
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 September 2017
Sec. Plant Breeding
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01593
 Aman Kumar1
Aman Kumar1 Monika Garg1*
Monika Garg1* Navneet Kaur1
Navneet Kaur1 Venkatesh Chunduri1
Venkatesh Chunduri1 Saloni Sharma1
Saloni Sharma1 Swati Misser1
Swati Misser1 Ashish Kumar1
Ashish Kumar1 Hisashi Tsujimoto2
Hisashi Tsujimoto2 Quan-Wen Dou3
Quan-Wen Dou3 Raj K. Gupta4
Raj K. Gupta4The protein content and its type are principal factors affecting wheat (Triticum aestivum) end product quality. Among the wheat proteins, glutenin proteins, especially, high molecular weight glutenin subunits (HMW-GS) are major determinants of processing quality. Wheat and its primary gene pool have limited variation in terms of HMW-GS alleles. Wild relatives of wheat are an important source of genetic variation. For improvement of wheat processing quality its wild relative Thinopyrum elongatum with significant potential was utilized. An attempt was made to replace Th. elongatum chromosome long arm (1EL) carrying HMW-GS genes related to high dough strength with chromosome 1AL of wheat with least or negative effect on dough strength while retaining the chromosomes 1DL and 1BL with a positive effect on bread making quality. To create chromosome specific translocation line [1EL(1AS)], double monosomic of chromosomes 1E and 1A were created and further crossed with different cultivars and homoeologous pairing suppressor mutant line PhI. The primary selection was based upon glutenin and gliadin protein profiles, followed by sequential genomic in situ hybridization (GISH) and fluorescent in situ hybridization (FISH). These steps significantly reduced time, efforts, and economic cost in the generation of translocation line. In order to assess the effect of translocation on wheat quality, background recovery was carried out by backcrossing with recurrent parent for several generations and then selfing while selecting in each generation. Good recovery of parent background indicated the development of almost near isogenic line (NIL). Morphologically also translocation line was similar to recipient cultivar N61 that was further confirmed by seed storage protein profiles, RP-HPLC and scanning electron microscopy. The processing quality characteristics of translocation line (BC4F6) indicated significant improvement in the gluten performance index (GPI), dough mixing properties, dough strength, and extensibility. Our work aims to address the challenge of limited genetic diversity especially at chromosome 1A HMW-GS locus. We report successful development of chromosome 1A specific translocation line of Th. elongatum in wheat with improved dough strength.
Wheat is one of the major cereal food crops with worldwide consumption. More than 700 million metric tons of wheat have been produced globally each year since 2013 which means it accounts for more than 30% of total cereal production and provides almost 20% of total calorific input to the world (USDA/ARS, 2017). Wheat (2n = 6X = 42) contains three closely related genomes: A, B, and D that are derived from three different diploid species (Feldman and Levy, 2015). Wheat flour protein combination distinguishes it from other cereals as it confers properties of viscoelasticity and extensibility to the dough (Wang et al., 2013). Gliadins and glutenins are two main components of seed storage proteins that build the gluten polymer and determine its properties (Shewry, 2009). Gliadins are monomeric proteins that impart extensibility to the dough. These can be categorized into α/β, γ, and ω gliadins. Glutenins are polymeric proteins that impart dough its viscoelasticity. Glutenins can be classified into high molecular weight glutenin subunits (HMW-GS) and low molecular weight glutenin subunits (LMW-GS). HMW-GS confer dough strength while the LMW-GS are responsible for dough extensibility (Wang et al., 2013). The loci encoding HMW-GS and LMW-GS are located on the long and short arms, respectively, of homoeologous group 1 chromosomes: 1A, 1B, and 1D (Sreeramulu and Singh, 1997). Each locus produces two subunits of different sizes, larger x-type and smaller y-type subunits. The variation in different HMW-GS is due to variation in size and number of cysteine residues (Shewry and Tatham, 1997). Subunits 1Bx, 1Dx, and 1Dy are expressed in all wheat cultivars while 1By and 1Ax subunits are expressed in some wheat cultivars. Different alleles of these subunits have different impact on end product quality and have accordingly been given different quality scores (Payne and Lawrence, 1983). 1Dx5+1Dy10 subunits from D genome and 1Bx17+1By18, 1Bx13+1By16, and 1Bx14+1By15 from B genome are associated positively with bread making quality as compared to other corresponding alleles (Mills et al., 2000; Branlard et al., 2001; Tohver, 2007). The gene coding 1Ay subunit generally remains silent in most of the wheat cultivars (Rasheed et al., 2014). Chromosome 1A (Glu-A1) is reported to be the least contributor (Zhang et al., 2016) and may affect processing quality negatively in some backgrounds (Garg et al., 2007). The Glu-A1 loci thus can be targeted for introgression of new alleles from different genetic resources. Wild species of wheat have rich HMW-GS variations (Wang et al., 2013) and may provide potential quality gene sources for this purpose. Being a polyploid, wheat has a highly buffered genome that is tolerant to alien introgressions. Wheat has been targeted for improvement in yield, disease resistance, abiotic stress tolerance, nutrition, and processing quality (Wulff and Moscou, 2014). To accomplish sufficient yield and resistance to diseases, many economically important genes have been transferred to wheat through chromosome translocations from many different relative genera (Feldman and Levy, 2015; Ma et al., 2015) such as, resistance genes Pm53, Yr40, and Yr42 from Aegilops spp. (Kuraparthy et al., 2009; Marais et al., 2009; Petersen et al., 2015), Pm21 from Haynaldia villosa (Chen et al., 1995), Pm40 and Yr50 from Thinopyrum (Luo et al., 2009; Liu et al., 2013) and YrSn0096 from Leymus mollis (Bao et al., 2012). Secale cereal (rye) specific translocations have been used to transfer both yield and resistance genes (Yr9, Pm8, Lr26, Sr31) into wheat (Mago et al., 2005; Ren et al., 2009, 2017). The rye-wheat 1RS.1BL translocation was used worldwide in wheat breeding programs and is well distributed as cultivars across the world (Rabinovich, 1998). It has substantially contributed to yield enhancement, productivity increase, and has affected agricultural economy (Rabinovich, 1998). Despite their wide use in conferring resistance traits to wheat cultivars not too many translocation lines have been developed for quality traits such as, dough strength and extensibility. T1BL.1RS is known to have a negative impact on bread making quality (Kumlay et al., 2003).
Thinopyrum elongatum (syn. Agropyron elongatum) also known as tall wheat grass belongs to family Poaceae. It has a diploid E genome (2n = 2X = 14) that can be utilized for increasing the genetic diversity of HMW-GS genes (Chen et al., 2013; Royal Botanic Gardens Kew, 2017). Th. elongatum has been used to transfer genes for salinity, drought, and disease resistance to wheat in many breeding programs (Dewey, 1960, 1962; Sharma and Gill, 1983; Colmer et al., 2006). Yield related genes from Thinopyrum have also been transferred in form 7DL.7Ag translocation (Monneveux et al., 2003). Wheat-Th. elongatum translocations have been used for conferring resistance to leaf rust and stem rust (Friebe et al., 1994). The disomic addition line of chromosome 1E (DAL1E) of Th. elongatum in Chinese spring (CS) wheat cultivar background have been reported to possess superior bread making quality (Garg et al., 2009a). The presence of whole alien chromosomes may result in reduced yield and the line may be genetically unstable (Zamir, 2001). Translocation lines are appropriate alternative to DALs/DSLs for introgression of wild alleles. Our previous work on Th. elongatum and Aegilops geniculata substitution lines have indicated that positive processing quality potential of wild species can be best utilized if translocation is carried out by replacing chromosome 1A specific HMW-GS rather than those coded by 1B and 1D (Garg et al., 2009b, 2016). The current study was aimed at replacing Glu-A1 loci with better quality genes from wheat related species. We developed chromosome 1A specific translocation line of Th. elongatum in wheat background. The effects of this translocation on processing quality were studied.
