- 1Department of Pharmacy, Changzheng Hospital, Second Military Medical University, Shanghai, China
- 2Department of Pharmaceutical Botany, School of Pharmacy, Second Military Medical University, Shanghai, China
- 3Development and Utilization Key Laboratory of Northeast Plant Materials, School of Traditional Chinese Materia Medica, Shenyang Pharmaceutical University, Shenyang, China
Lignans, such as lariciresinol and its derivatives, have been identified as effective antiviral ingredients in Isatis indigotica. Evidence suggests that the APETALA2/ethylene response factor (AP2/ERF) family might be related to the biosynthesis of lignans in I. indigotica. However, the special role played by the AP2/ERF family in the metabolism and its underlying putative mechanism still need to be elucidated. One novel AP2/ERF gene, named Ii049, was isolated and characterized from I. indigotica in this study. The quantitative real-time PCR analysis revealed that Ii049 was expressed highest in the root and responded to methyl jasmonate, salicylic acid (SA) and abscisic acid treatments to various degrees. Subcellular localization analysis indicated that Ii049 protein was localized in the nucleus. Knocking-down the expression of Ii049 caused a remarkable reduction of lignan/lignin contents and transcript levels of genes involved in the lignan/lignin biosynthetic pathway. Ii049 bound to the coupled element 1, RAV1AAT and CRTAREHVCBF2 motifs of genes IiPAL and IiCCR, the key structural genes in the lignan/lignin pathway. Furthermore, Ii049 was also essential for SA biosynthesis, and SA induced lignan accumulation in I. indigotica. Notably, the transgenic I. indigotica hairy roots overexpressing Ii049 showed high expression levels of lignan/lignin biosynthetic genes and SA content, resulting in significant accumulation of lignan/lignin. The best-engineered line (OVX049-10) produced 425.60 μg·g−1 lariciresinol, an 8.3-fold increase compared with the wild type production. This study revealed the function of Ii049 in regulating lignan/lignin biosynthesis, which had the potential to increase the content of valuable lignan/lignin in economically significant medicinal plants.
Introduction
Isatis indigotica Fort. (I. indigotica) is a well -known medicinal herb in the family Cruciferae. Its dry root (Ban-Lan-Gen) is frequently used as an anti-inflammatory and antiviral drug for treating hepatitis, influenza, and various kinds of inflammation (Ho and Chang, 2002; Kumagai et al., 2016). Lignans (e.g., pinoresinol, lariciresinol, and their derivatives) in I. indigotica have been identified as biologically active ingredients that can significantly inhibit different subtypes of human and avian influenza viruses (Li, 2003; Yang et al., 2013; Li et al., 2015). Additionally, as a typical representative of lignans, lariciresinol also plays an important role in treating cardiovascular diseases and some types of cancer (Adlercreutz et al., 1992; Saarinen et al., 2008). Despite its obvious benefits to human health, the widespread use and availability of lignans are limited by the low concentration (<1‰) in Ban-Lan-Gen (Li, 2003). In this case, improving lignan contents in I. indigotica has become an urgent problem to solve.
As a subgroup of phenolic compounds, lignans are derived from phenylalanine through the phenylpropanoid pathway (Figure 1; Nakatsubo et al., 2008; Chen et al., 2013; Nguyen et al., 2016). Genes encoding the enzyme cascade of the phenylpropanoid pathway have been widely characterized in I. indigotica, including phenylalanine ammonia-lyase (PAL) (Lu et al., 2006), cinnamate-4-hydroxylase (C4H), coumaroyl-CoAligase (4CL) (Zhang L. et al., 2016), coumarate 3-hydroxylase (C3H) (Chen et al., 2015), cinnamoyl-CoA reductase (CCR) (Hu et al., 2011), cinnamyl alcohol dehydrogenase (CAD), dirigent protein (DIR) (Li et al., 2014), and pinoresinol reductase (PLR) (Xiao et al., 2015). An understanding of lignan biosynthesis makes it possible to increase the production of such health-promoting compounds by metabolic engineering. In a previous study, additional expression of IiPLR led to 6.3-fold of lariciresinol accumulation in I. indigotica hairy roots (Xiao et al., 2015). The overexpression of IiC3H resulted in an engineered hairy root producing lariciresinol to an average 1.87-fold of that in the control (Chen et al., 2015). Plant metabolic pathways are known to be complex and usually involve multiple enzymes; thus, increasing the target metabolite production by overexpressing one or two enzymes in the host plant is often not highly efficient (Yu et al., 2012). Transcription factors can channel the metabolic flux by simultaneously regulating the transcription of related biosynthetic genes and manipulating the accumulation of secondary metabolism more obviously (Zhao et al., 2015). However, transcription factors involved in the lignan pathway in I. indigotica have not been reported; transcriptional regulation mechanisms of lignan biosynthetic pathway remain unclear.
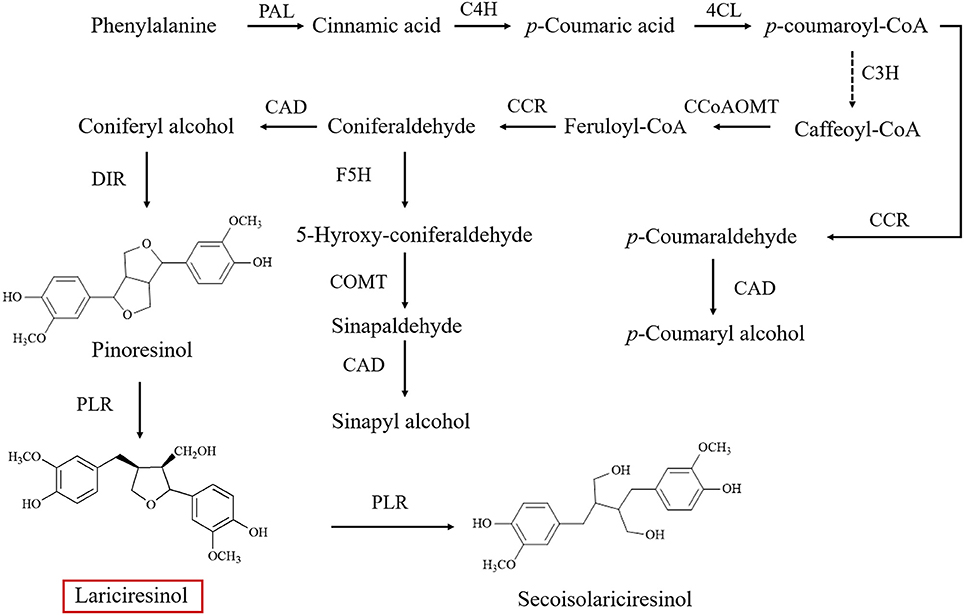
Figure 1. Biosynthetic pathway of lignins in Isatis indigotica (PAL, phenylalanine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate coenzyme A ligase; C3H, coumarate 3-hydroxylase; CCoAOMT, caffeoyl CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; F5H, ferulate 5-hydroxylase; COMT 1, caffeic acid O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase; DIR, dirigent protein; PLR, pinoresinol reductase).
The APETALA2/ethylene response factor (AP2/ERF) transcriptional factors are characterized by possessing one or two DNA-binding AP2 domains, which possess ~60 conserved amino acid residues (Sakuma et al., 2002; Nakano et al., 2006). This family plays various roles in plant development, stress responses and secondary metabolism in many plant species (Licausi et al., 2013). In recent years, AP2/ERF transcription factors have been reported to play an important role in lignin biosynthesis. For example, Eriobotrya japonica EjAP2-1 was a novel regulator of fruit lignification via interaction with EjMYB transcription factor (Zeng et al., 2015). Other study reported that the overexpression of Arabidopsis SHINE (an AP2/ERF gene) in rice showed a 45% reduction in lignin content compared with the wild type (WT) production (Ambavaram et al., 2011). These results suggested that AP2/ERF transcription factors had potential functions in lignin regulation. Whether AP2/ERF transcription factors could regulate the biosynthesis of lignan in I. indigotica would be interesting to explore, as lignan shares the same monolignols with lignin (Hano et al., 2006).
In a previous study, methyl jasmonate (MeJA) elicited I. indigotica hairy roots were used as resources to identify candidate lignan related AP2/ERF genes. Based on the results of transcriptome and metabolome, four AP2/ERF genes (Ii007, Ii049, Ii050, and Ii080, which were designated through query of a previously established I. indigotica transcriptome inventory by Chen) were identified to be significantly correlated with the biosynthesis of pharmaceutically valuable lignans in I. indigotica (Chen et al., 2015). Among these four candidates, Ii049, whose homologous gene in Arabidopsis thaliana (At4g13040) was reported to be a positive regulator for the accumulation of salicylic acid (SA), was an important signaling molecule in the secondary metabolite biosynthesis (Giri et al., 2014). Therefore, the present study, aimed to explore the function of Ii049 in the regulation of lignan biosynthesis, and also investigate the relationship among Ii049 expression, SA accumulation and lignan biosynthesis.
Materials and Methods
Plant Material and Treatments
I. indigotica plants were cultivated in a greenhouse with natural light at 20–25°C in the Second Military Medical University (SMMU), Shanghai, China. Species verification was performed by Professor Hanming Zhang of the School of Pharmacy, SMMU. Seeds of I. indigotica were pretreated with 75% alcohol for 5 min, followed by treatment with 0.1% HgCl2 for 10 min to obtain sterilized plants. After rinsing five times with sterile distilled water, the sterilized seeds were cultured on the Murashige and Skoog (MS) medium solidified with 0.6% agar for germination. The sterilized plants were grown in a controlled room at 25°C with a 16/8-h light/dark photoperiod cycle and a relative humidity of ~70%. Two-month-old I. indigotica plants, with 7–8 leaves, were used for genetic transformation and stress treatments. For stress treatment, plants in sealed boxes were sprayed with 0.1 mM MeJA, SA, and ABA, respectively, until the solution dropped off from the plants. The entire plants were harvested at 0-, 2-, 4-, 6-, 8-, 12-, and 24-h post-treatment. Three independent biological replicates for each group were performed. MeJA, SA, and ABA were purchased from Sigma-Aldrich (USA). All samples were immediately frozen in liquid nitrogen and stored at −80°C until required for quantitative real-time polymerase chain reaction (qRT-PCR) analysis.
