- 1College of Agronomy, Xinjiang Agricultural University, Urumqi, China
- 2Institute of Crop Science, National Wheat Improvement Center, Chinese Academy of Agricultural Sciences, Beijing, China
- 3International Maize and Wheat Improvement Center (CIMMYT), Beijing, China
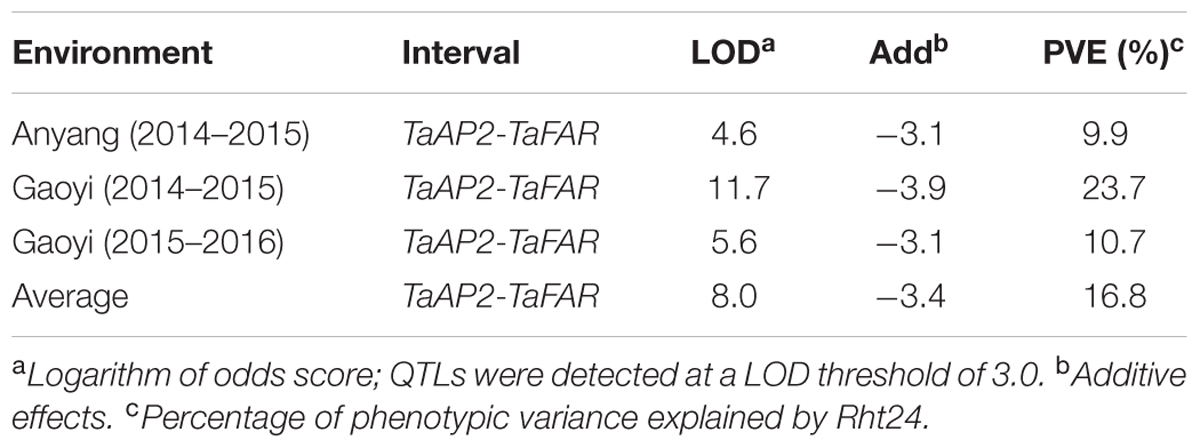
Height is an important trait related to plant architecture and yield potential in bread wheat (Triticum aestivum L.). We previously identified a major quantitative trait locus QPH.caas-6A flanked by simple sequence repeat markers Xbarc103 and Xwmc256 that reduced height by 8.0–10.4%. Here QPH.caas-6A, designated as Rht24, was confirmed using recombinant inbred lines (RILs) derived from a Jingdong 8/Aikang 58 cross. The target sequences of Xbarc103 and Xwmc256 were used as queries to BLAST against International Wheat Genome Sequence Consortium database and hit a super scaffold of approximately 208 Mb. Based on gene annotation of the scaffold, three gene-specific markers were developed to genotype the RILs, and Rht24 was narrowed to a 1.85 cM interval between TaAP2 and TaFAR. In addition, three single nucleotide polymorphism (SNP) markers linked to Rht24 were identified from SNP chip-based screening in combination with bulked segregant analysis. The allelic efficacy of Rht24 was validated in 242 elite wheat varieties using TaAP2 and TaFAR markers. These showed a significant association between genotypes and plant height. Rht24 reduced plant height by an average of 6.0–7.9 cm across environments and were significantly associated with an increased TGW of 2.0–3.4 g. The findings indicate that Rht24 is a common dwarfing gene in wheat breeding, and TaAP2 and TaFAR can be used for marker-assisted selection.
Introduction
Bread wheat (Triticum aestivum L.) is one of the most important food crops providing a significant proportion of the calories and protein consumed by humans (Mir et al., 2012; Chen et al., 2016). Grain yield in wheat largely depends on plant architecture, particularly plant height, which is significantly associated with biomass production and harvest index that ultimately determine yield potential (Law et al., 1978). Most importantly, appropriately reduced plant height reduces lodging and increases grain yield (Evans, 1998). The introduction of semi-dwarf varieties leading to the “Green Revolution” greatly enhanced crop yields globally (Peng et al., 1999).
To date, 23 dwarfing genes (Rht1–Rht23) have been cataloged in wheat (Supplementary Table S1) (McIntosh et al., 2015). Among them, only four homeologous genes Rht1 (Rht-B1b), Rht2 (Rht-D1b), Rht3 (Rht-B1c), and Rht10 (Rht-D1c) from the chromosome 4BS, 4DS, 4BS, and 4DS, respectively, had been cloned (Peng et al., 1999; Pearce et al., 2011; Wu et al., 2011), 13 were on chromosomes 2AS (2), 2BL (1), 2DL (1), 3BS (1), 5AL (1), 5DL (1), 6AS (3), 7AS (1), and 7BS (2), but the locations of six other dwarfing genes have not been determined yet (Peng et al., 1999, 2011; Ellis et al., 2005; Wu et al., 2011; McIntosh et al., 2013; Chen et al., 2015). So far, more than 50 quantitative trait loci (QTL) for plant height have also been identified on all wheat chromosomes in previous reports (Börner et al., 2002; Peng et al., 2003; Quarrie et al., 2005; Liu et al., 2011; Griffiths et al., 2012; Würschum et al., 2015). For example, Wei et al. (2010) identified quite a few stable plant height QTL located on chromosomes 2A, 2B, 2D, 3B, 4B, 5A, 5D, 7B, and 7D. Griffiths et al. (2012) found that all chromosomes except for 3D, 4A, and 5D, had plant height genes using meta-QTL analysis. However, only a few genes for reduced stature have been used in wheat breeding because most showed negative effects on grain yield (Law et al., 1978; Worland et al., 1998; Chen et al., 2015). Rht-B1b (4BS), Rht-D1b (4DS) and Rht8 (2DL) are extensively used in wheat breeding globally and molecular markers have been developed for their marker-assisted selection (MAS) (Korzun et al., 1998; Peng et al., 1999; Rebetzke and Richards, 2000; Ellis et al., 2002; Asplund et al., 2012). Rht-B1b (Rht1) and Rht-D1b (Rht2) were two major genes in the Green Revolution, and at present, approximately 70% of wheat varieties throughout the world contain at least one of them (Evans, 1998). Rht8 reduced plant height around 10% and it has not significant negative effect on grain yield (Worland et al., 1998).