The plant materials utilized in this study included (1) disomic addition line of Th. elongatum chromosome 1E and disomic substitution line of Th. elongatum chromosome 1E with chromosome 1D of wheat, both in the background of Chinese Spring (CS) {DSL-1E(1D)}, (2) genetic stocks nulli for chromosome 1A and tetra for chromosome 1D (N1AT1D), N1AT1B, N1BT1A, N1BT1D, and N1DT1B in the background of CS, and (3) four cultivars: AcDomain-hard Canadian, Norin 61 (N61)-soft Japanese, PBW343-hard Indian, Cham 6-hard Syrian.
Search for chromosome specific protein markers was accomplished from electrophoretic patterns of glutenin and gliadin proteins of Th. elongatum addition and substitution lines, and nulli tetra genetic socks of CS. For creation of translocation line, the DSL-1E(1D) was reciprocally crossed with N1AT1D to create double monosomics for chromosome 1E and 1A. The F1 generation was crossed/backcrossed with cultivars N61, CHAM6, PBW343, AcDomain, and with PhI mutant line (promote homoeologous pairing and recombination). Selection was based upon presence of HMW-GS and absence gliadins from Th. elongatum and absence of 1AS coded gliadins from wheat. Confirmation of translocation was carried out by GISH analysis. Plants with high vigor were selected and additional cycles of crossing/backcrossing and selfing were carried out.
Seed storage proteins were sequentially extracted according to Smith and Payne (1984) with modifications. Initially gliadin extraction was accomplished with 1.5 M DMF (dimethyl formamide), followed by glutenin extraction with 50% Isopropanol, 50 mM TrisHCl (pH 7.5), 2% DTT. Separation of glutenins was carried out on 10% polyacrylamide gel. Gliadins were separated on 15% polyacrylamide gels.
RP-HPLC of separated gliadin and glutenin proteins was carried out according to Mejias et al. (2014) with some modifications. The separation of proteins was carried out using C8 reversed phase analytical column Zorbax 300SB-C8 (Agilent Technologies) with 5 μm particle size and 300 Å particle diameter, 250 mm length, 4.6 mm internal diameter of Agilent 1260 Infinity Quaternary liquid chromatography system. The detector was diode array UV-vis detector and absorbance was detected at 210 nm. Injection volume was 20 μl. Linear gradient was set up using solvents A (0.1% TFA in MQ) and B (0.1% TFA in Acetonitrile) and column temperature was set up at 60°C. Glutenins were separated at a linear gradient 25–44% B for 120 min, 44–50% B for 5 min, and finally 50–65% B for 5 min. Gliadins were separated from 20 to 65% B for 120 min and then 65 to 75% B for 20 min.
Background screening of the translocation line was done by simple sequence repeat (SSR) markers. Total 536 deletion bin based primers (Sourdille et al., 2004; Carollo et al., 2005) were synthesized and used for background screening of BC4F3 plants (Supplementary Datasheet). N61, CS and 1E(1D) were used as controls. The genomic DNA of the selected samples was isolated using CTAB (Cetyl trimethyl ammonium bromide) method (Doyle and Doyle, 1987). The amplification was carried out in 20 μl of reaction mixture containing genomic DNA, PCR Mastermix (Thermo Fisher Scientific), and 0.4 μM primers using a thermal cycler (Eppendorf). Thermal cycling program involved an initial denaturation at 95°C for 5 min, followed by 32 cycles of denaturation at 95°C for 60 s, annealing at 5°C below the Tm of respective primers for 30 s, primer extension at 72°C for 30 s, followed by a final extension at 72°C for 10 min. The amplified PCR products were size fractioned by electrophoresis on 2.5% agarose gel and/or 10% polyacrylamide gel and visualized by staining with ethidium bromide (0.5 μg/ml) in a gel documentation system (Biospectrum UVP).
Developing seeds of N61 and translocation line were collected at four different stages (7DAA, 18DAA, 25DAA, and 28DAA). For SEM, seeds were embedded in the optimal cutting temperature (OCT) media (Leica biosystems) and kept for freezing for half an hour. Then the cryosectioning of the embedded seeds was carried out at −20°C in a cryomicrotome (Leica CM1950 cryostat). For carrying out scanning electron microscopy, the cryosections of 40 μm thickness were collected on the adhesive carbon strip and coated with gold particles. After coating, the samples were mounted on the scanning electron microscope and observed. For the mature seeds, instead of performing cryosectioning, the seeds were cracked under liquid nitrogen. The cracked seeds were then adhered onto the adhesive carbon strip. The rest of the procedure was same as in the case of the developing seeds.
The Acetocarmine squash method (Tsuchiya and Nakamura, 1979) was utilized for the preparation of mitotic chromosome spreads. Th. elongatum genomic DNA was isolated by CTAB method (Doyle and Doyle, 1987). Genomic in situ hybridization (GISH) and fluorescent in situ hybridization (FISH) were carried out for screening and confirmation of translocation line. Total genomic DNA of Th. elongatum was labeled with fluorescein-12-dUTP (Roche) or tetramethyl-rhodamine-5-dUTP (Roche) by random primer labeling method and was used as a probe to identify the translocation (GISH). pAS1 and AAG sequences labeled with cyanine (Cy3) and 6-fluorescein amidite (6-FAM). respectively, were used as probes for FISH to identify specific chromosomes. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Roche) and observed under the fluorescence microscope.
The moisture, protein content, dry gluten, and wet gluten of milled flour were determined according to standard American Association of Cereal Chemists (AACC, 1990) methods. Sodium dodecyl sulfate sedimentation (SDSS) values were measured on a small scale using 1 g of flour by the method of Takata et al. (1999). Dough extensibility tests (Totosaus et al., 2013) were performed on texture analyzer (Stable Microsystems) using Kieffer dough and gluten extensibility rig. The values of peak positive force, stretching distance, area to positive peak and force at the target were calculated for different dough samples. Solvent retention capacity (SRC) tests were performed according to AACC International Method 56-11.02 for deionized water, sucrose solution (50% w/w), sodium carbonate solution (5%w/w), and lactic acid solution (5%w/w). To determine mixing properties of flour a 10 g mixograph (National Mfg. Co., Lincoln, NE, USA) was used. Mixing tests were performed in triplicates using AACC method 54-40A. Results were analyzed using MixSmart software (AEW Consulting, Lincoln, NE, USA).
To determine the level of significance of different parameters, analysis of variance (ANOVA) was performed using the stat view program.
For generation of chromosome specific translocation line Th. elongatum substitution line DSL-1E(1D) and N1AT1D were utilized. As both are in the genetic background of CS, their glutenin, and gliadin seed storage protein electrophoretic profiles were studied for identification of protein markers (Supplementary Figure 1). Based on glutenin profile Th. elongatum specific HMW-GS were identified and further utilized as markers for screening of 1EL chromosome. From gliadin profile Th. elongatum specific gliadins were identified and utilized as 1ES specific marker. The HMW-GS profiling of CS indicated that it has a null allele at Glu-A1 locus. Therefore, LMW glutenin and gliadin profiles were explored for chromosome 1A specific protein subunits by utilizing its nullitetra stocks N1AT1B, N1AT1D, N1BT1A, N1BT1D, and N1DT1B. One, chromosome 1AS specific gliadin was identified and used as a protein marker for screening IAS chromosome (Supplementary Figure 2).
