- 1Consiglio Nazionale delle Ricerche – Institute for Agricultural and Forest Systems in the Mediterranean, Perugia, Italy
- 2Consiglio Nazionale delle Ricerche – Institute of Biosciences and Bioresources, Perugia, Italy
- 3Department of Agricultural, Food and Environmental Sciences, Università degli Studi di Perugia, Perugia, Italy
Germplasm collections of tree crop species represent fundamental tools for conservation of diversity and key steps for its characterization and evaluation. For the olive tree, several collections were created all over the world, but only few of them have been fully characterized and molecularly identified. The olive collection of Perugia University (UNIPG), established in the years’ 60, represents one of the first attempts to gather and safeguard olive diversity, keeping together cultivars from different countries. In the present study, a set of 370 olive trees previously uncharacterized was screened with 10 standard simple sequence repeats (SSRs) and nine new EST-SSR markers, to correctly and thoroughly identify all genotypes, verify their representativeness of the entire cultivated olive variation, and validate the effectiveness of new markers in comparison to standard genotyping tools. The SSR analysis revealed the presence of 59 genotypes, corresponding to 72 well known cultivars, 13 of them resulting exclusively present in this collection. The new EST-SSRs have shown values of diversity parameters quite similar to those of best standard SSRs. When compared to hundreds of Mediterranean cultivars, the UNIPG olive accessions were splitted into the three main populations (East, Center and West Mediterranean), confirming that the collection has a good representativeness of the entire olive variability. Furthermore, Bayesian analysis, performed on the 59 genotypes of the collection by the use of both sets of markers, have demonstrated their splitting into four clusters, with a well balanced membership obtained by EST respect to standard SSRs. The new OLEST (Olea expressed sequence tags) SSR markers resulted as effective as the best standard markers. The information obtained from this study represents a high valuable tool for ex situ conservation and management of olive genetic resources, useful to build a common database from worldwide olive cultivar collections, also based on recently developed markers.
Introduction
The cultivated olive (Olea europaea, subsp. europaea, var. europaea, Green, 2002) is one of the most important oil crops in the world and 95% of total olive oil production derives from the Mediterranean basin (Marra et al., 2013; Trujillo et al., 2014). The olive crop counts a very rich varietal heritage, represented by more than 1,200 named cultivars, over 3,000 minor cultivars and an uncertain number of genotypes including pollinators, local ecotypes and centennial trees (El Bakkali et al., 2013; Hosseini-Mazinani et al., 2014; Mazzitelli et al., 2015; Laroussi-Mezghani et al., 2016; Mousavi et al., 2017). Since time of ancient Greece, olive cultivars have been vegetatively propagated, either by cutting or grafting, allowing the accurate reproduction of the best-performing genotypes, leading to the present varietal assortment (Breton et al., 2009; Kaniewski et al., 2012). Thus, most cultivars represent ancient pre-bred genotypes, and the limited and sporadic genetic improvement initiatives, with classical or biotechnological approaches, forced the retention of numerous traditional cultivars despite their agronomical limitations. Among these, only a few have a large area of cultivation and a clear impact on the production of oil and table olives (Ilarioni and Proietti, 2014). But the availability of a large set of well characterized and highly different cultivars is critical to increase the ability to face new agronomical challenges (De Gennaro et al., 2012; Larbi et al., 2015) and future climatic constrains (Moriondo et al., 2013; Proietti et al., 2014; Tanasijevic et al., 2014), diversifying the gene pools, preserving unique genetic traits currently available (Bracci et al., 2011; Corrado et al., 2011; Potts et al., 2012; Klepo et al., 2013) and offering different sensory profiles of extra-virgin olive oils.
Several reasons make it difficult to ensure the identification of cultivars, as the joint cultivation of native and foreign cultivars, the ambiguous plant naming, seedlings or wild plants, or the interchange of plant material over the centuries (Marra et al., 2013; Lazović et al., 2016). Furthermore, the large number of cultivars, the high degree of kinship among many of them, mainly in cases of geographic proximity, and the possible appearance of clonal variation, have raised additional identification problems (Belaj et al., 2007; Caruso et al., 2014; Ipek et al., 2015).
Olive collections represent the main tool to preserve and certify germplasm resources (Belaj et al., 2012; Caruso et al., 2014), mainly when recent trends toward establishing modern orchards exclusively based on a few highly producing and low-vigor cultivars, may potentially lead to the erosion of this germplasm. More than 100 collections of olive genetic resources have been established at international, national and regional levels for conservation and evaluation purposes (Trujillo et al., 2014). A first World Olive Germplasm Bank (WOGB) was established since the years’ 70 at IFAPA (Cordoba, Spain), with about 500 accessions from 21 countries (Belaj et al., 2011; Trujillo et al., 2014). In 2003, a second WOGB was created at INRA (Marrakech, Morocco), including 560 accessions originating from 14 Mediterranean countries (Haouane et al., 2011). An international olive collection built by CNR (ISAFOM) and planted in Zagaria (Enna, Italy), includes about 400 cultivars collected worldwide (Las Casas et al., 2014). A national collection has been built by CREA-OLI (Cosenza, Italy), consisting of approximately 500 cultivars from Italy, corresponding to 85% of total Italian olive germplasm (Muzzalupo et al., 2014). In Turkey, a national olive germplasm collection in Izmir contains 96 genotypes (Kaya et al., 2013), whereas the Greek National Olive Germplasm Collection counts on 47 olive cultivars (Xanthopoulou et al., 2014). Also new olive growing countries, such as the United States of America, have organized important olive collections (NCGR-Davis, CA, United States) (Zelasco et al., 2012), as well as Argentina, Chile, Uruguay, Australia, China, and South Africa (Trujillo et al., 2014). In addition to these important gene banks, many other minor collections were set up along the time to preserve dedicated pools of genotypes, such as cultivars with specific characteristics, wild plants, segregating progenies or core collections (Belaj et al., 2011, 2012; Díez et al., 2011; Marchese et al., 2016). Among these, the UNIPG (Perugia University, Italy) collection, established 50 years ago, represents one of the first attempts to collect and conserve ex situ a large number of olive cultivars. It contains genotypes of different geographical origin (although with prevalence of Italian cultivars), and holds great potential for the complete agronomic and exhaustive evaluation of cultivars, as reported by numerous previous works on agronomical, morphological or biological varietal performance (Breton et al., 2014; Portarena et al., 2015).
Simple sequence repeats were the main molecular markers used to characterized the olive germplasm collection (Haouane et al., 2011; Muzzalupo et al., 2014; Trujillo et al., 2014). In fact, SSRs represent the most popular markers for olive genotyping, due to the high polymorphism, extraordinary abundance and fast transferability (Sarri et al., 2006; Baldoni et al., 2009; Díez et al., 2011; Belaj et al., 2012; Hosseini-Mazinani et al., 2014; Mousavi et al., 2014, 2017). However, all SSR loci published so far, characterized by dinucleotide repeat motifs, have demonstrated several drawbacks due to the difficult discrimination among alleles (Baldoni et al., 2009). On the contrary, EST-SSRs derive from expressed regions of the genome, have a greater transferability among species and, since they are located within genes, their variation could find correlation with the phenotype (Duran et al., 2009). However, EST-SSRs may reveal less variations and lower polymorphic information than standard SSRs, eventhough sufficient for population genetic analysis and for genotyping purpose (Yang et al., 2013). For this reason, new trinucleotidic EST-SSR loci recently identified (Mariotti et al., 2016) should now be widely applied for a more clear varietal characterization.