Simple sequence repeat (SSR) markers were favored for research and breeding in the last two decades due to high polymorphism, good repeatability, co-dominance and a simple polymerase chain reaction (PCR)-based system for analysis (Röder et al., 1998). With rapid advancements in sequencing technology, the quality of wheat genome assembly has been significantly improved and large numbers of high-quality scaffolds to facilitate gene isolation are available (Rogers, 2014). Simultaneously, many single nucleotide polymorphisms (SNPs) were identified for developing SNP chips, providing high-throughput genotyping platforms that have been extensively used for genome-wide association studies (GWAS) and QTL mapping at high resolution (Colasuonno et al., 2014; Wang et al., 2014; Sukumaran et al., 2015; Gao et al., 2016; Wu et al., 2016). Many QTL for agronomic traits, grain and industrial quality and disease resistance have been identified in wheat (Buerstmayr et al., 2009; Wang et al., 2014; Naruoka et al., 2015; Jin et al., 2016; Liu et al., 2016; Zhai et al., 2016). Thus, the availability of high quality wheat genome data and high-throughput SNP genotyping platforms greatly advanced wheat genetics and breeding.
We previously identified a plant height major QTL QPH.caas-6A between Xwmc256 and Xbarc103, which explained 8.0–10.4% of the phenotypic variance across eight environments using an F2:4 population derived from the Jingdong 8/Aikang 58 cross (Li et al., 2015). In addition, QPH.caas-6A probably was consistent with QTL_height_6A_1, which also linked with Xwmc256 on chromosome 6AL and explained 6.3–29.1% of the phenotypic variance (PVE) (Griffiths et al., 2012). Furthermore, QPH.caas-6A was identified to reduce TGW 6.5–8.2%, increase kernel number per spike and number of spike by 2.4–3.5% and NS by 2.0–4.6%, respectively, which is similar to “green revolution” gene Rht2 (Li et al., 2015). Thus QPH.caas-6A was a potentially useful dwarfing locus in wheat breeding. Here we designate QPH.caas-6A as Rht24. The previously closest markers flanking Rht24 were Xbarc103 and Xwmc256, which were 8.2 cM apart. The aim of the present study was to identify markers more closely linked to Rht24 using the wheat genomic database and a 660K SNP chip and thereby establish a more efficient MAS system for wheat breeding.
Materials and Methods
Plant Materials
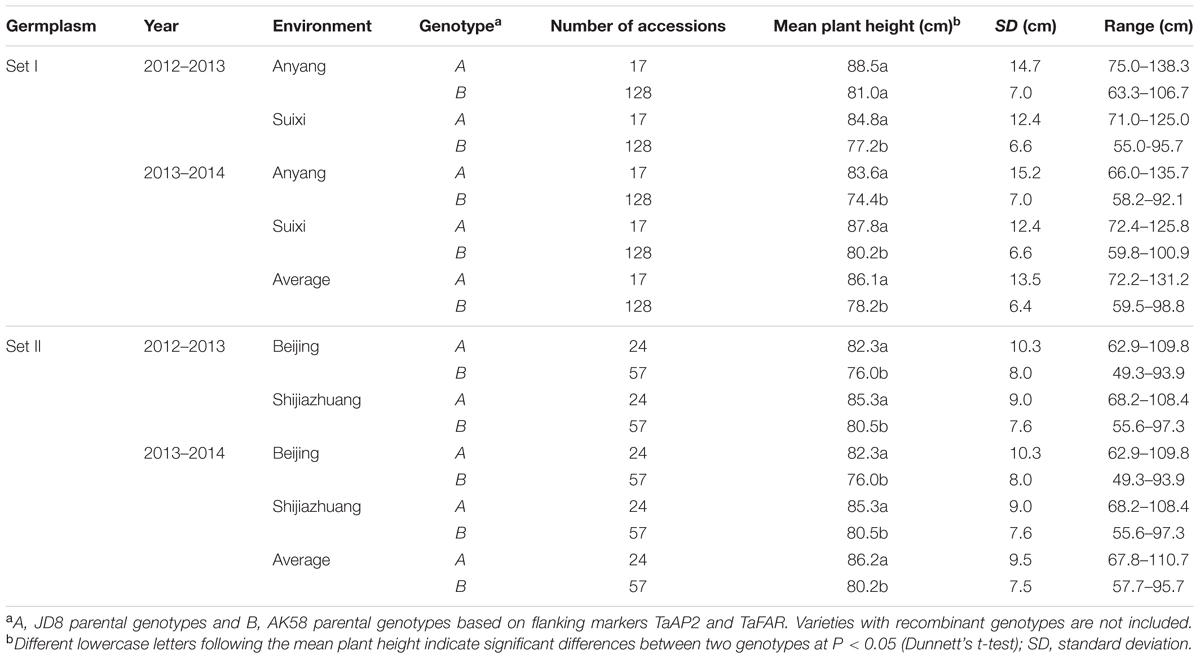
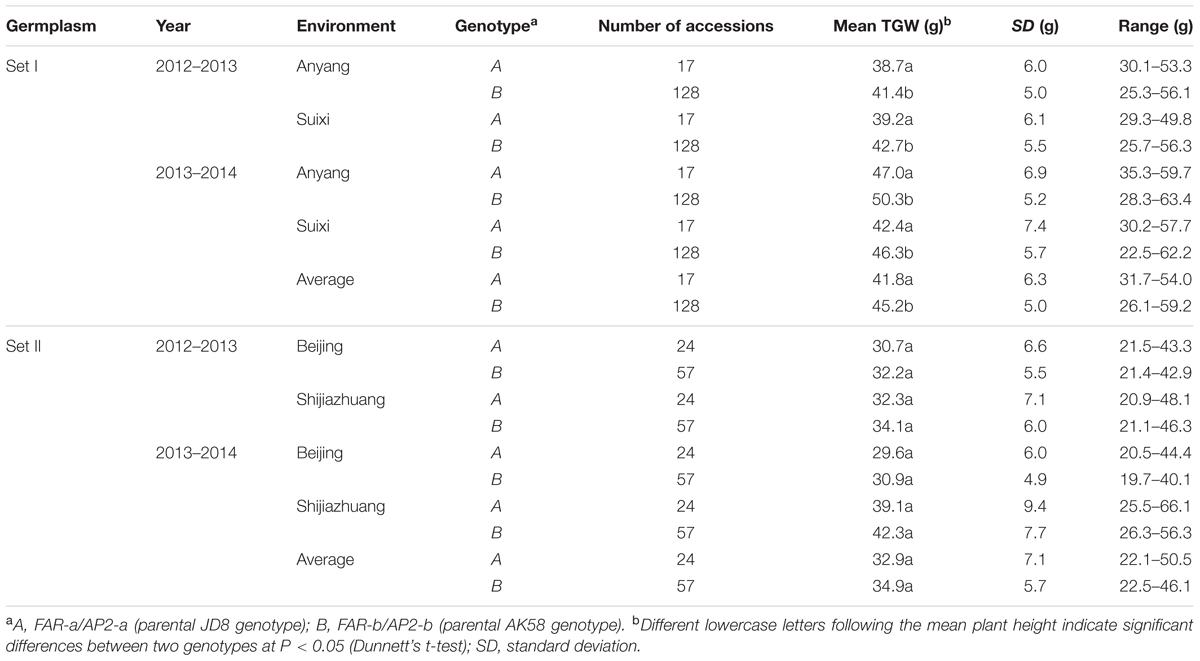
Two hundred and fifty-six recombinant inbred lines (RILs, F2:6) from a cross between Aikang 58 (AK58) × Jingdong 8 (JD8) were used for genetic analysis. AK58, a leading variety occupying more than one million ha in the Yellow and Huai Valley, has a short plant height and excellent lodging resistance. Jingdong 8, with relatively tall plant height and excellent resistance to heat during the grain filling stage, was an elite variety in the Northern China Plain Region. Two sets of Chinese and introduced elite varieties (Supplementary Tables S4, S5), comprising 154 (Set I) and 88 (Set II) varieties, were used to validate the efficacy of MAS for Rht24 and to investigate allelic distributions.
