- State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, China
The first YTH domain-containing RNA binding protein (YTP) was found in rat, where it was related to oxidative stress. Unlike characterizations in yeast and animals, functions of plant YTPs are less clear. Malus hupehensis (Pamp.) Rehd. YTP1 and YTP2 (MhYTP1 and MhYTP2) are known to be active in leaf senescence and fruit ripening. However, no research has been published about their roles in stress responses. Here, we investigate the stress-related functions of MhYTP1 and MhYTP2 in Arabidopsis thaliana. Both of the two genes participated in salicylic acid (SA), jasmonic acid (JA), and abscisic acid (ABA) signaling and play roles in plant responses to oxidative stress, chilling, high temperature, high salinity, and mannitol induced physiological drought stress. Moreover, MhYTP1 plays leading roles in SA and ABA signaling, and MhYTP2 plays leading roles in JA signaling and oxidative stress responses. These results will fill a gap in our knowledge about plant YTPs and stress responses and provide a foundation for future attempts to improve stress tolerance in apple.
Introduction
Phytohormones such as, abscisic acid (ABA), jasmonic acid (JA), and salicylic acid (SA) play vital roles in stress responses (Reje et al., 2014). For example, ABA is active when plants are exposed to heavy metals, drought, extremes in temperature, high salinity, or radiation. It also functions in various developmental processes, including seed germination and dormancy, and the closure of stomata (Vishwakarma et al., 2017). Both JA and SA are involved in responses to biotic stresses. The JA-signaling pathway is critical to the development of resistance to necrotrophic pathogens that derive nutrients from dead or dying cells, while the SA-signaling pathway is mainly activated against biotrophic pathogens that obtain nutrients from living host tissues (Glazebrook, 2005). In plants resistant to pathogen infections, the JA- and SA-defense pathways generally interact antagonistically (Verhage et al., 2010).
When plants are stressed, their hormone signaling is altered, which then changes the expression of responsive genes. In the gene regulation network, RNA binding proteins (RBPs) participate at the transcriptional and post-transcriptional levels, which is a powerful strategy of gene expression regulation. The most common post-transcriptional modifications are alternative splicing and polyadenylation, which change RNA sequences, protein functioning, and the availability of RNA-binding sites (Filichkin et al., 2010; Di Giammartino et al., 2011). The RBPs bind with target RNAs via their RNA-binding domains. To date, approximately 40 types of RNA-binding domains have been identified from various RBPs (Kishore et al., 2010). The novel YTH (for YT521-B homology) domain and YTH domain-containing RNA binding proteins (YTPs) have been described in yeast, animals (including humans; Homo sapiens), and plants (Stoilov et al., 2002).
The first identified YTP was YT521, found in the astrocytes of rat (Rattus norvegicus). The mRNA level of YT521 can be depressed under oxygen-deficient conditions but recovered after reoxygenation. Proteins that interact with YT521 are also RNA-binding proteins, e.g., RA301, SC35, and SF2, which indicates that YT521 is involved in post-transcriptional regulation (Imai et al., 1998). Another YTP gene, YT521-B, cloned from the brain library of rat, can alter the selection of alternative splicing sites in a concentration-dependent manner (Hartmann et al., 1999). Mmi1, a YTP in the model system Schizosaccharomyces pombe (fission yeast), is important in the molecular regulation of meiosis and controls sexual differentiation in fission yeast (Harigaya et al., 2006; Chen et al., 2011; Hiriart et al., 2012). However, meiosis-specific mRNAs are eliminated by binding to Mmi1 during the vegetative growth phase (Harigaya et al., 2006). Four YTPs occur in humans, and various YTHDF2 loci are associated with human longevity (Cardelli et al., 2006). Recently, researchers are focusing on functions of YTHDF2 and YTHDF3 in binding with the methylated N6 position of selected internal adenines in mRNAs and non-coding RNAs, which affect the translation status and lifetime of mRNA (Li F. et al., 2014; Zhu et al., 2014; Shi et al., 2017). These studies with yeast and animal YTPs have primarily focused on interactions between YTPs and target RNAs, and have not examined their roles in stress responses.
However, the information about plant YTPs are limited in contrast with researches on animals and yeast. The first plant YTP—cleavage and polyadenylation specificity factor 30 (CPSF30)—was discovered in Arabidopsis, where it participates in defense and oxidative-stress responses, as well as cellular signaling (Addepalli and Hunt, 2007; Chakrabarti and Hunt, 2015). Li D. et al. (2014) have identified YTP family members in Arabidopsis and analyzed their functions in abiotic-stress responses. Fifteen members of the Malus YTP family have been reported and their expression has been investigated via quantitative real-time PCR (qRT-PCR) experiments during leaf senescence and under various abiotic-stress conditions (Wang et al., 2014). Although Malus hupehensis (Pamp.) Rehd. YTP1 and YTP2 (MhYTP1 and MhYTP2) have functions in leaf senescence and fruit ripening (Wang et al., 2017), their roles in stressed plants have not been confirmed.
Here, we investigated the responses of MhYTP1 and MhYTP2 to various environmental stimuli in Arabidopsis thaliana, based on their promoter cis-elements. After fusion with the β-glucuronidase (GUS) gene, their promoters were transferred into Arabidopsis plants. We also treated plants that over-expressed MhYTP1 or MhYTP2 and tested the responses of those transgenics to SA, methyl jasmonate (MeJA), ABA, methyl viologen (MV), exposure to 4 or 40°C, salt (NaCl), or mannitol.