For a generation of translocation line the DSL-1E (1D) was reciprocally crossed with N1AT1D (Figure 1). The F1 was double monosomic for 1E and 1A and was supposed to produce a translocation line at very low frequency during next meiosis. The F1 plants were selfed and crossed with cultivars N61, Cham6, PBW343, AcDomain. The endosperm halves of selfed (F2 of N1ATID cross) and F1generations from crosses with cultivars were analyzed for HMW-GS and gliadins profile for the screening of translocation line (Supplementary Figure 3). Out of 243 analyzed seeds 108 carried Th. elongatum HMW-GS. Expression of CS specific chromosome 1A coded gliadins was not observed in 87 of total analyzed seeds. Total 21 seeds which showed expression of Th. elongatum HMW-GS and absence of CS specific chromosome 1A coded gliadins were selected for further study. Embryo halves of these seeds were sown in petri plates, their two roots were collected and the rest of the seedling was sown in the pot to raise plant. Collected roots were fixed and analyzed by GISH with genomic DNA of Th. elongatum, AAG and pAS1 sequences. Nineteen seeds including double monosomics and disomic substitution 1E(1A) were selected. Selection included six plants from AcDomain cross, two from Cham 6 cross, two from Norin61 cross, showing monosomic substitution (double monosomics), and nine from self (F2) showing disomic substitution. No translocations could be identified at this stage. We could recover two healthy plants with healthy spikes from AcDomain cross, two from self and one from Norin61 cross. As we could not identify any translocation line, to increase the chance of homoeologous recombination, these plants were crossed with PhI positive line. These plants were also crossed with AcDomain, N61 depending on the cross. Total 174 BC1/F1 seeds were screened for HMW-GS and gliadin profiles from the endosperm half while embryo part was used to get roots for GISH/FISH and raise the plants. Out of 76 seeds analyzed from AcDomain (BC1), four seeds having HMW-GS of Th. elongatum and missing gliadins of Th. elongatum were selected and used for further studies (Table 1). Similarly six out of 88 analyzed F1 seed from PhI positive line cross from the initial AcDomain crosswere selected. We could select only three seeds out of 20 analyzed seeds from N61 cross (BC1). These 13 selected seeds were checked by GISH analysis. Out of this one BC1 seed from N61 cross and one F1seed from PhI positive line cross from AcDomain cross were found to be translocation lines with 1EL from Th. elongatum and 1AS from wheat. Apart from translocation, we observed broken chromosomes, and missing Th. elongatum chromosomes in other 11 seeds. Translocation line development frequency was 1.1% from AcDomain/PhI cross and 5% from N61 backcross (Table 1). But screening efficiency by GISH was increased to 16.7% in AcDomain/PhI cross and 33.3% in N61 backcross after first screening by protein markers. These selected seeds were grown and further backcrossed with N61 or AcDomain depending on the cross, and selected by HMW-GS profile. AcDomain/PhI cross specific translocation line could not be carried forward due to poor plant growth and seed setting. The PhI selfed progeny could not produce viable plants and could not be used for screening of translocation line, thus only one translocation from N61 cross was selected. N61 cross specific translocation line was backcrossed four times and selfed. HMW-GS specific marker was used in each generation. Homozygous line was selected in the F3 generation by screening at least 10 seeds from individual F2 plants. One healthy F2 plant with all seeds showing 1E specific HMW-GS was considered homozygous. Individual F3 plants raised from these seeds were again checked for homozygosity and one healthy plant was selected for generation advancement and selfed until F6 generation. These BC4F6 seeds were used for further analysis.
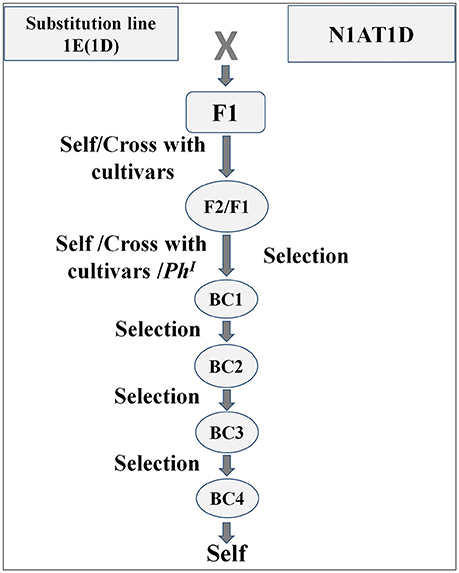
Figure 1. Schematic representation of crossing scheme involved in the generation of chromosome specific translocation line [1EL (1AS)]. Double monosomics were created by carefully selecting the breeding material and screening was carried out by combination of protein markers and genomic in situ hybridization. Multiple rounds of backcrossing were carried out to recover the background of recipient cultivar. N1AT1D = Genetic stock nullisomic for chromosome 1A and tetrasomic for chromosome 1D.
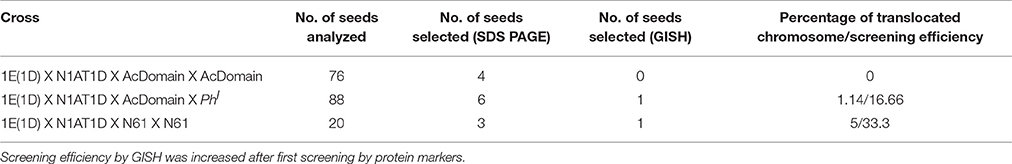
Table 1. Frequency of generation of chromosome specific translocation [1EL(1AS)] in different crosses.
The seed storage protein profile via SDS-PAGE indicated that DTL-1EL(1AS) had six HMW-GSs, four from N61 (1Dx2.2, 1Dy12, 1Bx7, 1By8), and two from Th. elongatum (1Ex and 1Ey; Figure 2). Th. elongatum specific 1Ex band was located between 1Bx7 and 1By8. The 1Ey band was located in the LMW glutenin region. Expression of 1Ax2* observed in N61 was not found in DTL-1EL(1AS). No differences were observed in the gliadin pattern of parent cultivar N61 and translocation line 1EL(1AS).
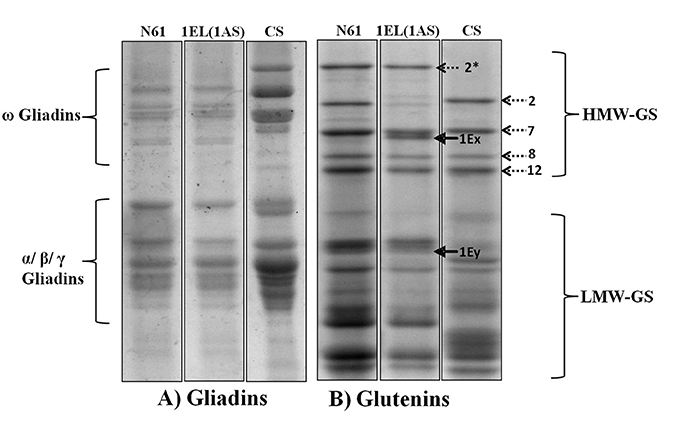
Figure 2. Seed storage protein electrophoretic profile of N61, CS, and DTL-(1EL1AS) (A) Gliadins profile; (B) Glutenins profile; arrow showing Th. elongatum specific protein band (1Ex). Electrophoretic profile indicated replacement of N61 chromosome 1AL coded HMW-GS with those of Th. elongatum. No difference was observed in the gliadin protein profiles of N61 and DTL-(1EL1AS).
RP-HPLC of glutenin fraction was done using a linear gradient (Figure 3). The proteins separated between RT-values 30–105 min. Total 40 major peaks were observed in the chromatogram. HMW-GS specific peaks could be detected between RT-values of 30–62 min. In HMW specific region 14 peaks were identified. There was one additional Th. elongatum specific peak in the glutenin fraction of translocation line 1EL(1AS) in the HMW GS region between peaks 7 and 8. Second peak was overlapping with peak 10. LMW-GS specific peaks could be detected between RT-values of 62–105 min. A total number of 26 peaks were identified in the LMW region. No differences were observed in LMW-GS specific region of glutenin fraction of translocation line and recipient cultivar N61.