Field Trials and Phenotype Evaluation
The RILs and parents were planted at Gaoyi in Hebei province and Anyang in Henan during the 2014–2015 cropping season, and at Gaoyi during 2015–2016 cropping season. The experimental design was randomized complete blocks and three replications. Each plot was a single 2 m row with 25 cm between rows.
Set I varieties were planted at Anyang in Henan province and Suixi in Anhui, respectively, during the 2012–2013 and 2013–2014 cropping seasons. Set II varieties were grown at Shijiazhuang in Hebei and Beijing, respectively, during the 2012–2013 and 2013–2014 cropping seasons. These were grown in randomized complete blocks with three replications. Each plot consisted of four 2 m rows spaced 30 cm apart and approximately 50 plants in each row. Field management was according to local practice. During the whole wheat growth period in our field trial, there is no extreme weather causing serious damages, such as cold spell in later spring, dry and hot wind in filling stage, and fertilization (bottom fertilizer including 150 kg/h2 Urea plus 400 kg/h2 Diammonium Phosphate; 300 kg/h2 Urea just before elongation stage) is enough for wheat growth and development.
Plant height was measured from the ground to spike (awns excluded) at grain-filling. For each plot, five representative primary tillers on different plants in the middle of each row were selected to measure the plant height, and the averaged value was used for subsequent analysis. TGW was determined by weighing triplicate 200 grain samples.
Genotypic Analysis Using Simple Sequence Repeat (SSR) Markers
Genomic DNA was isolated from young leaves using the modified CTAB method (Saghai-Maroof et al., 1984). Twenty-two SSR markers on chromosome 6AL were chosen for genetic analyses1 (Supplementary Table S7).
Polymerase chain reaction was performed in a 15 μl reaction system containing 7.5 μl of 2 × Taq PCR Mix (Tianwei Biotechnology Co., Ltd., Beijing2), 5 pmol of each primer and 100 ng of genomic DNA. Amplification was performed at 94°C for 5 min, followed by 35 cycles at 94°C for 20 s, 50–60°C (depending on specific primers) for 30 s and 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR products were separated in 6% polyacrylamide or 2% agarose gels.
Gene-Specific Marker Development by Genome Mining Approach
To confirm the target sequences of the SSR mentioned above, PCR products were purified using the TIANgel MIDI Purification Kit (Tiangen, Biotechnology Co., Ltd., Beijing3). The purified PCR products were ligated with cloning vector pEASY-T5 Zero and then transformed into Trans1-T1 competent cells by the heat shock method (TransGen Biotech Co., Ltd., Beijing4). At least three positive clones from each transformation were randomly selected and sequenced at Shanghai Sangon Biotech Co., Ltd5. The SSR target sequences were used as BLAST queries6 to identify desirable scaffolds. The genes of interest in the target scaffold were isolated and sequenced following the above procedure. Sequence sites polymorphic between the parents were identified by alignments with DNAMAN software7 for designing gene-specific markers. Cleaved amplified polymorphic sequences (CAPS) or derived CAPS (dCAPS) marker for each target gene was developed following Thiel et al. (2004).
SNP Chip-Based Screening
Wheat 660K SNP chip was developed by the Institute of Crop Science, Chinese Academy of Agricultural Sciences, synthesized by Affymetrix and commercially available at CapitalBio Corporation8. Recently, the chip has been efficiently used in our lab (Jin et al., 2016).
To identify more molecular markers linked to Rht24 the 660K SNP chip was used to test two parents and two contrasting bulks of the RIL population, comprising equal amounts of genomic DNA from 10 tall and 10 short RILs, respectively. Genotyping was performed at CapitalBio Technology Company9. SNPs polymorphic between parents and between two bulks were selected to develop CAPS markers.
Genetic Linkage Map Construction and Statistical Analysis
Quantitative trait locus IciMapping 4.010 was used for linkage map construction and QTL analysis using a LOD score threshold of 3.0 (Lincoln et al., 1992). Plant height was reformed with an algorithm SERiation and criterion SARF (sum of adjacent recombination frequencies). Variance analysis and t-test (Duncan method) were conducted with SAS 9.411. The broad-sense heritability (hB2) of the corresponding traits was calculated using the following formula:
where σ2g, σ2ge, and σ2ε were estimates of lines, line × environment interactions and residual error variances, respectively, and e and r represented the numbers of environments and replicates, respectively (Supplementary Table S6; Nyquist and Baker, 1991). T-test was used to perform the association analysis between Rht24 and plant height as well as TGW.
Results
Confirmation of Rht24
In our previous study, Rht24 was identified with LOD score of approximately 12, explaining 8.0–10.4% of the phenotypic variance; it was mapped within an 8.2 cM interval flanked by Xbarc103 and Xwmc256 (Li et al., 2015). To further verify Rht24, 22 SSR markers were selected from the targeted region of chromosome 6AL (Supplementary Table S7). Among them, only Xbarc103 and Xwmc256 were polymorphic between the parental lines and bulk lines built by 10 tall and 10 short lines (Supplementary Figure S1). Rht24 was located between the two markers, which were 19.46 cM apart. Although the genetic distance was greater than reported previously, the QTL for reduced plant height was confirmed.