Materials and Methods
Cloning and Sequence Analysis of Genes and Promoters
We previously determined the sequences of MhYTP1 (MDP0000124048) and MhYTP2 (MDP0000488588) from the Genome Database for Rosaceae (GDR; Wang et al., 2017). For promoter cloning, we used a CTAB-based method (Porebski et al., 1997) to isolate genomic DNA from young leaves of 1-year-old M. hupehensis seedlings. The promoter regions were cloned by PCR, based on the region upstream of MhYTP1 (GDR: contig MDC008177.673; NCBI: MF370872) or MhYTP2 (GDR: contig MDC011033.286; NCBI: MF370873). All primers are listed in Supplementary Table 1. The functions of cis-acting elements in those promoters were predicted from the PlantCARE database (Rombauts et al., 1999; Postel et al., 2002).
Production of Transgenic Arabidopsis thaliana Plants
To construct our GUS expression vectors, we inserted the promoter regions of MhYTP1 and MhYTP2 into 433 vectors (Gateway; attR1-attR2-GUS-TNOS). For construction of the overexpression (OE) vectors, the open reading frame of MhYTP1 or MhYPT2 was inserted into the pCambia2300 vector. These vectors were then transferred to Agrobacterium tumefaciens. Transformation and selection of transgenic Arabidopsis plants were performed by the floral-dip procedure (Zhang et al., 2006). Before being spread on wet soil in pots, seeds of the wild type (WT) A. thaliana (Columbia, or Col-0, ecotype) were suspended in re-distilled water and kept in the dark for 3 days at 4°C. After the seeded pots were placed in a growth chamber under short-day conditions (8-h photoperiod) for 3–4 weeks, they were moved to long-day conditions (16-h photoperiod) to induce flowering.
A positive single Agrobacterium colony was inoculated into a liquid YEB medium containing appropriate antibiotics and incubated at 28°C. After the OD600 reached 1.5–2.0, Agrobacterium cells were collected by centrifugation at 4,000 g for 10 min and re-suspended in a fresh 5% (w:v) sucrose solution with 0.02% (v:v) Silwet L-77. The Arabidopsis plants were inverted and their aerial parts dipped into this cell suspension for 10 s. Afterward, those plants were kept under darkness for 16–24 h before being returned to the growth chamber.
To select transgenic plants, we sterilized T0 seeds with 70% ethanol for 1 min and 5% bleach for 10 min. After being rinsed three times with sterile water, the seeds were re-suspended in 0.05% agarose and spread on an MS selection agar medium supplemented with kanamycin. Plates were held at 4°C for 3 days and then transferred to 22°C under short-day conditions. Transgenic plants were distinguishable after 7–10 days. Preliminarily confirmed T0 transgenic plants were transferred to fresh selection plates and grown for another 1–2 weeks. Afterward, fully verified plantlets were transferred to soil in pots, and then moved to a growth chamber for seed development. Homozygous T3 seeds were produced by T2 transgenic plants that had survived on the selection media. Homozygous lines were further confirmed by qRT-PCR.
Quantitative Real-Time PCR
Total RNA from snap-frozen Arabidopsis seedlings was extracted by a modified CTAB method (Gambino et al., 2008). The cDNA was reversed-transcribed from total RNA using a PrimeScript® RT reagent Kit with gDNA Eraser (Perfect Real Time; Takara). Quantitative real-time PCR was performed according to protocols for SYBR® Premix Ex Taq™ II (Tli RNaseH Plus; Takara) and the Real Time PCR System machine (Bio-Rad iQ™ 5; Bio-Rad). The gene-specific primers are shown in Supplementary Table 1. Expression levels were calculated relative to the expression of AtTublin8 mRNA.
Stress Treatments
For testing the responses of Arabidopsis to environmental inductions, seeds of the WT and homozygous T3 transgenic plants were surface-sterilized and germinated on a standard MS medium. After 7 days, seedlings of all lines were transferred to either a standard MS medium or one that was supplemented with SA (10 μM), MeJA (5 μM), ABA (2 μM), MV (1 μM), NaCl (200 mM), or mannitol (200 mM). After 7 days of stress treatment, GUS expression was investigated in both WT and transgenic plants. Fresh weights and root lengths were recorded from WT and OE plants of each treatment.
The response to chilling (4°C) or heat (40°C) was examined using 14-day-old seedlings grown for 4 days on standard MS media, and was compared with the performance of plants under normal (control) conditions, i.e., 25°C. Again, GUS expression was monitored for both WT plants and those transformed with MhYTP1 or MhYTP2 promoters. Relative electrolyte leakage (REL) and the concentration of total chlorophyll were calculated for leaf samples from WT and OE plants under normal and treatment conditions. The REL was measured with a conductivity meter (Leci; DDS-307). Chlorophyll was extracted with 80% acetone and concentrations were determined spectrophotometrically according to the methods of Lichtenthaler and Wellburn (1983).
Statistical Analysis
Data were subjected to one-way ANOVA, and mean differences were assessed by Duncan analyses (statistically significant at P < 0.05).
Results
MhYTP1 and MhYTP2 Participate in SA, JA, and ABA Signaling
Because the MhYTP1 promoter region contains a SA signaling-responsive cis-element (TCA-Element), we first investigated whether our two promoters could be induced by exogenously applied SA (Supplementary Figure 1). After they were fused with the GUS reporter gene in the expression vector, the constructs were transferred to Arabidopsis plants (Supplementary Figure 2). Our results indicated that SA treatment led to a decline in GUS expression levels in those transgenics (Figure 1A). We also over-expressed those YTP genes separately in Arabidopsis and obtained Lines OE-1, OE-2, OE-7, and OE-15 for MhYTP1; and Lines OE-3, OE-4, OE-6, and OE-10 for MhYTP2 (Supplementary Figure 3). After growing on either normal MS or an MS medium supplemented with 10 μM SA for 7 days, values for fresh weights were higher for all OE plants than for the WT. This indicated that the latter genotype was more sensitive to SA (Figures 1B,C). In contrast, we found no significant differences in root lengths between WT and OE plants.