In this work, we have provided the first molecular identification of the accessions present in the UNIPG olive varietal collection. The identification of all olive trees was performed by standard SSRs and, for the first time in olive collections, by EST-SSRs. We intended to reach numerous important goals: (1) the identification of all accessions, including those closely related or morphologically similar, (2) the evaluation of discrimination power between EST-SSRs and dinucleotide standard SSRs, and (3) establishing the level and wideness of the genetic variability inside a germplasm collection, in order to make available this important source of well-defined genotypes to all interested stakeholders and researchers.
Materials and Methods
Sample Collection and Archival Records
The Olive Varietal Collection of the University of Perugia – Department of Agricultural, Food and Environmental Sciences (UNIPG) – is located in Prepo, Perugia (43°04′ 53.94″ N – 12°22′ 53.25″ E, altitude about 400 m asl), on a clay soil, with medium content of organic matter, phosphorus and potassium, temperate-Mediterranean climate, average annual temperatures of 12.8°C and annual rainfall of about 900 mm. Planting distance is 5 × 5 m and trees are grown polyconic vase-shaped. Regular agricultural practices are applied to the olive plants, without irrigation. The collection, established in 1965, has been duplicated in 1984 and enlarged by adding further local, national and international cultivars. Based on the UNIPG archive, the collection consisted of 370 olive plants, where each genotype was represented at least by three replications, randomly distributed in a single block, although, some cultivars (Carolea, Maurino, Moraiolo, Leccino, Frantoio, San Felice, Nostrale di Rigali and Manzanilla de Sevilla) were represented by at least 20 trees per cultivar, distributed in four randomized blocks, allowing for their agronomical and morpho-bio-phenological evaluation. No information was available on the original source of plant material.
DNA Extraction and Molecular Analysis
Leaf samples were collected from each plant, for a total of 370 accessions, and plant position of each tree was recorded. For each accession, total DNA was extracted from fresh leaves following the standard manufacturer’s instructions of GeneElute Plant Genomic DNA Miniprep Kit (Sigma–Aldrich).
All samples were analyzed by using nine best ranked EST-SSR markers (OLEST1-7-9-12-14-16-20-22-23) recently developed (Mariotti et al., 2016). Double step polymerase chain reactions (PCR) were performed in a volume of 25 μl containing 25 ng of DNA, 10× PCR buffer, 200 μM of each dNTP, 10 pmol of primer forward (with 18 bp tail in 5′) and reverse, and 2 U of DNA Polymerase (Q5 High Fidelity DNA Polymerase, New England Biolabs). In the second step, fluorescent tail (10 pmol) was annealed to the forward primer using a double step PCR: the first step consisting in an initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 25 s, the second step (for tail annealing) made up of 20 cycles, with the same conditions of the first step except for annealing temperature (Tm = 52°C), a final elongation at 72°C for 40 min closed the second step PCR.
In order to verify the identity of cultivars present in the collection, all samples were genotyped by using standard dinucleotide SSRs markers, widely applied for cultivar characterization in most olive germplasm collections (Haouane et al., 2011; Muzzalupo et al., 2014; Trujillo et al., 2014). Ten high polymorphic markers were applied, including DCA3-5-9-16-18, EMO90, GAPU71B-101-103A and UDO-043 (Sefc et al., 2000; Carriero et al., 2002; Cipriani et al., 2002), previously selected as best performing loci (Baldoni et al., 2009) and common to the other genotyping works. Forward primers carried VIC, FAM, PET, or NED labels at their 5′-end. Standard PCR amplifications were performed in a reaction volume of 25 μl containing 25 ng of DNA, 10× PCR buffer, 200 μM of each dNTP, 10 pmol of each forward and reverse primer, and 2 U of Q5 High-Fidelity DNA Polymerase (New England Biolabs), with an initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, annealing temperature as suggested by authors (50–60°C) for 30 s and 72°C for 25 s, followed by a final elongation at 72°C for 40 min.
Polymerase chain reactions products were loaded on an ABI 3130 Genetic Analyzer (Applied Biosystems-Hitachi) using the internal GeneScan 500 LIZ Size Standard (Thermo Fisher Scientific). Output data were analyzed by GeneMapper 3.7 (Applied Biosystems).
In order to verify the match of the 370 olive samples with previously characterized cultivars, the data obtained for the 10 standard SSR markers were compared to those available in the database of olive SSR profiles established at CNR-IBBR of Perugia (Italy), including more than 1,000 worldwide olive cultivars, and to other available datasets (Baldoni et al., 2009; Trujillo et al., 2014), allowing to establish cultivar identity and determine all cases of identical profiles, presumably corresponding to clonal genotypes with undetermined presence of mutationsclonal replicates (Baldoni et al., 2009, 2011; Bartolini, 2009; Mousavi et al., 2017).
Allele Frequency and Diversity Analysis
Number of alleles per locus (Na), number of effective alleles (Ne), Shannon’s information index (I), observed (Ho) and expected heterozygosity (He), and fixation index (F) were calculated at each locus for novel and standard SSRs by the use of GenAlEx 6.501 software (Peakall and Smouse, 2012). Pairwise relatedness was performed on standard and OLEST SSR markers to calculate the allelic similarity for codominant data using GenAlEx 6.501 following the LRM = Lynch and Ritland (1999) estimator – Mean multiplied by 2 to give max of 1.00. The software FreeNA (Chapuis and Estoup, 2007) was applied to detect the presence of possible null alleles (Fnull), to determine the genetic uniqueness of each accession and to quantify redundancy. Polymorphic information content (PIC) was calculated for each microsatellite locus using CERVUS v.3.0 software (Marshall et al., 1998). We calculated the probabilities of identity for unrelated individuals [P(ID)] at each locus and across loci, as described by Waits et al. (2001), by using GenAlEx for both OLEST and standard SSR markers. Cumulative P(ID) was calculated by ranking the PIC values at each locus from high to low. We used the criterion of P(ID) lower than 0.001 for the estimation of the minimum number of loci required for individual identification in the study species (Waits et al., 2001).
A model-based Bayesian clustering method was applied to infer the genetic structure of 59 cultivars and to define the number of clusters in the dataset (gene pools) using the software STRUCTURE v.2.3 (Pritchard et al., 2009), for the same sample set separately for OLEST and standard SSRs. Tests were based on an admixture model with independent allele frequencies. No prior information was used to define clusters. Independent runs were done by setting the number of clusters (k) from 1 to 10. Each run comprised a burn-in length of 100,000 followed by 100,000 MCMC (Monte Carlo Markov Chain) replicates. An ad hoc statistic ΔK, based on the rate of change in the log probability of data between successive K values, as described by Evanno et al. (2005), was calculated through Structure Harvester v.0.9.93 website (Earl, 2012) and used to estimate the most likely number of clusters (k). In order to verify the breakdown of cultivars present in the Perugia collection to the Mediterranean groups previously observed (Sarri et al., 2006), their profiles for ten standard SSRs were analyzed with those of 281 most widely cultivated cultivars of Mediterranean from the CNR-IBBR database by using the same Structure parameters. Data of 281 cultivars were already published (Baldoni et al., 2009, 2011; Mousavi et al., 2017).