DNA and RNA Preparation
Genomic DNA was isolated from 2-month-old I. indigotica sterilized plants using the cetyltrimethyl ammonium bromide (CTAB) method (Doyle, 1990). The total RNAs of I. indigotica plants were extracted using the RNAiso Plus (Cat. #9108, TaKaRa, Japan). The DNase I (Cat. #GD201-01, Tiangen Biotech Co., China) was used to remove all DNAs from the RNA samples according to the protocol suggested by the manufacturer. The quality and concentration of DNA and RNA samples were examined by ethidium bromide-stained agarose gel electrophoresis and spectrophotometer analysis using a NanoDrop 2000C Spectrophotometer (Thermo Scientific, USA).
Gene/Promoter Isolation and Analysis
The Ii049 gene was isolated based on the sequencing result from transcription profiling of I. indigotica (Chen et al., 2015) with gene-specific primers Ii049-F and Ii049-R (Supplementary Table S1). The full-length coding region of Ii049 was obtained by PCR using the Pfu DNA Polymerase (Cat. #AP221-12, TransGen Biotech Co., China) and the first-strand cDNA as a template. PCR was performed under the following condition: denaturation at 94°C for 1 min, followed by 35 cycles, each one consisting of 94°C for 20 s, 45°C for 20 s, and 72°C for 1 min, followed by a final extension at 72°C for 5 min. Genomic DNA sequence of Ii049 was obtained by the same reaction system using genomic DNA as the template and the extension time at 72°C in the amplification cycles was prolonged to 3 min. The amplified PCR products were purified and cloned into the PMD18-T vector and then sequenced.
Searching for open reading frame (ORF) and prediction of nucleotide translation products were performed using the ORF Finder tool (https://www.ncbi.nlm.nih.gov/orffinder/). The molecular weight (MW) and theoretical isoelectric point of Ii049 were predicted using the Vector NTI Advance 11 software. An analysis of protein structure was performed using the Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/). Multiple alignment analysis was performed using the ClustalX software (version 1.80). A phylogenetic tree of Ii049 and various heterologous AP2/ERF members was constructed using the MEGA5.0 software by the neighbor-joining method (1,000 bootstrap replicates). Gene sequences used for multiple alignment analysis and phylogenetic tree are listed in Supplementary Table S3.
The 5′-upstream region of IiPAL was isolated using the Genome Walking Kit (TaKaRa, Japan) according to the manufacturer's instruction with primers IiPAL-SP1-R and IiPAL-SP2-R. The promoter of IiCCR was cloned based on the sequence obtained from Hu with primers IiCCR-F and IiCCR-R (Hu et al., 2011). All sequences are listed in Supplementary Table S1. PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and NSITE-PL(http://linux1.softberry.com/berry.phtml?topic=nsitep&group=help&subgroup=promoter) were used to predict the characteristics of the promoters (Lescot et al., 2002; Solovyev et al., 2010).
Quantitative Real-Time PCR (qRT-PCR)
The qRT-PCR analysis was performed to determine the transcript abundance of Ii049. High quality total RNA (1 μg) was used to prepare the first strand cDNA using the TransScript First-Strand cDNA Synthesis SuperMix Kit (Cat. #AT301-03, TransGen Biotech Co., China) following the manufacturer's protocol. QRT-PCR was performed on a Thermal Cycler Dice Real Time PCR machine (TaKaRa, Japan) using the SYBR-Green PCR Master Mix Kit (Cat. #RR820, TaKaRa, Japan) according to the manufacturer's instruction. The expression levels were normalized with the actin control gene using the 2−ΔΔCt method (Vandesompele et al., 2002; Udvardi et al., 2008). All the primers for qRT-PCR are listed in Supplementary Table S2.
Subcellular Localization in Protoplast
The encoding region without the stop codon of Ii049 was cloned into the pCAMBIA1301-GFP vector by Nco I and Spe I sites to generate Ii049-GFP to determine the subcellular localization of Ii049. The primers used for subcloning are listed in Supplementary Table S1. The sequence and fusion of GFP under the control of the cauliflower mosaic virus (CaMV) 35S promoter were confirmed by DNA sequencing. Plasmid Ii049-GFP was transiently expressed into rice protoplasts using the polyethylene glycol-mediated transformation for the observation of subcellular localization. Transfected protoplasts were incubated overnight at room temperature and the transient expressions were visualized using a confocal laser scanning microscope (Nikon, Japan) as described by Tan et al. (2015).
Construction of Ii049 RNAi Vector and I. indigotica transformation
The RNA interference (RNAi) approach was used to knock down Ii049 to determine the essential role of Ii049 in the regulation of lignan biosynthesis in I. indigotica. A less conserved region at the C-terminus of Ii049 (371 bp) was used to interfere with the expression of Ii049. Gene specific primers Ii049-RNAi-F and Ii049-RNAi-R (listed in Supplementary Table S1) were used to amplify the fragment with two restriction sites at both ends. After sequence confirmation, the PCR product was inserted into the modified pC1300-pHANNIBAL vector (Tan et al., 2015) upstream and downstream of the pyruvate orthophosphate dikinase (PDK) intron with opposite orientations to generate vector pIi049-RNAi (see Supplementary Figure S3).
Generation of Transgenic Hairy Root
Hairy root cultures of I. indigotica were initiated by infecting the 2-month-old sterilized leaf-disk with Agrobacterium tumefaciens strain C58C1 as described in a previous study (Chen et al., 2015; Xiao et al., 2015). Plasmids pIi049-RNAi together with pC1300-pHANNIBAL (Control check 1, CK1) were separately introduced into A. tumefaciens strain C58C1 for plant transformation. The infected leaf-disk was placed on the surface of 1/2 MS solid medium and supplemented with 500 mg·l−1 cefotaxime. When the transgenic hairy roots were 2–4 cm, single roots were isolated from leaves and cultivated on 1/2 MS solid medium containing 300 mg·l−1 cefotaxime. The hairy roots were transferred to fresh 1/2 MS solid medium with a gradual decrease in cefotaxime concentrations (300, 100, and 0 mg·l−1) every 2 weeks. The hairy root lines generated were screened using 10 mg·L−1 hygromycin for antibiotic-resistant roots. pIi049-RNAi was named as Ri049 lines. The hairy root lines generated through transformation with the blank C58C1 were named as WT line. Approximately 100 mg transgene hairy roots that had the same growth rates as WT lines were cultured in a 250 ml shake flask containing 200 ml of fresh liquid hormone-free half-strength MS medium at 110 rpm and 25°C in the dark. Clonal hairy root cultures were routinely subcultured every 9 days and harvested at 45 days. The fresh weights of root tissues were recorded at days 9, 18, 27, 36, and 45 for studying the biomass growth rate. After culturing for 45 days, the hairy roots were harvested for DNA and RNA extraction, phloroglucinol-HCl staining and metabolite analysis.
Analyses of Ii049-RNAi Hairy Roots
Genomic DNA was extracted from 45 days transgene hairy roots using the CTAB method. Primers JDPDK-F and JDPDK-R based on the sequence of vector pC1300-pHANNIBAL were used to detect the presence of inserted Ii049 fragment in Ri049 lines. The hygromycin resistance gene (hpt) and hairy root rolb gene were also checked using primers hpt-F/hpt-R and rolb-F/rolb-R. All primers are listed in Supplementary Table S1.
The expression levels of genes involved in lignan/lignin biosynthesis were analyzed using qRT-PCR in all positive lines. Primers used for qRT-PCR are listed in Supplementary Table S2. At least three independent control lines were tested in these experiments, and the mean value was shown as the control. For phloroglucinol-HCl staining, fresh hairy roots were immediately immersed for 2 min in 5% phloroglucinol (dissolved in 100% ethanol) and then incubated with concentrated HCl for 10 min.
Extraction and Determination of Lignin, Lignan, and SA
The extraction and determination of Klason lignin (acid-insoluble) and acid-soluble lignin were performed according to the protocol of Ma (2007). Lignan contents of transgenic I. indigotica hairy roots were measured as previously described with slight modification (Xiao et al., 2015). Hairy roots were collected and dried at 45°C until a constant dry weight (DW) was obtained. Dry hairy roots were ground into a fine powder (2-mm mesh). Then, 100 mg of dry power was extracted with 5 mL of methanol under sonication for 30 min. The supernatant was taken and the power was extracted again with 5 ml methanol. The supernatant was combined and further centrifuged at 4,000 rpm for 5 min at room temperature. The extract was diluted with methanol to 10 ml total volume. The extract (2 ml) of hairy root was evaporated to dryness and the residue was dissolved in 1 mL of 15% acetonitrile. The final solution was filtered through a 0.22-μm organic membrane filter prior to analysis.
HPLC/MS/MS was performed on an Agilent 1200 series coupled with an Agilent 6410 Triple Quadrupole Mass Spectrometer and an electrospray ionization source (Agilent, USA). The mobile phase was acetonitrile (Cat.1.00030.4008, # HPLC grade, Merck, Germany, eluent A) and 5 mM ammonium acetate solution (Cat. #218365000, HPLC grade, Merck, Germany, eluent B). The solvent gradient was used as follows: 0–4.00 min, 14% A; 4.00–4.50 min, 50% A, 4.50–8.50, 85% A. The negative-ion mode and multiple reaction monitoring mode for MS analyses were selected, under the following conditions: a flow rate of 0.3 mL·min−1, an Agilent ZORBAX SB-C18 (3.5 μm, 100 × 2.1 mm i.d.) at a column temperature of 35°C and injection with 5 μL samples. Characteristic m/z ions were 357→151 for pinoresinol, 359→329 for lariciresinol, 361→165 for secoisolariciresinol and 137→93 for SA. The HPLC/MS/MS data was acquired and processed using the MassHunter Workstation software provided by the manufacturer. All standards were purchased from Sigma-Aldrich (USA).
