins were involved in protein processing and degradation upon palmella formation (Figure 9H and Tables 1–3). The phosphorylation at S768 and S773 of prefoldin β subunit (PFDb) was decreased upon palmella formation (Figure 9H and Table 3). PFD is a cofactor of the group II chaperonins for capturing and transferring the unfolded actin and tubulin for correct folding (Millán-Zambrano and Chávez, 2014). Furthermore, the decrease of cytosolic chaperonin T-complex protein 1 β subunit (TCP) was examined upon palmella formation (Figure 9H and Table 2), which assists in the folding of newly synthesized actin, tubulin, and other polypeptides in vivo (Sternlicht et al., 1993). Previous proteomics results have showed that the cold-decreased PFD in cold-tolerant Zoysia japonica (Xuan et al., 2013) and salt-decreased TCP in Thellungiella halophila (Pang et al., 2010), indicating the protein folding is inhibited under stress. In our studies, the decrease of PFDb phosphorylation and the abundance of TCP might be related with microtubule organization upon palmella formation.
Besides, three endoplasmic reticulum-located chaperones, calreticulin (CALR), protein disulfide isomerase 1, and luminal binding protein Bip1, were all obviously increased upon palmella formation in D. salina (Figure 9H and Table 1). CALR is involved in the folding and quality control of newly synthesized proteins and glycoproteins, which is highly conserved and crucial for plant development and stress response (Garg et al., 2015). In addition, protein disulfide isomerase aids in the formation of proper disulfide bonds during protein folding in the endoplasmic reticulum (Appenzeller-Herzog and Ellgaard, 2008), which were salinity-increased in salt-tolerant Medicago sativa (Rahman et al., 2015), barley (Mostek et al., 2015), rice (Ghaffari et al., 2014), and halotolerant yeast (Rhodotorula mucilaginosa) (Lahav et al., 2004). Additionally, luminal binding protein Bip1 functions in precursor protein import and translocation (Wang et al., 2004b), which was high salt-induced in halotolerant yeast (Lahav et al., 2004). All these imply that salinity shock might enhance the protein folding and importation for maintaining the correct conformation and subcellular location of the oxidative proteins upon palmella formation.
In addition, two cytoplasmic heat shock protein 70s (HSP70) were decreased, but one chloroplast HSP70 was increased upon palmella formation (Figure 9H and Tables 1, 2). HSP70s assists the folding of de novo synthesized polypeptides and the translocation of precursor proteins in response to osmotic stress (Wang et al., 2004b), which was reported to increase in salinity-treated D. salina (Liska et al., 2004; Katz et al., 2007), and upon oxidative stress-induced early transition from green vegetative cells to red cysts in H. pluvialis (Wang et al., 2004a). This implies that the transportation and processing of certain newly synthesized peptide into chloroplasts are enhanced upon palmella formation of D. salina. Moreover, the chloroplast-located preprotein translocase SecY subunit is a component of the SecYEG translocon, being driven by the chloroplast ATPase SecA. Both SecY and SecA are essential for protein transportation from cytoplasm to chloroplasts (du Plessis et al., 2011). Their abundance changes may imply that the nuclear encoding proteins are necessary for chloroplasts in salinity shock-stressed D. salina (Figure 9H and Tables 1, 2). The increase of cyclophilin in D. salina would enhance the protein folding upon palmella formation (Figure 9H and Table 1; Zhang et al., 2012). The decrease of a mitochondrial GroEL and three chloroplastic chaperonin 60s would inhibit protein folding of RuBisCO, and the abundance of chaperonin 60s correlate positively with RuBisCO in plant cells (Figure 9H and Table 2; Holland et al., 1998).
Ubiquitin-dependent selective degradation of proteins is crucial upon palmella formation in D. salina (Ciechanover, 1998). In this study, salinity-induced abundance of 26S proteasome regulatory complex, as well as phosphorylation at S6 of UBC indicate that ubiquitin-dependent protein degradation in cytoplasm is enhanced upon palmella formation (Figure 9I and Tables 1, 3). Pervious proteomic study reported the increased 26S proteasome regulatory complex in D. salina under long-term salt stress (Liska et al., 2004). The increased phosphorylation at S6 of UBC located in the kinase motifs of CK2 implies that the conjugation of ubiquitin to the target protein may be enhanced (Supplemental Table S6).
The damaged proteins in chloroplasts need to be degraded through an efficient proteolytic system during chloroplast biogenesis, maintenance, and stress response (Ramundo et al., 2014). Upon palmella formation, the abundance and phosphorylation level of FtsH, as well as the abundance of ATP-dependent Clp protease proteolytic subunit were salinity-increased in chloroplasts in D. salina (Figure 9I and Tables 1, 3). FtsH can efficiently degrade proteins with low thermodynamic stability, and was increased in short-term salinity-stressed D. salina, osmotic shock-treated Oenococcus oeni (Bourdineaud et al., 2003), and heat/light-stressed cyanobacterium Synechocystis PCC 6803 (Silva et al., 2003). Additionally, FtsH was tightly connected with photosynthesis-related proteins in PPI network (Figure 8B). Therefore, the increase of T304 phosphorylation in AAA+ domain of FtsH would facilitate its assembly/disassembly with photosynthetic proteins to perform its peptidase function (Figures 7G, 9I and Table 3). Besides, the chloroplast gene encoding Clp protease is essential for cell viability (Ramundo et al., 2014), which was increased in D. salina under long-term salt stress (Katz et al., 2007). All these would enhance the removal of abnormal, modified, and mistargeted proteins in chloroplasts.