Results
Polymorphisms Detected at EST and Standard SSR Loci
The nine OLEST markers analyzed were easily scored, showed low stuttering and clear differentiation among alleles (Table 1, Supplementary Table S1 and Figure S1). Mean Na amounted to 7.9, ranging between 5 (OLEST9) and 15 (OLEST16). Ne was 4.466 on average, while the mean I value was 1.636. He (0.760) was in general higher than Ho (0.718), unless for OLEST22 and 23, where Ho was significantly higher than He. F values were positive on average, excluding OLEST22 and 23, and a negligible or moderate amount of null alleles was observed, with no effect on their discrimination power. PIC values were higher than 0.5 at all OLEST loci, with an average value of 0.726 and the maximum discrimination power for OLEST16 (0.848) and OLEST1 (0.804).
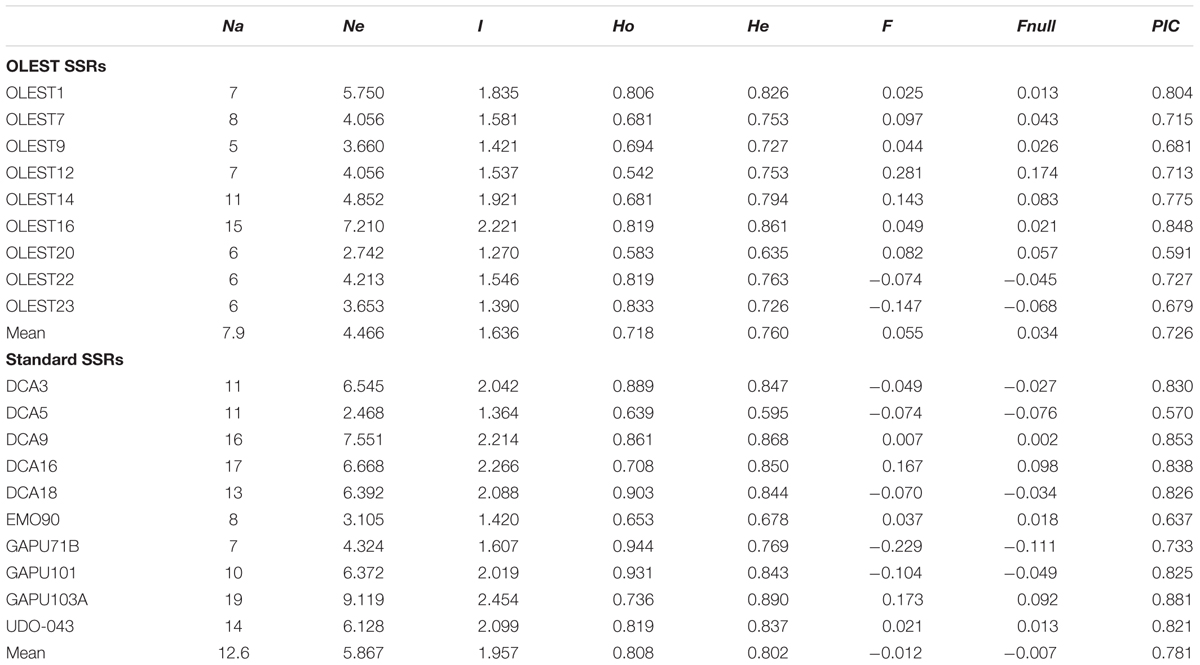
TABLE 1. Indices of genetic diversity at 72 cultivars for each SSR locus: number of alleles (Na), number of effective alleles (Ne), Shannon’s information index (I), observed heterozygosity (Ho), expected heterozygosity (He), fixation index (F) and presence of null alleles (Fnull), Polymorphism Information Content (PIC).
Total number of alleles for standard SSRs (Table 1) was considerably higher than for OLESTs, with 12.6 alleles per locus. Mean Ho was similar to He (0.808 and 0.802, respectively), and three out of 10 loci (DCA18, GAPU71B and GAPU101) with Ho higher than 0.9. F and Fnull were slightly negative, showing -0.012 and -0.007, respectively, whereas the mean value of PIC was 0.781. Cumulative probability of identity values (Figure 1) showed that a minimum of three loci was required for OLEST markers and only two for standard SSRs to reach P(ID) < 0.001. Therefore, only four and three loci were needed to distinguish all genotypes for OLEST and standard SSR markers, respectively. Nine OLEST [cumulative P(ID) = 2.5e-10] or 10 standard SSRs [cumulative P(ID) = 7.3e-14] allow for the unequivocal individual identification for this sample set with a high statistical confidence.
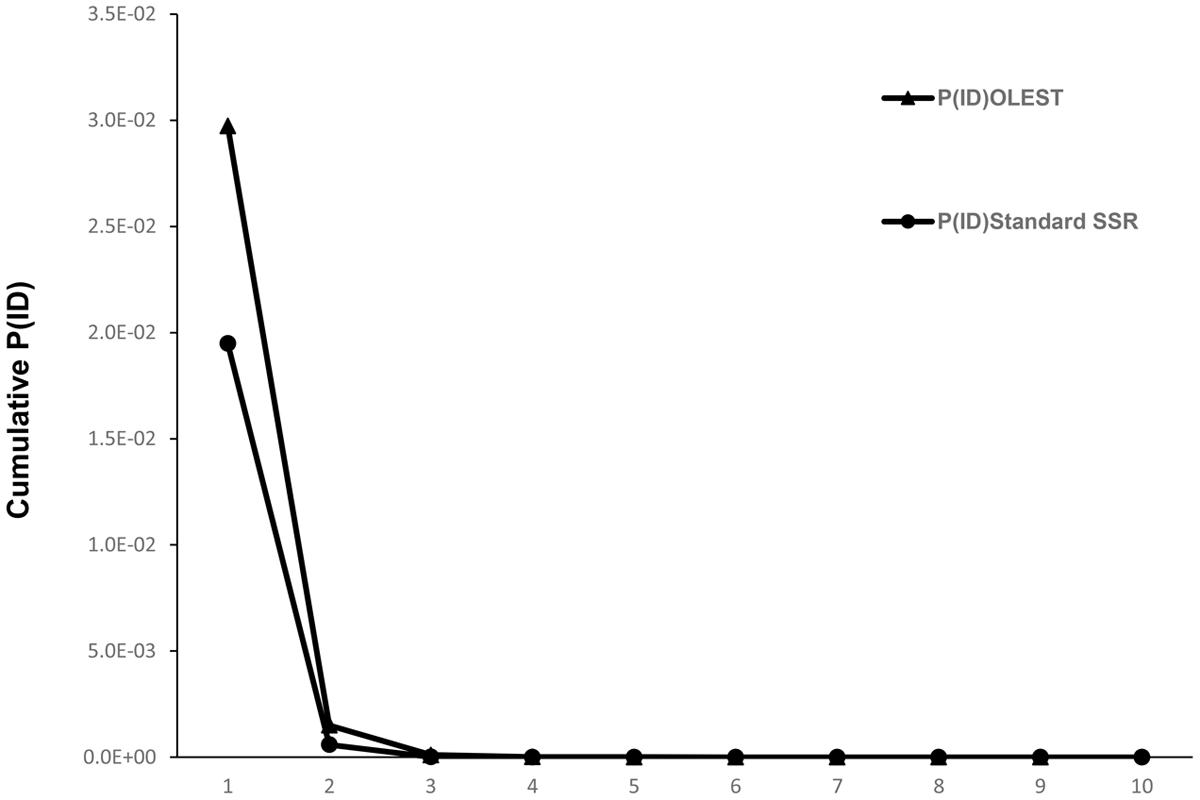
FIGURE 1. Cumulative probability of identity for OLEST and standard SSRs for unrelated individuals [P(ID)].