Electrophoretic Mobility Shift Assay
The full length coding sequence of Ii049 was amplified with the primers Ii049-pET-F and Ii049-pET-R (Supplementary Table S1), and inserted into the expression vector pET32a (Novagen, Denmark) between the Nco I and Hind III sites for the generation of Ii049 recombinant protein. The resulting Ii049 recombinant protein was expressed in Escherichia coli strain BL21(DE3) (Novagen, Denmark). E. coli cells harboring Ii049 were broken by an ultrasonic disintegrator (Cat. #BILON-650Y, Shanghai Bilon Instrument Co. Ltd., China) in a crushed-ice bath at an output level of 5 and a 50 duty cycle for 1 min with 0.5 s interval every second. After centrifugation (19,000 × g, 20 min, at 4°CC), the supernatants were applied to a 5 mL nickel column (Bio-Scale Mini Profinity IMAC Cartridges) for Ii049 recombinant protein purification according to the manufacturer's recommendation (Cat. #732-4612, Bio-Rad Laboratories, USA). Protein integrity was checked by western blotting using primary anti-His-Tag antibody (Cat. #12698, Cell Signaling Technology, USA) and horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Cat. #7074, Cell Signaling Technology, USA). The triple tandem copies of the coupled element 1 (CE1) motif (5′-TTCCACCGCCGTTCCACCGCCGTTCCACCGCCG-3′), RAV1AAT (RAA) motif (5′-AGCAACATATAAGCAACATATAAGCAACATATA-3′) and CRTAREHVCBF2CBF2 (CBF2) motif (5′-GCCGTCGATGTTGCCGTCGATGTTGCCGTCGATGTT-3′) were labeled with biotin at the 3' end. The specificity of binding was examined by competition with the unlabeled probes (10×, 50×, 100×). Electrophoretic mobility shift assay (EMSA) was carried out according to the protocol included in the EMSA/Gel-Shift Kit (Cat. #GS009, Beyotime Biotechnology, China).
Yeast One-Hybrid Assay
According to the results of EMSA, yeast one-hybrid assay (Y1H) was performed to verify physical binding of Ii049 with target promoters. The coding sequence of Ii049 was amplified by PCR and inserted into the pGADT7 vector to form the prey using primers Ii049-Y1H-F and Ii049-Y1H-R (Supplementary Table S1). The triple tandem copies of CE1, RAA and CBF2 were inserted into pHIS2 between EcoR I and Sal I as baits. The probe sequences were the same as those used in EMSA. The prey and baits were co-transformed into yeast strain AH109. The interactions were examined on the SD/-Leu-Trp-His containing 100 mM 3-amino-1,2,4-triazole (3-AT). Yeast cells carrying the blank pGADT7 and pHIS2 plasmids were used as negative controls. Yeast cells were incubated at 28°C and observed after 3 days.
Measurement of Lignan Contents after Exogenous SA Treatment
The WT hairy roots in the exponential phase (~18 days) were prepared for SA induction. Then, 100 mM SA was added to 200 mL of 1/2 MS liquid medium while untreated hairy roots were used as a control. Hairy roots were harvested at 0, 1, 3, 5, 7, 10, and 12 h after treatment. All samples were immediately frozen in liquid nitrogen and kept at −80°C until RNA extraction and compound analysis by HPLC/MS/MS. All treatments were performed in triplicate.
Generation and Analyses of Ii049 Overexpression Hairy Roots
A full-length cDNA coding sequence of Ii049 was cloned into the binary vector PHB-flag using BamH I/Spe I restriction sites under the control of two CaMV 35S promoter (for primers Ii049-OVX-F and Ii049-OVX-R, see Supplementary Table S1) to construct the Ii049 overexpression vector (pIi049-OVX, see Supplementary Figure S3). The construct was transferred into Agrobacterium C58C1 and introduced into leaf explant of I. indigotica. The plants that carried the PHB-flag vector were used as control (CK2). The analysis of hairy roots was performed following the aforementioned methods.
Results
Isolation and Sequence Analysis of Ii049
The full length genomic sequence and cDNA sequence of Ii049 were isolated from I. indigotica by RT-PCR. The full-length Ii049 was 1,352 bp in size and consisted of five introns and six extrons (Supplementary Figure S1). The ORF of Ii049 was 684 bp, encoding a putative AP2/ERF protein of 227 amino acids. The predicted protein had a calculated MW of 27.37 kDa with an isoelectric point of 9.37. Amino acid analysis indicated that the putative protein contained a conserved 58-residue AP2 domain at the 121st-178th amino acids. As MRG and HLG in the AP2 domain, the gene was deemed to be the member of Soloist subfamily (Zhuang et al., 2009), a subfamily of AP2/ERF transcription factors.
Sequence alignment was used to further analyze the structural relationship of Ii049 and other Soloist-like proteins from various plant species. The AP2 domain was highly conserved as it shared an 84–93% amino acid identity with other members of Soloist-like proteins from different species (Figure 2A). Moreover, the phylogenetic analysis showed that Ii049 was structurally closely related to AT4g13040 in Arabidopsis (Figure 2B). As both I. indigotica and Arabidopsis belonged to Cruciferae family, Ii049 and AT4g13040 might have similar biological functions.
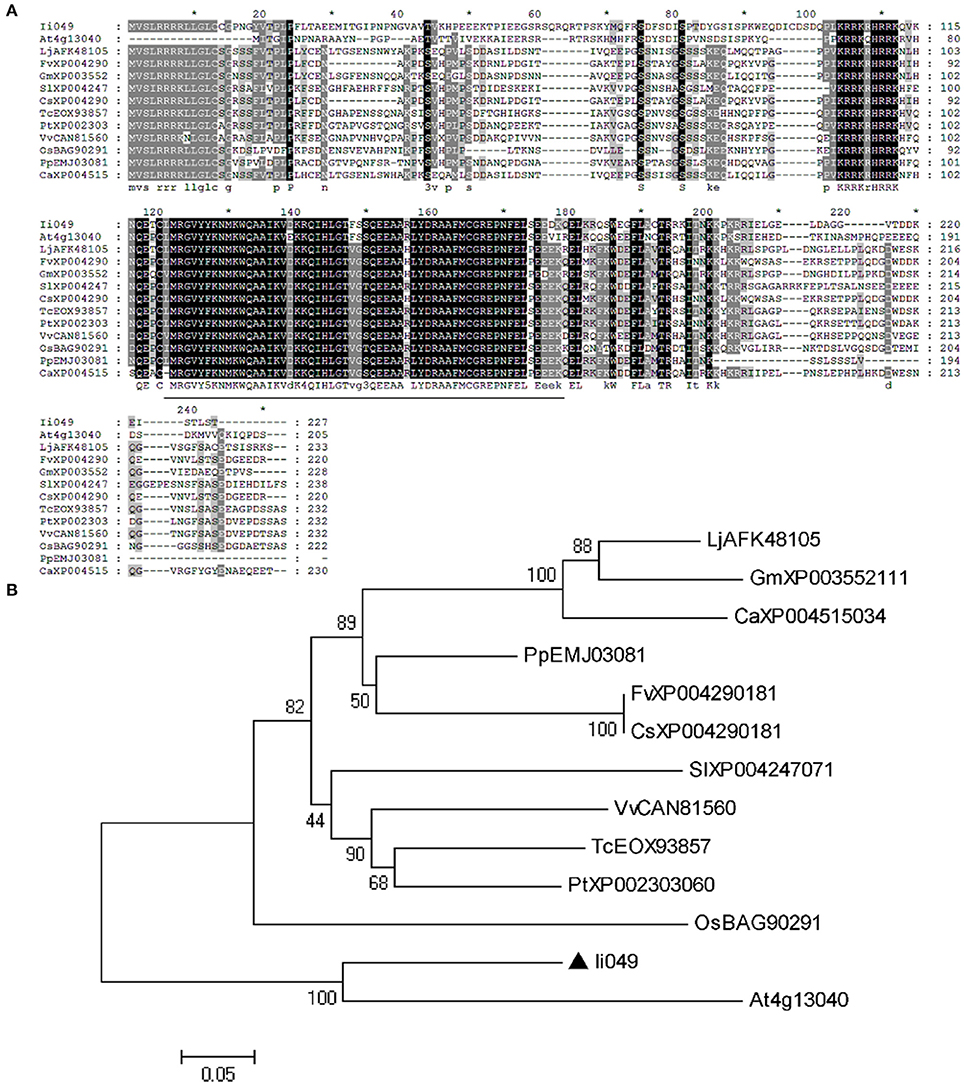
Figure 2. Sequence comparison between Ii049 and related AP2/ERF family protein. (A) Amino acid alignment of Ii049 with several Soloist-like proteins in different plant species, the AP2 domain has been underlined. (B) Phylogenetic tree of Ii049 and other plant Soloist-like proteins using MEGA 5.0 software based on the neighbor-joining method. Gene names with their accession numbers are listed in Supplementary Table S2.
Expression and Induction Patterns of Ii049
The total RNAs were isolated from roots, stems and leaves of 2-month-old sterilized I. indigotica plants to analyze the expression pattern of Ii049. The transcript of Ii049 could be detected in the roots, stems, and leaves. The maximum expression was found in the roots followed by stems, and the least expression was found in the leaves (Figure 3A). Next, the transcripts of Ii049 were detected in response to MeJA, SA and ABA by qRT-PCR. The result showed that the transcript level of Ii049 was regulated by MeJA, SA and ABA with significant variations depending on the time and/or the phytohormones (Figures 3B–D). After treatment with MeJA, the expression of Ii049 slightly declined at 2 h, then increased quickly. The high expression of Ii049 presented during the 4- to 6-h period (Figure 3B). In response to SA, Ii049 induction peaked at 6 h, with an increase of ~3.11-fold, and subsequently declined (Figure 3C). For the ABA treatment, the transcript level of Ii049 was reached a maximum at 2 h (1.85-fold) followed by a decline and was up-regulated again after 12 h treatment (Figure 3D).