The mitochondria peptidase M16 was increased in palmella cells (Figure 9I and Table 1) and in D. salina cells under long-term salt stress (Liska et al., 2004). The peptidase M16 was also increased in suspension cells of A. thaliana (Ndimba et al., 2005), gametophore of Physcomitrella patens (Wang et al., 2008), and roots of O. sativa (Li et al., 2010) under salt stress. These results suggest the proteolytic reaction in mitochondrial processing may be important for stress-induced palmella formation.
The palmella formation is regulated by a sophisticate signaling network in D. salina. The salinity-decreased protein abundance and/or phosphorylation level imply that several crucial salinity-responsive signaling pathways (e.g., mitogen-activated protein kinase (MAPK) signaling, brassinosteroid (BR) signaling, and Snf1-like protein kinase (SnRK) signaling) tend to be inhibited upon salinity shock-induced palmella formation (Figure 9K and Tables 2, 3). Among them, MAPKs are highly conserved serine/threonine kinases in combination with their upstream activators, which convey osmotic stress signals to appropriate effectors and contribute to adaptation to the high salt stress (Moustafa et al., 2014). Additionally, SH2 domains are crucial for protein docking to phosphorylated tyrosine residues on other proteins, which are common in adapter proteins that aid in the signal transduction of receptor tyrosine kinase pathways (Koytiger et al., 2013). The decrease of MAPK and SH2 domain containing protein indicated that the active salt-responsive MAPK signaling might be inhibited in the dormant palmelloid cells. Moreover, in the MAPK signaling pathway, GSK3 can phosphorylate MAP1B to regulate flagellar length and assembly (Wilson and Lefebvre, 2004). The phosphorylation of Y218 in C. reinhardtii GSK3 regulated its active states (Wilson and Lefebvre, 2004). The GSK3 in D. salina shared the conserved phosphorylation site with that in C. reinhardtii. This conserved phosphorylation site was localized in protein kinase domain (Figure 7H). Thus, the decreased phosphorylation of Y251 in GSK3 of D. salina would inhibit its activity to reduce the flagellar stability upon palmella formation.
The salinity-decreased calmodulin-like protein and KCBP would contribute to inhibition of microtubule-binding activity of motor domain during palmella formation of D. salina (Shi et al., 2013). The decreased phosphorylation level at S173 and S174 of SnRK indicates that SnRK-related signaling pathway for regulation of ion homeostasis and ROS production may be reduced upon palmella formation (Gong et al., 2002; Diedhiou et al., 2008; Mao et al., 2010; Kulik et al., 2012). The decreased phosphrylation level at S497 of Bsu 1 phosphatase would reduce its activity, leading to the inhibition of nuclear transcription factors to repress BR-responsive gene expression upon palmella formation (Mora-García et al., 2004).
In the life cycle of unicellular algae, palmella stage is critical for cell surviving in various stress conditions. Salinity-induced palmella formation is a fine-tuned cellular process. By integrating analysis of physiological, quantitative proteomics, and phosphoproteomics data, we revealed the specific molecular mechanisms upon palmella formation (Figure 9). They mainly include (1) cell membrane curvature and cytoskeleton dynamics are modulated for cell morphological changes, (2) accumulations of glycerol and EPSs are enhanced for protection of membrane system, (3) SOD and GPX pathways are specific for ROS scavenging, (4) the activities of photosynthesis oxygen evolution and PSII are inhibited, but the CET and C4 pathway are enhanced, (5) nuclear and chloroplastic gene expression are regulated in response to salinity, and (6) chloroplast and cytoplasm protein processing and turnover are enhanced. All these provide novel insights into the underlying salinity shock-induced palmella formation.
SW and SD designed research. SW, YB, QZ, CS, KC, ZX, CZ, and SD performed research. HZ, MY, and WM contributed new reagents or analytic tools. SW, QZ, JM, and SD analyzed data. SW, SD, and SC wrote the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The project was supported by the National Natural Science Foundation of China (No. 31270310) and The Fundamental Research Funds for the Central Universities (No. 2572014EA04) to SD, and the China State Key Research Program (2016YFA0501402), the National Natural Science Foundation of China (21535008, 21525524) to MY.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00810/full#supplementary-material
3D, three-dimensional; APX, ascorbate peroxidase; AsA, ascorbate; BR, brassinosteroid; CALR, calreticulin; CAT, catalase; CET, cyclic electron transport; DAS, days after salinity shock; DBH, DEAD-box helicases; DHA, hydroascorbate; DHAR, dehydroascorbate reductase; DTT, dithiothreitol; EF, elongation factor; eIF, eukaryotic initiation factor; EPSs, exopolysaccharides; FtsH, membrane AAA-metalloprotease; GPDH, glycerol-3-phosphate dehydrogenase; GPX, glutathione peroxidase; GR, glutathione reductase; GSH, reduced glutathione; GSK3, glycogen synthase kinase 3; GSSG, oxidized glutathione; GST, glutathione S-transferase; HAS, hours after salinity shock; HSP70, heat shock protein 70; KCBP, kinesin-like calmodulin binding protein; IMAC, Ti4+-immobilized metal ion affinity chromatography; LC, liquid chromatography; LHCb, light harvesting chlorophyll a/b binding proteins of photosystem II; LHCII, light harvesting chlorophyll a/b binding proteins; MAPK, mitogen-activated protein kinase; MDHAR, monodehydroascorbate reductase; MS, mass spectrometry; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PEP, phosphoenolpyruvate; PFDb, prefoldin β subunit; PGM, phosphoglucomutase 1; PM, plasma membrane; POD, peroxidase; PPDK, pyruvate phosphate dikinase; PRP1, PRP1 splicing factor; PrxR, 2-Cys peroxiredoxin; PS, photosystem; PsbH, photosystem II protein H; RNA pol, RNA polymerases; ROS, reactive oxygen species; RP, reversed-phase; RPB8, RNA polymerase subunit 8; RPS3, ribosomal protein S3; RPS3a, ribosomal protein S3a; RPS6, ribosomal protein S6; S, serine; SCX, strong cation exchange chromatography; SnRK, Snf1-like protein kinase; SOD, superoxide dismutase; SRPPs, salinity-responsive phosphoproteins; SRPs, salinity-responsive proteins; T, threonine; TCP, T-complex protein 1 β subunit; UBC, ubiquitin-conjugating enzyme E2; RuBisCO, ribulose-1,5- isphosphate carboxylase/oxygenase; RPL12, ribosomal protein L12; Y, tyrosine.