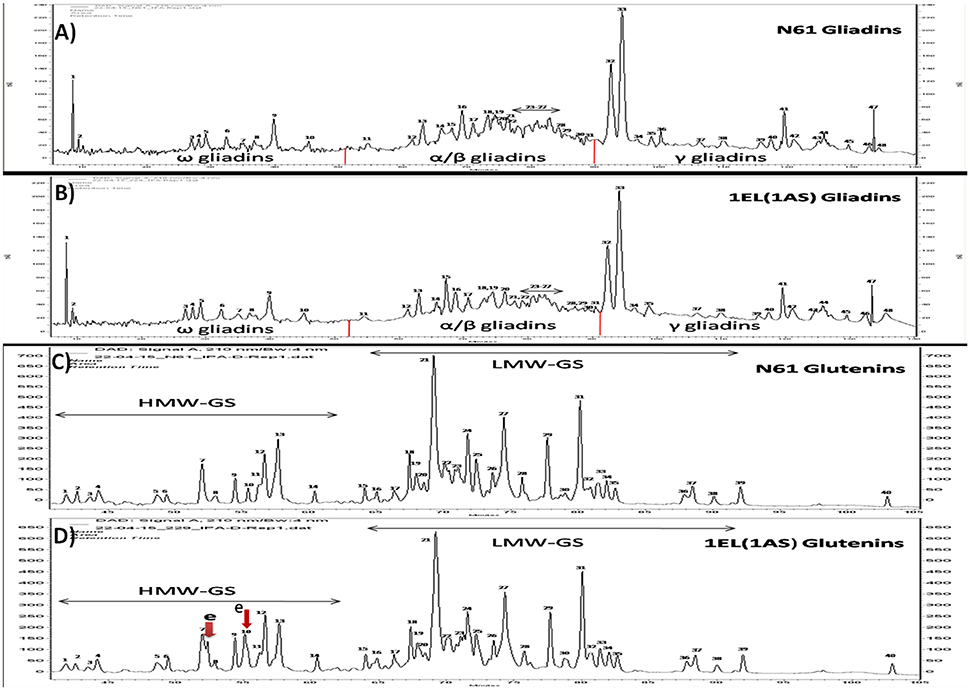
Figure 3. RP-HPLC analysis of gliadins fraction of (A) N61, (B) DTL-1EL(1AS) and glutenin fractions of (C) N61, (D) DTL-1EL(1AS). Two additional peaks were observed in the HMW-GS part of the HPLC profile (indicated by red arrows). Gliadin peak patterns of N61 and translocation line were similar.
The separation of gliadins fraction occurred between RT-values of 7–140 min. Three regions could be identified in the chromatogram: ω gliadins specific region between RT-values of 7–52 min, α/β gliadins specific region between 53 and 91 min, and γ gliadin specific region between 92 and 140 min. Ten peaks were observed in ω gliadins region while 21 peaks could be identified in α/β gliadins region. Total 17 major peaks were detected in γ gliadins region. No differences were observed in gliadin fractions of both N61 and 1EL(1AS) translocation line.
Sequential GISH and FISH analysis was carried out to confirm the translocation. First GISH with Th. elongatum genomic DNA was carried out (Figure 4C) and then both Th. elongatum genomic DNA and GAA sequences were used (Figure 4B). It was followed by FISH with GAA and pAS1 probes (Figure 4A). GISH analysis using Th. elongatum genomic DNA as probe revealed that Th. elongatum chromosome was translocated to one of the wheat chromosomes. It showed presence of Robertsonian translocation i.e., a whole arm of wheat chromosome was replaced with that of Th. elongatum (Figures 4C,D). The long arm of Th. elongatum was transferred to short arm of one of wheat chromosomes. Identity of wheat chromosomes was revealed by pAS1 and GAA FISH probes. These sequences give specific banding signals on individual chromosomes which can be utilized to identify them. All the 14 D-genome chromosomes were identified with pAS1 signals (Figure 4A) indicating that D-genome was not involved in translocation. GAA sequences could identify all the 14 B-genome chromosomes so B-genome involvement was also ruled out. Only 12 A-genome chromosomes (2A to 7A) could be identified from the GAA pattern (Figure 4A). It demonstrated that the long arm of Th. elongatum chromosome was attached to short arm of chromosome 1A of wheat.
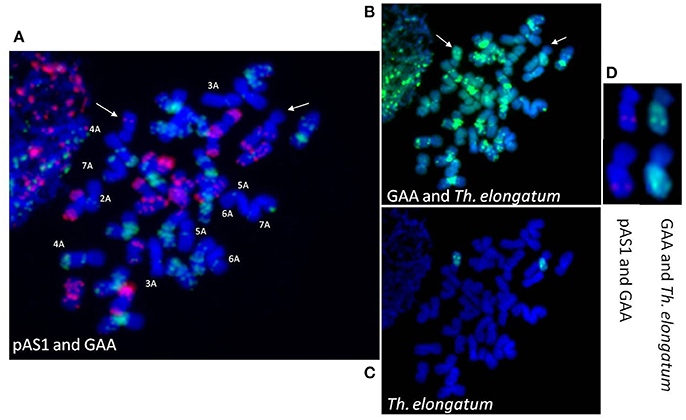
Figure 4. Genomic in situ hybridization for detection of Thinopyrum elongatum specific translocation in Triticum aestivum; (A) pAS1 (red) and GAA (green) signals for screening of individual wheat chromosomes; (B) GAA (green) and Th. elongatum (green marked by arrow) signals for screening of individual wheat chromosomes; (C) Th. elongatum specific signals (green) for screening of translocation line; (D) pAS1 (red), GAA (green), and Th. elongatum signals (green) detecting 1EL(1AS). Chromosomes were counterstained with DAPI (blue).
The BC4F3 derivatives of the confirmed translocation line in the background of Japanese cultivar Norin 61 were screened for background recovery. Total 536 SSR markers were utilized for this purpose. Out of 536, total 119 markers were found to be polymorphic. Background screening of the lines revealed that the translocation lines had mostly Norin 61 background (Figure 5; Supplementary Table 1). For chromosomes 1B, 1D, 2A, 2B, 2D all the polymorphic SSRs showed N61. CS pattern was observed in a very less number of cases; 3A, 3B, and 7B in one SSR each. Rest for all the chromosomes, N61 pattern was observed indicating good recovery of parent background. For chromosome 1A, total 41 SSR markers were used out of which 19 were found to be polymorphic. We observed N61 pattern using short arm region specific primers and Th. elongatum banding pattern using long arm specific primers (Figure 6). The markers which showed Th. elongatum pattern were WMC469, BARC17, WMC312, and CFA2219.
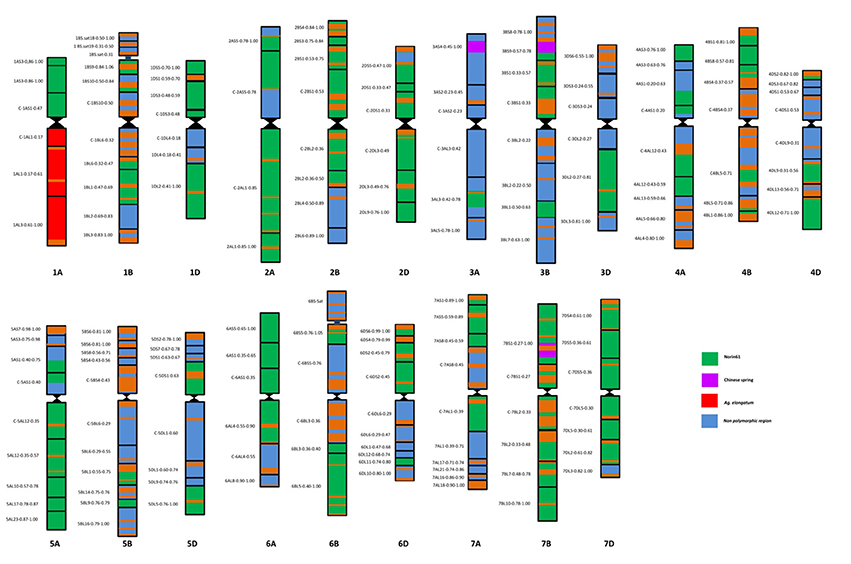
Figure 5. Schematic microsatellite marker based map of DTL-1EL (1AS) translocation lines; map showing N61 (green), CS (purple), and Th. elongatum (red) backgrounds in DTL-1EL(1AS). Good background recovery of N61 indicated generation of almost near isogenic line.