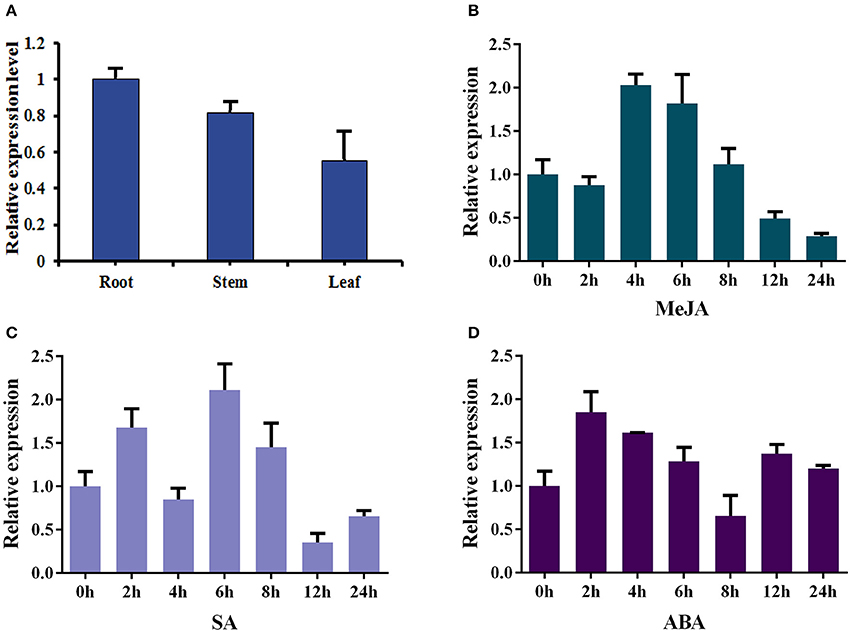
Figure 3. Expression profiles of Ii049 in different I. indigotica tissues (A) and under the induction of phytohormones [MeJA (B), SA (C), and ABA (D)].
Subcellular Localization of Ii049
The subcellular localization of Ii049 was examined in rice protoplasts using polyethylene glycol-mediated transformation to provide further evidence for the potential role of Ii049 in transcriptional regulation. The empty vector construct served as control. As shown in Figures 4A–D, the Ii049-GFP fusion protein was localized exclusively to the nucleus of the rice cells. In contrast, the free GFP was found in the cytoplasm (Figures 4E–H). Just as expected for a transcription factor, Ii049 was localized in the nucleus.
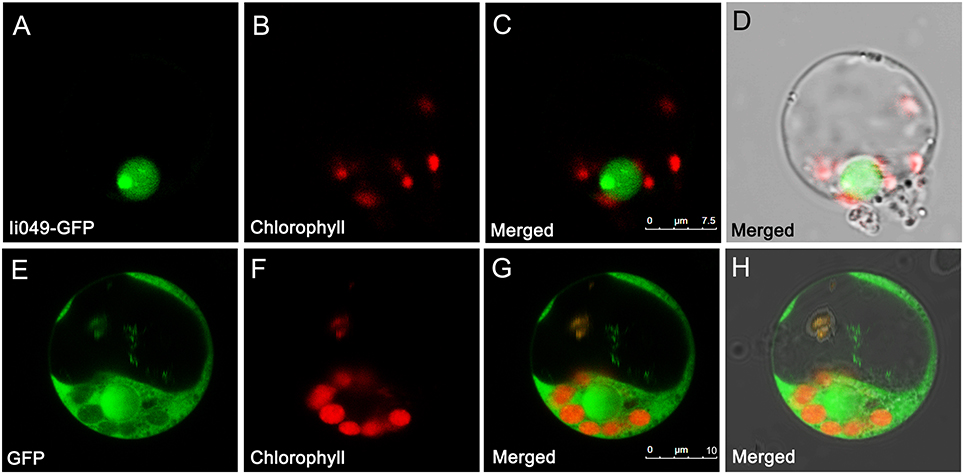
Figure 4. Subcellular localization of Ii049 in the rice cell. (A) A rice protoplast expressing Ii049-GFP showing green fluorescent signals in the nucleus. (B) The same protoplast cell of (A) showing the chlorophyll autofluorescence signal in the plastids. (C) The merged signal of (A,B). (D) The same protoplast of (C) under bright-field. Bars = 7.5 μm. (E) A rice protoplast expressing GFP showing green fluorescent signals. (F) The same protoplast cell of (E) showing the chlorophyll autofluorescence signal in the plastids. (G) The merged signal of (E,F). (H) The same protoplast of (G) under bright-field. Bars = 10 μm. This experiment was repeated three times with similar results.
Silencing of Ii049 Affected the Production of Lignans in I. indigotica
RNAi was used to knock down the expression of Ii049 to analyze the role of Ii049 in I. indigotica. All types of hairy root lines had the same growth rate and the highest growth rate was at 18 days after inoculation. Although the biomass of each hairy lines had little variations, no significant difference was observed after 45 days (P > 0.05) (Figure 5A). The PCR analysis of genomic DNA confirmed the presence of rolb, hpt genes and exogenous Ii049 fragment in Ri049 lines and the absence of exogenous Ii049 fragment in WT and CK1 lines (Figure 5B). In transgenic plants, five independent lines (Ri049-1, Ri049-2, Ri049-8, Ri049-9, and Ri049-13) with 22–67% observably down-regulated expression of Ii049 (Figure 5C) were selected for further study.
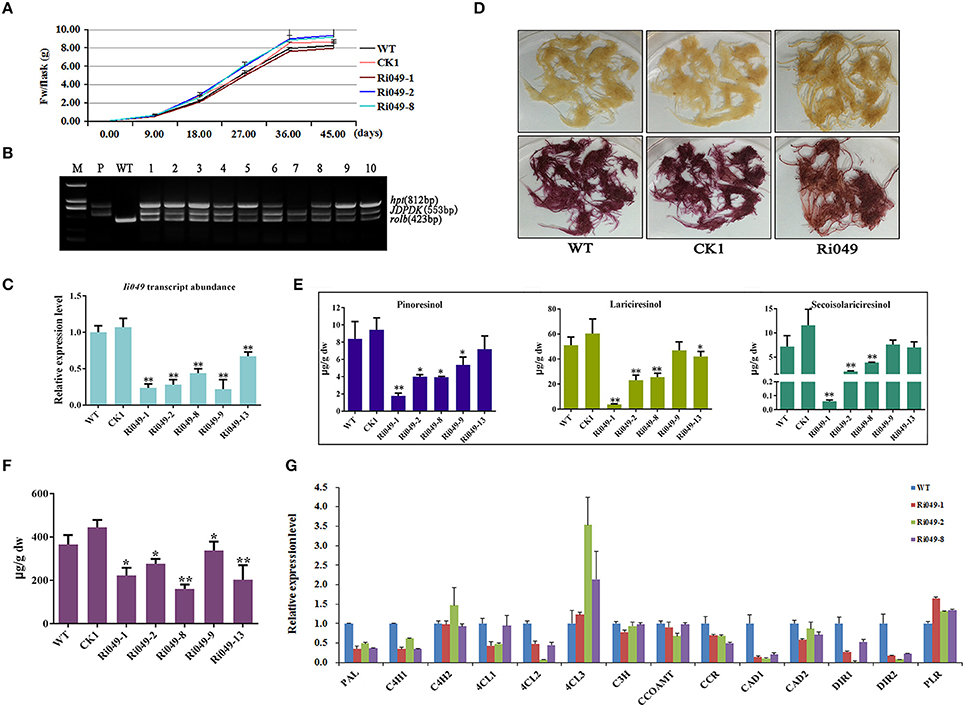
Figure 5. Characterization of transgenic hairy roots silencing Ii049. (A) Time course of biomass accumulation of hairy root lines WT, CK1, Ri049-1, Ri049-2, and Ri049-8. (B) Representative PCR analyses for the rolb and hpt genes and the specific gene in transgenic hairy root lines. M, DL-2000 Marker; P, the corresponding engineered bacteria (positive control); and WT, the wild-type hairy root (negative control). (C) Ii049 transcript levels. Bars indicate standard deviation. The level of significance obtained using the Student t-test is marked by the following: *P < 0.05, **P < 0.01. (D) The phenotype of Ri049 lines before (upper) and after (lower) phloroglucinol-HCl staining. (E) HPLC-MS/MS analysis of pinoresinol, lariciresinol and secoisolariciresinol of different Ri049 lines. (F) SA content in Ri049 lines. (G) Relative expression of lignan/lignin biosynthetic genes in hairy root lines WT, Ri049-1, Ri049-2, and Ri049-8. Each data point is the average of three biological replicates. Bars indicate standard deviation. The level of significance obtained using the Student t-test is marked by the following: *P < 0.05, **P < 0.01.
The hairy roots were stained with phloroglucinol-HCl (Weisner reagent) to reveal any differences of lignan/lignin accumulations between the transgenic lines and WT lines. The color intensity was associated with the presence of lignin and/or wall-bound or secreted phenolic derivatives (Hano et al., 2006). As shown in Figure 5D, WT and CK1 lines presented a similar purple-red color after staining with phloroglucinol-HCl, whereas the Ri049 line displayed a brown-red color after staining, which was weaker than that presented by the WT and CK1 lines. Such a color shift roughly indicated the decrease in lignan/lignin in Ri049 lines (Xiao et al., 2015).
LC-MS/MS was used to confirm further how lignans were modified among the Ri049, WT and CK1 lines. The productions of pinoresinol, lariciresinol and secoisolariciresinol reduced to 21.22–59.71%, 9.51–51.97%, 0.83–67.92%, respectively in lines Ri049-1, Ri049-2, and Ri049-8 (Figure 5E) compared with that in the WT control. Although Ri049-9 and Ri049-13 lines also had lesser lignan contents compared with that in control, no significant difference was found between them (P > 0.05).
The homologous gene of Ii049 in A. thaliana plays a role in the SA signaling pathways and is associated with the biosynthesis of SA (Giri et al., 2014). SA contents in transgenic hairy roots were measured to test whether Ii049 was involved in SA biosynthesis in I. indigotica. The contents of SA in transgenic hairy roots decreased to 43.76–75.61% of the WT control as predicted (Figure 5F). The reduction of lignan contents was consistent with the decrease in SA content. Moreover, the transcript levels of IiPAL, SA biosynthetic pathway gene, in Ri049 lines were reduced to 35.36–48.75% of the control level (Figure 5G). Therefore, the biosynthesis of lignans in I. indigotica might be related to the accumulation of SA.
Transcript analyses were performed on the lignan/lignin biosynthesis genes in transgenic hairy roots using qRT-PCR. Ri049-1, Ri049-2 and Ri049-8 were chosen as three biological replicates. The transcript levels of IiPAL, IiC4H1, Ii4CL1, Ii4CL2, IiCCR, IiCAD1, IiDIR1, and IiDIR2 significantly decreased in Ri049 lines. In particular, the expression levels of IiC4H2, Ii4CL3, and IiPLR were up-regulated while IiC3H, IiCCOAMT, and IiCAD2 were of the same level as the control (Figure 5G). These results demonstrated that Ii049 was an important positive regulator in the pathway of lignan biosynthesis, and it might regulate the expression levels of lignan/lignin biosynthetic genes resulting in a change in the lignan contents.