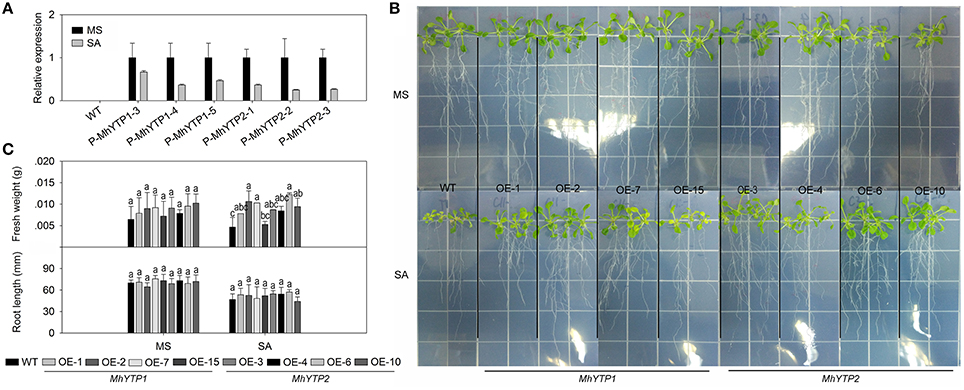
Figure 1. (A) Relative expression levels of GUS in WT and transgenic plants under normal conditions or salicylic acid (SA) treatment. Data are means ± SD for 5 replicates (at least 20 seedlings each). (B) Phenotype comparisons between WT and MhYTP1-overexpression plants, and between WT and MhYTP2-overexpression plants, in standard MS medium and under SA treatment. (C) Fresh weights and root lengths of OE and WT Arabidopsis plants grown in standard MS medium or MS medium supplemented with 10 μM SA. Data are means of 15 replicates ± SD. Values not followed by same letter are significantly different according to Duncan's multiple range test (P < 0.05).
Both JA signaling and SA signaling participate in plant defenses against pathogen infection, and their pathways interact with each other. We noted that GUS expression was induced by MeJA treatment, especially in plants transformed with the MhYTP2 promoter region (Figure 2A). Transcript levels of GUS were up to nine-fold higher in the treated transgenics than in the untreated plants. Separately, OE plants were generally more resistant to exogenously applied MeJA, as evidenced by their heavier fresh weights and longer root lengths when compared with the WT (Figures 2B,C). However, fresh weight of MhYTP1 overexpression line OE-1 and OE-7 are the same with that of WT; root lengths of WT, MhYTP1 overexpression line OE-2, MhYTP2 overexpression line OE-3 have no significant differences.
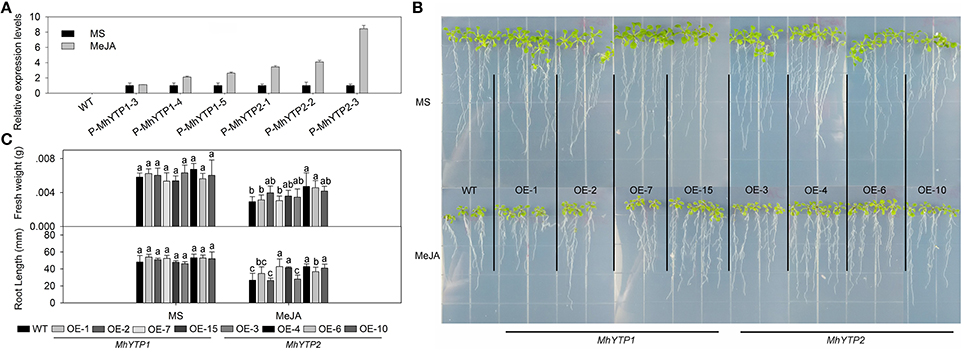
Figure 2. (A) Relative expression levels of GUS in WT and transgenic plants under normal conditions or methyl jasmonate (MeJA) treatment. Data are means ± SD for 5 replicates (at least 20 seedlings each). (B) Phenotype comparisons between WT and MhYTP1-overexpression plants, and between WT and MhYTP2- overexpression plants, in standard MS medium and under MeJA treatment. (C) Fresh weights and root lengths of OE and WT Arabidopsis plants grown in standard MS medium or MS medium supplemented with 5 μM MeJA. Data are means of 15 replicates ± SD. Values not followed by same letter are significantly different according to Duncan's multiple range test (P < 0.05).
The ABA-treated plants transformed with the MhYTP1 promoter also show induced GUS expression, whereas the level of expression in plants transformed with the MhYTP2 promoter did not differ significantly from their corresponding untreated plants (Figure 3A). After 7 days of growth on standard MS or the medium supplemented with 2 μM ABA, values for fresh weights and root lengths were higher for both MhYTP1-OE and MhYTP2-OE plants than for the WT (Figures 3B,C), indicating that the non-transformed plants were more sensitive to ABA.