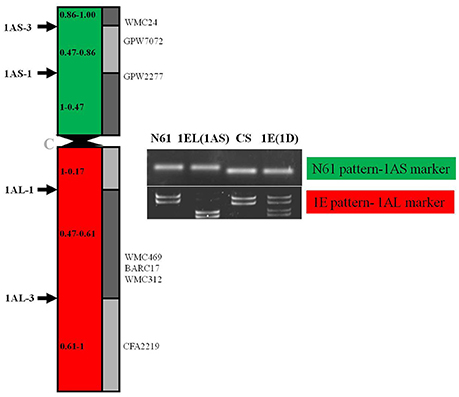
Figure 6. Schematic representation of translocated chromosome 1EL(1AS) in the DTL-1EL(1AS) to indicate number and position of SSR markers; the 1AS region showed N61 pattern, while long arm specific markers showed Th. elongatum specific pattern.
Morphologically the translocation line 1EL(1AS) plants were similar to parent N61 (Figure 7). We measured plant height, spike length, awn length, number of spikelets, flag leaf length, and width for N61 and DTL-1EL(1AS; Table 2). Spike length of DTL-1EL(1AS) was higher as compared to N61 but no differences were observed in a number of spikelets. There were no significant differences observed in plant height, awn length, flag leaf length, and width.
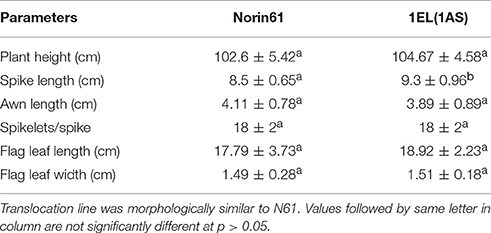
Table 2. Morphological data of translocation line (BC4F6-NIL) in comparison to recipient cultivar N61.
The scanning electron microscopy was performed on four different developmental stages (7 days after anthesis {DAA}, 18 DAA, 25 DAA, and 28 DAA) and mature seeds {Figures 8A–E for N61, Figures 8F–J for DTL-1EL(1AS)}. The results showed that the translocation line showed slow development at 7 DAA as compared to N61 (Figures 8A,F). But for the lateral developmental stages, there were no significant differences observed. Seed development of translocation line and N61 were similar at 18 DAA (Figures 8B,G), 25 DAA (Figures 8C,H), 28 DAA (Figures 8D,I). The SEM images of mature seeds, indicated similar packing of starch granules in translocation line and N61 (Figures 8E,J).
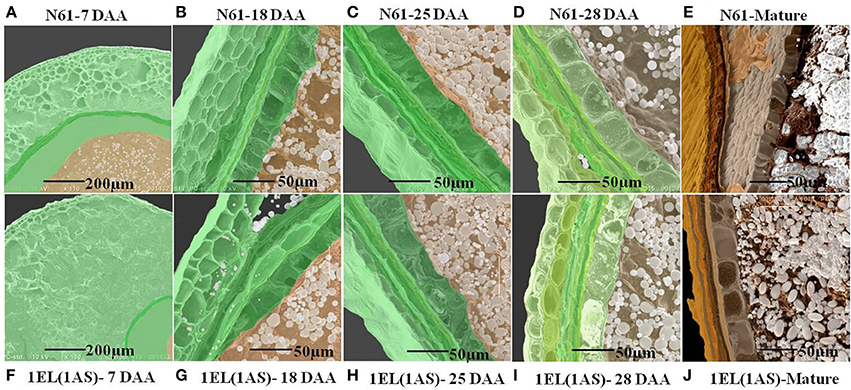
Figure 8. Scanning electron microscopy of different seed development stages starting from 7 days after anthesis (7 DAA) to 28 DAA of N61 and DTL-1EL(1AS). SEM images indicated that the translocation line showed slow development at 7 DAA as compared to N61. But for the later development stages there was no significant difference observed. (A) N61(7 DAA), (B) N61(18 DAA), (C) N61(25 DAA), (D) N61(28 DAA), (E) N61(mature seeds), (F) 1EL(1AS)(7 DAA), (G) 1EL(1AS) (18 DAA), (H) 1EL(1AS)(25 DAA), (I) 1EL(1AS) (28 DAA), (J) 1EL(1AS) (mature seeds).
We did not see any significant differences in total protein content between N61 and DTL-1EL(1AS) over a period of 3 years (2014–2016; Table 3). Similarly, dry gluten and wet gluten content remained similar in both N61 and DTL-1EL(1AS). However, variations were observed in values of different years, which might be contributing to different environmental conditions at different times. The values of grain hardness also remained similar for both N61 and DTL-1EL(1AS). The thousand kernel weight (TKW) for translocation line was less as compared to N61. The value of test weight remained same in both N61 and DTL-1EL(1AS). There was a significant increase in SDS sedimentation values for DTL-1EL (1AS) as compared to parent N61. Dough extensibility tests showed an increase in peak positive force, stretching distance and area to positive peak in the case of DTL-1EL(1AS) as compared to parent N61 (Table 4). The results were consistent for 2 years (2015, 2016). These results indicated improvement in dough extensibility properties of translocation line as compared to parent N61. Mixograph analysis also showed increases in midline peak time, midline peak value and midline peak integral in case of DTL-1EL(1AS) as compared to N61 (Table 4). There were no significant differences observed in values of midline peak width. Thus, overall mixing parameters for DTL-1EL (1AS) were shown to improve indicating improvement in dough strength. Solvent retention capacity test was carried out to confirm dough strength (Figure 9). In this test four different solutions of lactic acid, sucrose, water, and sodium carbonate were utilized for providing information on physical and chemical aspects of wheat samples. No difference was observed in the sodium carbonate retention capacity for translocation lines and parent N61 in the year 2015. The value increased in the year 2016. There was a significant increase in lactic acid SRC in DTL-1EL(1AS) in both years. Reverse trend was observed in case of sucrose SRC where the values decreased in DTL-1EL(1AS) in both years. There was an increase in MQ-SRC values of the translocation line in both years. The SRC values were used to calculate gluten performance index (GPI) of the flour. The results of SRC showed an increase in GPI of translocation line when compared to parent N61. Dough strength of translocation line was better than N61. Similar results were observed for both the studied years.
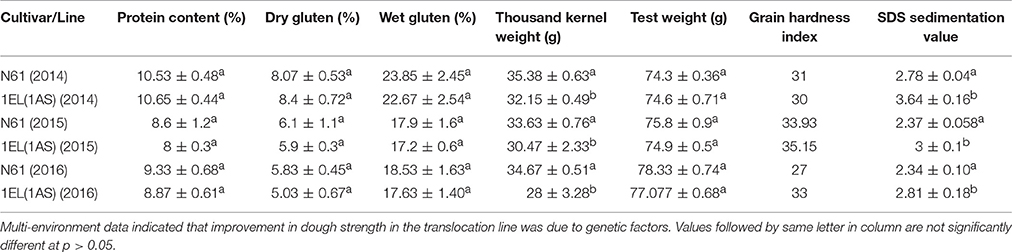
Table 3. Quality parameters across three years of translocation line in comparison to recipient cultivar.
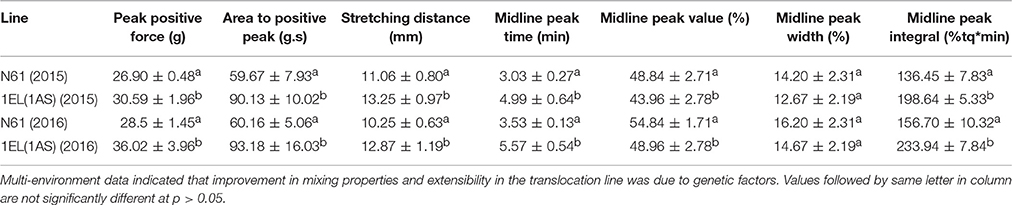
Table 4. Dough extensibility and mixing properties of translocation line in comparison to recipient cultivar.