Alkayal, F., Albion, R. L., Tillett, R. L., Hathwaik, L. T., Lemos, M. S., and Cushman, J. C. (2010). Expressed sequence tag (EST) profiling in hyper saline shocked Dunaliella salina reveals high expression of protein synthetic apparatus components. Plant Sci. 179, 437–449. doi: 10.1016/j.plantsci.2010.07.001
Appenzeller-Herzog, C., and Ellgaard, L. (2008). The human PDI family: versatility packed into a single fold. Biochim. Biophys. Acta 1783, 535–548. doi: 10.1016/j.bbamcr.2007.11.010
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Baker, M. A., Cerniglia, G. J., and Zaman, A. (1990). Microtiter plate assay for the measurement of glutathione and glutathione disulfide in large numbers of biological samples. Anal. Biochem. 190, 360–365. doi: 10.1016/0003-2697(90)90208-Q
Ban, Y., Kobayashi, Y., Hara, T., Hamada, T., Hashimoto, T., and Takeda, S. (2013). Alpha-tubulin is rapidly phosphorylated in response to hyperosmotic stress in rice and Arabidopsis. Plant Cell Physiol. 54, 848–858. doi: 10.1093/pcp/pct065
Ben-Amotz, A., and Avron, M. (1973). The role of glycerol in the osmotic regulation of the halophilic alga Dunaliella parva. Plant Physiol. 51, 875–878. doi: 10.1104/pp.51.5.875
Ben-Amotz, A., and Avron, M. (1990). The biotechnology of cultivating the halotolerant algae Dunaliella. Trends Biotechnol. 8, 121–126. doi: 10.1016/0167-7799(90)90152-N
Bian, Y., Li, L., Dong, M., Liu, X., Kaneko, T., Cheng, K., et al. (2016). Ultra-deep tyrosine phosphoproteomics enabled by a phosphotyrosine superbinder. Nat. Chem. Biol. 12, 959–966. doi: 10.1038/nchembio.2178
Bian, Y., Ye, M., Song, C., Cheng, K., Wang, C., Wei, X., et al. (2012). Improve the coverage for the analysis of phosphoproteome of HeLa cells by a tandem digestion approach. J. Proteome Res. 11, 2828–2837. doi: 10.1021/pr300242w
Boersema, P. J., Raijmakers, R., Lemeer, S., Mohammed, S., and Heck, A. J. (2009). Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494. doi: 10.1038/nprot.2009.21
Borowitzka, M. A., and Siva, C. J. (2007). The taxonomy of the genus Dunaliella (Chlorophyta, Dunaliellales) with emphasis on the marine and halophilic species. J. Appl. Phycol. 19, 567–590. doi: 10.1007/s10811-007-9171-x
Bourdineaud, J. P., Nehme, B., Tesse, S., and Lonvaud-Funel, A. (2003). The ftsH gene of the wine bacterium Oenococcus oeni is involved in protection against environmental stress. Appl. Environ. Microbiol. 69, 2512–2520. doi: 10.1128/AEM.69.5.2512-2520.2003
Boussiba, S., Bing, W., Yuan, J. P., Zarka, A., and Chen, F. (1999). Changes in pigments profile in the green alga Haeamtococcus pluvialis exposed to environmental stresses. Biotechnol. Lett. 21, 601–604. doi: 10.1023/A:1005507514694
Buchheim, M. A., Kirkwood, A. E., Buchheim, J. A., Verghese, B., and Henley, W. J. (2010). Hypersaline soil supports a diverse community of dunaliella (Chlorophyceae)1. J. Phycol. 46, 1038–1047. doi: 10.1111/j.1529-8817.2010.00886.x
Burda, K., Kruk, J., Borgstädt, R., Stanek, J., Strzalka, K., and Schmid, G. H. (2003). Mössbauer studies of the non-heme iron and cytochrome b559 in a Chlamydomonas reinhardtii PSI- mutant and their interactions with alpha-tocopherol quinone. FEBS Lett. 535, 159–165. doi: 10.1016/S0014-5793(02)03895-4
Büttner, M., and Sauer, N. (2000). Monosaccharide transporters in plants: structure, function and physiology. Biochim. Biophys. Acta 1465, 263–274. doi: 10.1016/S0005-2736(00)00143-7
Carpaneto, A., Koepsell, H., Bamberg, E., Hedrich, R., and Geiger, D. (2010). Sucrose-and H+-dependent charge movements associated with the gating of sucrose transporter ZmSUT1. PLoS ONE 5:e12605. doi: 10.1371/journal.pone.0012605
Chastain, C. J., Fries, J. P., Vogel, J. A., Randklev, C. L., Vossen, A. P., Dittmer, S. K., et al. (2002). Pyruvate, orthophosphate dikinase in leaves and chloroplasts of C(3) plants undergoes light-/dark-induced reversible phosphorylation. Plant Physiol. 128, 1368–1378. doi: 10.1104/pp.010806