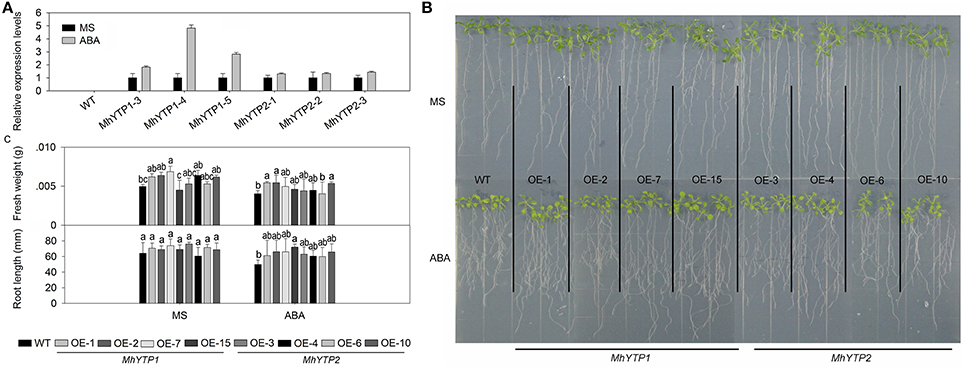
Figure 3. (A) Relative expression levels of GUS in WT and transgenic plants under normal conditions or abscisic acid (ABA) treatment. Data are means ± SD for 5 replicates (at least 20 seedlings each). (B) Phenotype comparisons between WT and MhYTP1-overexpression plants, and between WT and MhYTP2-overexpression plants, in standard MS medium and under ABA treatment. (C) Fresh weights and root lengths of OE and WT Arabidopsis plants grown in standard MS medium or MS medium supplemented with 2 μM ABA. Data are means of 15 replicates ± SD. Values not followed by same letter are significantly different according to Duncan's multiple range test (P < 0.05).
MhYTP1 and MhYTP2 Can Be Induced by MV, and Overexpression Plants Are More Resistant to MV Treatment
The presence of one ARE (anaerobic induction element) in the MhYTP1 promoter region as well as ARE and GC-motif (anoxic specific induction) elements in the MhYTP2 promoter region demonstrated their roles in the hypoxia-stress response (Supplementary Figure 1). Hypoxic conditions can cause oxidative stress in plants due to the generation of reactive oxygen species (ROS). Here, MV treatment resulted in greater GUS expression in plants carrying either the MhYTP1 or MhYTP2 promoter, and the response was especially strong when GUS was fused with the latter (Figure 4A).
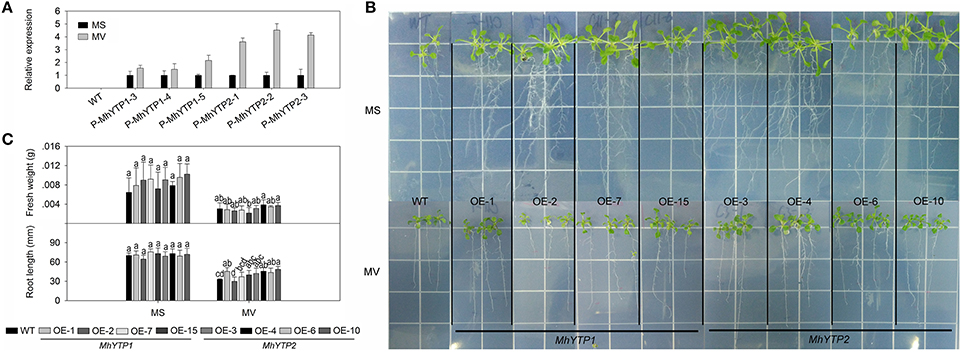
Figure 4. (A) Relative expression levels of GUS in WT and transgenic plants under normal conditions or methyl viologen (MV) treatment. Data are means ± SD for 5 replicate samples (at least 20 seedlings each). (B) Phenotype comparisons between WT and MhYTP1-overexpression plants, and between WT and MhYTP2-overexpression plants, in standard MS medium and under MV treatment. (C) Fresh weights and root lengths of OE and WT Arabidopsis plants grown in standard MS medium or MS medium supplemented with 1 μM MV. Data are means of 15 replicates ± SD. Values not followed by same letter are significantly different according to Duncan's multiple range test (P < 0.05).
After 7 days of growth on normal MS or MS media containing 1 μM MV, the roots were generally longer from OE plants than from the WT (Figures 4B,C), indicating that the latter was more sensitive to this stress treatment. The exception was for the roots of MhYTP1-OE 2, which were shorter than those of either MhYTP2-OE plants or the WT. MhYTP2 overexpression plants grow better than MhYTP1 overexpression and WT plant. A comparison of fresh weight values revealed no significant difference among genotypes, although MhYTP1-OE 15 was a bit lighter and MhYTP2-OE 4 and 10 were slightly heavier than the WT plants.
MhYTP1 and MhYTP2 Play Roles in Chilling and Heat Stress Responses
A HSE (heat stress response element) was found in the promoter region for MhYP2 but not for MhYTP1 (Supplementary Figure 1). Consistent with this, GUS expression was induced by heat stress when fused with the MhYTP2 promoter but not when fused with the MhYTP1 promoter (Figure 5A). In contrast, exposure to chilling induced expression when GUS was fused with the promoter of either MhYTP1 or MhYTP2 (Figure 5A).
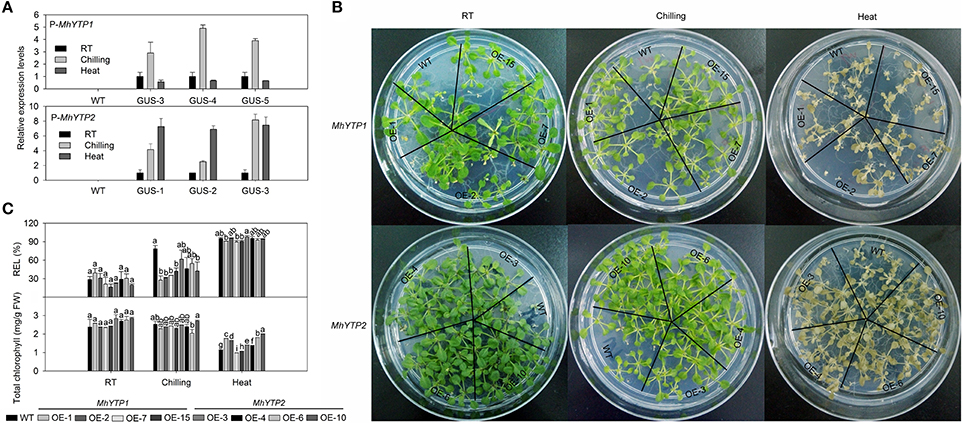
Figure 5. (A) Relative expression levels of GUS in WT and transgenic plants under normal (25°C), chilling (4°C), or heat-stress (40°C) conditions. Data are means ± SD for 5 replicates (at least 20 seedlings each). (B) Phenotype comparisons between WT and MhYTP1-overexpression plants, and between WT and MhYTP2-overexpression plants under normal, chilling, or heat treatment. (C) Relative electrolytic leakage (REL) in leaves and concentrations of total chlorophyll in OE and WT Arabidopsis plants grown under normal, chilling, or heat treatment. Data are means of 5 replicates ± SD. Values not followed by same letter are significantly different according to Duncan's multiple range test (P < 0.05).