Ii049 Directly Bound to the Promoter of IiPAL and IiCCR in I. indigotica
Transcription factors recognize and regulate target genes, which can be measured by EMSA and Y1H (Yu et al., 2012; Tan et al., 2015). Previous reports showed that IiPAL and IiCCR had the same expression patterns as those of Ii049. Both of them had the strongest expression in roots—a pattern where roots of I. indigotica (Ban-Lan-Gen) were used as a traditional Chinese medicine. Moreover, a consistent pattern was observed associating the phytohormone induction of lignification and the expression of Ii049, IiPAL, and IiCCR (Lu et al., 2006; Hu et al., 2011). Based on the reported binding sites of AP2/ERF transcription factor (Wu et al., 2007; Zhu et al., 2010; Yu et al., 2012), CE1 and RAA motifs were found in IiPAL promoter (814 bp, Supplementary Figure S2), and CE1, RAA, and CBF2 motifs in IiCCR promoter (GenBank accession no. HM636437.1), which were the preferred core binding sites of the AP2/ERF family.
EMSA was performed using the purified recombinant Ii049 protein (Supplementary Figure S4) and biotin-modified promoter fragments containing three repeated CE1, RAA, and CBF2 motifs, respectively. As presented in Figures 6A–C, the migration of biotinylated probe coding for CE1, RAA, and CBF2 motifs was retarded in the presence of Ii049 recombinant protein, whereas the His control was not. A competition assay was performed to further investigate the specific binding. The binding activity decreased along with the increase of competitor and when the ratio of the unlabeled to labeled probes was 100:1, almost all CE1-, RAA-, and CBF2-labeled probe were bound (Figures 6A–C, line 6).
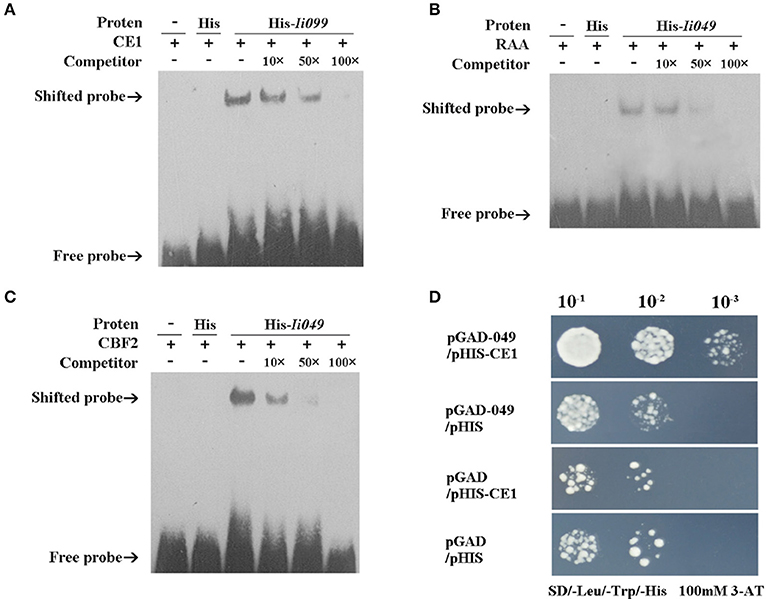
Figure 6. Ii049 protein bound to CE1, RAA, and CBF2 motifs. (A–C) The EMSA assay of Ii049 protein using biotinylated double-stranded CE1 (A), RAA (B), and CBF2 (C) probes, respectively. The detection of probes after reaction with His protein was taken as a negative control. Then, 10 μg of biotin-labeled were incubated with fusion protein at 25°C for 20 min to compete with cold probes (0, 10, 50, 100×) and then analyzed using EMSA. The bands were clarified with a solid black arrow. (D) Y1H assay for the interaction between His-Ii049 with CE1 motif. Triple CE1 motifs were used as bait. Yeast cells carrying pGADT7-Ii049 and pHIS2-CE1 grew normally on SD-Trp-Ura-His with 100 mM of 3-AT when diluted 1,000 times. Blank pGADT7 and PHIS2 were used as negative controls.
Y1H was further performed to verify the physical binding of Ii049 and CE1, RAA, and CBF2 motifs. The growth of yeast transformants on SD/-Leu-Trp-His containing 100 mM 3-AT was used to confirm the interactions. Only the combination of pGAD7-Ii049 and pHIS2-CE1 could have a normal growth when diluted 1,000 times (Figure 6D). The results of pGAD7-Ii049 and pHIS2-RAA/CBF2 were the same as that of pHIS2-CE1 (data not shown). Taken together, these results suggested that Ii049 was able to interact with RAA, CE1, and CBF2 elements physically.
SA-Induced Changes in the Lignan Accumulation and Transcript Profile
SA signaling is highly important for the synthesis of secondary metabolite. Accumulation of lariciresinol after SA treatment was monitored using LC-MS/MS in I. indigotica. As shown in Figure 7A, the content of lariciresinol significantly (P < 0.05) increased at 7 h after the treatment and the maximum contents (about 2.41-fold of the control) were observed at 10 h post-inoculation, followed by a gradual decline. Consequently, the genes encoding enzymes of the lignan/lignin biosynthetic pathway were further analyzed. All detected genes had a significant improvement in the transcription level when subjected to SA treatment except IiCAD1 (Figure 7B). The expression of IiPAL increased rapidly and peaked at 5 h. The transcription values of IiC4H2, Ii4CL1, Ii4CL2, Ii4CL3, IiC3H, IiCCOAMT, IiCCR, IiCAD2, and IiDIR2 gradually stimulated and reached the highest level at 7 h, followed by a reduction. The transcript levels of IiC4H1, IiDIR1, and IiPLR peaked at 10 h after SA treatment. These results indicated SA might play a role in the elicitation process and/or the signal transduction leading to gene activation and finally to the accumulation of lignan in I. indigotica.
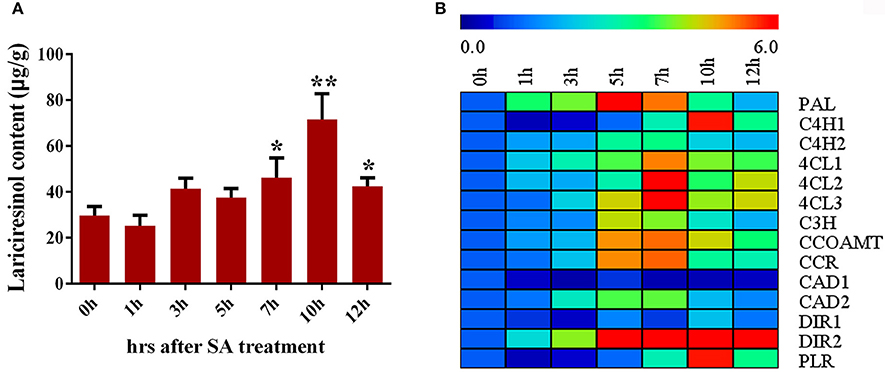
Figure 7. Regulation of SA on lignan biosynthesis in I. indigotica. (A) Lariciresinol contents under SA treatment (asterisks indicate values that are significantly different) *P < 0.05, **P < 0.01, from 0 h (Student t-test). Error bars indicate SD of three biological replicates. (B) Relative expression level of lignan biosynthetic genes under SA treatment.
Overexpression of Ii049 Improved Lignan Contents
Ii049 driven by double CaMV 35S promoter in vector PHB-flag was introduced into I. indigotica by Agrobacterium C58C1. Just as Ri049 lines, the morphological and growth rates between overexpression (OVX) lines and control lines did not show any difference (Figure 8A). PCR analyses confirmed the integration of the exogenous Ii049 gene in the transgenic lines and the absence of exogenous Ii049 in the WT and positive control lines (Figure 8B). The qRT-PCR analysis was conducted to examine the expression levels of the endogenous Ii049 in the OVX hairy roots. The transcript levels of Ii049 in all the independent OVX049 lines were significantly enhanced by 78.18- to 138.98-fold compared with the WT lines (Figure 8C). The expression of genes involved in lignan/lignin and SA biosynthesis remarkably improved, especially IiPAL and IiCCR (Figure 8G). Moreover, the Ii049 transcript level elevated lines (OVX049) had more SA accumulation compared with that in WT lines (Figure 8F).
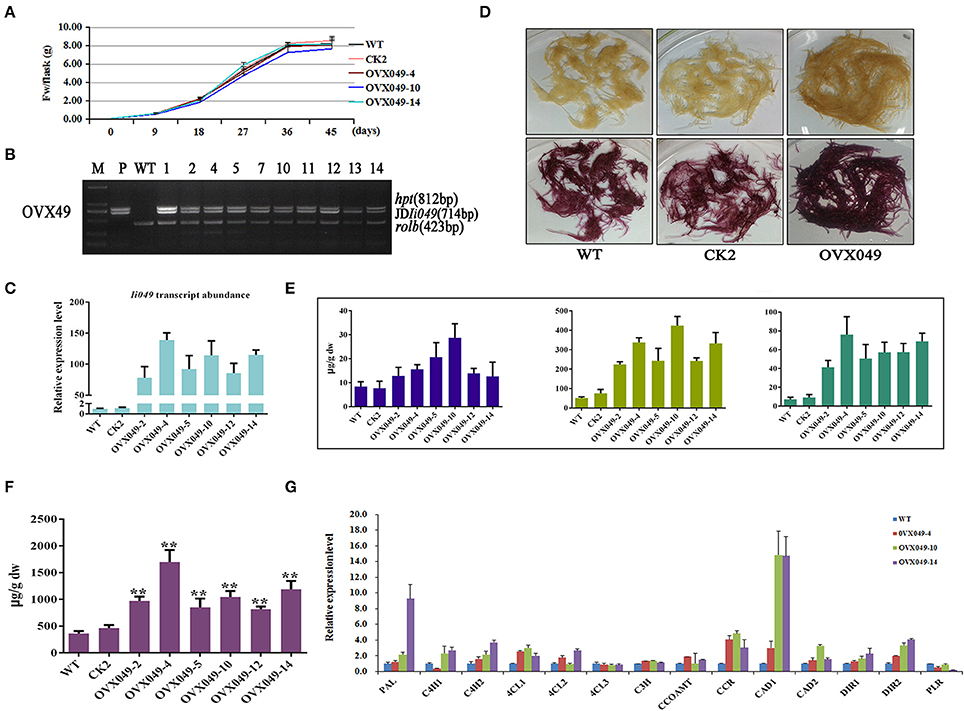
Figure 8. Characterization of transgenic hairy roots overexpressing Ii049. (A) Time course of biomass accumulation of hairy root lines WT, CK2, OVX049-4, OVX049-10 and OVX049-14. (B) Representative PCR analyses for the rolb and hpt genes and the specific gene in transgenic hairy root lines. M, DL-2000 Marker; P, the corresponding engineered bacteria (positive control); and WT, the wild-type hairy root (negative control). (C) Ii049 transcript levels. Bars indicate standard deviation. The level of significance obtained using the Student t-test is marked by the following: *P < 0.05, **P < 0.01. (D) Phenotype of OVX049 lines before (upper) and after (lower) phloroglucinol-HCl staining. (E) HPLC-MS/MS analysis of pinoresinol, lariciresinol and secoisolariciresinol of different OVX049 lines. (F) SA content in OVX049 lines. (G) Relative expression of lignan/lignin biosynthetic genes in hairy root lines WT, OVX049-4, OVX049-10, and OVX049-14. Each data point is the average of three biological replicates. Bars indicate standard deviation. The level of significance obtained using the Student t-test is marked by the following: *P < 0.05, **P < 0.01.