Narrow-Down of Rht24 by the Genome Mining Approach
To confirm the chromosomal location of Rht24, we cloned and sequenced the differential fragments of Xbarc103 and Xwmc256 in the tall and short lines, respectively (Supplementary Figure S1). Sequencing results showed that they were consistent with the reported sequences12. Using the sequences as queries to BLAST against International Wheat Genome Sequence Consortium database13, a super scaffold of approximately 208 Mb (approximately 1200 genes) spanning Rht24 was identified (Figure 1). Twenty genes involved in plant morphogenesis and hormone metabolism were selected from the scaffold and sequenced (Supplementary Table S8). Polymorphisms between AK58 and JD8 were detected at loci TaGA3, TaFAR, and TaAP2 (Supplementary Figures S2A–C and Table S2). Based on these polymorphisms, three gene-specific CAPS or dCAPS markers were developed (Figures 2A–C and Supplementary Figures S2A–C). Upon genotyping the RIL, the location of Rht24 was narrowed to a 1.85 cM interval flanked by TaFAR and TaAP2 (Figure 3).
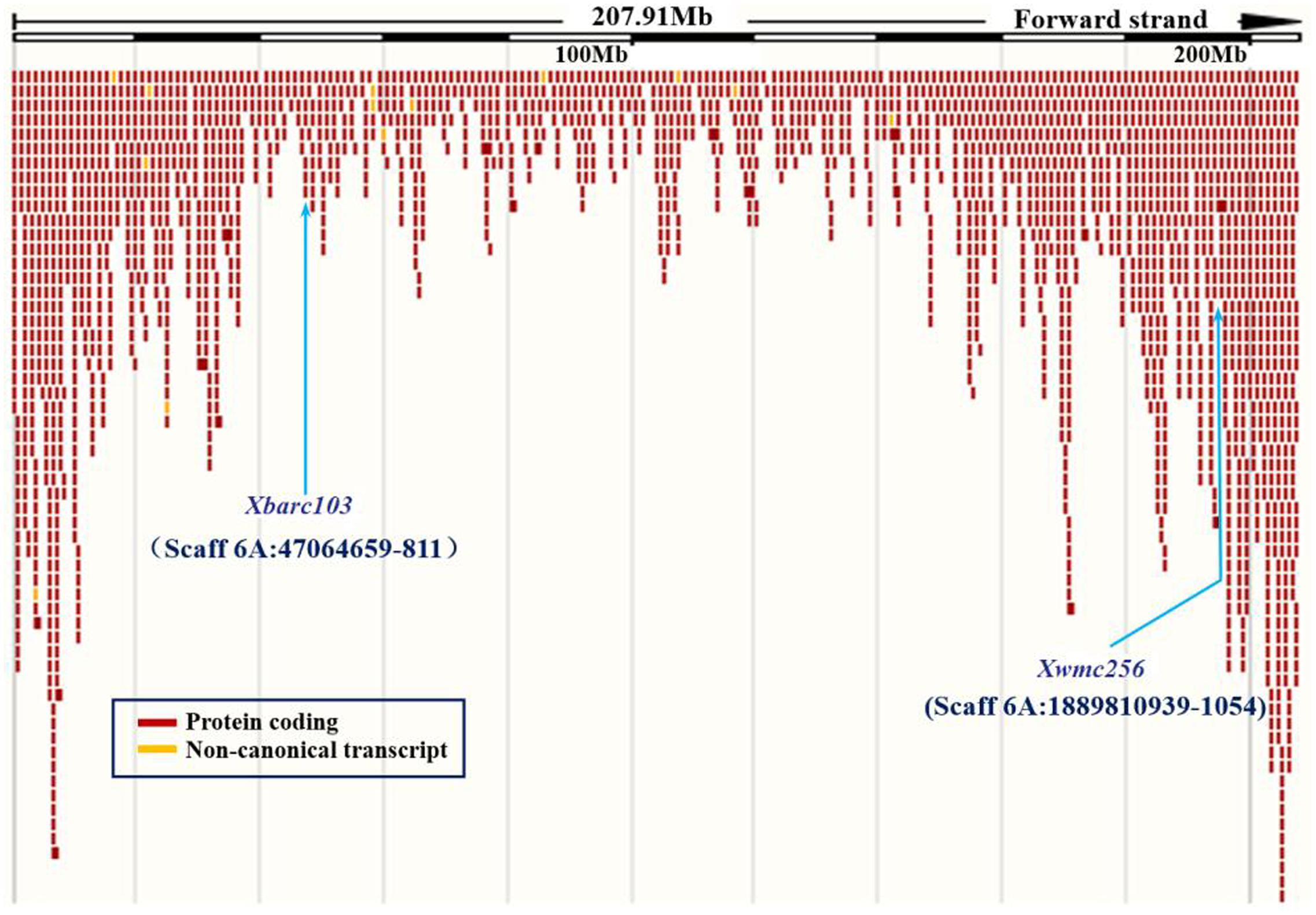
FIGURE 1. A super scaffold spanning Rht24. Locations of SSR loci Xbarc103 and Xwmc256 are shown by arrows.