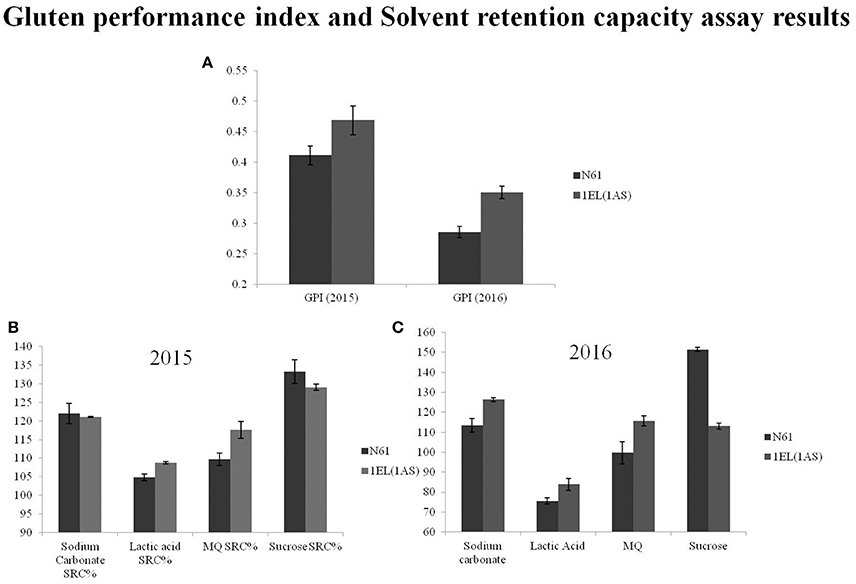
Figure 9. SRC data and GPI-values for Norin 61 and DTL-1EL(1AS); (A) Gluten performance index (GPI)-values for the year 2015 and 2016; (B) SRC-values for the year 2015; (C) SRC-values for the year 2016. SRC-values and GPI indicated improved dough strength in translocation line.
Wheat is one of the most important staple cereal crops consumed by billions across the world. Wide introgression in wheat to broaden its genetic base has resulted in significant improvement in productivity, disease resistance, and agricultural economy (Rabinovich, 1998). The conventional approach for development of alien introgression is through generation of amphiploids by crossing wheat directly with wild species and then screening and selecting subsequent generations for individual alien chromosome additions or substitutions (Qi et al., 2007; Wang et al., 2013). Whole chromosome introgression in form of additions or substitutions is usually associated with linkage drag causing unwanted agronomic traits (Zamir, 2001). Better alternative is to transfer small chromosome segment by means of translocations. Many chromosome engineering approaches have been used to transfer small segments of alien chromosomes without linkage drag (Mukai et al., 1993; Lukaszewski, 1997; Qi et al., 2007). Radiation treatment and gametocidal genes have been utilized for development of wheat-rye (Ahmad et al., 2000; Masoudi-Nejiad et al., 2002), - Aegilops umbellulata (Sears, 1956), and wheat-Leymus racemosus translocations (Chen et al., 2005). Both ionizing treatment and gametocidal genes induce random chromosome breakage and thus random translocations with often undesirable agronomic traits (Qi et al., 2007). The availability of nulli-terta genetic stocks (Sears, 1965) of hexaploid wheat makes it possible to create chromosome specific translocation using addition and substitution lines. Marais et al. (2009) have used addition line of Aegilops neglecta and crossed it with CS followed by repeated backcrossing and screening for development of translocations. Similar methodology was adopted by Kuraparthy et al. (2009) where they used substitution line 5Mg(5D) and crossed with CS(PhI) stocks for development of translocations. We utilized N1AT1D genetic stock along with DSL-1E(1D) as a targeted approach to replace chromosome 1AL with chromosome 1EL. The cross between N1AT1D and DSL-1E(1D) produced F1 generation which was double monosomic for both chromosomes 1A and 1E. Double monosomics were supposed to produce rare translocation in the next generation. We also selected for Disomic substitution DSL-1E(1A) in the next generation that was expected to produce double monosomics in next cross. The chances of homoeologous pairing between 1A and 1E were thus increased by selecting initial genetic material.
The chromosome specific translocation line of wheat generated in the current manuscript [1EL(1AS)] thus is a unique example of targeted wide introgression achieved by carefully selecting the breeding material and the screening technologies. To the best of our knowledge this is the first, unique approach targeted to improve wheat processing quality. Previously, Wang et al. (2013) and Garg et al. (2014) developed Aegilops longissima specific disomic substitution lines 1Sl(1B) and 1Sl(1A), which were reported to possess superior bread making quality. Similarly disomic addition lines of Aegilops searsii in CS background have been reported to possess improved dough strength (Garg et al., 2009b). But linkage drag and genetic instability makes use of addition/substitution lines as direct cultivar impractical. The basic concept was the utilization of Th. elongatum potential while retaining the existing wheat qualities. We targeted chromosome 1A as its HMW-GS have least contribution toward wheat dough strength. We wanted to retain both 1D and 1B specific HMW-GS due to their significant positive effect (Rogers et al., 1990; Kumlay et al., 2003; Garg et al., 2014). This involved our previous work on understanding the contribution of individual wheat and wild relative chromosomes by screening a large number of addition lines (Garg et al., 2009b, 2014).
Creation of translocation line is costly, long, and tedious process, involving screening of a large number of entries with very low success frequency (Ko et al., 2002). We have reduced the time, cost, and efforts involved by combining protein marker screening with GISH/FISH. GISH is the common technique to identify translocation that is costly, time consuming, and requires significant trained manpower hours. We first screened our carefully selected breeding material with chromosome specific protein markers from the endosperm half and raised the selected seedling from embryo part, collected first two roots for mitotic metaphase analysis and used rest of the seedling to raise plant. The selected seeds were screened by GISH. With this strategy, we were able to increase screening efficiency at the second stage to as high as 33.3%. Interestingly, we have developed this translocation from normal cross rather than homoeologous pairing suppressor mutant (PhI) cross. In the first screening cycle, we could not succeed in our efforts, so we involved PhI line in our crosses. Wheat is allohexaploid with three different genomes and pairing takes place between homologous pairs of chromosomes that makes wide introgression a difficult task. Wheat researchers have tried to circumvent this challenge by deleting the chromosome part that does not allow homoeologous recombination. Using this approach several translocation lines have been generated (Sears, 1976; Rey et al., 2015). Rather, our breeding material with PhI was poor in growth, seed setting and we could not use this material for screening. We expect that multiple deletion/translocation events might have had a detrimental effect on plant growth.
We have not only generated translocation line, but also recovered the background in one of the leading cultivars: N61 by several cycles of backcrossing/selfing (BC4F6) and thus developed almost near isogenic line (NIL). We have characterized it and carried out two year quality analysis. This line showed improved dough strength, mixing properties, and extensibility compared to N61 recipient parent. Our multi environment data indicated that improvement in the translocation line was due to genetic factors. This is a unique material that can be utilized in many breeding programs targeting grain processing quality. This material has been transferred to researchers and breeders for its utilization.
Conceived and designed the experiments: AmK, MG, and HT. Performed the experiments: AmK, MG, NK, SM, and QD. Analyzed the data: AmK, MG, and AsK. Contributed reagents/materials/analysis tools: VC, RG, and SS. Wrote the paper: AmK and MG.
The project was funded by Department of science and technology (DST), Govt. of India for the research project “Chromosome specific wide hybridization for the improvement of bread making quality of wheat.”