After 4 days of either chilling or heat stress, the phenotypes did not differ significantly among any of our transgenic or WT plants (Figure 5B). However, leaf REL values were lower for the transgenics (especially the MhYTP1-OE lines) than for the WT in response to chilling, while total chlorophyll concentrations were higher in the OE plants than in the WT under heat stress (Figure 5C). This showed that overexpression of the YTP genes conferred greater resistance to low and high temperatures. Based on the fact that leaves from MhYTP1-OE 7 and 15 contained less chlorophyll than did samples from any of the other genotypes under heat stress, MhYTP2-OE plants appeared to be more resistant to high temperatures. Therefore, these results indicated that MhYTP1 has the more dominant role in chilling resistance while MhYTP2 has a greater role in the heat stress response.
Overexpression of MhYTP1 and MhYTP2 Enhances Plant Resistance to Salinity and Drought
High salinity and drought are severe stresses commonly associated with the cultivation of apple. For high salinity, GUS gene can be increased in plants no matter transferred with MhYTP1 or MhYTP2 promoter under NaCl treatments (Figure 6A). Fresh weights were also higher in salinity-stressed OE plants than in similarly treated WT plants, meaning that the former were more resistant (Figures 6B,C). Simulated drought, also termed “physiological drought,” is imposed when plants are treated with high concentrations of mannitol that reduce their capacity to absorb water from the external environment. Although we did not find that mannitol regulated GUS expression in our transgenics (data not shown), plants over-expressing either MhYTP1 or MhYTP2 were more resistant to mannitol, i.e., drought, and produced longer roots when compared with the WT (Figure 7).
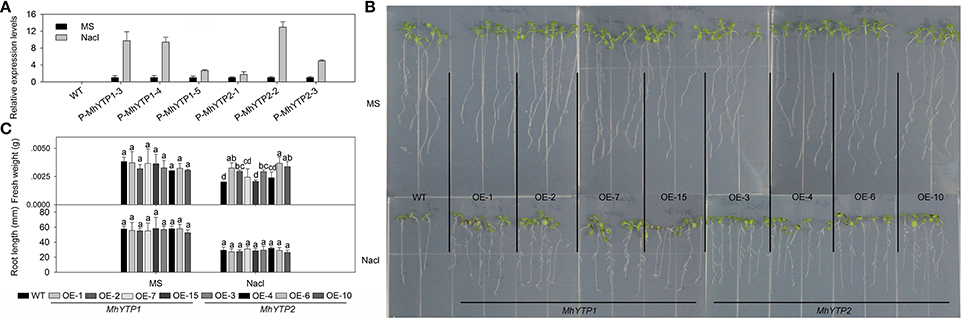
Figure 6. (A) Relative expression levels of GUS in WT and transgenic plants under normal or high salinity treatment. Data are means ± SD for 5 replicates (at least 20 seedlings each). (B) Phenotype comparisons between WT and MhYTP1-overexpression plants, and between WT and MhYTP2-overexpression plants, in standard MS medium or MS medium with 200 mM NaCl. (C) Fresh weights and root lengths of OE and WT Arabidopsis plants grown in standard MS medium or MS medium supplemented with 200 mM NaCl. Data are means of 15 replicates ± SD. Values not followed by same letter are significantly different according to Duncan's multiple range test (P < 0.05).
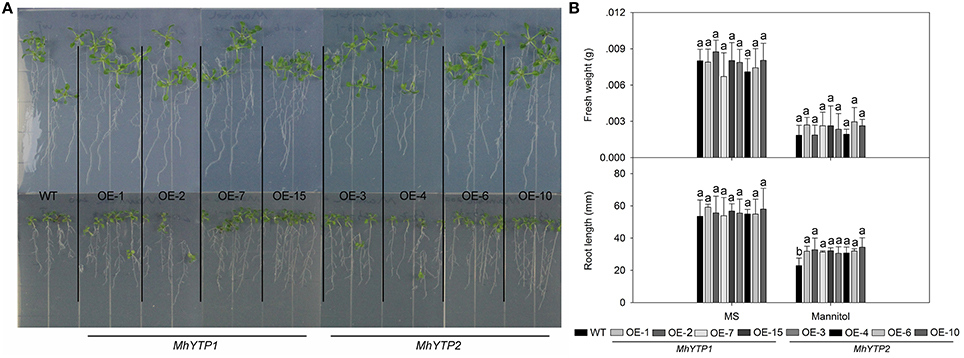
Figure 7. (A) Phenotype comparisons between WT and MhYTP1-overexpression plants, and between WT and MhYTP2-overexpression plants, in standard MS medium or MS medium with 200 mM mannitol. (B) Fresh weights and root lengths of OE and WT Arabidopsis plants under normal growing conditions or osmotic stress. Data are means of 15 replicates ± SD. Values not followed by same letter are significantly different according to Duncan's multiple range test (P < 0.05).