The OVX049 hairy roots presented a black-red color after staining with phloroglucinol-HCl (Figure 8D), which preliminarily predicting that OVX049 hairy roots accumulated more lignan/lignin compared with WT and CK2 lines. In agreement with the speculation derived from phloroglucinol-HCl staining, metabolism analysis showed that the OVX049 lines accumulated lariciresinol, which ranged from 4.41- to 8.32-fold of that in the control. Moreover, the contents of pinoresinol and secoisolariciresinol also significantly improved along with the increase in lariciresinol. The highest concentration was found in the OVX049-10 line, followed by OVX049-4 and OVX049-14 lines, with the mean levels of 425.60, 338.09 and 333.69 μg g−1 DW, respectively, for lariciresinol (Figure 8E). In comparison, lariciresinol content in the WT lines was 51.13 μg g−1 DW. This finding revealed the potential value of Ii049 in metabolic engineering of lignan production in I. indigotica.
Ii049 Promoted Lignin Biosynthesis in I. indigotica
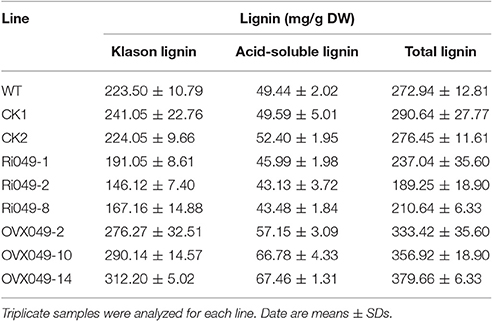
The total lignin (Klason lignin and acid-soluble lignin) content in the transgenic lines was detected. In the Ii049 RNAi lines (Ri049-1, Ri049-2, and Ri049-8), the Klason lignin concentration was 85.48, 65.37, and 74.79% compared with that in WT, and the levels of acid-soluble lignin and total lignin also decreased (Table 1). Meanwhile, a significant increase of lignin was found in the overexpression lines OVX049-2, OVX049-10, and OVX049-14. The contents of Klason lignin and acid-soluble lignin were increased by 23.61, 29.82, and 39.68% and 15.59, 35.07, and 36.45%, respectively (Table 1).
Discussion
Characterization of Ii049, an AP2/ERF Transcription Factor in I. indigotica
The AP2/ERF family is one of the largest superfamilies of plant-specific transcription factors (Dong et al., 2015). This family is involved in the control of plant growth and developmental programs, stress responses, and secondary metabolism (Licausi et al., 2013). To date, 112 putative AP2/ERF transcription factors have been identified in the I. indigotica transcriptome (Chen et al., 2015). However, none of the AP2/ERF family has been functionally characterized so far in I. indigotica. Interestingly, a gene encoding an AP2/ERF factor, namely Ii049, belonging to the Soloist subfamily was identified recently, which was a good candidate as a regulator of the lignan pathway. In this study, Ii049 was further characterized, and its role in lignan biosynthesis was analyzed in this study.
The Soloist subfamilies in Triticum aestivum L. (Zhuang et al., 2009) and Hevea brasiliensis Muell. Arg. (Du et al., 2013) were expressed in all the tested tissues with different expression levels. In I. indigotica, Ii049 also could be detected in all of the tissues with the highest expression in roots. This result was an indication that Ii049 might be positively correlated with the synthesis of lignans, as root is the main organ for the accumulation of lignans in I. indigotica (Chen et al., 2013).
The AP2/ERF family participates in varieties of signal transduction and is dramatically induced when subjected to environmental stress (Moffat et al., 2012; Cheng et al., 2013; Licausi et al., 2013; Zhang et al., 2015). Defense-related signaling molecules MeJA, SA, and ABA were chosen to examine the expression profile of Ii049. The results showed that the expression of Ii049 was up-regulated in response to MeJA, SA, and ABA. Ii049 was significantly induced by MeJA just like AaERF1 and AaERF2 in Artemisia annua. Ii049 was also proposed to play a major role in the regulation of secondary metabolism like AaERF1 and AaERF2 (Yu et al., 2012). Compared with MeJA and ABA, SA was shown to be the most effective in up-regulating Ii049. The results were fully consistent with a previous study on APD1, which was primarily regulated by SA (Giri et al., 2014). The expression of Ii049 under ABA treatment was also detected, as SA and ABA functioned cooperative or antagonistic in terms of plant defense (Xu et al., 2013; Alazem et al., 2014). As expected, ABA could effectively elevate the transcription level of Ii049, showing that SA and ABA might exert a synergistic action in I. indigotica. Taken together, as a transcriptional factor, Ii049 might play an important role through multiple signaling pathways in I. indigotica.
Ii049 Was a Key Regulator of Lignan Biosynthesis in I. indigotica
Lignans, such as lariciresinol, pinoresinol and secoisolariciresinol, are major phytoalexins in I. indigotica. When the expression of Ii049 was down-regulated by RNAi, the accumulation of lariciresinol, pinoresinol and secoisolariciresinol were significantly decreased in I. indigotica hairy roots compared with the WT lines. Moreover, Ii049 also could regulate the biosynthesis of SA just like its homologous in Arabidopsis (Giri et al., 2014). In addition, qRT-RCR analysis showed IiPAL, IiC4H1, Ii4CL1, Ii4CL2, IiCCR, IiCAD1, IiDIR1, and IiDIR2 involved in lignan/lignin biosynthesis were repressed in RNAi lines (Figure 5G). IiPAL, IiCAD1, IiC3H, IiCCR, and IiDIR1 were indicated to be the most possible genes involved in lignan biosynthesis (Chen et al., 2015). Interesting, IiPAL was also the first key enzyme in SA biosynthesis. All of these suggested that Ii049 might act as a regulator in lignan accumulation by regulating the pathway genes in lignan/lignin biosynthesis and SA biosynthesis.
Both PAL and CCR are key enzymes in the biosynthesis of lignan/lignin monomers and PAL is also a key enzyme in SA biosynthesis. PAL primarily links primary and secondary metabolism by catalyzing the conversion of L-phenylalanine into cinnamic acid, which is also a rate-limiting step of the phenylpropanoid metabolism (Jones, 1984). CCR plays a key regulatory role in lignan/lignin biosynthesis by catalyzing the nicotinamide adenine dinucleotide phosphate-dependent reduction of cinnamoyl-CoA esters to their corresponding cinnamaldehydes (Jones et al., 2001; Hu et al., 2011). The present study also showed that IiPAL and IiCCR were two of the most possible genes involved in lignan/lignin biosynthesis in I. indigotica (Chen et al., 2015).
AP2/ERF family can be divided into five subfamilies: AP2, DREB, ERF, RAV, and Soloist (Sakuma et al., 2002; Nakano et al., 2006). Members of different subfamilies were reported to display distinct DNA-binding activities. Members of the AP2 subfamily could recognize the sequence GCAC (A/G) N (A/T) TCCC (A/G) ANG (C/T) (Krizek, 2003; Yan et al., 2012). Many DREB proteins had been shown to bind to a dehydration-responsive element (DRE)/C-repeat (CRT) element [A/G)CCGAC] (Park et al., 2001; Zhang et al., 2014). The ERF proteins were first isolated as GCC-box binding proteins from tobacco (Ohme-Takagi and Shinshi, 1995). The RAV proteins bound specifically in vitro to the CAACA domain, namely RAA motif (Kagaya et al., 1999; Matías-Hernández et al., 2014). However, information about the DNA-binding properties of Soloist proteins is still lacking. Despite the generalization, evidence accumulated over the years have proved that ERF proteins bind not only to the GCC box but also to the DER/CRT, CE1, CBF2, and RAA motifs (Wu et al., 2007; Zhu et al., 2010; Yu et al., 2012) and some DREF proteins have been reported to bind to GCC box element (Wan et al., 2011). Based on these, whether Ii049 could bind to CE1, RAA, and CBF2 motifs in IiPAL and IiCCR promoters was verified.
The results of EMSA indicated that Ii049 could interact with the promoters of IiPAL and IiCCR by binding to the CE1, RAA, and CBF2 motifs (Figures 6A–C). Similar results had been observed in Y1H assay where Ii049 interacted with the promoter directly (Figure 6D). These results indicated that the change in lignan contents was due to the presence of Ii049-binding sites at the promoter of lignan/lignin and SA biosynthetic pathway genes such as IiPAL and IiCCR.
SA Activates Lignan Biosynthesis in I. indigotica
The phytohormone SA is a key regulator of plant development and stress responses, including, drought, cold and salinity stress, which are partly achieved by enhancing biosynthesis of secondary metabolites (Rivas-San and Plasencia, 2011; Khan et al., 2015). SA was widely used as an elicitor to improve active compounds in some plants. For example, the foliar sprays of 50 ppm SA could improve the contents of polyphenols, tannins, alkaloid and flavonoid and ameliorate water stress in Simarouba glauca (Awate and Gaikwad, 2014). In S. miltiorrhiza, SA induced the expression of tanshinone biosynthetic genes, such as SmIPPI, SmHMGR, SmDXS II, SmGGPPS, and SmCPS, coinciding with the induction of SA on improving the tanshinone production (Hao et al., 2015). In Vitis vinifera L, SA induced the accumulation of PAL mRNA, a key enzyme in phenylpropanoid metabolism, leading to a significant accumulation of phenolic and the development of thermotolerance (Wen et al., 2008). The accumulation of lignan in I. indigotica was also induced by SA treatment. Moreover, the expression of all other key genes in the lignan/lignin biosynthesis pathway significantly increased except for IiCAD1. This might explain the increased accumulation of lignan.