Genetic Identity and Differentiation
The comparison of standard SSR profiles with the CNR-IBBR dataset and previous published data allowed for the identification of UNIPG collection’s samples. Fifty nine distinct genotypes were identified, corresponding to 72 olive cultivars reported in the UNIPG archive. In fact, some samples called in the archive by different names, showed in our work identical genetic profiles (Supplementary Table S1 and Table 2). Among genotypes with identical profiles, the first group included the Portuguese cultivars Azeteira and Negrinha, eight cultivars resulted identical to Frantoio (Frantoio Corsini, Razzola, Casaliva, Razza, Taggiasca, Raja Sabina, and Ogliarola di Bitonto) (Group 2), Ogliarola Salentina, Mignola and Cima di Mola formed the third group, Moraiolo, Moraiolo Corsini and Corniolo the fourth, and Dritta di Moscufo and San Felice were the last case of identity. The OLEST markers showed exactly the same results, confirming all cases of identical profiles (Supplementary Table S1).
Eight different countries are represented in the collection, including Italy with 37 cultivars, Spain with nine, Greece with four, Portugal and France with three each, Morocco, Syria, and Tunisia with one each. Thirteen out of the 59 olive genotypes (Dolce d’Andria, Dritta di Loreto, Laurina, Morellona di Grecia, Negrera, Nostrale di Rigali, Olivago, Olivone, Orbetana, Pasola di Andria, Pocciolo, Santagatese, Tendellone) resulted exclusive to this collection and absent in the main WOGBs. Pairwise allelic relatedness performed by GenAlEx showed 100 percent of similarity between the synonymous cultivars (LRM = 1.00) for both set of markers. Comparing OLEST and standard SSRs for allelic similarity the highest values for non-synonymous cultivars were 0.67 and 0.57 respectively, while the minimum LRM values were -0.43 for OLEST and -0.31 for standard SSR markers.
Population Genetic Structure
From the Structure analysis of data derived from 10 standard SSR loci on the 59 UNIPG cultivars run with 281 Mediterranean representative cultivars (Supplementary Figure S2), the stabilization, in terms of log-likelihood values of ΔK values was observed at K = 3 and, assigning individuals to a population for values above 70%, it was observed that 16 cultivars clustered into the Western Mediterranean group, 35 in the Central one and 12 in the Eastern population, only nine genotypes showed high levels of admixture among two or three groups.
The Structure analysis within the cultivars of the collection performed on OLEST and standard SSRs showed the most probable grouping at K = 4 (Figures 2A,B). Most of the 59 cultivars resulted assigned to two of the four groups for standard SSRs while for OLEST the four structure population were well balanced. In fact, the proportion of membership for OLEST markers was from 0.158 (Pop2) to 0.406 (Pop1), while for standard SSRs the lowest value was 0.054 (Pop2) and for the Pop1 and Pop4 membership value were 0.423 and 0.414 respectively. Only 20 cultivars were assigned to the same population by both set of markers (Figures 2A,B). The expected heterozygosity individuated by Bayesian analysis within the same population was on average higher for standard than for OLEST markers (0.84 and 0.76, respectively). Furthermore, the level of population assignment for OLEST markers was lower than standard SSRs (0.75 and 0.88, respectively).
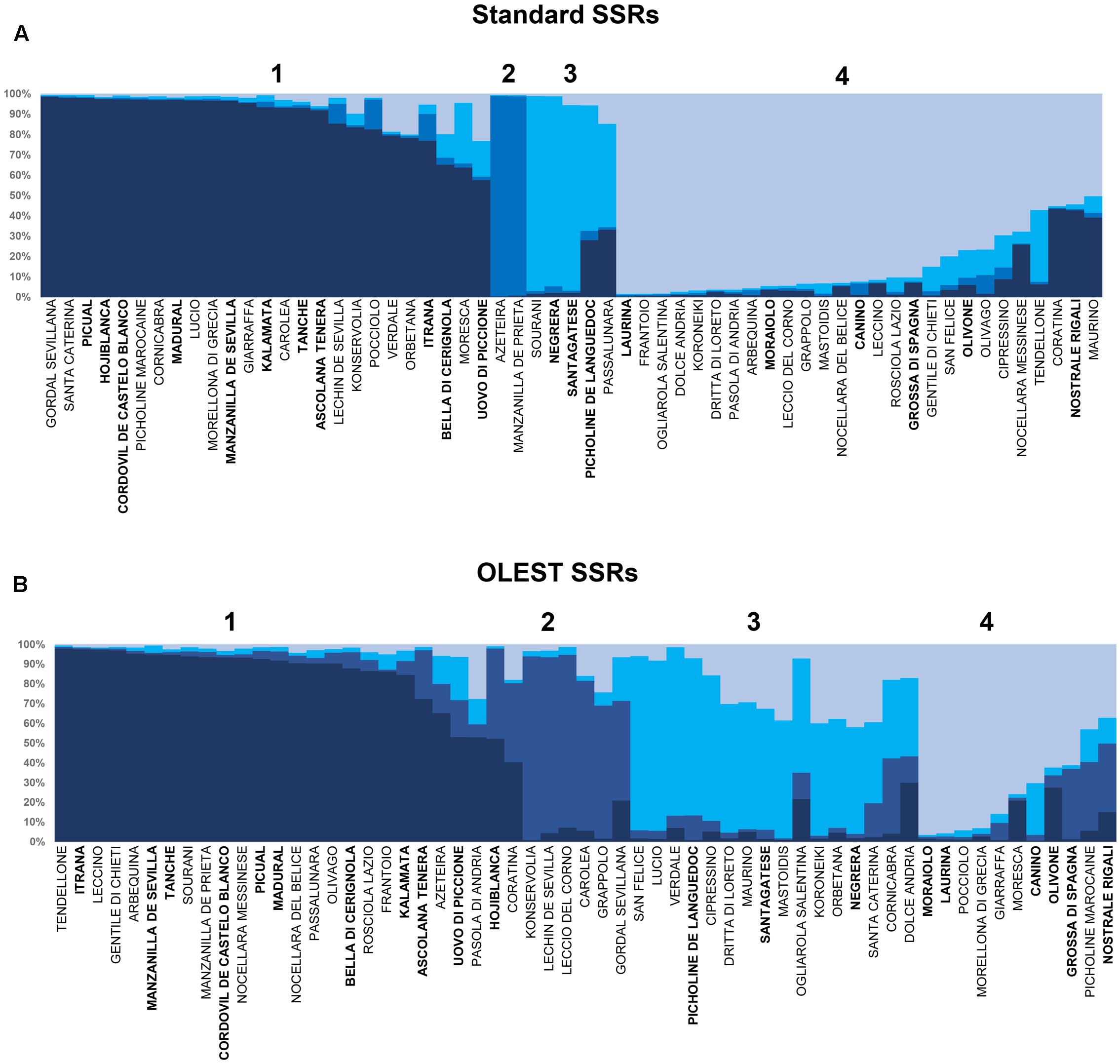
FIGURE 2. Genetic structure of the 59 cultivars identified at the UNIPG Olive Collection based on data derived from standard (A) and new OLEST (B) SSR markers. Each vertical bar represents single accessions and colors distinguish the four detected groups. Olive samples with more than one color indicate admixture in their genetic composition. Cultivars assigned to the same group by both kind of markers are reported in bold below every population (from Pop 1 to Pop 4).