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are thankful to the Department of Biotechnology, Govt. of India for the fellowship provided to AmK. We are grateful to National Agri-Food Biotechnology Institute, Mohali for providing the facilities for experimental work.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01593/full#supplementary-material
Ahmad, F., Comeau, A., Chen, Q., Collin, J., and St-Pierre, C. A. (2000). Radiation induced wheat-rye chromosomal translocations in triticale: optimizing the dose using fluorescence in situ hybridization. Cytologia 65, 1–6. doi: 10.1508/cytologia.65.1
Bao, Y., Wang, J., He, F., Ma, H., and Wang, H. (2012). Molecular, cytogenetic, identification of a wheat (Triticum aestivum) -American dune grass (Leymus mollis) translocation line resistant to stripe rust. Genet. Mol. Res. 11, 3198–3206. doi: 10.4238/2012.May.22.2
Branlard, G., Dardevet, M., Saccomano, R., Lagoutte, F. L., and Gourdon, J. (2001). Genetic diversity of wheat proteins and bread wheat quality. Euphytica 119, 59–67. doi: 10.1023/A:1017586220359
Carollo, V., Matthews, D. E., Lazo, G. R., Blake, T. K., Hummel, D. D., Lui, N., et al. (2005). GrainGenes 2.0. An improved resource for the small-grains community. Plant Physiol. 139, 643–651. doi: 10.1104/pp.105.064485
Chen, F. G., Liu, W., Yuan, J., Wang, X., Zhou, B., Wang, S., Zhang, S., et al. (2005). Development and characterization of wheat Leymus racemosus translocation lines with resistance to Fusarium head blight. Theor. Appl. Genet. 111, 941–948. doi: 10.1007/s00122-005-0026-z
Chen, P. D., Qi, L. L., Zhou, B., Zhang, S. Z., and Liu, D. J. (1995). Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 91, 1125–1128. doi: 10.1007/BF00223930
Chen, S., Huang, Z., Dai, Y., Qin, S., Zhang, L., Gao, Y., et al. (2013). The development of 7E chromosome specific molecular markers for Thinopyrum elongatum based on SLAF-seq technology. PLoS ONE 8:e65122. doi: 10.1371/journal.pone.0065122
Colmer, T. D., Flowers, T. J., and Munns, R. (2006). Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 57, 1059–1078. doi: 10.1093/jxb/erj124
Dewey, D. R. (1960). Salt tolerance of twenty five strains of Agropyron. Agron. J. 52, 631–635. doi: 10.2134/agronj1960.00021962005200110006x
Dewey, D. R. (1962). Breeding crested wheat grasses for salt tolerance. Crop Sci. 2, 403–407. doi: 10.2135/cropsci1962.0011183X000200050012x
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Feldman, M., and Levy, A. A. (2015). “Origin and evolution of wheat and related Triticeae species,” in Alien Introgression in Wheat, eds M. Molnar-Lang, C. Carla, and D. Jaroslav (Cham: Springer International Publishing), 21–76.
Friebe, B., Jiang, J., Knott, D. R., and Gill, B. S. (1994). Compensation indices of radiation-induced wheat-Agropyron elongatum translocations conferring resistance to leaf rust and stem rust. Crop Sci. 34, 400–404. doi: 10.2135/cropsci1994.0011183X003400020018x
Garg, M., Dhaliwal, H. S., Chhuneja, P., Kumar, D., Dou, Q. W., Elamein, H. M. M., et al. (2007). Negative effect of chromosome 1A on dough strength shown by modification of 1D addition in durum wheat (Triticum durum). Theor. Appl. Genet. 114, 1141–1150. doi: 10.1007/s00122-007-0506-4
Garg, M., Kumar, R., Singh, R. P., and Tsujimoto, H. (2014). Development of an Aegilops longissima substitution line with improved bread making quality. J. Cereal Sci. 60, 389–396. doi: 10.1016/j.jcs.2014.05.006
Garg, M., Tanaka, H., Ishikawa, N., Takata, K., Yanaka, M., and Tsujimoto, H. (2009a). Agropyron elongatum HMW-glutenins have the potential to improve wheat end product quality through targeted chromosome introgression. J. Cereal Sci. 50, 359–363. doi: 10.1016/j.jcs.2009.06.012
Garg, M., Tanaka, H., Ishikawa, N., Takata, K., Yanaka, M., and Tsujimoto, H. (2009b). A novel pair of HMW glutenin subunits from Aegilops searsii improves quality of hexaploid wheat. Cereal Chem. 86, 26–32. doi: 10.1094/CCHEM-86-1-0026
Garg, M., Tsujimoto, H., Gupta, R. K., Kumar, A., Kaur, N., Kumar, R., et al. (2016). Chromosome specific substitution lines of Aegilops geniculata alter parameters of bread making quality of wheat. PLoS ONE 11:e0162350. doi: 10.1371/journal.pone.0162350
Ko, J. M., Seo, B. B., Suh, D. Y., Do, G. S., Park, D. S., and Kwack, Y. H. (2002). Production of a new wheat line possessing the 1BL.1RS wheat-rye translocation derived from Korean rye cultivar Paldanghomil. Theor. Appl. Genet. 104, 171–176. doi: 10.1007/s00122-001-0783-2
Kumlay, A. M., Baenziger, P. S., Gill, K. S., Shelton, D. R., Graybosch, R. A., Lukaszewski, A. J., et al. (2003). Understanding the effect of rye chromatin in bread wheat. Crop Sci. 43, 1643–1651. doi: 10.2135/cropsci2003.1643
Kuraparthy, V., Sood, S., and Gill, B. S. (2009). Molecular genetic description of the cryptic wheat-Aegilops geniculata introgression carrying rust resistance genes Lr57 and Yr40 using wheat ESTs and synteny with rice. Genome 52, 1025–1036. doi: 10.1139/G09-076
Liu, J., Chang, Z., Zhang, X., Yang, Z., Li, X., Jia, J., et al. (2013). Putative Thinopyrum intermedium-derived stripe rust resistance gene Yr50 maps on wheat chromosome arm 4BL. Theor. Appl. Genet. 126, 265–274. doi: 10.1007/s00122-012-1979-3
Lukaszewski, A. J. (1997). Construction of midget chromosomes in wheat. Genome 40, 566–569. doi: 10.1139/g97-074
Luo, P. G., Luo, H. Y., Chang, Z. J., Zhang, H. Y., Zhang, M., and Ren, Z. L. (2009). Characterization and chromosomal location of Pm40 in common wheat: a new gene for resistance to powdery mildew derived from Elytrigia intermedium. Theor. Appl. Genet. 118, 1059–1064. doi: 10.1007/s00122-009-0962-0
Ma, P., Xu, H., Xu, Y., Li, L., Qie, Y., Luo, Q., et al. (2015). Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor. Appl. Genet. 128, 613–622. doi: 10.1007/s00122-015-2457-5
Mago, R., Miah, H., Lawrence, G. J., Wellings, C. R., Spielmeyer, W., Bariana, H. S., et al. (2005). High-resolution mapping and mutation analysis separate the rust resistance genes Sr31, Lr26 and Yr9 on the short arm of rye chromosome 1. Theor. Appl. Genet. 112, 41–50. doi: 10.1007/s00122-005-0098-9
Marais, F., Marais, A., McCallum, B., and Pretorius, Z. (2009). Transfer of leaf rust and stripe rust resistance genes Lr62 and Yr42 from Aegilops neglecta Req. ex Bertol. to common wheat. Crop Sci. 49, 871–879. doi: 10.2135/cropsci2008.06.0317
Masoudi-Nejiad, A., Nasuda, S., McIntosh, R. A., and Endo, T. R. (2002). Transfer of rye chromosome segments to wheat by a gametocidal gene. Chromosome Res. 10, 349–357. doi: 10.1023/A:1016845200960
Mejias, J. H., Lu, X., Osorio, C., Ullman, J. L., Wettstein, D., and Rustgi, S. (2014). Analysis of wheat prolamins, the causative agent of celic sprue, using reversed phase high performance liquid chromatography (RP-HPLC) and matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF-MS). Nutrients 6, 1578–1597. doi: 10.3390/nu6041578
Mills, E. N. C., Field, J. M., Kauffman, J. A., Tatham, A. S., Shewry, P. R., and Morgan, M. R. A. (2000). Characterization of monoclonal antibody specific for HMW subunits of glutenin and its use to investigate glutenin polymers. J. Agric. Food Chem. 48, 611–617. doi: 10.1021/jf9909499
Monneveux, P., Reynolds, M. P., Gonzalez, J., and Singh, R. P. (2003). Effects of 7DL.7Ag translocation from Lophopyrum elongatum on wheat yield and related morphophysiological traits under different environments. Plant Breed. 122, 379–384. doi: 10.1046/j.1439-0523.2003.00856.x
Mukai, Y., Friebe, B., Hatchett, J. H., Yamamoto, M., and Gill, B. S. (1993). Molecular cytogenetic analysis of radiation-induced wheat-rye terminal and intercalary chromosomal translocations and the detection of rye chromatin specifying resistance to Hessian fly. Chromosoma 102, 88–95 doi: 10.1007/BF00356025
Payne, P. I., and Lawrence, G. J. (1983). Catalogue of alleles for the complex gene loci Glu-A1, Glu-B1 and Glu-D1, which code for the high-molecular-weight subunits of glutenin in hexaploid wheat. Cereal Res. Commun. 11, 29–35.