Discussion
There are limited researches on plant YTPs. Although, MhYTP1 and MhYTP2 could promote leaf senescence and fruit ripening (Wang et al., 2017), their roles in stressed plants have not been confirmed. Each of their promoter regions contains several stress-related cis-elements indicating their roles in stress responses. MhYTP1 and MhYTP2 are homologous and share more than 90% similarity (Wang et al., 2017), but they differ in their responses to various environmental stimuli.
The TCA-Element occurs in the MhYTP1 promoter region but not in the MhYTP2 promoter. However, the latter contains a Box-W1 element (fungal elicitor responsive element). When we compared the responses to exogenous applied SA, expression was reduced when GUS was fused to either the MhYTP1 or MhYTP2 promoter, and all OE lines proved resistant to treatment with 10 μM SA. This indicated that the two genes are active in SA signaling. Another important participant in the network of plant pathogen defenses is JA. Here, GUS expression was induced by treatment with MeJA, especially in plants transformed with the MhYTP2 promoter. Moreover, gene overexpression plants, particularly the MhYTP2-OE lines, are more resistant to MeJA. All of these results led us to conclude that MhYTP1 plays a more important role in SA signaling and while MhYTP2 has a greater function in JA signaling.
Three phytohormones—SA, JA, and ethylene (ET)—are crucial to the regulation of signal transduction pathways. However, such control is not achieved through the isolated activation of only a single hormonal pathway but by a complex regulatory network that connects the different pathways. This enables each hormone to either assist or antagonize the others as required to fine-tune the defense response to an individual pathogen. Whereas, the SA-signaling pathway is mainly activated against biotrophic pathogens, the JA-signaling pathway is associated with the development of resistance to necrotrophic pathogens (Glazebrook, 2005; Adie et al., 2007; Verhage et al., 2010). Thus, our findings suggest that MhYTP1 has a leading role in defenses against biotrophic organisms while MhYTP2 functions in resistance to necrotrophic pathogens. The presence of an ET-responsive element (ERE) in the MhYTP2 promoter provides further evidence of a relationship between that gene and hormone signaling, again implying cooperation between JA- and ET-signaling in protecting plants against pathogen infections.
Under various biotic and abiotic stresses, e.g., drought, cold, salinity, and pathogen attacks, ABA signaling regulates the genes responsible for seed germination and stomatal movement (Adie et al., 2007; Beck et al., 2007; Hirayama and Shinozaki, 2007). The promoter regions of MhYTP1 or MhYTP2 do not contain any ABA- or drought-related cis-elements. Nevertheless, GUS expression levels of ABA treated plants transferred with MhYTP1 promoter were induced. However, the GUS expression levels of ABA treated plants transferred with MhYTP2 promoter were not significantly different from that of untreated plants. Overexpression plants are more resistant to ABA treatment. Thus, both of the two genes participated in ABA signaling and MhYTP1 may play a leading role. Then MhYTP1 and MhYTP2 regulated ABA signaling related stress responses.
Almost all promoter regions for Malus domestica YTP genes contain at least one anaerobic stress-related cis-element (Wang et al., 2014). We found that the promoter region of MhYTP1 has an ARE while that region of MhYTP2 have two ARE and a GC-motif. In examining how their expression is regulated when plants are under hypoxia stress due to elevated ROS production, we found that expression of GUS fused with either of those promoters increased after MV treatment. This was particularly true for the MhYTP2 promoter. Likewise, expression of the rat homolog YT521-B is inhibited under oxygen-deficient conditions but is recovered after re-oxygenation (Imai et al., 1998). The first YTP identified in a plant, AtCPSF30, also participates in the response to oxidative stress (Chakrabarti and Hunt, 2015). All of these reports suggest that YTPs are functionally homologous in plants and animals. After growing on normal MS or MS media supplemented with 1 μM MV for 7 days, MhYTP1-OE and, especially, MhYTP2-OE plants performed better that the WT. This difference might have been a result of the MhYTP2 promoter region carrying more anaerobic stress-related cis-elements. Therefore, we can conclude that both genes function in the oxidative stress response, with MhYTP2 having the dominant role.
The presence of HSEs in the MhYTP2 promoter region implies that expression of that gene is induced by changes, especially an increase, in temperature. Consistently, the GUS gene fused with the MhYTP2 promoter was induced by exposure to heat, i.e., 40°C, but not when fused with the MhYTP1 promoter. In contrast, expression of GUS, when fused with the promotor of either gene, was induced by chilling at 4°C. After 4 days of treatment at either temperature, overexpression of these YTP genes conferred resistant to both chilling and heat. This demonstrated that the function of a gene is determined not only by its promoter but also by its structure and that of its protein. As RBPs, the functions of MhYTP1 and MhYTP2 are also determined by their target RNAs. In addition, our transgenic plants also showed greater resistance to high salinity and mannitol (simulated drought) when compared with similarly treated WT plants.
From these results stated above, we can see that MhYTP1 and MhYTP2 participate in multiple stress responses. While these sources of stress can be present simultaneously, so also do MhYTP1 and MhYTP2 interact with each other, making it difficult to identify individual stress responses. One possible outcome of being exposed to multiple stresses is that plants are better able to develop resistance to more than one type of stress (Bowler and Fluhr, 2000). This phenomenon is called cross-tolerance, which is achieved through a powerful regulatory system that allows plants to adapt quickly to a changing environment (Bowler and Fluhr, 2000; Capiati et al., 2006; Fujita et al., 2006; Suzuki et al., 2012). JA, SA, and ET signaling interact with each other and all play roles in responses to both biotic and abiotic stresses. Miura and Tada (2014) have described the importance of SA in the drought response. This hormone also improves the resistance of Hordeum vulgare to water deficits (Bandurska and Stroiński, 2005). The pathogen related (PR) proteins are crucial for resistance against pathogens, and their expression is strongly up-regulated when plants are attacked (Seo et al., 2010). Moreover, many PR genes are induced when plants are exposed to abiotic stress (Seo et al., 2010).