Discussion
The application of highly effective and discriminant markers may allow the correct identification of all accessions, establishing their representativeness of the species variability and justifying their conservation in ex situ collections. This step is crucial to avoid redundancy in germplasm repositories, reducing management costs, distributing true-to-type genotypes for propagation, ratifying reliable genetic sources for breeding programs. The management of germplasm collections, in fact, requires attention and mistakes may be introduced at many stages, from the origin of plant material, that may derive from other collections, private orchards or unreliable sources, to propagation and field planting, and each accession needs correct identification and passport data (Kato et al., 2012; Potts et al., 2012; Trujillo et al., 2014). A thorough and accurate genotype profiling represents a crucial prerequisite to assist breeding programs, perform comparative studies and assess innovative researches.
The collection of olive cultivars established at the University of Perugia represents one of the first efforts to converge into a single set deeply diverse genotypes, deriving from areas with highly different climatic and growing conditions, in order to preserve the variation of cultivated olives and evaluate their characteristics. The genetic identity of genotypes at the UNIPG olive collection was never ascertained before and we were committed to achieve a complete genotyping of all accessions.
Simple sequence repeats markers have become the preferred tool for the identification of olive cultivars, due to their high discrimination power and straightforward data reading (Haouane et al., 2011; Trujillo et al., 2014), however, the largely used dinucleotide SSRs have shown problems related to difficult discrimination between neighboring alleles and low comparability of data among different labs, severely reducing their applicability for large-scale screening (Baldoni et al., 2009) and for comparing the molecular profiles of accessions distributed in different collections (Diez et al., 2015; Torkzaban et al., 2015). For this reason, we decided to apply both, the best ranked dinucleotide SSRs and the recently developed trinucleotide EST-SSRs (OLEST) (Mariotti et al., 2016), in order to also evaluate their reliability in genotyping germplasm repositories.
To establish cultivar identity and determine all clonal replicates, 10 standard dinucleotide SSR markers were preliminarly applied and allele profiles were compared with previously published data (Baldoni et al., 2009, 2011; Hosseini-Mazinani et al., 2014; Trujillo et al., 2014; Mousavi et al., 2017), or included in the CNR-IBBR database. Results derived from these analyses highlighted the presence of 59 distinct genotypes, including five groups of cultivars sharing identical SSR profiles (Bartolini, 2009; Trujillo et al., 2014), but coming from different areas of cultivation and carrying different names.
The same results were obtained when the analysis was independently performed with the new OLEST SSRs: 59 genotypes were distinguished and identified, and the same groups with identical profiles were displayed. Also the values of diversity parameters resulted quite similar to those of best ranked dinucleotide SSRs, particularly for the discrimination power and observed heterozygosity values, with a negligible presence of null alleles. The pairwise relatedness analysis demonstrated the same single-profile groups and highlighted that OLEST markers were more efficient to discriminate among the most polymorphic genotypes, showing the minimum values of allelic similarity.
The occurrence of cases of identical genotype under different cultivar names represents a primary source of problems for identification and a major challenge to the management of germplasm collections (Belaj et al., 2007; Abdessemed et al., 2015). In the olive case (Bradai et al., 2016), as for many other long living trees (Vezzulli et al., 2012; Urrestarazu et al., 2012; Fresnedo-Ramírez et al., 2013; Jiao et al., 2013; Frank and Chitwood, 2016), it can not be theoretically excluded that plant genotypes clonally propagated and living for thousands of years, may accumulate somatic mutations, over the time or as a result of environmental shocks. But these mutations could not be easily revealed by the use of a restricted set of SSR markers and, for this reason, we decided to leave the original names of cultivars, even if they showed the same SSR profile, making them available for future in-depth genomic analyses that would highlight eventual polymorphisms otherwise undetectable (Wu et al., 2014).
By using only three OLEST markers it was possible to discriminate 96.6% of all genotypes. Moreover, OLEST SSRs resulted more easily scorable than dinucleotide SSRs, and didn’t show stuttering problems due to the higher distance among similar alleles and lower slippage during replication. Using the three OLEST markers with the highest PIC values (OLEST1, OLEST14 and OLEST16), 57 out of 59 genotype were discriminated, whereas applying the three most discriminant standard SSRs (DCA09, DCA16 and GAPU103A), all 59 genotypes were completely recognized. In fact, the individual identification estimator [P(ID)] indicates two different accessions may have the same genotype at one specific locus in a population by chance rather than through inheritance, we found that both set of markers were able to clearly distinguish all 59 olive genotypes in the Perugia olive collection.
The Bayesian structure analysis of genotypes present in the Perugia assortment with the wide set of other important cultivars of Mediterranean basin, has shown that the collection well represents the groups in which the cultivated Mediterranean olives were previously splitted (Haouane et al., 2011; Diez et al., 2015), with a higher membership to the Central Mediterranean group, likely due to the prevalence of Italian cultivars. Furthermore, this repository owns 13 cultivars not present in the main international olive germplasm banks (Haouane et al., 2011; Trujillo et al., 2014), strengthening its relevant function for conservation, evaluation and protection of specific genotypes potentially endangered.
When the same analysis was exclusively performed on the UNIPG genotypes, 34% of cultivars resulted assigned to the same population by both sets of markers. The Bayesian results clearly highlighted the differences between OLEST and standard SSRs in the cultivar’s assignment into the structure populations. These dissimilarity was evidenced by the values of expected heterozygosity, the overall proportion of membership and admixture level. Therefore, the results of the present study suggest that, for phylogenetic studies, by using different set of markers could achieve unbalanced assignments. The different ability of both kinds of markers to group cultivars into different clusters could be explained by the nature of OLEST markers as mutations residing in the sequence of transcribed genes, and their alleles could display a higher frequency at regional level, where cultivars were selected based on common characteristics (Biton et al., 2015; Mariotti et al., 2016). Considering that olive domestication process has implied a selection of cultivars for certain agronomic characters, resulting in a loss of genetic variation due to genetic bottlenecks and, in some cases, episodes of founder effect (Cao et al., 2014; Hosseini-Mazinani et al., 2014; Mousavi et al., 2017), EST-SSRs could be related to agronomical traits more than neutral standard SSRs. The very long history of olive growing with several trading events, introduction of alien cultural practices and changes of dietary habits, may have blurred the fingerprints of independent domestication events and led to complex relationships among cultivars (Sarri et al., 2006; Soleri et al., 2010; Díez et al., 2011; Koehmstedt et al., 2011).