Chen, H., Jiang, J. G., and Wu, G. H. (2009). Effects of salinity changes on the growth of Dunaliella salina and its isozyme activities of glycerol-3-phosphate dehydrogenase. J. Agric. Food Chem. 57, 6178–6182. doi: 10.1021/jf900447r
Chen, H., Lao, Y. M., and Jiang, J. G. (2011). Effects of salinities on the gene expression of a (NAD+)-dependent glycerol-3-phosphate dehydrogenase in Dunaliella salina. Sci. Total Environ. 409, 1291–1297. doi: 10.1016/j.scitotenv.2010.12.038
Chen, L. Z., Li, D. H., Song, L. R., Hu, C. X., Wang, G. H., and Liu, Y. D. (2006). Effects of salt stress on carbohydrate metabolism in desert soil alga Microcoleus vaginatus Gom. J. Integr. Plant Biol. 48, 914–919. doi: 10.1111/j.1744-7909.2006.00291.x
Ciechanover, A. (1998). The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J. 17, 7151–7160. doi: 10.1093/emboj/17.24.7151
Contreras, L., Moenne, A., Gaillard, F., Potin, P., and Correa, J. A. (2010). Proteomic analysis and identification of copper stress-regulated proteins in the marine alga Scytosiphon gracilis (Phaeophyceae). Aquat. Toxicol. 96, 85–89. doi: 10.1016/j.aquatox.2009.10.007
Cox, J., and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Cox, J., Neuhauser, N., Michalski, A., Scheltema, R. A., Olsen, J. V., and Mann, M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. doi: 10.1021/pr101065j
Diedhiou, C. J., Popova, O. V., Dietz, K. J., and Golldack, D. (2008). The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 8:49. doi: 10.1186/1471-2229-8-49
Dittami, S. M., Gravot, A., Renault, D., Goulitquer, S., Eggert, A., Bouchereau, A., et al. (2011). Integrative analysis of metabolite and transcript abundance during the short-term response to saline and oxidative stress in the brown alga Ectocarpus siliculosus. Plant Cell Environ. 34, 629–642. doi: 10.1111/j.1365-3040.2010.02268.x
Doidy, J., Grace, E., Kühn, C., Simon-Plas, F., Casieri, L., and Wipf, D. (2012). Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 17, 413–422. doi: 10.1016/j.tplants.2012.03.009
du Plessis, D. J., Nouwen, N., and Driessen, A. J. (2011). The Sec translocase. Biochim. Biophys. Acta 1808, 851–865. doi: 10.1016/j.bbamem.2010.08.016
Dymek, E. E., Goduti, D., Kramer, T., and Smith, E. F. (2006). A kinesin-like calmodulin-binding protein in Chlamydomonas: evidence for a role in cell division and flagellar functions. J. Cell Sci. 119, 3107–3116. doi: 10.1242/jcs.03028
Ellis, R. J. (1972). Control of palmelloid formation in the green alga Pediastrum. Plant Cell Physiol. 13, 663–672. doi: 10.1093/oxfordjournals.pcp.a074778
Franqueira, D., Orosa, M., Torres, E., Herrero, C., and Cid, A. (2000). Potential use of flow cytometry in toxicity studies with microalgae. Sci. Total Environ. 247, 119–126. doi: 10.1016/S0048-9697(99)00483-0
Garg, G., Yadav, S., and Yadav, G. (2015). Key roles of calreticulin and calnexin proteins in plant perception under stress conditions: A review. Adv. Life Sci. 5, 18–26. doi: 10.5923/j.als.20150501.03
Ghaffari, A., Gharechahi, J., Nakhoda, B., and Salekdeh, G. H. (2014). Physiology and proteome responses of two contrasting rice mutants and their wild type parent under salt stress conditions at the vegetative stage. J. Plant Physiol. 171, 31–44. doi: 10.1016/j.jplph.2013.07.014
Gong, D., Gong, Z., Guo, Y., Chen, X., and Zhu, J. K. (2002). Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. J. Biol. Chem. 277, 28340–28350. doi: 10.1074/jbc.M107719200
Grieco, M., Tikkanen, M., Paakkarinen, V., Kangasjarvi, S., and Aro, E. M. (2012). Steady-state phosphorylation of light-harvesting complex II proteins preserves photosystem I under fluctuating white light. Plant Physiol. 160, 1896–1910. doi: 10.1104/pp.112.206466