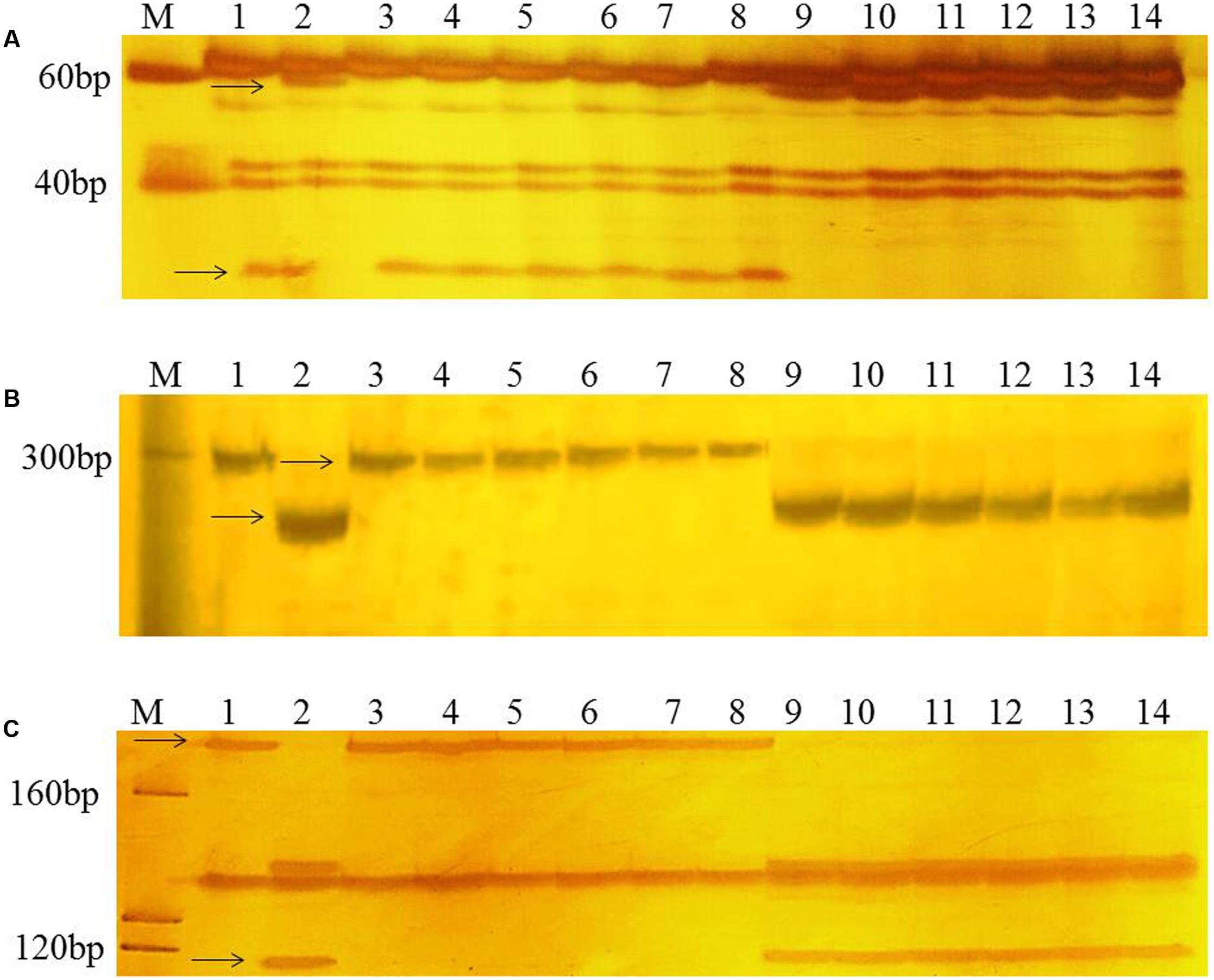
FIGURE 2. PCR patterns of polymorphic markers TaGA3 (A), TaFAR (B), and TaAP2 (C). Polymorphic fragments associated with height differences between short and tall RILs are shown by arrows. M, Marker (20 bp DNA ladder, Takara Bio Company). Lanes 1 and 2 are the parents, AK58 and JD8, respectively; lanes 3–8 are short RILs; lanes 9–14 are tall RILs.
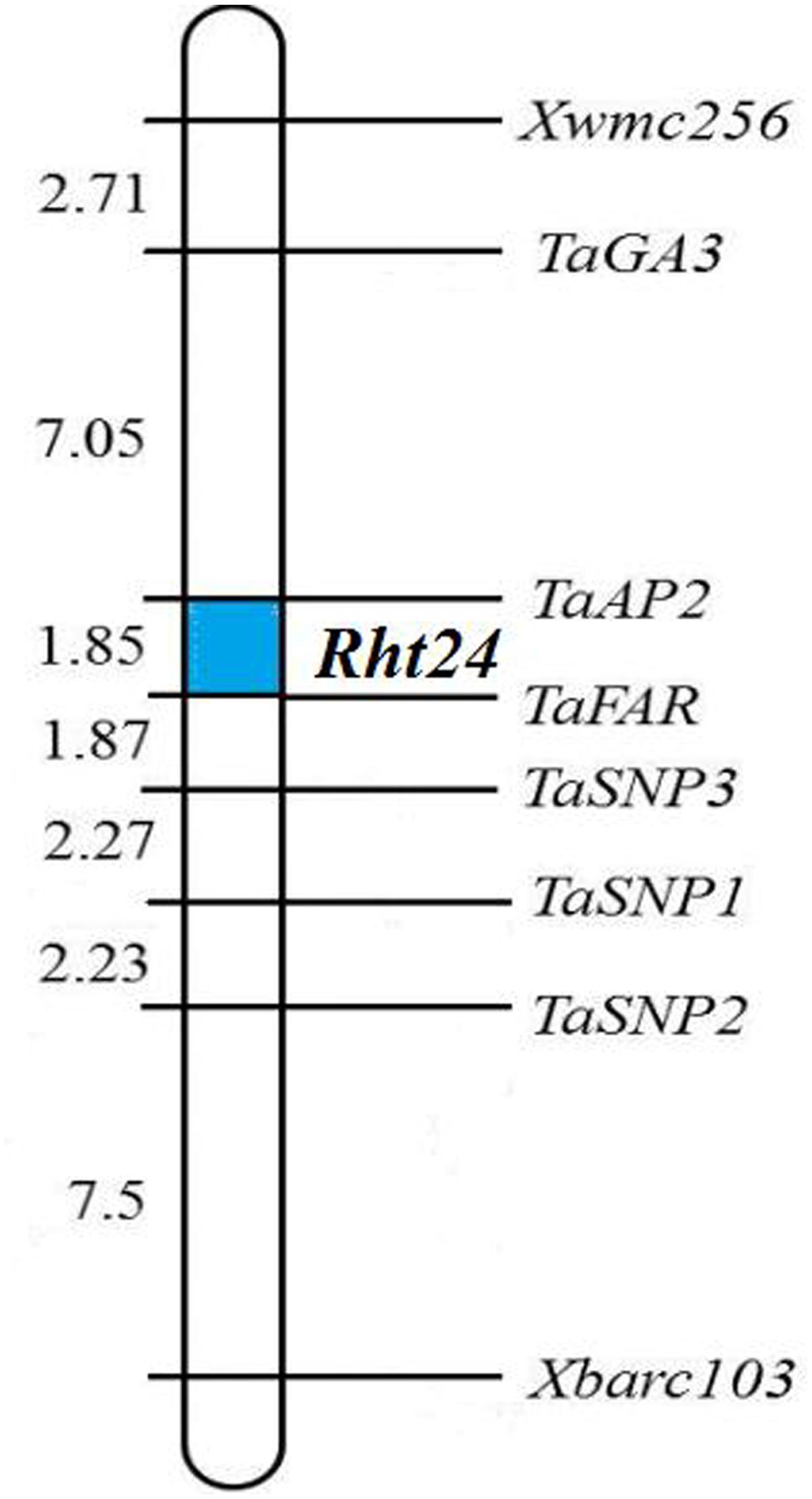
FIGURE 3. Linkage map of Rht24. The names and corresponding locations of all markers are indicated on the right side of the map. Genetic distance (cM) is shown on the left side of the map. The region of Rht24 is highlighted in blue.