We propose that MhYTP1 and MhYTP2 participate in such a variety of stress responses because of their YTH domains. This type of domain can bind with a short, degenerated, single-stranded RNA sequence motif that can be described only by a weight matrix (Zhang et al., 2010). However, an accurate motif sequence has not yet been confirmed. Thus, target RNAs of MhYTP1 and MhYTP2 are numerous, which may be the reason of their wide-spectrum of cellular functions. The way in which RBPs function in stress responses is determined by their promoter cis-elements, gene structures, protein structures, and target RNAs. Here, we analyzed the promoter regions of MhYTP1 and MhYTP2 and focused only on how each is induced by stresses. More research involving gene and protein structures, as well as target RNAs, is needed if we aim to improve our understanding of the exact roles of these proteins in stress responses.
In summary, we investigated the responses of MhYTP1 and MhYTP2 to various environmental stimuli by studying their promoter cis-elements. Although, MhYTP1 and MhYTP2 are homologous genes in apple, they still differ in their responses to various treatments. Both genes are active in SA, JA, and ABA signaling and play roles in plant responses to oxidative stress, chilling, heat, salinity, and mannitol-induced physiological drought stress. While MhYTP1 has the more dominant role in SA and ABA signaling, MhYTP2 is the main gene involved in JA signaling and oxidative stress responses.
Author Contributions
NW: Experiment design and implementation, manuscript composition; TG, PW, XS, YS, XJ, and BL: Experiment implementation and assistance; XG: Purchase and management of reagents, maintenance of laboratory apparatus; FM: Experiment design and implementation, composition and review of manuscript, financial support for experiments and laboratory apparatus.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Key Program of the National Natural Science Foundation of China (31330068), and by the earmarked fund for the China Agriculture Research System (CARS-28).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01367/full#supplementary-material
Supplementary Table 1. Sequences of primers for vector construction and quantitative real-time PCR.
Supplementary Figure 1. Main stress-related cis-elements in MhYTP1 and MhYTP2 promoter region. TCA-element, SA signaling-responsive cis-element; TC-rich repeat, cis-acting element involved in defense and stress responsiveness; ARE, anaerobic induction element; TGA-element, auxin-responsive element; HSE, heat stress response element; ERE, ET-responsive element; Box-W1, fungal elicitor responsive element; GC-motif, anoxic specific induction.
Supplementary Figure 2. Expression levels of GUS gene in Arabidopsis transferred with MhYTP1 or MhYTP2 promoter regions and WT plants.
Supplementary Figure 3. Expression levels of MhYTP1 and MhYTP2 in WT and transgenic Arabidopsis plants.
Abbreviations
ABA, Abscisic acid; CPSF30, cleavage and polyadenylation specificity factor 30; ET, ethylene; GDR, Genome Database for Rosaceae; GUS, β-glucuronidase; JA, jasmonic acid; MeJA, methyl jasmonate; MhYTP1, Malus hupehensis YTP1; MhYTP2, Malus hupehensis YTP2; MV, methyl viologen; NCBI, National Center for Biotechnology Information; qRT-PCR, quantitative real-time PCR; RBP, RNA binding protein; REL, relatively electrolyte leakage; SA, salicylic acid; YTH domain, YT521-B homology domain; YTP, YTH domain containing RNA binding protein.
References
Addepalli, B., and Hunt, A. G. (2007). A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res. 35, 4453–4463. doi: 10.1093/nar/gkm457
Adie, B. A., Pérez-Pérez, J., Pérez-Pérez, M. M., Godoy, M., Sánchez-Serrano, J. J., Schmelz, E. A., et al. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19, 1665–1681. doi: 10.1105/tpc.106.048041
Bandurska, H., and Stroiński, A. (2005). The effect of salicylic acid on barley response to water deficit. Acta Physiol. Plant. 27, 379–386. doi: 10.1007/s11738-005-0015-5
Beck, E. H., Fettig, S., Knake, C., Hartig, K., and Bhattarai, T. (2007). Specific and unspecific responses of plants to cold and drought stress. J. Biosci. 32, 501–510. doi: 10.1007/s12038-007-0049-5
Bowler, C., and Fluhr, R. (2000). The role of calcium and activated oxygens as signals for controlling cross-tolerance. Trends Plant Sci. 5, 241–246. doi: 10.1016/S1360-1385(00)01628-9
Capiati, D. A., Pais, S. M., and Tellez-Iñon, M. T. (2006). Wounding increases salt tolerance in tomato plants: evidence on the participation of calmodulin-like activities in cross-tolerance signaling. J. Exp. Bot. 57, 2391–2400. doi: 10.1093/jxb/erj212
Cardelli, M., Marchegiani, F., Cavallone, L., Olivieri, F., Giovagnetti, S., Mugianesi, E., et al. (2006). A polymorphism of the YTHDF2 gene (1p35) located in an Alu-rich genomic domain is associated with human longevity. J. Gerontol. A Biol. Sci. Med. Sci. 61, 547–556. doi: 10.1093/gerona/61.6.547
Chakrabarti, M., and Hunt, A. G. (2015). CPSF30 at the interface of alternative polyadenylation and cellular signaling in plants. Biomolecules 5, 1151–1168. doi: 10.3390/biom5021151
Chen, H., Futcher, B., and Leatherwood, J. (2011). The fission yeast RNA binding protein mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS ONE 6:e26804. doi: 10.1371/journal.pone.0026804