Engineering Lignan/Lignin Biosynthesis in I. indigotica
Lignans are pharmaceutically active compounds in I. indigotica for anti-virus, anti-inflammation, anti-tumor growth and angiogenesis (Li, 2003; Saarinen et al., 2008; Yang et al., 2013; Li et al., 2015). However, their utilization was limited due to the low yield in the roots of I. indigotica. The overexpression of Ii049 in hairy roots of I. indigotica increased the expression of lignan/lignin biosynthesis genes and SA content in this study, thereby improving lignan/lignin accumulation. The average production of lariciresinol was 5.9-fold of that in the control (Figure 8E). The results of this study indicated that the regulation of the expression of Ii049 was a promising option for increasing the accumulation of lignans. Otherwise, the overexpression of Ii049 significantly enhanced total lignin content in I. indigotica hairy roots, almost 1.22- to 1.39-fold of that in WT (Table 1). This study yielded similar results with the MYB transcription factor, which triggered lignin biosynthesis to 1.21- and 1.29-fold in Salvia miltiorrhiza and 1.48-fold of the control in loquat (Zhang et al., 2010; Zhang J. et al., 2016). The research might potentially manipulate the amount of lignin in other plant products based on the needs. Moreover, the transcription level of IiPLR decreased in the OVX049 lines, which might be due to the complexity in transcriptional regulation (Figure 8G). The overexpression of IiPLR resulted in an engineered I. indigotica hairy root ovx-2 producing about 353.5 μg g−1 DW of lariciresinol, a 6.3-fold increase compared with the WT production (Xiao et al., 2015). This study proposed that the co-expression of Ii049 and IiPLR might shed new light on substantially elevating lignan levels in I. indigotica, which might be used in the large-scale commercial production in the future.
Conclusions
This study showed that Ii049, a transcription factor from the AP2/ERF family, acted as a positive regulator of the biosynthesis of lignan/lignin in I. indigotica. Ii049 was mainly expressed in the roots and specially localized in the nucleus. The contents of lignan/lignin and SA significantly decreased in the RNAi transgenic lines. Moreover, the transcription of lignan/lignin and SA biosynthesis genes was also regulated by Ii049. EMSA and Y1H assays showed that Ii049 might trigger the expression of lignan/lignin and SA pathway genes by binding to CE1, RAA, and CBF2 motifs in their promoters. Also, SA could induce the accumulation of lignan and the expression of lignan/lignin biosynthetic pathway genes. Therefore, Ii049 controls lignan biosynthesis in two ways: by regulating the genes involved in lignan/lignin biosynthesis and by regulating SA biosynthesis, thus inducing lignan accumulation (Figure 9). In conclusion, this study provided strong evidence for genetic engineering of lignan/lignin production by overexpression AP2/ERF transcription factor.
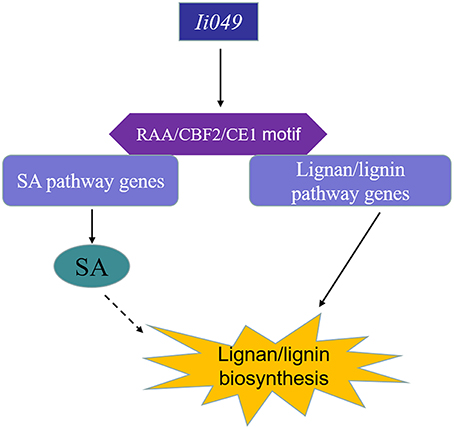
Figure 9. Schematic model of lignan/lignin biosynthesis regulated by Ii049. Ii049, an AP2/ERF transcription factor in I. indigotica; RAA/CBF2/CE1 motif, cis-element in genes promoter region.
Author Contributions
RM, YX, JY, LZ, and WC conceived and designed this study. RM, YX, and ZL performed the experiments. HT conducted the experiment of subcellular localization. RM, RC, and QL contributed to data analysis and bioinformatics analysis. JC and YW analyzed the accumulation of compounds through HPLC-MS/MS. RM and YX wrote the final manuscript and all authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the National Science Foundation of China (nos. 81325024, 31670292, 31100221, and 8160322).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01361/full#supplementary-material
Supplementary Figure S1. Nucleotide sequence and the deduced amino acid sequence of Ii049.
Supplementary Figure S2. Sequences of IiPAL promoter.
Supplementary Figure S3. Schematic diagram of constructed plant expression vectors.
Supplementary Figure S4. The purified recombinant Ii049 protein was stained with Coomassie Brilliant Blue (lane 1) or visualized after western blotting (lane 2).
Supplementary Table S1. PCR primers used for Ii049 isolation and vector construction.
Supplementary Table S2. Primers used in qRT-PCR.
Supplementary Table S3. Sequence data for the genes described in this study.
Abbreviations
4CL, 4-coumarate coenzyme A ligase; ABA, Abscisic acid; C3H, Coumarate 3-hydroxylase; C4H, Cinnamic acid 4-hydroxylase; CAD, Cinnamyl alcohol dehydrogenase; CCoAOMT, Caffeoyl-CoA O-methyltransferase; CCR, Cinnamoyl-CoA reductase; DIR, Dirigent protein; EMSA, Electrophoretic Mobility Shift Assay; MeJA, methyl jasmonate; MS, Murashige and Skoog medium; OVX049, overexpression of Ii049; qRT-PCR, quantitative real-time quantitative PCR; Ri049, knockdown the expression of Ii049; RNAi, RNA interference; SA, Salicylic acid; WT, Wild-type line.
References
Adlercreutz, H., Mousavi, Y., Clark, J., Hockerstedt, K., Hamalainen, E., and Wahala, K. (1992). Dietary phytoestrogens and cancer: in vitro and in vivo studies. J. Steroid Biochem. Mol. Biol. 41, 331–337 doi: 10.1016/0960-0760(92)90359-Q
Alazem, M., Lin, K. Y., and Lin, N. S. (2014). The abscisic acid pathway has multifaceted effects on the accumulation of Bamboo mosaic virus. Mol. Plant Microbe Interact. 27, 177–189. doi: 10.1094/MPMI-08-13-0216-R
Ambavaram, M. M., Krishnan, A., Trijatmiko, K. R., and Pereira, A. (2011). Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 155, 916–931. doi: 10.1104/pp.110.168641
Awate, P. D., and Gaikwad, D. K. (2014). Influence of growth regulators on secondary metabolites of medicinally important oil yielding plant Simarouba glauca DC. under water stress conditions. J. Stress Physiol. Biochem. 10, 222–229.
Chen, J. F., Dong, X., Li, Q., Zhou, X., Gao, S. H., and Chen, R. B. (2013). Biosynthesis of the active compounds of Isatis indigotica based on transcriptome sequencing and metabolites profiling. BMC Genomics 14:857. doi: 10.1186/1471-2164-14-857
Chen, R. B., Li, Q., Tan, H. X., Chen, J. F., Xiao, Y., and Ma, R. F. (2015). Gene-to-metabolite network for biosynthesis of lignans in MeJA-elicited Isatis indigotica hairy root cultures. Front. Plant Sci. 6:952. doi: 10.3389/fpls.2015.00952
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Dong, L., Cheng, Y. X., Wu, J. J., Cheng, Q., Li, W. B., and Fan, S. J. (2015). Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66, 2635–2647. doi: 10.1093/jxb/erv078
Du, L., Zhang, D. C., Deng, Z., Liu, X. H., and Li, D. J. (2013). Cloning and expression analysis of an AP2/EREBP gene in Hevea brasiliensis Muell. Arg. Plant Physiol. J. 49, 1238–1244
Giri, M. K., Swain, S., Gautam, J. K., Singh, S., Singh, N., and Bhattacharjee, L. (2014). The Arabidopsis thaliana At4g13040 gene, a unique member of the AP2/EREBP family, is a positive regulator for salicylic acid accumulation and basal defense against bacterial pathogens. J. Plant Physiol. 171, 860–867. doi: 10.1016/j.jplph.2013.12.015
Hano, C., Addi, M., Bensaddek, L., Cronier, D., Baltora-Rosset, S., and Doussot, J. (2006). Differential accumulation of monolignol-derived compounds in elicited flax (Linum usitatissimum) cell suspension cultures. Planta 223, 975–989. doi: 10.1007/s00425-005-0156-1
Hao, X., Shi, M., Cui, L., Xu, C., Zhang, Y., and Kai, G. (2015). Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol. Appl. Biochem. 62, 24–31. doi: 10.1002/bab.1236
Ho, Y. L., and Chang, Y. S. (2002). Studies on the antinociceptive, anti-inflammatory and anti pyretic effects of Isatis indigotica root. Phytomedicine 9, 419–424. doi: 10.1078/09447110260571661
Hu, Y. S., Di, P., Chen, J. F., Xiao, Y., Zhang, L., and Chen, W. S. (2011). Isolation and characterization of a gene encoding cinnamoyl-CoA reductase from Isatis indigotica Fort. Mol. Biol. Rep. 38, 2075–2083. doi: 10.1007/s11033-010-0333-6
Jones, D. H. (1984). Phenylalanine ammonia-lyase: regulation of its induction, and its role in plant development. Phytochemistry 23, 1349–1359. doi: 10.1016/S0031-9422(00)80465-3
Jones, L., Ennos, A. R., and Turner, S. R. (2001). Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J. 26, 205–216. doi: 10.1046/j.1365-313x.2001.01021.x
Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. doi: 10.1093/nar/27.2.470
Khan, M. I., Fatma, M., Per, T. S., Anjum, N. A., and Khan, N. A. (2015). Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 6:462. doi: 10.3389/fpls.2015.00462
Krizek, B. A. (2003). AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic Acids Res. 31, 1859–1868. doi: 10.1093/nar/gkg292
Kumagai, T., Aratsu, Y., Sugawara, R., Sasaki, T., Miyairi, S., and Nagata, K. (2016). Indirubin, a component of Ban-Lan-Gen, activates CYP3A4 gene transcription through the human pregnane X receptor. Drug Metab. Pharmacokinet. 31, 139–145. doi: 10.1016/j.dmpk.2016.01.002
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., and Van de Peer, Y. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, B. (2003). Studies on Active Constituents and Quality Evaluation of Banlangen. Ph.D. thesis, Shanghai: Second Military University.