Petersen, S., Lyerly, J. H., Worthington, M. L., Parks, W. R., Cowger, C., Marshall, D. S., et al. (2015). Mapping of powdery mildew resistance gene Pm53 introgressed from Aegilops speltoides into soft red winter wheat. Theor. Appl. Genet. 128, 303–312. doi: 10.1007/s00122-014-2430-8
Qi, L. L., Friebe, B., Zhang, P., and Gill, B. S. (2007). Homoeologous recombination, chromosome engineering and crop improvement. Chromosome 15, 3–19. doi: 10.1007/s10577-006-1108-8
Rabinovich, Y. (1998). Importance of wheat-rye translocations for breeding modern cultivars of Triticum aestivum L. Euphytica 100, 323–340. doi: 10.1023/A:1018361819215
Rasheed, A., Xianchun, X., Yan, Y., Appels, R., Mahmood, T., and He, Z. (2014). Wheat seed storage proteins: advances in molecular genetics, diversity and breeding applications. J. Cereal Sci. 60, 11–24. doi: 10.1016/j.jcs.2014.01.020
Ren, T. H., Yang, Z. J., Yan, B. J., Zhang, H. Q., Fu, S. L., and Ren, Z. L. (2009). Development and characterization of a new 1BL.1RS translocation line with resistance to stripe rust and powdery mildew of wheat. Euphytica 169, 207–213. doi: 10.1007/s10681-009-9924-5
Ren, T., Tang, Z., Fu, S., Yan, B., Tan, F., Ren, Z., et al. (2017). Molecular cytogenetic characterization of novel wheat-rye T1RS.1BL translocation lines with high resistance to diseases and great agronomic traits. Front. Plant Sci. 8:799. doi: 10.3389/fpls.2017.00799
Rey, M. D., Calderon, M. C., and Prieto, P. (2015). The use of ph1B mutant to induce recombination between the chromosomes of wheat and barley. Front. Plant Sci. 6:160. doi: 10.3389/fpls.2015.00160
Rogers, W. J., Rickatson, J. M., Sayers, E. J., and Law, C. N. (1990). Dosage effect of chromosomes of homoeologous groups 1 and 6 upon bread-making quality in hexaploid wheat. Theor. Appl. Genet. 80, 281–287. doi: 10.1007/BF00224399
Royal Botanic Gardens Kew (2017). Seed Information Database (SID). Version 7.1. Available online at: http://data.kew.org/sid/ (Accessed on 29 August 2017).
Sears, E. R. (1956). “The transfer of leaf rust resistance from Aegilops umbellulata to wheat,” in Genetics in Plant Breeding. Brook-Haven Symposia in Biology No. 9 (Upton, NY: Brookhaven National Laboratory), 1–22.
Sears, E. R. (1965). “Nullisomic-tetrasomic combinations in hexaploid wheat,” in Chromosome Manipulations and Plant Genetics, eds R. Riley and K. R. Lewis (Edinburgh: Oliver and Boyd), 29–45.
Sears, E. R. (1976). Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10, 31–51. doi: 10.1146/annurev.ge.10.120176.000335
Sharma, H. C., and Gill, B. S. (1983). Current status of wide hybridization in wheat. Euphytica 32, 17. doi: 10.1007/BF00036860
Shewry, P. R., and Tatham, A. S. (1997). Disulphide bonds in wheat gluten proteins. J. Cereal Sci. 25, 207–227. doi: 10.1006/jcrs.1996.0100
Smith, D. B., and Payne, P. I. (1984). A procedure for routine determination of Electrophoretic band patterns of barley and malt endosperm proteins. J. Nat. Ins. Agric. Bot. 16, 487–498.
Sourdille, P., Gandon, B., Chiquet, V., Nicot, N., Somers, D., Murigneux, A., et al. (2004). Wheat génoplante SSR mapping data release: a new set of markers and comprehensive genetic and physical mapping data. Available online at: https://wheat.pw.usda.gov/ggpages/SSRclub/GeneticPhysical
Sreeramulu, G., and Singh, N. K. (1997). Genetic and biochemical characterization of novel low molecular weight glutenin subunits in wheat (Triticum aestivum L.) Genome 40, 41–48. doi: 10.1139/g97-006
Takata, K., Yamauchi, H., Iriki, N., and Kuwabara, T. (1999). Prediction of bread-making quality by prolonged swelling SDS-sedimentation test. Breed Sci. 49, 221–223. doi: 10.1270/jsbbs.49.221
Tohver, M. (2007). High molecular weight (HMW) glutenin subunit composition of some nordic and middle European wheats. Genet. Resour. Crop. Evol. 54, 67–81. doi: 10.1007/s10722-005-1885-5
Totosaus, A., López, H., and Vera, N. G. (2013). Effect of lupinus (Lupinus albus) and jatropha (Jatropha curcas) protein concentrates on wheat dough texture and bread quality: optimization by a D-optimal mixture design. J. Text. Stud. 44, 424–435. doi: 10.1111/jtxs.12031
Tsuchiya, T., and Nakamura, C. (1979). Acetocarmine squash method for observing sugar beet chromosomes. Euphutica 28, 249–256. doi: 10.1007/BF00056582
[USDA/ARS] (2017). USDA, Foreign Agricultural Service, Production, Supply, and Distribution Database. Available online at: http://www.ers.usda.gov/data-products/wheat-data.aspx
Wang, S., Yu, Z., Cao, M., Shen, X., Li, N., Li, X., et al. (2013). Molecular mechanisms of HMW glutenin subunits from 1Sl genome of Aegilops longissima positively affecting wheat breadmaking quality. PLoS ONE 8:e58947. doi: 10.1371/journal.pone.0058947
Wulff, B. B. H., and Moscou, M. J. (2014). Strategies for transferring resistance to wheat: from wide crosses to GM cassettes. Front. Plant Sci. 5:692. doi: 10.3389/fpls.2014.00692
Zamir, D. (2001). Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2, 983–989. doi: 10.1038/35103590
Keywords: wheat, translocation line, HMW glutenin, wide hybridization, bread making quality, Thinopyrum elongatum, genomic in situ hybridization (GISH), marker assisted selection (MAS)
Citation: Kumar A, Garg M, Kaur N, Chunduri V, Sharma S, Misser S, Kumar A, Tsujimoto H, Dou Q-W and Gupta RK (2017) Rapid Development and Characterization of Chromosome Specific Translocation Line of Thinopyrum elongatum with Improved Dough Strength. Front. Plant Sci. 8:1593. doi: 10.3389/fpls.2017.01593
Received: 08 June 2017; Accepted: 30 August 2017;
Published: 14 September 2017.
Edited by:
Rodomiro Ortiz, Swedish University of Agricultural Sciences, SwedenReviewed by:
Galina Suvorova, All-Russia Research Institute of Legumes and Groat Crops, RussiaCopyright © 2017 Kumar, Garg, Kaur, Chunduri, Sharma, Misser, Kumar, Tsujimoto, Dou and Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monika Garg, mgarg100@yahoo.com; monikagarg@nabi.res.in
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.