Gu, W., Li, H., Zhao, P., Yu, R., Pan, G., Gao, S., et al. (2014). Quantitative proteomic analysis of thylakoid from two microalgae (Haematococcus pluvialis and Dunaliella salina) reveals two different high light-responsive strategies. Sci. Rep. 4:6661. doi: 10.1038/srep06661
Haghjou, M. M., Shariati, M., and Smirnoff, N. (2009). The effect of acute high light and low temperature stresses on the ascorbate–glutathione cycle and superoxide dismutase activity in two Dunaliella salina strains. Physiol. Plant. 135, 272–280. doi: 10.1111/j.1399-3054.2008.01193.x
Han, D., Wang, J., Sommerfeld, M., and Hu, Q. (2012). Susceptibility and protective mechanisms of motile and non motile cells of Haematococcus pluvialis (chlorophyceae) to photooxidative stress1. J. Phycol. 48, 693–705. doi: 10.1111/j.1529-8817.2012.01147.x
Holland, N., Belkind, A., Holland, D., Pick, U., and Edelman, M. (1998). Stress-responsive accumulation of plastid chaperonin 60 during seedling development. Plant J. 13, 311–316. doi: 10.1016/j.bbrc.2012.04.137
Ilag, L. L., Videler, H., McKay, A. R., Sobott, F., Fucini, P., Nierhaus, K. H., et al. (2005). Heptameric (L12) 6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc. Natl. Acad. Sci. U.S.A. 102, 8192–8197. doi: 10.1073/pnas.0502193102
Iwasa, K., and Murakami, S. (1968). Palmelloid formation of Chlamydomonas. Physiol. Plant. 21, 1224–1233. doi: 10.1111/j.1399-3054.1968.tb07353.x
Jackson, R. J., Hellen, C. U., and Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. doi: 10.1038/nrm2838
Kampfenkel, K., Van Montagu, M., and Inze, D. (1995). Extraction and determination of ascorbate and dehydroascorbate from plant tissue. Anal. Biochem. 225, 165–167. doi: 10.1006/abio.1995.1127
Kasamo, K. (1988). Inhibition of Tonoplast and Plasma Membrane H+-ATPase Activity in Rice (Oryza sativa L.) Culture Cells by Local Anesthetics. Plant Cell Physiol. 29, 215–221. doi: 10.1093/oxfordjournals.pcp.a077483
Katz, A., and Avron, M. (1985). Determination of intracellular osmotic volume and sodium concentration in dunaliella. Plant Physiol. 78, 817–820. doi: 10.1104/pp.78.4.817
Katz, A., Waridel, P., Shevchenko, A., and Pick, U. (2007). Salt-induced changes in the plasma membrane proteome of the halotolerant alga Dunaliella salina as revealed by blue native gel electrophoresis and nano-LC-MS/MS analysis. Mol. Cell. Proteomics 6, 1459–1472. doi: 10.1074/mcp.m700002-mcp200
Komis, G., Apostolakos, P., and Galatis, B. (2002). Hyperosmotic stress induces formation of tubulin macrotubules in root-tip cells of Triticum turgidum: their probable involvement in protoplast volume control. Plant Cell Physiol. 43, 911–922. doi: 10.1093/pcp/pcf114
Koytiger, G., Kaushansky, A., Gordus, A., Rush, J., Sorger, P., and MacBeath, G. (2013). Phosphotyrosine Signaling Proteins that Drive Oncogenesis Tend to be Highly Interconnected. Mol. Cell. Proteomics 12, 1204–1213. doi: 10.1074/mcp.M112.025858
Kulik, A., Anielska-Mazur, A., Bucholc, M., Koen, E., Szymanska, K., Zmienko, A., et al. (2012). SNF1-related protein kinases type 2 are involved in plant responses to cadmium stress. Plant Physiol. 160, 868–883. doi: 10.1104/pp.112.194472
Lahav, R., Nejidat, A., and Abeliovich, A. (2004). Alterations in protein synthesis and levels of heat shock 70 proteins in response to salt stress of the halotolerant yeast Rhodotorula mucilaginosa. Antonie Van Leeuwenhoek 85, 259–269. doi: 10.1023/b:anto.0000020361.81006.2b
Lechtreck, K. F., Rostmann, J., and Grunow, A. (2002). Analysis of Chlamydomonas SF-assemblin by GFP tagging and expression of antisense constructs. J. Cell Sci. 115, 1511–1522. Available online at: https://apps.crossref.org/titleList/
Len, A. C., Harty, D. W., and Jacques, N. A. (2004). Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150, 1339–1351. doi: 10.1099/mic.0.27008-0
Li, H. S., Sun, Q., Zhao, S. J., and Zhang, W. H. (2000). Principles and Techniques of Plant Physiological Biochemical Experiment. Beijing: Higher Education Press.