Enriching Molecular Markers in the Rht24 Region by a Combination of SNP Genotyping Assays and Genomic Identification
To enrich molecular markers in the Rht24 region, the 660K SNP chip was used in genotyping assays. In total, more than 400 polymorphic SNPs on chromosome 6A from wheat 660K SNP chips were identified between two contrasting bulks (data not shown). Among them, the target sequences of polymorphic SNPs anchored in the super scaffold above were used to develop site-specific CAPS markers (Supplementary Table S9). Among them, three polymorphic loci, TaSNP1, TaSNP2, and TaSNP3, were mapped in the region of Rht24 (Supplementary Figures S3A–C, S4A–C, Table S2 and Figure 3). However, they did not further narrow down the Rht24 locus.
Validation of Molecular Markers Closely Linked to Rht24
Among 242 wheat varieties genotyped by markers TaFAR and TaAP2 (Supplementary Figures S5A,B), Rht24 was present in 185 (76.4%) varieties and absent in 41 (17%) (Supplementary Table S3). The average plant height for the varieties containing Rht24 was 78.2 and 80.2 cm in Sets I and II, respectively, whereas for those without Rht24 the comparative values were 86.1 and 86.2 cm, respectively, indicating that varieties with Rht24 were generally shorter than those without (PI = 0.029, PII = 0.003, Table 1). Rht24 was also significantly associated with thousand grain weight (TGW) in Set I (P = 0.014, Table 2). Although Rht24 had no significant association with TGW in Set II, the average TGW of entries with Rht24 was 2 g higher than those without (P = 0.196, Table 2). On average Rht24 reduced plant height by 6.0–7.9 cm and increased TGW by 2.0–3.4 g in all varieties used in this study (Tables 1, 2).
Discussion
Rht24 Is an Important Locus Affecting Plant Height and Grain Weight in Wheat
Previous studies showed that loci controlling plant height were present in all 21 wheat chromosomes (Mccartney et al., 2005; Gale and Law, 1973; Griffiths et al., 2012). The QTL QTL_height_6A_1 linked with Xwmc256 on chromosome 6AL explained 6.3–29.1% of the phenotypic variance (PVE) (Griffiths et al., 2012). Rht24, previously designated as QPH.caas-6A, was flanked by SSR markers Xwmc256 and Xbarc103 (Li et al., 2015), indicating that it could be the same as QTL_height_6A_1. Here we developed two markers, TaFAR and TaAP2 that closely flanked Rht24. The two markers were used to test the genetic effect of Rht24 and observed similar result with the previously reported (Table 3; Griffiths et al., 2012; Li et al., 2015). Rht24 genotyped by TaAP2 and TaFAR as markers showed a significant association with reduced height among elite varieties. It was present in about 76% of elite varieties indicating that it was positively selected in wheat breeding programs (Supplementary Table S3). In our previous study, Rht24 reduced TGW by 6.9–8.5%. However, Rht24 had a positive effect on thousand kernel weight in the present study (Table 2). We speculate that two reasons probably account for this case. The AK58/JD8 RIL and natural populations have better hereditary stability than the previous F2:4 mapping population and are used to define Rht24 with more accurate effect analyses. Additionally, the interval of Rht24 was narrowed down and thus more interference effects of other genes were excluded using the closest flanking markers, TaFAR and TaAP2. In fact, it still cannot be ruled out that other genes disturb the effect of Rht24 on TGW based on our existing mapping information. Thus it is necessary to further narrow down the interval of Rht24. Thus, Rht24 is an important QTL for plant height and TGW in wheat and MAS mediated by selection for appropriate TaAP2 and TaFAR alleles should be effective.
Distribution of Rht-B1b, Rht-D1b, and Rht8 in Chinese germplasm was detected by molecular markers, with frequencies of 24.5, 45.5, and 46.8%, respectively (Zhang et al., 2006). In addition, Rht-B1b and Rht-D1b were tested by KASP assays in current Chinese leading varieties with frequencies of 36.9 and 38.6%, respectively (Rasheed et al., 2016). Our study showed that Rht24 was present in about 76% of elite Chinese varieties, a much higher frequency than either Rht-B1b or Rht-D1b (Supplementary Table S3). The results indicated that Rht24 had the highest frequency distribution compared with the other three loci, and it was frequently present in combination with Rht-D1b or Rht8. It was reported that Rht8 not only to reduce plant height but also significantly increase TGW (Zhang et al., 2016), however, Rht-B1b and Rht-D1b showed negative effects on TGW (Li et al., 1998). In this study, Rht24 increased TGW by 2.0–3.4 g in all varieties used (Table 2).
In all, Rht24 was important and extensively used in Chinese wheat breeding programs. Further study is needed to understand the origin of Rht24 in Chinese wheat and investigate Rht24 effect on the other traits, such as tiller number, flowering date, spike length and so on.
Combination of SNP Chip-Based Screening and Genome Mining Was an Effective Approach for Fine Mapping of Rht24
As the development of genome sequencing technology, high-quality wheat genome sequencing data has been continuously updated and released14. Based on BLAST against wheat genome databases, the markers from preliminary mapping can be used as entry points to mine new markers linked to the loci of interest. In the present study we used SSR markers flanking Rht24 as queries and identified a super scaffold. According to gene annotations, we developed three gene-specific markers and narrowed the region of Rht24 from a 19.46 cM interval (8.2 cM in our previous study) to 1.8 cM. A 660K SNP chip was recently developed and released15. The 660K chip has several advantages compared to the 90K SNP chip, such as higher marker density and higher resolution as well as better distribution on chromosomes. We used this chip to screen candidate markers linked to Rht24 and a significant number of polymorphic SNPs were obtained (data not shown). It was an obviously effective approach to take advantage of constantly updated genome information and high-throughput SNP platforms.
Ethics Statement
We declare that these experiments comply with the ethical standards in China.
Author Contributions
XT, LX, and DX performed the experiments. XT, WW, LF, CF, DW, XC, and QC did the field trials. SC and ZH designed the experiment. XT, SC, XX, and ZH wrote the paper.