Di Giammartino, D. C., Nishida, K., and Manley, J. L. (2011). Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866. doi: 10.1016/j.molcel.2011.08.017
Filichkin, S. A., Priest, H. D., Givan, S. A., Shen, R., Bryant, D. W., Fox, S. E., et al. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58. doi: 10.1101/gr.093302.109
Fujita, M., Fijita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Gambino, G., Perrone, I., and Gribaudo, I. (2008). A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 19, 520–525. doi: 10.1002/pca.1078
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Harigaya, Y., Tanaka, H., Yamanaka, S., Tanaka, K., Watanabe, Y., Tsutsumi, C., et al. (2006). Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442, 45–50. doi: 10.1038/nature04881
Hartmann, A. M., Nayler, O., Schwaiger, F. W., Obermeier, A., and Stamm, S. (1999). The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59fyn. Mol. Biol. Cell 10, 3909–3926. doi: 10.1091/mbc.10.11.3909
Hirayama, T., and Shinozaki, K. (2007). Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12, 343–351. doi: 10.1016/j.tplants.2007.06.013
Hiriart, E., Vavasseur, A., Touat-Todeschini, L., Yamashita, A., Gilquin, B., Lambert, E., et al. (2012). Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 31, 2296–2308. doi: 10.1038/emboj.2012.105
Imai, Y., Matsuo, N., Ogawa, S., Tohyama, M., and Takagi, T. (1998). Cloning of a gene, YT521, for a novel RNA splicing-related protein induced by hypoxia/reoxygenation. Brain Res. Mol. Brain Res. 53, 33–40. doi: 10.1016/S0169-328X(97)00262-3
Kishore, S., Luber, S., and Zavolan, M. (2010). Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief Funct. Genomics 9, 391–404. doi: 10.1093/bfgp/elq028
Li, D., Zhang, H., Hong, Y., Huang, L., Li, X., Zhang, Y., et al. (2014). Genome-wide identification, biochemical characterization, and expression analyses of the YTH domain-containing RNA-binding protein family in Arabidopsis and rice. Plant Mol. Biol. Rep. 32, 1169–1186. doi: 10.1007/s11105-014-0724-2
Li, F., Zhao, D., Wu, J., and Shi, Y. (2014). Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 24, 1490–1492. doi: 10.1038/cr.2014.153
Lichtenthaler, K., and Wellburn, A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 11, 591–592. doi: 10.1042/bst0110591
Miura, K., and Tada, Y. (2014). Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 5:4. doi: 10.3389/fpls.2014.00004
Porebski, S., Bailey, L. G., and Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. doi: 10.1007/BF02772108
Postel, D., Vanlemmens, P., Gode, P., Ronco, G., and Villa, P. (2002). PlantCARE, a database of Plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Reje, I. B., Pastor, V., and Mauch-Mani, B. (2014). Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants 3, 458–475. doi: 10.3390/plants3040458
Rombauts, S., Déhais, P., van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Seo, P. J., Kim, M. J., Park, J. Y., Kim, S. Y., Jeon, J., Lee, Y. H., et al. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61, 661–667. doi: 10.1111/j.1365-313X.2009.04091.x
Shi, H., Wang, X., Lu, Z., Zhao, B., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Stoilov, P., Rafalska, I., and Stamm, S. (2002). YTH: a new domain in nuclear proteins. Trends Biochem. Sci. 27, 495–497. doi: 10.1016/S0968-0004(02)02189-8
Suzuki, N., Koussevitzky, S., Mittler, R., and Miller, G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Verhage, A., van Wees, S. C., and Pieterse, C. M. (2010). Plant immunity: it's the hormones talking, but what do they say? Plant Physiol. 154, 536–540. doi: 10.1104/pp.110.161570
Vishwakarma, K., Upadhyay, N., Kumar, N., Yadav, G., Singh, J., Mishra, R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8:161. doi: 10.3389/fpls.2017.00161
Wang, N., Guo, T., Wang, P., Sun, X., Shao, Y., Liang, B., et al. (2017). Functional analysis of apple MhYTP1 and MhYTP2 genes in leaf senescence and fruit ripening. Sci. Hortic. 221, 23–32. doi: 10.1016/j.scienta.2017.04.018
Wang, N., Yue, Z., Liang, D., and Ma, F. (2014). Genome-wide identification of members in the YTH domain-containing RNA-binding protein family in apple and expression analysis of their responsiveness to senescence and abiotic stresses. Gene 538, 292–305. doi: 10.1016/j.gene.2014.01.039
Zhang, X., Henriques, R., Lin, S., Niu, Q., and Chua, N. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhang, Z., Theler, D., Kaminska, K. H., Hiller, M., de la Grange, P., Pudimat, R., et al. (2010). The YTH domain is a novel RNA binding domain. J. Biol. Chem. 285, 14701–14710. doi: 10.1074/jbc.M110.104711
Keywords: YTH domain, promoter, phytohormone, stress, Malus
Citation: Wang N, Guo T, Wang P, Sun X, Shao Y, Jia X, Liang B, Gong X and Ma F (2017) MhYTP1 and MhYTP2 from Apple Confer Tolerance to Multiple Abiotic Stresses in Arabidopsis thaliana. Front. Plant Sci. 8:1367. doi: 10.3389/fpls.2017.01367
Received: 17 May 2017; Accepted: 21 July 2017;
Published: 04 August 2017.
Edited by:
Jianhua Zhu, University of Maryland, College Park, United StatesReviewed by:
Yanmin Zhu, Agricultural Research Service (USDA), United StatesJi-Hong Liu, Huazhong Agricultural University, China
Copyright © 2017 Wang, Guo, Wang, Sun, Shao, Jia, Liang, Gong and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengwang Ma, fwm64@sina.com; fwm64@nwsuaf.edu.cn
 Na Wang
Na Wang Tianli Guo
Tianli Guo Ping Wang
Ping Wang Xun Sun
Xun Sun Xin Jia
Xin Jia Fengwang Ma
Fengwang Ma