Li, X. J., Yang, M. F., Chen, H., Qu, L. Q., Chen, F., and Shen, S. H. (2010). Abscisic acid pretreatment enhances salt tolerance of rice seedlings: proteomic evidence. Biochim. Biophys. Acta 1804, 929–940. doi: 10.1016/j.bbapap.2010.01.004
Liang, Y. (1999). Effects of silicon on enzyme activity and sodium, potassium and calcium concentration in barley under salt stress. Plant Soil 209, 217–224. doi: 10.1023/A:1004526604913
Lichtenthaler, H. K., and Wellburn, A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 11, 591–592. doi: 10.1042/bst0110591
Liska, A. J., Shevchenko, A., Pick, U., and Katz, A. (2004). Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella as revealed by homology-based proteomics. Plant Physiol. 136, 2806–2817. doi: 10.1104/pp.104.039438
Longworth, J., Noirel, J., Pandhal, J., Wright, P. C., and Vaidyanathan, S. (2012). HILIC-and SCX-based quantitative proteomics of Chlamydomonas reinhardtii during nitrogen starvation induced lipid and carbohydrate accumulation. J. Proteome Res. 11, 5959–5971. doi: 10.1021/pr300692t
Lurling, M., and Beekman, W. (2006). Palmelloids formation in Chlamydomonas reinhardtii: defence against rotifer predators?. Ann. Limnol. Int. J. Lim. 42, 65–72. doi: 10.1051/limn/2006010
Lützelberger, M., Bottner, C. A., Schwelnus, W., Zock-Emmenthal, S., Razanau, A., and Kaufer, N. F. (2010). The N-terminus of Prp1 (Prp6/U5-102 K) is essential for spliceosome activation in vivo. Nucleic Acids Res. 38, 1610–1622. doi: 10.1093/nar/gkp1155
Ma, W., Shi, D., Wang, Q., Wei, L., and Chen, H. (2005). Exogenous expression of the wheat chloroplastic fructose-1, 6-bisphosphatase gene enhances photosynthesis in the transgenic cyanobacterium, Anabaena PCC7120. J. Appl. Phycol. 17, 273–280. doi: 10.1007/s10811-005-4850-y
Ma, W., Wei, L., and Wang, Q. (2008). The response of electron transport mediated by active NADPH dehydrogenase complexes to heat stress in the cyanobacterium Synechocystis 6803. Sci. China Life Sci. 51, 1082–1087. doi: 10.1007/s11427-008-0139-0
Mao, X., Zhang, H., Tian, S., Chang, X., and Jing, R. (2010). TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 61, 683–696. doi: 10.1093/jxb/erp331
Mastrobuoni, G., Irgang, S., Pietzke, M., Aßmus, H. E., Wenzel, M., Schulze, W. X., et al. (2012). Proteome dynamics and early salt stress response of the photosynthetic organism Chlamydomonas reinhardtii. BMC Genomics 13:215. doi: 10.1186/1471-2164-13-215
Mendes, L. F., Zambotti-Villela, L., Colepicolo, P., Marinho-Soriano, E., Stevani, C. V., and Yokoya, N. S. (2013). Metal cation toxicity in the alga Gracilaria domingensis as evaluated by the daily growth rates in synthetic seawater. J. Appl. Phycol. 25, 1939–1947. doi: 10.1007/s10811-013-0036-1
Millán-Zambrano, G., and Chávez, S. (2014). Nuclear functions of prefoldin. Open Biol. 4:140085. doi: 10.1098/rsob.140085
Montoya, H. T., and Olivera, A. G. (1993). Dunaliella salina from saline environments of the central coast of Peru. Hydrobiologia 155–161. doi: 10.1007/978-94-011-2076-0_11
Mora-García, S., Vert, G., Yin, Y., Caño-Delgado, A., Cheong, H., and Chory, J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18, 448–460. doi: 10.1101/gad.1174204
Mostek, A., Borner, A., Badowiec, A., and Weidner, S. (2015). Alterations in root proteome of salt-sensitive and tolerant barley lines under salt stress conditions. J. Plant Physiol. 174, 166–176. doi: 10.1016/j.jplph.2014.08.020
Moustafa, K., AbuQamar, S., Jarrar, M., Al-Rajab, A. J., and Tremouillaux-Guiller, J. (2014). MAPK cascades and major abiotic stresses. Plant Cell Rep. 33, 1217–1225. doi: 10.1007/s00299-014-1629-0
Ndimba, B. K., Chivasa, S., Simon, W. J., and Slabas, A. R. (2005). Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5, 4185–4196. doi: 10.1002/pmic.200401282
O'Connor, H. E., Ruffle, S. V., Cain, A. J., Deak, Z., Vass, I., Nugent, J. H., et al. (1998). The 9-kDa phosphoprotein of photosystem II. Generation and characterisation of Chlamydomonas mutants lacking PSII-H and a site-directed mutant lacking the phosphorylation site. Biochim. Biophys. Acta 1364, 63–72. doi: 10.1016/S0005-2728(98)00013-9
Pang, Q., Chen, S., Dai, S., Chen, Y., Wang, Y., and Yan, X. (2010). Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599. doi: 10.1021/pr100034f