Funding
This study was supported by the National Key Research and Development Program of China (2016YFD0101802, 2016YFE0108600) and the National Natural Science Foundation of China (31161140346, 31461143021, 31371623).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors are grateful to Prof. R. A. McIntosh, Plant Breeding Institute, University of Sydney, for critical review of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01379/full#supplementary-material
Footnotes
- ^http://wheat.pw.usda.gov
- ^http://www.tw-biotech.com
- ^http://tiangen.casmart.com.cn
- ^http://www.transgen.com.cn
- ^http://www.sangon.com/
- ^http://plants.ensembl.org/Triticum_aestivum/Info/Index
- ^http://www.lynnon.com
- ^http://wheat.pw.usda.gov/ggpages/topics/Wheat660_SNP_array_developed_by_CAAS.pdf
- ^http://www.capitalbiotech.com
- ^http://www.isbreeding.net/
- ^http://www.sas.com
- ^http://wheat.pw.usda.gov
- ^http://plants.ensembl.org/Triticum_aestivum/Info/Index
- ^http://plants.ensembl.org/Triticum_aestivum
- ^http://wheat.pw.usda.gov/ggpages/topics/Wheat660_SNP_array_developed_by_CAAS.pdf
References
Asplund, L., Leino, M. W., and Hagenblad, J. (2012). Allelic variation at the Rht8 locus in a 19th century wheat collection. Sci. World J. 2012, 146–151. doi: 10.1100/2012/385610
Börner, A., Schumann, E., Fürste, A., Cöster, H., Leithold, B., Röder, S., et al. (2002). Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 105, 921–936. doi: 10.1007/s00122-002-0994-1
Buerstmayr, H., Ban, T., and Anderson, J. A. (2009). QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed. 128, 1–26. doi: 10.1111/j.1439-0523.2008.01550.x
Chen, C., He, Z. H., Lu, J. L., Li, J., Ren, Y., Ma, C. X., et al. (2016). Molecular mapping of stripe rust resistance gene YrJ22 in Chinese wheat cultivar Jimai 22. Mol. Breed. 36:118. doi: 10.1007/s11032-016-0540-5
Chen, S. L., Gao, R. H., Wang, H. Y., Wen, M. X., Xiao, J., Bian, N. F., et al. (2015). Characterization of a novel reduced height gene (Rht23) regulating panicle morphology and plant architecture in bread wheat. Euphytica 203, 583–594. doi: 10.1007/s10681-014-1275-1
Colasuonno, P., Gadaleta, A., Giancaspro, A., Nigro, D., Giove, S., Incerti, O., et al. (2014). Development of a high-density SNP-based linkage map and detection of yellow pigment content QTLs in durum wheat. Mol. Breed. 34, 1563–1578. doi: 10.1007/s11032-014-0183-3
Ellis, M., Spielmeyer, W., Gale, K., Rebetzke, G., and Richards, R. (2002). “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor. Appl. Genet. 105, 1038–1042. doi: 10.1007/s00122-002-1048-4
Ellis, M. H., Rebetzke, G. J., Azanza, F., Richards, R. A., and Spielmeyer, W. (2005). Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theor. Appl. Genet. 111, 423–430. doi: 10.1007/s00122-005-2008-6
Evans, L. T. (1998). Crop evolution, adaptation and yield. Photosynthetica 34, 56–60. doi: 10.1023/A:1006889901899
Gale, M. D., and Law, C. N. (1973). Semi-dwarf wheats induced by monosomy and associated changes in gibberellin levels. Nature 241, 211–212. doi: 10.1038/241211a0
Gao, L., Turner, M. K., Chao, S., Kolmer, J., and Anderson, J. A. (2016). Genome wide association study of seedling and adult plant leaf rust resistance in elite spring wheat breeding lines. PLoS ONE 11:e0148671. doi: 10.1371/journal.pone.0148671
Griffiths, S., Simmonds, J., Leverington, M., Wang, Y., Fish, L. J., Sayers, L., et al. (2012). Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Mol. Breed. 29, 159–171. doi: 10.1007/s00122-009-1046-x
Jin, H., Wen, W. E., Liu, J. D., Zhai, S. N., Zhang, Y., Yan, J., et al. (2016). Genome-wide QTL mapping for wheat processing quality parameters in a Gaocheng 8901/Zhoumai 16 recombinant inbred line population. Front. Plant Sci. 7:1032. doi: 10.1007/s00122-006-0346-7
Korzun, V., Röder, M. S., Ganal, M. W., Worland, A. J., and Law, C. N. (1998). Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 96, 1104–1109. doi: 10.1007/s001220050845
Law, C. N., Snape, J. W., and Worland, A. J. (1978). The genetic relationship between height and yield in wheat. Heredity 40, 15–20. doi: 10.1038/hdy.1978.13
Li, X. M., Xia, X. C., Xiao, Y. G., He, Z. H., Wang, D. S., Trethowan, R., et al. (2015). QTL mapping for plant height and yield components in common wheat under water-limited and full irrigation environments. Crop Pasture Sci. 66, 660–670. doi: 10.1071/CP14236
Li, X. P., Jiang, C. Z., and Liu, H. L. (1998). Effects of different dwarfing genes on agronomic characteristics of winter wheat. Acta Agron. Sin. 24, 475–478.
Lincoln, S. E., Daly, M. J., and Lander, E. S. (1992). Constructing Genetic Maps with MapMaker/EXP3.0, 3rd Edn. Cambridge, MA: Whitehead Institute.