Li, J., Zhou, B. X., Li, C. F., Chen, Q. Y., Wang, Y. T., and Li, Z. T. (2015). Lariciresinol-4-O-beta-D-glucopyranoside from the root of Isatis indigotica inhibits influenza A virus-induced pro-inflammatory response. J. Ethnopharmacol. 174, 379–386. doi: 10.1016/j.jep.2015.08.037
Li, Q., Chen, J. F., Xiao, Y., Di, P., Zhang, L., and Chen, W. S. (2014). The dirigent multigene family in Isatis indigotica: gene discovery and differential transcript abundance. BMC Genomics 15:388. doi: 10.1186/1471-2164-15-388
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Lu, B. B., Du, Z., Ding, R. X., Zhang, L., Yu, X. J., and Liu, C. H. (2006). Cloning and characterization of a differentially expressed phenylalanine ammonialyase gene (IiPAL) after genome duplication from tetraploid Isatis indigotica fort. J. Integr. Plant Biol. 48, 1439–1449. doi: 10.1111/j.1744-7909.2006.00363.x
Ma, Q. H. (2007). Characterization of a cinnamoyl-CoA reductase that is associated with stem development in wheat. J. Exp. Bot. 58, 2011–2021. doi: 10.1093/jxb/erm064
Matías-Hernández, L., Aguilar-Jaramillo, A. E., Marín-Gonzílez E Suárez-Lápez, P., and Pelaz, S. (2014). RAV genes: regulation of floral induction and beyond. Ann. Bot. 114, 1459–1470. doi: 10.1093/aob/mcu069
Moffat, C. S., Ingle, R. A., Wathugala, D. L., Saunders, N. J., Knight, H., and Knight, M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7:e35995. doi: 10.1371/journal.pone.0035995
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nakatsubo, T., Mizutani, M., Suzuki, S., Hattori, T., and Umezawa, T. (2008). Characterization of Arabidopsis thaliana pinoresinol reductase, a new type of enzyme involved in lignan biosynthesis. J. Biol. Chem. 283, 15550–15557. doi: 10.1074/jbc.M801131200
Nguyen, T. N., Son, S., Jordan, M. C., Levin, D. B., and Ayele, B. T. (2016). Lignin biosynthesis in wheat (Triticum aestivum L.): its response to waterlogging and association with hormonal levels. BMC Plant Biol. 16:28. doi: 10.1186/s12870-016-0717-4
Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 7, 173–182. doi: 10.1105/tpc.7.2.173
Park, J. M., Park, C. J., Lee, S. B., Ham, B. K., Shin, R., and Paek, K. H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell. 13, 1035–1046. doi: 10.1105/tpc.13.5.1035
Rivas-San, V. M., and Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. doi: 10.1093/jxb/err031
Saarinen, N. M., Warri, A., Dings, R. P., Airio, M., Smeds, A. I., and Makela, S. (2008). Dietary lariciresinol attenuates mammary tumor growth and reduces blood vessel density in human MCF-7 breast cancer xenografts and carcinogen-induced mammary tumors in rats. Int. J. Cancer. 123, 1196–1204. doi: 10.1002/ijc.23614
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Solovyev, V. V., Shahmuradov, I. A., and Salamov, A. A. (2010). Identification of promoter regions and regulatory sites. Methods Mol. Biol. 674, 57–83. doi: 10.1007/978-1-60761-854-6_5
Tan, H. X., Xiao, L., Gao, S. H., Li, Q., Chen, J. F., and Xiao, Y. (2015). TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol. Plant. 8, 1396–1411. doi: 10.1016/j.molp.2015.04.002
Udvardi, M. K., Czechowski, T., and Scheible, W. R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell. 20, 1736–1737. doi: 10.1105/tpc.108.061143
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., and De Paepe, A. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Wan, L. Y., Zhang, J. F., Zhang, H. W., Zhang, Z. J., Quan, R. D., and Zhou, S. R. (2011). Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 6:e25216. doi: 10.1371/journal.pone.0025216
Wen, P. F., Chen, J. Y., Wan, S. B., Kong, W. F., Zhang, P., and Wang, W. (2008). Salicylic acid activates phenylalanine ammonia-lyase in grape berry in response to high temperature stress. Plant Growth Regul. 55, 1–10. doi: 10.1007/s10725-007-9250-7
Wu, L. J., Chen, X. L., Ren, H. Y., Zhang, Z. J., Zhang, H. W., and Wang, J. Y. (2007). ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta 226, 815–825. doi: 10.1007/s00425-007-0528-9
Xiao, Y., Ji, Q., Gao, S. H., Tan, H. X., Chen, R. B., and Li, Q. (2015). Combined transcriptome and metabolite profiling reveals that IiPLR1 plays an important role in lariciresinol accumulation in Isatis indigotica. J. Exp. Bot. 66, 6259–6271. doi: 10.1093/jxb/erv333
Xu, J., Audenaert, K., Hofte, M., and De Vleesschauwer, D. (2013). Abscisic acid promotes susceptibility to the rice leaf blight pathogen Xanthomonas oryzae pv oryzae by suppressing salicylic acid-mediated defenses. PLoS ONE 8:e67413. doi: 10.1371/journal.pone.0067413
Yan, X. H., Zhang, L., Chen, B., Xiong, Z. Y., Chen, C. L., and Wang, L. J. (2012). Functional identification and characterization of the Brassica napus transcription factor gene BnAP2, the ortholog of Arabidopsis thaliana APETALA2. PLoS ONE 7:e33890. doi: 10.1371/journal.pone.0033890
Yang, Z. F., Wang, Y. T., Zheng, Z. G., Zhao, S. S., Zhao, J., and Lin, Q. (2013). Antiviral activity of Isatis indigotica root-derived clemastanin B against human and avian influenza A and B viruses in vitro. Int. J. Mol. Med. 31, 867–873. doi: 10.3892/ijmm.2013.1274
Yu, Z. X., Li, J. X., Yang, C. Q., Hu, W. L., Wang, L. J., and Chen, X. Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant. 5, 353–365. doi: 10.1093/mp/ssr087
Zeng, J. K., Li, X., Xu, Q., Chen, J. Y., Yin, X. R., and Ferguson, I. B. (2015). EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 13, 1325–1334. doi: 10.1111/pbi.12351
Zhang, J., Ge, H., Zang, C., Li, X., Grierson, D., Chen, K. S., et al. (2016). Ejodo1, a myb transcription factor, regulating lignin biosynthesis in developing loquat (Eriobotrya japonica) fruit. Front. Plant Sci. 7:1360 doi: 10.3389/fpls.2016.01360
Zhang, L., Chen, J. F., Zhou, X., Chen, X. F., Li, Q., and Tan, H. X. (2016). Dynamic metabolic and transcriptomic profiling of methyl jasmonate-treated hairy roots reveals synthetic characters and regulators of lignan biosynthesis in Isatis indigotica Fort. Plant Biotechnol. J. 14, 2217–2227. doi: 10.1111/pbi.12576
Zhang, P., Yang, P. Z., Zhang, Z. Q., Han, B., Wang, W. D., and Wang, Y. F. (2014). Isolation and characterization of a buffalograss (Buchloe dactyloides) dehydration responsive element binding transcription factor, BdDREB2. Gene 536, 123–128. doi: 10.1016/j.gene.2013.11.060
Zhang, Y., Li, D., Zhang, H., Hong, Y., Huang, L., Liu, S., et al. (2015). Tomato histone H2B monoubiquitination enzymes SlHUB1 and SlHUB2 contribute to disease resistance against Botrytis cinerea through modulating the balance between SA- and JA/ET-mediated signaling pathways. BMC Plant Biol. 15:252. doi: 10.1186/s12870-015-0614-2
Zhang, Y., Yan, Y. P., and Wang, Z. Z. (2010). The arabidopsis PAP1 transcription factor plays an important role in the enrichment of phenolic acids in salvia miltiorrhiza. J. Agric. Food Chem. 58, 12168–12175 doi: 10.1021/jf103203e
Zhao, S. J., Zhang, J. J., Tan, R. H., Yang, L., and Zheng, X. Y. (2015). Enhancing diterpenoid concentration in Salvia miltiorrhiza hairy roots through pathway engineering with maize C1 transcription factor. J. Exp. Bot. 66, 7211–7226. doi: 10.1093/jxb/erv418
Zhu, Q., Zhang, J. T., Gao, X. S., Tong, J. H., Xiao, L. T., and Li, W. B. (2010). The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 457, 1–12. doi: 10.1016/j.gene.2010.02.011
Keywords: AP2/ERF transcription factor, Isatis indigotica, lignan/lignin, metabolic regulation, salicylic acid, secondary metabolism
Citation: Ma R, Xiao Y, Lv Z, Tan H, Chen R, Li Q, Chen J, Wang Y, Yin J, Zhang L and Chen W (2017) AP2/ERF Transcription Factor, Ii049, Positively Regulates Lignan Biosynthesis in Isatis indigotica through Activating Salicylic Acid Signaling and Lignan/Lignin Pathway Genes. Front. Plant Sci. 8:1361. doi: 10.3389/fpls.2017.01361
Received: 09 April 2017; Accepted: 20 July 2017;
Published: 04 August 2017.
Edited by:
Raúl Alvarez-Venegas, Center for Research and Advanced Studies of the National Polytechnic Institute (CINVESTAV), MexicoReviewed by:
Xiaoya Chen, Shanghai Institute of Plant Physiology and Ecology (SIBS, CAS), ChinaRenu Deswal, University of Delhi, India
Copyright © 2017 Ma, Xiao, Lv, Tan, Chen, Li, Chen, Wang, Yin, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Yin, yinjun2002@yahoo.com
Lei Zhang, zhanglei@smmu.edu.cn
Wansheng Chen, chenwanshengsmmu@aliyun.com
†These authors have contributed equally to this work.
 Ruifang Ma
Ruifang Ma