Pick, U. (2002). “Adaptation of the halotolerant alga Dunaliella to high salinity,” in Salinity: Environment-Plants-Molecules, eds A. Läuchli, and U. Lüettge (Dordrecht: Kluwer Academic Publishers), 97–112. doi: 10.1007/0-306-48155-3_5
Pistocchi, R., Guerrini, F., Balboni, V., and Boni, L. (1997). Copper toxicity and carbohydrate production in the microalgae Cylindrotheca fusiformis and Gymnodinium sp. Eur. J. Phycol. 32, 125–132. doi: 10.1080/09670269710001737049
Preetha, K., John, L., Subin, C. S., and Vijayan, K. K. (2012). Phenotypic and genetic characterization of Dunaliella (Chlorophyta) from Indian salinas and their diversity. Aquat. Biosyst. 8:1. doi: 10.1186/2046-9063-8-27
Rahman, M. A., Alam, I., Kim, Y. G., Ahn, N. Y., Heo, S. H., Lee, D. G., et al. (2015). Screening for salt-responsive proteins in two contrasting alfalfa cultivars using a comparative proteome approach. Plant Physiol. Biochem. 89, 112–122. doi: 10.1016/j.plaphy.2015.02.015
Ramundo, S., Casero, D., Mühlhaus, T., Hemme, D., Sommer, F., Crèvecoeur, M., et al. (2014). Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. The Plant Cell 26, 2201–2222. doi: 10.1105/tpc.114.124842
Rao, Y., and Haucke, V. (2011). Membrane shaping by the Bin/amphiphysin/Rvs (BAR) domain protein superfamily. Cell. Mol. Life Sci. 68, 3983–3993. doi: 10.1007/s00018-011-0768-5
Reinbothe, C., Pollmann, S., and Reinbothe, S. (2010). Singlet oxygen signaling links photosynthesis to translation and plant growth. Trends Plant Sci. 15, 499–506. doi: 10.1016/j.tplants.2010.05.011
Sánchez, B., Champomier-Vergès, M. C., Anglade, P., Baraige, F., de Los Reyes-Gavilán, C. G., Margolles, A., et al. (2005). Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187, 5799–5808. doi: 10.1128/JB.187.16.5799-5808.2005
Scibilia, L., Girolomoni, L., Berteotti, S., Alboresi, A., and Ballottari, M. (2015). Photosynthetic response to nitrogen starvation and high light in Haematococcus pluvialis. Algal Res. 12, 170–181. doi: 10.1016/j.algal.2015.08.024
Shanmuganathan, A., Avery, S. V., Willetts, S. A., and Houghton, J. E. (2004). Copper-induced oxidative stress in Saccharomyces cerevisiae targets enzymes of the glycolytic pathway. FEBS Lett. 556, 253–259. doi: 10.1016/S0014-5793(03)01428-5
Sharma, K., D'Souza, R. C., Tyanova, S., Schaab, C., Wiśniewski, J. R., Cox, J., et al. (2014). Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Tr-based signaling. Cell Rep. 8, 1583–1594. doi: 10.1016/j.celrep.2014.07.036
Sheng, G., Yu, H., and Yue, Z. (2006). Factors influencing the production of extracellular polymeric substances by Rhodopseudomonas acidophila. Int. Biodeterior. Biodegrad. 58, 89–93. doi: 10.1016/j.ibiod.2006.07.005
Shi, K., Cui, L., Jiang, H., Yang, L., and Xue, L. (2013). Characterization of the microtubule-binding activity of kinesin-like calmodulin binding protein from Dunaliella salina. Res. Microbiol. 164, 1028–1034. doi: 10.1016/j.resmic.2013.08.009
Shoji, T., Suzuki, K., Abe, T., Kaneko, Y., Shi, H., Zhu, J. K., et al. (2006). Salt stress affects cortical microtubule organization and helical growth in Arabidopsis. Plant Cell Physiol. 47, 1158–1168. doi: 10.1093/pcp/pcj090
Silva, P., Thompson, E., Bailey, S., Kruse, O., Mullineaux, C. W., Robinson, C., et al. (2003). FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp PCC 6803. The Plant Cell 15, 2152–2164. doi: 10.1105/tpc.012609
Song, C., Wang, F., Cheng, K., Wei, X., Bian, Y., Wang, K., et al. (2014). Large-scale quantification of single amino-acid variations by a variation-associated database search strategy. J. Proteome Res. 13, 241–248. doi: 10.1021/pr400544j
Song, C., Ye, M., Liu, Z., Cheng, H., Jiang, X., Han, G., et al. (2012). Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol. Cell. Proteomics 11, 1070–1083. doi: 10.1074/mcp.M111.012625
Sternlicht, H., Farr, G. W., Sternlicht, M. L., Driscoll, J. K., Willison, K., and Yaffe, M. B. (1993). The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc. Natl. Acad. Sci. U.S.A. 90, 9422–9426.
Sukegawa, Y., Yamashita, A., and Yamamoto, M. (2011). The fission yeast stress-responsive MAPK pathway promotes meiosis via the phosphorylation of Pol II CTD in response to environmental and feedback cues. PLoS Genet. 7:e1002387. doi: 10.1371/journal.pgen.1002387
Summer, E. J., Schmid, V. H., Bruns, B. U., and Schmidt, G. W. (1997). Requirement for the H phosphoprotein in photosystem II of Chlamydomonas reinhardtii. Plant Physiol. 113, 1359–1368.