Liu, G., Xu, S. B., Ni, Z. F., Xie, C. J., Qin, D. D., Li, J., et al. (2011). Molecular dissection of plant height QTLs using recombinant inbred lines from hybrids between common wheat (Triticum aestivum L.) and spelt wheat (Triticum spelta L.). Chin. Sci. Bull. 56, 1897–1903. doi: 10.1007/s11434-011-4506-z
Liu, J. D., He, Z. H., Wu, L., Bai, B., Wen, W. E., Xie, C. J., et al. (2016). Genome-wide linkage mapping of QTL for black point reaction in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 129, 2179–2190. doi: 10.1007/s00122-016-2766-3
Mccartney, C. A., Somers, D. J., Humphreys, D. G., Lukow, O., Ames, N., Noll, J. S., et al. (2005). Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 × ‘AC Domain’. Genome 48, 870–883. doi: 10.1139/g05-055
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Morris, C., Appels, R., and Xia, X. C. (2015). Catalogue of Gene Symbols for Wheat: 2015–2016. Supplement. Available at: http://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2015.pdf
McIntosh, R. A., Hart, G. E., and Gale, M. D. (2013). Catalogue of Gene Symbols for Wheat: 2013-2014 Supplement. Available at: http://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2013-2014.pdf
Mir, R. R., Kumar, N., Jaiswal, V., Girdharwal, N., Prasad, M., Balyan, H. S., et al. (2012). Genetic dissection of grain weight in bread wheat through quantitative trait locus interval and association mapping. Mol. Breed. 29, 963–972. doi: 10.1007/s11032-011-9693-4
Naruoka, Y., Garland-Campbell, K. A., and Carter, A. H. (2015). Genome-wide association mapping for stripe rust (Puccinia striiformis f. sp. tritici) in US Pacific Northwest winter wheat (Triticum aestivum L.). Theor. Appl. Genet. 128, 1083–1101. doi: 10.1007/s00122-015-2492-2
Nyquist, W. E., and Baker, R. J. (1991). Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 10, 235–322. doi: 10.1080/07352689109382313
Pearce, S., Saville, R., Vaughan, S. P., Chandler, P. M., Wilhelm, E. P., Alkaff, N., et al. (2011). Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiol. 157, 1820–1831. doi: 10.1104/pp.111.183657
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Peng, J. H., Ronin, Y., and Fahima, T. (2003). Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. U.S.A. 100, 2489–2494. doi: 10.1073/pnas.252763199
Peng, Z. S., Li, X., Yang, Z. J., and Liao, M. L. (2011). A new reduced height gene found in the tetraploid semi-dwarf wheat landrace Aiganfanmai. Genet. Mol. Res. 10, 2349–2357. doi: 10.4238/2011
Quarrie, S. A., Steed, A., and Calestani, C. (2005). High-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring x SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor. Appl. Genet. 110, 865–880. doi: 10.1007/s00122-004-1902-7
Rasheed, A., Wen, W. E., Gao, F. M., Zhai, S. N., Jin, H., Liu, J. D., et al. (2016). Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 129, 1843–1860. doi: 10.1007/s00122-016-2743-x
Rebetzke, G. J., and Richards, R. A. (2000). Gibberellic acid-sensitive dwarfing genes reduce plant height to increase kernel number and grain yield of wheat. Crop Pasture Sci. 51, 235–246. doi: 10.1071/AR99043
Röder, M. S., Korzun, V., Wendehake, K., Plaschke, J., Tixier, M. H., Leroy, P., et al. (1998). A microsatellite map of wheat. Genetics 149, 2007–2023.
Rogers, J. (2014). “The IWGSC survey sequencing initiative,” in Proceedings of the International Plant and Animal Genome Conference XXII, San Diego, CA.
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 24, 8014–8018.
Sukumaran, S., Dreisigacker, S., Lopes, M., Chavez, P., and Reynolds, M. P. (2015). Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor. Appl. Genet. 128, 353–363. doi: 10.1007/s00122-014-2435-3
Thiel, T., Kota, R., Grosse, I., Stein, N., and Graner, A. (2004). SNP2CAPS: a SNP and indel analysis tool for CAPS marker development. Nucl. Acids Res. 32:e5. doi: 10.1093/nar/gnh006
Wang, S. C., Wong, D., Forrest, K., Allen, A. L., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using the high-density 90,000 SNP array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wei, T. M., Chang, X. P., Min, D. H., and Jing, R. L. (2010). Analysis of genetic diversity and tapping elite alleles for plant height in drought-tolerant wheat varieties. Acta Agron. Sin. 36, 895–904. doi: 10.3724/SP.J.1006.2010.00895
Worland, A. J., Korzun, V., Röder, M. S., Ganal, M. W., and Law, C. N. (1998). Genetic analysis of the dwarfing gene Rht8 in wheat. Part II. The distribution and adaptive significance of allelic variants at the Rht8 locus of wheat as revealed by microsatellite screening. Theor. Appl. Genet. 96, 1110–1120. doi: 10.1007/s001220050846
Wu, J., Kong, X., Wan, J., Liu, X., Zhang, X., Guo, X., et al. (2011). Dominant and pleiotropic effects of a GAI gene in wheat results from a lack of interaction between DELLA and GID1. Plant Physiol. 157, 2120–2130. doi: 10.1104/pp.111.185272
Wu, Q. H., Chen, Y. G., Fu, L., Zhou, S. H., Chen, J. J., Zhao, X. J., et al. (2016). QTL mapping of flag leaf traits in common wheat using an integrated high-density SSR and SNP genetic linkage map. Euphytica 208, 337–351. doi: 10.1007/s10681-015-1603-0
Würschum, T., Langer, S. M., and Longin, C. F. (2015). Genetic control of plant height in European winter wheat cultivars. Theor. Appl. Genet. 128, 865–874. doi: 10.1007/s00122-015-2476-2
Zhai, S. N., He, Z. H., Wen, W. E., Jin, H., Liu, J. D., Zhang, Y., et al. (2016). Genome-wide linkage mapping of flour color-related traits and polyphenol oxidase activity in common wheat. Theor. Appl. Genet. 129, 377–394. doi: 10.1007/s00122-015-2634-6
Zhang, D. Q., Song, X. P., Feng, J., Ma, W. J., Wu, B. J., Zhang, C. L., et al. (2016). Detection of dwarf genes Rht-B1b, Rht-D1b and Rht8 in huang-huai valley winter wheat areas and the influence on agronomic characteristics. J. Triticeae Crops 36, 975–981. doi: 10.7606/j.ssn.1009-1041.2016.08.01
Keywords: CAPS marker, genome mining, Rht24, SNP chip, Triticum aestivum
Citation: Tian X, Wen W, Xie L, Fu L, Xu D, Fu C, Wang D, Chen X, Xia X, Chen Q, He Z and Cao S (2017) Molecular Mapping of Reduced Plant Height Gene Rht24 in Bread Wheat. Front. Plant Sci. 8:1379. doi: 10.3389/fpls.2017.01379
Received: 14 January 2017; Accepted: 24 July 2017;
Published: 08 August 2017.
Edited by:
Agata Gadaleta, Università Degli Studi di Bari Aldo Moro, ItalyReviewed by:
Elisabetta Mazzucotelli, Consiglio per la Ricerca in Agricoltura e l’Analisi dell’Economia Agraria (CREA), ItalyInes Yacoubi, Centre of Biotechnology of Sfax, Tunisia
Copyright © 2017 Tian, Wen, Xie, Fu, Xu, Fu, Wang, Chen, Xia, Chen, He and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuanghe Cao, shcao8@163.com Zhonghu He, zhhecaas@163.com
 Xiuling Tian
Xiuling Tian Weie Wen1,2
Weie Wen1,2 Xianchun Xia
Xianchun Xia Zhonghu He
Zhonghu He Shuanghe Cao
Shuanghe Cao