The Perugia collection represents the first study case of a real olive germplasm repository validated by standard SSRs and characterized by EST-SSRs. The work has allowed to confirm the OLEST markers as effective genotyping tools, as good as best standard markers for cultivar identification, allowing to avoid the application of other unreliable dinucleotide SSRs. The use of the OLEST markers on a wide set of olive cultivars will help establishing a common fingerprint database without miscalling and binning, exploitable for several molecular investigations, representing a valuable resource for comparative genomics, evolutionary analyses and population studies.
Author Contributions
SM, LB, LR, PP, MB, RM, LN, and SP contributed substantially to the conception and design of the work; SM, LR, LN, RM, and SP contributed to plant material collection; SM, LB, and RM, performed all molecular work and genotype scoring; SM, LB, LR, RM, LN, and SP interpretation of data; SM, LB, LR, PP, MB, RM, LN, and SP drafted the text; SM, LB, LR, PP, MB, RM, LN, and SP approved the version to be published; SM, LB, LR, PP, MB, RM, LN, and SP agreed to be accountable for all aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The study has been partially performed within the Project “BeFOre – Bioresources for Oliviculture,” 2015–2019, H2020-MSCA-RISE- Marie Skłodowska-Curie Research and Innovation Staff Exchange, Grant Agreement N. 645595.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01283/full#supplementary-material
References
Abdessemed, S., Muzzalupo, I., and Benbouza, H. (2015). Assessment of genetic diversity among Algerian olive (Olea europaea L.) cultivars using SSR marker. Sci. Hortic. 192, 10–20. doi: 10.1016/j.scienta.2015.05.015
Baldoni, L., Cultrera, N., Mariotti, R., Pandolfi, S., Blanco, A., Montemurro, C. (eds). et al. (2011). Tematica I - Manuale per L’identificazione e il Riordino del Patrimonio Varietale di Olivo. Bari: Università degli Studi di Bari.
Baldoni, L., Cultrera, N. G., Mariotti, R., Ricciolini, C., Arcioni, S., Vendramin, G. G., et al. (2009). A consensus list of microsatellite markers for olive genotyping. Mol. Breed. 24, 213–231. doi: 10.1007/s11032-009-9285-8
Bartolini, G. (2009). “Banca dati mondiale: germoplasma dell’olivo (www.oleadb.eu) 1,” in Banca Dati del Germoplasma di Olivo (Olea europaea L.): Cultivar, Sinonimi, Aree di Coltivazione, Descrittori, Collezioni, Vol. 5, ed. Accademia dei Georgofili (Florence: Atti della Accademia dei Georgofili), 699–710. doi: 10.7349/OLEA_databases
Belaj, A., del Carmen Dominguez-García, M., Atienza, S. G., Urdíroz, N. M., De la Rosa, R., Satovic, Z., et al. (2012). Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet. Genomes 8, 365–378. doi: 10.1007/s11295-011-0447-6
Belaj, A., León, L., Satovic, Z., and De la Rosa, R. (2011). Variability of wild olives (Olea europaea subsp. europaea var. sylvestris) analyzed by agro-morphological traits and SSR markers. Sci. Hortic. 129, 561–569. doi: 10.1016/j.scienta.2011.04.025
Belaj, A., Muñoz-Diez, C., Baldoni, L., Porceddu, A., Barranco, D., and Satovic, Z. (2007). Genetic diversity and population structure of wild olives from the north-western Mediterranean assessed by SSR markers. Ann. Bot. 100, 449–458. doi: 10.1093/aob/mcm132
Biton, I., Doron-Faigenboim, A., Jamwal, M., Mani, Y., Eshed, R., Rosen, A., et al. (2015). Development of a large set of SNP markers for assessing phylogenetic relationships between the olive cultivars composing the Israeli olive germplasm collection. Mol. Breed. 35, 107. doi: 10.1007/s11032-015-0304-7
Bracci, T., Busconi, M., Fogher, C., and Sebastiani, L. (2011). Molecular studies in olive (Olea europaea L.): overview on DNA markers applications and recent advances in genome analysis. Plant Cell Rep. 30, 449–462. doi: 10.1007/s00299-010-0991-9
Bradai, F., Pliego-Alfaro, F., and Sánchez-Romero, C. (2016). Somaclonal variation in olive (Olea europaea L.) plants regenerated via somatic embryogenesis: influence of genotype and culture age on phenotypic stability. Sci. Hortic. 213, 208–215. doi: 10.1016/j.scienta.2016.10.031
Breton, C., Farinelli, D., Shafiq, S., Heslop-Harrison, J. S., Sedgley, M., and Bervillé, A. (2014). The self-incompatibility mating system of the olive (Olea europaea L.) functions with dominance between S-alleles. Tree Genet. Genomes 10, 1055–1067. doi: 10.1007/s11295-014-0742-0
Breton, C., Terral, J. F., Pinatel, C., Médail, F., Bonhomme, F., and Bervillé, A. (2009). The origins of the domestication of the olive tree. C. R. Biol. 332, 1059–1064. doi: 10.1016/j.crvi.2009.08.001
Cao, K., Zheng, Z., Wang, L., Liu, X., Zhu, G., Fang, W., et al. (2014). Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol. 15, 415. doi: 10.1186/s13059-014-0415-1
Carriero, F., Fontanazza, G., Cellini, F., and Giorio, G. (2002). Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor. Appl. Genet. 104, 301–307. doi: 10.1007/s001220100691
Caruso, T., Marra, F. P., and Costa, F. (2014). Genetic diversity and clonal variation within the main Sicilian olive cultivars based on morphological traits and microsatellite markers. Sci. Hortic. 180, 130–138. doi: 10.1016/j.scienta.2014.10.019
Chapuis, M. P., and Estoup, A. (2007). Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 24, 621–631. doi: 10.1093/molbev/msl191
Cipriani, G., Marrazzo, M. T., Marconi, R., Cimato, A., and Testolin, R. (2002). Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor. Appl. Genet. 104, 223–228. doi: 10.1007/s001220100685
Corrado, G., Imperato, A., La Mura, M., Perri, E., and Rao, R. (2011). Genetic diversity among olive varieties of Southern Italy and the traceability of olive oil using SSR markers. J. Hortic. Sci. Biotechnol. 86, 461. doi: 10.1080/14620316.2011.11512789
De Gennaro, B., Notarnicola, B., Roselli, L., and Tassielli, G. (2012). Innovative olive-growing models: an environmental and economic assessment. J. Clean. Prod. 28, 70–80. doi: 10.1016/j.jclepro.2011.11.004
Díez, C. M., Trujillo, I., Barrio, E., Belaj, A., Barranco, D., and Rallo, L. (2011). Centennial olive trees as a reservoir of genetic diversity. Ann. Bot. 108, 797–807. doi: 10.1093/aob/mcr194
Diez, C. M., Trujillo, I., Martinez-Urdiroz, N., Barranco, D., Rallo, L., Marfil, P., et al. (2015). Olive domestication and diversification in the Mediterranean Basin. New Phytol. 206, 436–447. doi: 10.1111/nph.13181
Duran, C., Appleby, N., Edwards, D., and Batley, J. (2009). Molecular genetic markers: discovery, applications, data storage and visualisation. Curr. Bioinform. 4, 16–27. doi: 10.2174/157489309787158198
Earl, D. A. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
El Bakkali, A., Haouane, H., Moukhli, A., Costes, E., Van Damme, P., and Khadari, B. (2013). Construction of core collections suitable for association mapping to optimize use of Mediterranean olive (Olea europaea L.) genetic resources. PLoS ONE 8:e61265. doi: 10.1371/journal.pone.0061265
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Frank, M. H., and Chitwood, D. H. (2016). Plant chimeras: the good, the bad, and the ‘Bizzaria’. Dev. Biol. 419, 41–53. doi: 10.1016/j.ydbio.2016.07.003
Fresnedo-Ramírez, J., Martinez-Garcia, P. J., Parfitt, D. E., Crisosto, C. H., and Gradziel, T. M. (2013). Heterogeneity in the entire genome for three genotypes of peach [Prunus persica (L.) Batsch] as distinguished from sequence analysis of genomic variants. BMC Genomics 14:750. doi: 10.1186/1471-2164-14-750
Haouane, H., El Bakkali, A., Moukhli, A., Tollon, C., Santoni, S., Oukabli, A., et al. (2011). Genetic structure and core collection of the World Olive Germplasm Bank of Marrakech: towards the optimised management and use of Mediterranean olive genetic resources. Genetica 139, 1083–1094. doi: 10.1007/s10709-011-9608-7
Hosseini-Mazinani, M., Mariotti, R., Torkzaban, B., Sheikh-Hassani, M., Ataei, S., Cultrera, N. G., et al. (2014). High genetic diversity detected in olives beyond the boundaries of the Mediterranean Sea. PLoS ONE 9:e93146. doi: 10.1371/journal.pone.0093146
Ilarioni, L., and Proietti, P. (2014). “Olive tree cultivars,” in The Extra-Virgin Olive Oil Handbook, Vol. 5, ed. C. Peri (Hoboken, NJ: John Wiley & Sons, Ltd), 59–67.