Suo, J., Zhao, Q., Zhang, Z., Chen, S., Cao, J., Liu, G., et al. (2015). Cytological and proteomic analyses of Osmunda cinnamomea germinating spores reveal characteristics of fern spore germination and rhizoid tip-growth. Mol. Cell. Proteomics 14, 2510–2534. doi: 10.1074/mcp.M114.047225
Sztrum, A. A., Sabatini, S. E., and Rodríguez, M. C. (2012). Isocitrate lyase activity and antioxidant responses in copper-stressed cultures of Chlamydomonas reinhardtii (Volvocales, Chlorophyceae). Phycologia 51, 135–143. doi: 10.2216/10-56.1
Takouridis, S. J., Tribe, D. E., Gras, S. L., and Martin, G. J. (2015). The selective breeding of the freshwater microalga Chlamydomonas reinhardtii for growth in salinity. Bioresour. Technol. 184, 18–22. doi: 10.1016/j.biortech.2014.10.120
Tange, T. Ø., Nott, A., and Moore, M. J. (2004). The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 16, 279–284. doi: 10.1016/j.ceb.2004.03.012
Valášek, L. S. (2012). ‘Ribozoomin’–translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr. Protein Peptide Sci. 13, 305–330. doi: 10.2174/138920312801619385
Vizcaino, J. A., Deutsch, E. W., Wang, R., Csordas, A., Reisinger, F., Rios, D., et al. (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226. doi: 10.1038/nbt.2839
Voutsina, A., Riva, M., Carles, C., and Alexandraki, D. (1999). Sequence divergence of the RNA polymerase shared subunit ABC14. 5 (Rpb8) selectively affects RNA polymerase III assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 27, 1047–1055. doi: 10.1093/nar/27.4.1047
Wang, F., Dong, J., Jiang, X., Ye, M., and Zou, H. (2007). Capillary trap column with strong cation-exchange monolith for automated shotgun proteome analysis. Anal. Chem. 79, 6599–6606. doi: 10.1021/ac070736f
Wang, L., Piao, T., Cao, M., Qin, T., Huang, L., Deng, H., et al. (2013). Flagellar regeneration requires cytoplasmic microtubule depolymerization and kinesin-13. J. Cell Sci. 126, 1531–1540. doi: 10.1242/jcs.124255
Wang, M. C., Peng, Z. Y., Li, C. L., Li, F., Liu, C., and Xia, G. M. (2008). Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 8, 1470–1489. doi: 10.1002/pmic.200700569
Wang, S. B., Chen, F., Sommerfeld, M., and Hu, Q. (2004a). Proteomic analysis of molecular response to oxidative stress by the green alga Haematococcus pluvialis (Chlorophyceae). Planta 220, 17–29. doi: 10.1007/s00425-004-1323-5
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004b). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wilson, N. F., and Lefebvre, P. A. (2004). Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot. Cell 3, 1307–1319. doi: 10.1128/EC.3.5.1307-1319.2004
Xu, J., Zhang, X., Ye, N., Zheng, Z., Mou, S., Dong, M., et al. (2013). Activities of principal photosynthetic enzymes in green macroalga Ulva linza: functional implication of C(4) pathway in CO(2) assimilation. Sci. China Life Sci. 56, 571–580. doi: 10.1007/s11427-013-4489-x
Xuan, J., Song, Y., Zhang, H., Liu, J., Guo, Z., and Hua, Y. (2013). Comparative proteomic analysis of the stolon cold stress response between the C4 perennial grass species Zoysia japonica and Zoysia metrella. PLoS ONE 8:e75705. doi: 10.1371/journal.pone.0075705
Zhang, H., Han, B., Wang, T., Chen, S., Li, H., Zhang, Y., et al. (2012). Mechanisms of plant salt response: insights from proteomics. J. Proteome Res. 11, 49–67. doi: 10.1021/pr200861w
Zhang, T., Gong, H., Wen, X., and Lu, C. (2010). Salt stress induces a decrease in excitation energy transfer from phycobilisomes to photosystem II but an increase to photosystem I in the cyanobacterium Spirulina platensis. J. Plant Physiol. 167, 951–958. doi: 10.1016/j.jplph.2009.12.020
Zhao, Q., Suo, J., Chen, S., Jin, Y., Ma, X., Yin, Z., et al. (2016). Na2CO3-responsive mechanisms in halophyte Puccinellia tenuiflora roots revealed by physiological and proteomic analyses. Sci. Rep. 6:32717. doi: 10.1038/srep32717
Zhou, H., Ye, M., Dong, J., Corradini, E., Cristobal, A., Heck, A. J., et al. (2013). Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 8, 461–480. doi: 10.1038/nprot.2013.010
Zhou, H., Ye, M., Dong, J., Han, G., Jiang, X., Wu, R., et al. (2008). Specific phosphopeptide enrichment with immobilized titanium ion affinity chromatography adsorbent for phosphoproteome analysis. J. Proteome Res. 7, 3957–3967. doi: 10.1021/pr800223m
Keywords: palmella formation, Dunaliella salina, salinity stress, quantitative proteomics, phosphoproteomics
Citation: Wei S, Bian Y, Zhao Q, Chen S, Mao J, Song C, Cheng K, Xiao Z, Zhang C, Ma W, Zou H, Ye M and Dai S (2017) Salinity-Induced Palmella Formation Mechanism in Halotolerant Algae Dunaliella salina Revealed by Quantitative Proteomics and Phosphoproteomics. Front. Plant Sci. 8:810. doi: 10.3389/fpls.2017.00810
Received: 23 January 2017; Accepted: 30 April 2017;
Published: 23 May 2017.
Edited by:
Setsuko Komatsu, University of Tsukuba, JapanReviewed by:
Ramesh Katam, Florida A&M University, United StatesCopyright © 2017 Wei, Bian, Zhao, Chen, Mao, Song, Cheng, Xiao, Zhang, Ma, Zou, Ye and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingliang Ye, mingliang@dicp.ac.cn
Shaojun Dai, daishaojun@hotmail.com
†These authors have contributed equally to this work.
‡In deep memory of Hanfa Zou.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.