Ipek, M., Seker, M., Ipek, A., and Gul, M. K. (2015). Identification of molecular markers associated with fruit traits in olive and assessment of olive core collection with AFLP markers and fruit traits. Genet. Mol. Res. 14, 2762–2774. doi: 10.4238/2015.March.31.6
Jiao, W. B., Huang, D., Xing, F., Hu, Y., Deng, X. X., Xu, Q., et al. (2013). Genome-wide characterization and expression analysis of genetic variants in sweet orange. Plant J. 75, 954–964. doi: 10.1111/tpj.12254
Kaniewski, D., Van Campo, E., Boiy, T., Terral, J. F., Khadari, B., and Besnard, G. (2012). Primary domestication and early uses of the emblematic olive tree: palaeobotanical, historical and molecular evidence from the Middle East. Biol. Rev. 87, 885–899. doi: 10.1111/j.1469-185X.2012.00229.x
Kato, S., Matsumoto, A., Yoshimura, K., Katsuki, T., Iwamoto, K., Tsuda, Y., et al. (2012). Clone identification in Japanese flowering cherry (Prunus subgenus Cerasus) cultivars using nuclear SSR markers. Breed. Sci. 62:248. doi: 10.1270/jsbbs.62.248
Kaya, H. B., Cetin, O., Kaya, H., Sahin, M., Sefer, F., Kahraman, A., et al. (2013). SNP discovery by Illumina-based transcriptome sequencing of the olive and the genetic characterization of Turkish olive genotypes revealed by AFLP, SSR and SNP markers. PLoS ONE 8:e73674. doi: 10.1371/journal.pone.0073674
Klepo, T., De la Rosa, R., Satovic, Z., León, L., and Belaj, A. (2013). Utility of wild germplasm in olive breeding. Sci. Hortic. 152, 92–101. doi: 10.1016/j.scienta.2012.12.010
Koehmstedt, A. M., Aradhya, M. K., Soleri, D., Smith, J. L., and Polito, V. S. (2011). Molecular characterization of genetic diversity, structure, and differentiation in the olive (Olea europaea L.) germplasm collection of the United States Department of Agriculture. Genet. Res. Crop Evol. 58, 519–531. doi: 10.1007/s10722-010-9595-z
Larbi, A., Vázquez, S., El-Jendoubi, H., Msallem, M., Abadía, J., Abadía, A., et al. (2015). Canopy light heterogeneity drives leaf anatomical, eco-physiological, and photosynthetic changes in olive trees grown in a high-density plantation. Photosynth. Res. 123, 141–155. doi: 10.1007/s11120-014-0052-2
Laroussi-Mezghani, S., Le Dréau, Y., Molinet, J., Hammami, M., Grati-Kamoun, N., and Artaud, J. (2016). Biodiversity of Tunisian virgin olive oils: varietal origin classification according to their minor compounds. Eur. Food Res. Technol. 242, 1087–1099. doi: 10.1007/s00217-015-2613-9
Las Casas, G., Scollo, F., Distefano, G., Continella, A., Gentile, A., and La Malfa, S. (2014). Molecular characterization of olive (Olea europaea L.) Sicilian cultivars using SSR markers. Biochem. Syst. Ecol. 57, 15–19. doi: 10.1016/j.bse.2014.07.010
Lazović, B., Adakalić, M., Pucci, C., Perović, T., Bandelj, D., and Belaj, A. (2016). Characterizing ancient and local olive germplasm from Montenegro. Sci. Hortic. 209, 117–123. doi: 10.1016/j.scienta.2016.06.022
Lynch, M., and Ritland, K. (1999). Estimation of pairwise relatedness with molecular markers. Genetics 152, 1753–1766.
Marchese, A., Marra, F. P., Caruso, T., Mhelembe, K., Costa, F., Fretto, S., et al. (2016). The first high-density sequence characterized SNP-based linkage map of olive (Olea europaea L. subsp. europaea) developed using genotyping by sequencing. Aust. J. Crop Sci. 10, 857–863. doi: 10.21475/ajcs.2016.10.06.p7520
Mariotti, R., Cultrera, N. G. M., Mousavi, S., Baglivo, F., Rossi, M., Albertini, E., et al. (2016). Development, evaluation, and validation of new EST-SSR markers in olive (Olea europaea L.). Tree Genet. Genomes 12, 120. doi: 10.1007/s11295-016-1077-9
Marra, F. P., Caruso, T., Costa, F., Di Vaio, C., Mafrica, R., and Marchese, A. (2013). Genetic relationships, structure and parentage simulation among the olive tree (Olea europaea L. subsp. europaea) cultivated in Southern Italy revealed by SSR markers. Tree Genet. Genomes 9, 961–973. doi: 10.1007/s11295-013-0609-9
Marshall, T. C., Slate, J., Kruuk, L. E. B., and Pemberton, J. M. (1998). Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 7, 639–655. doi: 10.1046/j.1365-294x.1998.00374.x
Mazzitelli, O., Calleja, A., Sardella, D., Farrugia, C., and Zammit-Mangion, M. (2015). Analysis of the molecular diversity of Olea europaea in the Mediterranean Island of Malta. Genet. Res. Crop Evol. 62, 1021–1027. doi: 10.1007/s10722-014-0205-3
Moriondo, M., Trombi, G., Ferrise, R., Brandani, G., Dibari, C., Ammann, C. M., et al. (2013). Olive trees as bioindicators of climate evolution in the Mediterranean Basin. Glob. Ecol. Biogeogr. 22, 818–833. doi: 10.1111/geb.12061
Mousavi, S., Mariotti, R., Bagnoli, F., Costantini, L., Cultrera, N. G. M., Arzani, K., et al. (2017). The eastern part of the Fertile Crescent concealed an unexpected route of olive (Olea europaea L.) differentiation. Ann. Bot. 119, 1305–1318. doi: 10.1093/aob/mcx027
Mousavi, S., Mazinani, M. H., Arzani, K., Ydollahi, A., Pandolfi, S., Baldoni, L., et al. (2014). Molecular and morphological characterization of Golestan (Iran) olive ecotypes provides evidence for the presence of promising genotypes. Genet. Res. Crop Evol. 61, 775–785. doi: 10.1007/s10722-013-0071-4
Muzzalupo, I., Vendramin, G. G., and Chiappetta, A. (2014). Genetic biodiversity of Italian olives (Olea europaea) germplasm analyzed by SSR markers. Sci. World J. 2014:296590. doi: 10.1155/2014/296590
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Portarena, S., Farinelli, D., Lauteri, M., Famiani, F., Esti, M., and Brugnoli, E. (2015). Stable isotope and fatty acid compositions of monovarietal olive oils: implications of ripening stage and climate effects as determinants in traceability studies. Food Control 57, 129–135. doi: 10.1016/j.foodcont.2015.03.052
Potts, S. M., Han, Y., Khan, M. A., Kushad, M. M., Rayburn, A. L., and Korban, S. S. (2012). Genetic diversity and characterization of a core collection of Malus germplasm using simple sequence repeats (SSRs). Plant Mol. Biol. Rep. 30, 827–837. doi: 10.1007/s11105-011-0399-x
Pritchard, J., Wen, X., and Falush, D. (2009). Documentation for Structure Software: Version 2.3. Available at: http://pritch.bsd.uchicago.edu/structure.html
Proietti, S., Sdringola, P., Desideri, U., Zepparelli, F., Brunori, A., Ilarioni, L., et al. (2014). Carbon footprint of an olive tree grove. Appl. Energy 127, 115–124. doi: 10.1016/j.apenergy.2014.04.019
Sarri, V., Baldoni, L., Porceddu, A., Cultrera, N. G. M., Contento, A., Frediani, M., et al. (2006). Microsatellite markers are powerful tools for discriminating among olive cultivars and assigning them to geographically defined populations. Genome 49, 1606–1615. doi: 10.1139/g06-126
Sefc, K. M., Lopes, M. S., Mendonça, D., Santos, M. R. D., Machado, L. M., and Machado, A. D. C. (2000). Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol. Ecol. 9, 1171–1173. doi: 10.1046/j.1365-294x.2000.00954.x
Soleri, D., Koehmstedt, A., Aradhya, M. K., Polito, V., and Pinney, K. (2010). Comparing the historic olive trees (Olea europaea L.) of Santa Cruz Island with contemporaneous trees in the Santa Barbara, CA area: a case study of diversity and structure in an introduced agricultural species conserved in situ. Genet. Resour. Crop Evol. 57, 973–984. doi: 10.1007/s10722-010-9537-9
Tanasijevic, L., Todorovic, M., Pereira, L. S., Pizzigalli, C., and Lionello, P. (2014). Impacts of climate change on olive crop evapotranspiration and irrigation requirements in the Mediterranean region. Agric. Water Manage. 144, 54–68. doi: 10.1016/j.agwat.2014.05.019
Torkzaban, B., Kayvanjoo, A. H., Ardalan, A., Mousavi, S., Mariotti, R., Baldoni, L., et al. (2015). Machine learning based classification of microsatellite variation: an effective approach for phylogeographic characterization of olive populations. PLoS ONE 10:e0143465. doi: 10.1371/journal.pone.0143465
Trujillo, I., Ojeda, M. A., Urdiroz, N. M., Potter, D., Barranco, D., Rallo, L., et al. (2014). Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) using SSR and morphological markers. Tree Genet. Genomes 10, 141–155. doi: 10.1007/s11295-013-0671-3
Urrestarazu, J., Miranda, C., Santesteban, L. G., and Royo, J. B. (2012). Genetic diversity and structure of local apple cultivars from Northeastern Spain assessed by microsatellite markers. Tree Genet. Genomes 8, 1163–1180. doi: 10.1007/s11295-012-0502-y
Vezzulli, S., Leonardelli, L., Malossini, U., Stefanini, M., Velasco, R., and Moser, C. (2012). Pinot blanc and Pinot gris arose as independent somatic mutations of Pinot noir. J. Exp. Bot. 63, 6359–6369. doi: 10.1093/jxb/ers290
Waits, L. P., Luikart, G., and Taberlet, P. (2001). Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol. Ecol. 10, 249–256. doi: 10.1046/j.1365-294X.2001.01185.x
Wu, G. A., Prochnik, S., Jenkins, J., Salse, J., Hellsten, U., Murat, F., et al. (2014). Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662. doi: 10.1038/nbt.2906
Xanthopoulou, A., Ganopoulos, I., Koubouris, G., Tsaftaris, A., Sergendani, C., Kalivas, A., et al. (2014). Microsatellite high-resolution melting (SSR-HRM) analysis for genotyping and molecular characterization of an Olea europaea germplasm collection. Plant Genet. Resour. 12, 273–277. doi: 10.1017/S147926211400001X
Yang, J., Dai, P., Zhou, T., Huang, Z., Feng, L., Su, H., et al. (2013). Genetic diversity and structure of wintersweet (Chimonanthus praecox) revealed by EST-SSR markers. Sci. Hortic. 150, 1–10. doi: 10.1016/j.scienta.2012.11.004
Zelasco, S., Salimonti, A., Baldoni, L., Mariotti, R., Preece, J. E., Aradhya, M., et al. (2012). Efficiency of SSR markers for exploring olive germplasm diversity through a genetic comparison between the USDA-NCGR and the CRA-OLI olive collections. Acta Hortic. 1057, 585–592. doi: 10.17660/ActaHortic.2014.1057.75
Keywords: genetic variability, ex situ conservation, germplasm management, genotyping, EST-SSR, Olea europaea L.
Citation: Mousavi S, Mariotti R, Regni L, Nasini L, Bufacchi M, Pandolfi S, Baldoni L and Proietti P (2017) The First Molecular Identification of an Olive Collection Applying Standard Simple Sequence Repeats and Novel Expressed Sequence Tag Markers. Front. Plant Sci. 8:1283. doi: 10.3389/fpls.2017.01283
Received: 21 March 2017; Accepted: 07 July 2017;
Published: 19 July 2017.
Edited by:
Mariela Torres, Instituto Nacional de Tecnología Agropecuaria (INTA), ArgentinaReviewed by:
Rosario Muleo, Università degli Studi della Tuscia, ItalyRaul De La Rosa, Andalusian Institute of Agricultural and Fisheries Research and Training, Spain
Copyright © 2017 Mousavi, Mariotti, Regni, Nasini, Bufacchi, Pandolfi, Baldoni and Proietti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Primo Proietti, primo.proietti@unipg.it
 Soraya Mousavi
Soraya Mousavi Roberto Mariotti2
Roberto Mariotti2 Luca Regni
Luca Regni Luigi Nasini
Luigi Nasini Luciana Baldoni
Luciana Baldoni Primo Proietti
Primo Proietti